Abstract
The CRISPR-Cas system has transformed the field of gene-editing and created opportunities for novel genome engineering therapeutics. The field has significantly progressed, and recently, CRISPR-Cas9 was utilized in clinical trials to target disease-causing mutations. Existing tools aim to predict the on-target efficacy and potential genome-wide off-targets by scoring a particular gRNA according to an array of gRNA design principles or machine learning algorithms based on empirical results of large numbers of gRNAs. However, such tools are unable to predict the editing outcome by variant Cas enzymes and can only assess potential off-targets related to reference genomes. Here, we employ normal mode analysis (NMA) to investigate the structure of the Cas9 protein complexed with its gRNA and target DNA and explore the function of the protein. Our results demonstrate the feasibility and validity of NMA to predict the activity and specificity of SpyCas9 in the presence of mismatches by comparison to empirical data. Furthermore, despite the absence of their exact structures, this method accurately predicts the enzymatic activity of known high-fidelity engineered Cas9 variants.
Keywords: Normal mode analysis, CRISPR specificity, CRISPR activity, CRISPR computational modelling, In silico activity simulation, Structure function, CRISPR
1. Introduction
The CRISPR-Cas system is a prokaryotic adaptive immune system, conferring immunity against bacteriophages and plasmids based on nucleic acids recognition [1]. It has been employed as a gene-editing tool in eukaryotic cells owing to its unique RNA-guided targeting attributes. Class 2 CRISPR systems consist of a single Cas effector protein. Upon binding to a guide-RNA (gRNA) molecule, it directs the Cas protein toward its target sequence [2], [3], DNA or RNA, depending on the type and subtype of the Cas protein [4]. Target recognition is mediated by base pairing between the gRNA and the target sequence. For the commonly studied Streptococcus pyogenes (Spy)Cas9, the gRNA base-pairs with the target strand DNA (TS-DNA), a stage that drives a conformational transformation of Cas9, leading it to cleave the target DNA [5]. Recently, the field of gene-editing has entered a new era, as the CRISPR-Cas system was introduced into patients cells, ex vivo [6], [7], [8], [9] and in vivo [10], and reportedly contributing positive results in clinical outcomes. Understanding the accuracy and specificity of CRISPR-Cas9 is essential to better design and develop improved future gene-editing therapeutics [11]. Previous studies have revealed the protein structure of SpyCas9, paving the way to structural investigation of the protein and its functions [12], [13], [14]. Existing tools designated to predict on-target activity efficiency, specificity and off-targeting are based on a set of gRNA design principles derived from experimental observations [15]. Such tools and principles do not apply to the variety of Cas enzymes, engineered variants and gRNAs that do not conform with reference genomes. Simulating the enzymatic activity is of interest in assessing the editing outcome of various Cas enzymes, as well as for in silico design of novel variants. Normal mode analysis (NMA) is a computational method used to probe large-scale motions in biomolecules. Typical application is for the prediction of functional motions in proteins. It relies on the premise that the physics of a protein around an equilibrium position behave as an oscillating system. This in turn describes the flexible and thermodynamical movements close to an equilibrium point. Coarse-grained NMA overcomes the computationally limiting factor of analyzing large numbers of atoms in a protein by representing each residue using a single atom – the α carbon [16]. Recently, we have demonstrated how NMA can be utilized to explain the function and dysfunction of proteins with pathogenic mutations in several different clinical conditions [17], [18], [19], [20]. It is largely agreed that the function of the protein and its dynamics can be inferred from NMA [16], [21]. In a previous study, NMA was shown to provide structural and dynamic details on the mechanism of action of SpyCas9 [22]. Nevertheless, it was not utilized to study the sequence-dependent activity of CRISPR-Cas systems. NMA can be utilized to investigate the entropic profile of the whole structure, or only a part of it (i.e., specific residues, nucleotides, or distinct macromolecules such as a protein or an RNA molecule). The method described herein characterizes allosteric entropic changes made in a structure with several macromolecules. By changing one macromolecule in the complex, we witness entropic changes in other macromolecules of the complex that might affect the complex functionality. We hypothesized that NMA could predict Cas9 activity and likewise its specificity (Fig. 1). In this study, we have done computational replications of previously published experimental studies and compared the entropy scores we obtained from the NMA to the observed empirical data. Our results support the relevance of NMA to study the function of proteins, particularly SpyCas9, and imply its ability to predict on-target and off-target activity and specificity of CRISPR-Cas systems.
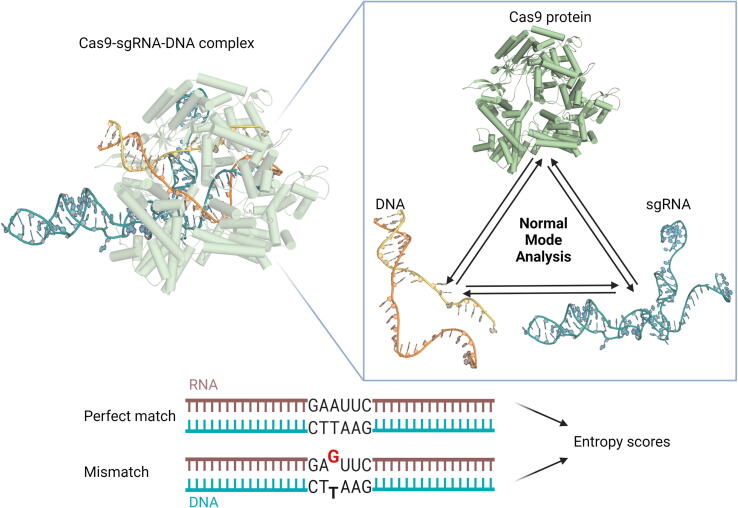
Fig. 1.
General scheme – NMA predicts the activity and specificity in a sequence-dependent manner. NMA yields entropy scores that correlate with empirical SpyCas9 activity data. Modifications were made to all parts of the structure: protein (high-fidelity variants mutations), DNA (four different EMX1 sites) and sgRNA (mismatches assay) while retaining high correlations. PDB: 5F9R [12].
2. Results
2.1. NMA accurately replicates empirical data of SpyCas9 activity and specificity profile
To test the relevance of NMA to predict the activity of SpyCas9, we performed an in silico replication of a previously published experiment by Hsu et al. [23]. The empirical data describe the specificity profile of SpyCas9 in four genomic loci within the EMX1 gene. The specificity was measured as the cleavage activity in the presence of mismatches between the single-guide RNA (sgRNA) and the target DNA, compared to a perfect-match sgRNA (Fig. 2a, left column) [23]. We hypothesized that the entropic changes caused by single-nucleotide mismatches would reflect the specificity patterns obtained from the experimental results. To examine our hypothesis, the structure of the SpyCas9 complex (bound to the sgRNA and the target DNA) was fetched from the Protein Data Bank (PDB, accession number: 5F9R [12]) and the RNA and DNA sequences were modified to match the four EMX1 loci. We then generated 57 modified structures per locus, where in each structure, one nucleotide of the sgRNA was changed, according to the original experiment (see Methods). In total, 232 structures were generated. The ΔG of each structure was measured using NMA. Since we have modified the sgRNA molecule in the structure, we sought to assess the single-nucleotide mismatch effect on the ΔG of the protein (chain B) and the DNA (chain C – target strand) separately (Fig. 2). Analysis of the ΔG measurements across the different EMX1 sites from both the protein and the DNA, unveils patterns that indeed resemble the empirical data (Fig. 2a). Moreover, the seed region can be clearly observed in the ΔG patterns, demonstrating the consistency of our results with the previous reports [3], [11], [23], [24]. The correlation for each combination of empirical results, ΔG of the protein and ΔG of the DNA, was calculated for all sites (Fig. 2b–d). The Pearson correlation coefficient (r) of the DNA entropy or the protein entropy with the empirical data is very similar and ranges from 0.6853 to 0.7875 (Fig. 2b) and 0.6577 to 0.7502 (Fig. 2c), respectively (absolute values). The high r values demonstrate the feasibility of in silico NMA to predict the activity outcome of SpyCas9, even when the DNA and sgRNA sequences of the structures are modified compared to the original structure. The r values are presented as absolute values since the direction of the correlation (positive or negative) does not affect the power of the correlation. Since the |r| values of the EMX1 site 3 are higher compared to the three other sites, we decided to perform the following analyses in this study on the EMX1 site 3. r values and p-values are summarized in Supplementary Table 1.
Fig. 2.
SpyCas9 empirical activity and structure-based entropy. a) Heatmap representations of previously reported empirical SpyCas9 activity (specificity measured as the ratio of mismatch/perfect match), the entropy of the DNA and the SpyCas9 protein () in the presence of single-base mismatches in four loci within the EMX1 gene. The color scale bar orientation is determined by the direction of the correlation (positive/negative). b) Correlations between the empirical activity (x) and the of the DNA (y). c) Correlations between the empirical activity (x) and the of the protein (y). d) Correlations between the of the DNA (x) and the of the protein (y). All correlation plots are shown with a 95% confidence interval and p-value <0.00005 (N = 57). The correlation values represent the Pearson correlation coefficient (r).
2.2. Residues’ entropy and empirical enzymatic activity correlate among different gRNAs with mismatches
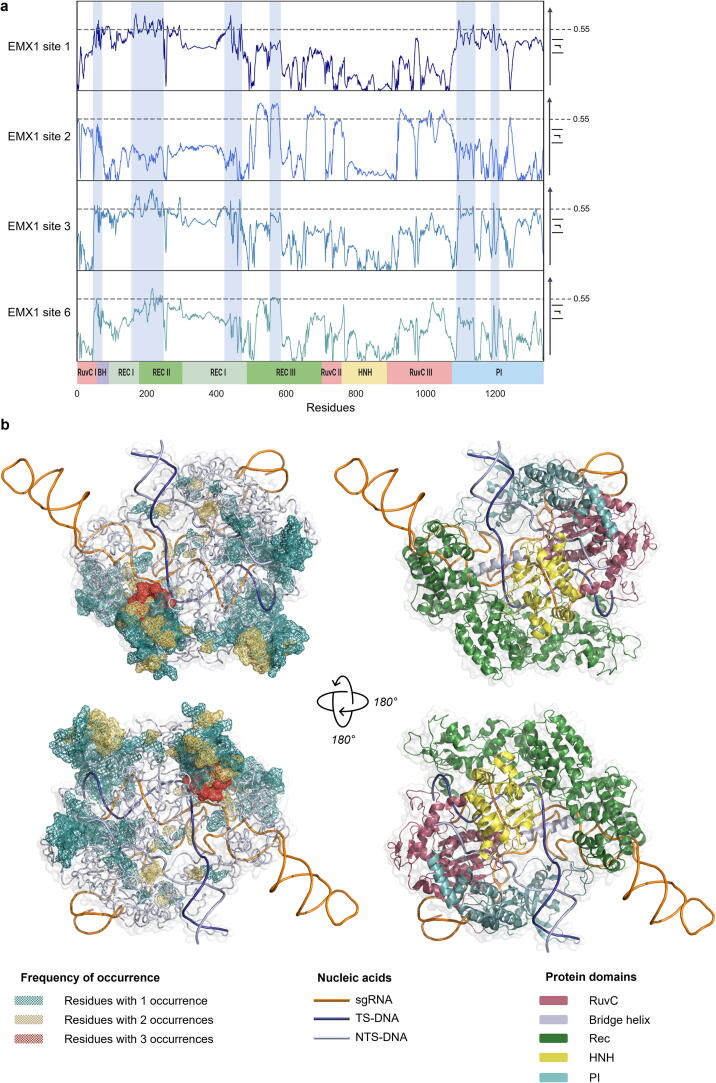
To examine which amino acids within the structure of SpyCas9 respond in the form of ΔG changes coordinately with activity rates in the presence of mismatches, we calculated the ΔG of each residue. The correlation between the ΔG and the activity was calculated (r) and plotted for each genomic site (Fig. 3a). It is apparent that the r values for each residue are highly consistent among the four EMX1 loci, indicating the coherence reactivity of the protein regions in varying genetic contexts. Noticeably, high r values were most abundant within the REC lobe (REC domains I-III) and the PAM interacting (PI) domain, as well as the bridge-helix (BH) that is known to confer mismatch sensitivity [25]. We set a tentative threshold of r = 0.55 and marked regions of residues that cross it in more than one EMX1 site, indicating protein regions where entropic response to mismatches harmoniously correlates with the empirical activity of the enzyme. Further to the 2D representation of the residues crossing the r = 0.55 threshold, we depicted the number of occurrences in which a residue crossed the threshold in a 3D representation to observe the structural relevance of such residues (Fig. 3b). Examination of the 3D structure confirms that residues that repeatedly have high r values are likely to interact directly with the nucleic acids within the structure. For instance, residues 164–174, which are part of the REC lobe (REC I domain), interact closely with the gRNA, stabilizing the R-loop (gRNA:TS-DNA heteroduplex), and cross the r threshold in two EMX1 sites. Remarkably, although the REC2 domain does not bind the gRNA and the DNA (despite residue D269), and SpyCas9 still retains its activity even after complete removal of the domain [13], it contains the most frequent residues (212–219 and 244–246). It is noteworthy that high r values of a certain residue do not implicate its role in specificity imparting. However, the ΔG of residues with high r values can be utilized to predict the enzymatic function.
Fig. 3.
The correlation between the empirical activity in the presence of mismatches and the entropy of each amino acid in the structure of SpyCas9 for each mismatch. a) Absolute values of the Pearson correlation coefficient r, measured in all amino acids along with the structure of SpyCas9 in the presence of mismatches in four genomic loci. The measured entropy relates to the α-carbon of each amino acid. The dashed line represents a threshold of r = 0.55. Regions containing residues with r greater than the threshold in more than one site are marked in light blue. The 2D representation of the protein domains shows the regions in which the entropy of the amino acids best correlate with the empirical activity data. Scale range 0 < r < 0.8. b) The structure of SpyCas9 highlighting the residues with r > 0.55 (mesh). Colors indicate the number of sites (1–3) in which the r value for this residue crossed the threshold (left). The right panel is a 3D representation of the protein domains. The target strand DNA (TS-DNA), non-target strand (NTS-DNA) and the sgRNA are represented as simplified lines, while the protein is visualized as a cartoon. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. NMA-based predictions of the activity and specificity of engineered SpyCas9 variants
As a modification of nucleic acids within the structure of SpyCas9 led to NMA-based results that were consistent with empirical data, we speculated whether NMA might also predict the outcome of amino acids modifications. Similar to the comparison of ΔG to the activity in the presence of mismatches (Fig. 2), the computationally modified protein should be compared to a priori empirical data of such variants. To that end, we obtained the specificity and activity scores of eight engineered SpyCas9 variants with improved specificity from a previously published study by Schmid-Burgk et al. This study provides high-throughput and uniformly collected data (using the TTISS method) of all eight variants, compared to the wildtype (WT) SpyCas9 [26]. The variants that were compared were eSpCas9(1.1) [27], SpCas9-HF1 [28], HypaCas9 [29], evoCas9 [30], Sniper-Cas9 [31], Hifi-Cas9 [32] and LZ3 Cas9 [26]. The authors tested 59 gRNAs to evaluate the on-target activity and specificity (genome-wide off-target activity), thus, generating comprehensive and robust data.
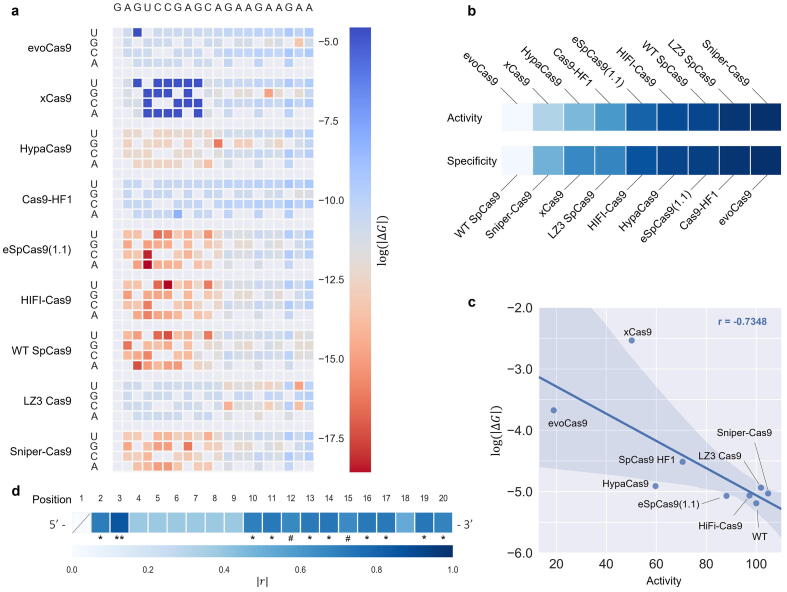
We focused on the protein structure with the altered nucleic acids corresponding to the EMX1 site 3 sequence and modified the amino acids according to the various engineered SpyCas9 variants. Thereafter, by generating structures of all the single mismatches for each variant (as previously described in this manuscript), we established a predicted specificity profile consisting of SpyCas9 and eight variants (Fig. 4a). The order of the variants was determined according to their activity, as measured by Schmid-Burgk and colleagues (Fig. 4b). The two most specific variants, evoCas9 and Cas9-HF1, exhibit highly specific entropy profiles compared to WT SpyCas9 and other less specific variants. Interestingly, the ΔG values are highly correlative with the average on-target activity scores (r = −0.7348; Fig. 4c). While most variants show ΔG patterns (Fig. 4a) that correlate with the empirical activity (Fig. 4b), xCas9 is seemingly not in line with the other variants. xCas9 is comprised of seven mutations and was initially screened as a PAM-modified variant that afterwards was found to have improved specificity. The inconsistent entropy pattern may be due to other molecular mechanisms underlying the specificity improvement and activity reduction of xCas9. We next calculated the correlation between the average ΔG of each position and each variant and the average activity score of each variant (Fig. 4d). High r values indicate the feasibility to predict the activity outcome based on the ΔG of a particular position. Surprisingly, the obtained r values pattern in the different positions of the gRNA resembles the seed region pattern, excluding positions two and three (PAM-distant region) that are thought to be the least stringent. These significantly correlative positions (2, 3, 10–17, 19 and 20) can be of great use in predicting the on-target activity of various Cas variants and serve as predictors for off-targets assessments.
Fig. 4.
NMA predicts and replicates specificity and activity of eight SpyCas9 variants with improved specificity. a) Entropy profile heatmaps of SpyCas9 variants in the presence of gRNA mismatches at the EMX1 – site3 locus ( measured at the DNA molecule (chain C – TS-DNA). b) Average activity and specificity scores as previously reported and determined by the TTISS method. c) Correlation between the activity score of each variant and its corresponding average entropy score (. The correlation plot is shown with a 95% confidence interval and p-value = 0.024123 (N = 9). d) The Pearson correlation coefficient (r) of each position within the gRNA, representing the feasibility of each position to predict the activity outcome (average per variant) using the entropy score (average per position per variant). #=0.05 < p-value < 0.06, *=p-value < 0.05, **=p-value < 0.005.
3. Discussion
The data presented in this study demonstrate the correlation between NMA and empirical enzymatic activity from experimental studies. The multicomponent complex of Cas9 protein, sgRNA and DNA (TS and NTS-DNA) allowed us to manipulate one or two elements (gRNA mismatches or protein mutations) and measure their influence on the constants (i.e., DNA). Strong correlations between the empirical enzymatic activity and NMA calculations were observed after changes were made to the original structure. Strikingly, after also changing the protein residues the correlation remained as strong. While examining different hypotheses, whether NMA correlates with WT SpyCas9 in the presence of mismatches and if SpyCas9 variants correlate with their reported activity, we utilized two independent datasets. One, by Hsu et al. characterizes the specificity profile of WT SpyCas9 in four loci within the EMX1 gene [23]. The other, by Schmid-Burgk et al. compares eight variants with improved specificity and attempts to find genome-wide off-targets and determine their on-target efficiency [26]. The consistent correlation between NMA and empirical experimental data from different studies provide strong evidence for the validity of NMA to predict the outcome of Cas9 activity. Although the first part of this work is focused on the EMX1 gene, we show four distinct gRNA sequences that were analyzed using NMA. Moreover, the predicted NMA scores were compared to empirical data of HF variants targeting 59 target loci and nevertheless, provided consistent correlation. Thus, the robustness of NMA has shown to be generalized and not restricted to a specific gRNA. The method presented herein may lay the groundwork for generating future gene-editing tools and technologies such as off-targets assessment tools and engineering of novel Cas variants. The latter can benefit from the NMA activity-based standard curve (Fig. 4) or a similar NMA specificity-based curve. Moreover, applying this method on other Cas enzymes (i.e., Cas9 orthologs, Cas12 or other Cas effector proteins) can lead to the development of novel effector proteins from different classes with unique functions. This is restraint to the limitations of the method, as it requires available structure of the protein of interest in association with related molecules (e.g., DNA, RNA), and detailed data that can be used for comparison and calibration. Any engineered protein candidate that was predicted using this method should be tested experimentally in a “wet lab”. Notably, actual experimental results may be subjected to variance resulted from multiple parameters. This may affect both the empirical data used for analysis and the validation experiment of the proteins of interest. Furthermore, structures depicting the protein (or complex) in different conformations might result in different conclusions. Taken together, this study demonstrates the feasibility and accuracy of NMA in the context of the CRISPR-Cas9 system. Future studies may make use of the method and data presented in this work to further improve its accuracy and conduct experimental validations of the computational predictions.
4. Methods
4.1. In silico analysis
The structure of the SpyCas9 complex was taken from the Protein Data Bank (PDB-101; accession numbers PDB: 5F9R [12]). Next, using the mutagenesis software X3dna- DSSR (https://x3dna.org/) Linux package [33], [34], [35], [36], we performed in silico bases mutagenesis of the given gRNA (chain A) and DNA (chain C – TS-DNA and chain D – NTS-DNA) to the gRNA and DNA sequences used in the study of Hsu et al. [23]. For the WT structure now modified with four new gRNA sequences, we created a structure for each of the possible mismatches in positions 1–19, using the aforementioned X3dna- DSSR software. WT and mismatched structures were analyzed by an ENCoM coarse-grained NMA method to evaluate the effect of the analyzed mismatch on the stability of the protein and the DNA. This method is based on an entropic considerations C package of ENCoM [37] available at the ENCoM development website (https://github.com/NRGlab/ENCoM), compiled and used on a Ubuntu platform (Canonical Group, UK). For each analyzed variant, we calculated the entropy difference (ΔG) by subtracting the NMA-based mismatched structure’s entropic profile from the entropic profile of the WT perfect-match structure model.
The calculation of the entropic difference (ΔG) was done using MATLAB software (MathWorks, Natick, MA).
Next, to build the nine high fidelity structures, the Mutagenesis plugin in PyMol Molecular Graphics System Version 1.8 (Schrödinger, LLC., Cambridge, MA) was used to perform the appropriate in silico point mutagenesis in the WT protein structure with changed gRNA (as mentioned above, the gRNA was modified using X3dna-DSSR) modelled structure (EMX1 site 3). Using this structure, in silico mutagenesis was performed for each variant to replace the amino acids in accordance with each of the eight variants. These in silico mutations were made only in chain B. All variants were also analyzed for mismatches in the gRNA by the same procedure as described above. Mismatched structures of all variants were analyzed by an ENCoM coarse-grained NMA method to evaluate the effect of the analyzed mismatch on the stability of the protein. For each analyzed variant, we calculated the ΔG by subtracting the NMA-based mismatched structure’s entropic profile from the entropic profile of the perfect-match structure model. The calculation of the ΔG was done using MATLAB software.
Author contributions
R.R. and O.S. conceived the study with input from D.O. and F.B. O.S. performed the NMA and generated the modified structures. O.S. and R.R. designed the in silico experiments and analyzed data. R.R. wrote the manuscript and prepared figures with input from O.S. D.O. and F.B. The manuscript was reviewed and approved by all co-authors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Fig. 1 was created using Biorender.com and using a protein structure from PDB (accession: 5F9R). R.R. is supported by external PhD scholarships from the “Dan David Prize” and “Teva BioInnovation Program”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.04.026.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Barrangou R., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (80-) 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Jinek M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (80-) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science (80-) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova K.S., et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2019;182(18):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang F., Doudna J. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 6.Frangoul H., et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 7.Lacey S.F., Fraietta J.A. First trial of CRISPR-edited T cells in lung cancer. Trends Mol Med. 2020;26:713–715. doi: 10.1016/j.molmed.2020.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Stadtmauer E.A., et al. CRISPR-engineered T cells in patients with refractory cancer. Science (80-) 2020;367 doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y., et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;265(26):732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 10.Gillmore J., et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med. 2021;385:493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 11.Rabinowitz R., Offen D. Single-base resolution: increasing the specificity of the CRISPR-Cas system in gene editing. Mol Ther. 2020 doi: 10.1016/j.ymthe.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang F., et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science (80-) 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimasu H., et al. Crystal structure of Cas9 in Complex with Guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M., et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343 doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haeussler M., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005;13:373–380. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Wilf-Yarkoni A., et al. Mild phenotype of wolfram syndrome associated with a common pathogenic variant is predicted by a structural model of wolframin. Neurol Genet. 2021;7 doi: 10.1212/NXG.0000000000000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helbig I., et al. A recurrent missense variant in AP2M1 impairs clathrin-mediated endocytosis and causes developmental and epileptic encephalopathy. Am J Hum Genet. 2019;104:1060–1072. doi: 10.1016/j.ajhg.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shauer A., et al. Novel RyR2 mutation (G3118R) is associated with autosomal recessive ventricular fibrillation and sudden death: clinical, functional, and computational analysis. J Am Heart Assoc. 2021;10 doi: 10.1161/JAHA.120.017128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fellner A., et al. In-silico phenotype prediction by normal mode variant analysis in TUBB4A-related disease. Sci Rep. 2022;12 doi: 10.1038/s41598-021-04337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahar I., Rader A. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng W. Probing the structural dynamics of the CRISPR-Cas9 RNA-guided DNA-cleavage system by coarse-grained modeling. Proteins. 2017;85:342–353. doi: 10.1002/prot.25229. [DOI] [PubMed] [Google Scholar]
- 23.Hsu P.D., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doench J.G., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bratovič M., et al. Bridge helix arginines play a critical role in Cas9 sensitivity to mismatches. Nat Chem Biol. 2020 doi: 10.1038/s41589-020-0490-4. [DOI] [PubMed] [Google Scholar]
- 26.Schmid-Burgk J.L., et al. Highly parallel profiling of Cas9 variant specificity. Mol Cell. 2020;1–7 doi: 10.1016/j.molcel.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slaymaker I.M., et al. Rationally engineered Cas9 nucleases with improved specificity. Science (80-) 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinstiver B.P., et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J.S., et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casini A., et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.K., et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vakulskas C.A., et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X., Olson W. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu X., Olson W. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng G., Lu X., Olson W. Web 3DNA–a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colasanti A., Lu X., Olson W. Analyzing and building nucleic acid structures with 3DNA. J Vis Exp. 2013 doi: 10.3791/4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frappier V., Chartier M., Najmanovich R. ENCoM server: exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res. 2015;43:W395–W400. doi: 10.1093/nar/gkv343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.