Abstract
Several studies have focused on using cell carriers to solve the problem of mesenchymal stem cell expansion on regenerative medicine. However, the disadvantages of using prolonged enzymatic treatment and low cell harvest efficiency still trouble researchers. In this study, PNIPAAm-immobilized gelatin microspheres (abbreviated as GNMS) were synthesized using a simple power-driven flow-focusing microinjection system. The developed thermosensitive GNMS can allow easier harvesting of cells from the microspheres, requiring only 10 min of low-temperature treatment and 5 min of trypsin treatment. The developed GNMS was characterized by Fourier-transform infrared spectroscopy, optical microscopy, and scanning electron microscopy. Further, live/dead staining, F-actin staining, and PrestoBlue cell viability assays were used to evaluate cytotoxicity, cell morphology, cell proliferation, and harvest efficiency. The gene expression of stem cell markers was determined by real-time quantitative PCR (Q-PCR) analysis to investigate the stemness and phenotypic changes in Wharton's jelly-derived mesenchymal stem cells. The results showed that the engineered cell-laden thermosensitive GNMS could significantly increase the cell harvest rate with over 99% cell survival rate and no change in the cell phenotype. Thus, the described strategy GNMS could be the suitable 3D cell carriers in the therapeutic application and opens new avenues for regenerative medicine.
Keywords: Thermosensitive cell carriers, Gelatin-PNIPAAm microspheres, Cell expansion, Stem cell therapy, Regenerative medicine
Graphical abstract
1. Introduction
Regenerative medicine is a promising therapy for the development of functional cells, tissues, and organs to repair or replace unhealthy organs in the body due to aging, illness, and damage. It can be applied in musculoskeletal disorders, oncology, wound care, dental, ocular disorders, etc. [[1], [2], [3]]. In most cases, stem cells are frequently used in these applications. Because stem cells self-renew and differentiate into specialized cell properties, stem cell-based therapy has emerged as an extremely promising technology [[4], [5], [6]]. Wharton's jelly-derived mesenchymal stem cells (WJ-MSCs) are MSCs that have been isolated from the human umbilical cord. Compared with other bone marrow-derived MSCs and adipose-derived MSCs, WJ-MSCs express higher levels of undifferentiated human embryonic stem cell markers and pluripotency markers [[7], [8], [9]].
In clinical practice, a successful regenerative medicine application often requires 108-10 [10] cells, which is difficult to achieve using traditional 2D culture [[10], [11], [12]]. Therefore, researchers have developed a 3D culture method using microcarriers for cell expansion, including polystyrene beads, dextran beads, and poly(lactic-co-glycolic acid) beads [[13], [14], [15]]. However, these polymers lack the integrin recognition sites for cell identification, limiting cell growth and attachment of cells [16,17]. To overcome these shortcomings, collagen beads and gelatin-based microcarriers have been developed. Cells can recognize the arginyl-glycyl-aspartic acid (RGD) peptide, which is abundant in collagen and gelatin for better biocompatibility, cell attachment efficiency, and low immunogenicity [18,19]. However, the non-uniform size of these beads may cause a heterogeneous distribution of cells on the beads, which results in limited applications [20].
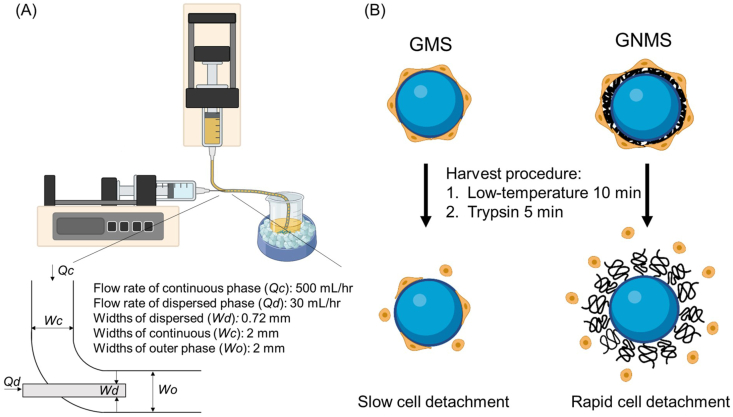
Although many studies have addressed stem cell expansion, the low cell collection efficiency remains a challenge due to the size of the microcarriers, cell-cell junctions, and abundant extracellular matrix formation in the 3D culture environment. Traditionally, treatments by proteolytic enzymes, such as dispase, collagen, and trypsin, were used for 15–30 min to detach the cells on the 3D carriers [[21], [22], [23]]. However, such enzymatic treatment may irreversibly harm the cells and denature the protein on the cell membrane, reducing the cell survival rate [24]. To overcome this, thermoresponsive poly(N-isopropylacrylamide) (PNIPAAm)-grafted materials have been developed and applied to cell culture. When the temperature of PNIPAAm is higher than the lower critical solution temperature (LCST) of approximately 32 °C, the polymer is in a relatively hydrophobic state, which is favorable for the attachment of cells [25]. However, when the temperature is lower than the LCST, PNIPAAm becomes more hydrophilic and forms a brush-like structure on the surface of the material, and the cells detach from the material [26,27]. Despite this, there remains a sophisticated relationship between cells and materials. The influence of temperature alone is often unreliable for achieving a high cell detachment efficiency, hence reducing to a lower temperature or extending the time of the low-temperature treatment to 120 min improves cell collection efficiency, which would also decrease the cell viability.
In this study, we aimed to develop a thermosensitive cell carrier and modify the current traditional cell harvesting procedure for 3D cell expansion. Gelatin microspheres (abbreviated as GMS) were prepared using a simple power-driven flow-focusing microinjection system. The prepared microspheres had a narrow bead size distribution. Subsequently, the thermoresponsive polymer PNIPAAm was immobilized onto GMS via EDC/NHS crosslinking. We hypothesized that the PNIPAAm-immobilized GMS (abbreviated as GNMS) would allow the cells to proliferate with high cell viability on the carriers, requiring only 10 min of low-temperature treatment and 5 min of trypsin treatment to harvest the WJ-MSCs. The developed cell-laden thermosensitive GNMS can be applied in the therapeutic application and open new avenues for regenerative medicine. The developed GNMS was characterized by Fourier-transform infrared (FTIR) spectroscopy, optical microscopy, and scanning electron microscopy (SEM) for functional group identification, sphere size measurement, and microstructure examination, respectively. The cell viability of the WJ-MSCs adhering to the gelatin microspheres was evaluated using a live/dead staining assay. Further, microfilaments were observed by confocal laser microscopy using F-actin staining. Cell proliferation ability was investigated using the PrestoBlue cell viability assay. In addition, the gene expressions of stem cell markers and pluripotency markers were determined by real-time quantitative PCR (Q-PCR) analysis to examine the stemness and phenotypic changes in WJ-MSCs. The overall design is illustrated in Scheme 1.
Scheme 1.
The Scheme of GNMS preparation and its applications for cell expansion. (A) Schematic representation of the power-driven flow-focusing microinjection system. (B) Schematic illustration of GNMS exhibiting a temperature-dependent cell adhesion and detachment for cell expansion.
2. Materials and methods
2.1. Materials
Gelatin from porcine skin, type A (Cat. No. G2500), glutaraldehyde, poly (N-isopropylacrylamide), amine-terminated (Cat. No. 724831, average molecular weight 5500), N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC), and N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Further, olive oil was purchased from Weiyi Enterprise Co., Ltd. (Taiwan). Alpha minimum essential medium, fetal bovine serum (FBS), and trypsin-EDTA (0.25%) were purchased from Gibco (Grand Island, NY, USA). Finally, the LIVE/DEAD viability kit, Alexa Fluor 488 phalloidin, and PrestoBlue cell viability reagent were purchased from Invitrogen (Carlsbad, CA, USA).
2.2. Preparation of gelatin microspheres using a power-driven flow-focusing microinjection system
Uniform gelatin microspheres were prepared using a power-driven flow-focusing microinjection system based on a water-in-oil emulsion, which consisted of three parts: two syringe pumps with a flow rate of 500 mL/h as the continuous phase, a syringe pump with a flow rate of 30 mL/h as the discontinuous phase, and a two-way channel. The two-way channel was fabricated by inserting a 26G needle into a silicone tube (2 mm inner diameter × 4 mm outer diameter), which was fixed with a hot-melt adhesive. A 10% (w/v) gelatin solution was used in the discontinuous phase and injected into the continuous phase via a 26G needle. Olive oil was used as the continuous phase and was injected through a silicone tube. Gelatin microspheres were formed in the channels and collected in the collecting tank. The prepared gelatin microspheres were hardened on ice for 15 min and then washed with cold acetone to remove excess oil. Thereafter, the resultant gelatin microspheres were added to 1% (v/v) glutaraldehyde at a volume ratio of 1.67 to obtain crosslinked gelatin microspheres. Finally, the crosslinked gelatin microspheres were washed several times and lyophilized for further analysis.
2.3. Preparation of PNIPAAm immobilized gelatin microspheres
GNMS were prepared by crosslinking the amine-terminated PNIPAAm to the free carboxyl groups on the gelatin microspheres using N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS), which is briefly described as follows. The crosslinked gelatin microspheres (10 g) prepared in Section 2.2 were added to a solution of EDC (1.92 g) and NHS (1.15 g) in 100 mL of 0.1 M MES buffer and stirred at 21 °C for 24 h. The amine-terminated PNIPAAm (146.7 mg) was added to the resultant solution and stirred at 21 °C for another 24 h to complete the crosslinking reaction. The GNMS were washed several times and lyophilized for further analysis.
2.4. FTIR spectrophotometry analysis
An FTIR spectrophotometer with an auto attenuated total reflection (ATR) system (Perkin Elmer) was used to identify the functional groups of the GMS and GNMS. The freeze-dried GMS and GNMS were first ground into a powder and scanned using the ATR method. Thereafter, the spectra were recorded in the wavelength range of 600–4000 cm−1 with a resolution of 8 cm−1.
2.5. Inoculation of WJ-MSCs with gelatin microspheres
WJ-MSCs were obtained from BCRC (Hsinchu, Taiwan) and grown in alpha minimum essential medium containing 20% FBS, 4 ng/mL hFGF, 100 units/mL penicillin, and 100 μg/mL streptomycin in a humidified atmosphere containing 5% CO2 maintained at 37 °C. Approximately 20 mg of GMS and GNMS was transferred to a low-attachment 24-well plate. For each well, 1 × 105 WJ-MSCs were seeded and the volume of the medium was 150 μL for the first 15 min and adjusted to 500 μL for 24 h. Following incubation of 24 h, the microspheres were removed from the new well plate and carefully washed with phosphate-buffered saline (PBS) to remove non-adherent cells. The gelatin microspheres were cultured for seven days, and the medium was changed every three days.
2.6. WJ-MSCs viability on the microspheres
The cell viability of the WJ-MSCs adhering to the GMS or GNMS was evaluated using a live/dead staining assay. On day 7 after incubation, the microspheres were gently washed twice with PBS and stained with calcein AM and propidium iodide in the dark for 15 min. The microspheres were observed under a fluorescent microscope, where the living cells and dead cells are shown in green and red, respectively, by the specific wavelength of the light excitation.
2.7. SEM examination of the microspheres
After being cultured for 1 day and 7 days, WJ-MSCs adhering to the GMS or GNMS, respectively, were observed under a scanning electron microscope (Hitachi TM-1000, Japan) at an accelerating voltage of 15 kV. The microspheres were washed twice with PBS and subsequently fixed with 4% formaldehyde for 15 min. The microspheres were then immersed step-by-step in the dehydration series, followed by a critical point dryer. Thereafter, the CPD-dried microspheres were coated with a thin gold (thickness = 5 nm) film to prepare the samples for SEM examination.
2.8. Microfilament observation of the WJ-MSCs on the microspheres
To observe the morphology and cytoskeleton of the WJ-MSCs adhered to the microspheres, F-actin staining was performed. On days 1 and 7, the cells were washed twice with PBS and then fixed with 4% formaldehyde for 15 min. Subsequently, the microspheres were stained with Alexa Fluor 488 phalloidin and Hoechst 33342 and observed under a confocal microscope, where green and blue fluorescence represented actin filaments and nuclei, respectively.
2.9. Cell harvest efficiency
After culturing for 7 days, a cell harvest experiment was performed. and GMS or GNMS were washed twice with PBS. The microspheres were then placed on ice for 10 min and subsequently treated with 0.25% trypsin at 21 °C for 5 min. The cell viability of the harvested cells was investigated using a live/dead staining assay using fluorescence microscopy. Further, the relative cell numbers were quantified and calculated using the ImageJ software. The cell harvest efficiency was determined as follows: [cell harvest efficiency (%)] = [number of harvested cells]/([number of harvested cells] + [number of residual cells on the microspheres]) × 100%.
2.10. Cell proliferation assessment
The cell proliferation ability of the WJ-MSCs adhered to the microspheres was assessed using the PrestoBlue cell viability assay. At day 0, the WJ-MSCs were inoculated with gelatin microspheres, as described in Section 2.5. On days 1, 4, and 7, the microspheres were washed with PBS and the culture medium was replaced with 10% PrestoBlue reagent in the dark for 30 min. Fluorescence was analyzed using an Enspire microplate reader (PerkinElmer, Massachusetts, USA) using a fluorescence excitation wavelength of 560 nm and fluorescence emission at 590 nm.
2.11. Gene expression of WJ-MSCs on the microspheres
At day 7 after WJ-MSCs were cultured on GMS or GNMS, the microspheres were washed with PBS and lysed with TRIzol reagent (Thermo Fisher). The supernatant was collected, and the total RNA was extracted using the Direct-zol™ RNA MiniPrep Kit (Zymo Research, USA) following the protocol established by the manufacturer. The RNA was mixed with KAPA SYBR® FAST One-Step qRT-PCR kit mixed (KAPA Biosystems, USA) and then detected via qPCR. The primers used in this analysis are listed in Supplemental Table S1. Further, the intensities were detected and recorded using a ViiA 7 real-time PCR instrument (Applied Biosystems, Foster City, CA, USA), while the mRNA levels of CD29, CD44, CD73, CD90, CD34, and CD45 were calculated using the comparative Ct method [28], where GAPDH was used as the calibrator to calculate the relative mRNA levels. Moreover, pluripotency genes, including SOX2, NANOG, REX1, and OCT4, were used to evaluate the stemness of WJ-MSCs. Relative gene expression was normalized to GAPDH gene expression and expressed as fold change relative to the control group. The WJ-MSCs cultured in the 2D culture plate served as the control group.
2.12. Statistics
The results obtained were expressed as the mean and standard deviation of at least three independent measurements. One-way analysis of variance (ANOVA) with Tukey's multiple comparisons test and student's t-test were used for all statistical evaluations. Differences were considered significant at a p-value of less than 0.05. (p < 0.05, ∗; p < 0.01, ∗∗; p < 0.001, ∗∗∗).
3. Results
3.1. FTIR functional groups analysis
The functional groups of PNIPAAm, GMS, and GNMS were analyzed using FTIR spectrophotometry (Fig. 1). The characteristic absorption bands at 1635 cm−1 and 1535 cm−1 corresponded to C O stretching and N–H bending, respectively, and were observed in both GMS and GNMS. The absorption bands at 851 cm−1 and 878 cm−1 correspond to the asymmetric and symmetric stretching vibrations of the aliphatic sulfide group on PNIPAAm, which were not observed in the GMS. In addition, the absorption band at 1170 cm−1 was attributed to the C–N stretching of amide groups; bands at 1084 cm−1 and 1036 cm−1 represent vibrations involving C–C(CH3)2 stretching of the isopropyl group from PNIPAAm. These data confirmed that PNIPAAm was successfully immobilized on the surface of the gelatin microspheres.
Fig. 1.
FTIR spectra of PNIPAAm, GMS, and GNMS.
3.2. Size measurement and SEM examination of developed gelatin microspheres
Gelatin microspheres were prepared using a power-driven flow-focusing microinjection system, as shown in Fig. 2. The freeze-dried microspheres were uniform with a diameter of 518.23 ± 27.09 μm and 522.72 ± 28.23 μm for GMS and GNMS, respectively. Further, the sizes of the microspheres were expanded to a mean value of 690.28 ± 21.07 μm and 717.32 ± 20.11 μm for GMS and GNMS respectively, owing to swelling in the medium No significant difference was observed in the morphology and bead size of the GMS and GNMS.
Fig. 2.
Morphology and microstructure of gelatin microspheres were observed via SEM and optical microscopy. (A) SEM image of GMS under 60× magnification, scale bar = 1 mm and (B) 250 magnification, scale bar = 300 μm. (C) Optical micrograph of GMS, scale bar = 400 μm. (D) SEM image of GNMS under 60× magnification, scale bar = 1 mm and (E) 250 magnification, scale bar = 300 μm. (F) Optical micrograph of GNMS, scale bar = 400 μm.
3.3. WJ-MSC viability on the microspheres
The cell viability of the WJ-MSCs on the microspheres was examined by a live/staining assay on day 7. The living cells were stained with calcein AM, whereas dead cells were stained with propidium iodide, which turned green and red, respectively. As shown in Fig. 3, more than 99% of the WJ-MSCs on the microspheres were alive on both GMS and GNMS on days 1 and 7. Moreover, the cell number on day 7 was much higher than that on day 1. It can be concluded that the developed GNMS microspheres were not cytotoxic to the WJ-MSCs and served as a good microenvironment for cell proliferation.
Fig. 3.
Evaluation of WJ-MSCs viability on the gelatin microspheres by live/dead staining on day 7. The living cells in green were stained with calcein AM, and the dead cells colored in red were stained with propidium iodide. Nuclei were stained with Hoechst 33342, which appeared blue under fluorescence microscopy. Scale bar = 500 μm.
3.4. Cell morphology and microfilament observation of the WJ-MSCs on the microspheres
The cell morphology and cytoskeleton of the WJ-MSCs on the microspheres were evaluated by SEM observation and F-actin staining, and on days 1 and 7. As shown in Fig. 4(A), the cells adhered and spread on the developed gelatin microspheres on day 1, and became much more well spread and flattened on the substrate on day 7. The microfilaments in the cytoskeleton were screened via F-actin staining (Fig. 4(B)). To investigate the distribution of the WJ-MSCs on the microspheres, F-actin stained GMS and GNMS were further reconstructed by z-stacking under confocal microscopy. The results showed that the cells could significantly proliferate on the microspheres and reach confluence after culturing for 7 days. Further, the cells were found to spread on the surface, as shown in the monolayer on the microspheres, as shown in Fig. 5.
Fig. 4.
Evaluation of WJ-MSCs morphology and cytoskeleton on the gelatin microspheres via SEM observation and F-actin staining. (A) After being cultured for 1 day and 7 days, the gelatin microspheres were fixed, series hydration, critical point dried, and observed under the SEM. (B) On day 1 and day 7, the gelatin microspheres were fixed, stained with dyes, and observed under confocal microscopy. Nuclei were stained with Hoechst 33342, and F-actin are stained with Phalloidin, which were shown in blue and green, respectively.
Fig. 5.
3D reconstruction of WJ-MSC on the gelatin microspheres by z-stacking under confocal microscopy. Nuclei were stained with Hoechst 33342, and F-actin were stained with Phalloidin, which are shown in blue and green, respectively.
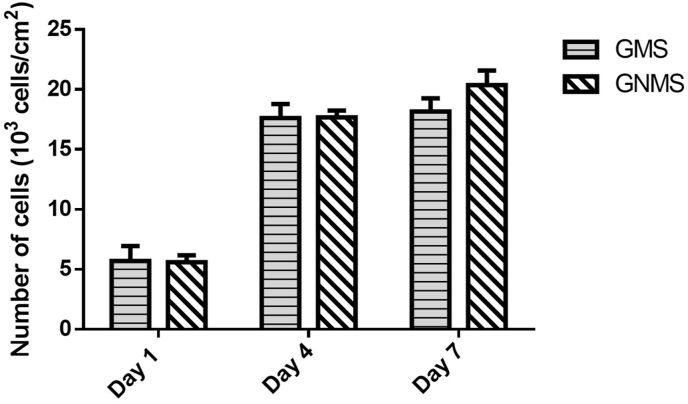
3.5. Cell proliferation of the WJ-MSC on the microspheres
The proliferation of WJ-MSCs adhered to the microspheres was determined by the PrestoBlue cell viability assay, wherein different cell numbers were measured for the standard curve, as shown in Fig. 6. After culturing for 7 days, GMS and GNMS showed approximately 3.2- and 3.6-fold cell growth from the initial cell density, respectively. Moreover, no significant difference was observed in the cell proliferation ability between GMS and GNMS, indicating that the PNIPAAm modification did not influence cell growth. This result suggested that GNMS could support the growth of WJ-MSCs and be a biocompatible cell carrier for cell expansion.
Fig. 6.
The proliferation of WJ-MSCs adhered on the gelatin microspheres as determined by Prestoblue cell viability assay on day 1, day 4, and day 7.
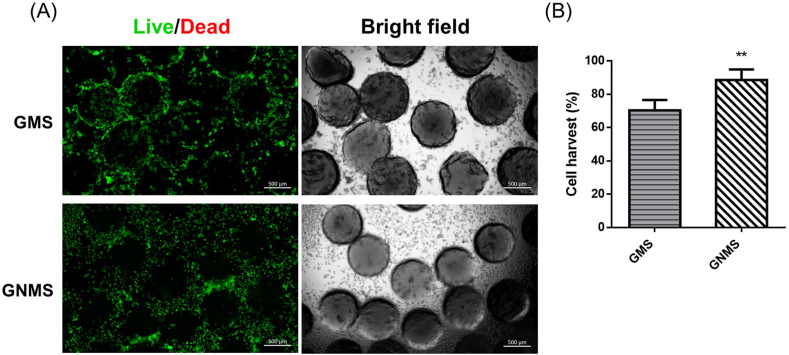
3.6. Cell harvest efficiency
The cell harvest efficiency and harvest cell viability were determined by live/dead staining and quantified using ImageJ software, as shown in Fig. 7. After culturing for 7 days, the microspheres were collected and harvested by placing them on ice for 10 min followed by trypsin for 5 min. In this protocol, almost all the cells were alive (green), and negligible number of cells were dead (red) (Fig. 7(A)). In addition, most of the cells were shown to be single-cell suspensions, with no cell aggregates being observed in the study. Quantification of the cell harvest efficiency was achieved by calculating the relative cell numbers on the microspheres and the number of detached cells in the supernatant (Fig. 7(B)). The harvesting efficiency reached nearly 90% in the GNMS group. These results indicated that the harvesting procedure could significantly increase the cell harvest efficiency in thermosensitive GNMS compared to GMS.
Fig. 7.
(A) Fluorescence images of gelatin microspheres and cell suspension after dissociation by the harvesting procedure. The cell viability of the harvested WJ-MSCs was evaluated by live/dead staining under fluorescence microscopy. The living cells were stained with calcein AM, and the dead were stained with propidium iodide, which are shown in green and red, respectively. (B) Quantification of cell harvest efficiency was determined by calculating relative cell numbers using ImageJ software. (p < 0.01, ∗∗ compared to GMS group; by student's t-test).
3.7. Gene expression of WJ-MSCs on the microspheres
To explore the effect of GMS and GNMS on the stemness properties and phenotype of WJ-MSCs, several stem cell and pluripotency markers were measured via qPCR analysis (Fig. 8). As shown in Fig. 8(A), all the positive stem cell markers, such as CD29, CD44, CD73, and CD90, were detected in WJ-MSCs at the mRNA level, whereas CD34 and CD45, the hematopoietic markers, were not detected. We further investigated classical embryonic pluripotent stem cell markers (SOX2, NANOG, REX1, and OCT4), as shown in Fig. 8(B). The results showed that the WJ-MSC culture on the 3D microspheres could express a certain degree of gene expression, which further indicated that GMS and GNMS did not change the stemness of the WJ-MSCs and maintained the cell phenotypes compared to the normal 2D cell culture method.
Fig. 8.
Gene expression of WJ-MSCs on the microspheres by Q-PCR at day 7. (A) Positive MSC markers, including CD29, CD44, CD73, CD90, and negative MSC markers, including CD34 and CD45 were used to identify the WJ-MSCs phenotype. The mRNA levels were expressed as relative levels normalized to GAPDH (defined as 1). (B) Pluripotency genes including SOX2, NANOG, REX1, OCT4 gene expression were used to evaluate the stemness of WJ-MSCs. Relative gene expression was normalized by GAPDH gene expression and expressed as fold change to the 2D culture.
4. Discussion
Gelatin is a natural polymer with good biocompatibility and biodegradability. As it can easily gel at low temperatures, it has been prepared into films, hydrogels, and beads for use in the food, pharmaceutical, and cosmetic industries [29]. More importantly, gelatin has many cell recognition domains that allow cells to spread, survive, and proliferate in gelatin-based cell carriers [30,31].
However, culturing cells with traditional cell carriers is primarily limited by the long time required for enzyme treatment to collect cells, which may affect the cell surface proteins and reduce the cell survival rates [24,32]. To overcome this shortcoming, certain researchers have successfully used external force disturbance and shear force to greatly shorten the trypsin time to 7 min [33]. Although certain researchers also used temperature-responsive materials to avoid enzyme treatment, they often require a long period of low-temperature treatment up to 2 h to achieve high collection efficiency [27].
PNIPAAm is a temperature-responsive polymer, where it shrinks from a hydrated state to a dehydrated state at a temperature higher than the LCST. PNIPAAm has been widely grafted onto the substrate and has been used as a smart material in tissue engineering and drug delivery [34,35]. In this study, the prepared gelatin microspheres were modified with a thin layer of PNIPAAm brush via amidation between the surface carboxyl groups on the gelatin microcarrier and the amine group of PNIPAAm (Fig. 1). The synthesized GNMS allows the cells to be easily collected from the microcarrier compared with the unmodified GMS, while maintaining the high survival rate of the collected cells. Thus, the developed GNMS reduced the trypsin reaction time to 5 min, which combined with the 10 min low-temperature treatment could yield a high harvest efficiency and over 99% cell survival rate (Fig. 7).
In addition, in the cell carrier, the bead size in the micron size range can reduce the diffusion rate of nutrition, oxygen, and waste, which can increase the proliferation and survival rate of the cells [17,36]. Traditionally, microcarriers are created via emulsification caused by mechanical force or shear force, but this method causes a nonhomogeneous size distribution. In contrast, other researchers formed microcarriers by applying a fixed force, including an electric field or hydrodynamic focusing, to interrupt the discontinuous phase [17,37,38]. The uniform bead size of the microspheres has the advantages of promoting cell proliferation, survival, and the surface-to-volume ratio in the homogeneous distribution with the best optimization [15,39,40]. In this study, we developed a simple power-driven flow-focusing microinjection system to prepare gelatin microspheres. A water-in-oil emulsion was formed in the silicone tube. The diameter of the prepared GNMS has a uniform bead diameter of 522.72 ± 28.23 μm, which after immersing in the medium swells to approximately 717.32 ± 20.11 μm. These were easily handled (Fig. 2), allowed cells to survive (Fig. 3), and provided a good environment to proliferate on the microspheres (Fig. 4, Fig. 5, Fig. 6).
Wharton's jelly is a gel-like connective tissue in the umbilical cord and is abundant in mesenchymal stem cells, often called WJ-MSCs. Unlike other MSCs, WJ-MSCs have an extraembryonic mesodermal origin and are considered to be more primitive and possess characteristics between embryonic and adult stem cells [41]. They can express the proteins that are considered pluripotency markers such as SOX2, NANOG, REX1, and OCT4, also known as embryonic stem cell markers [42,43]. In this study, the gene expression of pluripotency markers was slightly increased compared to that in 2D culture (Fig. 8(B)). It has been shown that 3D culture can increase pluripotent gene expression in mesenchymal stem cells [44,45]. In general, MSCs are defined as CD29+, CD44+, CD73+, CD90+, CD34−, and CD45− [46]; [47]. The WJ-MSCs on the GMS and GNMS had a phenotype consistent with the reference and showed small deviations (Fig. 8(A)). These results indicated that WJ-MSCs cultured on the developed GNMS did not change the cell phenotype, maintained their stemness, and could be a potential cell carrier for cell expansion.
3D cell culture scaffolds have been reported to promote stem cell behavior and extracellular matrix (ECM) formation. Stem cells and their niche are reciprocal in that stem cells can remodel niches and secrete ECM components [48,49]. In this study, cell-cell and cell-substrate interactions were found on the GNMS with abundant ECM formation. We believe that these cell-laden thermosensitive GNMS could be suitable carriers stem cell therapy and applications in tissue engineering. In clinical applications of regenerative medicine, the ideal cell carrier for cell expansion is based on the following criteria: (1) sufficient cell proliferation on the microcarriers, (2) ability to efficiently collect cells and maintain cell survival, and (3) lack of change in cell morphology and phenotype during culture [[50], [51], [52], [53]]. In this study, the cells were expanded 3.6-fold from the initial cell density after 7 days of culture, as shown in Fig. 6. Meanwhile, approximately 90% of the cells were collected through the thermosensitive GNMS microcarriers, with over 99% of the cells alive (Fig. 3). In addition, the results of gene expression revealed that the cells maintained their phenotype and pluripotent stemness when cultured on 3D GNMS microcarriers compared with 2D culture.
In this study, we showed that the GNMS is a promising gelatin-based cell microcarrier, which could be applied to cell expansion, stem cell therapy, and other applications in tissue engineering. The efficacy of the developed GNMS was determined by comparing it with GMS and 2D cultures. The developed GNMS can also be applied in the static or dynamic bioreactor at a 1-L scale for scale-up cell expansion to meet the clinical needs. The suspension of the medium and GNMS can easily separate by the density. The cells would be suspended in the suspension and collected by centrifugation. However, several investigations into the large-scale quantities still need to address. To begin with, the interconnected pores in the GNMS were believed not only to increase the specific surface area for cell attachment, which might enhance the cell expansion capacity, but also to facilitate the exchange of oxygen, nutrients, and waste, thereby increasing cell survival rate. The size of microcarriers and associated long-term cell survival rate should be looked into carefully and determined by controlling the needle size in a two-way channel, increasing the flow rate of the continuous phase, and the decrease the flow rate of the dispersed phase, and so on. Moreover, the fold of the cell growth is highly related to the seeding density and the dynamic cell culture, which should also be optimized to expand 7 to 10-fold as potential cell carriers. Besides, shortening the preparation time for the preparation of GNMS is also another issue for mass production and should be evaluated in the manufactory system based on pH value, temperature, the specific surface area of the materials, rotational speed of the reaction, etc.
5. Conclusion
In this study, we successfully synthesized thermosensitive PNIPAAm-immobilized gelatin microspheres using a simple power-driven flow-focusing microinjection system. The prepared microspheres had a narrow sphere size distribution. Further, the developed thermosensitive GNMS provided a good environment to allow the WJ-MSCs to spread, survive, proliferate, and maintain the cell phenotype and stemness without cytotoxicity. The immobilized PNIPAAm would allow the WJ-MSCs to be harvested more easily on the microspheres, requiring only 10 min of low-temperature treatment and 5 min of trypsin. These findings suggest that the engineered cell-laden thermosensitive GNMS can be potential cell carriers in the therapeutic application and opens new avenues for regenerative medicine.
Credit author statement
I-Hsuan Yang: Methodology, Formal analysis, Investigation, Visualization, Writing - original draft. Che-Yung Kuan: Methodology, Formal analysis, Investigation. Zhi-Yu Chen: Methodology, Validation. Chi-Han Li: Methodology, Validation. Chih-Ying Chi: Methodology, Validation. Yu-Ying Lin: Methodology. Ya-Jyun Liang: Methodology. Wei-Ting Kuo: Validation. Yi-An Li: Validation. Feng-Huei Lin: Conceptualization, Methodology, Formal analysis, Funding acquisition, Writing - review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the National Health Research Institutes (BN-110-PP-01, BN-110-GP-07, and BN-110-GP-09), and subsidized by Ministry of Science and Technology and National Taiwan University (NTU), Taiwan.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100266.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mao A.S., Mooney D.J. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112(47):14452. doi: 10.1073/pnas.1508520112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oie Y., Nishida K. BioMed Res. Int. 2013:2013. doi: 10.1155/2013/428247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatullo M., et al. Journal of tissue engineering and regenerative medicine. 2015;9(11):1205. doi: 10.1002/term.1899. [DOI] [PubMed] [Google Scholar]
- 4.Strauer B.E., Kornowski R. Circulation. 2003;107(7):929. doi: 10.1161/01.cir.0000057525.13182.24. [DOI] [PubMed] [Google Scholar]
- 5.Wang L.L.W., et al. Bioeng. Trans. Med. 2021;6(2) [Google Scholar]
- 6.El-Husseiny H.M., et al. Mater. Today Biol. 2022;13:100186. doi: 10.1016/j.mtbio.2021.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bharti D., et al. Cell Tissue Res. 2018;372(1):51. doi: 10.1007/s00441-017-2699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christodoulou I., et al. 2013. Stem Cells International; p. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nekanti U., et al. Stem Cell. Dev. 2010;19(1):117. doi: 10.1089/scd.2009.0177. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes A., et al. J. Biotechnol. 2007;132(2):227. doi: 10.1016/j.jbiotec.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 11.Moloudi R., et al. Sci. Rep. 2018;8(1):1. doi: 10.1038/s41598-018-31019-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng L., Gautrot J.E. Mater. Today Biol. 2021;12:100159. doi: 10.1016/j.mtbio.2021.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malda J., Frondoza C.G. Trends Biotechnol. 2006;24(7):299. doi: 10.1016/j.tibtech.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Muoio F., et al. Appl. Sci. 2021;11(3):925. [Google Scholar]
- 15.Seeto W.J., et al. Small. 2019;15(47):1902058. doi: 10.1002/smll.201902058. [DOI] [PubMed] [Google Scholar]
- 16.Rajzer I., et al. Mater. Sci. Eng. C. 2017;77:493. doi: 10.1016/j.msec.2017.03.306. [DOI] [PubMed] [Google Scholar]
- 17.Yang I.-H., et al. Bioact. Mater. 2021;6(6):1699. doi: 10.1016/j.bioactmat.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dosta P., et al. J. Biomed. Mater. Res. B Appl. Biomater. 2020;108(7):2937. doi: 10.1002/jbm.b.34624. [DOI] [PubMed] [Google Scholar]
- 19.Glattauer V., et al. The Japanese society for biomaterials, and the Australian society for biomaterials and the. Korean Soc. Biomater. 2010;92(4):1301. Journal of Biomedical Materials Research Part A: An Official Journal of The Society for Biomaterials. [Google Scholar]
- 20.Choi S.-W., et al. J. Mater. Chem. 2012;22(23):11442. [Google Scholar]
- 21.Gebb C., et al. Dev. Biol. Stand. 1983;55:57. [PubMed] [Google Scholar]
- 22.White L., Ades E. J. Clin. Microbiol. 1990;28(2):283. doi: 10.1128/jcm.28.2.283-286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., et al. Amb. Express. 2019;9(1):1. doi: 10.1186/s13568-019-0794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H.S., et al. Cell Transplant. 2010;19(9):1123. doi: 10.3727/096368910X516664. [DOI] [PubMed] [Google Scholar]
- 25.Nagase K., et al. Biomaterials. 2018;153:27. doi: 10.1016/j.biomaterials.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 26.Tamura A., et al. Acta Biomater. 2012;8(11):3904. doi: 10.1016/j.actbio.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Tamura A., et al. Biomaterials. 2012;33(15):3803. doi: 10.1016/j.biomaterials.2012.01.060. [DOI] [PubMed] [Google Scholar]
- 28.D'Angelo W., et al. Stem Cell Res. Ther. 2018;9(1):1. doi: 10.1186/s13287-018-1033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karim A.A., Bhat R. Trends Food Sci. Technol. 2008;19(12):644. doi: 10.1016/j.tifs.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidenko N., et al. J. Mater. Sci. Mater. Med. 2016;27(10):1. doi: 10.1007/s10856-016-5763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Afewerki S., et al. Bioeng. Trans. Med. 2019;4(1):96. doi: 10.1002/btm2.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirai H., et al. J. Biosci. Bioeng. 2002;94(4):351. doi: 10.1263/jbb.94.351. [DOI] [PubMed] [Google Scholar]
- 33.Nienow A.W., et al. Biochem. Eng. J. 2014;85:79. [Google Scholar]
- 34.Inomata H., et al. Macromolecules. 1990;23(22):4887. [Google Scholar]
- 35.Wei H., et al. Biomaterials. 2007;28(1):99. doi: 10.1016/j.biomaterials.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen L.T., et al. Sci. Rep. 2019;9(1):1. [Google Scholar]
- 37.Anna S.L., et al. Appl. Phys. Lett. 2003;82(3):364. [Google Scholar]
- 38.van der Graaf S., et al. J. Membr. Sci. 2005;251(1–2):7. [Google Scholar]
- 39.Choi S.W., et al. Small. 2010;6(14):1492. doi: 10.1002/smll.201000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi S.W., et al. Small. 2009;5(4):454. doi: 10.1002/smll.200801498. [DOI] [PubMed] [Google Scholar]
- 41.Stefańska K., et al. J. Clin. Med. 2020;9(4):1102. doi: 10.3390/jcm9041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlin R., et al. Reprod. Biol. Endocrinol. 2006;4(1):1. doi: 10.1186/1477-7827-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong C.K., et al. Cell Biol. Int. 2011;35(3):221. doi: 10.1042/CBI20100326. [DOI] [PubMed] [Google Scholar]
- 44.Cesarz Z., Tamama K. Stem Cell. Int. 2016:2016. doi: 10.1155/2016/9176357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y., et al. J. Cell Mol. Med. 2017;21(6):1073. doi: 10.1111/jcmm.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell J.B., et al. Stem cells. 2006;24(2):376. doi: 10.1634/stemcells.2005-0234. [DOI] [PubMed] [Google Scholar]
- 47.Tirpáková M., et al. Genes. 2021;12(3):431. [Google Scholar]
- 48.Guneta V., et al. J. Biomed. Mater. Res. 2016;104(5):1090. doi: 10.1002/jbm.a.35644. [DOI] [PubMed] [Google Scholar]
- 49.Gattazzo F., et al. Biochim. Biophys. Acta Gen. Subj. 2014;1840(8):2506. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eibes G., et al. J. Biotechnol. 2010;146(4):194. doi: 10.1016/j.jbiotec.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 51.Nienow A., et al. Stem Cell Manufacturing. Elsevier; 2016. Bioreactor engineering fundamentals for stem cell manufacturing; p. 43. [Google Scholar]
- 52.Rafiq Q.A., et al. Biotechnol. Lett. 2013;35(8):1233. doi: 10.1007/s10529-013-1211-9. [DOI] [PubMed] [Google Scholar]
- 53.Sun L.Y., et al. Cell Prolif. 2010;43(5):445. doi: 10.1111/j.1365-2184.2010.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.