Abstract
The severe pollution of bacterial resistance induced by the wide and even indiscriminate use of antibacterial agents has posed serious threats to human health and ecological safety. Furthermore, the combined effects of antibacterial agents have a closer relationship with the pollution of bacterial resistance than single antibacterial agent. However, there is little information regarding how multiple antibacterial agents interplay to induce bacterial resistance. Here, we developed a simple judgment method with five basic procedures for the joint action of antibacterial agents on bacterial resistance, involving toxicity determination, mutation frequency determination, conjugative transfer frequency determination, dose-response relationship fitting, key parameters obtaining, and joint resistance action judgment. This proposed approach was validated through investigating the joint resistance action between silver nanoparticle (AgNP) and 1-pyrrolidino-1-cyclohexene (1P1C, a kind of quorum sensing inhibitors). According to the procedures, the mutation unit and conjugative transfer unit for the AgNP-1P1C mixture were calculated to be 64.27 and 5.10, respectively, indicating the antagonism for their joint resistance action. This method can not only benefit the mechanistic explanation for how mixed antibacterial agents stimulate the bacterial resistance, but also guide the environmental risk assessment and clinical use of combined antibacterial agents in the related fields.
• We present a novel method to judge the joint resistance action of antibacterial agents, taking the emergence and dissemination of antibiotic resistance genes into account.
• Toxicity determination can help to design the mixtures of antibacterial agents and confirm the appropriate test concentration range of antibacterial agents used in mutation frequency and conjugative transfer frequency determination.
• The mutation unit and conjugative transfer unit were proposed according to the toxic unit in the judgment of joint toxic action.
Keywords: Bacterial resistance, Antibacterial agents, Joint action, Mutation unit, Conjugative transfer unit
Graphical abstract
Specifications table
| Subject Area: | Environmental Science |
| More specific subject area: | Environmental Toxicology |
| Method name: | Joint resistance action of antibacterial agents |
| Name and reference of original method: | Hormetic dose-responses for silver antibacterial compounds, quorum sensing inhibitors, and their binary mixtures on bacterial resistance of Escherichia coli, Shen et al. 2021 (doi: 10.1016/j.scitotenv.2021.147464) |
| Resource availability: | Non |
Method details
Background
Bacterial resistance, as a global health crisis, has aroused wide public concern over the last few decades [1]. The wide and even indiscriminate use of antibiotics and some other antibacterial compounds in medicine, animal husbandry, and family practice are considered to cause the severe pollution of bacterial resistance [2,3]. Bacteria that possess resistance to antibacterial agents make the prevention and control of infectious diseases difficult, which pose serious threats to human health and ecological safety [4]. Hence, it is necessary to investigate how antibacterial agents result in bacteria developing resistance.
One of the most important conditions for bacteria developing resistance is to obtain antibiotic resistance genes (ARGs). As emerging contaminants with environmental persistence, ARGs have been found in various environmental media, featured by high abundance and diversity, which are regarded to closely relate with the pollution of bacterial resistance [5]. Mutation and horizontal gene transfer are the main ways for bacteria acquiring ARGs, which are usually used to characterize the emergence and dissemination of ARGs [6,7]. Previous studies have observed the facilitation of many single antibacterial agents on the mutation and horizontal gene transfer, which indicated that these agents could induce bacterial resistance through triggering hormetic effects on bacteria acquiring ARGs [8,9]. For example, sulfonamides could significantly promote the mutation frequency and RP4 plasmid conjugative transfer frequency in Escherichia coli (E. coli) through acting on the SdiA protein in quorum sensing system of E. coli [8]. Furthermore, Lu et al. found that silver nanoparticle (AgNP) and triclosan at environmentally relevant concentrations could facilitate the conjugative transfer of plasmid-borne ARGs across bacterial genera via regulating the oxidative stress and cell membrane permeability in bacteria [10,11]. Currently, considerable interest has centered on hormesis-based bacterial resistance of single agent, and the potential drug resistance (whether the facilitation of mutation or horizontal gene transfer) has been a significant indicator in the environmental risk evaluation of antibacterial agent [12,13]. However, there is little information regarding how multiple antibacterial agents interplay to induce the emergence and dissemination of ARGs. Antibacterial agents usually co-exist in environment [14], thus the pollution of bacterial resistance probably relates to the joint selection pressure of multiple antibacterial agents on the bacterial acquisition of ARGs. Furthermore, the co-existence of antibacterial agents may change the resistance risk of single agent. Therefore, it is of importance to judge the joint action of antibacterial agents on bacterial resistance, which will benefit the environmental risk assessment of mixed antibacterial agents and the investigation of bacterial resistance pollution.
Here, bacterial mutation and conjugative transfer (as the main way of horizontal gene transfer) are, respectively used to characterize the emergence and dissemination of ARGs [15], [16], [17]. Through obtaining key parameters in the dose-response of antibacterial agents on the mutation frequency and conjugative transfer frequency, the method for judging joint action of antibacterial agents on bacterial resistance was established according to the judgment of joint toxic action. This method included five basic procedures, and the specific steps in each procedure were described and explained in detail. Furthermore, each procedure was validated using the judgement of the joint resistance action for AgNP and quorum sensing inhibitor (QSI) as a case study.
Method details: the five basic procedures (Fig. 1)
Fig. 1.
Schematic representation of the five basic procedures in the method for judging joint resistance action of antibacterial agents.
Procedure 1: toxicity determination
The single toxicity of each test antibacterial agent was investigated to obtain the toxicity parameters, which were used to design the mixture of antibacterial agents. Setting E. coli MG1655 K-12 (purchased from Biovector Co., Ltd. (Bejing, China)) as the model bacteria, the dose-response relationship of single antibacterial agent to the growth of E. coli was determined using microplate toxicity analysis.
Step 1: A preculture of E. coli was grown at 37 °C with 180 rpm shaking for 6 h (logarithmic growth period) in LB broth media (containing 1% (w/v) tryptone, 0.5% (w/v) yeast extract, and 1% (w/v) NaCl with the pH adjusted to 7.0–7.2), which was then diluted about 105 times and used as an inoculum (approximately 5 × 104 cell mL−1) in the following toxicity test. Tryptone was obtained from Sinopharm Chemical Reagent Company (Shanghai, China). Yeast extract (super pure) and NaCl (AR, purity 99.5%) were bought from Aladdin Industrial Corporation (Shanghai, China).
Step 2: Serial dilutions of the test antibacterial agent were set through fixing equal log dose interval between adjacent concentrations (usually 12–16 concentration points).
Note: The lowest and largest concentration of test agent should be set based on the result in the preliminary experiment, which guarantee that the dose-response relationship covers the no effect (0) and the maximum inhibition.
Step 3: 80 μL of test antibacterial agent, 80 μL of MH broth media (composed of 3% (w/v) beef infusion, 1.75% (w/v) casein hydrolysate, and 0.15% (w/v) water-soluble starch with the pH adjusted to 7.2–7.4), and 40 μL of the E. coli inoculum were added into each well of a transparent 100-well (or 96-well) microplate in order (total 200 μL for the treatment group). Meanwhile, the control group was obtained through substituting the 80 μL of test agent in the above 200 μL mixed solution with 80 μL of 1% (w/v) NaCl solution. All treatment and control groups had three or five parallels that independently carried out. Beef infusion, casein hydrolysate (approx. 70% free amino acids), and water-soluble starch (AR) were all purchased from Solarbio Life Sciences co., Ltd. (Beijing, China).
Step 4: The OD600 (optical density at 600 nm) of each well was determined on a microplate reader (Bioscreen C MBR, Growth Curves USA, Piscataway, NJ, USA) after the microplate was incubated at 37 °C with 180 rpm shaking for 24 h.
- Step 5: The toxicity of test antibacterial agent to E. coli growth was expressed as the Inhibition (%) of OD600 value (Eq. (1)):
(1)
where OD600,c and OD600,t were the average OD600 values of the control and treatment groups, respectively. Then, the dose-response relationship of agent concentration to the corresponding inhibition of E. coli growth was obtained, and the standard deviation was used to represent for the error bar.
Procedure 2: mutation frequency determination
Step 1: The above microplate toxicity analysis in toxicity determination step was repeated in mutation test.
Step 2: The 200 μL solution in three or five paralleled wells for each treatment or control group were collected into a 1.5 mL centrifuge tube. E. coli in this collected solution was washed thrice with 1% (w/v) NaCl solution and then suspended in the same saline solution.
Note: The process of washing E. coli included centrifuging for 3 min with 6000 rpm, removing the supernatant, shaking for 5 s using a vortex mixer, and adding 1 mL 1% (w/v) NaCl solution.
Step 3: Some of the E. coli suspension (usually at most 10 μL) was diluted about 106 times with 1% (w/v) NaCl solution, which was used to determine the number of total E. coli in the suspension. Some of the E. coli suspension (usually the rest solution) was concentrated about 3 times, which was used to determine the number of mutant E. coli in the suspension.
Step 4: The diluted suspension and concentrated suspension were, respectively incubated on antibiotic-free and rifampicin-added (25 mg L−1) solid plate media (LB broth with 1.5–2% agar) for 12–15 h, and the number of total E. coli and mutant E. coli were obtained by counting the colony-forming units per milliliter of culture (CFU mL−1) in these two kinds of solid plates. Rifampicin (purity 97%) was obtained from Aladdin Industrial Corporation (Shanghai, China). Agar was bought from Sinopharm Chemical Reagent Company (Shanghai, China).
Note: In Step 3 and 4, the diluted and concentrated ratio of suspension, the concentration of rifampicin, and the culture period of solid plate are not fixed, which could be adjusted according to the density of CFU in the preliminary experiment (convenient and clear to count).
- Step 5: The mutation frequency (MF) of E. coli in treatment and control groups were all obtained via calculating the ratio of CFU value for mutant E. coli to CFU value for total E. coli (Eq. (2)):
(2)
where CFUmE and CFUtE, respectively referred to the average CFU values of mutant E. coli and total E. coli in each group. There were at least two duplicates for each group. The influence of test antibacterial agent on the mutation frequency of E. coli was expressed as the Promotion (%) of MF value (Eq. (3)):
| (3) |
where MFt and MFc, respectively indicated the average MF values of the treatment and control groups. Then, the dose-response relationship of agent concentration to the corresponding promotion of MF value was obtained, and the standard deviation was used to represent for the error bar.
Procedure 3: conjugative transfer frequency determination
RP4 plasmid conjugative transfer between E. coli (RP4) strain and E. coli (Nal) strain was used as the research model for investigating the conjugative transfer of ARGs. The E. coli (RP4) strain that harbored the RP4 plasmid (carrying the ampicillin, kanamycin, and tetracycline resistance genes) was used as the donor, and the E. coli (Nal) strain that was resistant to nalidixic acid was used as the recipient.
Step 1:E. coli (RP4) and E. coli (Nal) were cultured at 37 °C with 180 rpm shaking for 6 h to logarithmic growth period in LB broth media that containing corresponding antibiotics.
Step 2: The cultured E. coli (RP4) and E. coli (Nal) were washed thrice with 1% (w/v) NaCl solution, the OD600 value of which were then both adjusted to 0.5 with the same saline solution.
Note: The process of washing E. coli (RP4) and E. coli (Nal) included centrifuging for 3 min with 6000 rpm, removing the supernatant, shaking for 5 s using a vortex mixer, and adding 1 mL 1% (w/v) NaCl solution.
Step 3:E. coli (RP4) and E. coli (Nal) were both shaken for 20 min with 180 rpm, and then mixed at a ratio of 1:2 to obtain a mixed inoculum. After 180 rpm shaking for 20 min, the final inoculum (approximately 5 × 106 cell mL−1) was obtained. Serial dilutions of the test antibacterial agent were set through fixing equal log dose interval between adjacent concentrations (usually 10–12 concentration points).
Note: The lowest and largest concentration of test agent should be set based on the result in the preliminary experiment, which guarantee that the dose-response relationship exhibits inverted U shape and the lowest concentration of test agent has no effect (0).
Step 4: 40 μL of test antibacterial agent, 100 μL of 2-fold LB broth media, and 60 μL of the inoculum for mixed bacteria were added into a 1.5 mL centrifuge tube in order (total 200 μL for the treatment group). Meanwhile, the control group was obtained through substituting the 40 μL of test agent in the above 200 μL mixed solution with 40 μL of 1% (w/v) NaCl solution.
Step 5: After a 12 h mating period at 37 °C, the suspension in the centrifuge tube was diluted about 105 times with 1% (w/v) NaCl solution. The diluted suspension was then incubated on both solid plate media (LB broth with 1.5–2% agar) containing 40 mg mL−1 of nalidixic acid and solid plate media (LB broth with 1.5–2% agar) containing 40 mg mL−1 of nalidixic acid and 40 mg mL−1 of kanamycin, and the number of recipients and transconjugants were measured by counting CFU mL−1 in these two kinds of solid plates. Nalidixic acid (purity 98%) and kanamycin (purity 98%) were both purchased from Aladdin Industrial Corporation (Shanghai, China).
Note: The diluted ratio of suspension and the concentration of nalidixic acid and kanamycin are not fixed, which could be adjusted according to the density of CFU in the preliminary experiment (convenient and clear to count).
- Step 6: The conjugative transfer frequency (CTF) of RP4 plasmid in treatment and control groups were all obtained via calculating the ratio of CFU value for transconjugants to CFU value for recipients (Eq. (4)):
(4)
where CFUt and CFUr, respectively represented the average CFU values of transconjugants and recipients. There were at least two duplicates for each group. The influence of test antibacterial agent on the conjugative transfer frequency of RP4 plasmid was expressed as the Promotion (%) of CTF value (Eq. (5)):
| (5) |
where CTFt and CTFc, respectively referred to the average CTF values of the treatment and control groups. Then, the dose-response relationship of agent concentration to the corresponding promotion of CTF value was obtained, and the standard deviation was used to represent for the error bar.
Procedure 4: data fitting, key parameters obtaining, and antibacterial mixture designing
- Step 1: The toxic dose-response relationship of single antibacterial agent to the growth of E. coli was fitted via two kinds of regression function: (a) the built-in DoseResp regression function in Origin software (version 86, OriginLab Corporation, Northampton, MA, USA) was used when there was no hormetic effect in the dose-response relationship; (b) the artificially constructed Hormesis regression function in Origin software was used when there was a hormetic effect in the dose-response relationship. The fitting equation for Hormesis regression function (Eq. (6)) was as follows [18]:
(6)
where x was the concentration of test antibacterial agent, and y was the corresponding inhibitory effect; C and D represented the inhibitory rates corresponding to the top asymptote and the bottom asymptote, respectively; m referred to the stimulatory rate corresponding to the stimulation asymptote; a and b indicated the first median and slope in the stimulatory region, respectively; p and q were the median and slope in the inhibitory region, respectively. In the mutation and conjugative transfer test, the observed dose-response data of single antibacterial agents were fitted through the inverted Hormesis regression function.
Note: The Hormesis regression function and its inverted one should be input into Origin software by users, and complied via the program with the corresponding parameters precisely defined. When conducting the fitting process, a certain value should be artificially given to each parameter in the Hormesis regression function based on the actual dose-response relationship of single agent.
Step 2: According to the fitted toxic dose-response curve of single antibacterial agent to the growth of E. coli, the median effective concentration of each agent (EC50) was obtained to characterize its toxicity. The mixture of antibacterial agents used in mutation and conjugative transfer test was designed based on the equitoxic ratio of EC50 for each single agent [19]. Serial dilutions of the mixed antibacterial agents were set through fixing equal log dose interval between adjacent concentrations.
Note: The design of antibacterial mixture is not fixed, and the concentration of each agent and their mixing ratio could be adjusted based on actual needs. The lowest and largest concentration of mixed agents should be set based on the result in the preliminary experiment, which guarantee that the dose-response relationship exhibits inverted U shape and the lowest concentration of mixed agents has no effect (0).
Step 3: Then, the impacts of mixed antibacterial agents on mutation frequency and conjugative transfer frequency of RP4 plasmid in E. coli were determined using the same method as mentioned in Procedure 1–3. The corresponding dose-response data were fitted through the inverted Hormesis regression function.
Note: When conducting the fitting process, a certain value should be artificially given to each parameter in the inverted Hormesis regression function based on the actual dose-response relationship of mixed agents.
Step 4: To characterize the facilitation of single and mixed antibacterial agents on the mutation and conjugative transfer frequency, some key parameters should be selected in the corresponding fitted dose-response curves. Here, the initial (minimum) concentrations that single and mixed agents began to promote the mutation and conjugative transfer frequency were set as the key parameters, which could be regarded as the risk thresholds in inducing the emergence or dissemination of ARGs. Compared with the concentration that provoked initial 0% or 5% promotion, the concentration that provoked initial 10% promotion in the actual regression curve was more appropriate to be the risk threshold because it was easier to observe and accurately determine. Thus, the concentrations that induced initial 10% promotion on the mutation and conjugative transfer frequency in the regression curves, i.e., MC10 and CTC10, were obtained to feature the facilitation of single and mixed antibacterial agents on bacterial resistance.
Procedure 5: judgement of joint resistance action
- Step 1: Toxic unit (TU) method has been widely used to judge the joint toxic action among the components in the mixed antibacterial agents, in which the TU value is calculated via the EC50 of the components and the concentrations of components when the mixture is at EC50 [20]. According to TU method, the joint resistance action of test antibacterial agents was judged via calculating the mutation unit (MU) and the conjugative transfer unit (CTU), and the corresponding Eqs. (7) and (8) were as follows:
(7) (8)
where ci and ci′ represented the concentrations of component i when the mixture of antibacterial agents were at the concentrations of MC10 and CTC10 (MC10mix and CTC10mix), respectively; MC10i and CTC10i referred to the MC10 and CTC10 values of component i when acting alone, respectively. Based on the 10% promotion effect and the fixed ratio of agents in the mixture, these values were obtained from the dose-response curves of single and mixed agents to mutation and conjugative transfer frequency.
Step 2: In TU method, the joint toxic action of antibacterial agents could be judged to be antagonism, addition, or synergism via comparing TU value with 0.8 and 1.2 [21,22]. Similarly, the joint resistance action of antibacterial agents could be judged as follows: when MU (CTU) < 0.8, the joint action of components on the promotion of mutation frequency (conjugative transfer frequency) was synergism; when 0.8 ≤ MU (CTU) ≤ 1.2, the joint action of components on the promotion of mutation frequency (conjugative transfer frequency) was addition; and when MU (CTU) > 1.2, the joint action of components on the promotion of mutation frequency (conjugative transfer frequency) was antagonism.
Method validation
As the possible antibiotic alternatives, AgNP and QSI have the great potential in preventing and treating microbial infections [23,24]. Over the past two decades, AgNP and QSI have been used in medicine, hygiene, cosmetics, and breeding industry [25,26]. Due to the possible coexistence of these two antibacterial agents in the environment, it is necessary to explore how AgNP and QSI interplay on the bacterial resistance, i.e., to judge their joint resistance action, which will benefit evaluating the resistance risk for the combined exposure of AgNP and QSI.
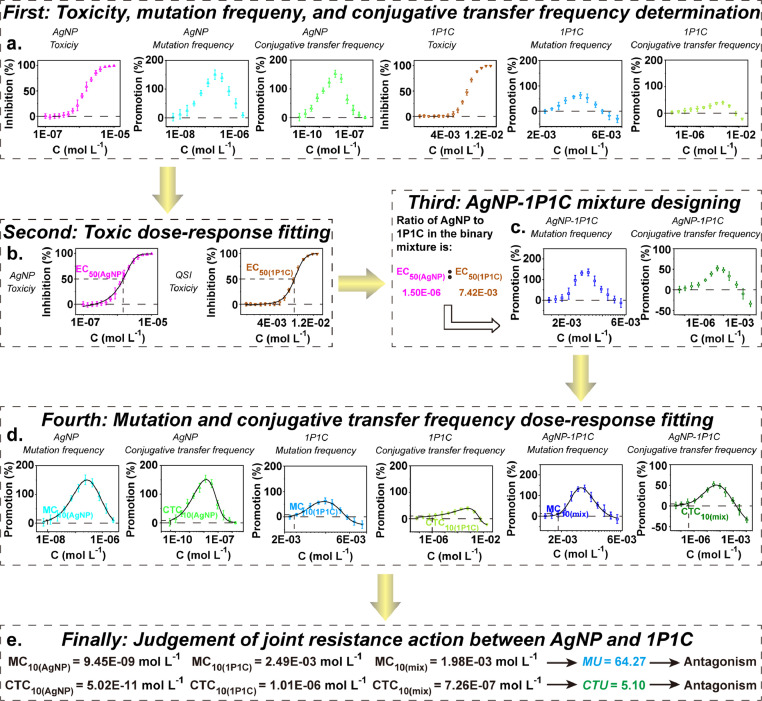
The joint resistance action of AgNP and QSI was judged using the above method (the data originated from Shen et al. 2021), and the five basic procedures were validated (Fig. 2) [15]. The test AgNP (5 nm) was purchased from Huzheng Nano Technology Co., Ltd (Shanghai, China). 1-pyrrolidino-1-cyclohexene (1P1C), as a typical QSI, which was all obtained from Sigma-Aldrich Co., Ltd. (St. Louis, MO, USA) which without repurification (purity ≥ 99%). First, the influence of single test AgNP and 1P1C on the growth, mutation frequency, and RP4 plasmid conjugative transfer frequency of E. coli were investigated according to Procedure 1–3, and the results were depicted in Fig. 2a. Second, because there was no hormetic phenomenon in the dose-response relationship of each test agent to E. coli growth, the DoseResp regression function was applied to fit these dose-response data (Procedure 4), as shown in Fig. 2b. It was obtained from these fitted curves that the EC50 values of AgNP and 1P1C were 1.50E-06 and 7.42E-03 mol L−1, respectively. Third, the binary mixture of AgNP and 1P1C was prepared, in which the ratio of component AgNP to 1P1C was equal to EC50(AgNP): EC50(QSI) (Procedure 4), and the impact of AgNP-1P1C mixture on the mutation frequency and RP4 plasmid conjugative transfer frequency was determined, and the results were displayed in Fig. 2c. Fourth, the observed dose-response data of AgNP, 1P1C, and their binary mixture on the mutation frequency and RP4 plasmid conjugative transfer frequency were fitted through the inverted Hormesis regression function (depicted in Fig. 2d). As described in Procedure 4, MC10 and CTC10 values for AgNP, 1P1C, and their binary mixture were obtained from the corresponding regression curves. Finally, the MU and CTU values for the binary mixture of AgNP and 1P1C were calculated on the basis of the equations in Procedure 5, as shown in Fig. 2e. It was obvious that the MU value (64.27) and CTU value (5.10) were all larger than 1.2, indicating that the joint resistance action between AgNP and 1P1C exhibited antagonistic effect. The results implied that although AgNP and QSI could facilitate the emergence and dissemination of ARGs, the coexistence of AgNP and QSI may largely mitigate the resistance risk induced by the single agent.
Fig. 2.
The judgment of joint resistance action of AgNP and QSI using the developed method. Source: Agathokleous et al. 2021 [13].
Conclusion
This study introduced five procedures for evaluating the joint resistance action of binary or multicomponent antibacterial agents under the laboratorial level. This method can help to fast assess the potential resistance risk for the mixture of antibacterial agents through experimental operation and mathematical calculation, which can be used in environmentology, toxicology, pharmacology, biology, and medicine. In addition, this method can also guide the mechanistic explanation for the facilitation of mixed antibacterial agents on bacterial resistance, the environmental risk assessment and clinical use of combined antibacterial agents.
Acknowledgments
We thank and Dr. Hongyan Shen (Hebei University of Science and Technology, China) for technical help with the equations of joint mutation and conjugative transfer action judgment. This work was supported by the National Natural Science Foundation of China (22006116 and 42107416).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Fair R.J., Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agyare C., Boamah V.E., Zumbi C.N., Osei F.B. Antibiotic use in poultry production and its effects on bacterial resistance. Antimicrob. Resist. Glob. Threat. 2018;3:33–50. doi: 10.5772/intechopen.79371. Chapter. [DOI] [Google Scholar]
- 3.Breijyeh Z., Jubeh B., Karaman R. Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules. 2020;25:1340. doi: 10.3390/molecules25061340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nat. News. 2017;543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 5.Martínez J.L. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 6.Martinez J.L., Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 2000;44:1771–1777. doi: 10.1128/AAC.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jutkina J., Rutgersson C., Flach C.F., Larsson D.G.J. An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance. Sci. Total Environ. 2016;548–549:131–138. doi: 10.1016/j.scitotenv.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 8.Sun H., Chen R., Jiang W., Chen X., Lin Z. QSAR-based investigation on antibiotics facilitating emergence and dissemination of antibiotic resistance genes: a case study of sulfonamides against mutation and conjugative transfer in Escherichia coli. Environ. Res. 2019;173:87–96. doi: 10.1016/j.envres.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Ma Q., Su B., Chen R., Lin J., Lin Z., Wang D., Yu Y. A study on the role that quorum sensing play in antibiotic-resistant plasmid conjugative transfer in Escherichia coli. Ecotoxicology. 2018;27:209–216. doi: 10.1007/s10646-017-1886-0. [DOI] [PubMed] [Google Scholar]
- 10.Lu J., Wang Y., Jin M., Yuan Z., Bond P., Guo J. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 2020;169 doi: 10.1016/j.watres.2019.115229. [DOI] [PubMed] [Google Scholar]
- 11.Lu J., Wang Y., Li J., Mao L., Nguyen S.H., Duarte T., Coin L., Bond P., Yuan Z., Guo J. Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environ. Int. 2018;121:1217–1226. doi: 10.1016/j.envint.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 12.Iavicoli I., Fontana L., Agathokleous E., Santocono C., Russo F., Vetrani I., Fedele M., Calabrese E.J. Hormetic dose responses induced by antibiotics in bacteria: a phantom menace to be thoroughly evaluated to address the environmental risk and tackle the antibiotic resistance phenomenon. Sci. Total Environ. 2021;798 doi: 10.1016/j.scitotenv.2021.149255. [DOI] [PubMed] [Google Scholar]
- 13.Agathokleous E., Wang Q., Iavicoli I., Calabrese E.J. The relevance of hormesis at higher levels of biological organization: hormesis in microorganisms. Curr. Opin. Toxicol. 2021;29:1–9. doi: 10.1016/j.cotox.2021.11.001. [DOI] [Google Scholar]
- 14.Kümmerer K. Antibiotics in the aquatic environment–a review–part I. Chemosphere. 2009;75:417–434. doi: 10.1016/j.chemosphere.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 15.Shen H., Liu Y., Liu Y., Duan Z., Wu P., Lin Z., Sun H. Hormetic dose-responses for silver antibacterial compounds, quorum sensing inhibitors, and their binary mixtures on bacterial resistance of Escherichia coli. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147464. [DOI] [PubMed] [Google Scholar]
- 16.Li X., Wang Y., Liu Y., Lin Z., Wang D., Sun H. Resistance risk induced by quorum sensing inhibitors and their combined use with antibiotics: mechanism and its relationship with toxicity. Chemosphere. 2021;265 doi: 10.1016/j.chemosphere.2020.129153. [DOI] [PubMed] [Google Scholar]
- 17.Li X., Shi J., Sun H., Lin Z. Hormetic dose-dependent response about typical antibiotics and their mixtures on plasmid conjugative transfer of Escherichia coli and its relationship with toxic effects on growth. Ecotoxicol. Environ. Saf. 2020;205 doi: 10.1016/j.ecoenv.2020.111300. [DOI] [PubMed] [Google Scholar]
- 18.Deng Z., Lin Z., Zou X., Yao Z., Tian D., Wang D., Yin D. Model of hormesis and its toxicity mechanism based on quorum sensing: a case study on the toxicity of sulfonamides to Photobacterium phosphoreum. Environ. Sci. Technol. 2012;46:7746–7754. doi: 10.1021/es203490f. [DOI] [PubMed] [Google Scholar]
- 19.Sun H., Pan Y., Gu Y., Lin Z. Mechanistic explanation of time-dependent cross-phenomenon based on quorum sensing: a case study of the mixture of sulfonamide and quorum sensing inhibitor to bioluminescence of Aliivibrio fischeri. Sci. Total Environ. 2018;630:11–19. doi: 10.1016/j.scitotenv.2018.02.153. [DOI] [PubMed] [Google Scholar]
- 20.Bundschuh M., Goedkoop W., Kreuger J. Evaluation of pesticide monitoring strategies in agricultural streams based on the toxic-unit concept – experiences from long-term measurements. Sci. Total Environ. 2014;484:84–91. doi: 10.1016/j.scitotenv.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Sprague J.B., Ramsay B.A. Lethal levels of mixed copper – zinc solutions for juvenile salmon. J. Fisheries Board Can. 1965;22:425–432. doi: 10.1139/f65-042. [DOI] [Google Scholar]
- 22.Long X., Wang D., Lin Z., Qin M., Song C., Liu Y. The mixture toxicity of environmental contaminants containing sulfonamides and other antibiotics in Escherichia coli: differences in both the special target proteins of individual chemicals and their effective combined concentration. Chemosphere. 2016;158:193–203. doi: 10.1016/j.chemosphere.2016.05.048. [DOI] [PubMed] [Google Scholar]
- 23.Hamad A., Khashan K.S., Hadi A. Silver nanoparticles and silver ions as potential antibacterial agents. J. Inorg. Organomet. Polym. Mater. 2020;30:4811–4828. doi: 10.1007/s10904-020-01744-x. [DOI] [Google Scholar]
- 24.Rémy B., Mion S., Plener L., Elias M., Chabrière E., Daudé D. Interference in bacterial quorum sensing: a biopharmaceutical perspective. Front. Pharmacol. 2018;9:203. doi: 10.3389/fphar.2018.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahamed M., AlSalhi M.S., Siddiqui M.K.J. Silver nanoparticle applications and human health. Clin. Chim. Acta. 2010;411:1841–1848. doi: 10.1016/j.cca.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Kumar P., Patel S.K.S., Lee J.K., Kalia V.C. Extending the limits of Bacillus for novel biotechnological applications. Biotechnol. Adv. 2013;31:1543–1561. doi: 10.1016/j.biotechadv.2013.08.007. [DOI] [PubMed] [Google Scholar]