Abstract
The lymph node (LN) is a vital organ of the lymphatic and immune system that enables timely detection, response, and clearance of harmful substances from the body. Each LN comprises of distinct substructures, which host a plethora of immune cell types working in tandem to coordinate complex innate and adaptive immune responses. An improved understanding of LN biology could facilitate treatment in LN-associated pathologies and immunotherapeutic interventions, yet at present, animal models, which often have poor physiological relevance, are the most popular experimental platforms. Emerging biomaterial engineering offers powerful alternatives, with the potential to circumvent limitations of animal models, for in-depth characterization and engineering of the lymphatic and adaptive immune system. In addition, mathematical and computational approaches, particularly in the current age of big data research, are reliable tools to verify and complement biomaterial works. In this review, we first discuss the importance of lymph node in immunity protection followed by recent advances using biomaterials to create in vitro/vivo LN-mimicking models to recreate the lymphoid tissue microstructure and microenvironment, as well as to describe the related immuno-functionality for biological investigation. We also explore the great potential of mathematical and computational models to serve as in silico supports. Furthermore, we suggest how both in vitro/vivo and in silico approaches can be integrated to strengthen basic patho-biological research, translational drug screening and clinical personalized therapies. We hope that this review will promote synergistic collaborations to accelerate progress of LN-mimicking systems to enhance understanding of immuno-complexity.
Keywords: Lymph node, Biomaterials, Computational models, Lymphatic system, Immunotherapy
Abbreviations: ABM, agent-based model; APC, antigen-presenting cell; BV, blood vessel; CPM, Cellular Potts model; DC, dendritic cell; ECM, extracellular matrix; FDC, follicular dendritic cell; FRC, fibroblastic reticular cell; LEC, lymphatic endothelial cell; LN, lymph node; LV, lymphatic vessel; ODE, ordinary differential equation; PDE, partial differential equation; PDMS, polydimethylsiloxane
Graphical abstract
1. Introduction
The lymph node (LN) is a pea-sized (0.1–2.5 cm long), bean-shaped organ of the lymphatic and immune system. A human possesses approximately 500–600 LNs, with clusters in the neck, chest, and abdomen, that are connected by a network of lymphatic vessels (LVs). The lymph is filtered through the LNs, which remove some pathogens and serve as the meeting site for antigen-presenting cells (APCs) and intra-LN immune cells [[1], [2], [3]]. Here, with sufficient APC-interaction and stimulation, lymphocytes (T/B cells) undergo activation and proliferation, to produce immune cells that can defend against foreign pathogens. This response is an important part of our adaptive immune response. LN-associated diseases such as lymphadenopathy, lymphedema, and lymphoma substantially compromise the immune defense system. Some of these diseases occur with high incidences and inflict long-lasting effects but are often neglected in the medical field. For instance, lymphedema, a swelling (edema) of limbs due to impaired lymphatic transport, is an undesired side effect of cancer therapeutic intervention, such as radiotherapy and LN dissection [4]. There is a high prevalence of cancer-associated lymphedema (∼83%) following complete inguinal LN dissection during melanoma treatment [4], and ∼60% of the cases of lymphedema in patients with gynecological cancers are persistent [5]. Currently, there are limited treatment options for these LN-associated diseases including a lack of proven drugs to efficiently alleviate lymphedema. Management instead primarily consists of physiotherapeutic interventions such as decongestive lymphatic therapy (stimulating lymphatic drainage manually through massages) and compression therapy (wearing compression garments) [6]. The LN may also play an important role in general cancer progression, as it has been demonstrated that tumor cells, like melanoma cells, often gain useful adaptations in the draining LNs, before metastasizing to distant organs [7]. These examples point to LNs as critical organs for maintaining health. Hence, there is an urgent necessity to conduct more in-depth studies on LN biology and the associated disease pathogenesis, to facilitate treatment development.
Recent advances in bioimaging techniques, such as cross-sectional imaging, functional imaging, tissue clearing, and interstitial contrast agent injection [[8], [9], [10]], have vastly improved understanding of LN anatomy and physiology. However, these tools allow observation of the histological appearances without enabling further verification of the biological hypotheses, which LN-mimicking models can provide. Traditionally, the study of LN function and associated diseases is performed using animal models [[11], [12], [13]], but this approach has several limitations including a lack of relevant immune system to humans, physiological differences, high costs, intensive labor, complex operations, low reproducibility, and ethical considerations preventing eligibility for selected studies [14]. For instance, in cancer research, less than 8% of treatments are successfully translated to clinical trials, even though they exhibit significant therapeutic effects on animal models [15]. It is also difficult to control the physiological parameters of animal models [16]. There is thus an urgent need for physiologically relevant LN models, which can mimic the architecture and function of healthy and diseased LN tissues without the limitations associated with animal models. LN mimicking-models can provide convenient and controllable platforms to investigate complex cell-cell/tissue interactions and assess the efficacy of proposed treatments.
Recently, several biomaterial-based models such as hydrogel, organoids and microfluidic devices have been developed to mimic LN architectures and immune functions. The entire LN or a specific functional area can be established on these synthetic models, with tailored modifications to achieve specific scientific aims [17]. Compared to current animal models, biomaterial-based models offer controllable bio-chemical and -physical features as inputs and easier access to specific outputs. With rapid progress in computer technology, mathematical and computational models are now better able to simulate sophisticated cell/tissue/organ activities in controlled physiological LN micro-environment and can offer massive amounts of data to support in vitro/vivo biomaterial works [18].
In this review, we first provide an overview on the anatomy, physiology, and role of LN in the immune system. This is followed by a summary on the utility of biomaterials and computational approaches to create LN-mimicking models. Next, we have an in-depth review of recent progress made in LN-mimicking models with biomaterial engineering, describing aspects of LN biology including LN structural features and functions, immune cell dynamics, lymph flow dynamics, as well as LN-associated disease and treatments. The potential roles and supports of mathematical and computational models are also explored, particularly in the research era of big data. Finally, we propose ways in which these existing in vitro/vivo and in silico systems can be integrated and exploited to serve as a powerful alternative to animal models while complementing each other to enhance basic, translational, and clinical studies. Besides providing a comprehensive review on latest LN-mimicking biomaterial models to explore LN bio-pathology, to the best of our knowledge, this is also the first study summarizing LN models spanning the fields of biomaterials and computational science. We are hopeful that this review will demonstrate the impact of integrative LN systems to spur exciting developments.
2. Lymph node biology
The lymphatic system plays key roles in mediating fluid homeostasis, lipid absorption, and immunity, and is comprised of primary and secondary lymphoid organs and a network of LVs (Fig. 1A) [19]. The primary organs (i.e., bone marrow and thymus) create immune cells called lymphocytes, and the secondary organs (i.e., LNs, spleen, tonsils, and specialized mucosal tissue) provide sites of immune response. The LVs collect and transport lymph, from the tissues, via the LNs, to the lymphatic or thoracic ducts located at right and left shoulder height respectively, where the fluid is returned to the blood circulation. The lymph is composed primarily of macromolecule-rich interstitial fluid that has extravasated from blood vessels (BVs), but also contains circulating APCs and lymphocytes. Lymphatic transport plays a critical role in the maintenance of fluid balance in the body, and by filtering lymph through LNs, also allows detection and immune response to harmful pathogens. The LNs contain a variety of stromal cells and macrophages that can fight infection by capturing and destroying pathogen directly [2]. They are also key sites for monitoring antigenic inflammation and inducing adaptive immunity after lymph-borne APCs and antigens/pathogens such as bacteria, viruses, and even tumor cells, drain into LNs and induce lymphocyte responses.
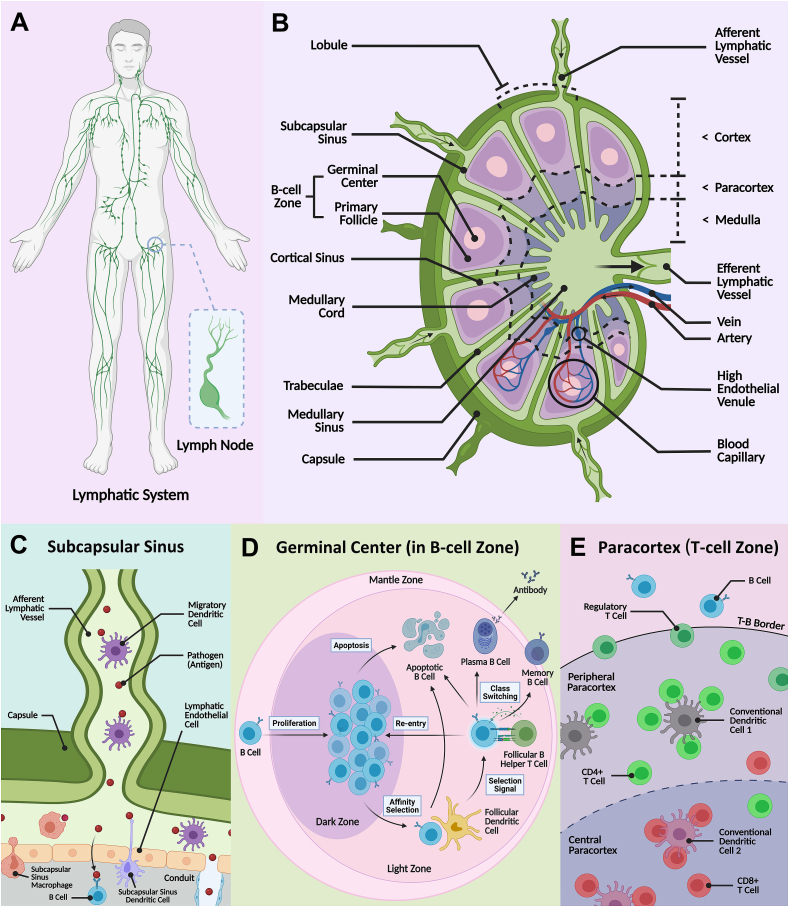
Fig. 1.
Schematic illustration of the lymphatic vessel network and the lymph node architecture with specific lymphocytes. A) Sampling of the contents of the lymphatic fluid within the lymph nodes is a crucial component in initiating immune response. B) The lymph node is mainly divided into three areas: cortex, paracortex, and medulla, and various immune cells reside in distinct microzones to orchestrate the adaptive immune system. C) Pathogens transported in the lymph are deposited via afferent vessels into the subcapsular sinus [21]. Free antigen may be phagocytosed by subcapsular sinus macrophages or drain into the cortex, containing the D) germinal center (in ‘B-cell zone’) and E) an inner paracortex (or ‘T-cell zone’). The dendritic cells also enter the cortex in a chemokine/cytokine-mediated manner. B cells and T cells can be activated by antigen-presenting cells (e.g., the migrating or resident dendritic cells that scavenge free antigen) and clonally expand and differentiate in the germinal center (within the B-cell zone) and T-cell zone, respectively. Effector T cells, and antibody-secreting plasma cells are produced that enter the medulla and exit the LN to defend against foreign pathogens.
As shown in Fig. 1B, the LN is typically comprised of multiple lymphoid lobules that are structurally supported by a three-dimensional (3D) reticular meshwork of specialized and spatially-characterized fibroblastic reticular cells (FRCs) and their secreted reticular fibers [[20], [21], [22]]. Each lymphoid lobule is served by a single afferent LV and surrounded by lymph-filled sinuses. The lobule is divided into a cortex region that contains the B follicles, where B cells reside, an inner paracortex, where T cells reside, and a medullary region at the base of the lobule. The stromal cells of the LN (FRCs and endothelial cells) secrete chemokines that guide incoming immune cells which express the corresponding receptors to the relevant compartments [23]. Lymph enters the LN through the afferent LVs, which deposit fluid into the subcapsular sinuses that cap the lobules. From here, approximately 90% of the fluid takes a peripheral path around the lobule, directly into the medullary sinuses and cords [24]. The remaining fluid percolates into the cortex and through the paracortex before being absorbed into paracortical blood vessels, or draining to medullary sinuses around the base of the lobule and into the efferent LV that leaves the LN.
The contents transported in the lymph take varying paths. The endothelial cells on the floor of the subcapsular sinuses form a continuous layer that excludes large particles (>70 kDa) from entering the LN cortex. This forces the particles to remain in the peripheral pathway, where larger pathogens can be phagocytosed by the macrophages in the sub-capsular or medullary sinuses, or shunted to the next LN (Fig. 1C) [21]. Macrophages can also capture and present lymph-born antigens to incoming immune cells. Other APCs such as dendritic cells (DCs), enter the LN in the afferent lymph, and cross the subcapsular sinus into the cortex and underlying paracortex in a chemokine-mediated manner [25]. Small free antigen can drain directly into the cortex but also enter conduits that fast track their journey to specialized blood vessels within the paracortex [26,27]. The LN is highly vascularized with blood capillaries branching from the hilus of the LN to form a dense network within the lobules, enabling efficient nutrient and oxygen delivery to cells. The naïve lymphocytes (B/T cells) migrate into the LN, from blood circulation, via specialized blood vessels in the paracortex called high endothelial venules, and return to the blood via the efferent LVs [28]. Most DCs remain trapped in LN parenchyma and undergo apoptosis within the LN after fulfilling their antigen-presentation role [29]. A small number of DCs have been detected in efferent lymph, but their origin remains unknown [30].
Chemokines play an important role in the localization of the immune cells to the correct compartments of the LN. Expression of CCL21-scavenging atypical chemokine receptor ACKR4 on endothelial cells of the subcapsular sinus ceiling, allows formation of a CCL21 gradient, to guide deposited DCs, which express complementary receptor CCR7, to the subcapsular sinus floor and cortex border [26]. Within the paracortex, FRCs secrete CCL21 and CCL19, another ligand for the CCR7 receptor. This allows chemotactic guidance of the DCs that cross the subcapsular sinus floor, and the naïve T cells that enter from paracortical blood vessels that also express CCR7 [23]. Stromal cell expression of CXCL13 by marginal reticular cells and follicular dendritic cells (FDCs) within the B follicles allows chemotactic localization of B cells, which express corresponding receptor, CXCR5 [31,32]. Further roles involving CCR8, CXCR4 and CCR3 are the subject of in-depth reviews [33].
Coordination of immune cell localization is crucial as it is within the cortex and paracortex that lymphocytes can interact with APCs and/or free antigen, to induce the adaptive immune response to defend against foreign pathogens [34,35]. Antigen recognition occurs through crosstalk between T cell or B cell receptors and peptide-MHC presented, and approximately 1 in 1 × 104–106 lymphocytes will express the complementary receptor [36]. B cells reside in primary follicles (B-cell zones), that are interspersed with a FDC network that organizes the B cells into well-defined follicles (Fig. 1D). The FDCs are also the predominant APCs that capture and present antigen to B cells, to induce B cell activation and the formation of a germinal center (secondary follicle) [37]. Activated B cells migrate to the follicle border where they present antigen to a T cell subset called T helper cells, and receive co-stimulation, before returning to the germinal center. Here, B cells can undergo clonal expansion, and differentiate into short-lived high-affinity antibody-secreting plasma cells and memory B cells, further supported in the process by the FDCs, that provide survival and affinity maturation signals [38]. Both plasma and memory cells exit the LN in efferent lymph and re-enter the blood circulation. When encountering the antigen source, the plasma cells secrete antibodies that neutralize or flag the pathogen for destruction. Longer-lasting memory B cells circulate to enable rapid response in future encounters with the same antigen. After entry, T cells primarily home to the paracortical area (T-cell zone) (Fig. 1E). Here they may encounter their cognate antigen presented by antigen-bearing migratory DCs that enter in afferent lymph, or LN resident DCs that capture free-antigen [39]. There are several subsets of T cells, with the majority composed of cytotoxic T cells (expressing co-receptor CD8+), that will directly attack pathogens, and helper T cells (expressing co-receptor CD4+), that help to activate both B cells and CD8+ T cells [40]. If T cells are sufficiently stimulated, they undergo activation, differentiation and clonal expansion to produce effector T cells [41]. These cells exit the LN in efferent lymph, enter blood circulation and migrate to peripheral tissues where they can generate vast numbers of effector cytokines upon antigen challenge. Circulating memory CD4+ and CD8+ T cells are also produced. Regulatory T cells (CD4+ CD25+) are an additional subtype of suppressive T cells involving in homeostasis maintenance and self-tolerance. Regulatory T cells can suppress effector T cell activities to prevent autoimmunity and have also recently been identified as key factors in tumor progression, thereby becoming a hot research target in development of new-generation cancer therapy [[42], [43], [44]].
The LN is a complex organ, with distinct regions and sub-structures adapted to perform different functions, that plays a crucial role in orchestrating a coordinated immune response. Understanding LN function is key to be able to manipulate immune response including to enhance vaccine design and safety, develop cancer immunotherapy and prevent auto-immune response, such as in graft failure following organ transplant [45]. However, limited understanding of the sophisticated LN architecture, cell interactions and functions continue to impede development in these areas. For example, use of biomolecule-induced immunotherapies, that aim to induce T/B cell activation in the cortex, is hindered by the fact that the majority of lymph fluid travels around the cortex, therefore new strategies to direct biomolecules to LN substructures are needed [46]. Furthermore, in clinical trials, after injection of primed DCs designed to act as anti-tumor T cell-activating vaccines, patient survival rate remained relatively low, despite prior observation of T cell expansion and differentiation in pre-clinical animal models. In humans, although T cell activation occurred in the draining LNs, the antigen-loaded DCs may not effectively sustain the effector phase of the T cell immune response [47]. It is therefore invaluable to develop physiologically relevant LN-mimicking models by integrating technologies in biomaterial engineering with mathematical and computational approaches.
3. Advanced biomaterial approaches for mimicking and studying lymph node
Designing LN-mimicking models will allow reconstruction of the lymphatic architecture to gain a better understanding of LN biology and assist the development of targeted immunotherapy. Currently, animal models such as mice and pigs are widely adopted in LN experiments [11,12]. To better simulate the LN microenvironment, decellularized animal-derived LNs have been developed to engineer LN-like scaffolds that preserve intrinsic biological cues and architectural structure, and maintain immunological function [48]. These immune cell-loaded decellularized LN scaffolds have been found to induce both in vitro cytokine production and in vivo antitumor immune responses [49]. However, animal LN-centered models are not ideal candidates for clinical research and application due to the wide physiological differences of lymphoid organs and the immune system between humans and animals [50]. Taking laboratory mice as an example, despite many similarities, there remain important differences between mouse and human LNs, such as immune cell composition (e.g., proportion of lymphocytes) [51], gene expression [52], LN geometry (e.g., size) and LN microstructures (e.g., lymphatic vasculature system) [52]. The use of animal platforms is accompanied by ethical complications and generally require high cost, time, and effort for maintenance. In recent years, technological advancements have led to a rise in alternative artificial biomaterial-based in vitro models for LN tissue engineering. These include the use of hydrogel scaffolds, organoid systems, and microfluidic devices (i.e., LN-on-a-chip). These artificial models can be modified by altering chemical composition and structure/architecture of biomaterials to obtain appropriate experimental subjects [17]. Importantly, in a biomaterial model, it is feasible to isolate and study a single factor in the complex biosystem and focus research on a specific pathway within an organ or body part, which provides us with new avenues for LN tissue engineering that is difficult to achieve with animal models,. Table 1 presents a comparison of the various models and their features including biophysical properties, physiological relevance, and application prospects.
Table 1.
Comparisons of different lymph node-mimicking models.
| Dish culture | Animal model | Hydrogel | Organoid | Microfluidic device | |
|---|---|---|---|---|---|
| Spatial feature | 2D | 3D | 3D | 3D | 3D |
| Cost | Low | High | Low | Medium | Medium |
| Operation time | Short | Long | Short | Medium | Medium |
| Scalability | High | Medium | Medium | Medium | Medium |
| Organ-level function | Absent | High | Low | Medium | High |
| Physiological relevance | Low | High | Medium | Medium | High |
| Structural variability | Low | Low | High | High | Medium |
| Cell-cell/tissue communication | Low | High | Low | Medium | High |
| Dynamic flow | Absent | High | Absent | Absent | High |
| Real-time read-out | High | Low | Medium | Medium | High |
| Environment control | High | low | Medium | Medium | High |
| Portability | Medium | Hard | Medium | Medium | Easy |
| Ethical issuea | Possible | Yes | Possible | Possible | Possible |
Ethical considerations of cell experiment are largely dependent on the source of cells.
Hydrogel is a type of soft material with interpenetrating 3D natural/synthetic networks swollen in water [47,53,54]. As hydrogels can be programmed to exhibit nature biomechanical and biochemical properties similar to natural tissues and organs, they have therefore, been extensively used to model human organs for drug development and screening [[55], [56], [57]]. Its 3D architecture enables hydrogel to offer more physiologically relevant conditions than alternative 2D/planar substrates, and also enables tuning of biochemical and biophysical parameters (e.g., mechanical stiffness) to better mimic the microenvironment and regulate cell behaviors and interactions [58]. Currently, a variety of hydrogel-based LN-mimicking platforms have been created to study LN patho-biology such as LN regeneration and immune cell behaviors [[59], [60], [61], [62]].
An organoid is a miniaturized version of any human tissue or organ in vitro that models the architecture and function of the corresponding organ, and is another key biomaterial model for LN engineering. Encapsulated tissues and cell clusters can form 3D organ-like structure via a self-organizing process [63,64]. Compared to hydrogel-based scaffolds, organoids provide a more comprehensive multicellular system that can incorporate organ-specific cell types and appropriate matrices for cell interaction/function, allowing better insight by mimicking whole organ physiology and dysfunction in vitro. Compared to animal models, in vitro organoid model may offer more convenient and informative read-outs of drug tests and mechanistic insight [59]. Furthermore, organoids often resemble the phase of early embryonic development of tissues and organs [[65], [66], [67]]. Therefore, creating an ex vivo LN organoid is a promising approach to allow study of the process and crucial factors of adaptive immune response, particularly in early tissue development. This provides unique opportunities for the study of LN-associated immune development, lymphoid diseases and their corresponding treatments. In the future, immune-functionalized organoids may be transplanted into the human body to replace or repair damaged lymphoid organs (e.g., lymphadenectomy and lymphodepletion) [59].
Organ-on-a-chip models exploit the remarkable developments in microfluidic techniques to offer new human tissue/organ in vitro models for drug screening and disease modeling [68,69]. An organ-on-a-chip is a novel micro-scale system, that mimics human organ function as well as microenvironment, and consists of various microstructures with live cells/tissues [70]. Compared to the static organ-mimicking models (e.g., hydrogel scaffold and organoid-based system), organ-on-a-chip devices can better simulate the actual LN physiological microenvironment and related immune function under input and output flow, more accurately mimicking the dynamic physiological conditions in vivo. Some key parameters of the LN system can also be realized in this 3D dynamic model, such as continuous fluid flow, shear force (i.e., dynamic mechanical stress), concentration gradients, drug/molecule diffusion, high cell density and activity, and cell-cell/tissue interfaces [59,70,71]. Recently, a library of LN-on-a-chip models have been reported to recapitulate several specific functional zones and pathological processes in the LN [59,60]. Incorporation of the minimal functional unit of a part of, or a whole LN, and their dynamics on a small portable chip, allows us to conveniently investigate the targeted bioprocesses. The microfluidic device combined with live LN tissues and cells can be utilized to capture the LN dynamic signaling events and understand how LN-associated diseases occur and metastasize. Microfluidic platforms can enable crosstalk between separate modules through dynamic fluid flow, therefore they can better replicate the sophisticated LN microzones and intricate interactions among different immune cells, compared to organoid or hydrogel models [59]. Features like regulation of recirculating flow rate and external mechanical force/stress allow fine environmental control and study of any one specific factor/variable within the model. Control of these factors allows for precise result analysis through direct visualized read-outs with the help of macroscopic imaging equipment [72,73]. Unlike animal models or traditional in vitro models, microfluidic platforms can be designed to enable real-time read-outs and continuous measurements without disturbance. The monitoring probes (e.g., integrated biosensors) and intelligent control system (e.g., fluid flow control) make it possible for long-period experiments and data collection with fully automated fashion [74].
In summary, biomaterial-based models provide a reliable platform to simulate LN architecture and conduct patho-biological research while mitigating some of the problems associated with use of animal models. Nevertheless, limitations of LN-mimicking biomaterial models continue to exist, which include limited matrix similarity (i.e., artificial extracellular matrix (ECM) generally cannot represent the actual lymphoid tissue matrix), absence of microarchitecture and vasculature, as well as lack of inter-organ communication and homeostasis [63,75,76]. As such, significant efforts have been invested to alleviate existing limitations. For example, to better study the interplay between LN immune cells and lymphoid matrix, ECM-derived macromolecules such as collagen and hyaluronic acid could be used. These materials replace the traditional synthetic polymers which do not represent the actual matrix due to lacking specific peptide sequences that are helpful for cell growth. Based on the traditional synthetic hydrogels, some research groups incorporated a series of biofunctional peptide sequences into hydrogels, including cell-adhesion sequence (e.g., RGD [77,78]), specific peptide for cell growth (e.g., laminin sequence IKVAV [79,80]) and matrix metalloproteinases-sensitive sequences (e.g., Ac-GCRD-GDQGIAGF-DRCG [81]) to mimic native ECM while keeping costs affordable (generally ECM macromolecules such as collagen are relatively high-cost) [82]. Furthermore, integration of the LN microfluidic model with hydrogels and organoids, in particular with other organ models on a single chip (i.e., body or human-on-a-chip), can allow for more accurate studies of the immune response. For instance, to improve the low cellular fidelity of biochips and the lack of dynamic control, such as input/output fluid flow (e.g., vasculature system), within organoids, a good strategy is to synergize organoids with biochips. We call these organoid-on-a-chip models, with the aim of offsetting the drawbacks of each individual approach [83].
Novel adjustments and improvements to existing biomaterial related techniques are also improving the development of LN-mimicking models. The fabrication of complex LN microstructures (e.g., capillary-like structures) in hydrogel models is challenging to replicate using traditional microfabrication technologies. However, new methods like polydimethylsiloxane (PDMS) micromolding template enable hydrogel microfabrication [84], which has already been achieved on other complex organ/tissue constructs such as polyethylene glycol (PEG)-based hydrogel microvascular networks [85]. There are also emerging 3D printing approaches including inkjet, extrusion-based and light-based technologies, as well as the layer-by-layer high-precision construction, to pattern cell-laden biomaterial model with precise control over their composition, spatial distribution and architecture [86,87]. This provides great opportunity to achieve complex LN structures and composition of immune cells in one model. To improve the use of 3D printing and bioinks in tissue regeneration [88], nanogels encapsulating cell clusters can be integrated into 3D printed gel models. Compared to the traditional bioinks, the nanogel can prevent unexpected cell behaviors (e.g., aggregation, dispersion, and sedimentation), and fabricate 3D models with higher precision and less cell damages during the printing process [89,90]. Laser ablation (e.g., multi-photon ablation) and electrospun techniques are also conducive to fabricate sophisticated LN biomaterial models as well as improved reproducibility and larger-scale applications [91].
4. Mathematical and computational models as support tools for biomaterial studies
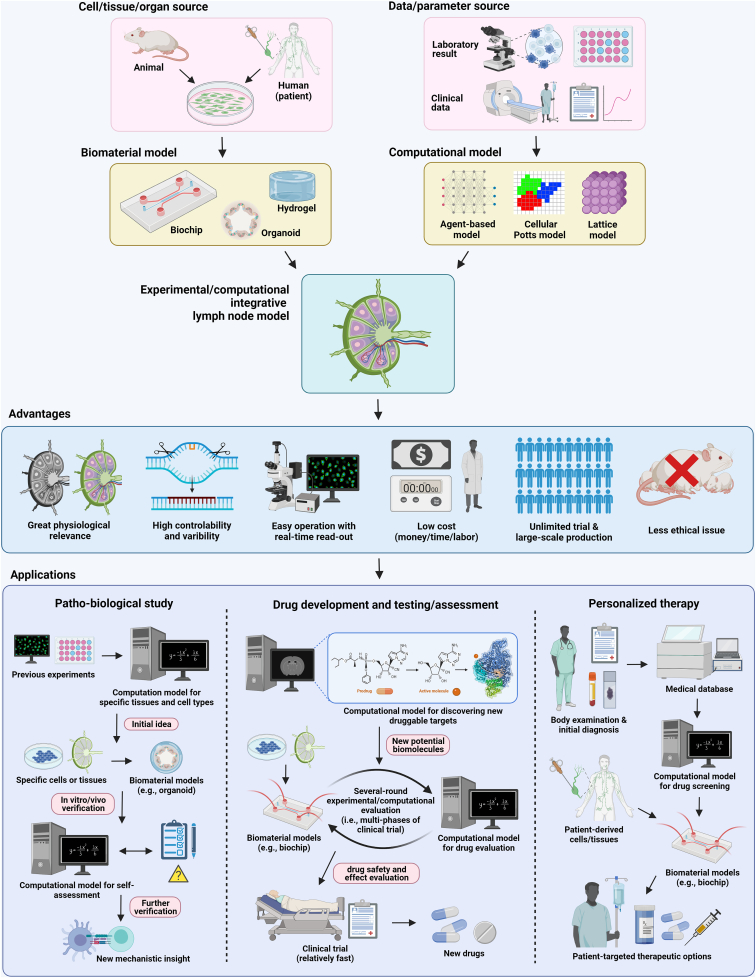
Mathematical and computational models can complement deficiencies of current biomaterial models. The in silico models can theoretically study complex biological systems, complement experimental data, and focus future work. In many published biomaterial-related works, computational models are widely adopted to analyze and verify the results of in vitro experiments and aid in standardization. Even when the mechanisms are not fully elucidated, or when new hypotheses remain difficult to test experimentally, in silico models can be built using assumptions and constraints to explore and explain phenomena [92]. Unlike in vitro/vivo experiments, researchers can perturbate parameters of interest without inadvertently interfering with other biological pathways in the system. A range of network or regulatory connections can also be easily manipulated. Furthermore, the number of trials using in silico methods are not limited by the number of animals available, or ethical considerations, increasing the feasibility of high numbers of repeated trials and often allowing a broader range of experiments at less cost. Besides biological studies, mathematical and computational tools are largely adopted for biomaterial model design to optimize physical/chemical properties, assess therapeutic effect, as well as predict potential bio-application.
The first mathematical model of the lymphatic system, to the best of our knowledge, was established by Reddy in 1974, consisting of a one-dimensional discrete model using Navier-Stokes equations of fluid mechanics [93]. Nowadays, with the development of new research approaches (e.g., algorithm application, computer simulation, and 3D bioimaging), a greater variety of mathematical and computational models describing the lymphatic system and lymphoid organs have been built. Currently, models based on systems of differential equations, agent-based models (ABMs), and hybrid multiscale models are typically applied to study and simulate tissue/cell-level processes (summarized in Table 2) [18]. Behaviors such as growth and decay, signal integration and receptor expression are easily described using ordinary differential equations (ODEs), but the geometry of the system is not accounted for. Incorporating more computationally expensive partial differential equations (PDEs) can improve spatial description, by describing diffusion and chemotaxis. Lattice based models, such as Cellular Potts models (CPMs) or ABMs, where the modelling environment is divided into discreet grid compartments, allow an extensive description of the environment, while hybrid multiscale models combine different model types to best describe different aspects of the system. Based on these, a large aspects of LN biology including LN structure and geometry, immune cell dynamics and immune response, immune cell chemotaxis, lymph flow dynamics, as well as LN-associated disease could be described on mathematical and computational models (summarized in Table 3), serving as an outstanding supplemental tool for biomaterial studies.
Table 2.
Primary mathematical approaches and computational models for simulating biological systems.
| Method/Modela | Definition | Application/Simulation |
|---|---|---|
| Agent-based model (ABM)b | Model contains autonomous decision-making entities termed agents. Each agent makes behavioral decisions individually based on pre-defined probabilistic rules considering agent internal state, surrounding agents, and the environment. | (1) Simulate many interactions at the individual level to uncover emergent behavior at the whole-population level; (2) Stochastic gene expression [94]; (3) Tissue formation and morphogenesis [95]; (4) Mammary stem cell subpopulation dynamics [96]; (5) Inflammation pathways [97,98]; (6) Immune system dynamics [99,100]; (7) Tumor models [101]; (8) Cell migration [102,103]; (9) Chemotaxis [104,105] |
| Boolean network | A discrete set of boolean variables which can be presented by a graph of linked nodes | (1) Gene regulatory networks [[106], [107], [108]]; (2) Interaction between pathogens and different cytokines [109] |
| Cellular automatonc | A discrete spatio-temporally extended dynamical system that includes identically programmed automaton cells and interacting units with a finite number of discrete states. | (1) 3D multicellular tissue growth [110]; (2) Cell behaviors and activities (e.g., neurons and fibroblasts); (3) Cardiac model [111]; (4) Tissue growth [112] |
| Cellular Potts model (CPM) | A lattice-based model where a cell can be described using a cluster of points, allowing capture of cell shape changes. The movement of the points/cluster around the lattice is governed by calculation of force equations and energy of the system therefore considering cell state and the grid environment. | (1) Simulate individual and collective cell behavior [[113], [114], [115], [116]]; (2) Tissue morphogenesis [113,117]; (3) Cancer development [118]; (4) Chemotaxis [114]; (5) Vasculogenesis and angiogenesis [119] |
| Differential equation modeld | Model containing differential equations that relates functions and their derivatives, to describe the dynamic aspects of biosystems. Inclusion of partial-differential equations can include some spatially dependent characteristics such as diffusion. | (1) Cancer diseases biology [120,121]; (2) Immunology and immunotherapy [[122], [123], [124], [125], [126], [127], [128]]; (3) Virus infection [129]; (4) Pharmacodynamics [130]; (5) Organ disease [131]; (6) Vascular network [132] |
| Hybrid multiscale model | Integrative model combining several kinds of computational models. | (1) Vascular network [[133], [134], [135]]; (2) Cancer and therapy [113,[136], [137], [138], [139]]; (3) Disease model [140]; (4) Immune system (cell and organ) [141] |
| Lattice modele | Model established on a 2D/3D lattice (or grid), as opposed to the continuum of spatial or spatio-temporal coordinates (off-lattice model). | (1) Predict protein structure [142]; (2) Tissue differentiation [143]; (3) Cell migration [102,116]; (4) Tumor growth [144] |
| Petri net formalism | Directed bipartite graph with two types of elements- places and transitions, to describe discrete-event dynamical systems. | (1) System biology [[145], [146], [147]]; (2) Gene network [148] |
The open source toolkits of computational approaches can be found in Ref. [149].
Agent-based model is also called individual-based model in some fields [150].
To some extent, cellular automaton model is an agent-based model with finite grids and limited degrees of freedom.
Ordinary differential equation and partial differential equation models are the main differential model types used in the biological study.
A large number of agent-based models and Cellular-Potts models are established on lattice models.
Table 3.
Mathematical and computational models for lymph node architectures, features, functions, and diseases.
| OBJECT | CONTENT | REFERENCE |
|---|---|---|
| LYMPH NODE STRUCTURE AND GEOMETRY | Reticular network | [141,169,[263], [264], [265]] |
| Microvascular network and conduit system | [168,170] | |
| B-cell follicle | [266] | |
| Cortical sinus | [267] | |
| Whole lymph node geometry | [171] | |
| Body-wide lymphatic system | [172] | |
| Subcapsular sinus | [171] | |
| IMMUNE CELL DYNAMICS AND IMMUNE RESPONSE | T cell motility and dynamics | [103,116,188,[190], [191], [192],196,268,269] |
| T cell activation and proliferation | [188,[270], [271], [272]] | |
| Dendritic cell-T cell interaction (immune response) | [202,271,[273], [274], [275], [276], [277], [278], [279]] | |
| Stromal cell-immune cell interaction (fibroblastic reticular cells) | [104,141,169,278,280] | |
| B cell proliferation and activation | [281] | |
| IMMUNE CELL CHEMOTAXIS | T cell motility | [104] |
| T cell-dendritic cell contact | [105,202,277] | |
| Tumor metastasis | [282,283] | |
| Chemokine gradient formation | [201] | |
| Stromal cell/fibroblastic reticular cell network | [141] | |
| LYMPH FLOW DYNAMICS | Intra-lymph node flow | [24,201,214,215] |
| Lymphatic vessel flow (lymphangion) | [[217], [218], [219], [220], [221], [222], [223],271] | |
| lymphocyte recirculation | [215] | |
| LYMPH NODE-ASSOCIATED DISEASE | Tumor metastasis through lymph node | [[284], [285], [286]] |
| Tumor-immune interplay in lymph node | [139,287,288] | |
| Lymphoma | [255,256] | |
| Lymphedema | [260,271,289] | |
| Lymph node degeneracy | [261] | |
| Infection | [262,263,290] |
5. Recent progress on lymph node-mimicking models
Designing LN-mimicking models that reconstruct the lymphatic architecture and biology is important to enable better development of targeted immunotherapy. In recent years, production of lymphatic-associated biomaterial models has significantly increased, with some designs having already completed the bench-to-business process and become commercialized in the tissue regeneration and drug discovery industry [151]. This largely advances our understanding of LN as well as lymphatic immune system, and promotes the development of immunotherapy. Plus, with rapid progress in computer technology, mathematical and computational models are now better able to simulate sophisticated cell/tissue/organ activities in controlled physiological LN micro-environment and offer massive amounts of data to support biomaterial in vitro/vivo works [18]. In this section, we will mainly focus on biomaterial engineering describing aspects of LN biology including LN structural feature and function, immune cell dynamics, lymph flow dynamics, as well as LN-associated disease and treatment. Mathematical and computational approaches are discussed (summarized in Table 3), to explore how they can support and supplement the biomaterial LN-mimicking models.
5.1. Lymph node structural feature and function
The structure of LN is complex, comprised of multiple lymphoid lobules and separated into microzones (Fig. 1). The geometrical organization of the LN is essential for effective immune responses [61], necessitating the building of in vitro models that account for structural features to investigate many key LN functions. Currently, animal models offer some feasible models to study LN architecture, however, the physiological differences of lymphoid organs and the immune system between humans and animals typically make them inappropriate for further clinical research and application [50]. In addition, it is often difficult to carry out complex experimental processes that target specific small microzones. Biomaterial models are great alternatives to simulate LN architecture in vitro.
Researchers have mimicked various LN macro/microstructures using biomaterial models, to learn how LN physiological conditions affect lymphatic cell behaviors. For example, a biochip that replicated the T-cell zone in the LN paracortex was fabricated to study the T cell/DC interaction and physiological conditions (Fig. 2A) [152]. Researchers observed different attachment and detachment behaviors of T cells to DCs under varying flow shear stresses, which could potentially impact the effect of antigen presentation and corresponding immune responses. To highlight the importance of interstitial flow to T-cell zone fibroblastic reticular cells (TRCs), Tomei et al. developed a platform that demonstrated how fluid flow facilitated TRCs organization in the matrix (Fig. 2B) [153]. The B-cell follicles are additional critical zones for LN adaptive immune responses [64]. Using continuous perfusion of fluid, Goyal and colleagues assembled T cells and B cells to form a germinal center-like lymphoid follicle on the microfluidic organ chip (Fig. 2C) [154]. The follicle exhibited several functions including plasma cells induction, immunoglobulin class switching, and cytokine production. This microfluidic device presents as a valuable tool to assess vaccine responses and the effects of immunotherapies in vitro, especially in the early phase of adaptive immune response.
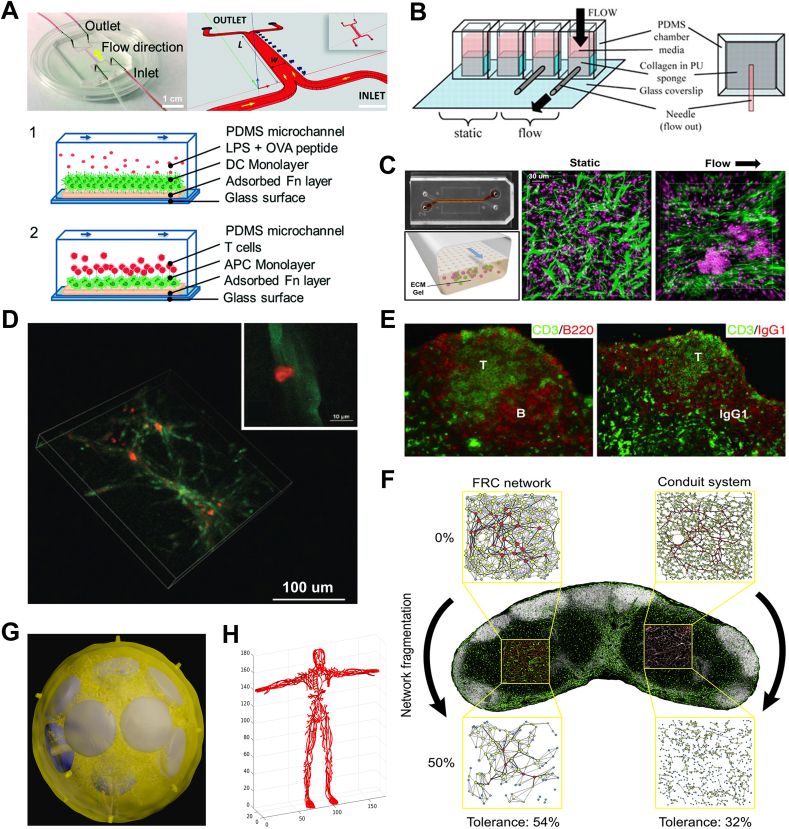
Fig. 2.
Biomaterial models and computational tools for simulating lymph node architectures and functions. The lymph node functional zones recapitulated on microfluidic devices including A) paracortex with T cell/dendritic cell interaction (Reproduced with permission [152]. Copyright 2016, The Royal Society of Chemistry), B) paracortex with T-cell zone fibroblastic reticular cells (Reproduced with permission [153]. Copyright 2009, The American Association of Immunologists, Inc.), and C) B-cell follicle (Reproduced with permission [154]. Copyright 2022, Wiley-VCH). D) Confocal images showing T cell (red) interaction and attachment with stromal network (green). Reproduced with permission [164]. Copyright 2015, Wiley-VCH. E) Clearly-segregated T-/B-cell zones and antigen-specific secondary IgG responses are observed in transplanted lymphoid organoid in NP-OVA-preimmunized BALB/c mice. Reproduced with permission [166]. Copyright 2007, American Society for Clinical Investigation. F) Topological structure and tolerance of FRC network and conduit system using small-world network models. Reproduced with permission [170]. Copyright 2020, Elsevier. G) 3D model of the whole lymph node geometry using confocal images and a FRC lattice network model. Reproduced with permission [171]. Copyright 2015, MDPI. H) Body-wide lymphatic network graph model. Reproduced with permission [172]. Copyright 2018, MDPI. (FRC: fibroblastic reticular cell).
Another advantage of a biomaterial model is that, through tuning macromolecular chemical features and material physical properties, the matrix profile can be programmed to desired properties [155,156]. This is useful for experiments which require specific tissue features. For example, in some studies of aging, scientists are able to programme ‘old’ matrix profiles on models by tailoring material biochemical and biomechanical properties, such as stiffening the matrix and reducing its porous structures [157,158]. Although possible to replicate aging tissue with animal models, this would be extremely expensive and time-consuming as several years of raising animal would be needed. Furthermore, by modulating the size and shape of the biomaterial, it is possible to better test conservative treatments (e.g., manual compression devices), which is difficult to achieve with animal models because of the physical differences between animals and humans. Laco et al. engineered collagen/nanofiber hydrogel composites to study topographical cues for growth, migration, elongation, and vessel formation of human lymphatic microvascular endothelial cells around LN [159]. This findings suggest a possibility of connecting implanted or grafted lymphoid organs onto existing LV network as a form of therapy. Additionally, to realize regeneration of LNs and ambient LVs, some hydrogels have been developed to understand critical factors affecting LN regeneration and this has been reviewed elsewhere [156].
Besides enabling the creation of tailored microstructures and matrix properties, biomaterial models also allow regulation of cell types in specific zones, which is hard to achieve with traditional animal models. For example, stromal cells, like FRCs and lymphatic endothelial cells (LECs), can be easily included in LN models to investigate T cell activity. The stromal network provides structural basis to the lymphoid organs, but recent studies have also shown that stromal network interaction with lymphocytes could greatly contribute to the construction of different microzones, affecting immune reactions [160], such as determining the type of immune response [161] and silencing self-reactive CD8+ T cells [162,163]. As such, Kim et al. encapsulated stromal cells in a hydrogel scaffold to remodel the stromal network and LN microenvironment [164]. After T cells were added to the newly formed stromal networks (designed to replicate the T-cell zone environment), the presence of T cell migration and interaction with the stromal network was verified through confocal microscopy (Fig. 2D). The replication of in vivo behavior indicated that the described stromal cell-contained hydrogel model could help to identify cellular factors regulating T cell behaviors. Suematsu and Watanabe also constructed LN-like organoids by incorporating stromal cells into collagenous scaffolds before transplanting the material into mice [165,166]. Separated B-cell and T-cell zones, endothelial venule-like vessels, germinal centers, and FDCs networks were clearly observed in the organoid model. After transplanting the organoid into an immunocompromised animal model, antibody production could be successfully induced (Fig. 2E), suggesting similar techniques may offer promising solutions in the treatment of immuno-deficiency.
Due to the constraints of current material fabrication technology, some sophisticated structures are still difficult to replicate in the biomaterial models, such as microvascular network and reticular network of FRCs in the LN. For example, despite the advent of high-precision 3D printing techniques, the appropriate high-performance bioinks required for tiny feature resolution are lacking [167]. Advanced mathematical and computational approaches can provide us with another way to recreate LN structure and support in vitro/vivo models. Attributing to recent advances in imaging technology and processing techniques, in silico models can now include descriptions of the LN microzones/subregions when modelling the immune response, improving model resolution and system description, and providing potentially personalized modelling frames. For example, Kelch et al. provided visualization of the entire LN microvascular network topology, to study vascular function in different LN subregions, by combining the use of advanced confocal microscopy, with new processing techniques [168]. The LN reticular network of FRCs has been described using a small-world network model by Ludewig et al. [169] By removing nodes in the network, and carrying out in vivo validation, it was shown that above a 50% destruction of the FRC network resulted in significant changes in immune cell recruitment, T cell migration and activation of CD8+ T cells. When the conduit system was added to the model and similarly disrupted [170], the FRC network showed higher robustness and tolerance to perturbation than the conduit system network (Fig. 2F). Apart from simulating LN microzone/subregion models, computational models can also simulate changes in the whole LN, helping us to understand how macroscopic alterations may affect cell (immune or tumor) interactions, cell migration and drug diffusion. Bocharov's group constructed 3D models of the subcapsular sinus, B-cell follicle, T-cell zone, and vascular system based on confocal imaging of the LN macroscopic structures. These models were then integrated with a FRC latticed network model, built using structural properties, to produce an idealized 3D whole LN geometric model (Fig. 2G) [171]. The research group has also developed a body-wide lymphatic system model, using network graph modelling, to gain insight into the topological properties and regulation of the system that can impact aspects such as tumor dissemination, systemic infection and fluid regulation (Fig. 2H) [172]. In contrast, a body-wide approach is difficult to achieve with current biomaterial models because of fabrication limits and extremely high cost.
5.2. Immune cell dynamics
After phagocytosing pathogens and presenting antigen fragments on MHCs, APCs, such as DCs, migrate from peripheral tissue and enter the initial LVs. These blind-ended vessels converge into collecting LVs, which transport DCs to draining LNs in the lymph [173,174]. The DCs are deposited into the LN subcapsular sinus within the lymph before migrating to the paracortex, while lymphocytes (T/B cells), enter directly into the paracortex as described in Section 2. A successful immune response involves sufficient antigen-recognition via cross talk between lymphocyte receptors and APCs to allow clonal expansion of pathogen-destroying immune cells. Therefore, it is paramount to understand how T cells migrate and communicate in the LNs to facilitate, for example, immunotherapy or vaccine design. As such, recently, a series of biomaterial models regarding T cell migration and immune behaviors have been engineered. Stachowiak et al. developed inverse opal hydrogel/collagen-fiber composite scaffolds with interconnected pores to explore scaffold/microenvironment conditions that generate rapid LN-like T cell migration [175,176]. Combined with lymphoid tissue chemokine (i.e., CCL21), unconstrained T cell motility was observed in the 80 μm-porous scaffolds (Fig. 3A), which indicated successful fabrication of the scaffold supporting T cell migration and the possibility of using CCL21 to induce lymphocyte motility. Based on this finding, Pérez del Río et al. designed a PEG-based 3D hydrogel loaded with CCL21, to provide structural and biological functions similar to LN tissues for studying T cell migration and proliferation in vitro [177].
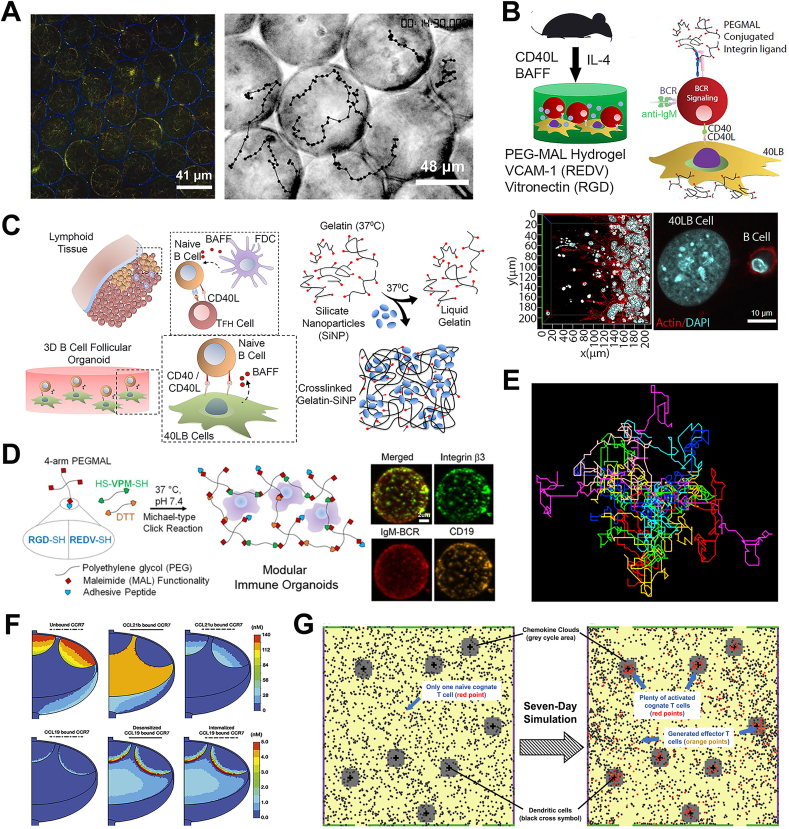
Fig. 3.
Biomaterial models and computational tools for studying immune cell dynamics and corresponding immune responses in the LN. A) Confocal image showing hydrogel-scaffold/collagen-fiber composite (scaffolds are shown in blue and fibers are in green) and brightfield image showing T cell migration paths in 80 μm-pore-sized composite scaffolds. Reproduced with permission [175]. Copyright 2008, Wiley-VCH. B) Schematic illustration of 3D maleimide/polyethylene glycol-based hydrogel for mimicking germinal center-like B cells. Reproduced with permission [178]. Copyright 2019, Elsevier. C) Overview of the cell interaction within the germinal center reaction and the formation of gelatin hydrogel/silicate nanoparticles-combined organoid scaffold. Reproduced with permission [180]. Copyright 2015, Elsevier. D) Schematic illustration of preparing maleimide/polyethylene glycol-based organoid with integrin-specific ligands and confocal images of various protein biomarker expressions on immune organoid. Reproduced with permission [184]. Copyright 2016, American Chemical Society. E) Simulated motion trajectories of twenty T cells in the lymph node paracortex using a 2D agent-based model. Reproduced with permission [103]. Copyright 2008, Australian and New Zealand Society for Immunology Inc. F) CCR7 concentration contours (in nM) of desensitized or internalized CCR7 showing variations in the different regions. Reproduced with permission [201]. Copyright 2017, The American Association of Immunologists, Inc. G) Snapshot of an agent-based model describing T cell and dendritic cell interaction. Adapted with permission [202]. Copyright 2019, Frontiers Media S.A.
In addition to T cells, B cells are another important lymphocyte type in the LN. Several studies involving LN-like hydrogel or organoid models that simulate the B cell behaviors and development of the germinal center (i.e., B-cell zone) have been reported. Purwada et al. reported the use of maleimide(MAL)/PEG-based hydrogel to develop a 3D synthetic immune tissue platform that regulated B cell differentiation and promoted B cell enrichment based on antigen affinity (Fig. 3B) [178]. This model not only recreates the LN germinal center area in vitro, but also provides a platform to study antigen-specific B cell immune response. Using peptide RGD-presenting gelatin hydrogel and silicate nanoparticles, Singh's group developed a B-cell follicle organoid that encapsulated B cells and 40LB stromal cells (i.e., stromal cells with CD40 and BAFF, which was reported in previous work [179]) (Fig. 3C) [180]. It accelerated the germinal center response ex vivo and induced robust antibody class switching. Compared to results using 2D culture systems, higher expression of CD40L (i.e., ligand for B cell maturation and activation) surface marker was found in this B cell-centered organoid system, highlighting the significance of 3D-structural follicular niche. This protocol was then optimized with a >90% success rate of fabricating organoid droplets [181]. Recently, to better identify LN microenvironment factors modulating germinal center responses, Singh's group further developed a PEG-based immune organoid for B cell proliferation and germinal center-like induction [182]. Interestingly, this work investigated the mode of Klebsiella pneumoniae (antibiotic-resistant bacteria) antigen presentation on B cell differentiation ex vivo, offering a powerful platform for vaccine development. Similarly, Béguelin et al. fabricated a hydrogel/nanoparticle combined organoid that modeled the induction of the germinal center. They elucidated that EZH2 histone methyltransferase regulated germinal center B cell expansion by suppressing CDKN1A (i.e., cyclin-dependent kinase inhibitor) [183]. Nevertheless, due to the abundance of RGD ligands on gelatin, it is difficult to investigate the role of specific integrin-ligand interaction during germinal center response in the abovementioned organoid model. A MAL/PEG-based B-cell follicle organoid was later engineered with specific integrins (i.e., α4β1 and αvβ3) and ligands (i.e., RGD and REDV) respectively (Fig. 3D) [184]. In this LN-mimicking model, it was observed that α4β1-based organoid led to a more robust early-germinal center phenotype with REDV ligands, while αvβ3 could regulate the two ligands similarly in the niche, suggesting that the different integrin-ligand interactions had distinct influences on the germinal center responses. This observation will be a potential entry point for modulating the germinal center reaction in the future immunotherapy. Besides the germinal center area, other zones such as the T-cell zone and FDCs network are also valuable to mimic in organoid models [64].
Biomaterial models offer researchers valuable in vitro platform to study immune responses, but they cannot include all complex architectures and factors (e.g., physical obstacle, stromal cell network, high cellular density, and intercellular interactions that results in change in T cell behavior) that may influence cell motion in the LN. For example, following activation, T cell mobility varies in three distinct phases including swarming and pausing with varying durations [185]. Cell type and stage of activation has also been shown to influence whether T cells follow directed or random patterns during migration [[186], [187], [188], [189]], as has location, fluid flow and exposure to signaling molecules [27]. Cell migration is therefore difficult to replicate in biomaterial models and assess by single parameters such as speed or mean displacement [[190], [191], [192]]. With the support of mathematical and computational models, these limitations on biomaterial models can be easily overcome. Biophysics-based in silico models have been used to describe T cell movement by ascribing a predetermined motion that best-matches in vivo observations, for example, by describing two T cell subsets, one subset moving with Brownian motion and the other with a random-walk with persistence [193]. T cell migration has also been described by applying a heavy-tailed distribution of step lengths, a Lévy walk, and Brownian motion, with the latter also considering geometric parameters, such as through a sphere and cylinder [190,194,195].
Computational models that allow a sophisticated description of the LN geometry, by dividing the environment with lattices, provide ideal platforms to unravel the pattern of T cell movement in differing micro-environments [196]. For example, a lattice-based CPM was created by Beltman et al. to study how LN topology dictates naïve T cell migration behaviors [116]. Several groups have developed ABMs of T cell motility in the LN, where the environment is divided into grid-compartments and cells are described as discrete agents that follow probabilistic rules. The agents can store internal properties, access information regarding the surrounding agent and grid-compartment states, and move in 2D or 3D space. Bogle and Dunbar developed a 3D ABM describing T cell motility in the LN paracortex region, capturing the characteristics of random motion by allowing agents to make discrete movements along the lattice sites, with movement probability determined using in vivo-informed probability distributions (Fig. 3E) [103]. This model also allowed LN swelling and contraction by altering grid-compartment number proportionally to T cell number, which is infeasible in current in vitro models.
Another advantage of the in silico model is enabling inclusion of the chemokine gradient in the LN, which is known to play an important role in aiding immune response in the LN, but has also been associated with LN metastasis. Although a rough gradient condition can be achieved across biomaterial models on some multiple chamber-based chemotaxis kit (e.g., Boyden Chamber [197], Zigmond Chamber [198], Dunn Chamber [199], μ-Slide chemotaxis [200], etc.), the current technology is still far from replicating actual physiological environments. Harnessing the computational simulation, some mechanisms of LN chemokine gradient formation and regulation in the LN are gradually being unraveled. For example, Moore's group built a computational model to investigate CCL19 and CCL21 transport and corresponding gradient formation in the LN using ordinary and partial differential equations [201]. The model demonstrated that intra-LN CCL21 gradients and distribution were substantially controlled by lymph flow and that a large CCL19 gradient was formed that served as a key regulator of B cell behavior at the boundary between the T-cell zone and B-cell follicle (Fig. 3F). An ABM developed by Azarov et al. showed that the reduced unique total contacts may be due to the swarm behavior as T cells flocked to DCs, that could physically restrict access to DCs by new T cells (Fig. 3G) [202]. The same model also showed that attraction of activated T cells to DCs could accelerate time to initiation of immune response. Dunbar et al. developed an agent-based T cell model to explore how chemotaxis affects T cell migration in LN paracortex by using vector addition [104]. In this model, the subtle bias of T cell movement towards chemokine sources was observed while a largely random-walk cell motion was maintained. These features were previously difficult to detect occurring simultaneously when using conventional in vitro models, highlighting the benefits that modelling in silico can provide.
5.3. Lymph flow dynamics
The LVs transport lymph, mainly composed of water and proteins that leak from blood vessels into interstitial spaces, but also antigens and APCs, that is absorbed into the initial LVs. These vessels are blind ended structures with walls one cell thick. The initial lymphatics converge into collecting lymphatics, thicker vessels with a layer of contracting muscle cells, that help pump fluid, and contain uni-directional valves to reduce backflow [203]. The collecting lymphatics drain to, and through, LNs, that provide an environment for sampling of lymph content and initiation of adaptive immune response, before being eventually deposited into venous circulation via junctions with the subclavian veins. This uptake and return of fluid to the blood is critical to prevent collection of fluid in interstitial spaces that can result in lymphoedema. Due to the role in transporting immune cells, it is vital to measure intra-LN flow dynamics and understand how LN internal structure affects the fluid flow, which could be key parameters in the immune response. Microfluidic techniques can be used to describe intra-LN dynamics through LN structures (i.e., flow-through microchannels) [204] and lymphatic flow can be established using external hydrostatic pressure setups [205]. In vitro models such as hydrogel and organoid systems are unable to capture these fluid-associated features. The addition of fluid flow to the cell-loaded matrix, to mimic in vivo lymph flow, can also increase the formation of stromal network and expression of chemokine ligands [153].
Several LN-centered fluid systems using microfluidic chips have been built to investigate the in vivo relationship between LN and lymphatic fluid flow. To understand how shear flow in the LN affects immune cell homing and lymphatic metastasis, Birmingham et al. developed a LN subcapsular sinus-on-a-chip that recreated the LN hydrodynamic microenvironment and the effect of wall shear stress (Fig. 4A) [206]. They found that cell motility and adhesion are critical to immune cell recruitment and cancerous metastasis in the LN. Based on these findings, by altering the fluid flow profile, the cell motility and adhesion, and therefore downstream effects, have the potential to be manipulated. Scope for improved experimental design remains. For example, to better simulate the in vivo situation, lymphatic flow could be set as variable rate instead of constant value. Furthermore, the inflow angle between afferent LV and subcapsular sinus should be inconstant and variable, rather than simply fixed at 90°. Fathi et al. designed a gravity-driven microfluidic device to mimic the normal flow condition (0.92 dyn/cm2) and disease flow condition (6.70 dyn/cm2) in the LVs (Fig. 4B) [207]. Interestingly, without requiring an external pump, this biochip operated with gravitational forces to simulate the cyclic fluid flow, and a range of flow shear stress could be achieved by periodically rotating the platform. To evaluate BV-LV interaction and lymphatic return rate, blood vascular endothelial cells and LECs were co-cultured on a microfluidic platform to recreate intervascular permeability [208]. By integrating distinct compartments of LN, simulation of the spatial dynamic LN microenvironment and complex immune cell interactions can be performed on one chip, which will be helpful to allow a comprehensive evaluation of the LN. Diverse whole-LN-mimicking microfluidic devices, including IG-Device™ [209], HIRIS™ III (Fig. 4C) [[209], [210], [211]], and multi-compartment 3D LN-on-a-chip (Fig. 4D) [212], have also been fabricated and included several key LN microstructures (e.g., subcapsular sinus, follicle, paracortex, reticular network, etc.) as well as immune cells (e.g., T/B cells, DCs, macrophages, etc.) to assist drug development with lower labor and time costs.
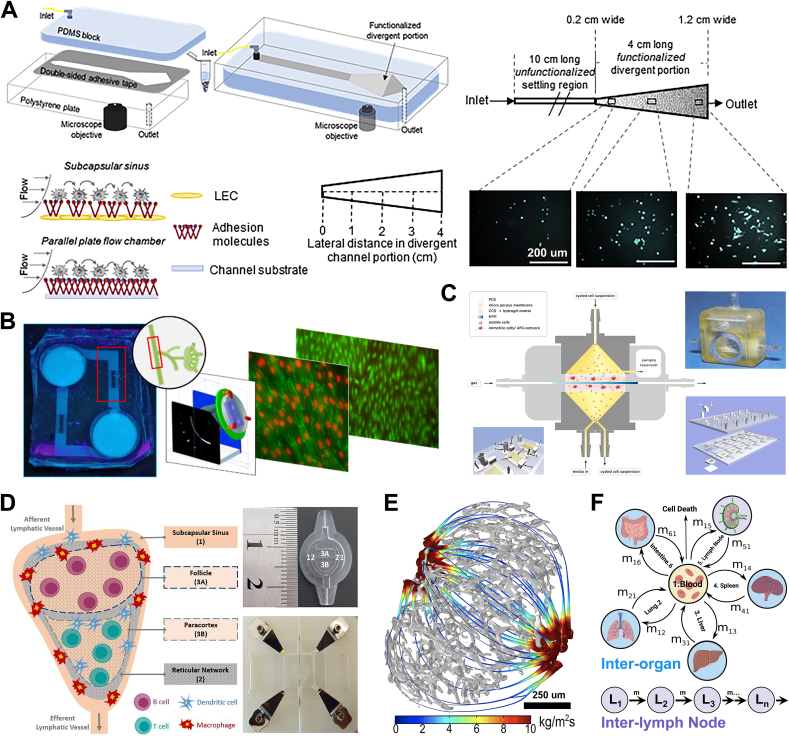
Fig. 4.
Biomaterial models and computational tools for studying lymph flow dynamics. A) Schematic illustration of a subcapsular sinus microenvironment-mimicking, divergent parallel-plate microfluidic perfusion with an adhesion-molecule-functionalized, divergent portion. Reproduced with permission [206]. Copyright 2020, Elsevier. B) The gravity-driven microfluidic device mimics the cyclical lymphatic flow under normal and disease conditions. Reproduced with permission [207]. Copyright 2020, American Chemical Society. C) Whole-LN-mimicking microfluidic devices (Reproduced with permission [209]. Copyright 2010, Elsevier) and D) multi-compartment 3D lymph node-on-a-chip (Reproduced with permission [212]. Copyright 2020, MDPI) for inflammatory responses and drug development. E) Lymph flow flux in stream tubes when efferent lymphatic pressure is 0 Pa. Reproduced with permission [214]. Copyright 2016, Springer Nature. F) Mathematical ordinary differential equation-based models informed by experimental data to describe and predict lymphocyte recirculation in the lymphatic system between lymph nodes and between different organs. Inspired by [215].
To achieve a more accurate replication of LN, there are multiple challenges that need to be addressed when developing the next iteration of improvements in LN representative microfluidic devices. These include the capability to describe complex organ architecture with location-specific cell seeding, corresponding cellular interaction, and interorgan communication via the transport of cells and signaling molecules both inter and intra model. Fluid dynamics within the LN have proven difficult to characterize, but determination of afferent and efferent flow is another important requirement to allow description of molecular and cellular transport. Biomaterial strategies such as fluorophore-labelled microparticles have been previously used to track flow pathways in the LN but capture of the different intra-LN flow rates proved difficult. This was because small-size particles (∼10 kDa) were able to cross the subcapsular sinus floor either into B-cell follicles and T-cell cortex area or into conduits that rapidly transported them to the surfaces of high endothelial venule, while larger particles (∼2000 kDa) were restricted to the subcapsular sinus and took a peripheral route to medullary sinuses then into efferent vessels [27].
Intra-LN flow mathematical and computational models can support and inform the in vitro experiments to achieve more insight into LN flow parameters. An in silico model was constructed that considered LN geometry and structure and calculated lymphatic flow through the LN using input generated with selective plane illumination microscopy [213,214]. By modelling flow rates under different lymphatic pressures, it was demonstrated that the fluid flow in stream tubes, curves that velocity vectors are tangential to with area proportional to fluid flux, was dependent on efferent lymphatic pressure (Fig. 4E). Moore's group also designed a 3D computational fluid dynamics model to describe lymphatic flow and fluid exchange in the LN [24]. Moreover, intra-LN flow greatly influenced cytokine transport and immune cell mobility in the LN, such as lymphocyte residence time. This insight shows that factors such as fluid flow, may affect lymphocyte activity, and therefore are an important target to consider when designing immunotherapy on biomaterial models. An overview of mathematical models that estimate how lymph flow impacts retention of lymphocytes in the LN can be found in a recent review [215].
In addition, some specific LV segments cannot be accurately described with simple microfluidic channels. Collecting LVs, unlike initial LVs, are not simple tubes, but contain unidirectional valves, with the lymphatic chambers between valves termed a “lymphangion”, and a layer of contracting smooth muscle that provides an intrinsic pumping mechanism [216]. This behavior is not easy to replicate with current biomaterial approaches. Consequently, several computational and mechanical models describing lymphangions have been developed to include lymphatic valves [[217], [218], [219]], lymphangion pumping behavior [220,221], and passive/active contraction [[221], [222], [223]]. Lymphocyte recirculation in the lymphatic system between LNs and between different organs has also been described and predicted by using experimental data to inform and parameterize ODE-based mathematical models (Fig. 4F) [215]. These lymphatic flow models provide new avenues to investigate the importance of factors, such as transport and recruitment of immune cells on a body-wide scale, on the immune response. Models of lymph flow dynamics could also be adjusted to support drug distribution and delivery research in the lymphatic system.
5.4. Lymph node-associated disease and treatment
Besides recreating LN architecture and immune cell behaviors, another important application of LN-like biomaterial platforms is to mimic lymphoid disease models. In recent years, several in vitro models have been developed to study lymphomas progression. Apoorva et al. developed controllable synthetic hydrogels that simulated the stiffness of healthy and neoplastic lymphoid tissue of diffuse large B-cell lymphoma [224]. Using this LN-mimicking model, they found that B-cell tumors showed maximum proliferation in the hydrogel with medium stiffness (∼2000 Pa) when compared to those grown in higher or lower stiffness models. The tissue stiffness was found to regulate tumor expansion via mechanical stimuli to surface receptors (e.g., B-cell antigen receptor and integrin). These results highlighted the role of lymphoid tissue stiffness in lymphoma progression and drug resistance, which had been largely neglected. Gravelle et al. fabricated a 3D follicular lymphoma-based organoid using a hanging-drop method in which cells were suspended in droplets of medium and then developed into 3D organoids [225]. Compared to growth in 2D cell suspension, over 600 different gene expressions, which correspond to several gene ontology biological processes (e.g., activation of NF-κB pathway and hypoxia) and lymphoma cell activities, were found in the 3D organoid culture system. This highlights the importance of 3D spatial organization in influencing cell-cell/matrix interaction in follicular lymphoma. Further ex vivo investigations are required to include the ECM cues of LN tissues and tumor heterogeneity, which were excluded in this work. To uncover the effect of integrin-specific ligands on malignant B/T-cell lymphoma, Tian et al. developed a lymphoid organoid embedded with malignant B/T cells and FDCs (which act as supporting stromal cell subtype) [226]. They found that B/T-cell lymphomas had different pro-survival signaling requirements, in that B-cell lymphomas depended on REDV ligand to promote expansion, while T-cell lymphomas required the presence of RGD ligand. The results link with separate findings that different integrin-ligand interactions induced distinct germinal center responses, identifying REDV and RGD ligands as critical markers to regulate B cell activity and related diseases [184]. This model also demonstrated that FDCs played a key role in the enhanced proliferation of lymphoma cells in 3D microenvironments, which pointed to FDCs as a potential avenue to target lymphoma.
As lymphoma is a type of cancer presented in the lymphatic system, it can quickly metastasize or spread throughout the body, and most commonly spreads to the liver, bone marrow, or lungs. When cancerous cells break away from the primary tumor, metastasis can occur via both the cardiovascular and the lymphatic system, with LNs presenting as a common metastasis site that is associated with worse prognosis in many types of cancers [227,228]. Recent studies suggest tumor cells like melanoma cells that metastasize through the lymphatic system possess a higher probability of survival than those that travel through the blood circulatory system because there are lower levels of oxidative products in the lymph [7,229]. Generally, tumor cells invade the surrounding LVs before migrating to adjacent LNs, followed by dissemination to other organs via the lymphatic circulation [230,231]. Development of a LN/LV-on-a-chip model could aid in understanding the mechanism of tumor metastasis through the lymphatic system. Additionally, compared to human cases, some recent studies found differences in the metastatic regions on currently used animal models, which could be caused by the physiological differences between human and animal LNs [232]. A microfluidic model is a reliable candidate to bridge this gap and verify results from animal experiments prior to clinical trials. During the first stage of the metastatic cascade, tumor cells secrete and overexpress pro-lymphangiogenic factors (e.g., vascular endothelial growth factor C) to induce LV sprouting. Kim et al. created a microfluidic platform to investigate how interstitial flow affects lymphangiogenesis (Fig. 5B) [233]. They observed that, synergized with proangiogenic factors, interstitial flow acted as a central regulator to direct lymphatic sprouting. The outcome was supported by results using a similar biochip from Swartz et al. [234] Notably, an immortalized human LEC has recently been developed, which stably maintains the biophysical features and genetic expression over 12 months and 53 passages [235]. In comparison, common primary LECs begin to deteriorate after 12–15 population doublings. When using this newly developed cell line in microfluidic chips, researchers will have a highly standardized platform to study lymphangiogenesis and LV/tumor cells interaction. Secondly, within the tumor microenvironment and under the influence of biophysical/biochemical cues, cancer cells migrate and invade into the LVs by penetrating LECs [236]. To explore potential targets for cancer therapy, a class of factors such as luminal and transmural flow (Fig. 5D) [237], surrounding matrix density (Fig. 5E) [238], and LECs/tumor cells crosstalk [239] have been replicated on microfluidic devices. Compared to hydrogel scaffolds and organoid-centered systems, microfluidic devices can be better platforms to investigate tumor transendothelial intravasation and migration because of the dynamic fluidic flow and direct visualization available via live-cell imaging. After cancer intravasation, tumor cells drain into the surrounding LNs and circulate throughout body via the lymphatics, invading a secondary site for tumorigenesis. As the viscosity of lymph fluid is lower than blood, due to the lack of platelets and red blood cells [240], the resulting lower shear rate makes the survival and metastasis of cancerous cells easier in the LVs [241]. A better understanding of tumor migration into the LVs, in order to design prevention is therefore warranted. Chen et al. created a single-cell migration biochip with choke points (6–30 μm) to investigate individual tumor cell migration in the LVs (Fig. 5C) [242]. This platform could describe the intrinsic differences in tumor cells responsible for chemotactic heterogeneity (i.e., the ability of tumor cells to invasion and migration), which makes it a great tool to help unlock the mechanism of tumor metastasis. Shim et al. combined LN and tumor slices on a two-compartment microfluidic platform to study LN/tumor crosstalk under recirculating flow (Fig. 5F) [243]. By detecting the secreted proteins/biomolecules from living tissues, use of this platform enabled the authors to understand the factors inducing immunosuppression and cancer metastasis. After draining into the LN, tumor cells can lodge and acclimatize to the LN microenvironment and suppress the function of immune cells [244]. This invites the development of LN-on-a-chip to mimic interactions between cancer cells and immune cells in LN. For instance, a LN micrometastasis-on-a-chip was created to simulate the deep cortical unit of LN and assess the efficacy of natural killer (NK) cells to kill lodged tumor cells [245].
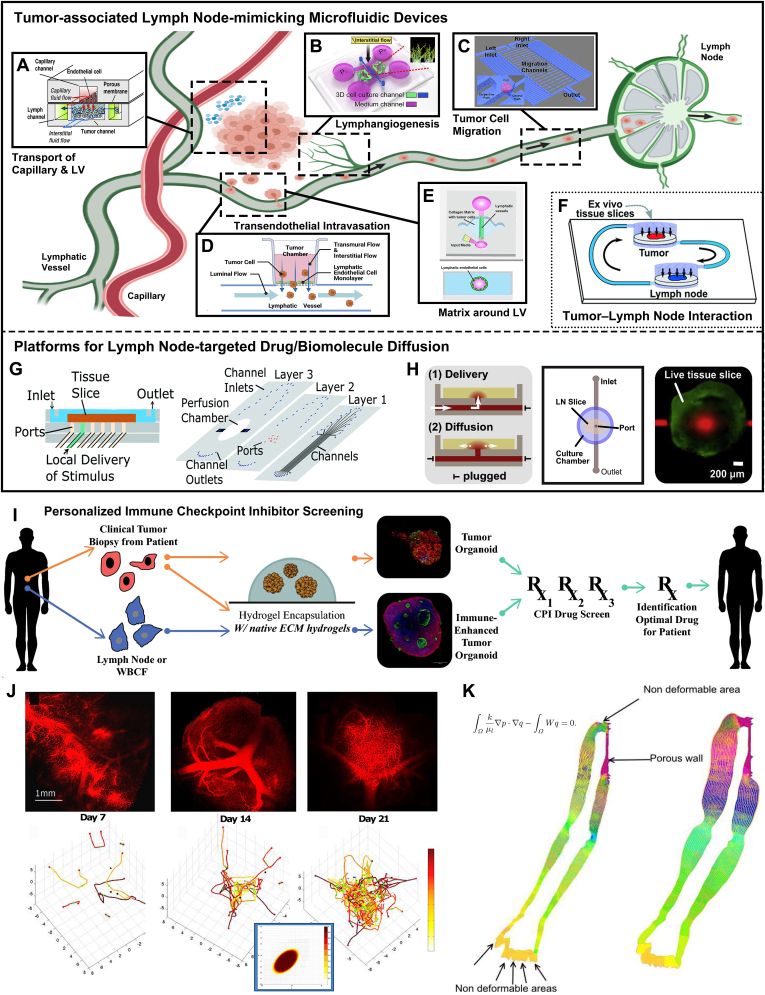
Fig. 5.
Biomaterial models and computational tools for mimicking lymphoid disease progression and developing corresponding treatments. Several tumor-associated lymph node-mimicking microfluidic devices have been developed, to stimulate different stages of the metastatic cascade and understand how tumor cells invade and spread via LV system, including A) transport of LV/BV and tumor (Reproduced with permission [252]. Copyright 2014, Elsevier), B) lymphangiogenesis (Reproduced with permission [233]. Copyright 2016, Elsevier), C) tumor cell migration in LV (Reproduced with permission [242]. Copyright 2015, Springer Nature), D) tumor intravasation into LV (Inspired by [237]), E) extracellular matrix around LV (Reproduced with permission [238]. Copyright 2020, The Royal Society of Chemistry), and F) LN/tumor on a chip (Reproduced with permission [243]. Copyright 2019, The Royal Society of Chemistry). Some microfluidic platforms have been created to mimic and study LN-targeted drug delivery and diffusion, such as G) LN local drug stimulation (Reproduced with permission [251]. Copyright 2017, The Royal Society of Chemistry) and H) LN-targeted delivery and diffusion of cytokine (reproduced with permission [73]. Copyright 2018, Elsevier). I) Overall concept of the lymph node/melanoma-combined organoid for studying personalized immunotherapy response. Immune cells or tumor cells are biopsied from patients and used to fabricate patient-specific tumor/immune cell organoids, to screen checkpoint inhibitor drug efficacy and identify optimal drugs for patients. Reproduced by permission [246]. Copyright 2019, Springer Nature. J) Comparison of vasculature and angiogenesis in lymphoma in animal model (above) and computational simulation (below) models. Reproduced with permission [255]. Copyright 2013, PLOS. K)In silico model of the lymph flow through the lymphatic system in a whole upper limb including the corresponding interstitial fluid exchanges (e.g., the pressure of the fluid is shown with the gradient of color) and domain deformation. Reproduced with permission [260]. Copyright 2017, EDP Sciences. (BV: blood vessel; LN: lymph node; LV: lymphatic vessel).
Advanced biomaterial models also provide great platforms to develop treatment to LN-related diseases such as immunotherapy. Votanopoulos et al. developed a series of LN/melanoma-combined organoid that contains cells from the same patient [246,247]. This platform allows for personalized immunotherapy, where drug responses can be screened in organoids prior to treatment (Fig. 5I). Compared to animal models, advantages include low expenses, short-time processing, and capability for mass-scale production, generating great potential to use this tool for other adaptive immunity applications such as DC vaccine development. To avoid immune complications (e.g., secondary lymphedema) following LN dissection, Lenti et al. engineered replacement artificial lympho-organoids using decellularized ECM-based scaffolds with LN stromal progenitors, which successfully recovered lymphatic function and induced antigen-specific immune responses upon transplantation into a mouse model [248]. In the context of humans, however, the progenitor cells seeded in these artificial systems must be human leukocyte antigen (HLA)-matched to reduce the risk of autoimmune responses such as graft-versus host disease which is often seen post-allogenic transplant [249]. Several microfluidic devices have been created to mimic and study LN-targeted drug delivery and diffusion [250]. To explore localized signaling on live LN tissue, Pompano et al. designed integration of live LN slices with microfluidic platform for locally controlled stimulation (Fig. 5G) [251]. By leveraging the modulation of flow rates through ports, researchers could simultaneously deliver drugs to two separate regions on the LN platform and compare immune responses and spatial dynamics. For instance, retention of delivered glucose-conjugated albumin was greater in B-cell zone than in the T-cell zone. The authors later developed a live LN tissue slice-contained biochip and integrated this platform with an optical imaging system (Micro-IOI) to facilitate analysis of the diffusion of fluorescent-labelled cytokines in the LN (Fig. 5H) [73]. These LN-on-a-chips are useful to deeply understand LN-targeted pharmacokinetics and efficacy and inform the design of cutting-edge immunotherapies. Moreover, to promote the study of drug efficacy, microfluidic platforms have also been developed to study the transport of nanoparticles to LV/BV (Fig. 5A) [252] and carry out rapid vaccine assessment [253].
Mathematical and computational models have also been developed to offer another way to unravel the pathology of LN-associated diseases and explore potential treatments, for example, metastases, lymphoma, and lymphedema. Despite lacking a description of all complex physiological conditions, models offer tight control over the simulated disease development allowing investigation of specific disease progression pathways, monitoring of specific outcomes of interest, and conduction of personalized treatment designs. By combining model-development techniques, such as use of single in vivo measured parameters and simplified assumptions, with reverse-engineering to fit the data/images and phenomena observed in animal or biomaterial in vitro models, the advantages of both methods can be used to create disease models that fill data gaps.
To describe the interplay between cancer progression and immune response, “prey-predator” models based on ordinary differential equations can simulate the “competition” between immune and cancer cells under no treatment [121] and chemotherapy conditions [126,127]. Factors such as cytokine diffusion, that can affect tumor progression, and tumor dormancy (where cancer cells exist without significant growth) [254], can also be integrated and their influence investigated using this framework. Furthermore, use of partial differential equations allow consideration of spatiotemporal aspects of tumor-immune cell interaction (e.g., cellular spatial density) [128]. Cellular automaton model, ABMs, continuous models, and hybrid models which can capture sophisticated LN geometries, have also been adopted to gain insight into immune cell and tumor cell interaction. Computational models have also been established to investigate lymphoma. For example, Frieboes et al. built a 3D continuum model describing non-Hodgkin's lymphoma using parameters obtained from murine in vivo experiments and murine immune-histological data [255]. When compared to an in vivo murine tumor experimental model, the computational model precisely predicted lymphoma growth and vascularization (Fig. 5J). Du et al. developed a model of a subtype of non-Hodgkin's lymphoma, called diffuse large B-cell lymphoma, using estimated molecular reaction kinetics to describe B-cell receptor signaling [256]. The resulting model can predict drug-induced tumor viability with applications such as optimization of drug combinatorial therapies. As well, the model has the potential to be used in investigating other B-cell malignancies related to aberrant B-cell receptor signaling (e.g., chronic lymphocytic leukemia) [257]. Plus, lymphedema is one of the most common post-surgical side-effects, with up to 20% of women reporting lymphedema of their operative side within 3 years post-surgery [258]. Effective therapies for, and pathological mechanisms underlying lymphedema, have typically been investigated using animal models, most commonly murine tail and hindlimb models, but only acute lymphedema can be effectively mimicked [259]. Current biomaterial in vitro models or larger animal models such as a monkey limb model, remain lacking and are costly to develop. Eymard et al. developed a 2D computational model to simulate clinical characteristics of lymphedema and explore reports physical compression therapy may reduce limb swelling post-LN dissection (Fig. 5K) [260]. Despite omission of key factors, such as the biophysical properties of patient tissues, this model presents as a powerful tool to design personalized compressive devices for lymphedema management. Other LN-associated disease models such as LN degeneracy [261] and infection [262,263] have also been designed.
6. Conclusion and perspectives
Recent advances in developing LN-mimicking models have offered researchers increasingly powerful tools to study LN anatomy and physiology to make new discoveries about the immune system. In the face of various LN-associated diseases, the models discussed earlier show promise for use as platforms for pathological studies and treatment, particularly personalized therapy. Input of anatomical images from microscopy and experiment-derived parameters can now be used to generate artificial models that accurately simulate pathological conditions. Significantly, the features in these artificial systems (e.g., microfluidic architecture of the biochip models and the equations in computational support models) can be readily modulated to meet specific requirements. Artificial systems also offer advantages such as lower costs, shorter operation times, reduced work effort, and fewer ethical issues. Nevertheless, limitations of LN-mimicking models remain. Generally, it is difficult to comprehensively capture the architectural complexity of the human tissue, as well as the high-ordered/random-combined cell activity that are key qualities of in vivo animal models, particularly as our biological understanding improves. For example, recently it was found that human LN LECs can be clustered into six transcriptomically distinct subpopulations that are differentially located in the LN and display different properties [291]. To accurately capture these features would require major improvements in cell seeding techniques within microfluidic devices. Location-specific LN models may also be required as LNs can differ in their unique marker expression depending on their anatomical localization. For example, in peripheral LNs, migration of lymphocytes through the high endothelial venules is controlled by peripheral node addressin (PNAd) whereas the same process in mucosal LNs is mediated by mucosal addressin cell adhesion molecule (MAdCAM) [292]. Furthermore, it is challenging to achieve a realistic scale of human organ size and cell numbers [250], and in most research studies, often only the tissues of interest are included in the model design. For example, to study a specific issue in the LN, the microfluidic platform is designed to describe only the LN subsection of interest [293]. This lack of a comprehensive simulation of the whole LN or body system means that the natural biological interactions between different LN substructures, other immune organs and the surrounding environment are excluded in the experimental setup, which may cause the outcomes to deviate from actual physiological observations. The same problem applies to computational models.
To establish more effective models, there is a need to promote synergy between in vitro/vivo experiments and in silico modeling where each model can be fully exploited for its strengths. Computational tools are be adopted to devise biomaterial models and assess corresponding properties [294,295]. For example, an in silico model can create the theoretical representation of polymer network to fabricate hydrogels with an in silico determined porous structure and mesh size, resulting in better control when the hydrogel is used for drug delivery [296]. Furthermore, properties such as the crosslinking condition, embrittlement phenomena, water absorption capacity, surface biophysical features, and degradation profiles can also be virtually emulated and tested in silico before hydrogel fabrication [[297], [298], [299], [300]]. The organoid developmental processes, such as self-assembled formation, can be studied and understood by computational multicellular modeling. By using in silico models to inform the processes from initial seeding of organoid models to maturation and organization, spatiotemporal predictions of organoid properties can be improved [301]. Computational methods can also provide insight when developing microfluidic devices, such as informing constructing microstructures based on organ-tissue specific architectures and features, as well as predicting fluid flow profile in the microchannels [302,303]. For example, through combining micro-computed tomography scan, artificial intelligence approach and high-resolution bioprinting, high accurate on-a-chip platforms which can mimic tiny complex organs are possible to be created [304]. The combinatorial strategy of biomaterial and computational tools is also promising when applied to tissue engineering [305]. A multi-scale computational investigation allows characterization of the effect of important material design parameters and can predict the corresponding biofunctions and therapeutic effects. In conjunction, the biomaterial system implements and validates the computational simulation and theory and can also provide new in vivo/vitro data to inform the in silico model. Several combinatorial models have recently been used within cardiac tissue engineering. For example, Wang et al. conceptualized the framework of integrating computational stimulation modelling with 3D printing material model for structural heart disease [306]. The image-based in silico models initially simulated cardiac tissue architecture and acquired data for 3D printing. The printed models then provided data such as fluid flow during device testing. This was then followed by analysis and optimization within an in silico model. Therapeutic drug-laden biomaterials for epicardial drug delivery have also been developed by Shirazi et al., using a combined iterative computational and in vitro modelling approach to optimize the biomaterial design [307] Through rapid iteration, the complex processes of drug release and transportation were mimicked on the model, and following, it optimized the biomaterial design and fabrication. Sulejmani et al. used computational modeling to evaluate biomaterials for transcatheter heart valve [308]. Similarly, this framework have also been adopted on investigating scaffold for bone grafting and cartilage-tissue repair [[309], [310], [311]].
Within the context of the LN, as shown in Fig. 6, we provide three examples to illustrate the impact such a combined system could deliver. First, mathematical and computational models can serve as “initiator or pioneer” at the beginning of the study by providing big data through large numbers of virtual, in silico simulations. Biomaterial models can then act as “verifiers” to confirm in silico results in vitro or in vivo. Cell behaviors and antigen presentation in the LN present as ideal subjects to apply such integrative framework. For instance, computational models like ABMs [202] can be used to study how DCs encounter intra-LN T cells by simulating the mobility patterns of these cells. A biomaterial model such as LN-on-a-chip can alternatively verify the ABM-produced observations through real-time microscopic read-outs and continuous measurements, which is difficult to achieve within animal models. Second, an integrative model is an alternative to drug testing using animals. It takes over $2.5 billion dollars and about 12–15 years on average, for full research, development, testing, assessment and US Food and Drug Administration (FDA) approval processing to develop one single drug [312,313]. Our proposed integrative LN model can perform repeated drug screening in high-throughput and/or high-content screening with shorter time and lower cost, to discover new druggable targets as well as evaluate their safety and efficacy. Third, the integrative framework can be a useful clinical tool for personalized medicine. For instance, data from lymphatic bioimaging and patient information can be fed into artificial intelligence system and risk-scoring classification computational models to predict high-risk groups of cancer recurrence of LN metastasis [[314], [315], [316]]. By developing biochips with patient-derived biological materials, combinatorial drugs can be identified to treat cancer metastasis involving LNs.
Fig. 6.
Overall concept of an experimental/computational integrative lymph node model system. An integrative system that combines biomaterial and mathematical and computational models can summate their individual strengths, and counteract individual drawbacks, to comprehensively mimic and study lymph node biology. Compared to traditional approaches such as animal models, integrative modelling has multiple advantages and offers promising improvements in biomedical applications including patho-biological study, drug development, and clinical therapy.
Apart from LN, this combinatorial strategy is appliable to other tissues and organs with complex structures and functions, such as heart, lung, kidney, and brain. This will offer qualitative/quantitative analytical approaches to investigate tissue growth and regeneration phenomena as well as pathological process comprehensively and precisely, based on hand-on in vitro/vivo experiments and virtually dynamic in silico models, promoting research and development understanding of path-biology. We envision that through this review, there will be increased collaboration between the materials and computational fields to design innovative integrated LN-mimicking models as well as other tissue and organ systems, to accelerate this promising area of research.
Author contributions
Y. S. and S. C. J. contributed equally to this work. All authors conceptualized the idea, wrote, and reviewed the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The research was supported by NUS Presidential Young Professorship and MOE Tier 1 grant. Y. S. is supported by the NUS Research Scholarship, and Y. J. Q. is supported by the A∗STAR Graduate Scholarship. Fig. 1, Fig. 6 are created with BioRender.com.
References
- 1.Gretz J.E., Anderson A.O., Shaw S. Immunol. Rev. 1997;156:11. doi: 10.1111/j.1600-065x.1997.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 2.Willard-Mack C.L. Toxicol. Pathol. 2006;34:409. doi: 10.1080/01926230600867727. [DOI] [PubMed] [Google Scholar]
- 3.Ohtani O., Ohtani Y. Arch. Histol. Cytol. 2008;71:69. doi: 10.1679/aohc.71.69. [DOI] [PubMed] [Google Scholar]
- 4.Rockson S.G., Keeley V., Kilbreath S., Szuba A., Towers A. Nat. Rev. Dis. Prim. 2019;5:22. doi: 10.1038/s41572-019-0072-5. [DOI] [PubMed] [Google Scholar]
- 5.Hayes S.C., Janda M., Ward L.C., Reul-Hirche H., Steele M.L., Carter J., Quinn M., Cornish B., Obermair A. Gynecol. Oncol. 2017;146:623. doi: 10.1016/j.ygyno.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Rockson S.G., Engl N. J. Med. 2018;379:1937. doi: 10.1056/NEJMcp1803290. [DOI] [PubMed] [Google Scholar]
- 7.Ubellacker J.M., Tasdogan A., Ramesh V., Shen B., Mitchell E.C., Martin-Sandoval M.S., Gu Z., McCormick M.L., Durham A.B., Spitz D.R., Zhao Z., Mathews T.P., Morrison S.J. Nature. 2020;585:113. doi: 10.1038/s41586-020-2623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luciani A., Itti E., Rahmouni A., Meignan M., Clement O. Eur. J. Radiol. 2006;58:338. doi: 10.1016/j.ejrad.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F., Niu G., Lu G., Chen X. Mol. Imag. Biol. 2011;13:599. doi: 10.1007/s11307-010-0421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian T., Yang Z., Li X. J. Anat. 2021;238:489. doi: 10.1111/joa.13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muhanna N., Chan H.H.L., Douglas C.M., Daly M.J., Jaidka A., Eu D., Bernstein J., Townson J.L., Irish J.C. BMC Med. Imag. 2020;20:106. doi: 10.1186/s12880-020-00507-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng H.Y., Zhang Y., Liu H.J., Dong X., Yang S.C., Lu Q., Meng F., Chen H.Z., Sun P., Fang C. Oncol. Lett. 2018;16:5179. doi: 10.3892/ol.2018.9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Servais E.L., Colovos C., Bograd A.J., White J., Sadelain M., Adusumilli P.S. J. Mol. Med. 2011;89:753. doi: 10.1007/s00109-011-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Norman G.A. JACC Basic Transl. Sci. 2019;4:845. doi: 10.1016/j.jacbts.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mak I.W., Evaniew N., Ghert M. Am. J. Transl. Res. 2014;6:114. [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Gill E., Shery Huang Y.Y. Futur. Sci. OA. 2017;3:FSO173. doi: 10.4155/fsoa-2016-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shou Y., Campbell S.B., Lam A., Lausch A.J., Santerre J.P., Radisic M., Davenport Huyer L. ACS Appl. Polym. Mater. 2021;3:1943. [Google Scholar]
- 18.Eftimie R., Gillard J.J., Cantrell D.A. Bull. Math. Biol. 2016;78:2091. doi: 10.1007/s11538-016-0214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaahtomeri K., Karaman S., Mäkinen T., Alitalo K. vol. 31. Cold Spring Harbor Laboratory Press; 2017. pp. 1615–1634. (Lymphangiogenesis Guidance by Paracrine and Pericellular Factors). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant S.M., Lou M., Yao L., Germain R.N., Radtke A.J. J. Cell Sci. 2020;133:1. doi: 10.1242/jcs.241828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jalkanen S., Salmi M. Nat. Rev. Immunol. 2020;20:566. doi: 10.1038/s41577-020-0281-x. [DOI] [PubMed] [Google Scholar]
- 22.Chang J.E., Turley S.J. Trends Immunol. 2015;36:30. doi: 10.1016/j.it.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamurty A.T., Turley S.J. Nat. Immunol. 2020;21:369. doi: 10.1038/s41590-020-0635-3. [DOI] [PubMed] [Google Scholar]
- 24.Jafarnejad M., Woodruff M.C., Zawieja D.C., Carroll M.C., Moore J.E. Lymphatic Res. Biol. 2015;13:234. doi: 10.1089/lrb.2015.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun A., Worbs T., Moschovakis G.L., Halle S., Hoffmann K., Bölter J., Münk A., Förster R. Nat. Immunol. 2011;12:879. doi: 10.1038/ni.2085. [DOI] [PubMed] [Google Scholar]
- 26.Ulvmar M.H., Werth K., Braun A., Kelay P., Hub E., Eller K., Chan L., Lucas B., Novitzky-Basso I., Nakamura K., Rülicke T., Nibbs R.J.B., Worbs T., Förster R., Rot A. Nat. Immunol. 2014;15:623. doi: 10.1038/ni.2889. [DOI] [PubMed] [Google Scholar]
- 27.Gretz J.E., Norbury C.C., Anderson A.O., Proudfoot A.E.I., Shaw S. J. Exp. Med. 2000;192:1425. doi: 10.1084/jem.192.10.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ager A. Front. Immunol. 2017;8:45. doi: 10.3389/fimmu.2017.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granucci F., Zanoni I. Cell Cycle. 2009;8:3816. doi: 10.4161/cc.8.23.9998. [DOI] [PubMed] [Google Scholar]
- 30.Alvarez D., Vollmann E.H., von Andrian U.H. Immunity. 2008;29:325. doi: 10.1016/j.immuni.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heesters B.A., Myers R.C., Carroll M.C. Nat. Rev. Immunol. 2014;14:495. doi: 10.1038/nri3689. [DOI] [PubMed] [Google Scholar]
- 32.Katakai T. Front. Immunol. 2012;3:200. doi: 10.3389/fimmu.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farnsworth R.H., Karnezis T., Maciburko S.J., Mueller S.N., Stacker S.A. Front. Immunol. 2019;10:518. doi: 10.3389/fimmu.2019.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura R., Sawada S.I., Mukai S.A., Sasaki Y., Akiyoshi K. Biomacromolecules. 2020;21:621. doi: 10.1021/acs.biomac.9b01351. [DOI] [PubMed] [Google Scholar]
- 35.Hommel M., Kyewski B. J. Exp. Med. 2003;197:269. doi: 10.1084/jem.20021512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins M.K., Moon J.J. J. Immunol. 2012;188:4135. doi: 10.4049/jimmunol.1102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Cho B., Suzuki K., Xu Y., Green J.A., An J., Cyster J.G. J. Exp. Med. 2011;208:2497. doi: 10.1084/jem.20111449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mesin L., Ersching J., Victora G.D. Immunity. 2016;45:471. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schudel A., Francis D.M., Thomas S.N. Nat. Rev. Mater. 2019;4:415. doi: 10.1038/s41578-019-0110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latif R., de Rosbo N.K., Amarant T., Rappuoli R., Sappler G., Ben-Nun A. Infect. Immun. 2001;69:3073. doi: 10.1128/IAI.69.5.3073-3081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pennock N.D., White J.T., Cross E.W., Cheney E.E., Tamburini B.A., Kedl R.M. Adv. Physiol. Educ. 2013;37:273. doi: 10.1152/advan.00066.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vignali D.A.A., Collison L.W., Workman C.J. Nat. Rev. Immunol. 2008;8:523. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira L.M.R., Muller Y.D., Bluestone J.A., Tang Q. Nat. Rev. Drug Discov. 2019;18:749. doi: 10.1038/s41573-019-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savage P.A., Klawon D.E.J., Miller C.H. Annu. Rev. Immunol. 2020;38:421. doi: 10.1146/annurev-immunol-100219-020937. [DOI] [PubMed] [Google Scholar]
- 45.Dietrich T., Bock F., Yuen D., Hos D., Bachmann B.O., Zahn G., Wiegand S., Chen L., Cursiefen C. J. Immunol. 2010;184:535. doi: 10.4049/jimmunol.0903180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimoto Y., Nagashima S., Nakajima K., Ohira T., Sato T., Izawa T., Yamate J., Higashikawa K., Kuge Y., Ogawa M., Kojima C. Int. J. Pharm. 2020;576:119021. doi: 10.1016/j.ijpharm.2020.119021. [DOI] [PubMed] [Google Scholar]
- 47.Singh A., Peppas N.A. Adv. Mater. 2014;26:6530. doi: 10.1002/adma.201402105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuzzone D.A., Albano N.J., Aschen S.Z., Ghanta S., Mehrara B.J. Lymphatic Res. Biol. 2015;13:186. doi: 10.1089/lrb.2013.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raje N., Berdeja J., Lin Y., Siegel D., Jagannath S., Madduri D., Liedtke M., Rosenblatt J., Maus M.V., Turka A., Lam L.P., Morgan R.A., Friedman K., Massaro M., Wang J., Russotti G., Yang Z., Campbell T., Hege K., Petrocca F., Travis Quigley M., Munshi N., Kochenderfer J.N. N. Engl. J. Med. 2019;380:1726. doi: 10.1056/NEJMoa1817226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morton J.J., Bird G., Refaeli Y., Jimeno A. Cancer Res. 2016;76:6153. doi: 10.1158/0008-5472.CAN-16-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mestas J., Hughes C.C.W. J. Immunol. 2004;172:2731. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 52.Xiang M., Grosso R.A., Takeda A., Pan J., Bekkhus T., Brulois K., Dermadi D., Nordling S., Vanlandewijck M., Jalkanen S., Ulvmar M.H., Butcher E.C. Front. Cardiovasc. Med. 2020;7:52. doi: 10.3389/fcvm.2020.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shou Y., Zhang J., Yan S., Xia P., Xu P., Li G., Zhang K., Yin J. ACS Biomater. Sci. Eng. 2020;6:3619. doi: 10.1021/acsbiomaterials.0c00545. [DOI] [PubMed] [Google Scholar]
- 54.Wang Q., Shi Z., Shou Y., Zhang K., Li G., Xia P., Yan S., Yin J. ACS Biomater. Sci. Eng. 2020;6:1715. doi: 10.1021/acsbiomaterials.0c00010. [DOI] [PubMed] [Google Scholar]
- 55.Mantha S., Pillai S., Khayambashi P., Upadhyay A., Zhang Y., Tao O., Pham H.M., Tran S.D. Materials. 2019;12:3323. doi: 10.3390/ma12203323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geckil H., Xu F., Zhang X., Moon S., Demirci U. Nanomedicine. 2010;5:469. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Z., Wang Q., Li G.-F., Shou Y.-F., Zong H.-J., Yan S.-F., Zhang K.-X., Yin J.-B. Chin. J. Polym. Sci. 2021;39:327. [Google Scholar]
- 58.Caliari S.R., Burdick J.A. Nat. Methods. 2016;13:405. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Najibi A.J., Mooney D.J. Adv. Drug Deliv. Rev. 2020;161–162:42. doi: 10.1016/j.addr.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shanti A., Hallfors N., Petroianu G.A., Planelles L., Stefanini C. Front. Pharmacol. 2021;12:711307. doi: 10.3389/fphar.2021.711307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andorko J.I., Hess K.L., Jewell C.M. AAPS J. 2015;17:323. doi: 10.1208/s12248-014-9708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graney P.L., Tavakol D.N., Chramiec A., Ronaldson-Bouchard K., Vunjak-Novakovic G. iScience. 2021;24:102179. doi: 10.1016/j.isci.2021.102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim J., Koo B.K., Knoblich J.A. Nat. Rev. Mol. Cell Biol. 2020;21:571. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shah S.B., Singh A. Acta Biomater. 2017;53:29. doi: 10.1016/j.actbio.2017.01.075. [DOI] [PubMed] [Google Scholar]
- 65.Ye W., Luo C., Li C., Huang J., Liu F. Cancer Lett. 2020;477:31. doi: 10.1016/j.canlet.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 66.Gatto D., Brink R. J. Allergy Clin. Immunol. 2010;126:898. doi: 10.1016/j.jaci.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 67.Drakhlis L., Biswanath S., Farr C.-M., Lupanow V., Teske J., Ritzenhoff K., Franke A., Manstein F., Bolesani E., Kempf H., Liebscher S., Schenke-Layland K., Hegermann J., Nolte L., Meyer H., de la Roche J., Thiemann S., Wahl-Schott C., Martin U., Zweigerdt R. Nat. Biotechnol. 2021;39:737. doi: 10.1038/s41587-021-00815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimura H., Sakai Y., Fujii T. Drug Metabol. Pharmacokinet. 2018;33:43. doi: 10.1016/j.dmpk.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 69.Radisic M., Loskill P. ACS Biomater. Sci. Eng. 2021;7:2861. doi: 10.1021/acsbiomaterials.1c00831. [DOI] [PubMed] [Google Scholar]
- 70.Wu Q., Liu J., Wang X., Feng L., Wu J., Zhu X., Wen W., Gong X. Biomed. Eng. Online. 2020;19:9. doi: 10.1186/s12938-020-0752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sosa-Hernández J.E., Villalba-Rodríguez A.M., Romero-Castillo K.D., Aguilar-Aguila-Isaías M.A., García-Reyes I.E., Hernández-Antonio A., Ahmed I., Sharma A., Parra-Saldívar R., Iqbal H.M.N. Micromachines. 2018;9:536. doi: 10.3390/mi9100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hennessy R., Koo C., Ton P., Han A., Righetti R., Maitland K.C. 2011. Multimodal Biomed. Imaging VI. 7892, 78920R. [Google Scholar]
- 73.Ross A.E., Pompano R.R. Anal. Chim. Acta. 2018;1000:205. doi: 10.1016/j.aca.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Mousavi Shaegh S.A., Massa S., Riahi R., Chae S., Hu N., Avci H., Zhang W., Silvestri A., Sanati Nezhad A., Manbohi A., De Ferrari F., Polini A., Calzone G., Shaikh N., Alerasool P., Budina E., Kang J., Bhise N., Ribas J., Pourmand A., Skardal A., Shupe T., Bishop C.E., Dokmeci M.R., Atala A., Khademhosseini A. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114:E2293. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He J., Zhang X., Xia X., Han M., Li F., Li C., Li Y., Gao D. J. Mol. Cell Biol. 2020;12:569. doi: 10.1093/jmcb/mjaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dahl-Jensen S. A. Grapin-Botton, Dev. 2017;144:946. doi: 10.1242/dev.143693. [DOI] [PubMed] [Google Scholar]
- 77.Liu S.Q., Tian Q., Wang L., Hedrick J.L., Hui J.H.P., Yang Y.Y., Ee P.L.R. Macromol. Rapid Commun. 2010;31:1148. doi: 10.1002/marc.200900818. [DOI] [PubMed] [Google Scholar]
- 78.Burdick J.A., Anseth K.S. Biomaterials. 2002;23:4315. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 79.Brun P., Zamuner A., Peretti A., Conti J., Messina G.M.L., Marletta G., Dettin M. Sci. Rep. 2019;9:5583. doi: 10.1038/s41598-019-42071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel R., Santhosh M., Dash J.K., Karpoormath R., Jha A., Kwak J., Patel M., Kim J.H. Polym. Adv. Technol. 2019;30:4. [Google Scholar]
- 81.Lutolf M.P., Lauer-Fields J.L., Schmoekel H.G., Metters A.T., Weber F.E., Fields G.B., Hubbell J.A. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:5413. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kyburz K.A., Anseth K.S. Ann. Biomed. Eng. 2015;43:489. doi: 10.1007/s10439-015-1297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takebe T., Zhang B., Radisic M. Cell Stem Cell. 2017;21:297. doi: 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 84.Billiet T., Vandenhaute M., Schelfhout J., Van Vlierberghe S., Dubruel P. Biomaterials. 2012;33:6020. doi: 10.1016/j.biomaterials.2012.04.050. [DOI] [PubMed] [Google Scholar]
- 85.Cuchiara M.P., Gould D.J., McHale M.K., Dickinson M.E., West J.L. Adv. Funct. Mater. 2012;22:4511. doi: 10.1002/adfm.201200976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.V Murphy S., Atala A. Nat. Biotechnol. 2014;32:773. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y.S., Yue K., Aleman J., Mollazadeh-Moghaddam K., Bakht S.M., Yang J., Jia W., Dell'Erba V., Assawes P., Shin S.R., Dokmeci M.R., Oklu R., Khademhosseini A. Ann. Biomed. Eng. 2017;45:148. doi: 10.1007/s10439-016-1612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramiah P., du Toit L.C., Choonara Y.E., Kondiah P.P.D., Pillay V. Front. Mater. 2020;7:76. [Google Scholar]
- 89.Zhang J., Xin W., Qin Y., Hong Y., Xiahou Z., Zhang K., Fu P., Yin J. Chem. Eng. J. 2022;430:132713. [Google Scholar]
- 90.Gungor-Ozkerim P.S., Inci I., Zhang Y.S., Khademhosseini A., Dokmeci M.R. Biomater. Sci. 2018;6:915. doi: 10.1039/c7bm00765e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valverde M.G., Mille L.S., Figler K.P., Cervantes E., Li V.Y., Bonventre J.V., Masereeuw R., Zhang Y.S. Nat. Rev. Nephrol. 2022 doi: 10.1038/s41581-021-00528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gunawardena J. BMC Biol. 2014;12:29. doi: 10.1186/1741-7007-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Margaris K.N., Black R.A. J. R. Soc. Interface. 2012;9:601. doi: 10.1098/rsif.2011.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thomas P. Sci. Rep. 2019;9:474. doi: 10.1038/s41598-018-35927-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tang J., Enderling H., Becker-Weimann S., Pham C., Polyzos A., Chen C.-Y., V Costes S. Integr. Biol. (Camb). 2011;3:408. doi: 10.1039/c0ib00092b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang J., Fernandez-Garcia I., Vijayakumar S., Martinez-Ruis H., Illa-Bochaca I., Nguyen D.H., Mao J.-H., V Costes S., Barcellos-Hoff M.H. Stem Cell. 2014;32:649. doi: 10.1002/stem.1533. [DOI] [PubMed] [Google Scholar]
- 97.Tang J., Ley K.F., Hunt C.A. BMC Syst. Biol. 2007;1:14. doi: 10.1186/1752-0509-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tang J., Hunt C.A. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baldazzi V., Castiglione F., Bernaschi M. Cell. Immunol. 2006;244:77. doi: 10.1016/j.cellimm.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 100.Chiacchio F., Pennisi M., Russo G., Motta S., Pappalardo F. BioMed Res. Int. 2014;2014:907171. doi: 10.1155/2014/907171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Siddiqa A., Niazi M., Mustafa F., Bokhari H., Hussain A., Akram N., Shaheen S., Ahmed F., Iqbal S. International Conference on Information and Communication Technologies. IEEE; 2009. pp. 134–139. 2009. [Google Scholar]
- 102.Manifacier I., Chauvière A., Verdier C., Chagnon G., Cheddadi I., Glade N., Stéphanou A. Comput. Methods Biomech. Biomed. Eng. 2020;23:S183. [Google Scholar]
- 103.Bogle G., Dunbar P.R. Immunol. Cell Biol. 2008;86:676. doi: 10.1038/icb.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bogle G., Dunbar P.R. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Riggs T., Walts A., Perry N., Bickle L., Lynch J.N., Myers A., Flynn J., Linderman J.J., Miller M.J., Kirschner D.E. J. Theor. Biol. 2008;250:732. doi: 10.1016/j.jtbi.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tran Q.N. Emerging Trends in Applications and Infrastructures for Computational Biology, Bioinformatics, and Systems Biology. Elsevier; 2016. pp. 107–120. [Google Scholar]
- 107.Macauley M., Jenkins A., Davies R. Algebraic and Combinatorial Computational Biology. Elsevier; 2019. pp. 89–146. [Google Scholar]
- 108.Murrugarra D., Aguilar B. Algebraic and Combinatorial Computational Biology. Elsevier; 2019. pp. 147–173. [Google Scholar]
- 109.Tay J.C., Tan P. International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2006. pp. 5315–5321. 2006. [Google Scholar]
- 110.Ben Youssef B. Emerging Trends in Applications and Infrastructures for Computational Biology, Bioinformatics, and Systems Biology. Elsevier; 2016. pp. 287–303. [Google Scholar]
- 111.Akdur G. Procedia - Soc. Behav. Sci. 2011;28:825. [Google Scholar]
- 112.Lehotzky D., Zupanc G.K.H. Dev. Neurobiol. 2019;79:497. doi: 10.1002/dneu.22686. [DOI] [PubMed] [Google Scholar]
- 113.A. F. M. Marée, V. A. Grieneisen, P. Hogeweg, In Single-Cell-Based Models in Biology and Medicine, Birkhäuser Basel, Basel, pp. 107–136.
- 114.Guisoni N., Mazzitello K.I., Diambra L. Front. Physiol. 2018;6:61. [Google Scholar]
- 115.Durand M., Guesnet E. Comput. Phys. Commun. 2016;208:54. [Google Scholar]
- 116.Beltman J.B., Marée A.F.M., Lynch J.N., Miller M.J., De Boer R.J. J. Exp. Med. 2007;204:771. doi: 10.1084/jem.20061278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hirashima T., Rens E.G., Merks R.M.H. Dev. Growth Differ. 2017;59:329. doi: 10.1111/dgd.12358. [DOI] [PubMed] [Google Scholar]
- 118.Szabó A., Merks R.M.H. Front. Oncol. 2013;3:87. doi: 10.3389/fonc.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.S. E. M. Boas, Y. Jiang, R. M. H. Merks, S. A. Prokopiou, E. G. Rens, 2018, pp. 279–310.
- 120.Ilea M., Turnea M., Rotariu M. Rev. Med.-Chir. Soc. Med. Nat. Iasi. 2013;117:572. [PubMed] [Google Scholar]
- 121.Michelson S., Miller B.E., Glicksman A.S., Leith J.T. J. Theor. Biol. 1987;128:233. doi: 10.1016/s0022-5193(87)80171-6. [DOI] [PubMed] [Google Scholar]
- 122.Ilea M., Turnea M., Arotăriţei D., Toma C.M. Rev. Med.-Chir. Soc. Med. Nat. Iasi. 2010;114:937. [PubMed] [Google Scholar]
- 123.Louzoun Y. Immunol. Rev. 2007;216:9. doi: 10.1111/j.1600-065X.2006.00495.x. [DOI] [PubMed] [Google Scholar]
- 124.Day J.D., Metes D.M., Vodovotz Y. Front. Immunol. 2015;6:484. doi: 10.3389/fimmu.2015.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sontag E.D. Cell Syst. 2017;4:231. doi: 10.1016/j.cels.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.d'Onofrio A. Math. Comput. Model. 2008;47:614. [Google Scholar]
- 127.foryś U., waniewski J., zhivkov P. J. Biol. Syst. 2006;14:13. [Google Scholar]
- 128.Matzavinos A. Math. Med. Biol. 2004;21:1. doi: 10.1093/imammb/21.1.1. [DOI] [PubMed] [Google Scholar]
- 129.Baleanu D., Mohammadi H., Rezapour S. Adv. Differ. Equ. 2020;2020:299. doi: 10.1186/s13662-020-02762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Banks H.T., Holm K., Wanner N.C., Cintrón-Arias A., Kepler G.M., Wetherington J.D. Math. Comput. Model. 2009;50:959. doi: 10.1016/j.mcm.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Friedman A., Hao W. Math. Biosci. Eng. 2017;14:143. doi: 10.3934/mbe.2017010. [DOI] [PubMed] [Google Scholar]
- 132.Myers T.G., Ribas Ripoll V., Sáez de Tejada Cuenca A., Mitchell S.L., McGuinness M.J. Math. Case Stud. 2017;8:2. doi: 10.1186/s40929-017-0011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Garbey M., Casarin S., Berceli S.A. Biomech. Model. Mechanobiol. 2019;18:29. doi: 10.1007/s10237-018-1065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Milde F., Bergdorf M., Koumoutsakos P. Biophys. J. 2008;95:3146. doi: 10.1529/biophysj.107.124511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Perfahl H., Hughes B.D., Alarcón T., Maini P.K., Lloyd M.C., Reuss M., Byrne H.M. J. Theor. Biol. 2017;414:254. doi: 10.1016/j.jtbi.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 136.Sfakianakis N., Madzvamuse A., Chaplain M.A.J. Multiscale Model. Simul. 2020;18:824. [Google Scholar]
- 137.Zangooei M.H., Habibi J. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oduola W.O., Li X. Cancer Inf. 2018;17 doi: 10.1177/1176935118790262. 117693511879026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Benchaib M.A., Bouchnita A., Volpert V., Makhoute A. Front. Bioeng. Biotechnol. 2019;7:104. doi: 10.3389/fbioe.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rikard S.M., Athey T.L., Nelson A.R., Christiansen S.L.M., Lee J.-J., Holmes J.W., Peirce S.M., Saucerman J.J. Front. Physiol. 2019;10:1481. doi: 10.3389/fphys.2019.01481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Novkovic M., Onder L., Cheng H.-W., Bocharov G., Ludewig B. Front. Immunol. 2018;9:2428. doi: 10.3389/fimmu.2018.02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mann M., Backofen R. Bio. Algorithm Med. Syst. 2014;10:213. [Google Scholar]
- 143.Checa S., Prendergast P.J. Ann. Biomed. Eng. 2009;37:129. doi: 10.1007/s10439-008-9594-9. [DOI] [PubMed] [Google Scholar]
- 144.Roose T., Chapman S.J., Maini P.K. SIAM Rev. 2007;49:179. [Google Scholar]
- 145.Bean D.M., Heimbach J., Ficorella L., Micklem G., Oliver S.G., Favrin G. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Reisig W. In: Modeling in Systems Biology. Koch I., Reisig W., Schreiber F., editors. Springer London; London: 2011. pp. 37–56. [Google Scholar]
- 147.Chaouiya C., Remy E., Thieffry D. J. Discrete Algorithm. 2008;6:165. [Google Scholar]
- 148.Imoto S., Miyano S., Matsuno H. Computational Systems Biology. Elsevier; 2006. pp. 205–228. [Google Scholar]
- 149.Metzcar J., Wang Y., Heiland R., Macklin P. JCO Clin. Cancer Inf. 2019;3:1. doi: 10.1200/CCI.18.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.DeAngelis D.L., Grimm V. F1000Prime Rep. 2014;6:39. doi: 10.12703/P6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang B., Radisic M. Lab Chip. 2017;17:2395. doi: 10.1039/c6lc01554a. [DOI] [PubMed] [Google Scholar]
- 152.Moura Rosa P., Gopalakrishnan N., Ibrahim H., Haug M., Halaas Ø. Lab Chip. 2016;16:3728. doi: 10.1039/c6lc00702c. [DOI] [PubMed] [Google Scholar]
- 153.Tomei A.A., Siegert S., Britschgi M.R., Luther S.A., Swartz M.A. J. Immunol. 2009;183:4273. doi: 10.4049/jimmunol.0900835. [DOI] [PubMed] [Google Scholar]
- 154.Goyal G., Prabhala P., Mahajan G., Bausk B., Gilboa T., Xie L., Zhai Y., Lazarovits R., Mansour A., Kim M.S., Patil A., Curran D., Long J.E., Sharma S., Junaid A., Cohen L., Ferrante T.C., Levy O., Prantil-Baun R., Walt D.R., Ingber D.E. Adv. Sci. 2022 doi: 10.1002/advs.202103241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mitragotri S., Lahann J. Nat. Mater. 2009;8:15. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li X., Shou Y., Tay A. Adv. NanoBiomed. Res. 2021;1:2000073. doi: 10.1002/anbr.202000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Phillip J.M., Aifuwa I., Walston J., Wirtz D. Annu. Rev. Biomed. Eng. 2015;17:113. doi: 10.1146/annurev-bioeng-071114-040829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.R. Akhtar, B. Derby, 2015, pp. 1–6.
- 159.Laco F., Grant M.H., Black R.A. J. Biomed. Mater. Res. 2013;101 A:1787. doi: 10.1002/jbm.a.34468. [DOI] [PubMed] [Google Scholar]
- 160.Roozendaal R., Mebius R.E. Annu. Rev. Immunol. 2011;29:23. doi: 10.1146/annurev-immunol-031210-101357. [DOI] [PubMed] [Google Scholar]
- 161.Molenaar R., Greuter M., van der Marel A.P.J., Roozendaal R., Martin S.F., Edele F., Huehn J., Förster R., O'Toole T., Jansen W., Eestermans I.L., Kraal G., Mebius R.E. J. Immunol. 2009;183:6395. doi: 10.4049/jimmunol.0900311. [DOI] [PubMed] [Google Scholar]
- 162.Lee J.-W., Epardaud M., Sun J., Becker J.E., Cheng A.C., Yonekura A., Heath J.K., Turley S.J. Nat. Immunol. 2007;8:181. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]
- 163.Gardner J.M., DeVoss J.J., Friedman R.S., Wong D.J., Tan Y.X., Zhou X., Johannes K.P., Su M.A., Chang H.Y., Krummel M.F., Anderson M.S. Science (80-.) 2008;321:843. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim J., Wu B., Niedzielski S.M., Hill M.T., Coleman R.M., Ono A., Shikanov A. J. Biomed. Mater. Res. 2015;103:2701. doi: 10.1002/jbm.a.35409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Suematsu S., Watanabe T. Nat. Biotechnol. 2004;22:1539. doi: 10.1038/nbt1039. [DOI] [PubMed] [Google Scholar]
- 166.Okamoto N., Chihara R., Shimizu C., Nishimoto S., Watanabe T. J. Clin. Invest. 2007;117:997. doi: 10.1172/JCI30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ashammakhi N., Ahadian S., Xu C., Montazerian H., Ko H., Nasiri R., Barros N., Khademhosseini A. Mater. Today Bio. 2019;1:100008. doi: 10.1016/j.mtbio.2019.100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kelch I.D., Bogle G., Sands G.B., Phillips A.R.J., LeGrice I.J., Rod Dunbar P. Sci. Rep. 2015;5:16534. doi: 10.1038/srep16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Novkovic M., Onder L., Cupovic J., Abe J., Bomze D., Cremasco V., Scandella E., Stein J.V., Bocharov G., Turley S.J., Ludewig B. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Novkovic M., Onder L., Bocharov G., Ludewig B. Cell Rep. 2020;30:893. doi: 10.1016/j.celrep.2019.12.070. [DOI] [PubMed] [Google Scholar]
- 171.Kislitsyn A., Savinkov R., Novkovic M., Onder L., Bocharov G. Computation. 2015;3:222. [Google Scholar]
- 172.Tretyakova R., Savinkov R., Lobov G., Bocharov G. Computation. 2017;6:1. [Google Scholar]
- 173.von Andrian U.H., Mempel T.R. Nat. Rev. Immunol. 2003;3:867. doi: 10.1038/nri1222. [DOI] [PubMed] [Google Scholar]
- 174.Förster R., Braun A., Worbs T. Trends Immunol. 2012;33:271. doi: 10.1016/j.it.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 175.Stachowiak A.N., Irvine D.J. J. Biomed. Mater. Res. 2008;85A:815. doi: 10.1002/jbm.a.31661. [DOI] [PubMed] [Google Scholar]
- 176.Stachowiak A.N., Bershteyn A., Tzatzalos E., Irvine D.J. Adv. Mater. 2005;17:399. [Google Scholar]
- 177.Pérez del Río E., Santos F., Rodriguez Rodriguez X., Martínez-Miguel M., Roca-Pinilla R., Arís A., Garcia-Fruitós E., Veciana J., Spatz J.P., Ratera I., Guasch J. Biomaterials. 2020;259:120313. doi: 10.1016/j.biomaterials.2020.120313. [DOI] [PubMed] [Google Scholar]
- 178.Purwada A., Shah S.B., Béguelin W., August A., Melnick A.M., Singh A. Biomaterials. 2019;198:27. doi: 10.1016/j.biomaterials.2018.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nojima T., Haniuda K., Moutai T., Matsudaira M., Mizokawa S., Shiratori I., Azuma T., Kitamura D. Nat. Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 180.Purwada A., Jaiswal M.K., Ahn H., Nojima T., Kitamura D., Gaharwar A.K., Cerchietti L., Singh A. Biomaterials. 2015;63:24. doi: 10.1016/j.biomaterials.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Purwada A., Singh A. Nat. Protoc. 2017;12:168. doi: 10.1038/nprot.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Graney P.L., Lai K., Post S., Brito I., Cyster J., Singh A. Adv. Funct. Mater. 2020;30:2001232. doi: 10.1002/adfm.202001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Béguelin W., Rivas M.A., Calvo Fernández M.T., Teater M., Purwada A., Redmond D., Shen H., Challman M.F., Elemento O., Singh A., Melnick A.M. Nat. Commun. 2017;8:877. doi: 10.1038/s41467-017-01029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Purwada A., Shah S.B., Beguelin W., Melnick A.M., Singh A. ACS Biomater. Sci. Eng. 2017;3:214. doi: 10.1021/acsbiomaterials.6b00474. [DOI] [PubMed] [Google Scholar]
- 185.T.R M., S.E H., U.H V.A. Nature. 2004;427:154. [Google Scholar]
- 186.Miller M.J. Science (80-.) 2002;296:1869. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 187.Cahalan M.D., Parker I., Wei S.H., Miller M.J. Curr. Opin. Immunol. 2003;15:372. doi: 10.1016/s0952-7915(03)00079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bajénoff M., Egen J.G., Koo L.Y., Laugier J.P., Brau F., Glaichenhaus N., Germain R.N. Immunity. 2006;25:989. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cyster J.G. Science (80-.) 1999;286:2098. doi: 10.1126/science.286.5447.2098. [DOI] [PubMed] [Google Scholar]
- 190.Textor J., Henrickson S.E., Mandl J.N., von Andrian U.H., Westermann J., de Boer R.J., Beltman J.B. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Beauchemin C., Dixit N.M., Perelson A.S. J. Immunol. 2007;178:5505. doi: 10.4049/jimmunol.178.9.5505. [DOI] [PubMed] [Google Scholar]
- 192.Mandl J.N., Liou R., Klauschen F., Vrisekoop N., Monteiro J.P., Yates A.J., Huang A.Y., Germain R.N. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18036. doi: 10.1073/pnas.1211717109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Banigan E.J., Harris T.H., Christian D.A., Hunter C.A., Liu A.J. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fricke G.M., Letendre K.A., Moses M.E., Cannon J.L. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Harris T.H., Banigan E.J., Christian D.A., Konradt C., Tait Wojno E.D., Norose K., Wilson E.H., John B., Weninger W., Luster A.D., Liu A.J., Hunter C.A. Nature. 2012;486:545. doi: 10.1038/nature11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Castro M., Lythe G., Molina-París C., Ribeiro R.M. Interface Focus. 2016;6:14. doi: 10.1098/rsfs.2015.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Boyden S. J. Exp. Med. 1962;115:453. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zigmond S.H. J. Cell Biol. 1977;75:606. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zicha D., Dunn G.A., Brown A.F. J. Cell Sci. 1991;99(Pt 4):769. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 200.Zengel P., Nguyen-Hoang A., Schildhammer C., Zantl R., Kahl V., Horn E. BMC Cell Biol. 2011;12:21. doi: 10.1186/1471-2121-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jafarnejad M., Zawieja D.C., Brook B.S., Nibbs R.J.B., Moore J.E. J. Immunol. 2017;199:2291. doi: 10.4049/jimmunol.1700377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Azarov I., Peskov K., Helmlinger G., Kosinsky Y. Front. Immunol. 2019;10:1289. doi: 10.3389/fimmu.2019.01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Moore J.E., Bertram C.D. Annu. Rev. Fluid Mech. 2018;50:459. doi: 10.1146/annurev-fluid-122316-045259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Scott S., Ali Z. Micromachines. 2021;12:319. doi: 10.3390/mi12030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mavrogiannis N., Ibo M., Fu X., Crivellari F., Gagnon Z. Biomicrofluidics. 2016;10 doi: 10.1063/1.4950753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Birmingham K.G., O'Melia M.J., Bordy S., Reyes Aguilar D., El-Reyas B., Lesinski G., Thomas S.N. iScience. 2020;23:101751. doi: 10.1016/j.isci.2020.101751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fathi P., Holland G., Pan D., Esch M.B. ACS Appl. Bio Mater. 2020;3:6697. doi: 10.1021/acsabm.0c00609. [DOI] [PubMed] [Google Scholar]
- 208.Sato M., Sasaki N., Ato M., Hirakawa S., Sato K., Sato K. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Giese C., Lubitz A., Demmler C.D., Reuschel J., Bergner K., Marx U. J. Biotechnol. 2010;148:38. doi: 10.1016/j.jbiotec.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 210.Giese C., Demmler C.D., Ammer R., Hartmann S., Lubitz A., Miller L., Müller R., Marx U. Artif. Organs. 2006;30:803. doi: 10.1111/j.1525-1594.2006.00303.x. [DOI] [PubMed] [Google Scholar]
- 211.Seifert M., Lubitz A., Trommer J., Könnig D., Korus G., Marx U., Volk H.-D., Duda G., Kasper G., Lehmann K., Stolk M., Giese C. Int. J. Artif. Organs. 2012;35:986. doi: 10.5301/ijao.5000118. [DOI] [PubMed] [Google Scholar]
- 212.Shanti A., Samara B., Abdullah A., Hallfors N., Accoto D., Sapudom J., Alatoom A., Teo J., Danti S., Stefanini C. Pharmaceutics. 2020;12:464. doi: 10.3390/pharmaceutics12050464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mayer J., Swoger J., Ozga A.J., Stein J.V., Sharpe J. Comput. Math. Methods Med. 2012;2012:1. doi: 10.1155/2012/128431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cooper L.J., Heppell J.P., Clough G.F., Ganapathisubramani B., Roose T. Bull. Math. Biol. 2016;78:52. doi: 10.1007/s11538-015-0128-y. [DOI] [PubMed] [Google Scholar]
- 215.Ganusov V.V., Tomura M. Mathematical, Computational and Experimental T Cell Immunology. Springer International Publishing; Cham: 2021. pp. 151–169. [Google Scholar]
- 216.van Helden D.F. J. Physiol. 2014;592:5353. doi: 10.1113/jphysiol.2014.286039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Galie P., Spilker R.L. J. Biomech. Eng. 2009;131:111004. doi: 10.1115/1.3212108. [DOI] [PubMed] [Google Scholar]
- 218.Contarino C., Toro E.F. Biomech. Model. Mechanobiol. 2018;17:1687. doi: 10.1007/s10237-018-1050-7. [DOI] [PubMed] [Google Scholar]
- 219.Tretyakova R.M., Lobov G.I., Bocharov G.A. Math. Model Nat. Phenom. 2018;13:45. [Google Scholar]
- 220.Jamalian S., Davis M.J., Zawieja D.C., Moore J.E. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Macdonald A.J., Arkill K.P., Tabor G.R., McHale N.G., Winlove C.P. Am. J. Physiol. Cell Physiol. 2008;295:H305. doi: 10.1152/ajpheart.00004.2008. [DOI] [PubMed] [Google Scholar]
- 222.Quick C.M., Venugopal A.M., Gashev A.A., Zawieja D.C., Stewart R.H. Am. J. Physiol. Integr. Comp. Physiol. 2007;292:R1510. doi: 10.1152/ajpregu.00258.2006. [DOI] [PubMed] [Google Scholar]
- 223.Venugopal A.M., Stewart R.H., Laine G.A., Dongaonkar R.M., Quick C.M. Am. J. Physiol. Cell Physiol. 2007;293:H1183. doi: 10.1152/ajpheart.01340.2006. [DOI] [PubMed] [Google Scholar]
- 224.Apoorva F.N.U., Tian Y.F., Pierpont T.M., Bassen D.M., Cerchietti L., Butcher J.T., Weiss R.S., Singh A. J. Biomed. Mater. Res. 2017;105:1833. doi: 10.1002/jbm.a.36031. [DOI] [PubMed] [Google Scholar]
- 225.Gravelle P., Jean C., Familiades J., Decaup E., Blanc A., Bezombes-Cagnac C., Laurent C., Savina A., Fournié J.J., Laurent G. Am. J. Pathol. 2014;184:282. doi: 10.1016/j.ajpath.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 226.Tian Y.F., Ahn H., Schneider R.S., Yang S.N., Roman-Gonzalez L., Melnick A.M., Cerchietti L., Singh A. Biomaterials. 2015;73:110. doi: 10.1016/j.biomaterials.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jones D., Pereira E.R., Padera T.P. Front. Oncol. 2018;8:36. doi: 10.3389/fonc.2018.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Stachura J., Wachowska M., Kilarski W.W., Güç E., Golab J., Muchowicz A. OncoImmunology. 2016;5 doi: 10.1080/2162402X.2016.1182278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Brown M., Assen F.P., Leithner A., Abe J., Schachner H., Asfour G., V Stein J., Uhrin P., Sixt M. Science (80-.) 2018;359:1408. doi: 10.1126/science.aal3662. [DOI] [PubMed] [Google Scholar]
- 230.Ji R.-C. Int. J. Mol. Sci. 2016;18:51. doi: 10.3390/ijms18010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hoshida T., Isaka N., Hagendoorn J., di Tomaso E., Chen Y.-L., Pytowski B., Fukumura D., Padera T.P., Jain R.K. Cancer Res. 2006;66:8065. doi: 10.1158/0008-5472.CAN-06-1392. [DOI] [PubMed] [Google Scholar]
- 232.Takahashi K., Saga Y., Mizukami H., Takei Y., Urabe M., Kume A., Suzuki M., Ozawa K. Cancer Sci. 2011;102:2272. doi: 10.1111/j.1349-7006.2011.02099.x. [DOI] [PubMed] [Google Scholar]
- 233.Kim S., Chung M., Jeon N.L. Biomaterials. 2016;78:115. doi: 10.1016/j.biomaterials.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 234.Bonvin C., Overney J., Shieh A.C., Dixon J.B., Swartz M.A. Biotechnol. Bioeng. 2010;105:982. doi: 10.1002/bit.22608. [DOI] [PubMed] [Google Scholar]
- 235.Frenkel N., Poghosyan S., Alarcón C.R., García S.B., Queiroz K., van den Bent L., Laoukili J., Rinkes I.B., Vulto P., Kranenburg O., Hagendoorn J. ACS Biomater. Sci. Eng. 2021;7:3030. doi: 10.1021/acsbiomaterials.0c01378. [DOI] [PubMed] [Google Scholar]
- 236.Valastyan S., Weinberg R.A. Cell. 2011;147:275. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pisano M., Triacca V., Barbee K.A., Swartz M.A. Integr. Biol. (United Kingdom) 2015;7:525. doi: 10.1039/c5ib00085h. [DOI] [PubMed] [Google Scholar]
- 238.Lugo-Cintrón K.M., Ayuso J.M., White B.R., Harari P.M., Ponik S.M., Beebe D.J., Gong M.M., Virumbrales-Muñoz M. Lab Chip. 2020;20:1586. doi: 10.1039/d0lc00099j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Polacheck W.J., Charest J.L., Kamm R.D. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:11115. doi: 10.1073/pnas.1103581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bouta E.M., Wood R.W., Brown E.B., Rahimi H., Ritchlin C.T., Schwarz E.M. J. Physiol. 2014;592:1213. doi: 10.1113/jphysiol.2013.266700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Greenlee J.D., King M.R. Biomicrofluidics. 2020;14 doi: 10.1063/1.5133970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Chen Y.-C., Allen S.G., Ingram P.N., Buckanovich R., Merajver S.D., Yoon E. Sci. Rep. 2015;5:9980. doi: 10.1038/srep09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Shim S., Belanger M.C., Harris A.R., Munson J.M., Pompano R.R. Lab Chip. 2019;19:1013. doi: 10.1039/c8lc00957k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lee C., Jeong S., Jang C., Bae H., Kim Y.H., Park I., Kim S.K., Koh G.Y. Science (80-.) 2019;363:644. doi: 10.1126/science.aav0173. [DOI] [PubMed] [Google Scholar]
- 245.Chandrasekaran S., McGuire M.J., King M.R. Lab Chip. 2014;14:118. doi: 10.1039/c3lc50584g. [DOI] [PubMed] [Google Scholar]
- 246.Votanopoulos K.I., Forsythe S., Sivakumar H., Mazzocchi A., Aleman J., Miller L., Levine E., Triozzi P., Skardal A. Ann. Surg Oncol. 2020;27:1956. doi: 10.1245/s10434-019-08143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Votanopoulos K.I., Skardal A. Ann. Surg Oncol. 2020;27:1968. doi: 10.1245/s10434-020-08351-7. [DOI] [PubMed] [Google Scholar]
- 248.Lenti E., Bianchessi S., Proulx S.T., Palano M.T., Genovese L., Raccosta L., Spinelli A., Drago D., Andolfo A., Alfano M., Petrova T.V., Mukenge S., Russo V., Brendolan A. Stem Cell Rep. 2019;12:1260. doi: 10.1016/j.stemcr.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Petersdorf E.W., Malkki M., Gooley T.A., Martin P.J., Guo Z. PLoS Med. 2007;4:e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shanti A., Teo J., Stefanini C. Pharmaceutics. 2018;10:278. doi: 10.3390/pharmaceutics10040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ross A.E., Belanger M.C., Woodroof J.F., Pompano R.R. Analyst. 2017;142:649. doi: 10.1039/c6an02042a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kwak B., Ozcelikkale A., Shin C.S., Park K., Han B. J. Contr. Release. 2014;194:157. doi: 10.1016/j.jconrel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Higbee R.G., Byers A.M., Dhir V., Drake D., Fahlenkamp H.G., Gangur J., Kachurin A., Kachurina O., Leistritz D., Ma Y., Mehta R., Mishkin E., Moser J., Mosquera L., Nguyen M., Parkhill R., Pawar S., Poisson L., Sanchez-Schmitz G., Schanen B., Singh I., Song H., Tapia T., Warren W., Wittman V. ATLA Altern. Lab. Anim. 2009;37:19. doi: 10.1177/026119290903701S05. [DOI] [PubMed] [Google Scholar]
- 254.Wilkie K.P., Hahnfeldt P. Interface Focus. 2013;3:20130010. doi: 10.1098/rsfs.2013.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Frieboes H.B., Smith B.R., Chuang Y.-L., Ito K., Roettgers A.M., Gambhir S.S., Cristini V. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Du W., Goldstein R., Jiang Y., Aly O., Cerchietti L., Melnick A., Elemento O. Cancer Res. 2017;77:1818. doi: 10.1158/0008-5472.CAN-16-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Burger J.A., Chiorazzi N. Trends Immunol. 2013;34:592. doi: 10.1016/j.it.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Clark B., Sitzia J., Harlow W. QJM An Int. J. Med. 2005;98:343. doi: 10.1093/qjmed/hci053. [DOI] [PubMed] [Google Scholar]
- 259.Hadrian R., Palmes D. Lymphatic Res. Biol. 2017;15:2. doi: 10.1089/lrb.2016.0015. [DOI] [PubMed] [Google Scholar]
- 260.Eymard N., Volpert V., Quere I., Lajoinie A., Nony P., Cornu C. Math. Model Nat. Phenom. 2017;12:180. [Google Scholar]
- 261.Andrews P.S., Timmis J. Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2006. pp. 164–177. [Google Scholar]
- 262.Mirsky H.P., Miller M.J., Linderman J.J., Kirschner D.E. J. Theor. Biol. 2011;287:160. doi: 10.1016/j.jtbi.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Grebennikov D., Bocharov G. IEEE Congress on Evolutionary Computation (CEC) IEEE; 2017. pp. 2653–2655. 2017. [Google Scholar]
- 264.Grebennikov D., van Loon R., Novkovic M., Onder L., Savinkov R., Sazonov I., Tretyakova R., Watson D., Bocharov G. Computation. 2016;5:3. [Google Scholar]
- 265.Bocharov G., Novkovic M., Onder L., Kislitsyn A., Savinkov R. International Workshop on Artificial Immune Systems (AIS) IEEE; 2015. pp. 1–2. 2015. [Google Scholar]
- 266.Meyer-Hermann M., Figge M.T., Toellner K.M. Trends Immunol. 2009;30:157. doi: 10.1016/j.it.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 267.Grigorova I.L., Panteleev M., Cyster J.G. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20447. doi: 10.1073/pnas.1009968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Baldazzi V., Paci P., Bernaschi M., Castiglione F. BMC Bioinf. 2009;10:387. doi: 10.1186/1471-2105-10-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Johnson S.C., Frattolin J., Edgar L.T., Jafarnejad M., Moore J.E. J. R. Soc. Interface. 2021;18 doi: 10.1098/rsif.2021.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Bouchnita A., Bocharov G., Meyerhans A., Volpert V. BMC Immunol. 2017;18:29. doi: 10.1186/s12865-017-0205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mozokhina A., Savinkov R. Mathematics. 2020;8:1467. [Google Scholar]
- 272.Ziraldo C., Gong C., Kirschner D.E., Linderman J.J. Front. Microbiol. 2016;6:1477. doi: 10.3389/fmicb.2015.01477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Celli S., Day M., Müller A.J., Molina-Paris C., Lythe G., Bousso P. Blood. 2012;120:3945. doi: 10.1182/blood-2012-01-408260. [DOI] [PubMed] [Google Scholar]
- 274.Zheng H., Jin B., Henrickson S.E., Perelson A.S., von Andrian U.H., Chakraborty A.K. Mol. Cell Biol. 2008;28:4040. doi: 10.1128/MCB.00136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bogle G., Dunbar P.R. Immunol. Cell Biol. 2010;88:172. doi: 10.1038/icb.2009.78. [DOI] [PubMed] [Google Scholar]
- 276.Huang W.Z., Hu W.H., Wang Y., Chen J., Hu Z.Q., Zhou J., Liu L., Qiu W., Tang F.Z., Zhang S.C., Ouyang Y., Ye Y.N., Xu G.Q., Long J.H., Zeng Z. Int. J. Biol. Sci. 2019;15:1396. doi: 10.7150/ijbs.33412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Linderman J.J., Riggs T., Pande M., Miller M., Marino S., Kirschner D.E. J. Immunol. 2010;184:2873. doi: 10.4049/jimmunol.0903117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Gong C., Mattila J.T., Miller M., Flynn J.L., Linderman J.J., Kirschner D. J. Theor. Biol. 2013;335:169. doi: 10.1016/j.jtbi.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Gong C., Linderman J.J., Kirschner D. Front. Immunol. 2014;5:57. doi: 10.3389/fimmu.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Graw F., Regoes R.R. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Erwin S., Childs L.M., Ciupe S.M. Sci. Rep. 2020;10:3935. doi: 10.1038/s41598-020-60316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yagawa Y., Robertson-Tessi M., Zhou S.L., Anderson A.R.A., Mulé J.J., Mailloux A.W. Sci. Rep. 2017;7:15996. doi: 10.1038/s41598-017-15924-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Shields J.D., Fleury M.E., Yong C., Tomei A.A., Randolph G.J., Swartz M.A. Cancer Cell. 2007;11:526. doi: 10.1016/j.ccr.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 284.Hartung N., Mollard S., Barbolosi D., Benabdallah A., Chapuisat G., Henry G., Giacometti S., Iliadis A., Ciccolini J., Faivre C., Hubert F. Cancer Res. 2014;74:6397. doi: 10.1158/0008-5472.CAN-14-0721. [DOI] [PubMed] [Google Scholar]
- 285.Liu T., Su X., Chen W., Zheng L., Li L., Yang A. Eur. Radiol. 2014;24:2827. doi: 10.1007/s00330-014-3335-2. [DOI] [PubMed] [Google Scholar]
- 286.Norton K.-A., Gong C., Jamalian S., Popel A. Processes. 2019;7:37. doi: 10.3390/pr7010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.DePillis L., Gallegos A., Radunskaya A. Front. Oncol. 2013;3:56. doi: 10.3389/fonc.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Jain R.K., Tong R.T., Munn L.L. Cancer Res. 2007;67:2729. doi: 10.1158/0008-5472.CAN-06-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Campanholi L.L., Duprat Neto J.P., Fregnani J.H.T.G. Int. J. Surg. 2011;9:306. doi: 10.1016/j.ijsu.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 290.Jenner A.L., Aogo R.A., Davis C.L., Smith A.M., Craig M. Curr. Pathobiol. Rep. 2020;8:149. doi: 10.1007/s40139-020-00213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Takeda A., Hollmén M., Dermadi D., Pan J., Brulois K.F., Kaukonen R., Lönnberg T., Boström P., Koskivuo I., Irjala H., Miyasaka M., Salmi M., Butcher E.C., Jalkanen S. Immunity. 2019;51:561. doi: 10.1016/j.immuni.2019.06.027. [DOI] [PubMed] [Google Scholar]
- 292.Roosenboom B., van Lochem E.G., Meijer J., Smids C., Nierkens S., Brand E.C., van Erp L.W., Kemperman L.G.J.M., Groenen M.J.M., Horjus Talabur Horje C.S., Wahab P.J. Cells. 2020;9:891. doi: 10.3390/cells9040891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Morsink M., Willemen N., Leijten J., Bansal R., Shin S. Micromachines. 2020;11:849. doi: 10.3390/mi11090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Hutmacher D.W., Little J.P., Pettet G.J., Loessner D. J. Mater. Sci. Mater. Med. 2015;26:185. doi: 10.1007/s10856-015-5518-z. [DOI] [PubMed] [Google Scholar]
- 295.V. B. Nguyen, C. H. Le, Z. Zhang, R. Lee, T. A. Nguyen, M. S. Packianather, 2018, pp. 655–660.
- 296.Campbell K.T., Wysoczynski K., Hadley D.J., Silva E.A. ACS Biomater. Sci. Eng. 2020;6:308. doi: 10.1021/acsbiomaterials.9b01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hajikhani A., Marino M., Wriggers P. Proc. Appl. Math. Mech. 2019:19. [Google Scholar]
- 298.Barrera O., Cocks A.C.F. Philos. Mag. A. 2013;93:2680. [Google Scholar]
- 299.Labus K., Radosinski L., Kotowski P. Int. J. Mol. Sci. 2021;22:9909. doi: 10.3390/ijms22189909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Amor N., Geris L., Vander Sloten J., Van Oosterwyck H. Acta Biomater. 2011;7:779. doi: 10.1016/j.actbio.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 301.Norfleet D.A., Park E., Kemp M.L. Curr. Opin. Biomed. Eng. 2020;13:113. [Google Scholar]
- 302.Carvalho V., Rodrigues R.O., Lima R.A., Teixeira S. Micromachines. 2021;12:1149. doi: 10.3390/mi12101149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Yazdian Kashani S., Keshavarz Moraveji M., Bonakdar S. Sci. Rep. 2021;11:12130. doi: 10.1038/s41598-021-91616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Zhu Z., Ng D.W.H., Park H.S., McAlpine M.C. Nat. Rev. Mater. 2021;6:27. [Google Scholar]
- 305.Almouemen N., Kelly H.M., O'Leary C. Comput. Struct. Biotechnol. J. 2019;17:591. doi: 10.1016/j.csbj.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Wang D.D., Qian Z., Vukicevic M., Engelhardt S., Kheradvar A., Zhang C., Little S.H., Verjans J., Comaniciu D., O'Neill W.W., Vannan M.A. JACC Cardiovasc. Imaging. 2021;14:41. doi: 10.1016/j.jcmg.2019.12.022. [DOI] [PubMed] [Google Scholar]
- 307.Shirazi R.N., Islam S., Weafer F.M., Whyte W., Varela C.E., Villanyi A., Ronan W., McHugh P., Roche E.T. Adv. Healthc. Mater. 2019;8:1900228. doi: 10.1002/adhm.201900228. [DOI] [PubMed] [Google Scholar]
- 308.Sulejmani F., Caballero A., Martin C., Pham T., Sun W. J. Mech. Behav. Biomed. Mater. 2019;97:159. doi: 10.1016/j.jmbbm.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Chappard D., Kün-Darbois J.-D., Guillaume B. Micron. 2020;133:102861. doi: 10.1016/j.micron.2020.102861. [DOI] [PubMed] [Google Scholar]
- 310.Lehder E.F., Ashcroft I.A., Wildman R.D., Maskery I., Cantu L.R. Solid Freeform Fabrication 2019: Proceedings of the 30th Annual International Solid Freeform Fabrication Symposium - an Additive Manufacturing Conference. SFF; 2019. pp. 1954–1964. 2019. [Google Scholar]
- 311.Farooqi A.R., Zimmermann J., Bader R., van Rienen U. Comput. Methods Progr. Biomed. 2020;197:105739. doi: 10.1016/j.cmpb.2020.105739. [DOI] [PubMed] [Google Scholar]
- 312.Kinch M.S., Merkel J. Drug Discov. Today. 2015;20:920. doi: 10.1016/j.drudis.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 313.DiMasi J.A., Grabowski H.G., Hansen R.W. J. Health Econ. 2016;47:20. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 314.Jiang L., Yin K., Wen Q., Chen C., Ge M., Tan Z. Sci. Rep. 2020;10:710. doi: 10.1038/s41598-019-55991-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zhu J., Wang W., Xu S., Jia C., Zhang Q., Xia Y., Wang W., Wen M., Wang X., Wang H., Zhang Z., Cai L., Zhang L., Jiang T. Front. Oncol. 2020;10:563700. doi: 10.3389/fonc.2020.563700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Navarro J.G., Lee J.H., Kang I., Rho S.Y., Choi G.H., Han D.H., Kim K.S., Choi J.S. HPB. 2020;22:1411. doi: 10.1016/j.hpb.2020.01.009. [DOI] [PubMed] [Google Scholar]