Abstract
Models of heart disease and drug responses are increasingly based on human pluripotent stem cells (hPSCs) since their ability to capture human heart (dys-)function is often better than animal models. Simple monolayer cultures of hPSC-derived cardiomyocytes, however, have shortcomings. Some of these can be overcome using more complex, multi cell-type models in 3D. Here we review modalities that address this, describe efforts to tailor readouts and sensors for monitoring tissue- and cell physiology (exogenously and in situ) and discuss perspectives for implementation in industry and academia.
Keywords: Human induced pluripotent stem cells, Cardiac microtissue, Engineered heart tissue, Heart-on-chip, Cardiomyocyte maturation, Multicellular cell diseases and drug efficacy platform, Structural readout, Functional readout
Graphical abstract
1. Introduction
Animal models have been very useful in understanding how the heart develops and the molecular mechanisms underlying congenital heart defects. They have been less successful in identifying the genetic and other causes of cardiac dysfunction or predicting the effects of drugs, largely because of differences in animal and human physiology. The advent of human pluripotent stem cells (hPSCs) [1,2] has however opened new avenues to study the human heart and its pathologies. We review here state-of-the-art models now being used to investigate the human heart, how they are being used to monitor human heart (patho-)physiology, their shortcomings and the expectations for the future.
Cardiomyocytes (CM) were among the first cell types to be differentiated from hPSCs [3,4], specifically from human embryonic stem cells (hESCs). Differentiation protocols initially entailed culture of undifferentiated cells as aggregates in suspension, called “embryoid bodies”, in the presence of growth factors. Since these first reports, many groups have developed methods to differentiate CMs in 2D (reviewed in Ref. [5]) and to form three dimensional (3D) cardiac tissues (Table 1). Whilst strikingly evident because of their cyclic contraction, at most 5% of the differentiated cells were initially CMs. This meant that most analysis was based on methods suitable for single cells such as patch clamp electrophysiology and immunostaining of sarcomeric proteins. hPSC-derived CMs (hPSC-CMs) were also immature compared to primary adult CMs with respect to their morphology, contractility, electrophysiology, metabolism and gene expression [[6], [7], [8]]. Nevertheless, various studies have shown that hPSC-CMs can be valuable for disease modelling and drug screening even if the CMs are immature, especially if they are derived from human induced pluripotent stem cells (hiPSCs) from patients [[9], [10], [11], [12], [13]]. Notably, all cardiac ion channels are expressed in hPSC-CMs so that modelling channelopathies caused by mutations in ion channel genes has been particularly successful. hiPSCs carry the genetic background of the individuals from whom they are derived and thus allow diseased genomes or disease predisposition to be captured in the derivative cardiac cell types; they thus support more personalized approaches to disease modelling and drug screening.
Table 1.
Literature summary of microtissues/organoids.
| Study | Cell types | Application | Readouts |
|
|---|---|---|---|---|
| Structural | Functional | |||
| Microtissues/Organoids | ||||
| [80] Archer et al., 2018 | hiPSC-CM Primary human cardiac ECs Primary human cFBs |
Drug Screening | Histology Confocal Imaging – immunofluorescence |
Gene expression (qRT-PCR) Cell viability assay (cellular ATP content) |
| [14,17] Beauchamp et al., 2015; 2020 | hiPSC-CM human embryonic-cFBs |
Cardiac development Disease modelling -cardiac fibrosis |
Confocal imaging-immunofluorescence Electron microscopy Histology |
Cell viability assay (live-dead staining) Ca2+ imaging Pharmacological analysis Gene expression (qRT-PCR) Western Blot Motion analysis Patch clamp |
| [28] Drakhlis et al., 2021 | hECS-CM | Cardiac development | Histology Multiphoton microscopy –immunofluorescence Live Optical fluorescence imaging - reporter cells; DiI-Ac-LDL Cell composition analysis – Flow cytometry Electron microscopy |
Ca2+ imaging Patch clamp Gene expression (microarray, RNAseq) |
| [22] Giacomelli et al., 2017 | hiPSC or hECS-CM hiPSC or hESC-cardiac EC |
Cardiac development Cardiac Maturation |
Optical Microscopy Confocal Imaging-immunofluorescence |
Gene expression (qRT-PCR) FACS Motion analysis Patch clamp MEA analysis |
| [18] Giacomelli et al., 2020 | hiPSC-CM hiPSC-cardiac EC hiPSC-cFBs |
Cardiac development Cardiac maturation Disease modelling -arrtyhmogenic cardiomyopathy |
Confocal Imaging-immunofluorescence Electron microscopy |
Gene expression (qRT-PCR, RNA seq) Western Blot Motion analysis Patch clamp Sharp electrode electrophysiology Ca+2 imaging Mitocondrial stress test- seahorse ELISA-cAMP level Nuclear magnetic resonance spectroscopy |
| [[21], [79]] Pointon et al., 2015; 2017 | hiPSC-CM Primary human cardiac microvascular EC Primary human cFBs |
Drug Screening | Confocal imaging-immunofluorescence | Gene expression (qRT-PCR) Motion analysis Cell viability assay (cellular ATP content) |
| [77] Ravenscroft et al., 2016 | hESC-CM Primary human EC Primary human cFBs or FBs |
Drug Screening Toxicity Testing |
Confocal imaging-immunofluorescence | Gene expression (qRT-PCR) Motion analysis Ca+2 Imaging |
| [20] Richards et al., 2020 | hiPSC-CM HUVECs Primary human cFBs |
Disease modelling -myocardial infraction | Confocal imaging- immunofluorescence | Gene expression (qRT-PCR) Motion analysis Ca+2 imaging Mitochondrial stress test - seahorse |
| [30] Silva et al., 2020 | hiPSC-CM | Cardiac development Cardiac maturation |
Optical and light sheet microscopy – immunofluorescence Histology |
Ca+2 imaging Patch clamp Gene expression (RNAseq) |
| [57] Lee et al., 2019 | hESC-CM hESC-mesenchymal stem cells |
Disease modelling Cardiac Fibrosis |
Optical Microscopy Confocal imaging-immunofluorescence |
Gene expression (qRT-PCR, microarray) FACS Motion analysis MEA analysis |
| [29] Hofbauer et al., 2021 | hiPSC-CM hESC-CM Human cardiac microvascular EC Chicken embryo |
Cardiac development | Optical Microscopy Confocal imaging-immunofluorescence Histology Electron microscopy |
Ca+2 and voltage imaging Motion analysis Gene expression (RNAseq) Reporter cell lines Proteomics analysis |
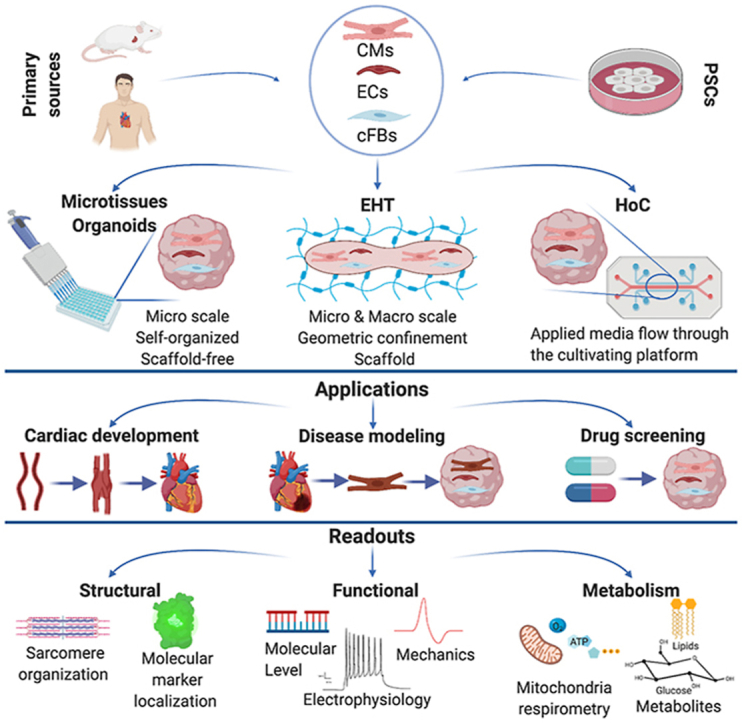
hPSC-CM immaturity has, however, been a challenge to modelling disease other than channelopathies and improving maturation state has been studied extensively over the last several years. Several reports have shown that culturing hPSC-CMs in 3D, mimicking the environment of cardiac tissue in vivo, improves the physiological and functional properties of CMs compared to 2D monolayers [[14], [15], [16]]. Moreover, if these 3D models also capture the complexity of native heart tissue, such as the communication between CMs and cardiac stromal- and vascular cells, then maturation is improved even further [[17], [18], [19], [20]]. Initially, primary endothelial cells (EC) such as those from human umbilical vein (HUVECs), or dermal fibroblasts (FB) were used in these 3D models but more recently it has been possible to generate ECs (specifically cardiac ECs) and cardiac FBs (cFB) from hPSC [18,21,22]. These have been combined in different ratios with CMs in microphysiological heart tissues (MHTs) (Table 1). The results broadly showed benefits for hPSC-CM maturation using cFB rather than dermal FBs and that this was mediated by junctional coupling with CMs [18]. 3D MHTs not only offer the advantage of being able to integrate multiple cell types but also microenvironmental cues, such as forced electrical or mechanical “pacing” or those mediated by the tissue scaffold, to closely mimic native heart. In this review, we first describe current MHT models, then summarize their applications with an overview of structural and functional readouts that can be used to report (patho-)physiology and drug/compound responses. Finally, we discuss how the field might move forward in refining and implementing these in the near future.
2. Current microphysiological tissue models
MHTs can be broadly divided into three types: those based on self-organized aggregates of cells (also referred to as spheroids, microtissues or organoids), those relying on biomaterials and bioengineering (engineered heart tissues (EHT)) and finally those in which perfusion can be integrated (Heart-on-chip). Human MHT models together with their cell sources, applications and readouts are summarized in Table 1, Table 2, Table 3. These tables are based on the selection from many studies in the field with a focus on more recent studies.
Table 2.
Literature summary of EHTs.
| Study | Cell types | Application | Readouts |
|
|---|---|---|---|---|
| Structural | Functional | |||
| EHTs | ||||
| [128] Dostanić et al., 2020 | hiPSC-CM hiPSC-cFBs |
Miniaturized cardiac tissues - Collagen/Matrigel hydrogel | Motion analysis Force measurement |
|
| [50,67,68,88] Nunes et al., 2013; Wang et al., 2019; Zhao et al., 2019; Feric et al., 2019 |
hiPSC-CM Human ventricular cFBs |
Cardiac maturation Disease modelling Drug screening - Collagen/Matrigel/Fibrin hydrogel |
Histology Confocal imaging- immunofluorescence Electron microscopy Second harmonic generation |
Force measurement Motion analysis Ca2+ imaging Pharmacological analysis Gene expression (qRT-PCR; RNAseq) Electrophysiological measurements Western Blot Viability assay Pharmacological analysis |
| [38,126] Huebsch et al., 2015; 2016 | hiPSC-CM Stromal cells |
Miniaturized cardiac tissues - geometric confinment | Confocal imaging- immunofluorescence | Force measurement Motion analysis MEA analysis Ca2+ Imaging Patch-clamp Pharmacological analysis |
| [40] Leonard et al., 2018 | hiPSC-CM | Cardiac Maturation - Fibrin hydrogel | Optical and confocal imaging- immunofluorescence Electron microscopy |
Force measurement Motion analysis Gene expression (qRT-PCR) Ca2+ Imaging Western Blot |
| [89] Lind et al., 2017 | hiPS-CM HUVEC Neonatal Rat Ventricular CM |
Drug screening | Confocal imaging- immunofluorescence | Motion analysis + sensors |
| [87] Lu et al., 2017 | hiPSC-CM | Drug screening - Fibrin hydrogel | Confocal imaging- immunofluorescence Electron microscopy |
Cell viability assay (MTS) Gene expression (qRT-PCR) Motion analysis Pharmacological analysis |
| [33,34] Ma et al., 2014; 2015 | hESC-CM hiPSC-CM |
Geometric confinement Disease modeling |
Confocal imaging- immunofluorescence | Motion analysis MEA analysis Gene expression (qRT-PCR) Cell proliferation assay (EdU) Cell migration imaging Pharmacological analysis |
| [70] Macqueen et al., 2018 | Neonatal rat ventricular CM hiPSC-CM | Disease modelling - Polycaprolactone (PCL)/gelatin nanofibres scaffold | Confocal imaging- immunofluorescence | Ca2+ Imaging Pressure-volume measurements Echocardiology |
| [85,86] Mannhardt et al., 2016; 2020 | hiPSC-CM | Drug screening - Fibrin hydrogel | Confocal imaging- immunofluorescence Histology Electron microscopy |
Motion analysis Pharmacological analysis Gene expression (RNAseq) |
| [66] Prondzynski et al., 2019 | Human primary CM hiPSC-CM | Disease modelling - Fibrin/Matrigel hydrogel | Confocal imaging- immunofluorescence | Gene expression (RNAseq, nanoString nCounter) Western Blot Motion analysis Action potential measurement (electrodes and patch clamp) |
| [41] Ronaldson-Bouchard et al., 2018 | hiPSC-CM human FBs |
Cardiac maturation - Fibrin/hydrogel | Confocal imaging- immunofluorescence Electron microscopy |
Force measurement Path-clamp Motion analysis Ca2+ imaging Gene expression (qRT-PCR) Pharmacological analysis Mitochondrial respirometry - Seahorse |
| [78] Takeda et al., 2018 | hiPSC-CM | Drug cytotoxicity | Confocal imaging- immunofluorescence Histology |
Gene expression (qRT-PCR) MEA analysis Cell viability assay (cellular ATP content) LDH assay Ca2+ imaging Motion analysis |
| [55] Tiburcy et al., 2017 | hPSC-CM human FBs |
Cardiac maturation Disease modelling - Collagen hydrogel |
Cell composition analysis – Flow cytometry; Confocal imaging- immunofluorescence | Force measurement Motion analysis Gene espression (qRT-PCR, RNAseq) Action potential measurement – electrodes |
| [19,53] Mills et al., 2017, 2019 | hESC-CM Neonatal Rat Ventricular CM |
Cardiac Maturation Drug screening - Collagen/Matrigel Hydrogel |
Optical and Confocal imaging - Immunofluorescence Cell composition analysis – Flow cytometry Electron microscopy |
Motion analysis Ca2+ imaging Patch-clamp Gene expression (qRT-PCR, RNAseq) Mass Spectrometry - proteomic analysis; Central carbon metabolite analysis Mitochondrial respirometry - Seahorse |
| [63] Bailey et al., 2021 | hiPSC-CM hiPSC-fibroblasts Primary macrophages |
Disease modelling - SARS-CoV-2 | Confocal imaging- immunofluorescence Histology Electron microscopy |
Gene expression (qRT-PCR; RNAseq; RNAscope) Motion analysis Cell viability assay (TUNEL) |
Table 3.
Literature summary of HoCs.
| Study | Cell types | Applications | Readouts |
|
|---|---|---|---|---|
| Structural | Functional | |||
| HoCs | ||||
| [83] Lee at al., 2021 | HER-2-overexpressing human breast cancer cell line hiPSC – CM | Disease modeling | Optical microscopy – brightfield Confocal microscopy – immunofluorescence |
Biosensors on micro-electrodes |
| [121] Maoz et al., 2017 | hiPSC – CM | Drug screening | Electron microscopy Confocal microscopy – immunofluorescence |
MEA analysis |
| [45] Marsano et al., 2016 | neonatal rat ventricular CM hiPSC – CM |
Disease modelling | Optical microscopy – brightfield Confocal microscopy – immunofluorescence |
Motion analysis |
| [95] Mastikhina et al., 2020 | human cFBs hiPSC – CM |
Disease modelling | Optical microscopy – brightfield Confocal microscopy – immunofluorescence |
Atomic force microscopy Gene expression (RNAseq) Pharmacological analysis |
| [46] Mathur et al., 2015 | hiPSC – CM | Drug cytotoxicity | Optical microscopy – brightfield Confocal microscopy – immunofluorescence |
Motion analysis Ca2+ imaging Pharmacological analysis |
| [47] Schneider et al., 2019 | hiPSC – CM | Optical microscopy – brightfield Confocal microscopy – immunofluorescence |
Motion analysis | |
| [154] Schneider et al., 2022 | hiPSC – CM human primary dermal fibroblasts |
Optical microscopy – brightfield Confocal microscopy – immunofluorescence |
Motion analysis Ca2+ imaging O2 sensors |
|
| [69] Wang et al., 2014 | hiPSC -CM | Disease modelling | Confocal microscopy – immunofluorescence | Motion analysis |
| [82] Skardal et al., 2017 | human hepatic stellate cells hiPSC – CM human lung microvasculature ECs human airway stromal mesenchymal cells human bronchial epithelial cells |
Drug screening | Optical microscopy - brightfield Confocal microscopy - immunofluorescence | LC-MS metabolic profiling electromechanical biosensors |
| [153] Tanumihardja et al., 2021 | hiPSC – CM | Electron microscopy | pH and O2 sensors | |
| [48] Xiao et al., 2014 | primary neonatal rat CM hESC-CM |
Drug screening | Optical microscopy - brightfield Confocal microscopy - Immunofluorescence Electron microscopy |
Motion analysis |
| [[125], [155]] Zhang et al., 2017; 2021 | human primary hepatocytes hiPSC – CM |
Drug screening | Brightfield microscopy - cellular morphology Fluorescence microscopy – cardiac markers |
Electromechanical biomarkers sensors Motion analysis |
| [140] Shen et al., 2017 | Murine ESC-CM hESC-CM | Cardiac maturation | Confocal microscopy – immunofluorescence | Ca2+ imaging RAMAN spectroscopy |
Microtissues (or spheroids) and organoids commonly refer to 3D tissue models although these terms are not always used as originally described. Lancaster and Knoblich defined organoids as “self-organized tissues that develop from PSCs or progenitor cells by recapitulation of embryonic development” [23]. The term “spheroid” has traditionally been used to describe aggregates of terminally differentiated cells. Subsequently though, many groups used the word “organoid” to describe any collection of PSCs or adult SCs that were differentiated into region specific cells of the organ with the help of growth factors (reviewed in Ref. [24]). For the heart, all three terms have been used as well as “cardioids” to describe beating aggregates of differentiated hPSCs whether they have formed spontaneously or have been created by mixing multiple pre-differentiated heart cell types. These beating aggregates can be formed entirely scaffold-free, e. g. on a non-adhesive surface (see reviews [[25], [26], [27]] where cFBs produce their own extracellular matrix (ECM), or by integration of natural ECM or substitute scaffolds (e.g. hydrogels). These models generally have different applications and some are useful for studying early heart development [[28], [29], [30]] but it is of note that unlike for many organs, organoids cannot be formed from stem cells in the adult heart since despite earlier claims, it does not seem that such a population is present to any extent after birth [31]. This may in part explain why the adult mammalian heart repairs poorly after damage.
Integration of biomaterials and tissue engineering principles provides environmental cues that further benefit tissue organization, cell-cell communication and results in elasticity properties close to the human heart [[32], [33], [34], [35]]. One of the first EHTs was generated by seeding a mixture of cells and extracellular matrix around Velcro-coated glass posts which allowed contraction of tissues against resistance. These structures resembled in vivo cardiac muscle fibers and allowed direct measurement of contraction force from deflection of the posts [36]. Several groups adopted and improved this simple yet effective system to quantify the change in contractile force of tissues, for example upon external metabolic or electromechanical stimulus (reviewed in Ref. [37]).
Two main strategies were employed to generate the EHTs - geometric confinement and mechanical load. Providing appropriate geometric confinement to the tissue is important for tissue morphology and functionality. For example, a rectangular frame induced CMs to exhibit a more rod-shape morphology, uniaxial calcium flow and improved sarcomeric organization compared to square frames or monolayer cultures in multi-well plates [6,38]. In addition, physiological mechanical stretching is important for tissue development and maturation [[39], [40], [41]].
Microtissues and EHTs can recapitulate several physiological features of heart muscle tissue (multiple cell types, typical contraction rates, 3D structure). However, they lack perfusable vessel-like structures, which in vivo transport oxygen, nutrients and cells. Organ-on-chip (OoC) technologies offer opportunities to integrate perfusion through microfluidics and study neovasculogenesis of the tissue constructs; vessel structures and transport processes are thus created much like those in vivo (reviewed in Refs. [[42], [43], [44]]).
Heart-on-chip systems (HoCs) are typically devices made of polymer modules featuring microscale networks of channels and chambers. The chambers commonly house the microtissues, while the channels act as ducts that supply the tissues with nutrients, growth factors, gases, soluble compounds and (blood or immune) cells. The tissue chambers can be separated by a semi-permeable barrier (e.g. micro-channels or porous membranes) from the direct flow of fluid to protect the tissues from sheer stress, yet allow perfusion of nutrients [[45], [46], [47], [48]]. Some HoCs allow detection of contractile forces by introducing cantilevers that moved with CM contraction [49]. They integrate cyclic mechanic stimulation mimicking contraction of the heart which in turn induces the pulsating flow [45] or electrical pacing [50]. Depending on the targeted research question and experimental requirements, chip designs might differ from one another to suit the specific purpose but ultimately tailored or in-house designs create challenges for standardization. There are a number of systems now commercially available that address this to some extent by ensuring that at least the manufacturing process is standardized.
Collectively, MHT models are poised to advance the cardiovascular field as their potential to study CM maturation and disease models and carry out drug screening begins to be exploited.
3. Applications
Aside from supporting CM maturation, MHTs are already being implemented in several areas including some mentioned above: studying cardiogenesis, investigating disease mechanisms, identifying which cardiac cell types are responsible for pathophysiology, assessing cardiotoxicity and identifying potential drug therapies. For all applications, models from any of the three MHT categories might be most suitable depending on the specific research questions. The model of choice should be as simple as possible but as complex as necessary.
3.1. Mediating cardiomyocyte maturation
One application of MHT models is to study intercellular communication in the heart which is important for heart development and maturation. Integration of CMs with ECs into microtissues improved structure and functionality of hPSC-CM [51]. Inclusion of cFBs enhanced CM maturation in their contraction force and frequency, gene expression, Ca2+ handling, sarcomere assembly and β-adrenergic receptor signaling proteins [17,18]. In addition, 3D tissue formats prevented cFBs forming stress fibers compared to monolayer cultures and eliminated profibrotic growth factors for cFB activation and trans-differentiation into myofibroblasts [17]. Furthermore, combining all three cell types (CMs, ECs and cFBs) in these tissues increased intracellular cAMP levels in CMs through CM-cFB crosstalk; this was further enhanced by Endothelin-mediated signaling from ECs to CMs [18]. Together, this resulted in better assembly of Connexin43 (Cx43) gap junctions and further promoted maturation of CMs. This was in line with in vivo studies [52]. Crosstalk between CMs and FBs and resulting maturation was not seen when dermal FBs replaced cFBs [18]. Dermal FBs express low or negligible levels of Cx43, depending on the particular batch which might explain why they are less effective in mediating maturation compared to cFBs. However, other tissue sources of FBs may express Cx43 and promote maturation. In addition, any paracrine effects of FBs may be independent of Cx43.
In addition to co-culture, other approaches to tissue maturation have used EHTs, in which systematic optimization of ECM components and regulation of exogenous mechanical- and metabolic factors (including energy substrates) have resulted in significant benefit [41,[53], [54], [55]].
These studies demonstrated that multiple cues must be carefully orchestrated to govern distinct aspects of the adult CM phenotype. Therefore, although much progress has been made and there are multiple ways to induce a postnatal CM phenotype, none is yet definitive and obtaining fully mature/adult CMs in MHTs (or any other model) remains a challenge.
3.2. Disease modelling
2D hPSC-CMs are easy to generate and use and are adequate for modeling ion channelopathies and cardiomyopathies caused by specific mutations, as mentioned earlier [51]. However, for more complex disorders, MHTs provide better structural, functional and metabolic maturation as well as allowing integration of multiple cell types and simulation of environmental conditions. These 3D models have also been used for studying inflammation subsequent to myocardial infarction or other injury [20,56]; or fibrosis [57,58].
Bacterial or viral infection can affect the heart. Severe acute respiratory syndrome coronavirus (SARS-CoV-2) infection causes coronavirus disease (COVID-19). Many groups worldwide are investigating viral infection and disease mechanisms using human stem cell models. Although the predominant pathology is observed in the respiratory system, several studies have shown that the cardiovascular system is also affected by this virus [59,60]. 20–30% of all COVID-19 patients suffer from severe cardiac symptoms and increased levels of cardiac protein troponin I in their blood which indicates heart damage and is linked to increased risk of morbidity [61,62]. 3D culture systems discussed here are also increasingly used to investigate how inflammation causes heart damage and the effects of therapy on the heart [[63], [64], [65]].
Taking advantage of the ability to integrate differentiated cardiac cells from healthy- (genetically corrected) and patient derived hiPSCs, multiple cardiac pathologies such as hypertrophy [[66], [67], [68]], arrhythmogenic cardiomyopathy [18], long QT syndrome type 3 (LQT3) [33] and Barth syndrome [69] have been modelled using different MHTs. Aside from providing insights into disease mechanisms, they have also been used to identify potential therapeutics [69,70].
These models are potentially useful for providing insights into disease mechanism but to date they have lacked immune/inflammatory cells, involved in triggering or sustaining these pathological processes [71], or nerve cells that can invade heart excessively and cause arrhythmias after myocardial infarction [72].
3.3. Cardiotoxicity assessment
Cardiotoxicity is still a major reason that drugs in development fail to reach the clinic or are labelled as high risk after market entry. Rodents do not always accurately predict drug effects on humans because the physiological parameters like contraction kinetics such as the beat rate of the human heart differ significantly from those in widely used laboratory animals (reviewed in Ref. [73]). In addition, direct translation of drug efficacy and toxicology from rodents to humans is limited due to differences in pharmacokinetic properties. Large animals such as non-human primates or dogs are more predictive but have financial and ethical burdens. Alternative models to animals are thus of growing interest.
One of the first major milestones towards using hiPSC-CMs among these alternatives is the Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative, a collaboration between academia, pharmaceutical industries, regulatory authorities and stem cell companies [74]. This showed a good correlation between the assay outcome in hiPSC-CMs and known toxicity in humans. In a later study, simultaneous measurement of contraction kinetics, action potentials and Ca2+ transients using voltage and calcium-sensitive dyes, was shown to be even more predictive of cardiac toxicity [75,76]. It is an ongoing effort to assess the drug-induced proarrhythmic risk using hiPSC-CMs and emerging in vitro/in silico assays. Collectively, these studies provided evidence that assays based on hiPSC-CMs could eventually replace traditional toxicology studies with the combination of 3D in vitro models and in silico tools.
MHT platforms are also being used to test functional toxicity of a number of drugs [21,77,78]. In these studies, tissues exhibited alterations in cardiac contraction in response to inotropic or non-inotropic drugs with an ability to predict in vivo outcome with high sensitivity and specificity [21]. Moreover, these models were able to distinguish between negative and positive inotropic compounds as opposed to previous studies on hiPSC-CM monolayer cultures [79]. This confirmed improved accuracy of risk assessment for a spectrum of compounds. Structural cardiotoxicity of FDA approved compounds was also assessed [80]. Morphological assessment of drug-exposed MHTs showed changes in structural integrity and cellular viability. Together, these studies revealed the potential value of MHTs in drug screening even though, because of their size and formats, they are not yet suitable for large scale compound screens. Pharmaceutical companies are increasingly adopting MHTs and more specifically OoC platforms for drug effectivity assessment and disease modelling [81] with current efforts on more standardized protocols.
Addressing drug metabolization and multi-organ effects, especially in OoC platforms integrating multiple organ systems with perfusion, are providing new opportunities to evaluate possible side effects of drug treatment and off-target tissues. For example, a heart-liver-lung multi-organ on chip platform showed that bleomycin, an anti-cancer drug that causes lung inflammation, also induced cardiotoxicity [82]. Another example was described by Lee at al. (2021), who connected HoC to a breast-cancer-on-chip to examine chemotherapy induced cardiotoxicity [83]. Combining cancer patient-derived hiPSC-CMs in this model could in the future aid prediction of individual cardiotoxic risk.
3.4. Development of cardiac therapeutics
Complementary studies also used hiPSC-CMs to predict drug-induced electrophysiological- and contractile changes (reviewed in Ref. [84]). 2D hiPSC-CMs and 3D EHTs [85,86] were compared for their (blinded) response to a list of known drugs. This resulted in highest predictivity and sensitivity from 3D-EHTs based on measuring force of contraction although combined measurement of contraction, action potential and calcium transients in 2D hiPSC-CM monolayer culture also gave high predictivity as mentioned above [76,84].
Several EHT and HoC drug screening platforms are scalable and have shown drug responses comparable to in vivo clinical observations [19,[87], [88], [89]]. Scalability (and corresponding cost) is important for pharma in large compound screens for therapeutics. EHTs and HoCs have additional advantages for some drug screens in providing an endothelial (or vascular wall) barrier (in Transwell® inserts). Drugs can be added on the basal side of the endothelial barrier whilst the cardiac tissue is on the apical side [89]. These models allow assessment of drug availability through the vasculature. Together, these platforms are increasingly used to develop de novo cardiac therapeutics.
4. Readouts
How cell responses can best be measured in situ preferably in real-time, is a crucial question for their utility. Readouts constitute an integral part of any model and are necessary to validate MHTs prior to implementation. Two important features of these heart models are relevant to translation: cardiac structure and function. In this section, we summarize the commonly used assays, both in situ in real time and terminal; these are summarized in Table 1, Table 2, Table 3
4.1. Microscopy-based analysis of MHT assembly and composition
CMs in the heart are tightly coupled to create the structure necessary for highly synchronized, repetitive contraction and electrical conduction. This is evidenced as aligned sarcomeres that form characteristic “bands” in the heart. Thus, proper physiology requires defined 3D structure in the tissue. MHTs may thus include architectural guidelines to shape the in vitro model e.g. scaffolds, spatial constraints or anchoring points. However, the structure of cardiac microtissues can vary from spheroid-like to extended fibers. Analysis of the tissue structure is thus highly relevant to demonstrate enhanced maturation or quantify pathological changes in disease. Here, we summarize microscopic methods and their readouts used for this purpose.
When designing MHTs, the material properties, such as transparency, refractive index and organic solvent compatibility, must be considered. In addition, the thickness of the material should be taken into account for imaging with short or long-distance objectives. If the platform does not have the same footprint as microscope slides or plates, tailored holders are needed that fit on the microscope stage. Moreover, when live imaging is preferred, an on-stage incubator for climate control is required.
The main ways to visualize MHT structure and its cellular components are label free imaging, immunofluorescence and use of fluorescent reporter cell lines. Label-free [89] and reporter cells [90] do not require additional steps such as antibody staining before imaging. On the other hand, immunofluorescent labelling may have higher specificity for protein detection and allow multiple proteins to be imaged simultaneously. Another post-fixation labelling technique is standard histology where various cellular and intracellular structures are visible and can be characterized [50]. In addition to labelling, tissue clearing techniques are increasingly used to improve image quality. 3D tissue imaging might be limited due to the scattered light by the lipids, intracellular or extracellular fluids present in the tissue. Tissue clearance eliminates these components and allows penetrate of light into the tissue by keeping refractive indices equal [91,92]. As a result, thicker tissue samples can be visualized by different microscopic techniques while preserving the 3D integrity of the sample.
Optical microscopy is the gold standard to image tissue structures. It includes a wide range of techniques from widefield to high-resolution microscopy. Transmitted light microscopy, such as bright-field or phase contrast, provides information on microtissue compaction, shape and motion (see Table 1, Table 2, Table 3) without requiring additional labeling or fixation, and is suitable for live imaging. However, transmitted light microscopy is usually unable to distinguish specific cell types and their intracellular structures. Confocal microscopy and super-resolution microscopy overcome this. They also provide high spatial resolution with the possibility of optical sectioning, imaging in the Z-direction and 3D-reconstruction although this adds additional labor and time in the post-imaging process [93]. However, photo damage remains a limitation of these techniques. In addition, higher magnification objectives and corresponding short working distances (<1 mm) may require removing the microtissue from the platform [94] which may be challenging and affect structural integrity of the microtissue. Two-photon microscopy can also provide 3D information. In comparison to confocal microscopy, it has better penetration depth due to greater wavelengths of photons used for imaging allowing visualization of deeper regions. It does not require optical clearance of the tissue and reduces photobleaching. Additionally, second harmonic generation allows label-free imaging of specific cell structures, like collagen deposition [67,95]. Light sheet microscopy based on illuminating an entire plane of tissue by laser in the form of a light sheet, has proven particularly useful for imaging 3D objects in particular because photo-bleaching is lower than conventional confocal imaging. Although not yet broadly applied to MHTs, first reports indicate advantages for tissue imaging on microfluidic chips [96].
Electron microscopy uses electron beams instead of light to generate a projection image. Compared to optical microscopy, electron microscopy provides much higher resolution. However, it requires fixed samples and often tissue slices, which make it a terminal assay. It also presents tissue collection challenges for HoC platforms [38,97].
4.2. Functional readouts
Functional analysis of cardiac tissue models can broadly be divided into four approaches: molecular (e.g. protein or gene expression), electrophysiological, mechanical and metabolic readouts.
4.2.1. Molecular analysis
Molecular analysis is not generally regarded as a direct functional readout but it does reveal expression profiles and pathways relevant for functionality. Commonly used assays for both in vivo and in vitro studies is high-throughput next generation sequencing (NGS) for transcriptome, genome and epigenome analysis. Besides well-known techniques like RT-PCR, RNA sequencing (RNAseq) is used increasingly either on bulk cell populations, where it reports average signal expression in cell populations, or on single cells. Bulk measurements on thousands to millions of cells is straightforward but does not distinguish different cell populations thereby potentially missing rare events that could be highly relevant to a disease mechanism [[98], [99], [100]]. By contrast, single cell analysis allows tracking of individual cell populations and detection of rare events (reviewed in Refs. [[101], [102], [103], [104], [105]]). RNAseq has been used for microtissues, organoids and EHTs but not so far for HoCs, probably due to difficulties in extracting the tissues from chips. Whilst highly informative, RNAseq lacks the corresponding protein expression profiles and require disruption of tissues and cells. Disruption itself can upregulate stress genes [106] and thereby introduce artefacts and spatial information is lost. Batch-to-batch and line-to-line variability may in the future be addressed by automation of procedures. Alternative high content imaging methods e. g Co-detection by indexing (CODEX) [107], imaging mass cytometry [108], MACSima imaging cycling staining (MICS) [109] or RNAscope [110] complement the toolbox for protein and RNA expression profiling. Spatial transcriptomics, although not yet commonly applied to MHTs offer next generation methodology for in situ tissue profiling [111]. Other approaches include collecting tissue-conditioned media for biomarker detection or recent impedimetric immunosensors with antibody coated electrodes which transmit electrical signals when biomarkers or pathogens bind to them [112]. These methods are currently used for quality control of embryos derived from in vitro fertilization where invasive cell sampling carries high risk [[113], [114], [115], [116]]. Clinical cardiac biomarkers [117,118] can also be used on conditioned media and all of these non-invasive methods allow long-term monitoring of live tissues.
4.2.2. Electrophysiology
Electrophysiological properties such as the action potential (AP) profile of the heart reflect functionality of CMs. Rapid ion transfer in and out of membrane channels alters the membrane voltage which results in AP changes. The AP starts with depolarization of the membrane by sodium ion influx and continues with initial repolarization by potassium (K+) ion efflux and a plateau phase in which calcium ions (Ca+2) move inwards. Further repolarization by K+ currents bring the membrane potential back to the resting level of −85 mV. Intracellular recordings of electrophysiological parameters are obtained using sharp electrodes or single cell patch clamp; these remain the benchmark for quantification of functionality in microtissues and EHTs. Voltage sensitive dyes [119] and integration of (non-invasive) electrodes are alternatives for extracellular recordings of field potential. Voltage sensitive dyes can be used in any platform and is suitable for whole tissue analysis [120]. However, they can be cytotoxic and are sensitive to photobleaching. Multielectrode arrays (MEAs) are culture plates with embedded electrodes [121] which are widely used for real-time estimation of QT-prolongation (an important safety parameter) [122] and conduction velocity [123]. They are mostly used for 2D cell cultures as it is a challenge to record the whole tissue electrophysiology considering their limited surface interface between the electrodes and the tissue. Adopting a similar strategy, self-rolling biosensor arrays which wraps around the 3D microtissue/organoid [124], or microelectrodes integrated in EHT and HoC platforms allow continuous and multiplexed recording of electrophysiological signals [88,121,125].
4.2.3. Mechanics
Excitation-contraction coupling is one of the key indications of a functional cardiac tissue and Ca2+ is one of the key ions of this process. Sarcoplasmic reticulum releases Ca2+ ions in response to electrical excitation during the AP plateau phase which increases the level of intracellular Ca2+. Coupling and uncoupling of intracellular Ca2+ with troponin C in myofilaments activates and deactivates contraction, respectively. In order to reflect the physiological properties of the 3D models with respect to their Ca2+ level changes, fluorescent dyes with a variety of excitation ranges and Ca2+ affinities are used. Although this technique is real-time, fast and efficient, it is not suitable for long-term analysis and sensitivity of recordings depends highly on the type of the dye. Genetically modified hPSC lines that encode a calcium indicator (GECI) circumvents limitations of Ca2+ dyes and offer user-independent possibilities to measure and analyze Ca2+ flux and contractility [126].
Optical tracking of cells, tissues, beads [127], pillars or cantilevers around which tissues have compacted [88,128] is another way to retrieve contraction parameters as relative pixel displacement or absolute contraction force. Traction force microscopy, fractional shortening of sarcomere length and micropost displacement are examples of such techniques mainly used for single cells [54,129]. Software such as MuscleMotion [130] and OpenHeartWare [47] are user-friendly and open access tools for optical tracking of live contracting tissue from video recordings or image stacks. This type of automated contraction analysis enables quantification of contraction changes in diseased and healthy states or in response to drugs and can also be used for MHTs discussed earlier (Table 1, Table 2, Table 3 [47,131]). However, precise and consistent tracking becomes a challenge with increased heterogeneity in contrast and texture of 2D or 3D models; this may create measurement artefacts. In addition, preload dependent mechanics cannot be well analyzed with optical tracking. Therefore, classical isometric force measurements remain the gold standard in biomechanical characterizations of the heart muscle. Integrated sensors in MHTs allow continuous electronic examination of contractile stress and beat rate [89]. Such analysis does not require invasive methodology and is user-friendly.
4.2.4. Metabolic readouts
The adult heart has a low energy reserve. To meet the high energy demand, CMs produce ATP from several metabolic substrates (e.g. glucose, pyruvate, triglycerides, glycogen, lactate, ketone bodies, fatty acids (FAs) and some amino acids) based on environmental conditions [132,133]. During early heart development, glucose and lactate are the major energy sources and glycolysis the major metabolic pathway to produce ATP. Postnatally though, the mitochondrial oxidative capacity increases and FA beta oxidation becomes the major energy source [134,135]. Recapitulating this “metabolic switch” from glycolysis to mitochondrial oxidative phosphorylation is an important reflection of maturity in MHTs [53].
4.2.4.1. Oxygen consumption and glycolysis measurements
Several commercially available kits and instruments (e.g. BioProfile Analyzers and Biochemistry Analyzers) allow quantification of extracellular glucose and lactate levels by colorimetric and fluorometric detection in cell culture media and are thus non-invasive. The Seahorse (extracellular flux) XF Analyzer is a fast and comprehensive approach that allows simultaneous recording of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of live cells and tissues. Three main metabolic assays can be performed with this technology: (I) mitochondrial stress; (II) glycolytic stress and (III) palmitate oxidation [136].
4.2.4.2. Quantification of metabolites and cell secretome
Conventional biochemical assays (RNAseq, LC-MS, ELISA) are widely used to detect metabolic changes in static cultures such as EHTs and microtissues/organoids and in a time-resolved manner for perfused systems. Secretome analysis can provide insight into disease mechanisms [137] and microenvironmental changes in the ECM, for example during hPSC differentiation into CMs [138]. However, proteome and secretome data acquisition and interpretation are challenging, even when applied to simple systems (e.g. monocultures). Low concentration of analytes, serum components in the media, protein splice variants can all result in false negatives and positives; this might be the reason why this technique has not been widely adopted in more complex cardiac systems (e.g. co-culture of different cell types).
Nuclear magnetic resonance spectroscopy (NMR) detects extra-/intra-cellular metabolites in MHTs in tissue supernatants and lysates. The technique can be used non-invasively and the data gives valuable input in the metabolic state of the tissues [18]. Alternatively, RAMAN spectroscopy provides non-invasive measurement of intracellular metabolites, and it is already in use to monitor hPSC-CM maturity [139,140] as well as ECM components in tissues [141,142].
5. Perspectives
MHTs have become much more robust and reproducible over the last several years but individually, each system still has issues to overcome before widespread application in the cardiovascular field. MHTs based on hPSCs can mimic aspects of embryonic development and integrate multiple (pre-differentiated) cell types. Cell types that have not yet been integrated into these models include (i) cardiac resident macrophages (CRM) that link with CMs and can regulate electrical activity and cardiac homeostasis [[143], [144], [145]] and (ii) the cardiac autonomic nervous system that mediates the electrical and mechanical signals from the brain to the heart [146]. Additionally, the intrinsic cardiac nervous system (ICNS) with its sensory, motor and interconnecting neurons regulates electrical conduction in the heart [147]. Several studies have indicated important roles of CRMs [145,148,149] and ICNS (reviewed in Ref. [150]) in cardiac pathophysiology in vivo. Moreover, many differentiation protocols, cell lines and platform designs mean little standardization between the systems, precluding the direct comparison of outcomes. One example of the challenge this presents is that identical EHTs made from different hiPSC lines exhibited different baseline contractile force, kinetics, and beating rate [86]. It is unclear whether this derives from different genetic backgrounds, line-specific disease associated SNPs or gene variants or just different state-of-maturity of the CMs used in the EHTs. In addition, efforts towards more head-to-head comparisons including batch-to-batch and clone-to-clone variability are needed.
Self-organized microtissues and organoids miss the full complement of micro-environmental cues that direct physiological maturation of CMs. EHTs can introduce natural or synthetic microenvironments that together with mechanical cues go some way to increasing physiological realism. Likewise, HoCs with microchannels have been developed which can integrate perfusion and fluidic flow, the critical feature of native tissues. In addition, most platforms are fabricated by academic labs which produce devices in small series and only few are at present commercially available. In addition, physical and chemical properties of materials may be variable with respect to absorption or retention of small molecules in the culture medium which could lead to misinterpretation of results. It is important to characterize these properties prior to use.
MHTs nevertheless offer remarkable opportunities in many applications including disease modelling and drug screening in the cardiac field particularly if populated with human cells. Data produced using current validation assays suggests that these systems can recapitulate many complex physiologies of heart tissue in healthy and diseased states with respect to structure, functionality and metabolism. However, most of these assays have not yet been used in HoCs, therefore lack options to mimic (blood and lymph) vascular flow. Most HoC designs are closed so tissues are not easily accessible for readouts. For this reason, terminal (or end-point) readouts are often preferred. Moving towards non-invasive assays and adapting these to be compatible with HoCs will be one way of addressing and providing alternatives for this issue. Integration of biosensors in chip design is an option. However, most commercial systems, for example, do not allow in-house integration of these sensors, perhaps explaining why many labs prefer their own in-house designs.
For structural characterization, high resolution imaging of 3D tissues along the Z-axis is still challenging in HoCs since most are still too thick to be imaged using conventional confocal microscopy. This means that the tissue must be removed from the platform, making high resolution imaging an end-point readout. Even though not often reported for HoCs, light-sheet microscopy and 2-photon microscopy can offer alternatives for direct on-chip imaging of live tissues and combination with transgenic cell lines expressing fluorescent marker proteins provide non-invasive structural readouts.
Metabolic studies are mainly focused on O2 consumption and glycolysis. However, most techniques are not suitable for real-time measurement in HoCs as they need either tissue lysis or effluent collection over several hours of perfusion. For example, although the Seahorse-XF is widely used on cell monolayers, microtissues and EHTs (Table 1, Table 2), most of the microfluidic devices are made of PDMS which is highly permeable for oxygen. Hence oxygen consumed by the tissues is replenished almost instantly downstream. In addition, they are mostly closed systems where the cell or tissue proximity is not easily accessible by instrumentation and the culture medium is delivered through continuous flow. The design flexibility however, allows, in principle for the integration of the sensors directly into the chip platform. Alternatively, non-invasive fluorescent-lifetime imaging microscopy (FLIM) allows characterization of mitochondrial metabolic oxidative changes by means of endogenous fluorescence in live samples [151].
Quantification techniques for metabolites and secretome (ELISA, MS, NMR and others reviewed in Ref. [133]) are used for microtissues and EHTs, they have not yet been adapted for HoC systems. However, some of these techniques have been applied in other OoCs, for example Raman spectroscopy in pancreas-on-chip [152].
Integrated or modular sensors offer in situ real-time automated tools for metabolic readouts. Integrated sensors include electrochemical sensors to measure oxygen and pH analytes as a difference of electrical current or potential [153]. However, they can also be adapted to measure non-electrically active species such as glucose and lactate. These sensors often require a transducer to detect signals. On the other hand, optical sensors can replace transducers and detect signals from a simple transparent window on top of the sensor. Integrated sensors can be positioned close to the tissue allowing real-time measurement [154]. However, they are in the system for entire culture periods which in some cases last multiple weeks while the measurement is only relevant when the system is challenged, for example with drugs. This means for enzyme-based sensors (but also others) that they might already be degraded before they are used. Integrated sensors are single use which significantly increases the cost per chip.
Modular multisensors, on the other hand, can be re-usable and thus cost efficient. They may contain several physical sensors for monitoring pH, oxygen and temperature or electrochemical sensors [155]. Modular multisensor systems in combination with modular OoC platforms can be useful to detect differential secretion of enzymes in different organs, for example in response to different compounds [82]. However, they are dependent on the flow rate and some measurements, such as oxygen consumption in PDMS chips can be impaired if not positioned properly [155]. Electrochemical and optical sensor alternatives and their integration in OoC systems have recently been extensively reviewed [156].
Emerging technologies like FLIM, Raman spectroscopy and integrated sensors may greatly contribute to non-invasive metabolic analysis of living tissues inside HoC platforms.
6. Conclusions
In sum, we reviewed here different 3D cardiac tissue generation platforms in combination with hiPSC-derived cardiac cell types together with their applications and readouts. These models are emerging tools to investigate cardiac development and pathophysiology. Further refinement in the generation and readout methods might eliminate hurdles that have been discussed here and allow closer mimicry of the adult heart with a possibility to standardize, reduce costs and adopt less labor-intensive methodology.
Declaration of competing interest
J.N and D.E. are employees of Miltenyi Biotec B.V. & Co. KG.
Acknowledgements
The authors' work is supported by the European Union's Horizon 2020 research and innovation programme under grant agreement No. 812954.
Contributor Information
Christine L. Mummery, Email: c.l.mummery@lumc.nl.
Dominik Eckardt, Email: dominike@miltenyi.com.
Peter Loskill, Email: peter.loskill@uni-tuebingen.de.
Valeria V. Orlova, Email: v.orlova@lumc.nl.
References
- 1.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007 doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Thomson J.A. Embryonic stem cell lines derived from human blastocysts. Science. 1998;80– doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Mummery C., Oostwaard D.W., Doevendans P., Spijker R., van den Brink S., Hassink R., van der Heyden M., Opthof T., Pera M., de la Riviere A.B., Passier R., Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.cir.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 4.Kehat I., Kenyagin-Karsenti D., Snir M., Segev H., Amit M., Gepstein A., Livne E., Binah O., Itskovitz-Eldor J., Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J. Clin. Invest. 2002;108:407–414. doi: 10.1172/jci12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G., Kamp T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ. Res. 2012;111:344–358. doi: 10.1161/circresaha.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G., Atsma D.E., Passier R., Mummery C.L. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:733–745. doi: 10.1016/j.celrep.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birket M.J., Mummery C.L. Pluripotent stem cell derived cardiovascular progenitors - a developmental perspective. Dev. Biol. 2015 doi: 10.1016/j.ydbio.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Ribeiro M.C., Tertoolen L.G., Guadix J.A., Bellin M., Kosmidis G., D'Aniello C., Monshouwer-Kloots J., Goumans M.J., li Wang Y., Feinberg A.W., Mummery C.L., Passier R. Biomaterials; 2015. Functional Maturation of Human Pluripotent Stem Cell Derived Cardiomyocytes Invitro - Correlation between Contraction Force Andelectrophysiology. [DOI] [PubMed] [Google Scholar]
- 9.Davis R.P., Casini S., Cathelijne W., Van Den Berg M.Hoekstra, Carol A., Remme C., Dambrot D., Salvatori Dorien W.-V., Oostwaard A.A.M., Wilde C.R., Bezzina A.O., Verkerk C., Mummery Freund C.L. 2012. Arrhythmia/Electrophysiology Cardiomyocytes Derived from Pluripotent Stem Cells Recapitulate Electrophysiological Characteristics of an Overlap Syndrome of Cardiac Sodium Channel Disease. [DOI] [PubMed] [Google Scholar]
- 10.Braam S.R., Tertoolen L., van de Stolpe A., Meyer T., Passier R., Mummery C.L. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4 doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Denning C., Borgdorff V., Crutchley J., Firth K.S.A., George V., Kalra S., Kondrashov A., Hoang M.D., Mosqueira D., Patel A., Prodanov L., Rajamohan D., Skarnes W.C., Smith J.G.W., Young L.E. Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform. BBA - Mol. Cell Res. 2016;1863:1728–1748. doi: 10.1016/j.bbamcr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang P., Lan F., Lee A.S., Gong T., Sanchez-Freire V., Wang Y., Diecke S., Sallam K., Knowles J.W., Wang P.J., Nguyen P.K., Bers D.M., Robbins R.C., Wu J.C. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127 doi: 10.1161/CIRCULATIONAHA.113.001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moretti A., Bellin M., Welling A., Jung C.B., Lam J.T., Bott-Flügel L., Dorn T., Goedel A., Höhnke C., Hofmann F., Seyfarth M., Sinnecker D., Schömig A., Laugwitz K.-L. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010;363:1397–1409. doi: 10.1056/nejmoa0908679. [DOI] [PubMed] [Google Scholar]
- 14.Beauchamp P., Moritz W., Kelm J.M., Ullrich N.D., Agarkova I., Anson B.D., Suter T.M., Zuppinger C. Development and characterization of a scaffold-free 3D spheroid model of induced pluripotent stem cell-derived human cardiomyocytes. Tissue Eng. C Methods. 2015 doi: 10.1089/ten.tec.2014.0376. [DOI] [PubMed] [Google Scholar]
- 15.Desroches B.R., Zhang P., Choi B.R., King M.E., Maldonado A.E., Li W., Rago A., Liu G., Nath N., Hartmann K.M., Yang B., Koren G., Morgan J.R., Mende U. Functional scaffold-free 3-D cardiac microtissues: a novel model for the investigation of heart cells. Am. J. Physiol. Heart Circ. Physiol. 2012 doi: 10.1152/ajpheart.00743.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soares C.P., Midlej V., de Oliveira M.E.W., Benchimol M., Costa M.L., Mermelstein C. 2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beauchamp P., Jackson C.B., Ozhathil L.C., Agarkova I., Galindo C.L., Sawyer D.B., Suter T.M., Zuppinger C. 3D Co-culture of hiPSC-derived cardiomyocytes with cardiac fibroblasts improves tissue-like features of cardiac spheroids. Front. Mol. Biosci. 2020 doi: 10.3389/fmolb.2020.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giacomelli E., Meraviglia V., Campostrini G., Cochrane A., Cao X., van Helden R.W.J., Krotenberg Garcia A., Mircea M., Kostidis S., Davis R.P., van Meer B.J., Jost C.R., Koster A.J., Mei H., Míguez D.G., Mulder A.A., Ledesma-Terrón M., Pompilio G., Sala L., Salvatori D.C.F., Slieker R.C., Sommariva E., de Vries A.A.F., Giera M., Semrau S., Tertoolen L.G.J., V Orlova V., Bellin M., Mummery C.L. Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell. 2020;26:862–879. doi: 10.1016/j.stem.2020.05.004. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills R.J., Parker B.L., Quaife-Ryan G.A., Voges H.K., Needham E.J., Bornot A., Ding M., Andersson H., Polla M., Elliott D.A., Drowley L., Clausen M., Plowright A.T., Barrett I.P., Wang Q.-D., James D.E., Porrello E.R., Hudson J.E. Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell. 2019;24:895–907. doi: 10.1016/j.stem.2019.03.009. e6. [DOI] [PubMed] [Google Scholar]
- 20.Richards D.J., Li Y., Kerr C.M., Yao J., Beeson G.C., Coyle R.C., Chen X., Jia J., Damon B., Wilson R., Starr Hazard E., Hardiman G., Menick D.R., Beeson C.C., Yao H., Ye T., Mei Y. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 2020;4:446–462. doi: 10.1038/s41551-020-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pointon A., Pilling J., Dorval T., Wang Y., Archer C., Pollard C. From the cover: high-throughput imaging of cardiac microtissues for the assessment of cardiac contraction during drug discovery. Toxicol. Sci. 2017;155:444–457. doi: 10.1093/toxsci/kfw227. [DOI] [PubMed] [Google Scholar]
- 22.Giacomelli E., Bellin M., V Orlova V., Mummery C.L. Co-differentiation of human pluripotent stem cells-derived cardiomyocytes and endothelial cells from cardiac mesoderm provides a three-dimensional model of cardiac microtissue. Curr. Protoc. Hum. Genet. 2017;95:21. doi: 10.1002/cphg.46. 9.1-21.9.22. [DOI] [PubMed] [Google Scholar]
- 23.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(80-) doi: 10.1126/science.1247125. 1247125–1247125. [DOI] [PubMed] [Google Scholar]
- 24.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 25.Giraud M.N., Armbruster C., Carrel T., Tevaearai H.T. Current state of the art in myocardial tissue engineering. Tissue Eng. 2007;13:1825–1836. doi: 10.1089/ten.2006.0110. [DOI] [PubMed] [Google Scholar]
- 26.Günter J., Wolint P., Bopp A., Steiger J., Cambria E., Hoerstrup S.P., Emmert M.Y. Microtissues in cardiovascular medicine: regenerative potential based on a 3D microenvironment. Stem Cell. Int. 2016:1–20. doi: 10.1155/2016/9098523. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirt M.N., Hansen A., Eschenhagen T. Cardiac tissue engineering : state of the art. Circ. Res. 2014;114:354–367. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- 28.Drakhlis L., Biswanath S., Farr C.M., Lupanow V., Teske J., Ritzenhoff K., Franke A., Manstein F., Bolesani E., Kempf H., Liebscher S., Schenke-Layland K., Hegermann J., Nolte L., Meyer H., de la Roche J., Thiemann S., Wahl-Schott C., Martin U., Zweigerdt R. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021;39 doi: 10.1038/s41587-021-00815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofbauer P., Jahnel S.M., Papai N., Giesshammer M., Deyett A., Schmidt C., Penc M., Tavernini K., Grdseloff N., Meledeth C., Ginistrelli L.C., Ctortecka C., Šalic Š., Novatchkova M., Mendjan S. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184:3299–3317. doi: 10.1016/j.cell.2021.04.034. e22. [DOI] [PubMed] [Google Scholar]
- 30.Silva A.C., Matthys O.B., Joy D.A., Kauss M.A., Natarajan V., Lai M.H., Turaga D., Alexanian M., Bruneau B.G., McDevitt T.C. Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Kretzschmar K., Post Y., Bannier-Hélaouët M., Mattiotti A., Drost J., Basak O., Li V.S.W., van den Born M., Gunst Q.D., Versteeg D., Kooijman L., van der Elst S., van Es J.H., van Rooij E., van den Hoff M.J.B., Clevers H. Profiling proliferative cells and their progeny in damaged murine hearts. Proc. Natl. Acad. Sci. U. S. A. 2018 doi: 10.1073/pnas.1805829115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouten C.V.C., Dankers P.Y.W., Driessen-Mol A., Pedron S., Brizard A.M.A., Baaijens F.P.T. Substrates for cardiovascular tissue engineering. Adv. Drug Deliv. Rev. 2011;63 doi: 10.1016/j.addr.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Ma Z., Koo S., Finnegan M.A., Loskill P., Huebsch N., Marks N.C., Conklin B.R., Grigoropoulos C.P., Healy K.E. Three-dimensional filamentous human diseased cardiac tissue model. Biomaterials. 2014;35 doi: 10.1016/j.biomaterials.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Z., Wang J., Loskill P., Huebsch N., Koo S., Svedlund F.L., Marks N.C., Hua E.W., Grigoropoulos C.P., Conklin B.R., Healy K.E. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun. 2015;6 doi: 10.1038/ncomms8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y., Feric N.T., Thavandiran N., Nunes S.S., Radisic M. The role of tissue engineering and biomaterials in cardiac regenerative medicine. Can. J. Cardiol. 2014;30 doi: 10.1016/j.cjca.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eschenhagen T., Fink C., Remmers U., Scholz H., Wattchow J., Weil J., Zimmermann W., Dohmen H.H., Schäfer H., Bishopric N., Wakatsuki T., Elson E.L. Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: a new heart muscle model system. Faseb. J. 1997;11:683–694. doi: 10.1096/fasebj.11.8.9240969. [DOI] [PubMed] [Google Scholar]
- 37.Mills R.J., Hudson J.E. Bioengineering adult human heart tissue: how close are we? APL Bioeng. 2019;3 doi: 10.1063/1.5070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huebsch N., Loskill P., Deveshwar N., Spencer C.I., Judge L.M., Mandegar M.A., Fox C.B., Mohamed T.M.A., Ma Z., Mathur A., Sheehan A.M., Truong A., Saxton M., Yoo J., Srivastava D., Desai T.A., So P.L., Healy K.E., Conklin B.R. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garoffolo G., Pesce M. Mechanotransduction in the cardiovascular system: from developmental origins to homeostasis and pathology. Cells. 2019;8:1607. doi: 10.3390/cells8121607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonard A., Bertero A., Powers J.D., Beussman K.M., Bhandari S., Regnier M., Murry C.E., Sniadecki N.J. Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J. Mol. Cell. Cardiol. 2018;118:147–158. doi: 10.1016/j.yjmcc.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L.J., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahadian S., Civitarese R., Bannerman D., Mohammadi M.H., Lu R., Wang E., Davenport-Huyer L., Lai B., Zhang B., Zhao Y., Mandla S., Korolj A., Radisic M. Organ-on-A-chip platforms: a convergence of advanced materials, cells, and microscale technologies. Adv. Healthc. Mater. 2018;7:1–53. doi: 10.1002/adhm.201700506. [DOI] [PubMed] [Google Scholar]
- 43.Browne S., Gill E.L., Schultheiss P., Goswami I., Healy K.E. Stem cell-based vascularization of microphysiological systems. Stem Cell Rep. 2021;16 doi: 10.1016/j.stemcr.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S., Wan Z., Kamm R.D. vol. 21. 2021. p. 473. (Lab on a Chip CRITICAL REVIEW Vascularized Organoids on a Chip: Strategies for Engineering Organoids with Functional Vasculature). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsano A., Conficconi C., Lemme M., Occhetta P., Gaudiello E., Votta E., Cerino G., Redaelli A., Rasponi M. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip. 2016;16:599–610. doi: 10.1039/c5lc01356a. [DOI] [PubMed] [Google Scholar]
- 46.Mathur A., Loskill P., Shao K., Huebsch N., Hong S.G., Marcus S.G., Marks N., Mandegar M., Conklin B.R., Lee L.P., Healy K.E. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 2015;5:1–7. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider O., Zeifang L., Fuchs S., Sailer C., Loskill P. User-friendly and parallelized generation of human induced pluripotent stem cell-derived microtissues in a centrifugal heart-on-a-chip. Tissue Eng. 2019;25:786–798. doi: 10.1089/ten.tea.2019.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Y., Zhang B., Liu H., Miklas J.W., Gagliardi M., Pahnke A., Thavandiran N., Sun Y., Simmons C., Keller G., Radisic M. Microfabricated perfusable cardiac biowire: a platform that mimics native cardiac bundle. Lab Chip. 2014;14 doi: 10.1039/c3lc51123e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal A., Goss J.A., Cho A., McCain M.L., Parker K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip. 2013;13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., Jiang J., Massé S., Gagliardi M., Hsieh A., Thavandiran N., Laflamme M.A., Nanthakumar K., Gross G.J., Backx P.H., Keller G., Radisic M., Biowire A platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giacomelli E., Mummery C.L., Bellin M. Human heart disease: lessons from human pluripotent stem cell-derived cardiomyocytes. Cell. Mol. Life Sci. 2017;74:3711–3739. doi: 10.1007/s00018-017-2546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brutsaert D.L. Cardiac endothelial-myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol. Rev. 2003;83:59–115. doi: 10.1152/physrev.00017.2002. [DOI] [PubMed] [Google Scholar]
- 53.Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L., Plowright A.T., Needham E.J., Wang Q.-D., Gregorevic P., Xin M., Thomas W.G., Parton R.G., Nielsen L.K., Launikonis B.S., James D.E., Elliott D.A., Porrello E.R., Hudson J.E. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114:E8372. doi: 10.1073/pnas.1707316114. –E8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribeiro A.J.S., Ang Y.S., Fu J.D., Rivas R.N., Mohamed T.M.A., Higgs G.C., Srivastava D., Pruitt B.L. Contractility of Single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12705–12710. doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tiburcy M., Hudson J.E., Balfanz P., Schlick S., Meyer T., Chang Liao M.-L., Levent E., Raad F., Zeidler S., Wingender E., Riegler J., Wang M., Gold J.D., Kehat I., Wettwer E., Ravens U., Dierickx P., van Laake L.W., Goumans M.J., Khadjeh S., Toischer K., Hasenfuss G., Couture L.A., Unger A., Linke W.A., Araki T., Neel B., Keller G., Gepstein L., Wu J.C., Zimmermann W.-H. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation. 2017;135:1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren L., Liu W., Wang Y., Wang J.C., Tu Q., Xu J., Liu R., Shen S.F., Wang J. Investigation of hypoxia-induced myocardial injury dynamics in a tissue interface mimicking microfluidic device. Anal. Chem. 2013;85:235–244. doi: 10.1021/ac3025812. [DOI] [PubMed] [Google Scholar]
- 57.Lee M., Jung K.B., Jo S., Hyun S., Moon K., Seo J., Kim S., Son M. Modelling cardiac fibrosis using three- dimensional cardiac microtissues derived from human embryonic stem cells. J. Biol. Eng. 2019;13:15. doi: 10.1186/s13036-019-0139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spencer T.M., Blumenstein R.F., Pryse K.M., Lee S.L., Glaubke D.A., Carlson B.E., Elson E.L., Genin G.M. Fibroblasts slow conduction velocity in a reconstituted tissue model of fibrotic cardiomyopathy. ACS Biomater. Sci. Eng. 2017;3:3022–3028. doi: 10.1021/acsbiomaterials.6b00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishiga M., Wang D.W., Han Y., Lewis D.B., Wu J.C. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020;17:543–558. doi: 10.1038/s41569-020-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020 doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey A.L., Dmytrenko O., Greenberg L., Bredemeyer A.L., Ma P., Liu J., Penna V., Winkler E.S., Sviben S., Brooks E., Nair A.P., Heck K.A., Rali A.S., Simpson L., Saririan M., Hobohm D., Stump W.T., Fitzpatrick J.A., Xie X., Zhang X., Shi P.Y., Hinson J.T., Gi W.T., Schmidt C., Leuschner F., Lin C.Y., Diamond M.S., Greenberg M.J., Lavine K.J. SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis, JACC basic to transl. Sci. 2021;6 doi: 10.1016/j.jacbts.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y., Wang P., Qin J. Microfluidic organs-on-a-chip for modeling human infectious diseases. Acc. Chem. Res. 2021;54 doi: 10.1021/acs.accounts.1c00411. [DOI] [PubMed] [Google Scholar]
- 65.Yiangou L., Davis R.P., Mummery C.L. Using cardiovascular cells from human pluripotent stem cells for COVID-19 research: why the heart fails. Stem Cell Rep. 2020;16:1–13. doi: 10.1016/j.stemcr.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prondzynski M., Lemoine M.D., Zech A.T., Horváth A., Di Mauro V., Koivumäki J.T., Kresin N., Busch J., Krause T., Krämer E., Schlossarek S., Spohn M., Friedrich F.W., Münch J., Laufer S.D., Redwood C., Volk A.E., Hansen A., Mearini G., Catalucci D., Meyer C., Christ T., Patten M., Eschenhagen T., Carrier L. Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol. Med. 2019;11:1–18. doi: 10.15252/emmm.201911115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang E.Y., Rafatian N., Zhao Y., Lee A., Lai B.F.L., Lu R.X., Jekic D., Davenport Huyer L., Knee-Walden E.J., Bhattacharya S., Backx P.H., Radisic M. Biowire model of interstitial and focal cardiac fibrosis. ACS Cent. Sci. 2019;5:1146–1158. doi: 10.1021/acscentsci.9b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Y., Rafatian N., Feric N.T., Cox B.J., Aschar-Sobbi R., Wang E.Y., Aggarwal P., Zhang B., Conant G., Ronaldson-Bouchard K., Pahnke A., Protze S., Lee J.H., Davenport Huyer L., Jekic D., Wickeler A., Naguib H.E., Keller G.M., Vunjak-Novakovic G., Broeckel U., Backx P.H., Radisic M. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell. 2019;176:913–927. doi: 10.1016/j.cell.2018.11.042. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L., Geva J., Roberts A.E., Ma Q., Ding J., Chen J., Wang D.Z., Li K., Wang J., Wanders R.J.A., Kulik W., Vaz F.M., Laflamme M.A., Murry C.E., Chien K.R., Kelley R.I., Church G.M., Parker K.K., Pu W.T. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macqueen L.A., Sheehy S.P., Chantre C.O., Zimmerman J.F., Pasqualini F.S., Liu X., Goss J.A., Campbell P.H., Gonzalez G.M., Park S.J., Capulli A.K., Ferrier J.P., Fettah Kosar T., Mahadevan L., Pu W.T., Parker K.K. A tissue-engineered scale model of the heart ventricle. Nat. Biomed. Eng. 2018;2:930–941. doi: 10.1038/s41551-018-0271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forte E., Furtado M.B., Rosenthal N. The interstitium in cardiac repair: role of the immune–stromal cell interplay. Nat. Rev. Cardiol. 2018;15:601–616. doi: 10.1038/s41569-018-0077-x. [DOI] [PubMed] [Google Scholar]
- 72.Tampakakis E., Gangrade H., Glavaris S., Htet M., Murphy S., Lin B.L., Liu T., Saberi A., Miyamoto M., Kowalski W., Mukouyama Y.-S., Lee G., Minichiello L., Kwon C. Heart neurons use clock genes to control myocyte proliferation. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abh4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milani-Nejad N., Janssen P.M.L. Small and large animal models in cardiac contraction research: advantages and disadvantages. Pharmacol. Ther. 2014;141:235–249. doi: 10.1016/j.pharmthera.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colatsky T., Fermini B., Gintant G., Pierson J.B., Sager P., Sekino Y., Strauss D.G., Stockbridge N. The comprehensive in vitro Proarrhythmia assay (CiPA) initiative — update on progress. J. Pharmacol. Toxicol. Methods. 2016;81:15–20. doi: 10.1016/j.vascn.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 75.Saleem U., Meer B.J.V., Katili P.A., Yusof N.A.N.M., Mannhardt I., Garcia A.K., Tertoolen L., De Korte T., Vlaming M.L.H., McGlynn K., Nebel J., Bahinski A., Harris K., Rossman E., Xu X., Burton F.L., Smith G.L., Clements P., Mummery C.L., Eschenhagen T., Hansen A., Denning C. Blinded, multicenter evaluation of drug-induced changes in contractility using human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2020;176 doi: 10.1093/toxsci/kfaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Meer B.J., Krotenberg A., Sala L., Davis R.P., Eschenhagen T., Denning C., Tertoolen L.G.J., Mummery C.L. Simultaneous measurement of excitation-contraction coupling parameters identifies mechanisms underlying contractile responses of hiPSC-derived cardiomyocytes. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-12354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ravenscroft S.M., Pointon A., Williams A.W., Cross M.J., Sidaway J.E. Cardiac non-myocyte cells show enhanced pharmacological function suggestive of contractile maturity in stem cell derived cardiomyocyte microtissues. Toxicol. Sci. 2016;152:99–112. doi: 10.1093/toxsci/kfw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takeda M., Miyagawa S., Fukushima S., Saito A., Ito E., Harada A., Matsuura R., Iseoka H., Sougawa N., Mochizuki-Oda N., Matsusaki M., Akashi M., Sawa Y. Development of in vitro drug-induced cardiotoxicity assay by using three-dimensional cardiac tissues derived from human induced pluripotent stem cells. Tissue Eng. C Methods. 2018;24:56–67. doi: 10.1089/ten.TEC.2017.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pointon A., Harmer A.R., Dale I.L., Abi-Gerges N., Bowes J., Pollard C., Garside H. Assessment of cardiomyocyte contraction in human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2015;144:227–237. doi: 10.1093/toxsci/kfu312. [DOI] [PubMed] [Google Scholar]
- 80.Archer C.R., Sargeant R., Basak J., Pilling J., Barnes J.R., Pointon A. Characterization and validation of a human 3D cardiac microtissue for the assessment of changes in cardiac pathology. Sci. Rep. 2018 doi: 10.1038/s41598-018-28393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vulto P., Joore J. Adoption of organ-on-chip platforms by the pharmaceutical industry. Nat. Rev. Drug Discov. 2021 doi: 10.1038/s41573-021-00323-0. [DOI] [PubMed] [Google Scholar]
- 82.Skardal A., Murphy S.V., Devarasetty M., Mead I., Kang H.W., Seol Y.J., Zhang Y.S., Shin S.R., Zhao L., Aleman J., Hall A.R., Shupe T.D., Kleensang A., Dokmeci M.R., Jin Lee S., Jackson J.D., Yoo J.J., Hartung T., Khademhosseini A., Soker S., Bishop C.E., Atala A. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017;7:1–16. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee J., Mehrotra S., Zare-Eelanjegh E., Rodrigues R.O., Akbarinejad A., Ge D., Amato L., Kiaee K., Fang Y., Rosenkranz A., Keung W., Mandal B.B., Li R.A., Zhang T., Lee H., Dokmeci M.R., Zhang Y.S., Khademhosseini A., Shin S.R. A heart-breast cancer-on-a-chip platform for disease modeling and monitoring of cardiotoxicity induced by cancer chemotherapy. Small. 2021;17:2004258. doi: 10.1002/smll.202004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Korte T., Katili P.A., Mohd Yusof N.A.N., van Meer B.J., Saleem U., Burton F.L., Smith G.L., Clements P., Mummery C.L., Eschenhagen T., Hansen A., Denning C. Unlocking personalized biomedicine and drug discovery with human induced pluripotent stem cell–derived cardiomyocytes: fit for purpose or forever elusive? Annu. Rev. Pharmacol. Toxicol. 2020;60:529–551. doi: 10.1146/annurev-pharmtox-010919-023309. [DOI] [PubMed] [Google Scholar]
- 85.Mannhardt I., Breckwoldt K., Letuffe-Brenière D., Schaaf S., Schulz H., Neuber C., Benzin A., Werner T., Eder A., Schulze T., Klampe B., Christ T., Hirt M.N., Huebner N., Moretti A., Eschenhagen T., Hansen A. Human engineered heart tissue: analysis of contractile force. Stem Cell Rep. 2016;7:29–42. doi: 10.1016/j.stemcr.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mannhardt I., Saleem U., Mosqueira D., Loos M.F., Ulmer B.M., Lemoine M.D., Larsson C., Améen C., de Korte T., Vlaming M.L.H., Harris K., Clements P., Denning C., Hansen A., Eschenhagen T. Comparison of 10 control hPSC lines for drug screening in an engineered heart tissue format. Stem Cell Rep. 2020;15:983–998. doi: 10.1016/j.stemcr.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu H.F., Leong M.F., Lim T.C., Chua Y.P., Lim J.K., Du C., Wan A.C.A. Engineering a functional three-dimensional human cardiac tissue model for drug toxicity screening. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa6c3a. [DOI] [PubMed] [Google Scholar]
- 88.Feric N.T., Pallotta I., Singh R., Bogdanowicz D.R., Gustilo M.M., Chaudhary K.W., Willette R.N., Chendrimada T.P., Xu X., Graziano M.P., Aschar-Sobbi R. Engineered cardiac tissues generated in the biowire II: a platform for human-based drug discovery. Toxicol. Sci. 2019;172:89–97. doi: 10.1093/toxsci/kfz168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lind J.U., Yadid M., Perkins I., O'Connor B.B., Eweje F., Chantre C.O., Hemphill M.A., Yuan H., Campbell P.H., Vlassak J.J., Parker K.K. Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab Chip. 2017;17:3692–3703. doi: 10.1039/C7LC00740J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ribeiro M.C., Slaats R.H., Schwach V., Rivera-Arbelaez J.M., Tertoolen L.G.J., van Meer B.J., Molenaar R., Mummery C.L., Claessens M.M.A.E., Passier R. A cardiomyocyte show of force: a fluorescent alpha-actinin reporter line sheds light on human cardiomyocyte contractility versus substrate stiffness. J. Mol. Cell. Cardiol. 2020;141:54–64. doi: 10.1016/j.yjmcc.2020.03.008. [DOI] [PubMed] [Google Scholar]
- 91.Kolesová H., Olejníčková V., Kvasilová A., Gregorovičová M., Sedmera D. Tissue clearing and imaging methods for cardiovascular development. iScience. 2021;24 doi: 10.1016/j.isci.2021.102387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Richardson D.S., Lichtman J.W. Clarifying tissue clearing. Cell. 2015;162 doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elliott A.D. Confocal microscopy: principles and modern practices. Curr. Protoc. Cytom. 2020;92 doi: 10.1002/cpcy.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.LoBiondo J., Abramowitz M., Friedman M.M. Microscope objectives. Curr. Protoc. Cytom. 2011;58:1–15. doi: 10.1002/0471142956.cy0202s58. [DOI] [PubMed] [Google Scholar]
- 95.Mastikhina O., Moon B.U., Williams K., Hatkar R., Gustafson D., Mourad O., Sun X., Koo M., Lam A.Y.L., Sun Y., Fish J.E., Young E.W.K., Nunes S.S. Human cardiac fibrosis-on-a-chip model recapitulates disease hallmarks and can serve as a platform for drug testing. Biomaterials. 2020;233 doi: 10.1016/j.biomaterials.2019.119741. 119741. [DOI] [PubMed] [Google Scholar]
- 96.Albert-smet I., Marcos-vidal A., Vaquero J.J., Desco M., Muñoz-barrutia A., Ripoll J. vol. 13. 2019. pp. 1–15. (Applications of Light-Sheet Microscopy in Microdevices). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ronaldson-Bouchard K., Vunjak-Novakovic G. Organs-on-a-Chip: a fast track for engineered human tissues in drug development. Cell Stem Cell. 2018;22:310–324. doi: 10.1016/j.stem.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cloonan N., Forrest A.R.R., Kolle G., Gardiner B.B.A., Faulkner G.J., Brown M.K., Taylor D.F., Steptoe A.L., Wani S., Bethel G., Robertson A.J., Perkins A.C., Bruce S.J., Lee C.C., Ranade S.S., Peckham H.E., Manning J.M., McKernan K.J., Grimmond S.M. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008 doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 99.Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008 doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 101.Deng Y., Finck A., Fan R. Single-cell omics analyses enabled by microchip technologies. Annu. Rev. Biomed. Eng. 2019;21 doi: 10.1146/annurev-bioeng-060418-052538. [DOI] [PubMed] [Google Scholar]
- 102.Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:96. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kolodziejczyk A.A., Kim J.K., Svensson V., Marioni J.C., Teichmann S.A. The technology and biology of single-cell RNA sequencing. Mol. Cell. 2015 doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 104.Olsen T.K., Baryawno N. Introduction to single-cell RNA sequencing. Curr. Protoc. Mol. Biol. 2018;122 doi: 10.1002/cpmb.57. [DOI] [PubMed] [Google Scholar]
- 105.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–643. doi: 10.1016/j.molcel.2017.01.023. e4. [DOI] [PubMed] [Google Scholar]
- 106.van den Brink S.C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., Baron C.S., Robin C., van Oudenaarden A. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 107.Black S., Phillips D., Hickey J.W., Kennedy-Darling J., Venkataraaman V.G., Samusik N., Goltsev Y., Schürch C.M., Nolan G.P. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 2021;16 doi: 10.1038/s41596-021-00556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chang Q., Ornatsky O., Hedley D. Staining of frozen and formalin-fixed, paraffin-embedded tissues with metal-labeled antibodies for imaging mass cytometry analysis. Curr. Protoc. Cytom. 2017;82 doi: 10.1002/cpcy.29. [DOI] [PubMed] [Google Scholar]
- 109.Kinkhabwala A., Herbel C., Pankratz J., Yushchenko D.A., Rüberg S., Praveen P., Reiß S., Rodriguez F.C., Schäfer D., Kollet J., Dittmer V., Martinez-Osuna M., Minnerup L., Reinhard C., Dzionek A., Rockel T.D., Borbe S., Büscher M., Krieg J., Nederlof M., Jungblut M., Eckardt D., Hardt O., Dose C., Schumann E., Peters R.-P., Miltenyi S., Schmitz J., Müller W., Bosio A. MACSima imaging cyclic staining (MICS) technology reveals combinatorial target pairs for CAR T cell treatment of solid tumors. Sci. Rep. 2022;12:1911. doi: 10.1038/s41598-022-05841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miao L., Lu Y., Nusrat A., Abdelnasser H.Y., Datta S., Zhou B., Schwartz R.J., Wu M. The spatiotemporal expression of notch1 and numb and their functional interaction during cardiac morphogenesis. Cells. 2021;10 doi: 10.3390/cells10092192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vickovic S., Eraslan G., Salmén F., Klughammer J., Stenbeck L., Schapiro D., Äijö T., Bonneau R., Bergenstråhle L., Navarro J.F., Gould J., Griffin G.K., Borg Å., Ronaghi M., Frisén J., Lundeberg J., Regev A., Ståhl P.L. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16 doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shin S.R., Kilic T., Zhang Y.S., Avci H., Hu N., Kim D., Branco C., Aleman J., Massa S., Silvestri A., Kang J., Desalvo A., Hussaini M.A., Chae S.K., Polini A., Bhise N., Hussain M.A., Lee H.Y., Dokmeci M.R., Khademhosseini A. Label-free and regenerative electrochemical microfluidic biosensors for continual monitoring of cell secretomes. Adv. Sci. 2017;4 doi: 10.1002/advs.201600522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abreu C.M., Thomas V., Knaggs P., Bunkheila A., Cruz A., Teixeira S.R., Alpuim P., Francis L.W., Gebril A., Ibrahim A., Margarit L., Gonzalez D., Freitas P.P., Conlan R.S., Mendes Pinto I. Non-invasive molecular assessment of human embryo development and implantation potential. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112144. [DOI] [PubMed] [Google Scholar]
- 114.Cortezzi S.S., Cabral E.C., Trevisan M.G., Ferreira C.R., Setti A.S., Braga D.P.de A.F., Figueira R. de C.S., Iaconelli A., Eberlin M.N., Borges E. Prediction of embryo implantation potential by mass spectrometry fingerprinting of the culture medium. Reproduction. 2013;145:453–462. doi: 10.1530/REP-12-0168. [DOI] [PubMed] [Google Scholar]
- 115.Iles R.K., Sharara F.I., Zmuidinaite R., Abdo G., Keshavarz S., Butler S.A. Secretome profile selection of optimal IVF embryos by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Assist. Reprod. Genet. 2019;36:1153–1160. doi: 10.1007/s10815-019-01444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Picton H.M., Elder K., Houghton F.D., Hawkhead J.A., Rutherford A.J., Hogg J.E., Leese H.J., Harris S.E. Association between amino acid turnover and chromosome aneuploidy during human preimplantation embryo development in vitro. Mol. Hum. Reprod. 2010;16:557–569. doi: 10.1093/molehr/gaq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jacob R., Khan M. Cardiac biomarkers: what is and what can Be. Indian J. Cardiovasc. Dis. Women WINCARS. 2018;3:240–244. doi: 10.1055/s-0039-1679104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McDonaugh B., Whyte M. EMJ Cardiol; 2020. The Evolution and Future Direction of the Cardiac Biomarker. [DOI] [Google Scholar]
- 119.Miller E.W. Small molecule fluorescent voltage indicators for studying membrane potential. Curr. Opin. Chem. Biol. 2016;33:74–80. doi: 10.1016/j.cbpa.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shea C.O., Holmes A.P., Yu T.Y., Winter J., Wells S.P., Correia J., Boukens B.J., De Groot J.R., Chu G.S., Li X., Ng G.A. vols. 1–13. 2019. (ElectroMap : High-Throughput Open-Source Software for Analysis and Mapping of Cardiac Electrophysiology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maoz B.M., Herland A., Henry O.Y.F., Leineweber W.D., Yadid M., Doyle J., Mannix R., Kujala V.J., Fitzgerald E.A., Parker K.K., Ingber D.E. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip. 2017;17:2294–2302. doi: 10.1039/c7lc00412e. [DOI] [PubMed] [Google Scholar]
- 122.Shinozawa T., Nakamura K., Shoji M., Morita M., Kimura M., Furukawa H., Ueda H., Shiramoto M., Matsuguma K., Kaji Y., Ikushima I., Yono M., Liou S.-Y., Nagai H., Nakanishi A., Yamamoto K., Izumo S. Recapitulation of clinical individual susceptibility to drug-induced QT prolongation in healthy subjects using iPSC-derived cardiomyocytes. Stem Cell Rep. 2017;8:226–234. doi: 10.1016/j.stemcr.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kehat I., Gepstein A., Spira A., Itskovitz-Eldor J., Gepstein L. High-resolution electrophysiological assessment of human embryonic stem cell-derived cardiomyocytes. Circ. Res. 2002;91:659–661. doi: 10.1161/01.RES.0000039084.30342.9B. [DOI] [PubMed] [Google Scholar]
- 124.Kalmykov A., Huang C., Bliley J., Shiwarski D., Tashman J., Abdullah A., Rastogi S.K., Shukla S., Mataev E., Feinberg A.W., Hsia K.J., Cohen-karni T. Science Advances; 2019. Organ-on-e-chip : Three-Dimensional Self-Rolled Biosensor Array for Electrical Interrogations of Human Electrogenic Spheroids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y.S., Aleman J., Arneri A. Biofabrication; 2021. Micro-electrode Channel Guide (ΜECG) Technology: an Online Method for Continuous Electrical Recording in a Human Beating Heart-On-Chip. [DOI] [PubMed] [Google Scholar]
- 126.Huebsch N., Loskill P., Mandegar M.A., Marks N.C., Sheehan A.S., Ma Z., Mathur A., Nguyen T.N., Yoo J.C., Judge L.M., Spencer C.I., Chukka A.C., Russell C.R., So P.L., Conklin B.R., Healy K.E. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng. C Methods. 2015 doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pasqualini F.S., Agarwal A., O'Connor B.B., Liu Q., Sheehy S.P., Parker K.K. Traction force microscopy of engineered cardiac tissues. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dostanić M., Windt L.M., Stein J.M., Van Meer B.J., Bellin M., Orlova V., Mastrangeli M., Mummery C.L., Sarro P.M. A miniaturized EHT platform for accurate measurements of tissue contractile properties. J. Microelectromechanical Syst. 2020;29 doi: 10.1109/JMEMS.2020.3011196. [DOI] [Google Scholar]
- 129.Ribeiro A.J.S., Schwab O., Mandegar M.A., Ang Y.-S., Conklin B.R., Srivastava D., Pruitt B.L. Multi-imaging method to assay the contractile mechanical output of micropatterned human iPSC-derived cardiac myocytes. Circ. Res. 2017;120:1572–1583. doi: 10.1161/CIRCRESAHA.116.310363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sala L., van Meer B.J., Tertoolen L.G.J., Bakkers J., Bellin M., Davis R.P., Denning C., Dieben M.A.E., Eschenhagen T., Giacomelli E., Grandela C., Hansen A., Holman E.R., Jongbloed M.R.M., Kamel S.M., Koopman C.D., Lachaud Q., Mannhardt I., Mol M.P.H., Mosqueira D., Orlova V.V., Passier R., Ribeiro M.C., Saleem U., Smith G.L., Burton F.L., Mummery C.L. Musclemotion: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ. Res. 2018;122 doi: 10.1161/CIRCRESAHA.117.312067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van Meer B.J., Sala L., Tertoolen L.G.J., Smith G.L., Burton F.L., Mummery C.L. Quantification of muscle contraction in vitro and in vivo using MUSCLEMOTION software: from stem cell-derived cardiomyocytes to zebrafish and human hearts. Curr. Protoc. Hum. Genet. 2018;99:e67. doi: 10.1002/cphg.67. [DOI] [PubMed] [Google Scholar]
- 132.Evans R.D., Heather L.C. Metabolic pathways and abnormalities. Surgery. 2016;34:266–272. doi: 10.1016/j.mpsur.2016.03.010. [DOI] [Google Scholar]
- 133.Taegtmeyer H., Young M.E., Lopaschuk G.D., Abel E.D., Brunengraber H., Darley-Usmar V., Des Rosiers C., Gerszten R., Glatz J.F., Griffin J.L., Gropler R.J., Holzhuetter H.-G., Kizer J.R., Lewandowski E.D., Malloy C.R., Neubauer S., Peterson L.R., Portman M.A., Recchia F.A., Van Eyk J.E., Wang T.J. Assessing Cardiac Metabolism, Circ. Res. 2016;118:1659–1701. doi: 10.1161/RES.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Breckenridge R.A., Piotrowska I., Ng K.-E., Ragan T.J., West J.A., Kotecha S., Towers N., Bennett M., Kienesberger P.C., Smolenski R.T., Siddall H.K., Offer J.L., Mocanu M.M., Yelon D.M., Dyck J.R.B., Griffin J.L., Abramov A.Y., Gould A.P., Mohun T.J. Hypoxic regulation of Hand1 controls the fetal-neonatal switch in cardiac metabolism. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lopaschuk G.D., Jaswal J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 136.Mdaki K.S., Larsen T.D., Weaver L.J., Baack M.L. Age related bioenergetics profiles in isolated rat cardiomyocytes using extracellular flux analyses. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kuhn T.C., Knobel J., Burkert-Rettenmaier S., Li X., Meyer I.S., Jungmann A., Sicklinger F., Backs J., Lasitschka F., Müller O.J., Katus H.A., Krijgsveld J., Leuschner F. Secretome analysis of cardiomyocytes identifies PCSK6 (proprotein convertase subtilisin/kexin type 6) as a novel player in cardiac remodeling after myocardial infarction. Circulation. 2020;141:1628–1644. doi: 10.1161/CIRCULATIONAHA.119.044914. [DOI] [PubMed] [Google Scholar]
- 138.Robert A.W., Pereira I.T., Dallagiovanna B., Stimamiglio M.A. Secretome analysis performed during in vitro cardiac differentiation: discovering the cardiac microenvironment. Front. Cell Dev. Biol. 2020;8:49. doi: 10.3389/fcell.2020.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Brauchle E., Schenke-Layland K. Raman microspectroscopy for the development and screening of recombinant cell lines. Biotechnol. J. 2017;12:1600412. doi: 10.1002/biot.201600412. [DOI] [PubMed] [Google Scholar]
- 140.Shen N., Knopf A., Westendorf C., Kraushaar U., Riedl J., Bauer H., Pöschel S., Layland S.L., Holeiter M., Knolle S., Brauchle E., Nsair A., Hinderer S., Schenke-Layland K. Steps toward maturation of embryonic stem cell-derived cardiomyocytes by defined physical signals. Stem Cell Rep. 2017;9:122–135. doi: 10.1016/j.stemcr.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Votteler M., Carvajal Berrio D.A., Pudlas M., Walles H., Stock U.A., Schenke-Layland K. Raman spectroscopy for the non-contact and non-destructive monitoring of collagen damage within tissues. J. Biophot. 2012;5:47–56. doi: 10.1002/jbio.201100068. [DOI] [PubMed] [Google Scholar]
- 142.Brauchle E., Schenke-Layland K. Raman spectroscopy in biomedicine – non-invasive in vitro analysis of cells and extracellular matrix components in tissues. Biotechnol. J. 2013;8:288–297. doi: 10.1002/biot.201200163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Harari E., Guo L., Smith S.L., Braumann R.E., Virmani R., Finn A.V. Heart-resident macrophages: are they involved in the rhythm of every beat? J. Thorac. Dis. 2017;9:2264–2267. doi: 10.21037/jtd.2017.07.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hulsmans M., Clauss S., Xiao L., Aguirre A.D., King K.R., Hanley A., Hucker W.J., Wülfers E.M., Seemann G., Courties G., Iwamoto Y., Sun Y., Savol A.J., Sager H.B., Lavine K.J., Fishbein G.A., Capen D.E., Da Silva N., Miquerol L., Wakimoto H., Seidman C.E., Seidman J.G., Sadreyev R.I., Naxerova K., Mitchell R.N., Brown D., Libby P., Weissleder R., Swirski F.K., Kohl P., Vinegoni C., Milan D.J., Ellinor P.T., Nahrendorf M. Macrophages facilitate electrical conduction in the heart. Cell. 2017 doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nicolás-Ávila J.A., Lechuga-Vieco A.V., Esteban-Martínez L., Sánchez-Díaz M., Díaz-García E., Santiago D.J., Rubio-Ponce A., Li J.L.Y., Balachander A., Quintana J.A., Martínez-de-Mena R., Castejón-Vega B., Pun-García A., Través P.G., Bonzón-Kulichenko E., García-Marqués F., Cussó L., A-González N., González-Guerra A., Roche-Molina M., Martin-Salamanca S., Crainiciuc G., Guzmán G., Larrazabal J., Herrero-Galán E., Alegre-Cebollada J., Lemke G., Rothlin C.V., Jimenez-Borreguero L.J., Reyes G., Castrillo A., Desco M., Muñoz-Cánoves P., Ibáñez B., Torres M., Ng L.G., Priori S.G., Bueno H., Vázquez J., Cordero M.D., Bernal J.A., Enríquez J.A., Hidalgo A. A network of macrophages supports mitochondrial homeostasis in the heart. Cell. 2020;183 doi: 10.1016/j.cell.2020.08.031. [DOI] [PubMed] [Google Scholar]
- 146.Ardell J.L., Armour J.A. Compr. Physiol. John Wiley & Sons; Hoboken, NJ, USA: 2016. Neurocardiology: structure-based function; pp. 1635–1653. [DOI] [PubMed] [Google Scholar]
- 147.Zipes D.P., Jalife J., Stevenson W.G. seventh ed. Circulation; 2017. Cardiac Electrophysiology: from Cell to Bedside. [Google Scholar]
- 148.Farache Trajano L., Smart N. Immunomodulation for optimal cardiac regeneration: insights from comparative analyses. Npj Regen. Med. 2021;6:1–11. doi: 10.1038/s41536-021-00118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nahrendorf M., Swirski F.K., Aikawa E., Stangenberg L., Wurdinger T., Figueiredo J.L., Libby P., Weissleder R., Pittet M.J. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J. Exp. Med. 2007 doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wake E., Brack K. Characterization of the intrinsic cardiac nervous system. Auton. Neurosci. 2016;199:3–16. doi: 10.1016/j.autneu.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 151.Marcek Chorvatova A., Cagalinec M., Chorvat D. Time-resolved imaging of mitochondrial flavin fluorescence and its applications fro evaluating the oxidative state in living cardiac cells. Mitochondrial Medicine. 2021 doi: 10.1007/978-1-0716-1262-0_26. [DOI] [PubMed] [Google Scholar]
- 152.Zbinden A., Marzi J., Schlünder K., Probst C., Urbanczyk M., Black S., Brauchle E.M., Layland S.L., Kraushaar U., Duffy G., Schenke-Layland K., Loskill P. Non-invasive marker-independent high content analysis of a microphysiological human pancreas-on-a-chip model. Matrix Biol. 2020;85–86:205–220. doi: 10.1016/j.matbio.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 153.Tanumihardja E., Slaats R.H., van der Meer A.D., Passier R., Olthuis W., van den Berg A. Measuring both pH and O 2 with a single on-chip sensor in cultures of human pluripotent stem cell-derived cardiomyocytes to track induced changes in cellular metabolism. ACS Sens. 2021;6:267–274. doi: 10.1021/acssensors.0c02282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schneider O., Moruzzi A., Fuchs S., Grobel A., Schulze H.S., Mayr T., Loskill P. Fusing spheroids to aligned μ-tissues in a Heart-on-Chip featuring oxygen sensing and electrical pacing capabilities. bioRxiv. 2022:2022. doi: 10.1101/2022.02.26.482011. 02.26.482011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Shaegh S.A.M., Massa S., Riahi R., Chae S., Hu N., Avci H., Zhang W., Silvestri A., Nezhad A.S., Manbohi A., De Ferrari F., Polini A., Calzone G., Shaikh N., Alerasool P., Budina E., Kang J., Bhise N., Ribas J., Pourmand A., Skardal A., Shupe T., Bishop C.E., Dokmeci M.R., Atala A., Khademhosseini A. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2293. doi: 10.1073/pnas.1612906114. –E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fuchs S., Johansson S., Tjell A.Ø., Werr G., Mayr T., Tenje M. In-line analysis of organ-on-chip systems with sensors: integration, fabrication, challenges, and potential. ACS Biomater. Sci. Eng. 2021 doi: 10.1021/acsbiomaterials.0c01110. [DOI] [PMC free article] [PubMed] [Google Scholar]


