Graphical abstract
Keywords: Transmembrane protein, Topology prediction, Transfer learning
Abstract
Transmembrane proteins (TMPs) are essential for cell recognition and communication, and they serve as important drug targets in humans. Transmembrane proteins' 3D structures are critical for determining their functions and drug design but are hard to determine even by experimental methods. Although some computational methods have been developed to predict transmembrane helices (TMHs) and orientation, there is still room for improvement. Considering that the pre-trained language model can make full use of massive unlabeled protein sequences to obtain latent feature representation for TMPs and reduce the dependence on evolutionary information, we proposed DeepTMpred, which used pre-trained self-supervised language models called ESM, convolutional neural networks, attentive neural network and conditional random fields for alpha-TMP topology prediction. Compared with the current state-of-the-art tools on a non-redundant dataset of TMPs, DeepTMpred demonstrated superior predictive performance in most evaluation metrics, especially at the TMH level. Furthermore, DeepTMpred could also obtain reliable prediction results for TMPs without much evolutionary feature in a few seconds. A tutorial on how to use DeepTMpred can be found in the colab notebook (https://colab.research.google.com/github/ISYSLAB-HUST/DeepTMpred/blob/master/notebook/test.ipynb).
1. Introduction
Biological membranes protect the vital contents of cells from the environment. Membrane proteins are an important part of biological membranes and play a diverse and important role in many processes such as molecule transport, immune system molecule recognition and metabolism [1], [2]. Membrane proteins are fully embedded into the membrane lipid bilayer, and often comprise a substantial fraction of the membrane total mass, ranging from 18% in the insulating myelin membrane of neurons to 75% in the inner membrane of mitochondria [3]. Membrane proteins are mainly divided into two structural classes: alpha-helical transmembrane proteins and beta-barrel transmembrane proteins. α-helical transmembrane proteins make up the majority of all known membrane proteins, which have been mostly found in the plasma membrane of eukaryotes and the inner membranes of bacterial cells. In humans, the importance of TMPs is reflected in the fact that they account for more than 50% of known drug targets even though they constitute a minority (between 20% and 30%) of all the proteins encoded in fully-sequenced genome [4], [5].
However, the structure determination of transmembrane proteins by X-ray crystallography and by NMR techniques is limited due to the difficulties in crystallizing them in an aqueous environment and by their relatively high molecular weight [6]. The known structures of transmembrane proteins (TMPs) only comprise about 1.8% (∼5800) of all structures in the Protein Data Bank [2], but there are more than 30,000,000 unique TMP sequences in the UniRef100 (release 2020.06). Considering the importance of transmembrane protein structures for drug development, automatic computational methods predicting topology of transmembrane proteins has been paid great attention. In particular, some low-resolution structural information about transmembrane proteins (TMPs) can be served as the constraint information for custom experiments [7] or modeling their 3D structures [8].
As far as we know, TopPred [9] is one of the earliest methods to predict the topology of TMPs, which was developed based on hydrophobicity analysis. Jones et al. [10] proposed a constrained dynamic programming to find the optimal location and orientation of transmembrane helices. In the following time, computational methods based on machine learning have been widely applied for predicting the topology of transmembrane proteins (TMPs) such as hidden Markov models [11], [12], [13], [14], SVMs [15], feed-forward neural network [16], [17], random forests [18], conditional random fields [19], [20], and K-nearest neighbor [21]. Besides, the prediction methods like TOPCONS [22], [23], CCTOP [24], and CNTOP [25] integrated the results of different computational methods into one consensus prediction and quantified the reliable TMHs based on the agreement between the different methods. Assaf et al. [26] proposed a graphical algorithm, called TopGraph, which was based on the minimum energy, the positive-inside rule and showed high accuracy on large transporters without structural homologues. Compared to traditional machine learning methods, computational methods based on deep learning can have significant advantages on complex biology problems [27], [28]. Recently, some methods based on deep learning have been developed to significantly improve the topology prediction of transmembrane proteins such as Membrain3.0 [29], DMCTOP [30]. Most of the above methods used evolutionary information as the input features such as sequence profile or multiple sequence alignments [31], [32], therefore they do not work when there are not enough homologous sequences.
Transfer learning aims at improving the performance of task on target domains by transferring the knowledge contained in different but related source domains, which can reduce the dependence on a large number of target domain data [33]. Transferring knowledge is particularly efficient when data are abundant in the source domain but scarce in the target domain. In the natural language processing (NLP) field, transfer learning in the form of pre-trained language models has become ubiquitous. Pre-trained language models mainly learn context-based word embeddings such as Bert [34]. Since there are more than billion protein sequences in databases, one efficient way is self-supervised language model to learn the latent information from unlabeled sequences. Recently, Bepler et al. [35] have reported competitive results with the help of transfer learning on secondary structure prediction and Rives et al. [36] trained the pre-trained language model called ESM based on Transformer which includes more than 680 million parameters to predict protein contact map. It is extremely enlightening for us to get a better feature representation of transmembrane proteins with the help of transfer learning.
In this study, we proposed a transfer learning method, DeepTMpred, using self-supervised pre-trained language models called ESM [36], convolutional neural networks, attentive neural network and conditional random fields for TMP topology prediction. Compared with the current state-of-the-art tools on an independent dataset, DeepTMpred can achieve superior results in most evaluation metrics. Furthermore, with the help of pre-trained language model, it could produce reliable topology prediction for TMPs in a few seconds. Finally, all source code can be freely available at https://github.com/ISYSLAB-HUST/DeepTMpred.
2. Materials and methods
2.1. Dataset
2.1.1. Data collection and preprocessing
We collected alpha-TMPs with known structures annotated in OPM(version: July 02, 2020) [37], which was an up-to-date experimental TMP structure database and provided spatial arrangements of membrane proteins with respect to the hydrocarbon core of the lipid bilayer. We first collected two classes of proteins from orientations of proteins in membranes (OPM) database such as the alpha-helical polytopic proteins and bitopic proteins. 8684 protein chains were collected in total. The protein chains were further chosen with the following conditions: (1) the chain is consecutive; (2) the length of the protein chain is less than 800 residues, and more than 20 residues; and (3) there is at least one TMH in the chain. Redundancy of the sequences was removed at 30% identity using CD-HIT and PSI-CD-HIT [38], [39]. We chose 40 test proteins used in Membrain 3.0 as the independent test dataset, where the similarity between them is less than 20%, and all of them are solved by either NMR or X-ray technique with resolution less than 4.5 Å. To guarantee a fair comparison on the independent test dataset, the collected sequences in the OPM that are similar to the independent test proteins at a threshold of 30% were dropped. By the above steps, the remaining TMPs from training dataset included 582 protein chains and was randomly divided into training set and validation set according to the ratio of 4:1. The annotation of TMHs and the orientations was taken from the OPM database (https://opm.phar.umich.edu/download). All training data and test data can be freely accessed at https://github.com/ISYSLAB-HUST/DeepTMpred. Table 1 lists a summary of the residues and TMHs located on TM and non-TM in the training set and test set. Besides, the training set and test set have approximately the same distributions with respect to the TMH length (Fig. S1). To explore whether the model can distinguish between soluble proteins and TMPs, we collected two types of soluble proteins from the TMSEG SP1441 test set, including 113 proteins with signal peptides and 173 proteins without signal peptides.
Table 1.
The summary of residues and TMHs located on TM and non-TM in training set and test set.
| Type |
Training Set |
Test Set |
||
|---|---|---|---|---|
| TM | non-TM | TM | non-TM | |
| Residues | 55,949 | 78,724 | 3126 | 3718 |
| TMHs | 2637 | – | 146 | – |
2.1.2. Construction of features based on evolutionary information
Position-specific scoring matrix (PSSM) is a commonly used protein sequence pattern representation. It contains evolutionary information and has been widely used in previous TMH prediction methods [15], [16], [40]. PSI-BLAST [41] is used to search against the non-redundant (NR) database with an iteration number equal to 3 and e-value threshold equal to 1e-3. For a protein sequence with length of L amino acids, the dimension of PSSM is .
Hidden Markov Model (HMM) profile is conducted by HHblits [42] to search against the Uniclust30 database with an iteration number equal to 3 and e-value threshold to 1e-3. The HMM profile is a matrix. For each residue column, it consists of 20 emission frequencies (EF), 7 transition probabilities, and 3 local diversities. PSSM and HMM profile was constructed for all training and test proteins.
2.1.3. Label of the topology
For the TMH prediction task, (0, 1) is assigned for labeling the residues in the sequence. ‘0’ indicates a non-transmembrane residue and ‘1’ indicates a membrane residue. For the N-terminus orientation prediction in the TMPs, ‘0’ denotes an inner membrane side and ‘1′ denotes an outer membrane side.
2.2. Model architecture
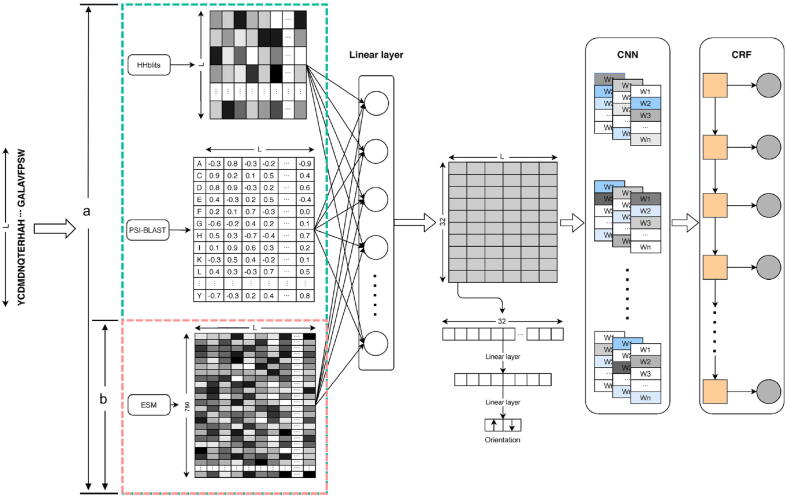
The model architecture shown in Fig. 1 consists of 6 parts, pre-trained self-supervised language model, evolutionary feature preprocessing, convolutional layer, two position-wise fully-connected layers, Conditional Random Fields, and attentive neural network. After the treatment of the evolutionary feature, the preprocessed feature vectors are concatenated with the output of the pre-trained language model. The integrated feature is fed into a linear layer to reduce the dimensionality. Using the convolution layer can obtain local features of the residues. In the next stage, two position-wise fully-connected layers are used for the classification of transmembrane protein helix. Finally, conditional random fields are used to optimize the distribution of sequence labeling.
Fig. 1.
The flowchart of DeepTMpred.
2.2.1. Pre-trained self-supervised language model
The development of pre-trained language models has brought the research in the protein representation to a new stage without artificial labels. Representation of protein sequences can be learned from massive unlabeled protein sequences, and downstream tasks can be significantly improved. Using pre-trained language model can reduce the risk of overfitting on small training data, which is equivalent to a kind of regularization method.
Here, we adopted a transformer-based self-supervised language model called ESM, which made full use of up to 250 million sequences of the Uniparc database [43] and included more than 85 million parameters (12-layer transformer) [36]. Here, the ESM model can accept a protein sequence and generate embedding matrices with .
2.2.2. Feature fusion and convolutional neural network
All embeddings are projected into the same feature space by using a fully connected neural layer. Given protein sequence PSSM feature , HMM profile feature , the embedding feature of the language model , we can obtain new embeddings via:
| (1) |
where is the concatenation and FC represents a fully connected neural layer ( represents the learnable parameters) which is applied to each position separately. As such, a nonlinear ReLU activation function can be applied to the fusion embedding. Considering the strong local correlation of the topological structure of transmembrane proteins, we use a convolutional layer based on sparse connectivity to obtain the local vector representations of adjacent residues.
2.2.3. Conditional random fields
Transmembrane protein topology prediction is a classical problem of sequence labeling. The aim is to determine whether the residue is embedded in the biological membrane or not. The residues are assigned by using Conditional Random Fields to strengthen the correlation between labels. Here, the use of a CRF layer is a helpful regularizer so that continuity of TMHs is encouraged. Conditional random fields were proposed by Lafferty et al. [44] for labeling sequence data. Given a protein sequence (length is ) of observations , the most probable topology sequence is , i.e. . By the fundamental theorem of a random field, the joint distribution over label sequence Y given X can be given by the following conditional probability:
| (2) |
where is the output of the convolutional neural network directly below the conditional random field, Z(h) is the normalization constant of the distribution , is a two fully-connected layers model which accepts as input and obtains outputs of m classes and is a learnable transition matrix with parameters.
2.2.4. Orientation prediction model based on the attentive neural network
The inside/outside orientation is only annotated at the N-terminus orientation of proteins in membranes (OPM) database. Therefore, this prediction is made for the entire protein, and then the orientation of the other non-transmembrane residues is inferred since they are supposed to switch after each TMH. Firstly, the parameters of the fully connected neural layer of feature fusion were fine-tuned with a small learning rate(1e-5). Since only the inside/outside position of the N-terminal is predicted, 5 residues before and after the TMP were chosen to classify the position which was taken based on the performance of the validation set. Then, an attention layer [45] with the ReLU activation function and a fully connected layer are used to map the features to the binary classification space. For the attention layer, the orientation representation of the TMP is formed by a weighted sum of these output vectors:
| (3) |
where is the embeddings of 5 residues before and after the TMP, is the dimension of the fusion layer, is a trained parameter vector andis a transpose. The dimension of , , is , 10, separately.
For model optimization, the label smoothing [46] method is adopted to avoid the overfitting of the prediction model.
2.3. Evaluation criteria
To evaluate our model performance, we adopted the same evaluation criteria that were also applied in previous studies [29].
For TMH prediction on residues level, it should be predicted whether each residue belongs to a membrane residue. PRE(r) is defined as:
| (4) |
The recall for residues level. REC(r) is defined as:
| (5) |
The F1 score for residues level. F1(r) is defined as:
| (6) |
The precision for TMH level. PRE(h) is defined as:
| (7) |
The recall for TMH level. REC(h) is defined as:
| (8) |
The F1 score for TMH level. F1(h) is defined as:
| (9) |
is the number of proteins with correctly predicted TMH, which is defined as:
| (10) |
A TMH segment is correctly predicted, if the segment satisfies both of the following criteria: (1) the endpoints of the predicted TMH does not deviate from those of the observed TMH by more than 5 residues; (2) the overlapped residues between the predicted TMH segment and the real TMH segment accounts for at least half of the longer one.
The inside/outside orientation prediction is a binary classification task. Thus, we adopt MCC, F1 score, and accuracy (ACC) criteria to evaluate prediction performance. These criteria are defined as:
| (11) |
| (12) |
| (13) |
where TP, TN, FP, FN represent the numbers of true positives, true negatives, false positives, and false negatives, respectively.
is used to evaluate the accuracy of soluble protein prediction, which is defined as:
| (14) |
3. Results
First of all, we showed how the hyperparameters of our model are tuned specifically. To make a comprehensive evaluation of transmembrane helix (TMHs) prediction, we compared DeepTMpred with three representative state-of-the-art methods including deep learning (MemBrain3.0), ensemble learning (CCTOP, TOPCONS), machine learning (MEMSAT-SVM) and graphical algorithm based on minimum energy (TopGraph) on the independent test data for TMH segments prediction. To demonstrate that DeepTMpred could work well even if TMPs are lack of evolutionary information, our proposed model was divided into two categories according to the input feature: (a) accepted the fusion feature of the evolutionary information and the output feature of ESM; (b) only accepted the output of ESM as input feature.
3.1. Hyperparameters tuning
DeepTMpred was constructed in Python 3.8 with PyTorch 1.5.0. Considering the limitations of GPU memory, we only finetuned 12-layer ESM with a small learning rate (1e-5). There are some hyperparameters needing to be optimized, including fusion embedding dimension, convolution filter setting, the numbers of convolution layers, and the fully connected layer dimension. All hyperparameters were tuned with Neural Network Intelligence (https://github.com/microsoft/nni). In this work, the optimized hyperparameters are as follows: fusion embedding dimension is 32, convolution filter is (16, 3) in which filter number is 16 and kernel size is 3, and the dimension of two fully connected layers are 16 and 2, respectively. For the inside/outside orientation prediction task, a small learning rate(1e-4) was applied for fine-tuning at the fully connected neural layer of feature fusion and the dimension of two fully connected layers for orientation classification are 8 and 2, respectively.
3.2. Comparison to the state-of-the-art methods for TMH segments prediction
To make a fair comparison with the three-representative state-of-the-art methods, the same test data has been adopted. Table 2 shows the prediction performance of different methods based on the 40 independent test proteins (the results of MemBrain3.0, MEMSAT-SVM, TOPCONS, TopGraph and CCTOP are from MemBrain3.0′s paper). DeepTMpred-a adopted the characteristics of evolutionary information of PSSM and HMM profile, while DeepTMpred-b does not include evolutionary information which is replaced by an all-zero matrix of . For DeepTMpred training, above process was equivalent that DeepTMpred-b had 50*32 less parameters than DeepTMpred-a.
Table 2.
Performance comparisons of DeepTMpred with the three representative state-of-the-art methods on the independent test set.
| Methods | PRE(r) | REC(r) | F1(r) | PRE(h) | REC(h) | F1(h) | V(p) |
|---|---|---|---|---|---|---|---|
| MemBrain3.0 | 0.892 | 0.904 | 0.898 | 0.808 | 0.819 | 0.814 | 21 |
| TopGraph | 0.873 | 0.717 | 0.787 | 0.504 | 0.464 | 0.483 | 12 |
| TOPCONS | 0.918 | 0.722 | 0.808 | 0.573 | 0.530 | 0.551 | 12 |
| MEMSAT-SVM | 0.926 | 0.710 | 0.804 | 0.591 | 0.547 | 0.568 | 10 |
| CCTOP | 0.931 | 0.679 | 0.785 | 0.511 | 0.477 | 0.493 | 10 |
| DeepTMpred-a | 0.874 | 0.918 | 0.895 | 0.857 | 0.863 | 0.860 | 26 |
| DeepTMpred-b | 0.889 | 0.911 | 0.900 | 0.864 | 0.870 | 0.867 | 28 |
From Table 2, we see that DeepTMpred-b including 12-layer Transformer achieved the highest F1(r) score of 0.900, followed by MemBrain3.0 (0.898), DeepTMpred-a (0.895), TOPCONS (0.808), MEMSAT-SVM (0.804), TopGraph (0.787), and CCTOP (0.785). CCTOP achieved the highest PRE(r) (0.931), followed by DeepTMpred-a (0.918), but CCTOP had the lowest REC(r) (0.679). It is interesting to be noted that all the PRE(h), REC(h) and F1(h) scores of DeepTMpred were significantly higher than those of the other three state-of-the-art methods. For the TMH segments prediction, the values of PRE(h), REC(h) and F1(h) of DeepTMpred-a are 0.857, 0.863 and 0.860, respectively, which are 6.1%, 5.4% and 5.7% higher than those of the MemBrain3.0 which performs the best at the TMH level among the three comparing state-of-the-art methods. Meanwhile, without including evolutionary information, DeepTMpred-b achieved PRE(h) of 0.864, REC(h) of 0.870 and F1(h) of 0.867, which are 6.9%, 6.2% and 6.5% higher than those of the MemBrain3.0.
Among the 40 test proteins, DeepTMpred-b correctly predicts all the TMHs for 28 proteins, i.e., 28 of 40 test proteins are assigned with the correct TMHs, while the highest V(p) of the three comparing state-of-the-art methods is 21. The second-best predictor on V(p) is DeepTMpred-a, which correctly predicts 26 out of the 40 test proteins. These data imply that DeepTMpred based the pre-trained model achieves better performance than other state-of-the-art methods.
The superior performance of DeepTMpred is mainly attributed to the following two reasons. Firstly, transfer learning which does not rely on large-scale training data is very suitable for TMP topology prediction with small scale data. Secondly, CRFs used in the last step of DeepTMpred can learn long-range correlations of TMHs and non-TMHs, which further improves the prediction performance of the method.
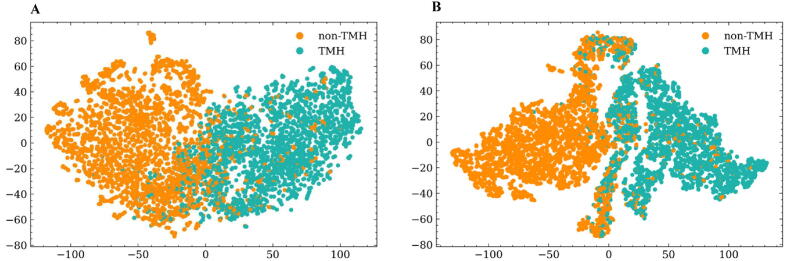
3.3. Representation of TMP residues in embedding space
DeepTMpred can be expected to capture meaningful patterns between TMH and non-TMH. To investigate whether the DeepTMpred model has learned to encode properties of TMH classification in its representations, we use 40 test TMPs from Membrain 3.0 and project the learned embedding of the fusion layer and convolutional layer of the DeepTMpred-b into two dimensions by applying the t-distributed stochastic neighbor embedding (t-SNE) algorithm. Fig. 2 demonstrates that the embedding is already meaningful after training in the convolutional layer. Furthermore, the clustering distribution of the embedding space of the convolutional layer is denser and more separable than the fusion layer and it shows that the convolutional layer enhances patterns in the embedding space. Based on the results, it is clearly concluded that the fusion layer and convolutional layer could encode and capture the different level feature from ESM embeddings and evolution information.
Fig. 2.
TMH properties of amino acids are represented in the output embeddings of the fusion layer(A) and convolutional layer (B), visualized here with t-SNE.
3.4. The inside/outside orientation prediction
Considering that other non-transmembrane positions can be inferred by the N-terminus position of a TMP, we only predicted the inside/outside orientation of the N-terminus by transfer learning. We adopt MCC, ACC, and F1 criteria to evaluate the effect of DeepTMpred. The performance of the two modules is shown in Table 3. From Table 3, we can see that DeepTMpred-a achieved F1 of 0.935, ACC of 0.900, and MCC of 0.722, which were better than those of DeepTMpred-b. The attentive neural network could efficiently learn and choose the import feature in 5 residues before and after the TMP. It also indicates that evolutionary information can significantly improve the performance of the orientation task and the latent information from ESM play a complementary role with evolutionary information, considering that the better performance of DeepTMpred-a than DeepTMpred-b. Regarding the prediction of orientation, we also compared DeepTMpred with the two tools (MEMSAT-SVM, TOPCONS). It can be seen from Table 3 that the scores of DeepTMpred-a and DeepTMpred-b in MCC, F1 and ACC were far higher than those of MEMSAT-SVM and TOPCONS. At present, our model only focuses on the orientation on the N-terminus, and it can be extended to the prediction of re-entrant loops in the future.
Table 3.
Performance of DeepTMpred for TMP orientation prediction with MEMSAT-SVM and TOPCONS on the independent test set.
| Methods | F1 | ACC | MCC |
|---|---|---|---|
| DeepTMpred-a | 0.935 | 0.900 | 0.722 |
| DeepTMpred-b | 0.915 | 0.875 | 0.680 |
| TOPCONS | 0.717 | 0.625 | 0.204 |
| MEMSAT-SVM | 0.769 | 0.700 | 0.406 |
3.5. Distinguishing TMPs with soluble proteins
Although DeepTMpred was not trained on soluble proteins, it is also important to distinguish TMPs with soluble proteins. 286 soluble proteins (two groups) from the SP1441 dataset were used. We mainly compared DeepTMpred with the other three methods (MEMSAT-SVM, TMHMM2.0 and TOPCONS) for this task. We adopted the same strategy as CCTOP and Phobius, that is, using SignalP software to predict the position information of signal peptides, which can greatly alleviate the problem of identifying signal peptides as TMH. In this study, SignalP 6.0 [47] was used. The prediction results are shown in Table 4. For the group A of 113 soluble proteins with signal peptides, TOPCONS achieved the highest ACC score of 0.929, followed by DeepTMpred (0.920), MEMSAT-SVM (0.699), and TMHMM2.0(0.655). For the group B of 173 soluble proteins without signal peptides, TMHMM2.0 achieved the highest ACC score of 0.977, followed by DeepTMpred (0.971), TOPCONS (0.971), and MEMSAT-SVM (0.965). Further comparison of the results showed that only DeepTMpred and TOPCONS could distinguish TMPs with soluble proteins well on both the two datasets.
Table 4.
Prediction performance of DeepTMpred and the other three methods on the SP1441 soluble protein test set (A: 113 proteins with signal peptides, B: 173 proteins without signal peptides.).
| Methods | ACCsp (A) | ACCsp (B) |
|---|---|---|
| DeepTMpred | 0.920 | 0.971 |
| MEMSAT-SVM | 0.699 | 0.965 |
| TMHMM2.0 | 0.655 | 0.977 |
| TOPCONS | 0.929 | 0.971 |
3.6. Analysis of time complexity
We also compared the time complexity for DeepTMpred and the other three methods on the 40 test proteins. TMHMM2.0, MEMSAT-SVM and TOPCONS were run as standalone package with default parameters and the pure running time for 40 test proteins was counted. Since the MemBrain3.0 model file cannot be run locally in our cluster, only the time of the feature generation step was computed. The results are shown in Table 5. MemBrain3.0 was the most time-consuming method, followed by MEMSAT-SVM, TOPCONS, DeepTMpred, and TMHMM2.0. Among the top three most accurate methods (DeepTMpred, MemBrain3.0 and TOPCONS), DeepTMpred required the least amount of time and it only took 8 s to predict the topology of 40 TMPs on the CPU device (6 s on GPU device). CCTOP did not provide a downloadable version, but its paper reported that a sequence took anywhere from a few minutes to 30 min. Overall, DeepTMpred had good performance in terms of both accuracy and running time.
Table 5.
The time complexity comparison between DeepTMpred and other four methods on the 40 test proteins.
| Methods | Running Time (s) | Database |
|---|---|---|
| TMHMM2.0 | 4 | – |
| MEMSAT-SVM | 12,557 | UniRef-50 |
| MemBrain3.0* | 38,303 | NR, UniClust-30 |
| TOPCONS | 869 | Pfam, CDD |
| DeepTMpred | 8/6 | – |
3.7. Case study
In Fig. 3, we showed an illustrative example from bacteriorhodopsin-I (PDB id:4pxkA) which is a kind of bacteriorhodopsin protein acting as a proton pump and including 7 TMHs. The prediction results of DeepTMpred-a and DeepTMpred-b were shown in Fig. 3A and Fig. 3B, respectively. From Fig. 3, it is easy to see that the first six TMHs of bacteriorhodopsin-I were correctly identified by both DeepTMpred-a and DeepTMpred-b. The predicted start and stop sites of the first six TMHs were all within the error range (<=5 residues) according to the annotation of the OPM database (Fig. 3C). Only the termination site of the seventh TMH predicted by DeepTMpred-a and DeepTMpred-b exceeded the error range. The errors were 6 and 7 residues, respectively, according to the annotation of the OPM database. While it is found that the starting and ending sites of the seventh TMH of bacteriorhodopsin-I are 208 and 229, respectively, based on the PDBTM database annotation which are a little difference from the annotation of the OPM database which are 205 and 219, respectively. The termination site of the seventh TMH predicted by DeepTMpred-a and DeepTMpred-b was also within the error range (<=5 residues), if judged by the annotation of the PDBTM database. Furthermore, the very similar performance of DeepTMpred-a and DeepTMpred-b on the case study of bacteriorhodopsin-I shows that DeepTMpred could also work well without using evolutionary information.
Fig. 3.
Case study of bacteriorhodopsin-I. (A)Molecular structure of bacteriorhodopsin-I (PDB entry: 4pxk) annotated according to DeepTMpred-a prediction: TMHs in green. (B)Molecular structure of bacteriorhodopsin-I (PDB entry: 4pxk) annotated according to DeepTMpred-b prediction: TMHs in red. (C)Comparison between the real topology (OPM database) and prediction of DeepTMpred. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We also compared the performance of three different tools (MEMSAT-SVM, MemBrain and TOPCONS) with DeepTMpred in hard-type TMH. For long-TMH prediction, 4b4aA was taken as an example, whose third TMH was of long type (>30). As can be seen from Table 6, both DeepTMpred, MemBrain, and MEMSAT-SVM could predict the long-TMH, but TOPCONS failed. Both MEMSAT-SVM and MemBrain showed a big error in the first TMH prediction, and DeepTMpred and TOPCONS also showed an error in the fourth TMH (more than 6 residues at the N-terminus). This example also showed that DeepTMpred was competitive with other tools in hard type TMH prediction.
Table 6.
Comparison between the true topology of 4b4aA (OPM database) and that predicted by DeepTMpred, MEMSAT-SVM, MemBrain, and TOPCONS.
| Methods | TMH-1 | TMH-2 | TMH-3(hard) | TMH-4 | TMH-5 | TMH-6 |
|---|---|---|---|---|---|---|
| OPM | (11–32) | (61–85) | (100–130) | (147–170) | (187–202) | (206–226) |
| MEMSAT-SVM | (15–40) | (59–87) | (101–130) | (150–173) | (190–205) | (210–225) |
| TOPCONS | (14–34) | (65–85) | (103–123) | (153–173) | (187–207) | (210–230) |
| MemBrain | (6–45) | (59–88) | (100–129) | (149–175) | (182–202) | (206–231) |
| DeepTMpred | (11–31) | (64–85) | (101–130) | (153–173) | (188–203) | (209–227) |
4. Discussion and conclusion
In this work, we proposed a transfer learning method, DeepTMpred, using pre-trained self-supervised language model called ESM, convolutional neural networks, conditional random field and attentive neural network for alpha-TMP topology prediction. Based on the comparison results, the proposed DeepTMpred showed superior performance in predicting the topology of TMP compared with the state-of-the-art methods.
Protein representation is an important problem for downstream tasks such as protein contact map prediction. Although evolutionary information is the most extensive method of protein representation, its disadvantages are obvious, relying too much on homologous sequences. Most of the state-of-art machine-learning based methods for TMP topology prediction made use of sequence profile as the input features, but they are time-consuming find enough homologous sequences. Further, some methods (such as Membrain 3.0) rely on the predicted structural features and biochemical properties, but these predicted features caused bias and calculations are time-consuming (more than dozens of minutes). DeepTMpred model which does not rely on the evolution information showed that the topology of proteins could be predicted only depending on the output feature of ESM within a few seconds.
Besides, after comparing DeepTMpred-a with DeepTMpred-b, DeepTMpred-b was slightly better at the TMH level, which implied that the combination of ESM and evolutionary information would bring noise and ESM model has already included most evolutionary information or adding parameters was difficult to optimize on training data sets. Intuitively, it is difficult to optimize a complex network (more parameters) for a small-scale data set, so this does not mean that adding evolutionary information will not lead to improvements in other tasks. In our final model, the fine-tuning ESM was used to replace the artificially constructed features in terms of evaluation performance. In the proposed model, CRFs also play an important role in TMH prediction and construct the correlation of labels. For the orientation prediction task, DeepTMpred can predict the orientation of transmembrane proteins through a transfer model with the attentive neural network which could capture the important contribution residues.
In addition to its best performance, DeepTMpred is also recommended due to its speed. The method is readily available for free: online via colab notebook, and as a standalone package from GitHub (https://github.com/ISYSLAB-HUST/DeepTMpred). Apart from that, as a stand-alone software package, DeepTMpred does not require any biological sequence database. A tutorial on how to use DeepTMpred can be found in the colab notebook (https://colab.research.google.com/github/ISYSLAB-HUST/DeepTMpred/blob/master/notebook/test.ipynb), mainly providing three different prediction modes (batch sequence prediction, single TMP sequence prediction, extra-long TMP sequence prediction).
As far as we know, this is the first time that transfer learning algorithm has been applied in the topology prediction of TMPs. Transfer learning provides new ideas for predicting the topology of transmembrane proteins lacking sufficient evolutionary information. In the future, we intend to improve the performance of TMPs topology prediction by developing new deep learning methods.
Funding
This work was supported by National Natural Science Foundation of China under Grant 61772217 and 62172172, Scientific Research Start-up Foundation of Binzhou Medical University under Grant BY2020KYQD01, and Fundamental Research Funds for the Central Universities under Grant 2016YXMS104 and 2017KFYXJJ225.
CRediT authorship contribution statement
Lei Wang: Software, Formal analysis, Data curation, Writing – review & editing. Haolin Zhong: Software, Formal analysis, Writing – review & editing, Visualization. Zhidong Xue: Conceptualization, Methodology, Writing – review & editing. Yan Wang: Conceptualization, Methodology, Writing – original draft, Writing – review & editing.
Acknowledgments
Acknowledgments
Thanks to the Facebook Research team for providing the pre-trained weights for the transformer protein language models.
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.04.024.
Contributor Information
Zhidong Xue, Email: zdxue@hust.edu.cn.
Yan Wang, Email: yanw@hust.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Heyden M., Freites J.A., Ulmschneider M.B., et al. Assembly and stability of alpha-helical membrane proteins. Soft Matter. 2012;8:7742–7752. doi: 10.1039/C2SM25402F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozma D., Simon I., Tusnady G.E. PDBTM: Protein Data Bank of transmembrane proteins after 8 years. Nucleic Acids Res. 2013;41:D524–529. doi: 10.1093/nar/gks1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan S., Tan H.T., Chung M.C. Membrane proteins and membrane proteomics. Proteomics. 2008;8:3924–3932. doi: 10.1002/pmic.200800597. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins A.L., Groom C.R. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 5.Wallin E., von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arora A., Tamm L.K. Biophysical approaches to membrane protein structure determination. Curr Opin Struct Biol. 2001;11:540–547. doi: 10.1016/s0959-440x(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 7.Das S., Hahn Y., Walker D.A., et al. Topology of NGEP, a prostate-specific cell:cell junction protein widely expressed in many cancers of different grade level. Cancer Res. 2008;68:6306–6312. doi: 10.1158/0008-5472.CAN-08-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., He Z., Zhang C., et al. Transmembrane protein alignment and fold recognition based on predicted topology. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0069744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 10.Jones D.T., Taylor W.R., Thornton J.M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 11.Krogh A., Larsson B., von Heijne G., et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 12.Kall L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Martelli P.L., Fariselli P., Casadio R. An ENSEMBLE machine learning approach for the prediction of all-alpha membrane proteins. Bioinformatics. 2003;19(Suppl 1):i205–211. doi: 10.1093/bioinformatics/btg1027. [DOI] [PubMed] [Google Scholar]
- 14.Tamposis I.A., Sarantopoulou D., Theodoropoulou M.C., et al. Hidden neural networks for transmembrane protein topology prediction. Comput Struct Biotechnol J. 2021;19:6090–6097. doi: 10.1016/j.csbj.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nugent T., Jones D.T. Transmembrane protein topology prediction using support vector machines. BMC Bioinf. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 17.Rost B., Fariselli P., Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernhofer M., Kloppmann E., Reeb J., et al. TMSEG: Novel prediction of transmembrane helices. Proteins. 2016;84:1706–1716. doi: 10.1002/prot.25155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H., Wang K., Lu L., et al. Deep conditional random field approach to transmembrane topology prediction and application to GPCR three-dimensional structure modeling. IEEE/ACM Trans Comput Biol Bioinform. 2017;14:1106–1114. doi: 10.1109/TCBB.2016.2602872. [DOI] [PubMed] [Google Scholar]
- 20.Lu W, Fu B, Wu H et al. CRF-TM: A conditional random field method for predicting transmembrane topology. Cham, 2015, p. 529-537. Springer International Publishing.
- 21.Shen H., Chou J.J. MemBrain: improving the accuracy of predicting transmembrane helices. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernsel A., Viklund H., Hennerdal A., et al. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsirigos K.D., Peters C., Shu N., et al. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43:W401–407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobson L., Remenyi I., Tusnady G.E. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408–412. doi: 10.1093/nar/gkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Zhang C., Shi X., et al. Improving transmembrane protein consensus topology prediction using inter-helical interaction. Biochim Biophys Acta. 2012;1818:2679–2686. doi: 10.1016/j.bbamem.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 26.Elazar A., Weinstein J.J., Prilusky J., et al. Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology. Proc Natl Acad Sci U S A. 2016;113:10340–10345. doi: 10.1073/pnas.1605888113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Q., Chen W.Y., Huang S.Q., et al. Deep learning for mining protein data. Briefings Bioinf. 2021;22:194–218. doi: 10.1093/bib/bbz156. [DOI] [PubMed] [Google Scholar]
- 28.Min S., Lee B., Yoon S. Deep learning in bioinformatics. Briefings Bioinf. 2017;18:851–869. doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- 29.Feng S.H., Zhang W.X., Yang J., et al. Topology prediction improvement of alpha-helical transmembrane proteins through helix-tail modeling and multiscale deep learning fusion. J Mol Biol. 2020;432:1279–1296. doi: 10.1016/j.jmb.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Wang H., Yang Y., Yu J., et al. 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2019. DMCTOP: topology prediction of alpha-helical transmembrane protein based on deep multi-scale convolutional neural network; pp. 36–43. [DOI] [PubMed] [Google Scholar]
- 31.Rost B., Casadio R., Fariselli P. Refining neural network predictions for helical transmembrane proteins by dynamic programming. Proc Int Conf Intell Syst Mol Biol. 1996;4:192–200. [PubMed] [Google Scholar]
- 32.Rost B., Casadio R., Fariselli P., et al. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang F.Z., Qi Z.Y., Duan K.Y., et al. A comprehensive survey on transfer learning. Proc IEEE. 2021;109:43–76. [Google Scholar]
- 34.Devlin J., Chang M.-W., Lee K., et al. Proceedings of the 2019 conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) 2019. Pre-training of deep bidirectional transformers for language understanding; pp. 4171–4186. [Google Scholar]
- 35.Bepler T., Berger B. International conference on learning representations. 2018. Learning protein sequence embeddings using information from structure. [Google Scholar]
- 36.Rives A., Meier J., Sercu T., et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. PNAS. 2021;118 doi: 10.1073/pnas.2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomize M.A., Lomize A.L., Pogozheva I.D., et al. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 38.Fu L., Niu B., Zhu Z., et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 40.Kall L., Krogh A., Sonnhammer E.L. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics. 2005;21(Suppl 1):i251–257. doi: 10.1093/bioinformatics/bti1014. [DOI] [PubMed] [Google Scholar]
- 41.Altschul S.F., Madden T.L., Schaffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remmert M., Biegert A., Hauser A., et al. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 43.Pundir S, Magrane M, Martin MJ et al. Searching and navigating UniProt databases, Curr Protoc Bioinformatics 2015;50:1 27 21-10. [DOI] [PMC free article] [PubMed]
- 44.Lafferty JD, McCallum A, Pereira FCN. Conditional random fields: probabilistic models for segmenting and labeling sequence data. International conference on machine learning. 2001, 282-289.
- 45.Zhou P., Shi W., Tian J., et al. Proceedings of the 54th annual meeting of the association for computational linguistics (volume 2: Short papers) 2016. Attention-based bidirectional long short-term memory networks for relation classification; pp. 207–212. [Google Scholar]
- 46.Müller R.R., Kornblith S., Hinton G. When does label smoothing help. Neural Inf Process Systems. 2019:4694–4703. [Google Scholar]
- 47.Teufel F., Almagro Armenteros J.J., Johansen A.R., et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022 doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.