Abstract
The actinobacterium Rhodococcus jostii RHA1 grows on a remarkable variety of aromatic compounds and has been studied for applications ranging from the degradation of polychlorinated biphenyls to the valorization of lignin, an underutilized component of biomass. In RHA1, the catabolism of two classes of lignin-derived compounds, alkylphenols and alkylguaiacols, involves a phylogenetically distinct extradiol dioxygenase, AphC, previously misannotated as BphC, an enzyme involved in biphenyl catabolism. To better understand the role of AphC in RHA1 catabolism, we first showed that purified AphC had highest apparent specificity for 4-propylcatechol (kcat/KM ∼106 M−1 s−1), and its apparent specificity for 4-alkylated substrates followed the trend for alkylguaiacols: propyl > ethyl > methyl > phenyl > unsubstituted. We also show AphC only poorly cleaved 3-phenylcatechol, the preferred substrate of BphC. Moreover, AphC and BphC cleaved 3-phenylcatechol and 4-phenylcatechol with different regiospecificities, likely due to the substrates’ binding mode. A crystallographic structure of the AphC·4-ethylcatechol binary complex to 1.59 Å resolution revealed that the catechol is bound to the active site iron in a bidentate manner and that the substrate’s alkyl side chain is accommodated by a hydrophobic pocket. Finally, we show RHA1 grows on a mixture of 4-ethylguaiacol and guaiacol, simultaneously catabolizing these substrates through meta-cleavage and ortho-cleavage pathways, respectively, suggesting that the specificity of AphC helps to prevent the routing of catechol through the Aph pathway. Overall, this study contributes to our understanding of the bacterial catabolism of aromatic compounds derived from lignin, and the determinants of specificity in extradiol dioxygenases.
Keywords: bacterial metabolism, actinobacteria, metalloenzyme, biotechnology, enzyme structure, lignin valorization, biocatalysis, guaiacol
Abbreviations: 4EC, 4-ethylcatechol; 6-phenyl HODA, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate; CatA, catechol 1,2-dioxygenase; MCP, meta-cleavage product; PCBs, polychlorinated biphenyls; RCF, reductive catalytic fractionation
The bacterial catabolism of aromatic compounds has long been studied for its potential in the bioremediation of pollutants such as polychlorinated biphenyls (PCBs) and, more recently, the valorization of lignocellulosic biomass, a critical feedstock for a bio-based economy. Studies in phylogenetically diverse bacteria that aerobically catabolize aromatic compounds, such as Pseudomonas and Rhodococcus, have revealed that the relevant pathways are organized in a modular, convergent fashion (1). More specifically, growth substrates are initially transformed via upper pathways to a limited number of shared intermediates (2). The latter, typically catechols including protocatechuate, are then transformed to central metabolites via lower pathways (2). The catecholic intermediates are subject to either meta-cleavage or ortho-cleavage, catalyzed by extradiol and intradiol dioxygenases, respectively. The convergent nature of bacterial aromatic catabolism provides an attractive approach for converting lignin depolymerization products, which typically occur as mixtures, to useful chemicals via biological funneling (3, 4). However, important aspects of aromatic catabolism have yet to be elucidated to realize this biocatalytic potential.
Rhodococcus jostii RHA1 is a model organism for a variety of physiological processes including the catabolism of aromatic compounds and steroids, the latter of which are also catabolized via a catecholic intermediate (5, 6, 7). RHA1 was isolated from lindane-contaminated soil for its remarkable ability to aerobically degrade PCBs (8). In bacteria such as RHA1, PCBs are cometabolized by the four-enzyme Bph pathway, which catabolizes biphenyl to benzoate and 2-hydroxypenta-2,4-dienoate (9). The potent PCB-degrading activity of RHA1 is due in part to its harboring multiple homologs of the Bph enzymes (8), including up to six homologs of BphC (10). BphC (EC 1.13.11.39) is the third enzyme of the Bph pathway, an extradiol dioxygenase that catalyzes the 2,3-cleavage of 3-phenylcatechol (also named 2,3-dihydroxybiphenyl) to 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (6-phenyl HODA), a meta-cleavage product (MCP). Although RHA1 contains 13 extradiol dioxygenases (11), the bacterium funnels many aromatic compounds to central metabolism via the β-ketoadipate pathway (2), an ortho-cleavage pathway. Among lignin-derived compounds, these include vanillate and guaiacol, which are cleaved by protocatechuate 3,4-dioxygenase and catechol 1,2-dioxygenase (CatA), respectively (12, 13), in the two convergent branches of the pathway.
RHA1 was recently shown to grow on two classes of compounds produced by the reductive catalytic fractionation (RCF) of lignin: alkylphenols and alkylguaiacols (13, 14). RCF has taken on new importance as the basis of a sustainable biorefining strategy to convert biomass (15). In this context, alkylphenol and alkylguaiacol catabolism are of interest to the development of microbial cell factories to valorize RCF lignin. In RHA1, alkylphenols are hydroxylated by AphAB, a two-component flavin-dependent monooxygenase, while alkylguaiacols are O-demethylated by AgcAB, a cytochrome P450 system (13, 14). In both cases, the products are 4-alkylcatechols, which are subject to meta-cleavage by AphC (Fig. 1), an extradiol dioxygenase that had initially been identified as BphC (10, 11) and for which a crystal structure has been deposited (PBD accession number 3LM4). AphC and BphC, of which the best characterized homolog is BphCLB400 from Paraburkholderia xenovorans LB400, are type I extradiol dioxygenases belonging to the VOC superfamily and utilize Fe2+ coordinated by a 2-His 1-carboxylate facial triad to catalyze catechol cleavage (16). Although RHA1 catabolizes 4-alkylcatechols via a meta-cleavage pathway and catechol via an ortho-cleavage pathway (13), it is unknown whether RHA1 utilizes the two pathways simultaneously. Indeed, the misrouting of catechols down the wrong pathway may be metabolically inefficient. For example, the ortho-cleavage of 4-methylcatechol leads to the formation of 4-methylmuconolactone, a dead-end metabolite in some bacteria (17).
Figure 1.
AphC reaction. AphC catalyzes the 2,3-cleavage of 4-alkylcatechols to 2-hydroxy-6-oxo-5-alkylhexa-2,4-dienoates (5-alkyl HODAs). 4-Alkylcatechols are intermediates in the catabolism of 4-alkylguaiacols (upper branch) and 4-alkylphenols (lower branch), generated in the reductive catalytic fractionation of lignin.
Herein, we studied AphC from RHA1 with the objectives of understanding the molecular basis of its activity and its contribution to the metabolism of aromatic compounds by RHA1. We evaluated the enzyme’s specificity (kcat/KM) for 4-substituted catechols and other substrates. To obtain insight into this specificity, we determined a crystal structure of AphC in complex with 4-ethylcatechol (4EC). Further insight was obtained by comparing the regiospecificities of AphC and BphCLB400 for 3-phenylcatechol and 4-phenylcatechol. Finally, we studied the ability of RHA1 to grow on a mixture of 4-ethylguaiacol and guaiacol, catabolized by meta-cleavage and ortho-cleavage pathways, respectively. Overall, the results provide insights into the molecular basis of specificity in extradiol dioxygenases and contribute to our understanding of the bacterial catabolism of aromatic compounds.
Results
Phylogenetic analysis
To better understand the relationship between AphC and its orthologs, we aligned the amino acid sequences of representative type I extradiol dioxygenases and analyzed their phylogeny. Conserved residues include His155, His217, and Glu268 (AphC numbering) that ligate the catalytically essential metal ion, as well as His200, His249, and Tyr258. In homologous enzymes, His249 and Tyr258 are proposed to deprotonate one of the hydroxyls of the Fe2+-bound catechol, and His200 is proposed to protonate an Fe2+-bound alkylperoxo intermediate (16, 18). The phylogenetic analysis (Fig. S1) revealed that AphC is most similar to PheC (49% amino acid sequence identity), an uncharacterized catechol 2,3-dioxygenase that occurs in Chloroflexi, an ancient phylum of bacteria. By contrast, AphC shares 25% amino acid sequence identity with XylE, a prototypical catechol 2,3-dioxygenase, and just over 15% identity with BphCLB400, a prototypical BphC-type enzyme. The phylogenetic relationship of AphC to PheC, together with the enzyme’s involvement in catabolizing 4-alkylcatechols (13) prompted us to characterize the enzyme further.
Preparation of AphC
To further characterize AphC, we heterologously produced the enzyme as a His-tagged protein in Escherichia coli. We purified AphC to >95% apparent homogeneity (Fig. S2) using immobilized metal affinity chromatography and proteolytically removed the affinity tag. This yielded ∼20 mg of purified AphC per liter of cell culture. Anaerobic reconstitution with Fe2+ followed by gel filtration yielded preparations of AphC containing ∼2 equivalents of iron. Nonreconstituted preparations contained similar amounts of iron, suggesting adventitious binding. In subsequent studies, the concentration of AphC was based on the concentration of protein determined by the BCA method.
The catechol-cleaving activities of AphC
To assess the substrate preference of AphC, we measured the activity of the enzyme using a range of catechols and an oxygraph assay (Table 1). Although AphC catalyzed the cleavage of a variety of catechols, two trends were apparent; it had significantly higher specific activities for alkylcatechols versus halocatechols, and it had higher activities for 4-substituted catechols than 3-substituted ones. This was perhaps most striking for the phenylcatechols. Indeed, AphC did not detectably consume O2 in the presence of 3-phenylcatechol despite the enzyme having been previously annotated as BphC (10, 11). To further assess the ability of AphC to cleave 3-phenylcatechol, we used a spectrophotometric assay to monitor the formation of the MCP at 428 nm. While some formation of an MCP was detected, the absorbance decayed rapidly (Fig. S3), complicating further studies. This is in contrast to 6-phenyl HODA, the MCP generated by the BphCLB400-catalyzed cleavage of 3-phenylcatechol, which has a half-life of ∼58 h under similar conditions (19). The AphC-catalyzed cleavage of 4-phenylcatechol, predicted to yield 5-phenyl HODA based on the Aph pathway (14), was also relatively stable. This MCP absorbed maximally at 383 nm, unlike 6-phenyl HODA, which absorbs maximally at 434 nm (19). Indeed, reaction of the MCP with ammonium yielded a stable pyridine derivative (20) whose NMR spectrum (1H NMR (D2O, 850 MHz): δ 8.47 (d, 1H, Ar-H), δ 8.23 (d, 1H, Ar-H), δ 8.05 (d, 1H, Ar-H), δ 7.52 to 7.78 (m, 5H, Ar-H) and mass spectrum (m/z = 198.05) are consistent with the MCP being 5-phenyl HODA. Finally, based on monitoring phenylcatechol depletion using HPLC, we estimated that the specific activity of AphC for 4-phenylcatechol is 30 × higher than for 3-phenylcatechol.
Table 1.
Specific activity of AphC with different catecholsa
| Substrate | Specific activity U/mg AphC |
|---|---|
| 4-propylcatechol | 240 (20) |
| 4-ethylcatechol | 290 (20) |
| 4-methylcatechol | 420 (20) |
| 4-phenylcatechol | 200 (10) |
| Catechol | 120 (20) |
| 3-phenylcatechol | 7 (5) |
| 3-methylcatechol | 120 (20) |
| 3-bromocatechol | 30 (10) |
| 3-fluorocatechol | 12 (5) |
| 4-chlorocatechol | 35 (7) |
The activity of AphC was measured using an oxygraph and 200 μM of each catechol (air-saturated Hepes (I = 0.1 M), pH 8, 25 °C) except for 3-phenylcatechol, for which specific activity was determined by monitoring substrate depletion using HPLC. The values represent the mean and standard deviations of three technical replicates.
To better characterize the MCPs resulting from the AphC-catalyzed cleavage of phenylcatechols, we compared them with those generated using BphCLB400. Incubation of 3-phenylcatechol and 4-phenylcatechol with each of the two enzymes yielded species with m/z values of 217.05 (Fig. S4), consistent with them all being phenyl HODAs. However, the four MCPs had different retention times (tR) on the HPLC column, suggesting that each arose from a different mode of cleavage. More specifically, the BphCLB400-catalyzed cleavage of 3-phenylcatechol (tR = 8.6 min) yields 6-phenyl HODA (21), and the cleavage of 4-phenylcatechol (tR = 8.1 min) is deduced to be 4-phenyl HODA. As noted above, the AphC-catalyzed cleavage of 4-phenylcatechol (tR = 7.4 min) yields 5-phenyl HODA. The MCP resulting from the AphC-catalyzed cleavage of 3-phenylcatechol (tR = 6.4 min) was obtained in too low yield and was too labile (Fig. S3) for NMR analysis. However, it is deduced to be 3-phenyl HODA. The structures of the four phenyl HODA products are shown in Fig. S5.
Substrate specificity
To establish AphC’s substrate specificity (kcat/KM) for select catechols, we used a spectrophotometric assay with the wavelengths and extinction coefficients listed in Table S1. In all cases, the dependence of the initial reaction rate on substrate concentration obeyed Michaelis–Menten kinetics (Fig. S6). As summarized in Table 2, AphC exhibited the highest apparent specificity for 4-propylcatechol (20 mM Hepes, I = 0.1 M, pH 8 at 25 °C) and cleaved 4-alkylcatechols in the following order of apparent specificity: propyl > ethyl > methyl > phenyl > no substituent. This is consistent with the trend determined apparent specificity of AgcAB for alkylguaiacols (13). AphC also displayed Michaelis–Menten behavior with respect to O2 concentration in the presence of saturating amounts of 4-propylcatechol (Fig. S6). Under these conditions, the enzyme’s KM O2 value was 70 ± 10 μM.
Table 2.
Steady-state kinetic parameters of AphCa
| Substrate |
kcat |
KM |
kcat/KM |
|---|---|---|---|
| s−1 | μM | mM−1 s−1 | |
| 4-propylcatechol | 0.205 (0.006) | 0.22 (0.02) | 920 (80) |
| 4-ethylcatechol | 0.246 (0.004) | 1.19 (0.06) | 206 (8) |
| 4-methylcatechol | 0.293 (0.009) | 2.8 (0.3) | 103 (9) |
| 4-phenylcatechol | 0.344 (0.005) | 7.4 (0.3) | 47 (1) |
| Catechol | 0.068 (0.002) | 47 (4) | 1.5 (0.1) |
| Oxygenb | 0.26 (0.02) | 70 (10) | 3.8 (0.5) |
Experiments were performed using air-saturated Hepes (I = 0.1 M), pH 8, at 25 °C and the spectrophotometric assay unless otherwise noted. Because the conditions were not saturating with respect to O2 concentration, the parameters are apparent. Parameters were calculated using a minimum of 15 data points at various substrate concentrations. Standard deviation is shown in parentheses.
Parameters evaluated using the oxygraph assay and 10 μM 4-propylcatechol.
The crystallographic structure of AphC
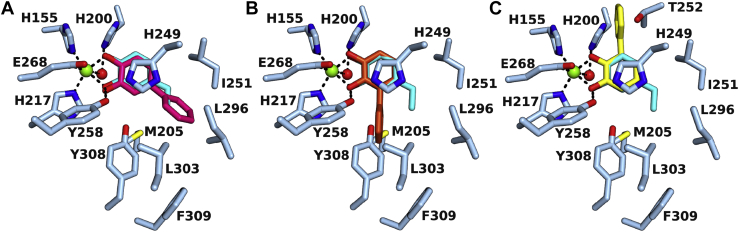
The crystal structure of AphC in complex with the substrate 4EC was solved by molecular replacement using the previously deposited crystal structure of AphC from a different crystal form, prepared aerobically and missing the last 30 amino acids (PBD ID 3LM4, (22)). There is no accompanying publication for this structure yet, and it is annotated as a catechol 2,3-dioxygenase, like BphC. In our anaerobic structure, 4EC is bound to the tetrameric AphC enzyme (Figs. 2A and S7). The iron is coordinated by His155, His217, and Glu268, consistent with the 2-His 1-carboxylate facial triad in homologous structures. In the complex, iron is six-coordinate in a distorted square bipyramidal geometry: the two oxygens from bidentate-bound 4EC and the Nε2 atoms of His155 and His217 are in the plane, while the Glu268 carboxylate and a water molecule occupy the axial positions (Fig. 2B). The 4EC hydroxyl-iron bond lengths are consistent in all protomers, with average distances of Fe-O1 ∼1.9 Å and Fe-O2 ∼2.2 Å. During catalysis, 4EC O2 is deprotonated, which is expected to shorten the Fe-O bond length. Indeed, several other published extradiol enzyme structures have an equivalent Fe-O2 bond length approximately 0.4 Å shorter than Fe-O1, consistent with this deprotonation. In BphCLB400·3-phenylcatechol, the equivalent Fe-O1 and Fe-O2 bond lengths are 2.4 Å and 2.0 Å, respectively (23). It is unclear why this is not the case in the AphC·4EC structure, but for this reason, 4EC was refined in its fully protonated form. His249 and Tyr258 are the conserved catalytic residues responsible for catechol deprotonation in homologous enzymes. In the structure, Tyr258 forms a hydrogen bond with the 4EC O2 hydroxyl group and His249 forms a stacking interaction with the catechol ring as well as a hydrogen bond with Tyr258. The other conserved catalytic residue, His200 (His195 in BphCLB400), forms a hydrogen bond with the iron coordinating water molecule. In the proposed mechanism, molecular oxygen replaces the water molecule at this site, and His200 protonates the alkylperoxo intermediate within the catalytic reaction (18). The 4-ethyl moiety from 4EC binds in a hydrophobic space defined by residues Trp192, Met205, Ile251, Leu296, Leu303, and Phe309, which can also accommodate larger side chains such as propyl and even phenyl groups (Figs. 2, B and C, 3).
Figure 2.
Structure of AphC active site and comparison of substrate-binding pocket with BphC and DoxG.A, AphC tetrameric assembly. One protomer is shown in cartoon presentation (blue) and the others are shown as surfaces. B, stereo view of the interactions of AphC active site residues (blue) with its substrate 4-ethylcatechol (cyan). The iron ion is shown in green with iron interactions indicated by dashed lines. Oxygen, nitrogen and sulfur atoms are shown in red, blue and yellow, respectively. C, substrate-binding cavity of AphC with 4-ethylcatechol. D, substrate-binding cavity of BphCLB400 with 3-phenylcatechol, shown in red (23) (PDB ID 1KMY). E, substrate-binding cavity of DoxG with 4-phenylcatechol, shown in red (PDB ID 2EI0).
Figure 3.
Models for 4-phenylcatechol and 3-phenylcatechol binding to AphC.A, modeling of 4-phenylcatechol (fuchsia) into the active site of the AphC·4-ethylcatechol (cyan) structure shows the phenyl ring is accommodated by the hydrophobic pocket contiguous to the catechol-binding site. Hydrogen bonding with 4-ethylcatechol and iron ligands are indicated by dashed lines. B, modeling of 3-phenylcatechol (orange) into the active site of the AphC·4-ethylcatechol (cyan) structure in an orientation as seen in the BphCLB400 crystal structure (PDB ID 1KMY). In this binding mode, the phenyl ring sterically clashes with AphC residues Met205 and Tyr308. C, an alternative binding mode with flipped 3-phenylcatechol (yellow) results in phenyl ring clashes with His200 and Thr252.
The structural basis of the substrate specificity and regiospecificity of AphC
To better understand the structural determinants of substrate specificity in AphC, the AphC·4EC structure was superimposed on those of BphCLB400·3-phenylcatechol (PBD ID 1KMY) and DoxG·4-phenylcatechol (PDB ID 2EI0). DoxG is a 1,2-dihydroxynaphthalene dioxygenase that efficiently cleaves 4-phenylcatechol (24). Unlike the tetrameric structure of AphC, BphCLB400 and DoxG crystallized as octamers. However, the tertiary structure of the three enzymes are highly similar (Fig. S8), with r.m.s.d values of 1.8 Å between αC atoms of AphC and BphC and 1.9 Å for AphC and DoxG. Comparison of the active sites of three enzymes revealed that the catecholic ring binds in the same bidentate configuration and that the architecture of the catalytic residues is very similar (Figs. 2, C–E and S8). By contrast, the proximal regions of the substrate-binding pocket differ in a manner consistent with the substrate specificities of the enzymes. In AphC, the hydrophobic binding pocket extends from the catechol C4-position away from the iron site, while in BphCLB400, the hydrophobic binding pocket extends away from the catechol C3 position, rotated by approximately 60° relative to AphC (Fig. 2, C and D). The deposited structure of the DoxG·4–phenylcatechol complex displays a more open proximal binding pocket consistent with the enzyme’s preference of 1,2-dihydroxynaphthalene, although it accommodates 4-phenylcatechol in an analogous position to AphC (Fig. 2E).
To better understand the basis of regiospecificity in AphC, 4-phenylcatechol and 3-phenylcatechol were modeled into the active site of AphC. In agreement with the enzyme’s substrate specificity, AphC easily accommodated 4-phenylcatechol with the phenyl group directed into the same hydrophobic pocket that supports the ethyl moiety of 4EC (Fig. 3A). This binding mode directs 2,3-cleavage to generate 5-phenyl HODA (Fig. S5). Steric hindrance between the phenyl group and Ile251 precludes 4-phenylcatechol binding with the catechol flipped 180°, swapping the positions of the C1 and C2 hydroxyls that would favor 1,6-cleavage. While AphC is able to cleave 3-phenylcatechols with much lower specificity to generate 3-phenyl HODA, steric clashes prohibit its modeling in either orientation. Docking in the same orientation as in BphC results in a clash with Met205 and Tyr308 (Fig. 3B). Docking in the flipped binding mode directs the phenyl moiety into a region occupied by His200 and Thr252 (Fig. 3C). In order to accommodate binding of 3-phenylcatechol, a conformational change is required.
Catabolism of 4-ethylguaiacol and guaiacol by RHA1
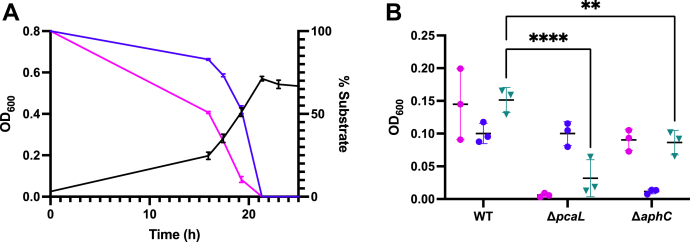
To investigate whether RHA1 efficiently utilizes meta-cleavage and ortho-cleavage pathways simultaneously, we grew the strain in a mixture of 0.5 mM guaiacol and 0.5 mM 4-ethylguaiacol. RHA1’s growth on this mixture was monophasic (Fig. 4A), and the culture reached the same final OD600 as cultures grown on 1 mM of either compound alone. Analysis of the culture supernatants further indicated that the two aromatic compounds were utilized concurrently throughout growth on the mixture, although guaiacol was depleted faster than 4-ethylguaiacol. To determine the pathways by which these substrates are catabolized, the growth of ΔaphC and ΔpcaL mutants, containing lesions in the meta-cleavage and ortho-cleavage pathways, respectively, was investigated on guaiacol, 4-ethylguaiacol, and a mixture of both. More specifically, PcaL functions as a β-ketoadipate enol-lactone hydrolase and γ-carboxymuconolactone decarboxylase in the β-ketoadipate pathway of RHA1 (2). The ΔaphC strain did not grow on 4-ethylguaiacol but grew on guaiacol (Fig. 4B). Conversely, the ΔpcaL strain grew on 4-ethylguaiacol but not guaiacol. Both mutants grew to less than 60% the culture density of WT RHA1 when grown on a mixture of 0.5 mM guaiacol and 0.5 mM 4-ethylguaiacol.
Figure 4.
Growth of WT and mutant RHA1 strains on guaiacols.A, growth curve of WT RHA1 (black) on a mixture of 4-ethylguaiacol (depletion shown in blue) and guaiacol (depletion shown in violet). Cells were grown on 0.5 mM of each substrate in M9 medium supplemented with Goodies (30 °C). Data represent the average and standard deviation of three biological replicates. B, growth yield of WT and mutant RHA1 strains on guaiacols. Strains were grown on either 1 mM guaiacol (violet), 1 mM 4-ethylguaiacol (blue), or a mixture of 0.5 mM guaiacol and 0.5 mM ethyl-guaiacol (teal) in M9 media supplemented with Goodies. Optical density was measured at 60 h postinoculation. Data represent the average and standard deviation of three biological replicates ∗∗∗∗p < 0.0001, ∗∗p < 0.01.
To further verify the activity of the pathways during growth, we measured the specific activities of the ortho-cleavage of catechol and the meta-cleavage of 4EC in cell lysates of RHA1 cells by spectrophotometrically measuring the production of muconate and 5-ethyl HODA, respectively (Table 3). In cells growing on guaiacol, the catechol ortho-cleavage activity, taken to represent CatA (2), was ∼40-fold higher than in cells growing on 4-ethylguaiacol. By contrast, the 4EC meta-cleavage was 85-fold higher in cells growing on 4-ethylguaiacol. This indicates that the β-ketoadipate and Aph pathways are specifically induced by substrates that are catabolized via these pathways. Importantly, both the ortho-cleavage and meta-cleavage activities were induced in cells growing on the mixture of guaiacol and 4-ethylguaiacol, albeit to a slightly lesser extent. Overall, these results demonstrate that RHA1 can simultaneously catabolize relatively similar substrates via ortho-cleavage and meta-cleavage pathways.
Table 3.
Specific activities of cell lysates from RHA1 growing on guaiacol and 4-ethylguaiacola
| Growth substrate | Assay substrate |
|
|---|---|---|
| Catechol U/mg | 4-Ethylcatechol U/mg | |
| Guaiacol | 19 (1) | 0.048 (0.006) |
| 4-Ethylguaiacol | 0.5 (0.1) | 4.1 (0.6) |
| Mixture | 15.0 (0.4) | 1.7 (0.4) |
Cells were grown on 1 mM of the indicated substrate or a mixture of the two and collected in the mid-log growth phase (OD600 ∼ 0.3). The ortho-cleavage of catechol and the meta-cleavage of 4-ethylcatechol were determined by spectrophotometrically following the production of muconate and 5-ethyl HODA, respectively. Data are averages from triplicate experiments with standard deviation shown in parentheses.
Discussion
This study establishes that AphC has a strong substrate specificity for 4-alkylcatechols, cleaving 4-propylcatechol with an apparent kcat/KM approximately three orders of magnitude higher than catechol. This specificity is consistent with the enzyme’s role in the catabolism of 4-alkylphenols and 4-alkylguaiacols by R. jostii RHA1 (13, 14). A crystal structure of AphC in complex with the substrate 4EC revealed that the enzyme’s catalytic machinery is essentially identical to that of other type I extradiol dioxygenases and that the alkyl substituent is accommodated by a large hydrophobic pocket that is contiguous with the catechol binding site. The catechol binding site is similar between AphC and BphCLB400, whose best substrate is 3-phenylcatechol, but the proximal pocket is extended either out from the catechol -C4 or -C3 carbon, respectively. The orientation of this proximal hydrophobic pocket appears to be a major determinant of substrate specificity.
The substrate specificity of AphC is unique among characterized extradiol dioxygenases (Table S2). For example, XylE (25) and AtdB (26), enzymes whose physiological role is also to cleave alkylcatechols, have highest specificity for unsubstituted catechol. Interestingly, AphC is as phylogenetically distant from these two enzymes as it is to BphC (Fig. S1) (27). Notably, the apparent specificities (kcat/KM) of these enzymes for their preferred substrates are one to two orders of magnitude higher than that of AphC for 4-propylcatechol. LapB, involved in the catabolism of 4-n-alkylphenols (C1–C5), has similar kinetic parameters for 4-methylcatechol, a physiological substrate, as those of AphC for 4-alkylcatechols (28). However, this enzyme is also phylogenetically distant from AphC, suggesting that the similar specificities of these enzyme may have arisen independently. Interestingly, AphC is closely related to a family of predicted catechol 2,3-dioxygenases that occur in Chloroflexi, exemplified in Fig. S1 by the sequence identified as PheCCa from Chloroflexus auranticus.
The basis for the different regiospecificities of AphC and BphC for 3-phenylcatechol and 4-phenylcatechol is unclear. In principle, the different regiospecificity of these enzymes could arise in one of two ways. In the case of 3-phenylcatechol, AphC would either have to accommodate the substrate in a flipped orientation versus BphC or direct O2 attack to C1 of the catecholic substrate instead of C2, as proposed for BphC (23). The architecture of the catalytic machinery is highly conserved in these homologs. The AphC iron ion, two His-carboxylate triad, His200, His247, and Tyr258 overlay with r.m.s.d values of 0.54 Å and 0.78 Å over all heavy atoms to BphC and DoxG, respectively. Given the conserved orientation of catalytic residues between AphC, BphC, DoxG and indeed other well-characterized enzymes such as 2,3-HPCD (18), particularly His249 and Tyr258 involved in catechol deprotonation and His200 involved in protonating the activated oxygen intermediate, the mode of O2 attack would almost certainly be conserved. This suggests that the differences in regiospecificity are due to 3-phenylcatechol binding in a flipped orientation. Our substrate modeling studies indicate that the binding of 3-phenylcatechol to AphC result in steric clashes regardless of its orientation, suggesting that some structural change occurs for activity. The low specific activity of AphC for 3-phenylcatechol is consistent with the poor fit of the substrate to the enzyme’s binding site. DoxG has an ∼260-fold higher specificity constant for 4-phenylcatechol over 3-phenylcatechol, and a much lower Km for 4-phenylcatechol (24). The DoxG substrate binding pocket is larger than that of AphC and BphCLB400, extending open from both the catechol C3 and C4 positions, so the preference for 4-phenylcatechol may be driven by favorable stacking of the phenyl against Tyr178 or other interactions in the hydrophobic pocket (Fig. 2E).
The presented data establish that RHA1 simultaneously catabolizes similar aromatic compounds by ortho-cleavage and meta-cleavage pathways, respectively. Thus, RHA1’s growth on a mixture of 4-ethylguaiacol and guaiacol was monophasic, and the two substrates were depleted with similar kinetics during this growth. Moreover, both pathways were required for growth on the mixture, and both intradiol and extradiol dioxygenases were present under these conditions. This simultaneous growth is somewhat unexpected given the potential of forming dead end metabolites such as 4-methylmuconolactone if 4-alkylcatechols are subject to ortho-cleavage (17). Our data suggest that the respective specificities of AphC and CatA play a major role in ensuring that the catechols are correctly routed. In this respect, it is noted that the relative specificity of AphC for 4EC over catechol (∼140) is less than that of BphCLB400 for 3-phenylcatechol over catechol (∼350 (27)) which are routed through meta-cleavage and ortho-cleavage pathways, respectively, in P. xenovorans LB400 and other biphenyl-degrading bacteria. However, it is possible that compartmentalization plays a role in correctly routing the catechols.
In conclusion, this study establishes the substrate specificity of a phylogenetically distinct alkylcatechol-cleaving extradiol dioxygenase and identifies a major structural determinant of that specificity. In addition, the enzyme’s specificity appears to play a major role in the correct routing of aromatic compounds in Rhodococcus.
Experimental procedures
Chemicals and reagents
All reagents were of analytical grade unless otherwise noted. Catechols were purchased from Sigma-Aldrich except for 3-bromocatechol (Alfa Aesar) and 4-propylcatechol, which was synthesized as described previously (13). Water for buffers was purified using a Barnstead Nanopure Diamond system to a resistivity of at least 18 MΩ.
Phylogenetic analysis
Amino acid sequences were aligned using MUSCLE (29). The phylogenetic tree was generated using the Maximum Likelihood method in MEGA X software (30) computing distances based on the JTT matrix (31), with 1000 bootstraps iterations.
Protein production and purification
AphC was produced heterologously as an N-terminal polyHis-tagged (Ht-) protein in E. coli BL-21 λ (DE3) containing pETAphC (13). Freshly transformed cells were grown at 37 °C at 200 rpm in LB supplemented with 50 μg ml−1 of kanamycin until the culture reached an optical density at 600 nm (OD600) ∼0.6. Expression of AphC was induced with 0.5 mM isopropyl β-D-thiogalactopyranoside, and the cells were incubated at 30 °C for an additional 16 h. Cells were harvested by centrifugation. Cells collected from 1 L of culture were suspended in 20 ml of 20 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 8 and lysed at 4 °C using an EmulsiFlex-C5 homogenizer (Avestin). Cellular debris was removed by centrifugation. Ht-AphC was purified from the cell extract using immobilized metal affinity chromatography (HisPur Ni-NTA Resin, ThermoFisher) according to the manufacturer’s protocol. The eluted Ht-AphC was exchanged into 20 mM Tris, 1 mM DTT, pH 8.0 using a 30 kDa centrifugal ultrafiltration device (Millipore). The affinity tag was removed by incubating the preparation with TEV protease (50:1 M ratio) for 2 h at room temperature followed by 16 h at 4 °C. The protease and uncleaved Ht-AphC were removed by passing the solution over the Ni-NTA resin. AphC was concentrated to ∼25 mg ml−1. BphCLB400 was produced and purified as described in Vaillancourt et al (27).
Anaerobic iron reconstitution and protein handling
AphC solution was briefly sparged with argon and brought inside a Labmaster Model 100 glovebox (Mbraun). The deoxygenated AphC was incubated with 3-fold molar excess of (NH4)2Fe(SO4)2·6H2O for ∼30 min at room temperature. The excess metal ion was removed by running the preparation over a 2.0 × 10 cm column of Sephadex G-25 resin (GE Healthcare) equilibrated with 20 mM Tris, pH 8.0. The reconstituted AphC was drawn into a syringe, removed from the glovebox, flash-frozen as beads in liquid N2, and stored at −80 °C until further use. Immediately prior to use, frozen beads of protein were brought into the glovebox and allowed to thaw anaerobically. Solutions of AphC were diluted as appropriate in 20 mM Hepes (I = 0.1 M, pH 8) and transferred to a glass vial. This was sealed with a rubber stopper and removed from the glovebox. Throughout the experiments, the vial was kept on ice, and aliquots were withdrawn using a gas-tight Hamilton syringe.
Protein analytical methods
Protein purity was evaluated using SDS-polyacrylamide gel stained with Coomassie Blue according to established procedures (32). Protein concentration was determined using a micro-BCA Protein Assay Kit (Pierce) using bovine serum albumin as a standard. Iron concentrations were determined colorimetrically using a Ferene-S-based assay and ferric chloride solution as a standard (33).
Steady-state kinetics assays
Enzymatic activities were measured using three different assays: monitoring the consumption of O2 using a Clark-type polarographic O2 electrode OXYG1 (Hansatech); monitoring the production of MCP using a spectrophotometer; and monitoring catechol depletion using an HPLC. Unless otherwise noted, assays were performed using air-saturated 20 mM Hepes (I = 0.1 M, pH 8) at 25 °C, and reactions were initiated by the addition of enzyme. Stock solutions of catechols were prepared fresh daily in the assay buffer, except for 3-phenylcatechol and 4-phenylcatechol, which were prepared in ethanol. One unit of activity (U) is defined as the amount of enzyme required to transform 1 μmol of substrate to product per min at 25 °C. Steady-state kinetic parameters were evaluated by fitting the Michaelis–Menten equation to the data using the least-squares fitting of LEONORA (34).
For the oxygraph assay, the electrode was calibrated daily according to the manufacturer’s instructions using air-saturated buffer and O2-depleted buffer prepared by adding sodium hydrosulfite. Specific activity reactions contained 1.4 μM AphC and 200 μM catechol. To evaluate the steady-state kinetic parameters for O2, the reaction buffer contained 10 μM 4-propylcatechol. Dissolved O2 concentrations were varied by equilibrating the reaction buffer with a gas mixture. Briefly, ultra-high purity O2 and prepurified N2 were mixed in the desired proportions with a stainless steel Concoa Model 561 gas proportioner and humidified by bubbling it through a 5 × 10 cm column of water. The concentration of dissolved O2 in the reaction mixture was determined using the O2 electrode.
Spectrophotometric assays were performed using 1 ml cuvettes and ∼0.1 μM AphC. In evaluating the steady-state kinetic parameters, the concentration of catechol was varied between 0.3 and 300 μM, depending on the catechol. Reaction progress was monitored at the λmax of the MCP. The extinction coefficients and wavelength used are listed in Table S1. The extinction coefficients were measured by determining the amount of O2 consumed by the AphC-catalyzed cleavage of the corresponding catechol using the oxygraph. These oxygraph reactions were initiated with the addition of excess AphC. Reactions were allowed to proceed to completion, and absorption spectra were recorded using a Cary5000 spectrophotometer equipped with a thermostatted cuvette holder to determine the λmax of each MCP. The amount of O2 consumed was taken to be the amount of MCP generated and was used to calculate the extinction coefficient from the absorbance and the Beer–Lambert equation.
HPLC analysis monitoring catechol depletion was used to determine specific activities when the MCP was not stable enough to be monitored by spectrophotometry or using the oxygraph. Reactions containing 0.4 μM AphC and 200 μM catechol were quenched with 10% acetic acid after 10 min and filtered through a 0.2 μm PTFE syringe filter. The run was conducted on a Waters 2695 separation HPLC module coupled to a Waters 2996 photodiode array detector. Samples were run over a Luna 5 μm C18(2) column at 0.7 ml min−1. Samples were eluted with a 16.8 ml linear gradient of 1% methanol +0.1% formic acid in H2O to 100% methanol and monitored at 280 nm.
For the activity assays of RHA1 cells growing in different compounds, 10 μl of lysates were reacted with 400 μM 4EC or catechol in 35 mM NaPO4, I = 0.1 M, pH 7.5 at 25 °C. The production of muconate and 5-ethyl HODA was measured on a Cary 60 UV-Vis spectrophotometer equipped with a thermostatted cuvette holder, and the specific activities were calculated using the extinction coefficients and wavelength listed in Table S1. The extinction coefficient of 5-ethyl HODA in the reaction buffer was calculated as described above, and the extinction coefficient of muconate was calculated using 100 μM of commercial standard.
Crystallography
AphC was additionally purified by size-exclusion chromatography, eluted into 20 mM Tris pH 7.5, 150 mM NaCl, 3 mM β-mercaptoethanol, and concentrated to 10 mg/ml. Sitting drop crystallization trials were set up with a Mosquito crystallization robot (sptlabtech) using SWISSCI 3-lens low profile crystallization plates. AphC crystals grew in condition D10 (40% PEG 300, 0.2 M calcium acetate, and 0.1 M sodium cacodylate pH 6.5) of the JCSG-plus screen (Molecular Dimensions). Crystals were further optimized in 240-well sitting drop plates and were microseeded from initial hits using a cat whisker. At this point, crystal trays were moved into an anaerobic glovebox (Belle Technology) and allowed to equilibrate for approximately 1 week and were then soaked in anaerobically prepared crystallization solution supplemented with 1 mM 4EC for 30 min. Soaked crystals were flash-frozen directly using a liquid nitrogen port on the glovebox, thereby minimizing exposure to oxygen. Diffraction data were collected on beamline CMCF-BM and processed with XDS (35) and STARANISO (36). The structure was solved by molecular replacement within CCP4 Cloud using Molrep (37) and one protein chain of PDB entry 3LM4 as a search model, resulting in a solution with four protein chains in the asymmetric unit. The 30 C-terminal residues missing from the search model were well-resolved in the electron density maps and between the four protomers all residues, excluding Gly330-Ala333 and the terminal Ser359-Leu360 were modeled. Model building was performed in Coot (38), and the structure was refined with REFMAC5 (39). 4EC was refined at 100% occupancy in the active site, and no significant Fo-Fc density was observed. MolProbity (40) was used to evaluate the final models and PyMOL (Schrödinger, LLC) for protein model visualizations. Data and refinement statistics are summarized in Table 4. The atomic coordinates have been deposited in the Protein Data Bank and are available under the accession code 7Q2A. Search for structural protein homologs were performed with the DALI server (41). Calculation of r.m.s.d values were done using GESMAT (42) for overall protein folds and LsqKab (43) for active site residues.
Table 4.
Crystallographic data and refinement statistics for AphC in complex with 4EC
| Parameter | AphC 4EC |
|---|---|
| Data collection | |
| Beamline | CMCF-BM |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 81.6/119.4/84.5 |
| α, β, γ (°) | 90.0/114.6/90.0 |
| Resolution (Å) | 47.14–1.59 |
| (1.74–1.59)a | |
| Rmerge [%] | 4.6 (97.5) |
| Rpim [%] | 2.9 (61.5) |
| <I/σI> | 12.4 (1.3) |
| Completeness (%)b | 94.0 (60.3) |
| Redundancy | 3.3 (3.5) |
| CC(1/2) | 0.999 (0.489) |
| Refinement | |
| Rwork/Rfree [%] | 17.7/20.9 |
| Ramachandran plot | |
| Most favored [%] | 97.5 |
| Allowed [%] | 2.5 |
| Disallowed [%] | 0.0 |
| No. atoms | |
| Protein | 11,110 |
| Water | 815 |
| Ligands | 50 |
| B-factors | |
| Protein | 33.6 |
| Water | 36.0 |
| Ligands | 37.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0132 |
| Bond angles (°) | 1.65 |
| pdb-code | 7Q2A |
Values in parentheses are for the highest-resolution shell.
Ellipsoidal completeness.
4-Ethylguaiacol and guaiacol catabolism by RHA1 and mutants
RHA1 was grown at 30 °C and, if in liquid media, shaking at 200 rpm. To assess growth substrate depletion and specific activities of cell lysates, single colonies of RHA1 were grown overnight in LB broth, harvested at 6000×g, washed twice with M9 minimal medium, and used to inoculate M9 medium (44) supplemented with an HCl-solubilized mineral solution described previously (27), and either 1 mM guaiacol, 1 mM 4-ethylguaiacol, or both at 0.5 mM each. To measure substrate depletion, 1 ml aliquots were sampled from 125 ml liquid culture, and the OD600 was measured using a Biochrom WPA Biowave II UV/Visible Spectrophotometer and a semimicro cuvettes (l = 1 cm). To measure the growth of the WT RHA1 and deletion mutants on media supplemented with various carbon sources, 150 μl were sampled from 3 ml cultures at endpoint (t = 60 h post inoculation), and OD600 was measured using a TECAN Spark Multimode Microplate Reader. Statistical significance of growth data was determined using a two-way ANOVA test with the Holm–Sidak multiple comparison test. Cells were harvested at OD600 0.2 to 0.5 by centrifugation (6000g for 20 min) and suspended in 10 mM sodium phosphate, pH 8. Cells were disrupted by bead-beading using a FastPrep-24 beat beater (MP Biomedicals) with 0.1 mm zirconia/silica beads and centrifuged to remove cell debris (4000g for 60 min). The specific activity in the supernatant was calculated as described above.
HPLC analysis was used to track the depletion of substrates during RHA1 growth. Culture supernatants (1 ml) were acidified by adding acetic acid to 10% final concentration, centrifuged (16,000g for 10 min), and filtered through a 0.2 μm PTFE membrane. The run was conducted as described above. Analytes were quantified by interpolation from a standard curve of 0 to 1 mM guaiacol or 4-ethylguaiacol. In assessing how compounds were degraded, RHA1 and its mutants (ΔaphC (14) and ΔpcaL (2) deletion strains) were grown in M9 media supplemented with Goodies and 5 mM succinate for 48 h, washed twice with M9 minimal media, and used to inoculate M9 supplemented with Goodies and either 1 mM guaiacol, 1 mM 4-ethylguaiacol, or both at 0.5 mM each.
LC/MS analysis
Reactions containing 3-phenylcatechol and 4-phenylcatechol treated with either AphC or BphCLB400 were quenched with one equivalent of methanol. LC-MS analysis was conducted on an Agilent 1290 Infinity II UHPLC in line with an Agilent 6546 Q-TOF equipped with a dual AJS ESI source operating in negative ionization mode. Sample (2 μL) was injected onto a Poroshell 120 PFP column (100 mm × 2.1 mm × 2.7 μM) and run on an 18 min linear gradient from 5 to 100% solvent B at 0.45 ml min−1. Solvent A was 0.1% formic acid in water, solvent B was 0.1% formic acid in acetonitrile. MS parameters in negative ionization mode were as follows: capillary voltage, 3500 V; nozzle voltage, 250 V; drying gas temp, 300 °C; drying gas flow rate, 10 L min−1; sheath gas temperature, 300 °C; sheath gas flow rate 12 L min−1, nebulizer pressure, 40 psi; fragmentor voltage, 100 V. MS/MS was collected on selected ions with 10, 20, and 40 V collision energies. Data were collected and analyzed using MassHunter Workstation Version 10.
NMR analysis
4-Phenylcatechol (2 mM) was combined with excess AphC in 20 mM Hepes (I = 0.1 M), pH = 8. The reaction was incubated at room temperature for 30 min, and the enzyme was removed by filtration through a 30 kDa molecular weight cut-off membrane. The filtrate was supplemented with 200 mM of NH4OH and incubated overnight at room temperature. The resultant reaction mixture was freeze dried and suspended in 0.5 ml of D2O for NMR analysis. NMR spectra were recorded using a Bruker AVANCE III 850 spectrometer. Data were analyzed using ACD NMR Processor Academic Edition.
Data availability
The atomic coordinates have been deposited in the Protein Data Bank and are available under the accession code 7Q2A.
Supporting information
This article contains supporting information (22, 24, 25, 26, 26).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Dr Mark Okon and Prof. Lawrence McIntosh assisted with the NMR spectroscopy. Angelé Arrieta assisted with the anaerobic crystallization. Part of the research described in this article was performed using CMCF-BM at the Canadian Light Source, a national research facility of the University of Saskatchewan, which is supported by the Canada Foundation for Innovation (CFI), the Natural Sciences and Engineering Research Council (NSERC), the National Research Council Canada (NRC), the Canadian Institutes of Health Research (CIHR), the Government of Saskatchewan, and the University of Saskatchewan.
Author contributions
L. E. N., M. Z., J. E. M., and L. D. E conceptualization; L. E. N., M. Z., H. B., J. C. G., and M. E. W. methodology; L. E. N., M. Zz., H. B., J. C. G., A. C. K. C., and M. E. W. formal analysis; L. E. N., M. Z., J. C. G., M. E. W., and L. D. E. writing-original draft; A. C. K. C., M. E. P. M., and J. E. M. writing-review & editing; M. E. P. M., J. E. M., and L. D. E. supervision; M. E. P. M., J. E. M., and L. D. E. funding acquisition.
Funding and additional information
This study was supported by the following grants from the Natural Sciences and Engineering Research Council of Canada: STPGP 506595 to 17 and DG 171359 to L. D. E.; and DG 04802 to M. E. P. M. L. D. E. is the recipient of a Canada Research Chair. M. Z. and J. E. M. acknowledge Research England for E3 funding.
Edited by Ruma Banerjee
Supporting information
Supplemental Figures S1–S8, Tables S1 and S2
References
- 1.Yam K.C., Geize R.V.D., Eltis L.D. Biology of Rhodococcus. Springer; Berlin/Heidelberg, Germany: 2010. Catabolism of aromatic compounds and steroids by Rhodococcus; pp. 133–169. [Google Scholar]
- 2.Patrauchan M.A., Florizone C., Dosanjh M., Mohn W.W., Davies J., Eltis L.D. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: Redundancies and convergence. J. Bacteriol. 2005;187:4050–4063. doi: 10.1128/JB.187.12.4050-4063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker J., Wittmann C. A field of dreams: Lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol. Adv. 2019;37:107360. doi: 10.1016/j.biotechadv.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Eltis L.D., Singh R. Lignin Valorization: Emerging Approaches. The Royal Society of Chemistry; Cambridge: 2018. Biological funneling as a means of transforming lignin-derived aromatic compounds into value-added chemicals; pp. 290–313. [Google Scholar]
- 5.McLeod M.P., Warren R.L., Hsiao W.W., Araki N., Myhre M., Fernandes C., Miyazawa D., Wong W., Lillquist A.L., Wang D., Dosanjh M., Hara H., Petrescu A., Morin R.D., Yang G., et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van der Geize R., Yam K., Heuser T., Wilbrink M.H., Hara H., Anderton M.C., Sim E., Dijkhuizen L., Davies J.E., Mohn W.W., Eltis L.D. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yam K.C., Okamoto S., Roberts J.N., Eltis L.D. Adventures in Rhodococcus - from steroids to explosives. Can J. Microbiol. 2011;57:155–168. doi: 10.1139/W10-115. [DOI] [PubMed] [Google Scholar]
- 8.Seto M., Masai E., Ida M., Hatta T., Kimbara K., Fukuda M., Yano K. Multiple polychlorinated biphenyl transformation systems in the gram-positive bacterium Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 1995;61:4510–4513. doi: 10.1128/aem.61.12.4510-4513.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa K. Biochemical and genetic bases of microbial degradation of polychlorinated biphenyls (PCBs) J. Gen. Appl. Microbiol. 2000;46:283–296. doi: 10.2323/jgam.46.283. [DOI] [PubMed] [Google Scholar]
- 10.Sakai M., Masai E., Asami H., Sugiyama K., Kimbara K., Fukuda M. Diversity of 2,3-dihydroxybiphenyl dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1. J. Biosci. Bioeng. 2002;93:421–427. doi: 10.1016/s1389-1723(02)80078-0. [DOI] [PubMed] [Google Scholar]
- 11.Goncalves E.R., Hara H., Miyazawa D., Davies J.E., Eltis L.D., Mohn W.W. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2006;72:6183–6193. doi: 10.1128/AEM.00947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H.P., Chow M., Liu C.C., Lau A., Liu J., Eltis L.D. Vanillin catabolism in Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2012;78:586–588. doi: 10.1128/AEM.06876-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fetherolf M.M., Levy-Booth D.J., Navas L.E., Liu J., Grigg J.C., Wilson A., Katahira R., Beckham G.T., Mohn W.W., Eltis L.D. Characterization of alkylguaiacol-degrading cytochromes P450 for the biocatalytic valorization of lignin. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25771–25778. doi: 10.1073/pnas.1916349117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy-Booth D.J., Fetherolf M.M., Stewart G.R., Liu J., Eltis L.D., Mohn W.W. Catabolism of alkylphenols in Rhodococcusvia a meta-cleavage pathway associated with genomic islands. Front. Microbiol. 2019;10:1862. doi: 10.3389/fmicb.2019.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y., Koelewijn S.F., Van den Bossche G., Van Aelst J., Van den Bosch S., Renders T., Navare K., Nicolai T., Van Aelst K., Maesen M., Matsushima H., Thevelein J.M., Van Acker K., Lagrain B., Verboekend D., et al. A sustainable wood biorefinery for low-carbon footprint chemicals production. Science. 2020;367:1385–1390. doi: 10.1126/science.aau1567. [DOI] [PubMed] [Google Scholar]
- 16.Vaillancourt F.H., Bolin J.T., Eltis L.D. The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 2006;41:241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 17.Catelani D., Fiecchi A., Galli E. Dextro-gamma-carboxymethyl-gamma-methyl-delta-alpha-butenolide. A 1,2-ring-fission product of 4-methylcatechol by Pseudomonas desmolyticum. Biochem. J. 1971;121:89–92. doi: 10.1042/bj1210089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fielding A.J., Lipscomb J.D., Que L., Jr. A two-electron-shell game: Intermediates of the extradiol-cleaving catechol dioxygenases. J. Biol. Inorg. Chem. 2014;19:491–504. doi: 10.1007/s00775-014-1122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seah S.Y., Labbe G., Nerdinger S., Johnson M.R., Snieckus V., Eltis L.D. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 2000;275:15701–15708. doi: 10.1074/jbc.275.21.15701. [DOI] [PubMed] [Google Scholar]
- 20.Dagley S., Evans W.C., Ribbons D.W. New pathways in the oxidative metabolism of aromatic compounds by microorganisms. Nature. 1960;188:560–566. doi: 10.1038/188560a0. [DOI] [PubMed] [Google Scholar]
- 21.Catelani D., Colombi A., Sorlini C., Treccani V. Metabolism of biphenyl. 2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate: The meta-cleavage product from 2,3-dihydroxybiphenyl by Pseudomonas putida. Biochem. J. 1973;134:1063–1066. doi: 10.1042/bj1341063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Syed Ibrahim B., Kumaran D., Burley S.K., Swaminathan S., New York SGX Research Center for Structural Genomics (NYSGXRC) Crystal structure of 2,3-Dihydroxy biphenyl dioxygenase from Rhodococcus sp. (strain RHA1) PDB. 2010 doi: 10.2210/pdb3lm4/pdb. [DOI] [Google Scholar]
- 23.Vaillancourt F.H., Barbosa C.J., Spiro T.G., Bolin J.T., Blades M.W., Turner R.F., Eltis L.D. Definitive evidence for monoanionic binding of 2,3-dihydroxybiphenyl to 2,3-dihydroxybiphenyl 1,2-dioxygenase from UV resonance Raman spectroscopy, UV/Vis absorption spectroscopy, and crystallography. J. Am. Chem. Soc. 2002;124:2485–2496. doi: 10.1021/ja0174682. [DOI] [PubMed] [Google Scholar]
- 24.Fortin P.D., MacPherson I., Neau D.B., Bolin J.T., Eltis L.D. Directed evolution of a ring-cleaving dioxygenase for polychlorinated biphenyl degradation. J. Biol. Chem. 2005;280:42307–42314. doi: 10.1074/jbc.M510456200. [DOI] [PubMed] [Google Scholar]
- 25.Cerdan P., Rekik M., Harayama S. Substrate specificity differences between two catechol 2,3-dioxygenases encoded by the TOL and NAH plasmids from Pseudomonas putida. Eur. J. Biochem. 1995;229:113–118. doi: 10.1111/j.1432-1033.1995.tb20445.x. [DOI] [PubMed] [Google Scholar]
- 26.Takeo M., Nishimura M., Shirai M., Takahashi H., Negoro S. Purification and characterization of catechol 2,3-dioxygenase from the aniline degradation pathway of Acinetobacter sp. YAA and its mutant enzyme, which resists substrate inhibition. Biosci. Biotechnol. Biochem. 2007;71:1668–1675. doi: 10.1271/bbb.70079. [DOI] [PubMed] [Google Scholar]
- 27.Vaillancourt F.H., Han S., Fortin P.D., Bolin J.T., Eltis L.D. Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J. Biol. Chem. 1998;273:34887–34895. doi: 10.1074/jbc.273.52.34887. [DOI] [PubMed] [Google Scholar]
- 28.Cho J.H., Jung D.K., Lee K., Rhee S. Crystal structure and functional analysis of the extradiol dioxygenase LapB from a long-chain alkylphenol degradation pathway in Pseudomonas. J. Biol. Chem. 2009;284:34321–34330. doi: 10.1074/jbc.M109.031054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 32.Ausubel F.M. 5th ed. Wiley; NY: 2002. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. [Google Scholar]
- 33.Haigler B.E., Gibson D.T. Purification and properties of ferredoxinNAP, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J. Bacteriol. 1990;172:465–468. doi: 10.1128/jb.172.1.465-468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornish-Bowden A. Oxford University Press; Oxford, U. K: 1995. Analysis of Enzyme Kinetic Data. [Google Scholar]
- 35.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tickle I.J., Flensburg C., Keller P., Paciorek W., Sharff A. Global Phasing Ltd; Cambridge, United Kingdom: 2018. STARANISO. [Google Scholar]
- 37.Vagin A., Teplyakov A. Molrep: An automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 38.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., Jain S., Lewis S.M., Arendall W.B., 3rd, Snoeyink J., Adams P.D., et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L. DALI and the persistence of protein shape. Protein Sci. 2020;29:128–140. doi: 10.1002/pro.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krissinel E. Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 2012;1:76–85. [PMC free article] [PubMed] [Google Scholar]
- 43.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. Sect. A. 1976;32:922–923. [Google Scholar]
- 44.Sambrock J., Mariatis T., Fritsch E. Cold Spring Harbor Laboratory; New York, NY: 1990. Molecular Cloning: A Manual Laboratory. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures S1–S8, Tables S1 and S2
Data Availability Statement
The atomic coordinates have been deposited in the Protein Data Bank and are available under the accession code 7Q2A.