Abstract
2-Ketopropyl-coenzyme M oxidoreductase/carboxylase (2-KPCC) is a member of the flavin and cysteine disulfide containing oxidoreductase family (DSOR) that catalyzes the unique reaction between atmospheric CO2 and a ketone/enolate nucleophile to generate acetoacetate. However, the mechanism of this reaction is not well understood. Here, we present evidence that 2-KPCC, in contrast to the well-characterized DSOR enzyme glutathione reductase, undergoes conformational changes during catalysis. Using a suite of biophysical techniques including limited proteolysis, differential scanning fluorimetry, and native mass spectrometry in the presence of substrates and inhibitors, we observed conformational differences between different ligand-bound 2-KPCC species within the catalytic cycle. Analysis of site-specific amino acid variants indicated that 2-KPCC-defining residues, Phe501-His506, within the active site are important for transducing these ligand induced conformational changes. We propose that these conformational changes promote substrate discrimination between H+ and CO2 to favor the metabolically preferred carboxylation product, acetoacetate.
Keywords: mass spectrometry, flavoenzyme, conformational change, carboxylation, ion mobility, native mass spectrometry, dynamics
Abbreviations: 2-KPC, 2-ketopropyl coenzyme M; 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; AlloSigMA, Allosteric Signaling and Mutation Analysis; CCS, collisional cross section; CoM, coenzyme M; DSF, differential scanning fluorimetry; DSOR, disulfide oxidoreductase; FAD, flavin adenine dinucleotide; GR, glutathione reductase; GSSG, glutathione disulfide; IM, ion mobility; m/z, mass to charge; NADPH, β-nicotinamide adenine dinucleotide phosphate; NMS, native mass spectrometry; WT, wildtype
Atmospheric carbon dioxide is a carbon source for building biomass in plants and microbes. In plants, RuBisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase), the most abundant enzyme on earth, catalyzes the first step in CO2 fixation (1, 2). A variety of lesser known, nonphotosynthetic microbial processes that fix CO2 or bicarbonate have also been described. However, how these processes contribute to the global carbon cycle and their catalytic mechanisms is not well understood (1).
2-ketopropyl-coenzyme M oxidoreductase/carboxylase (2-KPCC) is a unique CO2 fixing carboxylase which is part of the flavin/cysteine disulfide oxidoreductase (DSOR) family, members of which are best known for reducing metal ion or disulfide substrates (3). Glutathione reductase (GR) is a well characterized, paradigmatic DSOR that is responsible for cellular redox homeostasis (4) (Fig. 1A). Like GR, 2-KPCC is a homodimer with a redox active disulfide/FAD cofactor in each subunit. However, 2-KPCC has distinguishing features that appear to permit it to perform a carboxylation (3, 5, 6). Specifically, typical DSORs have a conserved His-Glu catalytic dyad (4, 7). The conserved His serves as a source of protons during the catalytic cycle and stabilizes an important reaction intermediate. The conserved Glu moderates the sidechain orientation and pKa of the conserved His (4, 7). In 2-KPCC, this catalytic dyad is substituted by a Phe-His pair. Previous studies have shown that the Phe-His in 2-KPCC produces a unique reduced active form and facilitates carboxylation (8). The reduced, reactive form of 2-KPCC reductively cleaves a thioether substrate 2-KPC (2-ketopropyl-coenzyme M). The product of this cleavage is an unstable enol-acetone anion, which as a nucleophile attacks enzyme-bound CO2 to form acetoacetate (Fig. 1B). In the absence of CO2 and in the presence of available protons, acetone forms as an undesirable side product. Therefore, 2-KPCC has a unique requirement, compared to GR, in that it must protect the enol-acetone intermediate from bulk solvent to produce the carboxylation product, acetoacetate (Fig. 1B) (1). Prior work showed that the Phe of the Phe-His dyad suppresses protonation of the enol-acetone intermediate (9), while the His helps stabilize acetoacetate (10), though whether their influence on the reaction is wholly electrostatic or something else is not known.
Figure 1.
Reactions of glutathione reductase and 2-KPCC showing the active site residues and intermediates.A, oxidized active site of GR is reduced by NADPH, the reduced reactive form then reacts with GSSG to make a mixed disulfide and one glutathione (GSH). Lastly, the active site returns to its oxidized form and releases two GSH. B, oxidized active site of 2-KPCC is reduced by NADPH, the reduced reactive form then reacts with 2-KPC to produce a mixed disulfide and the Ene-intermediate (blue). The Ene-intermediate needs to be protected from protons or it will form acetone (red) instead of acetoacetate (green). Lastly, after carboxylation is complete, the active site returns to its oxidized form and acetoacetate and CoM are released. 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; CoM, coenzyme M; GR, glutathione reductase; GSSG, glutathione disulfide.
The overall structures of 2-KPCC and GR are very similar. Both are homodimers of subunits that each contain FAD binding, NADPH binding, and interfacial domains (5, 6, 11). Unlike GR, 2-KPCC has extended N and C termini and a 13 amino acid insertion (Fig. 2) (5). The structure of 2-KPCC has been determined in the absence of added ligands and in several ligand-bound states (5, 6, 10, 11, 12). The structure of 2-KPCC in the presence of substrates and intermediates reveals conformational shifts in which the substrate access channel has collapsed. This is mainly due to differences in the conformation of the aforementioned extended N and C termini and 13 amino acid insertion not present in GR. In the presence of substrates and intermediates, structures of 2-KPCC exist with a smaller hydrophobic channel that we have proposed facilitates the access of CO2 but excluding other molecules including aqueous solvent. We have hypothesized that this arrangement and implied conformation change is of key importance in protecting reaction intermediates promoting carboxylation and the formation of acetoacetate relative to protonation and the formation of the unproductive product acetone (5, 6)
Figure 2.
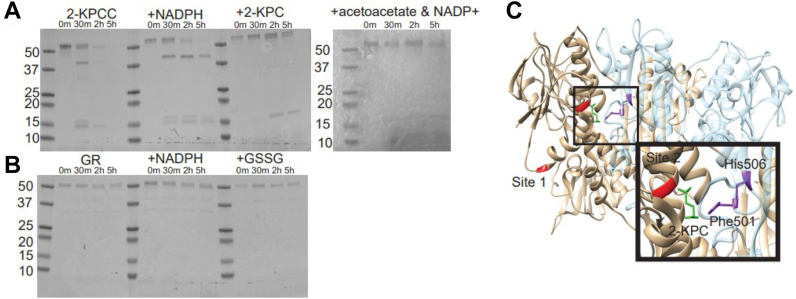
Structural comparison of 2-KPCC and glutathione reductase.A, structure of a glutathione reductase monomer (PDB: 1GRG). B, 2-KPCC (PDB: 1MO9) monomer highlighting extended regions, N terminus (red), C terminus (magenta), and extended interface domain (green). C, superimposed structures of 2-KPCC and glutathione reductase. D, dimeric structure of 2-KPCC. The regions unique to 2-KPCC surround the opening to the active site (black oval). 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; GR, glutathione reductase.
To test this hypothesis and gain insight into solution behavior, we have compared 2-KPCC and GR using a suite of biophysical techniques sensitive to conformational differences including ion mobility (IM)–native mass spectrometry (NMS), limited proteolysis, differential scanning fluorimetry (DSF), and the computational tool, Allosteric Signaling and Mutation Analysis (AlloSigMA). Experimental conditions include different ligand-bound and oxidation states of the enzyme. One condition of particular note is the oxidized enzyme and oxidized ligand complex, which was used to prevent product turnover and effectively explore the conformational changes reported in prior crystallographic work. Further, wildtype (WT) and amino acid–substituted variants of 2-KPCC in which one or both catalytic dyad residues (Phe501 and His506) are exchanged for the characteristic His-Glu pair found in GR were examined to determine whether the catalytic dyad, which promotes formation of the carboxylation product, might likewise play a role in producing conformational change. The evidence indeed indicates that 2-KPCC’s noncanonical Phe501-His506 active site dyad contributes to regulating dynamics associated with carboxylation.
Results and discussion
Addition of 2-KPC induced a more compact 2-KPCC structure
To begin a solution phase structural investigation of GR and 2-KPCC, NMS was conducted. In NMS, noncovalent interactions are maintained, facilitating an analysis of protein conformational changes upon ligand binding (13, 14). Solutions containing 5 μM GR or 2-KPCC with and without substrate (2 mM 2-KPC or glutathione disulfide (GSSG) intended to maximize the enzyme–substrate bound form) (15) were prepared in 300 mM ammonium acetate and analyzed by direct infusion NMS. The charge state distributions of 2-KPCC in the presence and absence of 2-KPC indicated that 2-KPC narrowed the distribution of charge states and shifted it to a higher mass to charge (m/z) ratio (Fig. 3A). This suggests a more compact protein fold when 2-KPC is present. In contrast, GR exhibited no change in charge state distribution upon GSSG binding (Fig. 3A).
Figure 3.
Mobiligrams and charge state distributions of WT, mutant 2-KPCC, and glutathione reductase. Charge state distribution of proteins in the native state is on the bottom of each panel. Ion mobility data for the specific charge species are directly above. The vertical axis is time for ion mobility with lower times indicating a smaller CCS. The color scale on the right shows relative intensity starting at 20%. A, from left to right WT 2-KPCC ± 2 mM 2-KPC and GR ± 2 mM GSSG. The comparison of WT 2-KPCC ± 2 mM 2-KPC depicts a change in ion mobility and charge state distribution based on 2-KPC binding. The comparison of GR ± 2 mM GSSG depicts no change in ion mobility or charge state distribution. B, from left to right Phe501His, His506Glu, Phe501His_His506Glu 2-KPCC ± 2 mM 2-KPC. The variants showed no change in ion mobility or charge state distribution based on 2-KPC binding. 2-KPC, 2-ketopropyl coenzyme M; 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; CCS, collisional cross section; GR, glutathione reductase; GSSG, glutathione disulfide.
IM coupled to NMS was then employed using the same experimental conditions. IM combines transfer time through an ion mobility cell and m/z to separate ions based on collisional cross section (CCS). Comparison of the observed CCS with values calculated from structural models provides information on protein conformation. Differences in protein conformation are detected as a change in CCS (13, 14). 2-KPCC in the absence of 2-KPC had a longer drift time than in its presence, corresponding to a larger CCS value for 2-KPCC in the absence of 2-KPC (Fig. 3A). Also, in the absence of substrate, 2-KPCC had a wider range of drift times, indicating a spread in the CCS which translates to a more diverse ensemble of conformations. This can be interpreted as greater conformational freedom in the absence of 2-KPC. In contrast, GR did not exhibit a shift in drift time with GSSG binding (Fig. 3A), giving strong evidence of no apparent difference in conformation.
Limited proteolysis revealed ligand-dependent protection of two protease cleavage sites
The IM-NMS data provided evidence for a substrate-dependent conformational change, but no information on its nature or location. To further our investigation, we used limited proteolysis to track the rate and location of kinetically favored sites of cleavage. The rate of proteolysis is expected to differ between dynamic and stable regions, with unstructured/dynamic regions of a protein exhibiting faster peptide bond hydrolysis (16, 17). Thermolysin was selected for this experiment because of the high frequency of potential cleavage sites in 2-KPCC (168) and GR (149) based on predicted sequence specificity (Fig. S1). Substrate binding can affect the rate and pattern of hydrolysis by changing accessibility to cleavable sites. Solutions of 2-KPCC and GR at 0.5 mg/ml were incubated with thermolysin at a 1:100 (wt:wt) ratio of protease to protein. To probe for substrate-induced conformational change, each enzyme was poised with different ligands (NADPH, 2-KPC, GSSG, and acetoacetate and NADP+). Aliquots were quenched for analysis at 0 min, 30 min, 2 h, and 5 h. The reaction was then visualized by SDS-PAGE.
For WT 2-KPCC, there were significant differences in the cleavage kinetics and SDS-PAGE gel banding pattern depending on the ligand present (Fig. 4A). 2-KPCC in the absence of ligands was completely digested within 2 h: more rapidly than any of the conditions where a ligand was present. This suggests that the ligand-free state of the enzyme has the greatest conformational freedom. Further, the banding pattern on the gel suggests there are two primary sites of cleavage in the 2-KPCC 57 kDa monomer, termed sites 1 and 2. Cleavage at site 1 generates a band with an apparent molecular weight just slightly smaller than the parent protein (51 kDa), while cleavage at site 2 gives rise to a 42 kDa band (Fig. 4A). To identify the main proteolysis cleavage sites on 2-KPCC, solution phase LC-MS and in-gel digestion were employed. Both sites are found on the N terminus, site 1 was identified as amino acid A26 and site 2, which gives rise to the formation of the 42 kDa band, is F142 (Fig. 4B). The expected 9 kDa peptide accompanying the 42 kDa fragment was too small to be resolved by the SDS-PAGE experiment.
Figure 4.
Limited proteolysis of WT 2-KPCC. The presence of substrate alters the kinetics and pattern of proteolysis. A, SDS-PAGE gel of limited proteolysis showing banding pattern of WT 2-KPCC under ligand-free, NADPH-bound, 2-KPC-bound, and acetoacetate & NADP+ conditions. B, SDS-PAGE gel of limited proteolysis showing banding pattern of GR under ligand-free, NADPH-bound, and GSSG-bound conditions. C, structure of 2-KPCC with two thermolysin cleavage sites shown in red. Inset, active site residues Phe501 and His506 (purple) and 2-KPC-bound (green) and proximity to the cleavage site (red) which produces the 42 KDa form. 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; GR, glutathione reductase; GSSG, glutathione disulfide.
In sharp contrast to the ligand-free state, the presence of 2-KPC dramatically slowed proteolysis. The intact protein was present throughout the 5 h incubation, with the 51 kDa band present after 2-h and the 42 kDa band not appearing at all. These data suggest that 2-KPC supplies substantial protection from proteolysis at both cleavage positions. Binding the products NADP+ and acetoacetate had very similar effects to 2-KPC, while in the NADPH present condition, the 42 kDa band appeared after 30 min and persisted to the 5-h time point. For GR, there was no significant change in cleavage kinetics or SDS-PAGE gel banding pattern upon substrate binding (Fig. 4C). In ligand-free, NADPH present, and GSSG present GR, the intact band was present at 5 h. GR did not exhibit change in proteolytic rate in the absence or presence of ligands, and this suggests little conformational flexibility regardless of ligand binding.
Collectively, the limited proteolysis data indicate that 2-KPCC underwent ligand-induced conformational changes that restricted protease cleavage in the presence of 2-KPC, NADPH, or the acetoacetate/NADP+ products, while GR did not. The ligand-induced conformational changes seen in 2-KPCC also depend on the ligand present, suggesting that the conformational change undergone during reaction may follow an ordered change which will help protect the reaction intermediate.
Global thermal stability is increased by substrate binding in 2-KPCC
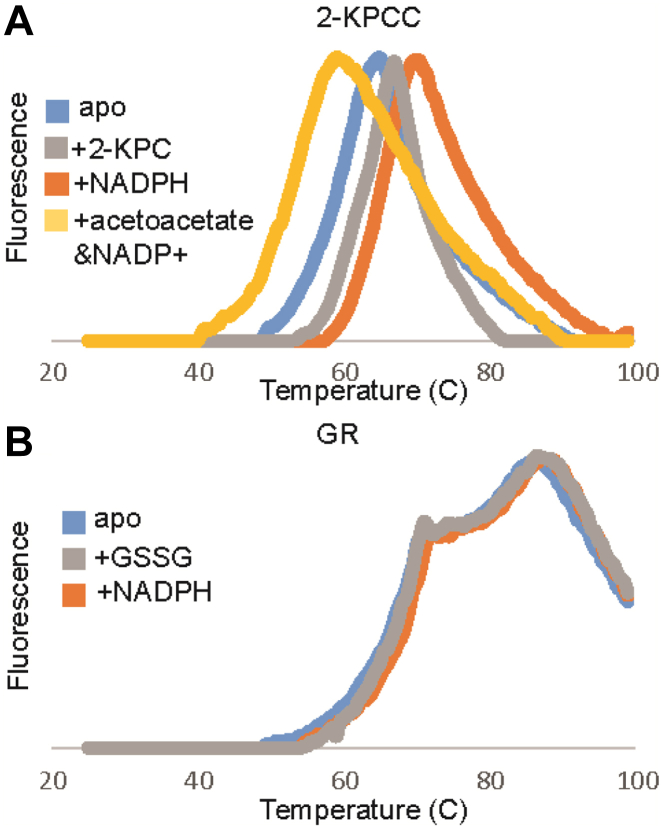
The limited proteolysis data revealed that substrate binding altered susceptibility to proteolysis. This inspired us to probe whether the same factors also influence global protein stability. To investigate this, we employed DSF to provide a read out of thermal stability of the proteins based on their melting profiles. A hydrophobic, fluorescent dye, which is quenched in a polar environment, is added to the protein solution. As the temperature is raised, the dye binds to hydrophobic pockets that become accessible as the protein structure denatures, producing a melting curve (18, 19, 20). DSF reports on local and global thermal stability. It has been shown that this technique can be utilized to investigate ligands that alter thermal stability (20, 21) and is particularly well-suited for protein complexes (22, 23). Therefore, we employed it to investigate the substrate binding effects on global thermal stability of 2-KPCC.
Reaction solutions of 0.25 mg/ml 2-KPCC or GR were incubated with 50× concentration of Sypro Orange dye with or without substrate (0–5 mM). 2-KPCC has a thermal melting point (Tm) of 65.5 °C (Fig. 5A). Binding 2-KPC and NADPH shifted the Tm to 67 °C and 70.5 °C, respectively. This indicates that the ligands have a stabilizing effect on 2-KPCC. Increased thermal stability is consistent with a model in which the substrate-bound conformation can exclude solvent and a more compact structure is present. The stability and compact form arise from enhanced noncovalent interactions in the protein–ligand complex (20, 21, 24). We would expect a more compact structure to have a higher Tm, like what is exhibited by binding of 2-KPC and NADPH. In contrast, binding of the reaction products acetoacetate and NADP+ lowered the Tm. This destabilization could characterize a product release form of 2-KPCC which was too small to be seen by limited proteolysis. Interestingly, GR has the highest Tm at 87 °C and exhibits no shift in Tm upon ligand binding but does exhibit a local destabilization at 72 °C (Fig. 5B). Together the IM-NMS, proteolysis, and thermal stability data reflect the crystallographic and catalytic models in which structural change is part of the catalytic cycle. The binding of different ligands produces different structural changes within 2-KPCC. These changes are not shared by GR, which has a high global thermal stability with none of the structural parameters interrogated here altered by substrate binding.
Figure 5.
Differential scanning fluorimetry curves of glutathione reductase and WT 2-KPCC.A, DSF curves showing substrate binding effects on melting curves of WT 2-KPCC. Apo (blue) Tm = 66.5 °C SD = 0.05, +2-KPC (gray) Tm = 67 °C SD = 0.34, +NADPH (orange) Tm = 70.5 °C SD = 0.74, +acetoacetate and NADP+ Tm = 60 °C SD = 0.071. B, DSF curves showing substrate binding effects on melting curves for GR. Apo (blue) SD = 0.013, +GSSG (gray) SD = 0.017, +NADPH (orange) SD = 0.017 Tm = 87.5 °C. Overall 2-KPCC exhibits lower melting temperatures than glutathione reductase. 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; DSF, differential scanning fluorimetry; GR, glutathione reductase; GSSG, glutathione disulfide.
Influence of 2-KPCC catalytic dyad on conformation
To better elucidate the cause for 2-KPCC’s conformational changes compared to GRs static behavior, we wanted to explore amino acid–substituted variants of 2-KPCC in which one or both catalytic dyad residues (Phe501 and His506) are substituted by the characteristic His-Glu pair found in GR (Phe501His and/or His506Glu). Prior work showed that the Phe of the Phe-His dyad suppresses protonation of the enol-acetone intermediate (9), while the His helps stabilize the formation of acetoacetate (10). We now sought to determine if the catalytic dyad, which is essential for promoting formation of the carboxylation product, might likewise play a role in the conformational changes that are proposed to exclude solvent from the enol-acetone intermediate.
To probe the structures of these variants, NMS and IM were employed in the presence and absence of 2-KPC. The charge state distributions of Phe501His, His506Glu, and Phe501His_His506Glu were lower than WT in the absence of 2-KPC, but not as low as WT in the presence of 2-KPC (Fig. 3B). In contrast to the WT enzyme, the variants did not have a shift in charge state upon addition of 2-KPC. The catalytic dyad of 2-KPCC promotes carboxylation and, when mutated, alters the product outcome (9, 10). Our results suggest a role for the unique catalytic dyad in carboxylation is manifested at least in part in transducing an essential conformational change. In the variants, the catalytic dyad stabilizes a more compact structure even without substrate. The 2-KPCC variants were also investigated in the presence and absence of 2-KPC (Fig. 3B). Importantly, they did not show a decrease in drift time with 2-KPC. However, they did exhibit a wider distribution of drift times. Taken with the NMS data, the variants appear to be in a conformation that closely resembles WT 2-KPCC in the presence of 2-KPC. This further implies that the Phe501His and His506Glu substitutions alter the conformational freedom of 2-KPCC.
Limited proteolysis indicates dyad-dependent protection of proteolytic sites
The 2-KPCC variants were also investigated by proteolysis. In the Phe501His variant, the ligand-free and NADPH present states exhibit lower proteolysis than WT (Fig. 6A), with the intact protein band persisting up to the 2 h time point. Addition of 2-KPC or NADPH extends the parent band lifetime to the 5 h mark and also stabilized the 42 kDa band so that it was likewise observable on this timescale. The His506Glu and the Phe501His_His506Glu reactions had intact protein remaining at the 2-h time point in all conditions and no observable 42 kDa species (Fig. 6, B and C). These results suggest that the His506Glu substitution stabilized the intact protein by limiting protease access to the helix that is cleaved to generate the 42 kDa form, but inhibited substrate-based protection.
Figure 6.
Limited proteolysis of 2-KPCC variants. The presence of substrate alters the kinetics and pattern of proteolysis. A, SDS-PAGE gel of limited proteolysis showing banding pattern of Phe501His 2-KPCC under ligand-free, 2-KPC-bound, NADPH-bound, and acetoacetate & NADP+-bound conditions. B, SDS-PAGE gel of limited proteolysis showing banding pattern of His506Glu 2-KPCC under ligand-free, 2-KPC-bound, NADPH-bound, and acetoacetate & NADP+-bound conditions. C, SDS-PAGE gel of limited proteolysis showing banding pattern of Phe501His_His506Glu 2-KPCC under ligand-free, 2-KPC-bound, NADPH-bound, and acetoacetate & NADP+-bound conditions. 2-KPC, 2-ketopropyl coenzyme M; 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase.
The active site residues Phe501 and His506 modulated protease access in 2-KPCC. When the active site was mutated to the canonical DSOR catalytic dyad residues, substrate-based protease protection was not observed in the variants with His506Glu substitution. This suggests that the protease-protective conformational changes could be the same as those needed for carboxylation. The His506Glu substitution appeared to alter conformational freedom more significantly than the Phe501His, as the substrate- and product-dependent modulation of protease access was not observed for this variant.
Active site dyad contributes to global thermal stability
The IM-NMS and limited proteolysis data demonstrated that the active site variants alter conformational freedom. We then employed DSF to investigate if the variants alter global protein stability. The ligand-free His506Glu variant has a decreased Tm relative to the WT and other variants, with a Tm of 57 °C (Fig. 7). The His506Glu variant exhibited small shifts to higher Tm upon binding of 2-KPC and NADPH and a slight shift to a lower Tm with acetoacetate and NADP+. This supports that the His506Glu substitution structure is possibly locked in a conformation not allowing for conformational freedom. Also, the melting temperatures of all conditions tested were similar to the acetoacetate and NADP+ condition of WT, which we interpret as the H506E substitution having a destabilizing effect on protein structure. The ligand free Phe501His and Phe501His_His506Glu variants have a similar Tm to WT with a Tm of 67 °C and 66 °C, respectively. However, the Phe501His variant did not have a large shift in Tm upon ligand binding except for with acetoacetate and NADP+ present, with a Tm of 71 °C. The Phe501His_His506Glu variant exhibited some hallmarks of both the single variants. A shoulder was present in some of the curves of Phe501His and Phe501His_His506Glu 2-KPCC. This shoulder suggests a local destabilization in the protein structure as the temperature is raised. Interestingly, GR shares this local destabilization.
Figure 7.
Differential scanning fluorimetry curves of 2-KPCC variants.A, DSF curves showing substrate binding effects on melting curves of Phe501His 2-KPCC. Apo (blue) Tm = 67 °C SD = 0.06, +2-KPC (gray) Tm = 67.5 °C SD = 0.02, +NADPH (orange) Tm = 68 °C SD = 0.09, +acetoacetate and NADP+ Tm = 71 °C SD = 0.05 n = 3. B, DSF curves showing substrate binding effects on melting curves for His506Glu 2-KPCC. Apo (blue) Tm = 57 °C SD = 0.04, +2-KPC (gray) Tm = 60 °C SD = 0.26, +NADPH (orange) Tm = 58 °C SD = 0.04, +acetoacetate and NADP+ Tm = 55.5 °C SD = 0.06 n = 3. C, DSF curves showing substrate binding effects on melting curves for Phe501His_His506Glu 2-KPCC. Apo (blue) Tm = 55 & 66 °C SD = 0.06, +2-KPC (gray) Tm = 57.5 °C SD = 0.02, +NADPH (orange) Tm = 50 °C SD = 0.06, +acetoacetate and NADP+ Tm = 56 °C SD = 0.017 n = 3. 2-KPC, 2-ketopropyl coenzyme M; 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; DSF, differential scanning fluorimetry.
Computational analysis implicates catalytic dyad in conformational change
To further elucidate how these active site residues influence conformational change, we turned to a computational analysis using AlloSigMA (25, 26). AlloSigMA uses a structure-based statistical mechanical model of allostery to calculate the per-residue change in free energy for all possible protein configurations due to interactions with ligands or mutations (27, 28). AlloSigMA defines the protein contact network of a protein by solving the structure-based statistical mechanical model of allostery for all possible local configurations in all available states. AlloSigMA defines mutations as either: UP-mutations, which cause over-stabilizing effects on the protein contact network, and DOWN-mutations, which cause destabilizing effects on a protein contact network (29). To test the impact of an amino acid substitution at a specified position, the user selects the residue to be probed and the type of mutation to be modeled, UP or DOWN. This information is used to predict changes in allosteric communication across a protein model. Allostery, in its broadest sense, is the functional change at one site on a protein caused by a change at a distant site (28, 29, 30). AlloSigMA interprets allostery as a change at any site in the structure that can lead to a shift in occupancy of the conformational states in which the protein can exist.
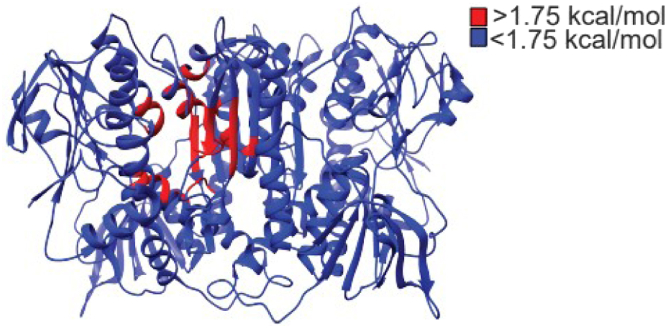
The crystal structure of 2-KPCC (PDB: 1MOK) was explored using AlloSigMA, where the Phe-His dyad was singly or doubly mutated to a DOWN-mutation (destabilizing residue). AlloSigMA then calculated the per-residue change in allosteric free energy (kcal/mol) caused by the mutation. A positive change in allosteric free energy indicates a change in force acting on the residue because of the mutation, and it can be interpreted as a possible increase in dynamics that could lead to a conformational change (31). A negative change in allosteric free energy indicates a stabilization of the residue caused by the mutation and possibly preventing conformational change or a decrease in dynamics. DOWN-mutations at Phe501, His506, and Phe501_His506 revealed an increase in free energy in the active site while the rest of the protein structure had a decrease in free energy. These values were color coded positive (red) or negative (blue) and displayed on the protein sequence or the structure (Fig. 8). These results are interpreted as an increase in conformational flexibility, where the number of possible conformational states is increased in the active site and a decrease in the rest of the protein. This was exhibited by all three variants, but the most prominent changes were seen in His506. A control experiment was conducted by mutating noncatalytically active residues in the active site and surface of the protein. These mutations produced very little change in free energy (Fig. S2).
Figure 8.
Change in free energy calculated by AlloSigMA as a result of mutations in the catalytic dyad plotted on structure of 2-KPCC (PDB:1MOK). Increase in free energy (red) indicates more perturbation and higher dynamics. Decrease in free energy (blue) indicates a stabilization and decrease in dynamics. 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; AlloSigMA, Allosteric Signaling and Mutation Analysis.
GR was also explored by AlloSigMA with mutations to the His-Glu catalytic dyad (Fig. S3). Interestingly, single substitutions at either residue produced similar behavior to 2-KPCC, where the active site showed an increase in free energy while the rest of the protein had a decrease. Tandem substitutions exhibited an increase in free energy near the active site. This further supports that the active site catalytic dyad has an important role in modulating conformational change in 2-KPCC and differences between the 2-KPCC and canonical DSOR enzymes such as GR.
Conclusion
Taken together, the DSF, limited proteolysis, IM-NMS data, and AlloSigMA analyses support a functional model of catalysis in which 2-KPCC transitions between multiple conformations, the equilibrium of which is controlled by substrate and product. The IM-NMS data showed a change in conformation of 2-KPCC when 2-KPC was present, and that active site substitutions altered the conformational ensemble of 2-KPCC. Limited proteolysis allowed us to localize conformational change upon binding of substrates and products to the N terminus. A change we believe serves to exclude solvent from the active site (see Fig. 2D). A key finding of this work is the role of the catalytic dyad (Phe-His) in the modulation of 2-KPCC’s conformational ensemble. DSF and AlloSigMA contributed significantly to this conclusion.
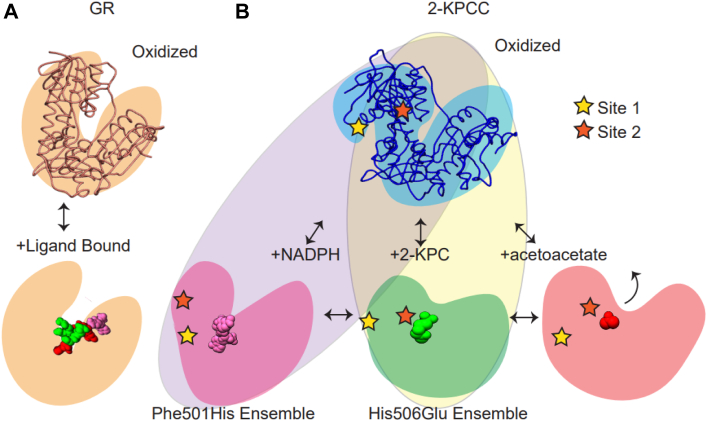
In recent years, our understanding of conformational change and allostery has shifted to a more nuanced model in which activity is best described in the context of conformational ensembles where population re-distributions influence catalysis (29, 32). Here we provide evidence that 2-KPCC is such a system, with a conformational ensemble that is influenced by substrate binding and modulated by the catalytic dyad. We propose the ligand free, oxidized state of 2-KPCC has full conformational freedom. During the catalytic cycle, binding of NADPH and reduction of the disulfide restricts the conformational ensemble to forms that support the binding of 2-KPC and formation of the mixed disulfide (Fig. 9). The binding of 2-KPC results in a more compact structure that presumably protects the active site, only allowing access of the second substrate CO2. Carboxylation then results in a form of 2-KPCC which supports the release of acetoacetate. In this scheme, changes to the conformational ensemble brought upon by substrate binding, help protect the enolacetone intermediate and promote carboxylation. Mutations of the catalytic Phe-His resulted in alterations to the conformational ensemble of 2-KPCC, and show the important role for the catalytic dyad in the conformational state of 2-KPCC.
Figure 9.
Conformational ensembles of GR and 2-KPCC. Schematic showing conformational changes associated with catalysis. One subunit of the dimer is shown. A, GR occupies a limited conformational space, remaining in the “open” conformation throughout the catalytic cycle (shown in tan). B, representative conformations of 2-KPCC highlighting the available conformational space sampled in solution. The oxidized, “open” conformations of 2-KPCC (blue) are shown with protease site 1 (yellow star) and site 2 (orange star). Binding of NADPH (pink) leads to altered access to site 2 and changes in conformational ensemble. 2-KPC binding (green) alters access to site 1 and the observed conformations of 2-KPCC. Binding of acetoacetate (red) represents yet another conformational ensemble favoring product release. The Phe501His variant restricts the available conformations which are populated in the ensemble (purple oval). Phe501His favors the oxidized and NADPH-bound conformations. The His506Glu variant restricts the accessible conformations (yellow oval) favoring oxidized and 2-KPC-bound conformations. 2-KPC, 2-ketopropyl coenzyme M; 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; GR, glutathione reductase.
2-KPCC’s mechanism is in contrast to catalysis by other DSOR proteins such as GR, which undergoes reduction, mixed disulfide formation, and product release with no detectable change in conformation (33). This suggests that the carboxylation is a derived function arising from adaptions by the active site chemistry and protein conformational stability.
Experimental procedures
Expression and purification of native 2-KPCC and amino acid–substituted variants
E. coli BL21 (DE3)pLysS cells were transformed with the pBAD plasmid harboring the WT or corresponding 2-KPCC variant gene from Xanthobacter autotrophicus Py2. The construct contained a His affinity tag, a thioredoxin tag, and a TEV-cleavage site. Cells were plated on LB agar + kanamycin (25 μg/ml) and grown overnight. A single colony from the plate was used to inoculate a 5 ml overnight culture in LB. The 5 ml overnight culture was used as the inoculum for a 500 ml baffled flask containing 500 ml of ZYP-rich medium + kanamycin (25 μg/ml). Cells were grown at 37 °C with agitation at 225 rpm until the A600 reached 0.6 to 1.0. The temperature was reduced to 25 °C, L-arabinose was added to 0.02%, and the cells were grown for an additional 16 to 18 h. The cells were pelleted by centrifugation, frozen, and stored at −80 °C. The cell pellet was re-suspended in three volumes of lysis buffer [20 mM Tris, pH 8.0, 300 mM NaCl, 5 mM imidazole, and protease inhibitor tablets (Pierce)] and thawed at 30 °C. All subsequent treatments were performed on ice or at 4 °C. The resuspended cells were lysed via multiple rounds of sonication (Branson Ultrasonifier). Cell lysates were clarified via centrifugation at 75,000g for 45 min. Clarified lysates were loaded onto a nickel–nitrilotriacetic–resin column (Bio-Rad) via a Bio-Rad FPLC, washed with lysis buffer, and eluted using a 0.005 to 0.4 M imidazole gradient in lysis buffer at 3 ml/min. The eluted 2-KPCC was diluted 6-fold into buffer A (20 mM Tris-Cl, pH 6.5, 5% w/v glycerol), applied to a Q-Sepharose ion-exchange column (Bio-Rad), and eluted using a 0 to 1 M NaCl gradient in buffer A. Fractions were screened using SDS-PAGE. Pure 2-KPCC protein was pooled and run on a desalting column (Bio-Rad) in 20 mM Tris, pH 7.4, 10% glycerol, and 200 mM NaCl. Protein was concentrated using a 30 kDa MWCO spin filter concentrator. To remove the 6x-His tag, 2-KPCC protein was incubated with TEV protease at a ratio of 1:100 (w/w) (Millipore) overnight at 4 °C. TEV protease was removed using a nickel–nitrilotriacetic–resin column via gravity. Fractions were screened using SDS-PAGE. Cleaved 2-KPCC protein was pooled and concentrated using 30kDaMWCO spin filter concentrator (Millipore). Total protein concentration was determined using a BCA protein assay (Thermo Scientific), and flavin concentration was determined from its UV-visible absorbance at 450 nm using Ɛ450 of 11,828 M−1 cm−1. All concentrations of protein cited in the text refer to flavin-containing protein.
NMS and IM of 2-KPCC and GR
Protein stock solutions were made anaerobic using a double-manifold Schlenk line with alternating cycles of argon gas purging and evacuation. Protein stocks were buffer exchanged into deoxygenated 300 mM ammonium acetate at pH 7.5 using 7.5 kDa mini-dialysis filters (Thermo-Fisher) overnight at 4 °C. Buffer exchanged proteins for bound conditions were then incubated for 15 min with either 2 mM 2-KPC, 2 mM GSSG, or 5 mM NADPH. Proteins were then loaded into in-house–prepared gold-coated borosilicate capillaries prepared as outlined in Luo et al. and analyzed using a Waters Synapt G2-Si with the following settings: capillary kV was set to 1.75; sampling cone, 95; source offset, 70; trap collision energy, 110 V; transfer collision energy, 105 V, source temperature, 30 °C; and trap gas, 7.0 ml/min. Data were analyzed using Waters MassLynx version 4.1. IM analysis was performed under the same conditions outlined above. The Synapt G2-Si instrument was set in IM mode, and data were analyzed by MassLynx 4.1 and DriftScope.
Limited proteolysis and peptide mapping of 2-KPCC and GR
2-KPCC (WT and variants) and GR samples at 0.5 mg/ml were prepared anaerobically (Mbraun anaerobic chamber Unilabpro) with deoxygenated buffer (200 mM Tris-HCl, 0.5 M CaCl2 pH 8) and incubated with either 2 mM 2-KPC, 2 mM GSSG, 5 mM NADPH, or 2 mM acetoacetate and 5 mM NADP+. Prepared reactions were then sealed using crimp seal vials and removed from the Mbraun. Using a gas-tight syringe, thermolysin (Promega) was added to reactions at a ratio of 1:100 (wt:wt) and incubated at 37 °C. At timepoints (0 min, 30 min, 2 h, 5 h) aliquots of 9 μl were removed from the reaction and immediately diluted into 36 μl of gel loading buffer, vortexed, and heated to 100 °C for 5 min. Following boiling, samples were frozen. Prior to gel loading, the frozen samples were simultaneously boiled at 100 °C for 5 min, centrifuged, vortexed, and maintained at 25 °C during gel loading. Samples were run on 4% to 20% (w/v) gradient SDS-PAGE gels (Bio-Rad) with tris-glycine buffer. Gels were stained with GelCode blue safe protein stain (Thermo).
Proteolysis reactions for mass spectrometry analysis were conducted identical to those for gel-based analysis except for quenching. Quenching of proteolysis was performed by multiple methods, these included dropping the pH, boiling, spotting directly onto a MALDI plate, and direct LCMS injection. LC-MS analysis of proteolysis reactions was completed on a 1200 infinity HPLC (Agilent) coupled to a MicroTOF LC mass spectrometer (Bruker). Peptides were separated on a size exclusion column (Phenomenex 150 × 2 mm) using a flow rate of 500 μl/min with solvent conditions 90% solvent A [0.1% FA (Sigma) in water (Thermo) and 10% solvent B (0.1% FA in acetonitrile) (Thermo)]. Data were acquired at 2 Hz over the scan range 50 to 1700 m/z in positive mode. Electrospray settings were as follows: nebulizer set to 3.7 bar, drying gas at 8.0 l/min, drying temperature at 350 °C, and capillary voltage at 3.5 kV. Data processing was carried out with Bruker DataAnalysis software. Unique peptides were identified by using Peptide Analysis Worksheet version 2000.6.8.0 with an accuracy of 100 ppm. Reactions were also analyzed by in-gel digestion and mass spectrometry. Protein bands from SDS-PAGE were cut and digested with trypsin (Promega) according to a standard protocol recommended by the manufacturer (1:50 wt:wt, overnight). Proteins were identified as described in (34) using a maXis Impact UHR-QTOF instrument (Bruker) interfaced with Dionex 3000 nano-uHPLC (Thermo). Data were analyzed and mapped by using Peptide Shaker, version 0.41.1, paired with Search GUI, version 1.30.1 (Compomics).
DSF of 2-KPCC and GR
Differential reactions containing 2 μg of protein in 200 mM Tris-HCl pH 8, a variety of ligands (2 mM 2-KPC, 2 mM GSSG, 5 mM NADPH, or 2 mM acetoacetate & 5 mM NADP+), and 1.2 μl of 50× stock SYPRO orange dye (Invitrogen) for a total reaction volume of 30 μl. The samples were loaded into a Roto-Gene Q instrument (Qiagen) with SYPRO orange absorbance excited at 470 nm and the fluorescence monitored at 570 nm as the temperature was ramped from 25 to 95 °C at 1 °C/min. All samples were run in triplicate.
AlloSigMA analysis of 2-KPCC and GR
The online computational tool AlloSigMA was used to explore the PDB files of both 2-KPCC and GR, (PDB: 1MOK, and PDB: 1GRG), respectively. Published protocols were followed for this analysis (31). Upon uploading the files, an allosteric signaling map was calculated to exhaustively compute the allosteric modulation of each residue due to mutation. For 2-KPCC, calculations were computed for DOWN-mutations at residues, Phe501, His506, Phe501_His506, Leu502, and Gly377. For GR, calculations were computed for DOWN-mutations of residues His467, Glu472, and His467_Glu472. The change in free energy calculated for each mutational study was then plotted on to the PDB structures 1MOK and 1GRG.
Source of glutathione reductase
Recombinant expressed in Escherichia coli, human GR was purchased from Sigma-Aldrich. Cas Number: 9001-48-3.
Data availability
Data will be shared upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
J. R. M. investigation; J. R. M. formal analysis; J. R. M. validation; J. R. M. visualization; J. R. M. data curation; J. R. M. writing-original draft; J. R. M., K. A. S., J. L. D., J. W. P., and B. B. writing-review & editing; K. A. S. resources; J. L. D., J. W. P., and B. B. conceptualization; J. L. D., J. W. P., and B. B. supervision; J. W. P. and B. B. methodology; J. W. P. project administration; J. W. P. funding acquisition.
Funding and additional information
This work was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award DE-FG02×04ER15563 to J. L. D., J. W. P., and B. B. Partial salary support for J. W. P. was supported by the USDA National Institute of Food and Agriculture, Hatch umbrella project #1015621. Funding for the Mass Spectrometry Facility used in this publication was made possible in part by the MJ Murdock Charitable Trust, the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers P20GM103474 and S10OD28650, and the MSU Office of Research, Economic Development and Graduate Education. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ruma Banerjee
Supporting information
Potential protease cleavage sites in GR and 2-KPCC.A, structure of GR (PDB: 1GRG) with potential thermolysin cleavage sites in red. B, structure of 2-KPCC (PDB: 1MO9) with potential thermolysin cleavage sites in red. 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; GR, glutathione reductase.
AlloSigMA control experiment mutating noncatalytically active residues.A, noncatalytically active site residue Leu502 was down mutated in AlloSigMA. Increase in free energy (red) indicates more perturbation and higher dynamics. Decrease in free energy (blue) indicates a stabilization and decrease in dynamics. No change in free energy (gray). B, noncatalytically active site residue Gly377 was down mutated in AlloSigMA. AlloSigMA, Allosteric Signaling and Mutation Analysis.
Change in free energy calculated by AlloSigMA plotted on structure of GR (PDB:1GRG). Increase in free energy (red) indicates more perturbation and higher dynamics. Decrease in free energy (blue) indicates a stabilization and decrease in dynamics. No change in free energy (gray). AlloSigMA, Allosteric Signaling and Mutation Analysis; GR, glutathione reductase.
References
- 1.Erb T.J. Carboxylases in natural and synthetic microbial pathways. Appl. Environ. Microbiol. 2011;77:8466–8477. doi: 10.1128/AEM.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey P., Hegeman A. Enzymatic Reaction Mechanisms. Oxford University Press; Oxford, UK: 2007. pp. 418–430. [Google Scholar]
- 3.Clark D.D., Allen J.R., Ensign S.A. Characterization of five catalytic activities associated with the NADPH:2-Ketopropyl-coenzyme M [2-(2-ketopropylthio)ethanesulfonate] oxidoreductase/carboxylase of the Xanthobacter strain Py2 epoxide carboxylase system. Biochemistry. 2000;39:1294–1304. doi: 10.1021/bi992282p. [DOI] [PubMed] [Google Scholar]
- 4.Rietveld P., Arscott L.D., Berry A., Scritton N.S., Deonarain M.P., Perham R.N., Williams C.H., Jr. Reductive and oxidative half-reactions of glutathione reductase from Escherichia coli. Biochemistry. 1994;33:13888–13895. doi: 10.1021/bi00250a043. [DOI] [PubMed] [Google Scholar]
- 5.Nocek B., Jang S.B., Jeong M.S., Clark D.D., Ensign S.A., Peters J.W. Structural basis for CO2 fixation by a novel member of the disulfide oxidoreductase family of enzymes, 2-ketopropyl-coenzyme M oxidoreductase/carboxylase. Biochemistry. 2002;41:12907–12913. doi: 10.1021/bi026580p. [DOI] [PubMed] [Google Scholar]
- 6.Pandey A.S., Nocek B., Clark D.D., Ensign S.A., Peters J.W. Mechanistic implications of the structure of the mixed-disulfide intermediate of the disulfide oxidoreductase, 2-ketopropyl-coenzyme M oxidoreductase/carboxylase. Biochemistry. 2006;45:113–120. doi: 10.1021/bi051518o. [DOI] [PubMed] [Google Scholar]
- 7.Deonarain R., Mahendra P., Berry A., Scrutton N.S., Perham R.N. Alternative proton donors/acceptors in the catalytic mechanism of the glutathione reductase of Escherichia coli: The role of histidine-439 and tyrosine-99. Biochemistry. 1989;28:9602–9607. doi: 10.1021/bi00451a008. [DOI] [PubMed] [Google Scholar]
- 8.Streit B.R., Mattice J.R., Prussia G.A., Peters J.W., DuBois J.L. The reactive form of a C-S bond-cleaving, CO2-fixing flavoenzyme. J. Biol. Chem. 2019;294:5137–5145. doi: 10.1074/jbc.RA118.005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prussia G.A., Gauss G.H., Mus F., Conner L., DuBois J.L., Peters J.W. Substitution of a conserved catalytic dyad into 2-KPCC causes loss of carboxylation activity. FEBS Lett. 2016;590:2991–2996. doi: 10.1002/1873-3468.12325. [DOI] [PubMed] [Google Scholar]
- 10.Prussia G.A., Shisler K.A., Zadvornyy O.A., Streit B.R., DuBois J.L., Peters J.W. The unique Phe–His dyad of 2-ketopropyl coenzyme M oxidoreductase/carboxylase selectively promotes carboxylation and S–C bond cleavage. J. Biol. Chem. 2021;297:100961. doi: 10.1016/j.jbc.2021.100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey A.S., Mulder D.W., Ensign S.A., Peters J.W. Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M oxidoreductase/carboxylase. FEBS Lett. 2011;585:459–464. doi: 10.1016/j.febslet.2010.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Kofoed M.A., Wampler D.A., Pandey A.S., Peters J.W., Ensign S.A. Roles of the redox-active disulfide and histidine residues forming a catalytic dyad in reactions catalyzed by 2-ketopropyl coenzyme M oxidoreductase/carboxylase. J. Bacteriol. 2011;193:4904–4913. doi: 10.1128/JB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tokmina-Lukaszewska M., Patterson A., Berry L., Scott L., Balasubramanian N., Bothner B. The role of mass spectrometry in structural studies of flavin-based electron bifurcating enzymes. Front. Microbiol. 2018;9:1–18. doi: 10.3389/fmicb.2018.01397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uetrecht C., Rose R.J., Van Duijn E., Lorenzen K., Heck A.J.R. Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 15.Janes W., Schulz G.E. The binding of the retro-analogue of glutathione disulfide to glutathione reductase. J. Biol. Chem. 1990;265:10443–10445. doi: 10.2210/pdb4gr1/pdb. [DOI] [PubMed] [Google Scholar]
- 16.Fontana A., Zambonin M., Polverino De Laureto P., De Filippis V., Clementi A., Scaramella E. Probing the conformational state of apomyoglobin by limited proteolysis. J. Mol. Biol. 1997;266:223–230. doi: 10.1006/jmbi.1996.0787. [DOI] [PubMed] [Google Scholar]
- 17.Bothner B., Taylor D., Jun B., Lee K.K., Siuzdak G., Schlutz C.P., Johnson J.E. Maturation of a tetravirus capsid alters the dynamic properties and creates a metastable complex. Virology. 2005;334:17–27. doi: 10.1016/j.virol.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Rayaprolu V., Kruse S., Kant R., Venkatakrishnan B., Movahed N., Brooke D., Lins B., Bennett A., Potter T., McKenna R.M., Agbandje-McKenna M., Bothner B. Comparative analysis of adeno-associated virus capsid stability and dynamics. J. Virol. 2013;87:13150–13160. doi: 10.1128/JVI.01415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayaprolu V., Kruse S., Kant R., Movahed N., Brooke D., Bothner B. Fluorometric estimation of viral thermal stability. Bio Protoc. 2014;4 doi: 10.21769/bioprotoc.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 21.Vedadi M., Niesen F.H., Allali-Hassani A., Fedorov O.Y., Finerty P.J., Jr., Wasney G.A., Yeung R., Arrowsmith C., Ball L.J., Berglund H., Hui R., Marsden B.D, Nordlund P., Sundstrom M., Weigelt J., et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright T.A., Stewart J.M., Page R.C., Konkolewicz D. Extraction of thermodynamic parameters of protein unfolding using parallelized differential scanning fluorimetry. J. Phys. Chem. Lett. 2017;8:553–558. doi: 10.1021/acs.jpclett.6b02894. [DOI] [PubMed] [Google Scholar]
- 23.Kopec J., Schneider G. Comparison of fluorescence and light scattering based methods to assess formation and stability of protein-protein complexes. J. Struct. Biol. 2011;175:216–223. doi: 10.1016/j.jsb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Matulis D., Kranz J.K., Salemme F.R., Todd M.J. Thermodynamic stability of carbonic anhydrase: Measurements of binding affinity and stoichiometry using thermofluor. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 25.Guarnera E., Tan Z.W., Zheng Z., Berezovsky I.N. AlloSigMA: Allosteric signaling and mutation analysis server. Bioinformatics. 2017;33:3996–3998. doi: 10.1093/bioinformatics/btx430. [DOI] [PubMed] [Google Scholar]
- 26.Guarnera E., Berezovsky I.N. Structure-based statistical mechanical model accounts for the causality and energetics of allosteric communication. PLoS Comput. Biol. 2016;12:1–27. doi: 10.1371/journal.pcbi.1004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurochkin I.V., Guarnera E., Wong J.H., Eisenhaber F., Berezovsky I.N. Toward allosterically increased catalytic activity of insulin-degrading enzyme against amyloid peptides. Biochemistry. 2017;56:228–239. doi: 10.1021/acs.biochem.6b00783. [DOI] [PubMed] [Google Scholar]
- 28.Swain J.F., Gierasch L.M. The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Gunasekaran K., Ma B., Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 30.Tee W.V., Guarnera E., Berezovsky I.N. Reversing allosteric communication: From detecting allosteric sites to inducing and tuning targeted allosteric response. PLoS Comput. Biol. 2018;14:1–26. doi: 10.1371/journal.pcbi.1006228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan Z.W., Guarnera E., Tee W.V., Berezovsky I.N. AlloSigMA 2: Paving the way to designing allosteric effectors and to exploring allosteric effects of mutations. Nucleic Acids Res. 2020;48:W116–W124. doi: 10.1093/nar/gkaa338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motlagh H.N., Wrabl J.O., Li J., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pai E.F., Schulz G.E. The catalytic mechanism of glutathione reductase as derived from x-ray diffraction analyses of reaction intermediates. J. Biol. Chem. 1983;258:1752–1757. [PubMed] [Google Scholar]
- 34.Yang Z.Y., Ledbetter R., Shaw S., Pence N., Tokmina-Lukaszewska M., Eilers B., Guo Q., Pokhrel N., Cash V.L., Dean D.R., Antony E., Peters J.W., Seefeldt L.C. Evidence that the Pi release event is the rate-limiting step in the nitrogenase catalytic cycle. Biochemistry. 2016;55:3625–3635. doi: 10.1021/acs.biochem.6b00421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Potential protease cleavage sites in GR and 2-KPCC.A, structure of GR (PDB: 1GRG) with potential thermolysin cleavage sites in red. B, structure of 2-KPCC (PDB: 1MO9) with potential thermolysin cleavage sites in red. 2-KPCC, 2-ketopropyl coenzyme M oxidoreductase/carboxylase; GR, glutathione reductase.
AlloSigMA control experiment mutating noncatalytically active residues.A, noncatalytically active site residue Leu502 was down mutated in AlloSigMA. Increase in free energy (red) indicates more perturbation and higher dynamics. Decrease in free energy (blue) indicates a stabilization and decrease in dynamics. No change in free energy (gray). B, noncatalytically active site residue Gly377 was down mutated in AlloSigMA. AlloSigMA, Allosteric Signaling and Mutation Analysis.
Change in free energy calculated by AlloSigMA plotted on structure of GR (PDB:1GRG). Increase in free energy (red) indicates more perturbation and higher dynamics. Decrease in free energy (blue) indicates a stabilization and decrease in dynamics. No change in free energy (gray). AlloSigMA, Allosteric Signaling and Mutation Analysis; GR, glutathione reductase.
Data Availability Statement
Data will be shared upon request.