Abstract
The issue of multidrug resistant bacteria is a worldwide health threat. To develop new antibacterial agents with new mechanisms of action is thus an urgent request to address this antibiotic resistance crisis. In the present study, a new thiophenyl-pyrimidine derivative was prepared and utilized as an effective antibacterial agent against Gram-positive strains. In the tests against MRSA and VREs, the compound showed higher antibacterial potency than that of vancomycin and methicillin. The mode of action is probably attributed to the effective inhibition of FtsZ polymerization, GTPase activity, and bacterial cell division, which cause bactericidal effects. The compound could be a potential candidate for further development as an effective antibiotic to combat drug-resistant bacteria.
F20 exhibits strong antibacterial activity through interacting with FtsZ.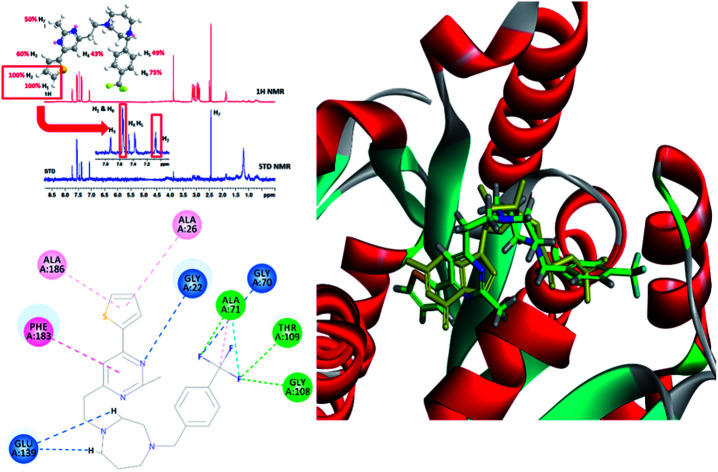
1. Introduction
In the past few decades, antibiotics have made a very significant contribution to the improvement of public health globally. Using antibiotics against bacteria was once considered the ultimate victory in fighting bacterial infections. Unfortunately, due to the improper use of antibiotics, bacteria can develop strong resistance to conventional antibiotics, which means that the battle is endless.1,2 Strong antibacterial resistance reduces the effectiveness for treatment of pathogenic bacterial infections and currently causes serious threats to public health worldwide. For example, both methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VRE) are representative of bacteria resistant to some of the clinical antibiotics currently used,3,4 such as methicillin and vancomycin. To combat the antibiotic resistance crisis, there is an urgent need to develop new antibiotics through the exploration of new chemotypes and targets of inhibition.
As far as current research is concerned, bacterial divisome is considered as a new area to develop new type antibiotics.5 Among the proteins in the divisome, FtsZ is considered as an attractive target for drug discovery since it is highly conserved in the bacterial kingdom. More importantly, FtsZ protein is not found in the mitochondria of higher eukaryotes.6,7 In addition, its biochemical activity is well-studied.6,7 FtsZ is an essential protein in bacterial cell division machinery because both GTP-dependent polymerization at midcell and the followed dynamic circular ring formation at the site of division are regulated by the protein. These biological activities are critically associated many other cell division proteins to produce bacterial daughter cells.8 The functional importance of FtsZ renders it as a good target for the discovery of new antibacterial agents.9
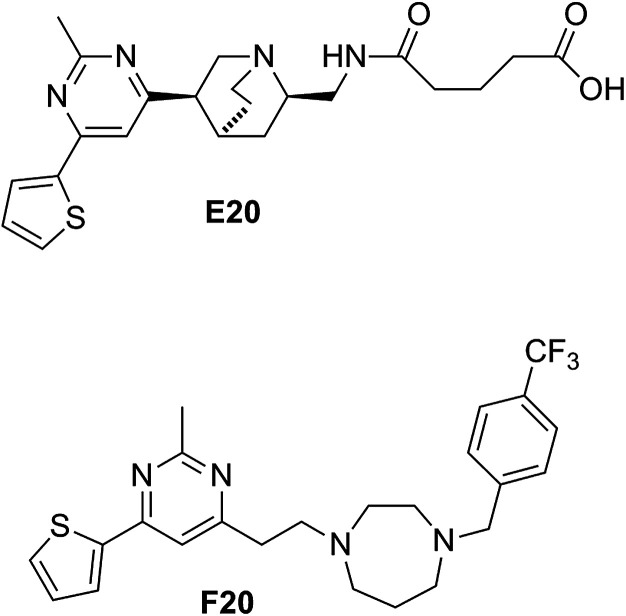
Some compounds have recently reported to inhibit the biological activity of the FtsZ protein and then to block the bacterial cell division.6,7,10,11 Mostly the GTP binding pocket or the H7 helix was found to be the binding target of the compounds. Nevertheless, no compound is able to move forward with a clinical trial. To expand the chemical diversity further with new chemotypes that target at the bacterial cell division may be a better choice for future development. In our previous study, we have identified a FtsZ inhibitor possessing a 2,4-disubstituted-6-thiophenyl-pyrimidine and a chiral amino quinuclidine moiety (E20, Fig. 1) through in silico virtual screening.12 This compound possesses weak inhibitory effects against bacteria and FtsZ protein. Nonetheless, the chiral quinuclidine moiety of the compound is structural complicated and thus impels further structural modification and investigation. In this study, the antibacterial activity of a new 2,4-disubstituted-6-thiophenyl-pyrimidine derivative (F20) shown in Fig. 1 was investigated and also studied its mode of action on FtsZ.
Fig. 1. Chemical structures of E20 and F20.
2. Materials and methods
2.1. Materials
The preparation of F20 was followed the previous report,10 and details of preparation and characterization of F20 can be referred to the ESI.† The pathogenic bacterial stains, Staphylococcus aureus (ATCC 29213, 29247, BAA-41, BAA-1717, BAA-1720, and BAA-1747), Staphylococcus epidermidis (ATCC 12228), Enterococcus faecium (ATCC 49624 and 700221), Enterococcus faecalis (ATCC 29212 and 51575), Escherichia coli (ATCC 25922 and BAA-2469), Klebsiella pneumoniae (ATCC BAA-2108) and Pseudomonas aeruginosa (ATCC BAA-1144) were ordered from American Type Culture Collection (ATCC, USA). Bacillus subtilis strain 168 was already available in our collection. Other materials were purchased from Sigma-Aldrich or Tin Hang Technology Limited.
2.2. Antimicrobial susceptibility assay
The antibacterial activities of F20 against the tested strains were estimated following the micro-dilution methods that are described in the Clinical and Laboratory Standards Institute (CLSI) guidelines.13 Different bacterial strains were cultured with appropriate nutrient broth solution,14 in the absence or presence of various concentration of F20. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of a compound causing no visible growth of bacteria. The minimum bactericidal concentration (MBC) was estimated by spreading 10 μL of bacterial culture from the MIC test onto Trypticase Soy Broth (TSB) agar plate, and MBC was defined the lowest concentration of a compound at which colonies were not observed after incubating at 37 °C for 24 h.13,15 Three independent assays were performed for MIC and MBC tests.
2.3. FtsZ polymerization assay
S. aureus and E. coli FtsZ protein was prepared by following the previously reported procedures.14,16 The polymerization of FtsZ was monitored through a light scattering assay at 37 °C, which was described in the literature.14 Briefly, FtsZ (6 μM) in 20 mM of Tris buffer (pH 7.4, containing 0.01%Triton X-100 to avoid compound aggregation) was placed in a 10 × 2 mm (excitation path) cell, the reaction was started by consecutive additions of 20 mM KCl, 5 mM MgCl2, 1 mM GTP and different concentrations of the test compound. 1% DMSO and 50 μg mL−1 methicillin were tested as vehicle and negative in this assay.
2.4. GTPase activity assay
An ATPase/GTPase Activity Assay Kit (Sigma MAK113) was used to detect the effect of F20 on the GTPase activity of SaFtsZ and EcFtsZ. This assay was performed by following the previously study.17 Briefly, FtsZ (6 μM) was preincubated with vehicle (1% DMSO) or different concentrations of each compound in 20 mM Tris buffer (pH 7.4, containing 0.01%Triton X-100 to avoid compound aggregation) for 10 min at 25 °C. Then 5 mM of MgCl2 and 200 mM of KCl were added. Reactions were started with the addition of 500 mM GTP and incubated at 37 °C. After 30 min, the reactions were quenched by adding 100 mL of Cytophos reagent for 10 min. Inorganic phosphate was quantified by measuring the absorbance at 650 nm with a microplate reader. Three independent tests were performed in this assay and the error bars were calculated for standard derivation.
2.5. Saturation transfer difference (STD) NMR study
The exchangeable protons of FtsZ were subjected to H–D exchange in a 50 mM sodium phosphate buffer, which contained 10% deuterium water, 50 mM NaCl and 5 mM MgCl2 (pH 6.5, uncorrected due to deuterium effects). The STD NMR experiment was performed on a Bruker AvanceIII 600 instrument equipped with a 5 mm QCI cryoprobe. Acquisitions were performed at 298 K using the standard STD pulse sequence with a train of 50 ms Gauss-shaped pulses, each separated by a 1 ms delay for selective protein irradiation, and an alternation between on and off resonances. A T1ρ spinlock filter (50 ms) was incorporated to suppress protein resonances. The NMR spectrum of 20 μM FtsZ and 1 mM F20 (molar ratio 1 : 50) was recorded using the standard pulse sequence. The details can be referred to the previously reported conditions.18 Group epitope mapping was carried out through the integration of the STD signal of the individual protons with respect to the strongest STD signal that assigned as 100%.
2.6. Visualization of bacterial morphology
The B. subtilis 168 or S. aureus ATCC 29213 cells were grown in Luria-Bertani (LB) medium for overnight. The cells were then diluted to approximately 1 × 105 CFU mL−1 in the same medium containing absence or MIC concentration of F20 and incubated at 37 °C for 4 h. The observation of bacterial morphology was carried out under a phase-contrast optical microscope with respect to the previous described conditions.19,20
2.7. Computational study
The molecular docking was conducted by using Discovery Studio (DS) 2016. A X-ray crystal structure of SaFtsZ was obtained from the PDB database (4DXD).21 FtsZ protein and F20 were prepared and minimized for docking using the tools in DS. The docking study was conducted with the DS-CDocker protocol.
2.8. Multistep resistance selection
A resistance selection experiment was performed to determine whether S. aureus could develop resistance to the compound F20.22 The broth microdilution assay was employed to obtain the MIC value of the compound treated to S. aureus ATCC 29213 over 21-passage for 21 days. As reported previously, resistance could be classified when an increase in the initial MIC was found greater than four-fold.23
3. Results
3.1. In vitro antibacterial activity of F20
A number of bacterial strains including some drug-resistant strains were used to examine the antibacterial activity of F20. Methicillin and E20 were examined as the reference compounds under the same conditions. The results showed that F20 can inhibit effectively the growth of the tested Gram-positive strains including MRSA and VRE strains, with the MIC values ranged from 24 μg mL−1 to 48 μg mL−1 (Table 1). When compared to the antibacterial activity of E20,12 the antibacterial potency of F20 is 10-time higher than that of E20. In addition, F20 can inhibit the growth of tested MRSA strains with a MIC value of 24 μg mL−1, which are 10 to 50 folds better than that of methicillin, and is slightly better than that of ampicillin. Moreover, F20 possesses inhibitory effects on the growth of vancomycin-resistant E. faecalis and E. faecium (VREFs) with MIC values of 48 μg mL−1, suggesting that F20 is more potent than that of vancomycin, which cannot inhibit the growth of above-mentioned VREFs at the concentration of 96 μg mL−1 (Table 1).16 For Gram-negative bacteria, F20 shows only moderate antibacterial activity against E. coli and its antibiotics resistant mutant with the MIC values of 96 μg mL−1. However, the compound does not have any effect on the other tested Gram-negative strains like K. pneumoniae and P. aeruginosa, at the concentration of 96 μg mL−1.
MIC of F20 against different bacterial strains.
| Organism | MIC (μg mL−1) | |||
|---|---|---|---|---|
| F20 | E20 | Met | Amp | |
| B. subtilis 168 | 24 | 384 | <1 | <1 |
| S. aureus ATCC 29213 | 24 | 384 | <1 | <1 |
| S. aureus ATCC 29247a | 24 | 384 | 6 | 48 |
| S. aureus ATCC BAA-41b | 24 | 384 | 1024 | 48 |
| S. aureus ATCC BAA-1717b | 24 | N.D. | 512 | 48 |
| S. aureus ATCC BAA-1720b | 24 | N.D. | 1024 | 48 |
| S. aureus ATCC BAA-1747b | 24 | N.D. | 256 | 48 |
| S. epidermidis ATCC 12228 | 24 | 384 | 1 | <1 |
| E. faecium ATCC 49624 | 48 | >384 | 4 | N.D. |
| E. faecium ATCC 700221c | 48 | >384 | 4 | N.D. |
| E. faecalis ATCC 29212 | 48 | >384 | 4 | N.D. |
| E. faecalis ATCC 51575c | 48 | >384 | 4 | N.D. |
| E. coli ATCC 25922 | 96 | 384 | 3 | 3 |
| E. coli ATCC BAA-2469d | 96 | 384 | >1024 | >96 |
| P. aeruginosa ATCC BAA-2108d | >96 | >384 | >256 | >96 |
| K. pneumoniae ATCC BAA-1144d | >96 | >384 | >256 | >96 |
Ampicillin-resistant.
MRSA.
Vancomycin-resistant.
Multidrug-resistant; Met = methicillin; Amp = ampicillin; N.D. = not determine.
3.2. Bactericidal test of F20 against selected bacterial strains
After determined the antibacterial activity of F20, we further investigated whether its antibacterial activity is bactericidal or bacteriostatic. In the test, MBC values of F20 against B. subtilis 168, S. aureus ATCC 29213, MRSA (ATCC BAA-41) and VRE (ATCC 700221) were determined (Table 2). According to the CLSI standards, an MBC/MIC ratio of 1 to 2 is considered indicative of bactericidal behaviour.13 The results show that the MBC/MIC ratios of F20 against the tested strains are equal or less than 2, indicating bactericidal activity.
MBC of F20 against tested bacterial strains (μg mL−1).
| Organism | MBC | MIC | MBC/MIC |
|---|---|---|---|
| B. subtilis 168 | 24 | 24 | 1 |
| S. aureus ATCC 29213 | 48 | 24 | 2 |
| S. aureus ATCC BAA-41 | 24 | 24 | 1 |
| E. faecium ATCC 700221 | 48 | 48 | 1 |
3.3. F20 disrupts FtsZ polymerization
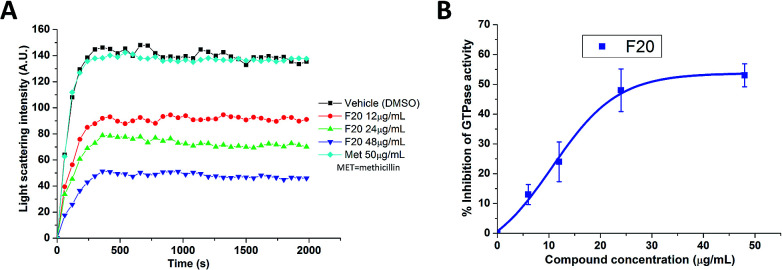
To confirm whether the FtsZ activity could be disturbed by F20, we first assessed the impacts of F20 on FtsZ polymerization dynamics. By using the light scattering in a fluorescence spectrometer, the changes in FtsZ polymerization can be determined by the corresponding absorbance changes at 600 nm. The time-dependent polymerization profiles of SaFtsZ and EcFtsZ in the absence and presence of F20 at a concentration range from 12 to 48 μg mL−1 was shown Fig. 2A and S2,† respectively. The results reveal that F20 inhibit the polymerization of FtsZ in a concentration-dependent manner that is similar to the reported FtsZ inhibitors.17,22,24 50 μg mL−1 methicillin is also examined as a non-FtsZ-targeting control antibiotic in the assay (Fig. 2A). As expected, it shows no effect in the FtsZ polymerization.
Fig. 2. (A) Time-dependent polymerization profiles of SaFtsZ with and without the use of F20. (B) Inhibition of GTPase activity of SaFtsZ by F20 at various concentrations (6, 12, 24 and 48 μg mL−1).
3.4. The inhibition of GTPase activity of FtsZ by F20
Since the dynamic FtsZ polymerization depends on the rate of GTP hydrolysis,25,26 we investigated further the inhibitory effect of F20 on the GTPase activity of SaFtsZ and EcFtsZ. The results indicate that F20 can inhibit the GTPase activity in a dose-dependent manner (Fig. 2B and S3†). For example, in the test of SaFtsZ, 6, 12, 24, and 48 μg mL−1 of F20 inhibited the GTPase activity of SaFtsZ by 13, 24, 48, and 53%, respectively (Fig. 2B). The result suggests that F20 inhibits the rate of GTP hydrolysis, which is probably due to the inhibition of FtsZ polymerization.
3.5. Saturation transfer difference (STD) NMR study of F20 with FtsZ
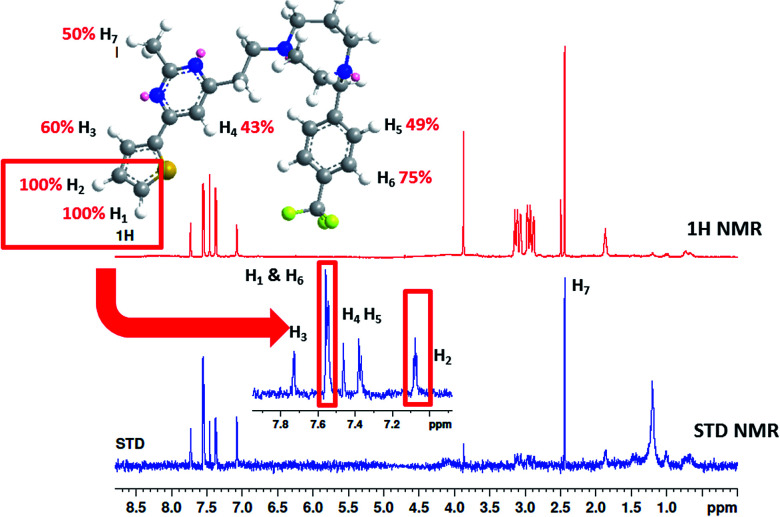
To understand more about the interaction between F20 and FtsZ protein, STD NMR spectroscopy was applied to characterize the binding properties and to identify the epitopes of compounds showing interactions with the receptor of protein. The relative degrees of saturation for individual protons of F20 are shown in Fig. 3 and the integral value of the largest signal is set as 100%. It was found that all of the protons of F20 displayed some degree of enhancement, which demonstrates that the interaction between the molecule and FtsZ has occurred (Fig. 3). The largest amount of saturation transfer was observed for H1 and H2, indicating that the thiophenyl group of the molecule closely contacts with the FtsZ protein.
Fig. 3. STD results of F20 interacting with FtsZ. Upper panel shows the chemical structure and proton assignments of F20; middle panel shows 1D (STD-off) spectrum (red); lower panel shows STD spectrum (blue). STD effects (ISTD/I0) for each proton are reported (upper panel).
3.6. Effect of F20 on the cell division of bacterial cells
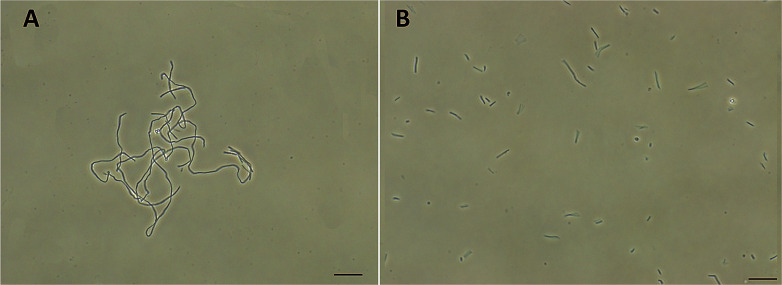
FtsZ inhibitors can disturb FtsZ activity and cause inhibitory effects on bacterial cell division. To investigate the mode of action of the antibacterial activity of F20, we observed the bacterial morphology of B. subtilis and S. aureus through an optical microscope under the conditions incubated with or without the compound. The treatment with F20 to the B. subtilis cells at a sublethal concentration significantly increased the cell length. The average length of the cell was longer than 20 μm (Fig. 4A) as compared with the control that cells treated with DMSO (cell length < 10 μm, Fig. 4B). On the other hand, treating spherical S. aureus with F20 caused the cells enlargement (Fig. S4†). The results obtained are consistent with the reported FtsZ inhibitors such as benzamide and quinolinium derivatives,27,28 suggesting that F20 could disrupt FtsZ activity and cause abnormal cell division.
Fig. 4. (A) Cells of B. subtilis 168 were grown in the presence of 24 μg mL−1 of F20. (B) Cells of B. subtilis 168 were grown in the absence of F20 (DMSO only). Scale bar = 15 μm.
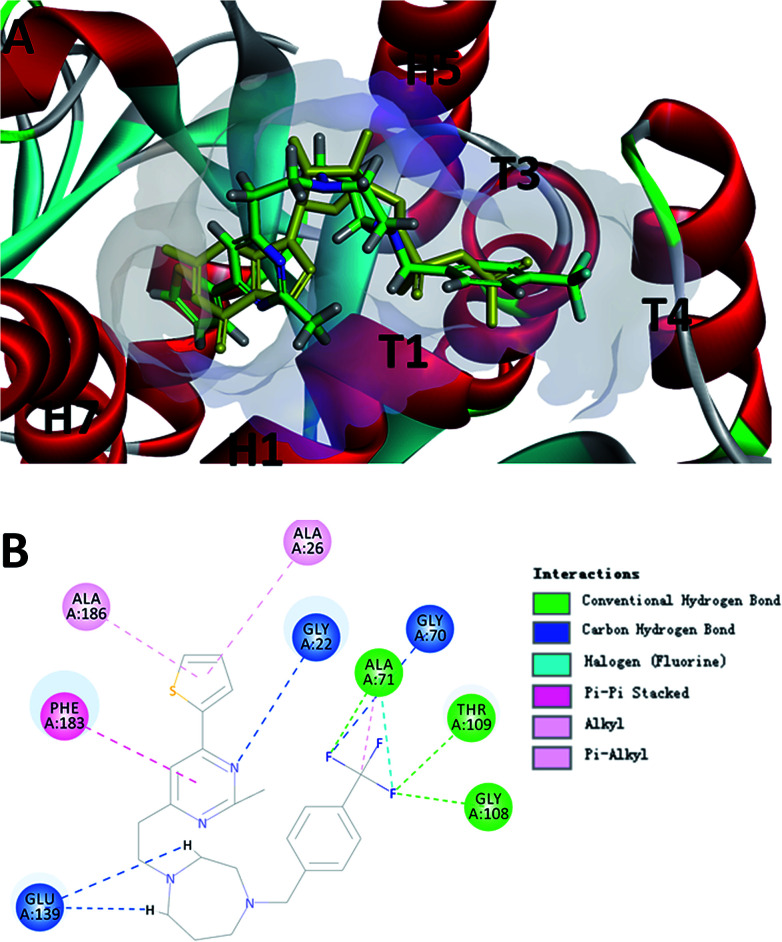
3.7. Computational studies of the binding mode of F20
To probe the potential binding mode of F20 interacted with FtsZ, we also performed molecular modeling study. Ten docking poses were generated in this study and the optimal docking pose, which has the highest score (-CDOCKER energy) was selected inspect visually. The result showed that F20 could bind into the GTP binding site and fully occupied the pocket where GDP is located (Fig. 5A). The 2-methyl-6-thiophenyl-pyrimidine moiety is located in a hydrophobic pocket found by the helices H1 and H7, whereas 4-trifluoromethyl-benzyl linked to the 7-member homo-piperazine cyclic ring that located at the hydrophilic pocket between the glycine rich loops T1, T3 and T4 (Fig. 5A). Three conventional hydrogen bond interactions between the trifluoromethyl group of F20 and the side chains of Ala71, Gly108 and Thr109 were predicted to be established. Moreover, the side chain of Gly22, Gly70 and Glu139 could interact with F20via carbon–hydrogen bond. A π–π stacking interaction can be observed between Phe183 and pyrimidine ring. In addition, amide–π interactions between Ala26/Ala186 and thiophenyl group was found (Fig. 5B). Several amino acids (such as Gly72, Ala73, Gly107, Thr133, Asp187, etc.) in this binding pocket can interact with F20via van der Waals force (data not shown). However, the most stable interactions of the natural GDP with the compound observed are shared five key binding residues including Gly22, Gly108, Thr109, Glu139 and Phe183.21
Fig. 5. (A) The prediction of F20 binds into the GTP binding site of FtsZ (F20: green; GDP: yellow). (B) The predicted interactions from F20 and the amino acids residues of FtsZ.
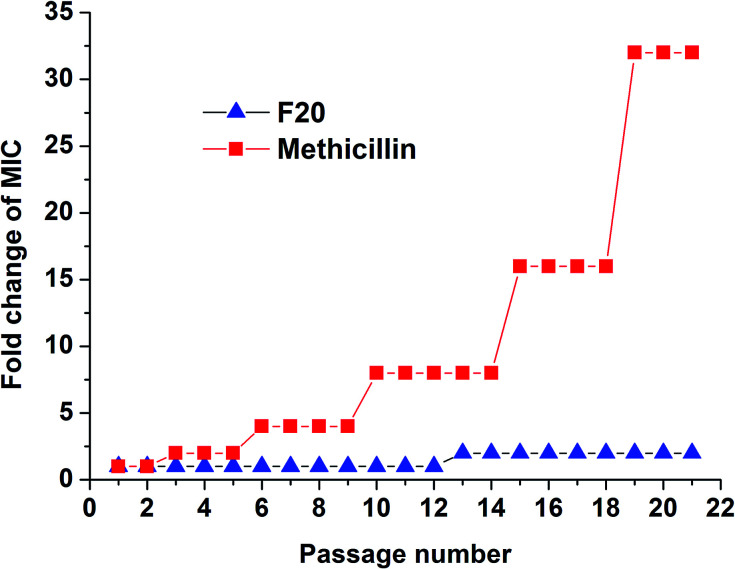
3.8. To investigate whether S. aureus develops resistance to F20
The results of resistance selection experiment on S. aureus ATCC 29213 revealed that the MIC value of F20 increases was found less than 2-fold after 21-passage. However, the MIC value of Methicillin increased 8-fold after 10 passages against S. aureus ATCC 29213 (Fig. 6). These results indicate that F20 seems not readily inducing the development of resistant mutants on S. aureus.
Fig. 6. Multi-step resistance selection of F20 and methicillin against S. aureus ATCC 29213.
4. Discussions
In this study, we showed that a thiophenyl-pyrimidine derivative F20 possesses strong antibacterial bacterial activity against Gram-positive strains with bactericidal effects. The results of biological assays also suggest that the mode of action may due to its inhibitory effect on the FtsZ activity. In the biochemical tests, F20 was found to inhibit the dynamic polymerization of FtsZ in a dose-dependent manner (Fig. 2A and S2†). Similar biological phenomena can also be observed in some FtsZ inhibitors such as cinnamaldehyde and quinoline derivatives.17,24 In addition, F20 is able to reduce the GTPase activity in a dose dependent manner (Fig. 2B and S3†). In the presence of F20 (24 μg mL−1, ∼50 μM), the inhibition of GTP hydrolysis activity was around 50%. Several FtsZ inhibitors were reported to suppress the GTPase activity of FtsZ,6 such as zantrins that inhibit GTP hydrolysis with IC50 values ranged from 20 to 100 μM (ref. 29) and the use of 9-phenoxyalkylberberine derivative at 40 μM was able to achieve 50% inhibition of GTP hydrolysis.16 Similarly, cinnamaldehyde derivative showed 50% inhibition at 30 μg mL−1.24 In our previous study, E20 is able to inhibit the GTPase activity with the IC50 value of 320 μM.12 Thus, the inhibition ability of F20 on GTPase activity is comparable with those of other known FtsZ inhibitors and is stronger than that of E20. Recent study revealed that the treadmilling dynamics of FtsZ was determined by its GTPase activity,25 suggesting that any interference on the GTPase activity of FtsZ may cause the disruption of FtsZ polymerization and cell division inhibition. This may be the reason that some reported FtsZ inhibitors16,24,28,30 cannot inhibit GTPase activity of FtsZ completely, but it can disrupt FtsZ polymerization effectively.
In the antibacterial test, F20 showed a stronger antibacterial activity against Gram-positive strains than Gram-negative strains (Table 1). The poor penetration ability of compound passing through the outer membrane of Gram-negative bacteria could be a possible reason. A similar antibacterial profile can be also found in other FtsZ inhibitors, such as 9-phenoxyalkylberberine and benzamide derivatives.16,27 The result of morphological study suggests that the possible mechanism of antibacterial could be the cell division inhibitory effect induced by F20 (Fig. 4A and S4†). Other inhibitors of FtsZ like quinoline and benzamide derivatives,17,27 can also effectively induce the cell elongation of B. subtilis or enlargement of S. aureus. These results obviously suggested that the inhibition ability of F20 on the GTPase activity resulted in the disruption of the FtsZ polymerization, and then causing the inhibition of cell division and finally resulted bacterial cell death. The findings of STD NMR study further confirm that F20 interacts with FtsZ directly. And its thiophenyl group shows more intimate contacts with FtsZ protein (Fig. 3). This result is in line with our molecular docking study, in which F20 was predicted to bind with the GTP binding site. And the thiophenyl group was predicted to interact with Ala 26 and Ala186 of FtsZ through the hydrophobic interactions (Fig. 5B). We also found that F20 is not readily to cause the development of resistant mutants on S. aureus. A possible reason could be attributed to the significance for recognizing the GTP molecules at the binding site. The mutation of amino acids at this site could also cause improper recognition of GTP and disrupting normal FtsZ activity.
5. Conclusion
In summary, F20 shows significant antibacterial activities against Gram-positive bacteria, which include the drug-resistant bacteria like MRSA and VREF. In addition, the antibacterial mechanism of F20 was studied through different biological assays. The results probably disclose that F20 is able to bind into the GTP binding site, inhibits the activity of GTPase, disrupts FtsZ polymerization and ultimately impairs bacterial cell division to cause cell death. In addition, F20 is not easy to induce the development of resistant mutant on the tested S. aureus. Therefore, the compound could be developed further as a potent and novel antibiotic.
Conflicts of interest
The authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgments
We acknowledge the support from the National Natural Science Foundation of China (81703333), Science and Technology Program of Guangzhou Municipal Health Commission (20192A011018), the Research Grants Council of Hong Kong (Grant No. 15100115), the Innovation and Technology Commission (K-BBX4) and The Hong Kong Polytechnic University (1-BBX8).
Electronic supplementary information (ESI) available: Experimental methods and materials. See DOI: 10.1039/c9ra01001g
Notes and references
- Zhang S. X. Zhou Y. M. Tian L. G. Chen J. X. Tinoco-Torres R. Serrano E. Li S. Z. Chen S. H. Ai L. Chen J. H. Xia S. Lu Y. Lv S. Teng X. J. Xu W. Gu W. P. Gong S. T. Zhou X. N. Geng L. L. Hu W. Infect. Dis. Poverty. 2018;7:53. doi: 10.1186/s40249-018-0427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D. Chem. Biol. 2012;19:3–10. doi: 10.1016/j.chembiol.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Li Y. Yang L. Fu J. Yan M. Chen D. Zhang L. Microb. Pathog. 2017;111:481–486. doi: 10.1016/j.micpath.2017.09.035. [DOI] [PubMed] [Google Scholar]
- Gould I. M. David M. Z. Esposito S. Garau J. Lina G. Mazzei T. Peters G. Int. J. Antimicrob. Agents. 2012;39:96–104. doi: 10.1016/j.ijantimicag.2011.09.028. [DOI] [PubMed] [Google Scholar]
- Sass P. Brotz-Oesterhelt H. Curr. Opin. Microbiol. 2013;16:522–530. doi: 10.1016/j.mib.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Hurley K. A. Santos T. M. Nepomuceno G. M. Huynh V. Shaw J. T. Weibel D. B. J. Med. Chem. 2016;59:6975–6998. doi: 10.1021/acs.jmedchem.5b01098. [DOI] [PubMed] [Google Scholar]
- Haranahalli K. Tong S. Ojima I. Bioorg. Med. Chem. 2016;24:6354–6369. doi: 10.1016/j.bmc.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser D. P. Margolin W. Nat. Rev. Microbiol. 2016;14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Ma S. Eur. J. Med. Chem. 2015;95:1–15. doi: 10.1016/j.ejmech.2015.03.026. [DOI] [PubMed] [Google Scholar]
- Chan K. F. Sun N. Yan S. C. Wong L. K. Lui H. K. Cheung K. C. Yuan J. Chan F. Y. Zheng Z. W. Chan W. C. Chen S. Leung Y. C. Chan T. H. Wong K. Y. ACS Omega. 2017;2:7281–7292. doi: 10.1021/acsomega.7b00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi F. Guo L. Wang Y. Venter H. Semple S. J. Liu F. Ma S. Bioorg. Med. Chem. Lett. 2017;27:958–962. doi: 10.1016/j.bmcl.2016.12.081. [DOI] [PubMed] [Google Scholar]
- Chan F. Y. Sun N. Neves M. A. C. Lam P. C. H. Chung W. H. Wong L. K. Chow H. Y. Ma D. L. Chan P. H. Leung Y. C. Chan T. H. Abagyan R. Wong K. Y. J. Chem. Inf. Model. 2013;53:2131–2140. doi: 10.1021/ci400203f. [DOI] [PubMed] [Google Scholar]
- Wikler M. A., Hindler J. F., Cookerill F. R., Patel J. B., Bush K. and Powell M., Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard, Clinical and Laboratory Standards Institute, 2009, vol. 29, pp. M07–A08 [Google Scholar]
- Sun N. Lu Y. J. Chan F. Y. Du R. L. Zheng Y. Y. Zhang K. So L. Y. Abagyan R. Zhuo C. Leung Y. C. Wong K. Y. Front. Microbiol. 2017;8:855. doi: 10.3389/fmicb.2017.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. Y. Du R. L. Cai S. Y. Liu Z. H. Fang Z. Y. Liu T. So L. Y. Lu Y. J. Sun N. Wong K. Y. Front. Microbiol. 2018;9:1937. doi: 10.3389/fmicb.2018.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N. Chan F.-Y. Lu Y.-J. Neves M. A. C. Lui H.-K. Wang Y. Chow K.-Y. Chan K.-F. Yan S.-C. Leung Y.-C. Abagyan R. Chan T.-H. Wong K.-Y. PLoS One. 2014;9:e97514. doi: 10.1371/journal.pone.0097514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N. Du R. L. Zheng Y. Y. Huang B. H. Guo Q. Zhang R. F. Wong K. Y. Lu Y. J. Eur. J. Med. Chem. 2017;135:1–11. doi: 10.1016/j.ejmech.2017.04.018. [DOI] [PubMed] [Google Scholar]
- Wang Y. Chan F. Y. Sun N. Lui H. K. So P. K. Yan S. C. Chan K. F. Chiou J. Chen S. Abagyan R. Leung Y. C. Wong K. Y. Chem. Biol. Drug Des. 2014;84:685–696. doi: 10.1111/cbdd.12361. [DOI] [PubMed] [Google Scholar]
- Fang Z. Ban L. Li Y. Yuan W. Liu Z. Liu T. Li X. Wong K. Y. Lu Y. Sun N. Yao X. J. Pharmacol. Sci. 2018;137:283–289. doi: 10.1016/j.jphs.2018.07.005. [DOI] [PubMed] [Google Scholar]
- Sun N. Ban L. Li M. Fang Z. Y. Li X. M. Yao W. X. Pan J. H. Lu Y. J. Liu Z. H. Wong W. L. J. Pharmacol. Sci. 2018;138:83–85. doi: 10.1016/j.jphs.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Tan C. M. Therien A. G. Lu J. Lee S. H. Caron A. Gill C. J. Lebeau-Jacob C. Benton-Perdomo L. Monteiro J. M. Pereira P. M. Elsen N. L. Wu J. Deschamps K. Petcu M. Wong S. Daigneault E. Kramer S. Liang L. Z. Maxwell E. Claveau D. Vaillancourt J. Skorey K. Tam J. Wang H. Meredith T. C. Sillaots S. Wang-Jarantow L. Ramtohul Y. Langlois E. Landry F. Reid J. C. Parthasarathy G. Sharma S. Baryshnikova A. Lumb K. J. Pinho M. G. Soisson S. M. Roemer T. Sci. Transl. Med. 2012;4:126ra135. doi: 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- Lui H. K. Gao W. Cheung K. C. Jin W. B. Sun N. Kan J. W. Y. Wong I. L. K. Chiou J. Lin D. Chan E. W. C. Leung Y.-C. Chan T. H. Chen S. Chan K.-F. Wong K.-Y. Eur. J. Med. Chem. 2019;163:95–115. doi: 10.1016/j.ejmech.2018.11.052. [DOI] [PubMed] [Google Scholar]
- Thangamani S. Mohammad H. Abushahba M. F. Hamed M. I. Sobreira T. J. Hedrick V. E. Paul L. N. Seleem M. N. Sci. Rep. 2015;5:16407. doi: 10.1038/srep16407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Sheng J. Huang G. Ma R. Yin F. Song D. Zhao C. Ma S. Eur. J. Med. Chem. 2015;97:32–41. doi: 10.1016/j.ejmech.2015.04.048. [DOI] [PubMed] [Google Scholar]
- Yang X. Lyu Z. Miguel A. McQuillen R. Huang K. C. Xiao J. Science. 2017;355:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho A. W. Hsu Y. P. Squyres G. R. Kuru E. Wu F. Jukes C. Sun Y. Dekker C. Holden S. VanNieuwenhze M. S. Brun Y. V. Garner E. C. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon D. J. Stokes N. R. Ure R. Galbraith G. Bennett J. M. Brown D. R. Baker P. J. Barynin V. V. Rice D. W. Sedelnikova S. E. Heal J. R. Sheridan J. M. Aiwale S. T. Chauhan P. K. Srivastava A. Taneja A. Collins I. Errington J. Czaplewski L. G. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- Sun N. Du R. L. Zheng Y. Y. Guo Q. Cai S. Y. Liu Z. H. Fang Z. Y. Yuan W. C. Liu T. Li X. M. Lu Y. J. Wong K. Y. J. Enzyme Inhib. Med. Chem. 2018;33:879–889. doi: 10.1080/14756366.2018.1465055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit D. N. Romberg L. Mets R. B. Hebert A. M. Mitchison T. J. Kirschner M. W. RayChaudhuri D. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11821–11826. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi F. Song D. Zhang N. Liu Z. Gu X. Hu C. Cai X. Venter H. Ma S. Eur. J. Med. Chem. 2018;159:90–103. doi: 10.1016/j.ejmech.2018.09.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.