Summary
Background
The adipocyte-hypertrophy associated remodeling of fat cell function is considered causal for the development of metabolic disorders. A better understanding of transcriptome and fatty acid (FA) related alterations with adipocyte hypertrophy combined with less-invasive strategies for the detection of the latter can help to increase the prognostic and diagnostic value of adipocyte size and FA composition as markers for metabolic disease.
Methods
To clarify adipocyte-hypertrophy associated transcriptomic alterations, fat cell size was related to RNA-Seq data from white adipose tissue and size-separated adipocytes. The relationship between adipocyte size and adipose tissue FA composition as measured by GC-MS was investigated. MR spectroscopy (MRS) methods for clinical scanning were developed to characterize adipocyte size and FA composition in a fast and non-invasive manner.
Findings
With enlarged adipocyte size, substantial transcriptomic alterations of genes involved in mitochondrial function and FA metabolism were observed. Investigations of these two mechanisms revealed a reciprocal relationship between adipocyte size and estimated thermogenic adipocyte content as well as depot-specific correlations of adipocyte size and FA composition. MRS on a clinical scanner was suitable for the in-parallel assessment of adipose morphology and FA composition.
Interpretation
The current study provides a comprehensive overview of the adipocyte-hypertrophy associated transcriptomic and FA landscape in both subcutaneous and visceral adipose tissue. MRS represents a promising technique to translate the observed mechanistic, structural and functional changes in WAT with adipocyte hypertrophy into a clinical context for an improved phenotyping of WAT in the context of metabolic diseases.
Funding
Competence network for obesity (FKZ 42201GI1128), ERC (No 677661, ProFatMRI; No 875488, FatVirtualBiopsy), Else Kröner-Fresenius-Foundation.
Keywords: White adipose tissue, Browning, Obesity, Transcriptomics, Fatty acids, Magnetic resonance
Research in context.
Evidence before the study
Fat cell size is related to measures of adiposity and obesity-related complications such as insulin resistance and cardiovascular risk factors. Despite adipocyte size being associated with obesity-related complications and markers thereof, the transcriptome-wide alterations with adipocyte-hypertrophy remain not well defined. Notwithstanding the relevance of adipocyte size and fatty acid composition for the development of cardiometabolic diseases, available techniques to assess the described parameters do rely on an invasive biopsy and therefore application in a large-scale and clinical context remains limited.
Added value of the study
Our data provides an in-depth characterization of the transcriptomic landscape associated with increased adipocyte size in a variety of human samples. Adipocyte hypertrophy manifests in the altered expression of genes involved in mitochondrial function and fatty acid metabolism. Increased adipocyte size was accompanied by a reduced predicted thermogenic adipocyte content and depot-specific alterations in fatty acid composition were revealed. Magnetic resonance spectroscopy demonstrated to be a suitable technique to translate the discovered structural and functional alterations with adipocyte-hypertrophy into a fast, non-invasive measurement procedure with potential applicability in large scale studies and routine clinical screenings.
Implications of all the available evidence
Our study sheds light on the transcriptome and fatty acid signatures associated with adipocyte hypertrophy and identifies transcripts and metabolic processes related to adipocyte size. As part of an actionable translational approach magnetic resonance spectroscopy represents a promising tool for the early and preventive detection of the described white adipose tissue phenotypes in a clinical setting.
Alt-text: Unlabelled box
Introduction
A higher body fat percentage and thus an expanded white adipose tissue (WAT) mass is a central hallmark of obesity. An increase in the number of small adipocytes (hyperplasia) is considered as a metabolically less harmful mechanism of WAT growth, than an expansion in volume (hypertrophy) of already existing fat cells which is recognized as a pathological form of WAT remodeling that is associated with adverse cardiometabolic outcomes.1,2 While hypertrophy in visceral adipose tissue (VAT) is associated with dyslipidemia and a pro-inflammatory state, both subcutaneous white adipose tissue (SAT) and VAT hypertrophy can contribute to systemic insulin resistance.3, 4, 5, 6, 7, 8 With progressing obesity, recent studies report a decrease in lipid turnover which can be attributed to elevated rates of triglyceride storage and decreased lipolytic activity.9,10 Additionally, differences in the secretion pattern of pro- and anti-inflammatory adipokines between small and large fat cells derived from the same individual were reported.11 The similarities and differences regarding measurement methods for adipocyte size and the pathophysiological implications of adipocyte hypertrophy have been extensively and systematically reviewed.12 Large-scale studies trying to unravel the causal transcriptional mechanisms behind adipocyte hypertrophy however, remain scarce.13
One of the largest public biobanks that includes both tissue histology together with concordant RNA-Seq data is the Genotype-Tissue expression (GTEx) project.14,15 Since our group previously estimated adipocyte size from over 500 SAT and VAT GTEx samples, we combined adipocyte size and RNA-Seq data from this cohort to investigate the transcriptomic changes that occur between different fat depots and depending on adipocyte hypertrophy.16
Considering the strong association of fatty acid (FA) composition and fat cell size with WAT (dys)-function and metabolic disease there is a need to develop, less invasive, yet highly accurate methods for the characterization of WAT.17 However, the feasibility of the simultaneous characterization of both FA composition and adipocyte size by magnetic resonance imaging (MRI) has not been shown yet and remains unclear. Furthermore, the motion sensitivity of magnetic resonance (MR) diffusion-based adipocyte size estimations appears to be challenging in a clinical context given the required long diffusion times.18,19 Thus, an MR relaxation-based characterization of adipocyte size – which is less sensitive to motion – combined with a chemical shift-based FA decomposition technique is desirable in the clinical context.
Therefore, the present study aimed to provide a comprehensive overview of gene expression changes associated with adipocyte hypertrophy in a variety of human samples. Further, we investigated two of the potential underlying mechanisms, including mitochondrial activity and FA metabolism. As the current study provides evidence for transcriptional, cellular and molecular alterations with adipocyte hypertrophy that could lay the foundation for the development of WAT dysfunction, an MRS-based method for the translational application towards an earlier and non-invasive diagnosis of WAT related cardiometabolic diseases was developed.
Methods
Ethics
All tissue donors gave written informed consent. The study protocols were approved by the ethics committee of the Technical University of Munich (Study №: 5716/13, 1946/07, 409/16s).
Clinical samples
All general study participants’ characteristics, cohorts and related outcome measures are summarized in Table S1.
GTEx
Parts of the data used in this study to investigate the relationship between adipocyte area and WAT transcriptome originate from the “Genotype-Tissue Expression (GTEx)” project.14,15 GTEx SAT samples originate from beneath the leg's skin sample located below the patella on the medial site. VAT was obtained from the greater omentum. Sex and age in 10 year brackets derived from the GTEx publicly available subject phenotypes were used to characterize the donors.
Samples for FA composition and MRS measurements
Larger SAT pieces (∼ 500 cm3) were obtained from abdominoplasty. Paired SAT and VAT samples were collected during general surgery or abdominal laparoscopic surgery. The described samples were used for MRS measurements, histology-based adipocyte sizing and to determine the FA composition by fatty-acid methyl ester (FAME) GC-MS. Collected phenotypic data included sex, age and BMI.
Samples for mature adipocyte isolation
Liposuction material was used to prepare and fractionate mature adipocytes for later isolation of RNA and transcriptomic characterization. Sex, age and BMI were given for the clinical characterization of the samples.
Cell culture
Adipocyte isolation and fractionation
Mature adipocytes were isolated from subcutaneous liposuction material originating from the abdominal region of four female donors based on collagenase-digestion as described previously.20 Briefly, 37.5 ml Krebs-ringer phosphate buffer (KRP) containing 4 % bovine serum albumin (BSA, heat shock fraction, Merck, Cat# A7906) and 100 u/ml collagenase (NB4 standard grade, Nordmark Biochemicals, Cat# S1745402) were added to 12.5 ml of liposuction material and incubated for one hour at 37°C under mild agitation in a water bath. The supernatant was filtered twice through meshes with pore sizes of 2000 µm and 200 µm and adipocytes were washed three times with Krebs-Ringer phosphate (KRP) buffer containing 0.1 % BSA. Mature adipocytes were fractionated based on buoyancy in a separating funnel.11,20 For size separation, 25 ml of mature adipocytes were gently mixed with 50 ml KRP 0.1 % BSA. After 45 s 25 ml were withdrawn from the funnel representing the small adipocyte fraction. The missing volume in the funnel was replaced with KRP 0.1 % BSA and this procedure was repeated 4 times. Similarly, an intermediate fraction was obtained with a flotation time of 20 s. Afterwards, only large cells remained in the funnel which were directly withdrawn.
Adipocyte quality control
All isolated fractions were visually inspected for the absence of lipid droplets and thus excess amounts of ruptured cells. Purity of the individual fractions was assured by direct comparisons of RNA-Seq data of mature adipocytes and isolated/cultured preadipocytes (day 0 undifferentiated & day 14 differentiated) from the stromal vascular fraction of the same donor (Fig. S1). Preadipocyte isolation and culture was performed as described earlier.21
Adipocyte size determination
Computationally derived adipocyte size estimates from histology samples
GTEx adipocyte area estimates originated from an earlier analysis and published method, the Adipocyte U-Net.16 In the initial publication by Glastonbury et al. mean adipocyte area ± SD was calculated for each individual and depot based on 500 unique sampled cells.16 For additional RNA-Seq analysis of the GTEx samples and to factorize adipocyte area individuals were assigned to one of four bins with equally spaced area intervals based on their mean adipocyte area (binsmall, binmedium, binlarge, binX-large). Summary statistics on bin-specific phenotypes and adipocyte areas are given in Table S2.
Histology-based adipocyte sizing of samples used for FA composition measurements and MRS
At least three 5 µm sections with a minimum distance of 100 µm between sections were cut from FFPE WAT. Sections were H&E stained and stitched high-definition range bright-field images at 200x magnification were taken (VHX 6000, Keyence). Adipocyte area was determined using the microscope's in-built image analysis software and diameter was calculated assuming spherical shape (VHX‐6000, Version 3.2.0.121). A lower bound threshold of 200 µm2 and an upper bound threshold of 16,000 µm2 was set to remove artefacts.16 Identified objects were inspected manually and artifacts were excluded from the analysis. Summary statistics on adipocyte area and diameter of the samples that were either used to correlate adipocyte size with FA levels or results derived from MRS are given in Tables S3 & S4, respectively.
Mature adipocytes
To measure mature adipocyte diameter a glass-slide was wetted with 200 µl of PBS and approximately 20 µl of adipocytes were pipetted onto the slide. Stitched, bright-field images in high-definition range were taken and adipocyte diameter was automatically determined using the microscope's software (VHX-6000). Identified objects were inspected manually and artifacts were excluded from the analysis.
RNA isolation, sequencing and differential expression analysis
Mature adipocyte RNA isolation
600 µl of adipocytes were pipetted into microtubes containing 600 µl of RLT (Qiagen, Cat # 79216) 1 % ß-mercaptoethanol (Merck, Cat#M6250), 200 mg of 0.5 mm ceramic beads (Carl-Roth, Cat# 11079105), inverted and immediately snap-frozen. Samples were allowed to thaw on ice, the RLT 1 % ß-Mercaptoethanol solution was removed and replaced with mirVana lysis buffer (Thermo Fisher Scientific, Cat# AM1560). Samples were lysed for 3 × 30 s in a homogenizer filled with dry ice set to 6.0 m/s (FastPrep, MP biomedicals, Cat# 116005500). Samples were centrifuged at 12,000 g for 10 min. at 4°C. Using a fine dosage syringe (B. Braun, Omnican 100, Cat# 9161502S) the adipocyte lysate located underneath the fat layer was transferred to a new tube. All remaining steps were carried out as described by the manufacturer.
RNA quality control of mature adipocytes
RNA concentration and 260/280 ratios from mature adipocytes were photometrically determined (Infinite 200 PRO, Tecan). The quality of isolated RNA was assessed based on RNA integrity number (RIN) obtained by using a Bioanalyzer RNA 6000 Nano chip (Agilent, Cat# 5067-1511). Across all samples, high RNA integrity was observed with an average RIN of 9.0 ± 0.8.
Mature adipocyte RNA sequencing
Library preparation, sequencing, and reference genome alignment of RNA isolated from mature adipocytes was carried out by the genomics services department of the Broad Institute. Reads were aligned to human genome assembly GRCh37 (hg19) using STAR.22 The number of raw read counts was determined using the feature counts function from the Rsubread package.23
GTEx RNA-Seq data
Raw gene read counts and gene transcripts per kilobase million (TPM) were obtained through the GTEx portal from GTEx analysis V8 (dbGap accession phs000424.v8.p2).14,15
Differential expression analysis
All RNA-Seq data was analyzed using edgeR.24, 25, 26 Multidimensional scaling (MDS) plots and dendrograms were generated to visualize clustering of samples dependent on experimental groups. EdgeR's quasi-likelihood pipeline was used to test for differential gene expression.26 Differential expression analysis was used to compare gene expression between GTEx-derived paired SAT and VAT samples (n = 99). A differential expression model with adipocyte size as a categorical variable (binsmall vs. binX-large) was applied to GTEx SAT (nbinsmall = 20, nbinX-large = 28) and VAT samples (nbinsmall = 41, nbinX-large = 11). Additionally, a model relating gene expression to SAT (n =153) or VAT (n = 141) adipocyte size expressed as a continuous variable was computed for the GTEx cohort. P-values from GTEx RNA-Seq analysis were FDR corrected and genes with an FDR < 0.05 and a log2 fold change > 1 were considered as significant.
In another differential expression analysis, the transcriptome of size-separated adipocytes (small fraction vs. large fraction) was compared. P-values from size-separated adipocytes were FDR corrected and genes with an FDR < 0.05 were considered as significant.
Gene set enrichment analysis
Gene set enrichment analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) was carried out using clusterProfiler.27 Data visualization in the form of pathway graphs was achieved using pathview.28
BATLAS analysis
SAT and VAT TPM expression values obtained from the GTEx portal were filtered for the relevant BATLAS marker genes.29 TPM expression values of marker genes that were not expressed in the GTEx cohort were set to zero. Brown and beige adipocyte content estimates per individual and respective WAT depot were obtained from the BATLAS web tools results sheet (https://shiny.hest.ethz.ch/BATLAS/).
Bisque marker analysis
Relative differences of cell-type abundances in GTEx samples, preadipocytes and size-separated adipocytes were estimated using Bisque marker gene-based decomposition (R-package: BisqueRNA: Decomposition of Bulk Expression with Single-Cell Sequencing).30 Marker genes for human cell-type clusters were derived from a recently published single cell atlas of human and mouse white adipose tissue.31 Relative differences in cell-type abundances predicted by Bisque were correlated with GTEx adipocyte area estimates. Group-wise comparisons were used to assess differences in cell type proportions between isolated mature adipocyte fractions and preadipocytes.
Adipose tissue fatty acid composition
Adipose tissue FA composition was determined based on FAME GC-MS in samples from abdominoplasty, general surgery or laparoscopy (nSAT = 22, nVAT = 12).32 10–20 mg of wet adipose tissue were snap-frozen on dry ice immediately after excision in tubes containing 700 mg lysing matrix D (MP biomedicals, Cat# 116913050-CF). Samples were weighed, allowed to thaw on wet ice, and concentration was set to 0.05 mg/µl using equal parts MeOH and water (Merck, Cat# 1060181000). Tissue was lysed using a homogenizer (FastPrep) set to 30 s and 6 m/s. Transesterification was carried out as described earlier and FAMEs were extracted using hexane (Merck, Cat# 1007951000).32,33 GC-MS based total FA analysis was performed as previously published.32 Individual FA species were presented as molar percentages of the total FA profile.
Magnetic resonance spectroscopy
Lipid droplet phantom studies
As a validation strategy and to further elucidate the interplay of lipid droplet size and the degree of FA unsaturation in the formation of MR signals, lipid droplet phantoms were manufactured.34 Preceding emulsification, the FA composition and degree of unsaturation of different vegetable oils were validated using FAME GC-MS. Sunflower seed oil and linseed oil were diluted 1:500 and 1:1000 in 3:1 isooctane isopropyl alcohol (Merck, Cat# 1047171000 & Cat# 1010401000). Transesterification and GC-MS were carried out similarly to WAT samples as described above. Lipid droplet phantoms were manufactured with the following ratios of sunflower and linseed oil: 1:0, 1:2, 2:1, 0:1. All lipid droplet phantoms were produced with a fat content of 80 % and a water content of 20 % closely resembling human WAT composition.35,36 For emulsification and conservation purposes the aqueous phase contained 2 % (v/v) tween 80 (Merck, Cat# P1754) and 0.5 % (m/m) sodium benzoate (Merck, Cat #71300). Emulsification was carried out using a colloid mill (Labor‐Pilot 2000/4, IKA‐Werke) set to 3000, 5000, 8000, and 12,000 rpm to obtain different lipid droplet sizes within the water matrix as described earlier.34,37,38 The generated lipid droplet size spectrum was determined using a laser diffraction particle size analyzer (Mastersizer 2000 with Hydro 2000S dispersing unit, Malvern Instruments).
Adipose tissue sample preparation
All human WAT samples originating from abdominoplasty, general surgery or laparoscopy used for MRS were fixed in 4 % formaldehyde (Histofix, Carl-Roth, Cat# P087) for 24 h and immediately measured afterwards (nSAT Donors = 16, nSAT Samples = 27; nVAT = 5).
Magnetic resonance spectroscopy
Lipid droplet phantoms and WAT were measured using a single-voxel short-TR multi-TI multi-TE (SHORTIE) stimulated echo acquisition mode (STEAM) MRS acquisition scheme (Fig. S5a).39 Both lipid droplet phantoms and WAT samples were measured using equivalent sequence parameters: Inversion time (TI) of 8, 83, 233, 458, 833, 1133 ms, echo time(TE) of 10, 15, 20, 25, 70 ms, mixing time (TM) of 16 ms, minimum repetition time (TR) of 801 ms, τ of 774 ms, 4 phase cycles in 4 averages, 2048 sampling points, spectral bandwidth of 3000 Hz, default voxel-size of 12 × 12 × 12 mm3 and a total scan time of 02:32 min. Spectra were obtained for all combinations of TI and TE. All measurements were performed at room temperature (21±1°C) on a clinical whole-body 3T MRI scanner (Ingenia Elition X, Philips Healthcare, The Netherlands). For signal reception, either the clinical 8-channel small extremity coil or the clinical 8-channel wrist coil was used depending on sample size. The acquired MRS data was processed using SVD-based coil combination followed by simple averaging and zero-order phase correction.40 Signal fitting was carried out using a time domain-based joint-series model fitting routine implemented in MATLAB (R2019b) using NL2SOL.41 The SHORTIE signal as a function of time S(t) was modeled as:
where ϕ represents a common phase term, ρi is the proton density, ωi is the precession frequency, di and gi are the Lorentzian and Gaussian damping factors, and T1,i and T2,i are the relaxation times of the ith frequency component, respectively. TI is the inversion time; TM is the mixing time and TE is the echo time. The proton MR-based FA profile characterization is based on the assumption that all MR-detectable FAs are present in the form of triglycerides and that all MR-detectable signals originate from water and triglycerides. Consequently, a 11-peak-signal-model was fitted with in total 21 degrees of freedom including constraints for a 10-peak-triglyceride-model and relaxation properties (Table S5).42 In course of the parametrization of the triglyceride model, the FA profile characterization results from three triglyceride characteristics: the mean number of double bounds per triglyceride (ndb), the mean number of methylene interrupted double bounds per triglyceride (nmidb) and the mean fatty acid carbon chain length (CL).
Statistical analysis
Data analysis was carried out using R.43 If not specified otherwise all results were given as mean ± SD. According to the sample distribution, variance, and experimental setting (paired vs. unpaired) parametric tests (paired samples t-test, independent samples t-test) or non-parametric tests (Mann-Whitney test, Wilcoxon signed-rank test) were used to test for/against differences between groups. Similarly, Pearson or Spearman correlation analysis was applied to investigate the association between continuous variables. One-sided tests were used within the BATLAS analysis to investigate whether a decrease in brown/beige adipocyte content was observed with larger fat cell size. Bonferroni correction was used to adjust for multiple testing in the FA composition analysis. Across all analyses p-values < 0.05 were considered significant. Linear regression analysis of the MRS data was carried out in python (V3.6.10) using the scipy package (V1.4.1).
Role of funders
The funding bodies were not involved in study design, data collection, and analysis, decision to publish, or manuscript preparation.
Results
SAT and VAT display different gene expression signatures
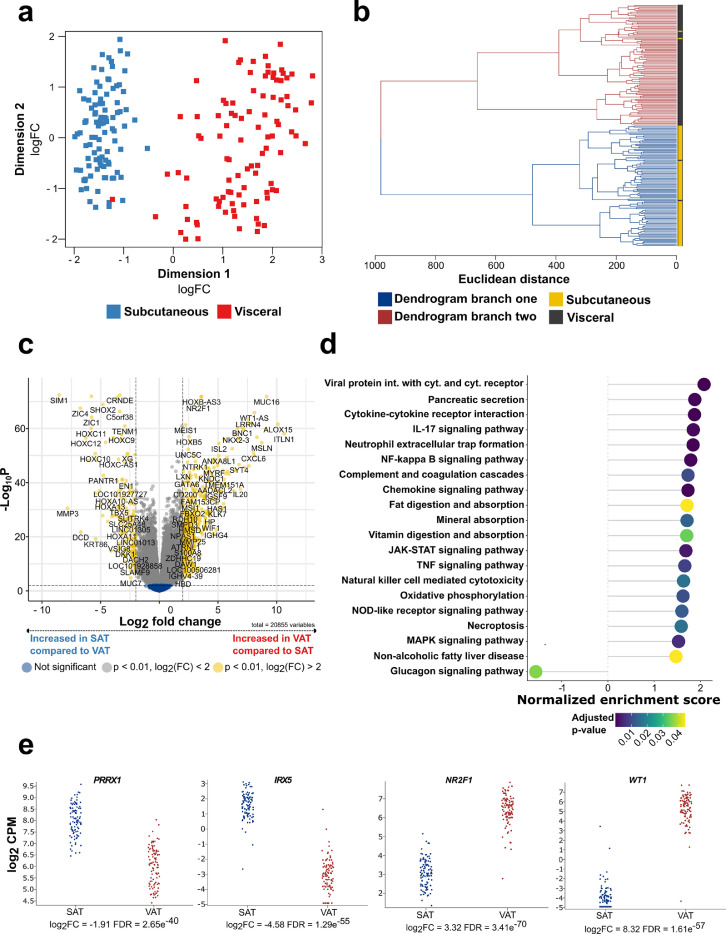
To assess the overall gene expression differences between SAT and VAT, GTEx derived WAT RNA-Seq data was used. As visible by two distinct clusters in the MDS plot and two separate branches in the dendrogram, global SAT and VAT gene expression were clearly distinguishable from each other (npaired = 99, Figure 1a, b). Differential gene expression analysis revealed a total of 689 genes characterized by positive fold changes (FC) and thus higher expression levels in SAT compared to VAT (FDR < 0.05, log2 (FC) > 1; Figure 1c, Table S6a), while 1437 genes were higher expressed in VAT compared to SAT (FDR < 0.05, log2 (FC) > 1; Figure 1c, Table S6a). MMP3 showed strong enrichment in SAT (log2(FC) = -7.82, FDR = 2.71*10−31, Figure 1c). On the contrary, ALOX15 expression was multiple fold higher in VAT compared to SAT and among the top 20 differentially expressed genes in the present analysis (log2(FC) = -10.11, FDR = 2.49*10−62, Figure 1c, Table S6a). KEGG gene set enrichment analysis revealed differences between the two depots revolving around chemokine and cytokine related signaling (hsa04060:Cytokine-cytokine receptor interaction, FDR = 1.63e−06; hsa04062:Chemokine signaling pathway, FDR = 3.44e−04), inflammatory processes (hsa04668:TNF signaling, FDR = 0.0084; hsa04064:NF-kappa b signaling, FDR = 7.51e−04) as well as energy metabolism (hsa00190:Oxidative phosphorylation, FDR = 0.010). An overview of the top 20 enriched KEGG pathways with an adjusted p-value < 0.05 is given in Figure 1d and all significantly enriched KEGG pathways are depicted in Table S6b. A targeted analysis of depot specific marker genes previously reported in literature indicated that SAT/VAT selective genes are expressed in a similar pattern in GTEx samples (Figures 1e). Further, both GTEx SAT and VAT displayed an adipogenic gene signature as shown by the expression of ADIPOQ, LEP, PPARG and FABP4 (Figure S2).
Figure 1.
SAT and VAT are characterized by different transcriptomic signatures
(a) MDS plot displaying the first two dimensions of the data (n = 99). (b) Hierarchical clustering of SAT and VAT samples based on euclidean sample distances (n = 99). (c) Volcano plot showing differentially expressed genes between SAT and VAT (n = 99). (d) Dotplot displaying the top 20 enriched KEGG pathways with an adjusted p-value < 0.05 from gene set enrichment analysis, thereby comparing the expression of KEGG pathway associated genes from SAT and to VAT depots (n =99). Positive scores indicate enrichment of gene-sets in VAT while negative scores indicate enrichment in SAT. (e) Log2 counts per million (CPM) of SAT and VAT lineage specific marker genes (n = 99).
The SAT and VAT transcriptome is altered with adipocyte hypertrophy
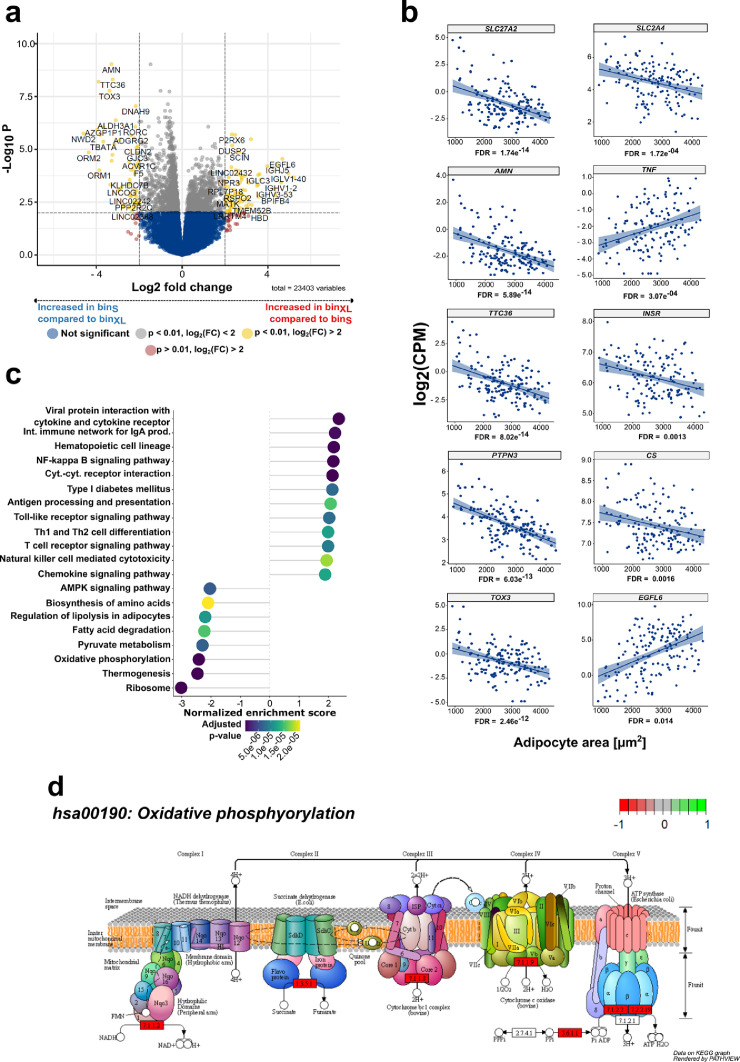
To evaluate the link between adipocyte area and gene expression profiles, individuals from GTEx were stratified into four equally spaced adipocyte area bins.16 A 2.75 fold change in SAT mean adipocyte area was observed between individuals assigned to either binsmall (n = 20, mean adipocyte area = 1,383 ± 241 µm2) or binX-large (n = 28, mean adipocyte area = 3,797 ± 235 µm2) (Table S2). Substantial changes in SAT gene expression were observed when comparing small with X-large adipocytes, including increased expression of 621 genes in binX-large compared to binsmall (FDR < 0.05, log2(FC) > 1; Figure 2a, Table S7a). 369 genes were characterized by higher expression in the small compared to the X-large bin (FDR < 0.05, log2(FC) > 1; Table S7a). Amongst the top-ranked genes, SLC27A2 exhibited significantly higher expression in the group with small adipocytes (FDR = 6.33e−09, log2(FC) = -3.90, Figure 2a, Table S7a).
Figure 2.
Adipocyte area is related to gene expression in SAT
(a) Volcano plot showing differential expression of genes between the binsmall (n = 20) and binX-Large (n = 28) in SAT. (b) Scatterplots displaying the results of the continuous model-based analysis that was applied to GTEx RNA-Seq data from SAT (n = 153). Log2 counts per million (CPM) are plotted against adipocyte area for 153 individuals. FDR is given for each gene below the plot. The left half of the figure shows 5 of the top-ranked genes from the continuous model-based RNA-Seq analysis (SLC27A2, AMN, TTC36, PTPN3, TOX3). On the right half 5 exemplary genes that are already known to be related to WAT biology and fat cell size that were also significant (FDR < 0.05) in our continuous model are depicted (SLC2A4, TNF, INSR, CS, EGFL6). A regression line including the standard error in a lighter shade of blue was added to the plots to visualize the relationship between gene expression and adipocyte area. (c) Dotplot displaying the top 20 enriched KEGG pathways with an adjusted p-value < 0.05 from gene set enrichment analysis in SAT. The “Human Diseases” category of the KEGG pathway database was not displayed on the graph. Positive scores indicate enrichment of gene-sets in binX-Large while negative scores indicate enrichment in binX-Small. (d) KEGG Pathview graph depicting the different complexes (I-V) of the respiratory chain (hsa00190: Oxidative phosphorylation). Rectangular red coloring around the EC number of the respective respiratory chain complex depicts mitochondrial complexes that are underrepresented in SAT from the binX-Large group (n = 28) compared to the binsmall group (n = 20). In contrast, green coloring (+1) displays complexes from the electron transport chain that are overrepresented in SAT from the binX-large group in relation to the binsmall samples.
Further SLC27A2 ranked highest by FDR in a differential expression analysis where SAT adipocyte area was set as a continuous variable (n = 153, FDR = 1.74e−14; Figure 2b, Table S7b). SLC27A1 (alias FATP1), the FA transporter that is mainly expressed in WAT and that has been previously associated with obesity was not found to be differentially expressed concerning SAT adipocyte area (FDRbinned = 0.70, FDRcontinuous = 0.33).44,45 Furthermore, EGFL6 was found to be higher expressed in individuals with larger SAT adipocyte areas (FDR = 2.83e−05, log2(FC) = 4.67, Figure 2a). In the SAT differential gene expression model with adipocyte area as a continuous variable SLC27A2 (FDR = 1.74e−14), AMN (5.89e−14), TTC36 (FDR = 8.02e−14), PTPN3 (FDR = 6.03e−13), and TOX3 (2.46e−12) were found as the top 5 genes by FDR (Figure 2b, left column; Table S7b). An overview of all genes with an FDR < 0.05 overlapping between the binned and continuous model is given in Table S7c and Figure S3a. The present work is consistent and expands on recent studies to link gene expression with obesity, adipogenesis and fat cell size.12,46 Amongst others, negative associations between SAT adipocyte area, SLC2A4 (alias GLUT4, FDR =1.72e−04), INSR (FDR = 0.0013) and CS (FDR = 0.0016) were found, while TNF (3.07e−04) and EGFL6 (0.014) were positively related with adipocyte area (Figure 2b, right column; Table S7b). KEGG gene set analysis in SAT revealed significant positive enrichment of genes involved in immune-related and inflammatory processes for individuals assigned to the binXL group (hsa04640:Hematopoietic cell lineage, FDR =1.05e−07; hsa04060:Cytokine-cytokine receptor interaction, FDR = 5.47e−09; hsa04064:NF-kappa B signaling pathway, FDR = 6.48e−08; Figure 2c; Table S7d). On the contrary, negative enrichment was seen for gene sets involved in key adipose tissue metabolic processes, such as hsa04923:Regulation of lipolysis in adipocytes (FDR = 1.13e−05; Figure 2c; Table S7d) and hsa00620:Pyruvate metabolism (FDR = 6.48e−06; Figure 2b; Table S7d). The analysis further revealed highly significant negative enrichment for hsa03010:Ribosome (FDR = 5.47e−09, Figure 2c; Table S7d) and hsa00190:Oxidative phosphorylation (FDR = 5.47e−09, Figure 2c; Table S7d). Pathway analysis confirmed this finding and revealed a consistent negative enrichment of subunits from the mitochondrial respiratory chain in SAT from individuals with enlarged adipocytes (Figure 2d). Differentially expressed SAT genes based on the continuous model, stratified by sex are given in Table S7f and S7g, respectively.
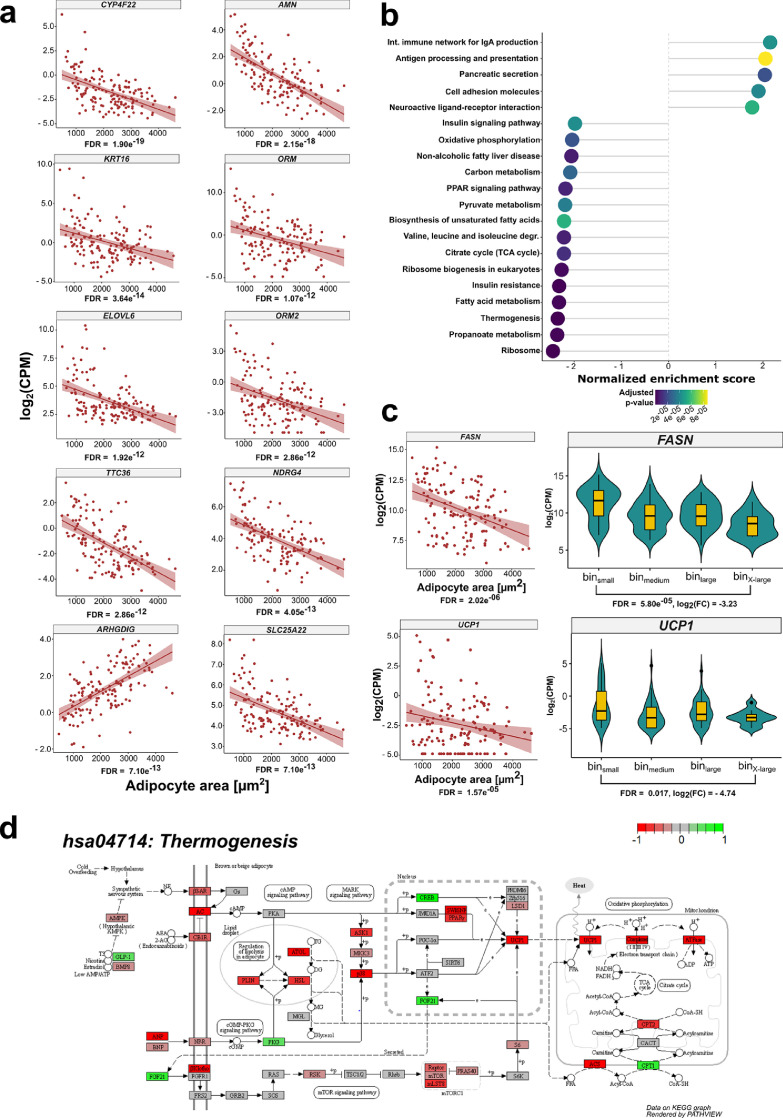
VAT adipocyte area binning resulted in a 3.54 fold change in fat cell size between the smallest and largest bin (Table S2). Differences in VAT adipocyte area were reflected by size-bin specific transcriptomic signatures with 500 genes showing higher expression in binX-large compared to binsmall and 327 genes showing higher expression in binsmall versus binX-Large (FDR < 0.05, log2(FC) > 1 or log2(FC) < -1, respectively; Table S8a). A differential gene expression model with adipocyte area as a continuous variable was applied to account for unequal group sizes and sex distributions. The continuous model revealed a higher number of significant genes with an FDR < 0.05 (continuous: 4,194 genes; categorical: 1,896 genes; Figure S3b, Table S8b) most likely due to higher sample sizes (ncont = 141). Differentially expressed genes from the categorical model were overlapping with the genes from the continuous model with only 208 differentially expressed genes specific to the categorical model (Figure S3b, Table S8c). An overview of ten exemplary genes that were found amongst the top 20 adipocyte size dependent genes in VAT by the continuous model is given in Figure 3a. All differentially expressed genes from the continuous model are listed in Table S8b. With both models a strong negative relationship between ELOVL6 and adipocyte area was found (FDRcontinuous = 1.92e−12, FDRcat = 9.88e−05, log2(FC)cat = -4.21, Figure 3a). As depicted in Figure 3b, KEGG gene set enrichment analysis revealed significant negative enrichment for genes involved in metabolic pathways (hsa00620:Pyruvate metabolism, FDR = 3.66e−05; hsa00020:Citric acid cycle (TCA), FDR = 1.17e−05; hsa01212:Fatty acid metabolism, FDR = 9.70e−07; Table S8d), signaling (hsa03320:PPAR signaling pathways, FDR = 8.80e−06; hsa04910:Insulin signaling, FDR = 4.21e−05; Table S8d) and energy homeostasis (hsa00190:Oxidative phosphorylation, FDR = 2.29e−05; hsa04714:Thermogenesis, FDR = 6.60e−09; Table S8d). In line with findings from KEGG enrichment analysis suggesting that the FA metabolism of individuals with large adipocytes is perturbed and an inverse relationship between adipocyte area and ELOVL6, a negative relationship between adipocyte area and FASN expression (FDRCat = 0.004, log2(FC)Cat = - 3.23; FDRCont = 2.02e−06) was observed in both models (Figure 3c). In agreement with the KEGG pathway analysis, the categorical and the continuous model revealed reduced expression of UCP1 in VAT from individuals with enlarged fat cells (FDRCat = 0.017, log2(FC)Cat = - 4.74; FDRCont = 1.57e−05, Figure 3c). A detailed visualization of the KEGG thermogenesis pathway indicated concordant negative enrichment of genes involved in lipolysis (PNPLA2 alias ATGL, LIPE alias HSL, PLIN), respiration and energy dissipation (UCP1) in VAT from individuals with hypertrophic adipocytes (Figure 3d). Differentially expressed VAT genes based on the continuous model, stratified by sex are given in Table S8f and S8g, respectively. Together, adipocyte hypertrophy in both SAT and VAT was associated with decreased marker gene expression of genes involved in processes crucial for WAT function and homeostasis, i.e. FA metabolism and mitochondrial activity.
Figure 3.
Adipocyte area is related to gene expression in VAT
(a) Scatterplots displaying the results of the continuous model-based analysis that was applied to GTEx RNA-Seq data from VAT. Log2 counts per million (CPM) are plotted against adipocyte area for 141 individuals. FDR is given for each gene below the plot. Displayed is the relationship between adipocyte area and gene expression for 10 exemplary genes that were ranked highest in the analysis according to their FDR. A regression line including the standard error in a lighter shade of red was added to the plots to visualize the relationship between gene expression and adipocyte area. (b) Dotplot displaying the top 20 enriched KEGG pathways with an adjusted p-value < 0.05 from gene set enrichment analysis in VAT (n = 141). The “Human Diseases” category of the KEGG PATHWAY database is not displayed on the graph. (c) Scatterplot and violin plot assessing the relationship between the expression of FASN, UCP1 and adipocyte area (nscatter = 141, nbin small = 41, nbin medium = 49, nbin large = 40, nbin xlarge = 11). (d) KEGG Pathview graph depicting the different complexes and genes involved in the canonical thermogenesis pathway (hsa04714:Thermogenesis). Red (- 1) indicates thermogenesis related genes that are underrepresented in VAT from the binX-Large (n = 11) group compared to the binsmall (n = 40) group. In contrast, green (+1) displays thermogenesis genes that are overrepresented in VAT from the binX-large group in relation to the binsmall samples. Genes depicted in grey (0) did not show differences between the two groups.
Adipocyte size is inversely associated with the thermogenic adipocyte content of SAT and VAT
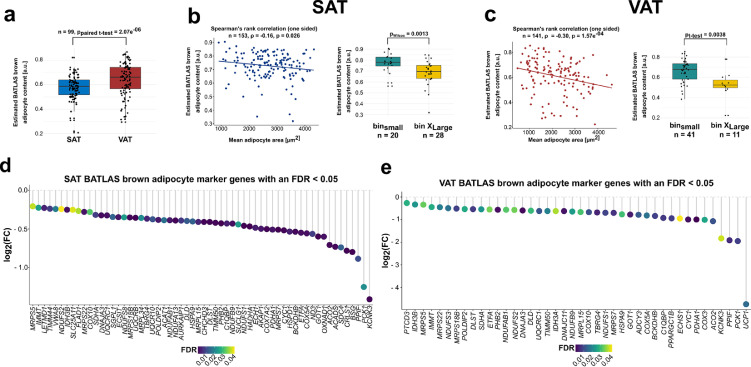
To further elucidate the link between adipocyte area and mitochondrial activity, the brown adipose tissue atlas (BATLAS) deconvolution tool was used to estimate brown and beige adipocyte content in GTEx WAT biopsies as a proxy of thermogenesis and browning capacity.29 BATLAS analysis revealed substantial differences in estimated thermogenic adipocyte content dependent on WAT depot and adipocyte area (Table S9). A 6.13 % higher brown/beige adipocyte content was predicted in paired VAT compared to SAT derived from 99 individuals (p = 2.07e−06, paired t-test, Figure 4a, Table S9). In both VAT and SAT, BATLAS predicted a decrease in brown/beige adipocyte content with hypertrophic (binX-Large) compared to small adipocytes (binsmall). An 8.79 % and 13.57 % reduction in brown/beige adipocyte content was observed for SAT and VAT, respectively (SC: nbinSmall = 20, nbinX-Large = 28, p = 0.0013, one-sided Wilcoxon test; VC: nbinSmall = 41, nbinX-Large = 11, p = 0.0038, one-sided t-test; Figure 4b, c, Table S9). Findings from the binned analysis were confirmed by correlating the estimated brown adipose tissue (BAT) content with adipocyte area across all samples from both adipose depots. In SAT, a negative relationship between brown/beige adipocyte content and adipocyte area was found (n = 153, ρ = -0.16, p = 0.026, one-sided spearman correlation, Figure 4b). Similar results were observed in VAT where adipocyte area was inversely associated with BAT content estimated by BATLAS (n = 141, ρ = -0.3, p = 1.57e−04, one-sided spearman correlation, Figure 4c). A slightly higher overall BAT content and a stronger negative relationship with adipocyte area were seen for VAT. In both depots, multiple of the 98 BAT marker genes published by Perdikari et al.29 were found to be significant with an FDR < 0.05 between the binsmall and binX-Large groups (Figure 4d, e). Among the significant genes, greater fold changes of BAT marker genes were observed in VAT compared to SAT (Figure 4d, e). From all BATLAS BAT marker genes KCNK3 showed the strongest downregulation in SAT from individuals with large adipocytes (Figure 4d; log2(FC) = -1.41, p = 2-54e−04). In VAT, UCP1 showed the strongest downregulation in individuals with large adipocytes (Figure 4e; log2(FC) = -4.74, p =0.017). Similarly, the continuous model indicated a significant negative relationship between UCP1 expression and VAT adipocyte area (FDR = 1.57e−05).
Figure 4.
BATLAS analysis indicates differences in brown adipocyte content with adipocyte hypertrophy
(a) Boxplot comparing the BATLAS estimated brown adipocyte content of paired SAT and VAT biopsies (n = 99, paired t-test). (b) Scatterplot depicting the correlation between estimated brown adipocyte content and adipocyte area in SAT (n = 153, one-sided spearman correlation). Boxplot comparing the estimated brown adipocyte content between individuals with small (binsmall, n = 20) and large subcutaneous adipocytes (binX-large, n= 28) by means of a one-sided wilcoxon test. (c) Scatterplot depicting the correlation between estimated brown adipocyte content and adipocyte area in VAT (n = 141, one-sided spearman correlation). Boxplot comparing the estimated brown adipocyte content between individuals with small (binsmall, n = 41) and large visceral adipocytes (binX-large, n = 11) by means of a one-sided t-test. (d) Dotplot depicting all BATLAS brown adipocyte marker genes that were found to be differentially expressed with a FDR < 0.05 in SAT group-wise comparisons between the binsmall (n = 20) and binX-Large (n = 28) group. (e) Dotplot depicting all BATLAS brown adipocyte marker genes that were found to be differentially expressed with a FDR < 0.05 in VAT group-wise comparisons between the binsmall (n = 41) and binX-Large (n =11) group.
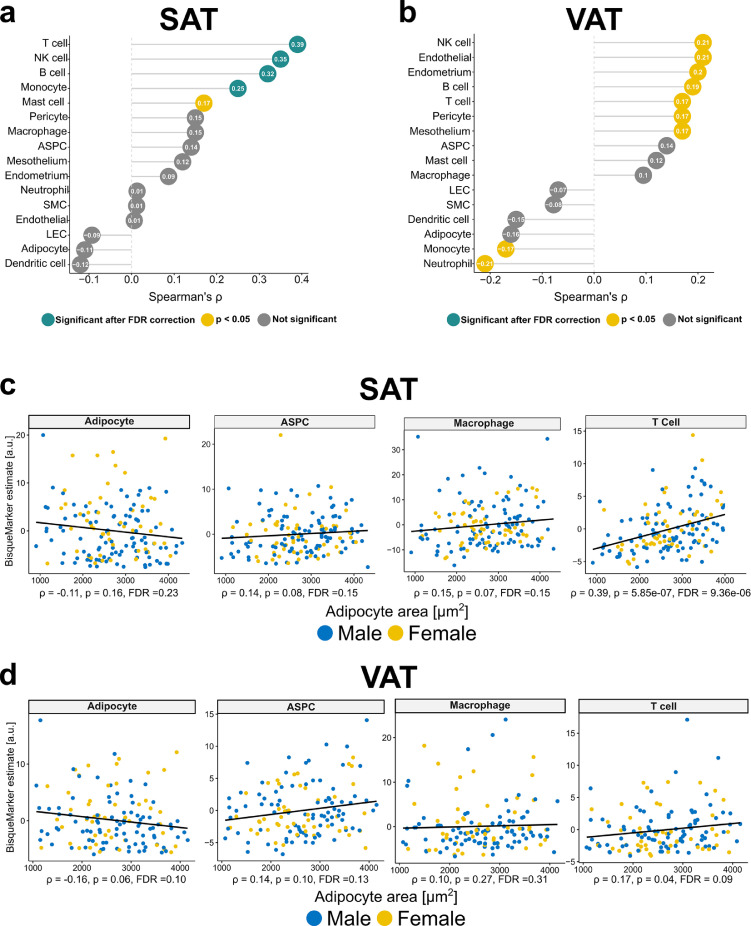
Associations between adipocyte area and predicted relative differences in abundances of cell types
Bisque marker gene based deconvolution in GTEx SAT suggested a positive spearman correlation between adipocyte area and myeloid/lymphoid lines (n = 153, Figure 5a, FDR < 0.05). In VAT no significant associations between adipocyte area and predicted relative differences in abundances of cell type clusters were seen based on spearman correlation after FDR correction (n = 141, Figure 5b). However, more cell types with a p-value < 0.05 were observed compared to the SAT analysis with predictions relating to immune (e.g. NK-cells, B-cells, T-cells), vascular (Endothelial cells, Pericytes) and other cell types (e.g. Mesothelial cells) (Figure 5b). In both depots, a non-significant, negative relationship between predicted relative differences in adipocyte content and adipocyte area was seen (Figure 5c, d). In contrast predicted adipose stem and progenitor cells (ASPCs) as well as macrophages tended to increase with adipocyte area (non-significant, Figure 5c, d). In SAT a strong positive association between adipocyte area and predicted relative differences in T-Cell abundance was seen (Figure 5c). Similarly, T-cells were positively associated with adipocyte area in VAT, but not significant after FDR correction (Figure 5d).
Figure 5.
Predicted relative differences in cell-type abundances in relation to adipocyte area.
(a) Dotplot displaying the spearman correlation between predicted relative differences in cell-type abundances and adipocyte area in SAT (n = 153). (b) Dotplot displaying the spearman correlation between predicted relative differences in cell-type abundances and adipocyte area in VAT (n=141). (c) Scatterplots displaying the spearman correlation between predicted relative differences of adipocytes, ASPCs, Macrophages and T cells with adipocyte area in SAT (n = 153). (d) Scatterplots displaying the spearman correlation between predicted relative differences of adipocytes, ASPCs, Macrophages and T cells with adipocyte area in VAT (n = 141).
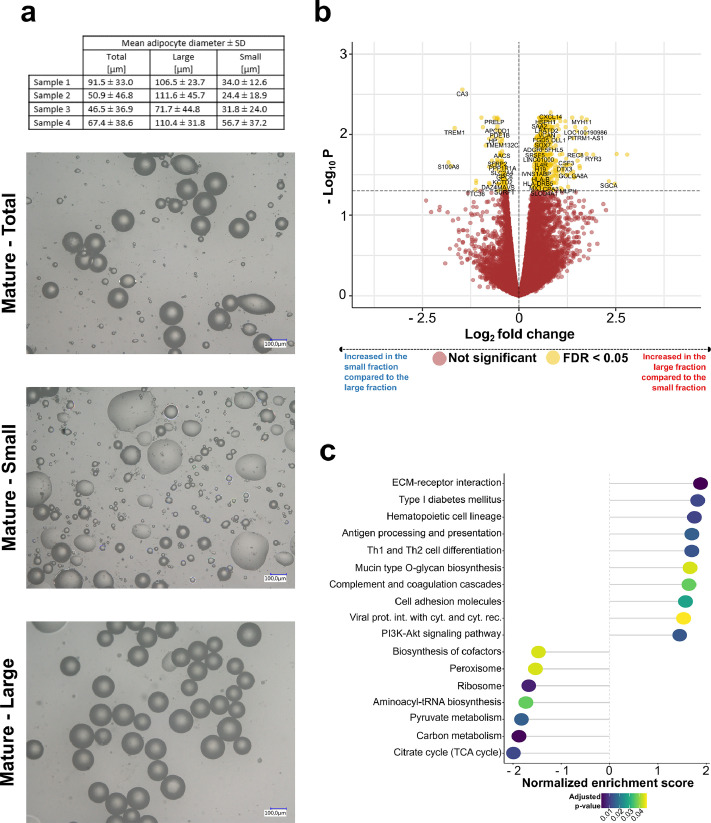
RNA-Seq reveals differences in the transcriptomic signature between small and large adipocytes from the same individual involving FA metabolism and inflammation
Results concerning the investigations on bulk adipose tissue from individuals with diverse genetic backgrounds (GTEx cohort) suggest substantial differences in transcriptomic signatures between individuals with large and small fat cells. To clarify, whether the observed differences in tissue can be explained by the transcriptomic signatures of large adipocytes themselves an investigation was carried out on the intra-individual gene expression patterns of size-separated mature adipocytes. Size separation based on buoyancy resulted in an average size difference of 2.98 fold between the small and large fraction (Figure 6a). A direct comparison with preadipocytes, isolated from the same donors showed clustering of samples with respect to cell type and high expression of adipocyte-specific marker genes in the mature adipocyte samples (Figure S1a, S1b).47 Enrichment and purity of the adipocyte cluster was confirmed by BisqueMarker gene based convolution of the data and contamination with other cell types was not significantly different between adipocyte size fractions (Figure S1c, d) RNA-Seq revealed 583 genes to be differentially expressed (FDR < 0.05) between small and large adipocytes (Table S10a). 480 genes were upregulated in the large fraction, while 103 genes were higher expressed in the small fraction (Figure 6b; Table S10a). SAA2 and TM4SF1, two genes previously identified to be differentially expressed in subcutaneous size-separated adipocytes by Jernas et al., reached significance in our RNA-Seq based analysis, as well (SAA1: log2(FC) = 0.38, FDR = 0.061; SAA2: log2(FC) = 0.53, FDR = 0.007; TM4SF1: log2(FC) = 0.75, FDR = 0.0062).48 KEGG pathway analysis revealed pathways analogous to the cross-sectional GTEx cohort to be enriched in the large (hsa04151:PI3K-Akt signaling, FDR = 0.014; hsa03010:Ribosome, FDR = 0.0049) and small fraction (hsa04512:Carbon metabolism, FDR = 0.0013; hsa00020:Citrate cycle, FDR = 0.011; hsa00620:Pyruvate metabolism, FDR = 0.015), respectively (Figure 6c; Table S10b). In contrast to the GTEx tissue-based analysis, OXPHOS and thermogenesis were not significantly enriched in the KEGG gene set enrichment analysis. Most of the KEGG pathways with an FDR < 0.05 in the adipocyte dataset were also found to be significant in binned GTEx-derived SAT. 12 out of 17 (71 %) KEGG pathways from the size-separated adipocyte dataset reached an FDR < 0.05 in the binned GTEx SAT analysis, too. Further, no major differences in BATLAS predicted thermogenic adipocyte content, most likely due to limited buoyancy/sedimentation of thermogenic cells/precursors and strong enrichment for unilocular mature adipocytes during isolation was observed (data not shown). The evidence for a decreased expression of genes related to respiratory chain function and thermogenesis was therefore limited to the GTEx RNA-Seq analysis assessing inter-individual differences in gene expression regarding adipocyte area.
Figure 6.
The transcriptomic signatures of size-separated mature adipocytes.
(a) Results from mature adipocyte size-separation experiments. Representative images of the total, small and large adipocyte fraction from one donor are shown in the top half of the graph. A tabular overview listing the mean adipocyte diameter ± SD per fraction across all four different donors is given in the bottom half. (b) Volcano plot depicting the DE of genes between the small and large adipocyte fraction (n = 4). (c) Dotplot displaying the top 20 enriched KEGG pathways with an adjusted p-value < 0.05 from gene set enrichment analysis comparing large and small fat cells that were size-separated based on buoyancy (n = 4). The “Human Diseases” category of the KEGG PATHWAY database is not displayed on the graph.
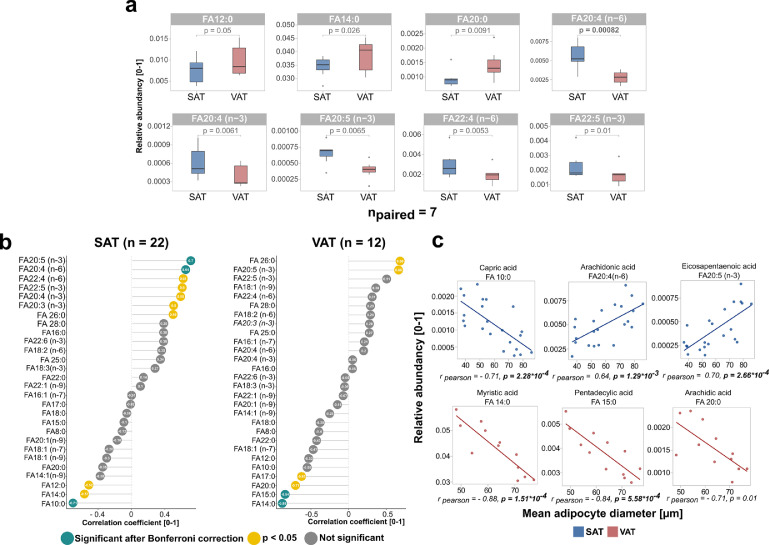
Adipose tissue FA composition is related to adipocyte size
To further understand the link between adipocyte area and FA metabolism that the data revealed, i.e. size-, and depot-dependent expression of de novo lipogenesis (DNL) and FA elongation related marker genes, the relationship between WAT FA composition and adipocyte diameter was investigated (Table S3). Comparisons of the FA composition between SAT and VAT revealed an increased relative abundance of the saturated FA species lauric acid (12:0, p = 0.05, paired t-test), myristic acid (14:0, p = 0.026, paired t-test) and arachidic acid (20:0, p = 0.0091, paired t-test) in VAT (Figure 7a). In contrast, ω-3 and ω-6 PUFAs with chain lengths of 20 or 22 carbon atoms were significantly higher in SAT (Figure 7a). Out of all FA species the ω-6 FA, arachidonic acid (ARA) showed the highest significance (FA20:4 (n-6), p = 0.00082, paired t-test, Figure 7a). Similar to depot-specific FA patterns the results indicate that FA composition is dependent on adipocyte diameter in both depots. In SAT (n = 22) a positive pearson correlation between C20 & C22 PUFAs (Figure 7b) and adipocyte diameter was observed, while an inverse association was revealed predominantly for medium-chain saturated FAs in VAT (n = 12, Figure 7b). Associations between SAT adipocyte diameter and capric acid (FA 10:0, r = -0.71, p = 2.28e−04, pearson correlation), eicosapentaenoic acid (FA20:5 (n-3); r = 0.70, p = 2.66e−04, pearson correlation) and ARA (FA20:4 (n-6); r = 0.70, p = 1.29e−03, pearson correlation) were observed (Figure 7b, c). Inverse relationships of VAT adipocyte diameter with myristic acid (FA14:0, r = -0.88, p = 1.51e−04, pearson correlation), pentadecylic acid (FA15:0, r = -0.84, p = 5.58e−04, pearson correlation) and arachidic acid (FA20:0, r = -0.71, p = 0.01, pearson correlation) were detected (Figure 7b, c).
Figure 7.
Differences in fatty acid composition with regard to adipose depot and fat cell size.
(a) Boxplots showing FA species that were found to show different abundancies in paired, SAT, and VAT with p-values originating from paired t-tests (n = 7). SAT samples are colored in red, while VAT samples are shown in blue. Out of the 28 FA species that were measured all FAs with a p-value < 0.05 are displayed. P-values written in bold are the ones that remained significant after Bonferroni adjustment for multiple testing (pbonf = 0.05/28 = 0.0018). (b) Dotplot showing the pearson correlation between adipocyte diameter and all 28 FA species assessed. SAT samples (n = 22) are displayed on the left, while VAT samples (n = n = 12) are shown on the right. (c) Scatterplots displaying the three FA species showing the strongest association with mean adipocyte diameter as assessed by pearson correlations in SAT (blue, n =22) and VAT (red, n =12).
MRS is suitable for the non-invasive and in-parallel characterization of WAT fat cell size and FA composition
To facilitate the translation of adipocyte size associated changes in FA and transcriptomic signatures into clinical practice an MRS-based technique for the simultaneous characterization of FA composition and adipocyte cell size-based relaxation properties was developed. As previously shown by our group, water-fat emulsions are a valuable model system to generate lipid droplets with highly defined size ranges for the MR-based morphological characterization of lipid-rich samples (water/fat content & droplet size).19,34
Independent of the ratio of sunflower to linseed oil, increasing lipid droplet diameters were observed with decreasing revolutions per minute of the colloid mill (Figure S4a–f, Table S11). At similar rpms a trend towards smaller lipid diameter with increasing linseed oil content was observed (Table S11). As published, linoleic acid (18:2 n-6) was the most abundant FA in sunflower oil (Figure S4g), while linolenic acid (18:3 n-3) was the most frequent FA in linseed oil (Figure S4h). ω-6 FAs (59 %) were predominant in sunflower oil, while linseed oil showed a high ω-3 FA (56 %) content (Table S12).49
The lipid droplet phantom experiment served as a control experiment and indicated that FA characteristics ndb (n = 16, r = 0.762, p < 0.001, pearson correlation) and nmidb (n = 16, r = 0.980, p<0.001, pearson correlation) can be adequately quantified independent from the median lipid droplet size (Figure S5b). The carbon chain length (CL) parameter showed a negative correlation between MRS and GC-MS (n = 16, r = -0.900, p < 0.001, pearson correlation). Furthermore, the lipid droplet phantom experiment revealed that the T2 relaxation of the triglyceride's methylene frequency was highly correlated with the median lipid droplet size (n = 16, r = 0.988, p < 0.001, pearson correlation) independently from the presence of double bonds (Figure S5c). Also, the statistics for T1 relaxation of the triglyceride's methylene frequency vs. median lipid droplet size showed a negative correlation (n = 16, r = -0.845, p < 0.001, pearson correlation). In contrast, the water component exhibited a positive correlation for the T1 relaxation (n = 16, r = 0.711, p < 0.001, pearson correlation) and a negative more likely exponential correlation for the T2 relaxation (n = 16, r = -0.849, p < 0.001, pearson correlation) vs. the median lipid droplet size, respectively.
The in vitro WAT MRS experiment was conducted with samples from abdominoplasty, general surgery, and laparoscopy. Matched MRS, histology and GC-MS data was available for 32 samples originating from 21 donors (Table S4). Similar to the lipid droplet phantom experiment, a positive but weaker correlation between MRS and GC-MS for the FA characteristics ndb (n = 32, r = 0.446, p = 0.011, pearson correlation) and nmidb (n = 32, r = 0.773, p < 0.001, pearson correlation) was found. No correlation between MRS and GC-MS was observed for FA chain length (Figure 8b).
Figure 8.
MRS-based characterization of fatty acid composition and fat cell size in vitro in human adipose tissue samples.
(a) Exemplary MRS spectra with matched histology for two female subjects with similar age (59 and 63 year) and BMI (36 and 34 kg/m2) but difference median fat cell size (49.8 vs.74.4 µm) in abdominal subcutaneous AT. MRS spectra display signal variations with TI (at TE = 10 ms) and TE (TI = 1133 ms), respectively. Fitted methylene T1 and T2 relaxation times are given in green and orange, respectively. (b) Linear regression plots (pearson correlation, n = 32) from the adipose tissue sample experiment: GC-MS-based vs. MRS-based quantification of the FA characteristics ndb, nmidb, CL. Sample origin is indicated by color. (c) Linear regression plots (pearson correlation, n = 32) from the adipose tissue sample experiment: GC-MS-based and MRS-based FA characteristics ndb, nmidb, CL vs. median fat cell size. Sample origin is indicated by color. (d) Linear regression plot (pearson correlation, n = 32) for the methylene T1 and T2 relaxation vs. median fat cell size. Sample ndb (binned) as measured by GC-MS is indicated by color encoding.
The linear regression analysis between the median fat cell size diameter and FA characteristics (Figure 8c) revealed significant moderate correlation for the GC-MS-based ndb (n = 32, r = 0.374, p = 0.035, pearson correlation), nmidb (n = 32, r = 0.512, p = 0.003, pearson correlation) and CL (n = 32, r = 0.419, p = 0.017, pearson correlation), but did not reach significance with the MRS-based ndb (n = 32, r = 0.192, p = 0.292, pearson correlation), nmidb (n = 32, r = 0.203, p = 0.265, pearson correlation) and CL (n = 32, r = 0.192, p=0.292, pearson correlation). In contrast, the median fat cell diameter significantly correlated with both T1 (n = 32, r = 0.484, p = 0.005, pearson correlation) and T2 relaxation (n = 32, r = 0.472, p = 0.006, pearson correlation) of the methylene frequency (Figure 8d).
The histology matched MRS example (Figure 8a) of SAT in comparison from two female donors with comparable age (59 and 63 years) and BMI (36 and 34 kg/m2) revealed between the subjects a larger median fat cell size diameter (49.8 vs. 74.4 µm) together with a longer T1 (225 vs. 244 ms) and T2 relaxation time (51 vs. 57 ms) of the methylene frequency.
Discussion
The depot-specific hypertrophic expansion of adipocytes has been identified as detrimental for the manifestation of metabolic disorders. Understanding the transcriptomic changes occurring with adipocyte hypertrophy and between different WAT depots is therefore of great relevance to identify the causal mechanisms underlying this relationship. Further, the translation of experimental findings into clinical practice remains a challenging task since morphology and composition analysis of WAT are laborious and dependent on biopsy-based techniques. To overcome this limitation and to translate the discovered mechanistic alterations accompanying adipocyte hypertrophy the potential of MRS as a biopsy-free method for the in-parallel assessment of adipocyte size and FA composition was explored.
Negative enrichment for KEGG ribosomal and mitochondrial pathways was observed in relation to adipocyte area in both depots. In contrast, immune and inflammatory pathways i.e. NF-κB signaling were positively enriched and the expression of pro-inflammatory marker genes (TNF-α & IL-6) was increased in individuals with large adipocytes. Mitochondrial dysfunction due to a reduced expression of respiratory chain genes could lead to elevated levels of ROS production impacting the endocrine and metabolic functions of the tissue.50 Together, the identified adipocyte-size associated gene expression patterns provide a transcriptomic foundation for the manifestation of obesity and its related metabolic disorders. In line with findings suggesting that fat distribution and adipocyte size are sexually dimorphic traits our data indicates that the relationship between gene expression and adipocyte size differs between males and females.16,51 This is most likely due to a complex, sex-specific interplay of genetic, epigenetic and hormonal differences.51 Limitations to the GTEx cohort should be acknowledged. Approximately 2/3 of the used GTEx data stems from males and differences in age and sex were observed with increasing adipocyte size. Further, comorbidities and causes of death differ between individuals. The validation of identified transcripts by means of independent cohorts and functional studies should therefore be part of future research. However, analysis was performed with a large number of individuals (n > 140) in a unique dataset that combines both transcriptomic data and histology-derived morphological information of SAT and VAT.
To rule out possible confounding effects of the stromal vascular fraction and different genetic backgrounds mature adipocytes were separated based on size and their transcriptome was characterized. Our RNA-Seq based approach confirms in an independent cohort the differential expression of SAA and TM4SF1 in different adipocyte size fraction that was previously reported by Jernas et al. based on qPCR validated microarray data.48 Together, pathways and genes that were significant in size-separated adipocytes imply that large adipocytes display a metabolically more harmful expression profile. Most of the size-separated adipocyte KEGG pathways with an FDR < 0.05 were also significant in the binned SAT analysis. It is therefore concluded that the detrimental transcriptomic profile of WAT from individuals with large fat cells is at least partially caused by enlarged mature adipocytes themselves. While multiple similar pathways were found, a large number of pathways was exclusively significant in the tissue dataset indicating alterations of other WAT-resident cells or non-adipocyte related pathways with hypertrophy. On the individual gene level, findings on size separated adipocytes are in good agreement with results from spatial transcriptomics suggesting that mature adipocytes from different size quartiles display distinct transcriptomic signatures.13 Less genes were differentially expressed between size-separated mature adipocytes when compared to the GTEx-derived differential expression analysis assessing the relationship between adipocyte size and gene expression in SAT. While different size fractions from a limited number of four individuals were compared, the bulk WAT analysis was conducted in more than 140 individuals. Further, in WAT next to mature adipocytes other cell types can contribute to gene expression.52 It should be acknowledged that other cell types (e.g. immune & endothelial) were present in isolated mature adipocytes at higher proportions compared to cultured and differentiated preadipocytes from the same donors on day 0 and day 14 (Suppl. Figure S1). This effect was however independent of the size fraction and can be explained by the fact that adipocytes are used immediately after isolation, while preadipocytes are plated, washed multiple times, undergo multiple proliferation cycles, and most importantly are enriched by highly selective cell culture media. Besides enrichment for a specific cell type (mature adipocytes) discrepancies in number and type of differentially expressed genes between the two datasets could also originate from different anatomical SAT sampling sites. A study assessing the transcriptome of 15 different adipose depots, comes to the conclusion that fat from the upper leg, upper arm, abdomen, interscapular and supraclavicular region are comparable while buccal, heel and carotid sheath deviated significantly.53 While our data indicates that SAT lineage specific markers and common adipogenic genes are abundantly expressed in GTEX SAT assumptions about similarities and differences between the two SAT sampling sites present in this study remain speculative as no direct comparison was available. Another layer of complexity is added as gene expression of adipocytes is dependent on progenitor lineage, spatial localization within the tissue and proximity to other cell types.13
SAT and VAT browning could display a possible protective mechanism against excessive lipid storage, adipocyte hypertrophy, and long-lasting metabolic complications. Pathway analysis revealed negative enrichment for lipolysis and FA degradation-related genes in large adipocytes from both depots, indicating that hypertrophic adipocytes favor lipid storage over lipid catabolism. In parallel, this phenomenon was accompanied by decreased expression of OXPHOS-related genes, indicating substantial mitochondrial aberrations with adipocyte hypertrophy. Evidence for an inverse relationship between brown/beige adipocyte content and adipocyte size, especially in VAT is provided. Despite large fold changes in UCP1 expression total UCP1 expression was low and accompanied by large individual variance. The BATLAS based deconvolution approach however provides evidence that there is concordant downregulation of numerous genes in the thermogenesis network with adipocyte hypertrophy. As UCP1 is constitutively inactive, the presence of mRNA in our study does not necessarily relate to thermogenic activity.54 A higher thermogenic adipocyte content in individuals with small adipocytes could be protective against excessive lipid accumulation, adipocyte hypertrophy and its associated metabolic complications. The stronger association between adipocyte area and predicted thermogenic adipocyte content in VAT compared to SAT is in good agreement with recent WAT single cell transcriptomics studies reporting the presence of beige-like adipocyte progenitors and thermogenic subpopulations predominantly in VAT.31,55
Bisque marker gene based deconvolution indicated changes in the relative differences in abundances of myeloid and lymphoid cell clusters with adipocyte area that only remained significant in SAT after FDR correction. The described associations are in good agreement with a recent study that reports positive associations between predicted myeloid/lymphoid immune cell proportions and BMI.31 Large parts of the stromal vascular fraction consist of immune cells and proportions are known to change with obesity contributing to a pro-inflammatory environment. Our data on T cells and adipocyte area relate well to studies reporting increased amounts, but also differing T cell subtypes with obesity.55,56 Similarly, T cell associated cytokines such as CCL5 (aka RANTES) are known to be elevated with obesity.57 While the negative association between adipocyte proportions and adipocyte area was not significant in GTEx samples the direction of effect was similar to a previous study that related estimated adipocyte proportions to BMI. Additionally, a positive association between fat cell size and ASPC content was reported by the authors.31 Associations not reaching significance in our dataset could be explained by a smaller number of samples and by the fact that different deconvolution algorithms and approaches were used. Decreasing proportions of mature adipocytes with higher BMI/adipocyte size could favor hypertrophic over hyperplasic expansion. Positive associations between adipocyte precursors and adipocyte area could be explained by the observation that WAT primarily expands through hypertrophy but that hyperplasia can be observed as well, especially with severe obesity.58 If the proportions of adipocyte precursor cells increase with fat cell size or BMI but an opposing trend is seen for mature adipocytes this could allow for the speculation that differentiation capacity of precursor cells is reduced with obesity. This mechanism would be in line with studies suggesting that (a) adipogenesis is crucial for healthy WAT expansion and that (b) hypertrophic obesity does not stem from a limited amount of precursor cells, but lacking commitment and differentiation and capacity of the latter.59,60
Up to date, numerous studies provide evidence for a strong association between hypertrophic WAT expansion and metabolic disease. The findings described here, combined with other recent approaches utilizing (single-cell) omics techniques and different model systems substantiate earlier observational studies and provide sound mechanistic evidence for the involvement of WAT hypertrophy in metabolic diseases. Considering the existing and continuously increasing more granular body of evidence, being able to determine WAT FA composition and adipocyte size in parallel as prognostic biomarkers for metabolic disease using non-invasive diagnostic tools would be highly desirable. Therefore, an MR-based technique for the simultaneous characterization of fat cell size and FA composition was developed to pave the way towards a non-invasive assessment of WAT morphology and composition that could be applicable in a clinical context. The lipid droplet phantom experiment demonstrated the feasibility of the characterization of the FA characteristics as described by ndb and nmidb, as previously reported.42 Beyond what was already known, the experiment empirically demonstrated for the first time the correlation of the methylene T2 relaxation with median lipid droplet size independent from the presence of double bounds in the considered range. Thus, the methylene T2 relaxation can be used as an indirect measure of lipid droplet size. With MR relaxation properties being temperature-dependent it should be acknowledged that in vivo relaxation times at body temperature will differ from the reported relaxation times of excised room temperature WAT samples. Interestingly, SHORTIE MRS-based ndb and nmidb measures correlated well with those from GC-MS, but not with the median adipocyte diameter. While we found a similar trend of increasing methylene T2 relaxation times with increasing lipid droplet diameter in WAT, the significance of the correlation flipped for methylene T1 relaxation showing an increase in T1 with increasing diameter. The differences between the phantom and the WAT sample experiment may be partly explained in consideration of a) the performed GC-MS and MRS are not characterizing the exact same pool of fatty acids and b) structural differences between emulsified lipid droplets and WAT microstructure. The relaxation-based characterization of the median fat cell size in WAT has several advantages compared to approaches based on the probing of restricted diffusion effects including reduced data acquisition times, reduced sensitivity to vibrational artefacts and reduced motion sensitivity.18,19,61 Furthermore, the reported MR parameter relationship pinpoints the possibility of a methodological translation of the proposed measurement framework into a quantitative imaging-based method allowing the spatially resolved probing of multiple AT depots. Thereby, an application towards an unperturbed in-vivo phenotyping of adipocyte size and FA composition is feasible and object of future investigations. Being able to simultaneously probe morphological and compositional parameters of WAT in a non-invasive manner would offer the opportunity to longitudinally monitor adipocyte size and FA composition in various depots of the body. Further, parallel MR-based measurements of FA patterns and adipocyte hypertrophy could serve as a potential diagnostic biomarker to detect early changes in WAT morphology and composition that might occur prior to the systemic manifestation of metabolic diseases. The reported MRS relaxation times could be affected by other tissue properties beyond adipocyte size (e.g. tissue oxygenation).62
The observed switch to an energy storing, pro-inflammatory transcriptome in individuals with large adipocytes, especially in the metabolically more harmful VAT provides important mechanistic insights on the transcriptional background of hypertrophic obesity. The identified reduction in thermogenic adipocyte content in individuals with large fat cells could display a mechanism facilitating hypertrophic WAT expansion and in turn the manifestation of metabolic disorders. Our MR-based proof-of-concept approach bridges from mechanistic insights to translational applicability and could become a promising tool for the non-invasive estimation of FA composition and adipocyte size in-vivo. An earlier and better diagnosis of metabolic derangements based on adipose tissue morphology and FA composition before systemic manifestations occur could substantially contribute to prevent cardiometabolic disorders. The present work is a first step towards this goal but needs confirmation in prospective human studies.
Contributors
Conceptualization: J.H.; S.R.; D.C.K.; M.C.; H.H.
Methodology: J.H.; S.R.; C.S.
Investigation: J.H.; S.R.; C.S.; S.L.; S.S.; P.P.; J.E.
Formal analysis: J.H.; S.R.; C.S.
Data verification: J.H.; S.R.; C.S.; M.C.; D.C.K.; H.H.
Resources: C.N.; C.M.L., U.K.; J.E.; D.C.K; M.C.; H.H.
Writing – original draft: J.H.; S.R.
Writing – review & editing, all co-authors
Supervision: D.C.K.; M.C.; H.H.
Data sharing statement
Results from the transcriptomic and gene set enrichment analysis are included in this article as Supplementary Information (Tables S6–S8, S10). SAT and VAT GTEx RNA-Seq data and phenotypes are publicly available on the GTEx Portal. GTEx SAT and VAT adipocyte area are available through a previous publication from our group (Glastonbury et al.; Plos Comp Biol; 2020; doi.org/10.1371/journal.pcbi.1008044). All remaining data; resources and reagents of this study are available from the corresponding authors upon reasonable request.
Declaration of interests
The authors have no conflicts of interest to disclose. C.M.L. has collaborated with Novo Nordisk and Bayer in research, and under a university agreement, did not accept any personal payment. M.C. has collaborated with Bayer without accepting personal payment. M.C. further serves as a member on the Scientific Advisory Board of Nestle and holds equity in the company Waypoint Bio. D.C.K acknowledges research grant support from Philips Healthcare for a different project outside of the present work.
Acknowledgement
This study was supported in part by a grant from the Kompetenznetz Adipositas (Competence Network for Obesity) funded by the Federal Ministry of Education and Research (FKZ 422 01GI1128) and by the European Research Council (Grant agreement No 677661, ProFatMRI and grant agreement No 875488, FatVirtualBiopsy). This study only reflects the authors’ view and the EU is not responsible for any use that may be made of the information it contains. Parts of the study were funded by the Else Kröner-Fresenius-Foundation, Bad Homburg. S.L. was funded by a Novo Nordisk Postdoctoral Research Fellowship. M.C. received funding from the Next Generation Fund at the Broad Institute of MIT and Harvard. C.M.L. is supported by the Li Ka Shing Foundation, Wellcome Trust, NIH, Gates Foundation, and the National Institute for Health Research Oxford Biomedical Research Centre. The funding bodies were not involved in study design, data collection, and analysis, decision to publish, or manuscript preparation.
We are grateful to Prof. T. Huettl, Dr. U.Schulze-Eilfing, Dr. C. Kleeberger, Dres. Hoffmann, Dr. O. Prokopchuk and their respective teams for obtaining adipose tissue samples. We thank S. Winkler for assisting with WAT sample collection, D. Weidlich for helpful discussions, M. Zamskiy, C. Held and L. Patzelt for helping with the logistics and execution of the MRS measurements.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104020.
Contributor Information
Julius Honecker, Email: julius.honecker@tum.de.
Hans Hauner, Email: hans.hauner@tum.de.
Appendix. Supplementary materials
References
- 1.Morigny P., Boucher J., Arner P., Langin D. Lipid and glucose metabolism in white adipocytes: pathways, dysfunction and therapeutics. Nat Rev Endocrinol. 2021;17(5):276–295. doi: 10.1038/s41574-021-00471-8. [DOI] [PubMed] [Google Scholar]
- 2.Laforest S., Labrecque J., Michaud A., Cianflone K., Tchernof A. Adipocyte size as a determinant of metabolic disease and adipose tissue dysfunction. Crit Rev Clin Lab Sci. 2015;52(6):301–313. doi: 10.3109/10408363.2015.1041582. [DOI] [PubMed] [Google Scholar]
- 3.Tandon P., Wafer R., Minchin J.E.N. Adipose morphology and metabolic disease. J Exp Biol. 2018;221(Pt Suppl 1) doi: 10.1242/jeb.164970. [DOI] [PubMed] [Google Scholar]
- 4.Verboven K., Wouters K., Gaens K., et al. Abdominal subcutaneous and visceral adipocyte size, lipolysis and inflammation relate to insulin resistance in male obese humans. Sci Rep. 2018;8(1):4677. doi: 10.1038/s41598-018-22962-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veilleux A., Caron-Jobin M., Noël S., Laberge P.Y., Tchernof A. Visceral adipocyte hypertrophy is associated with dyslipidemia independent of body composition and fat distribution in women. Diabetes. 2011;60(5):1504–1511. doi: 10.2337/db10-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffstedt J., Arner E., Wahrenberg H., et al. Regional impact of adipose tissue morphology on the metabolic profile in morbid obesity. Diabetologia. 2010;53(12):2496–2503. doi: 10.1007/s00125-010-1889-3. [DOI] [PubMed] [Google Scholar]
- 7.Weyer C., Foley J.E., Bogardus C., Tataranni P.A., Pratley R.E. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia. 2000;43(12):1498–1506. doi: 10.1007/s001250051560. [DOI] [PubMed] [Google Scholar]
- 8.Laforest S., Michaud A., Paris G., et al. Comparative analysis of three human adipocyte size measurement methods and their relevance for cardiometabolic risk. Obesity. 2017;25(1):122–131. doi: 10.1002/oby.21697. (Silver Spring, Md) [DOI] [PubMed] [Google Scholar]
- 9.Rydén M., Andersson D.P., Bernard S., Spalding K., Arner P. Adipocyte triglyceride turnover and lipolysis in lean and overweight subjects. J Lipid Res. 2013;54(10):2909–2913. doi: 10.1194/jlr.M040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arner P., Bernard S., Salehpour M., et al. Dynamics of human adipose lipid turnover in health and metabolic disease. Nature. 2011;478(7367):110–113. doi: 10.1038/nature10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skurk T., Alberti-Huber C., Herder C., Hauner H. Relationship between adipocyte size and adipokine expression and secretion. J Clin Endocrinol Metab. 2007;92(3):1023–1033. doi: 10.1210/jc.2006-1055. [DOI] [PubMed] [Google Scholar]
- 12.Ye R.Z., Richard G., Gévry N., Tchernof A., Carpentier A.C. Fat cell size: measurement methods, pathophysiological origins, and relationships with metabolic dysregulations. Endocr Rev. 2022;43(1):35–60. doi: 10.1210/endrev/bnab018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bäckdahl J., Franzén L., Massier L., et al. Spatial mapping reveals human adipocyte subpopulations with distinct sensitivities to insulin. Cell Metab. 2021;33(9):1869–1882.e6. doi: 10.1016/j.cmet.2021.07.018. [DOI] [PubMed] [Google Scholar]
- 14.Lonsdale J., Thomas J., Salvatore M., et al. The genotype-tissue expression (GTEx) PROJECT. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. (New York, NY) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glastonbury C.A., Pulit S.L., Honecker J., et al. Machine Learning based histology phenotyping to investigate the epidemiologic and genetic basis of adipocyte morphology and cardiometabolic traits. PLoS Comput Biol. 2020;16(8) doi: 10.1371/journal.pcbi.1008044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts R., Hodson L., Dennis A.L., et al. Markers of de novo lipogenesis in adipose tissue: associations with small adipocytes and insulin sensitivity in humans. Diabetologia. 2009;52(5):882. doi: 10.1007/s00125-009-1300-4. [DOI] [PubMed] [Google Scholar]
- 18.Lehnert A., Machann J., Helms G., Claussen C.D., Schick F. Diffusion characteristics of large molecules assessed by proton MRS on a whole-body MR system. Magn Reson Imaging. 2004;22(1):39–46. doi: 10.1016/j.mri.2003.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Weidlich D., Honecker J., Boehm C., et al. Lipid droplet-size mapping in human adipose tissue using a clinical 3T system. Magn Reson Med. 2021;86(3):1256–1270. doi: 10.1002/mrm.28755. [DOI] [PubMed] [Google Scholar]
- 20.Fischer B., Schottl T., Schempp C., et al. Inverse relationship between body mass index and mitochondrial oxidative phosphorylation capacity in human subcutaneous adipocytes. Am J Physiol Endocrinol Metab. 2015;309(4):E380–E387. doi: 10.1152/ajpendo.00524.2014. [DOI] [PubMed] [Google Scholar]
- 21.Sinnott-Armstrong N., Sousa I.S., Laber S., et al. A regulatory variant at 3q21.1 confers an increased pleiotropic risk for hyperglycemia and altered bone mineral density. Cell Metab. 2021;33(3):615–628. doi: 10.1016/j.cmet.2021.01.001. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47(8):e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Lun A.T., Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G., Wang L.G., Han Y., He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics A J Integr Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perdikari A., Leparc G.G., Balaz M., et al. BATLAS: deconvoluting brown adipose tissue. Cell Rep. 2018;25(3) doi: 10.1016/j.celrep.2018.09.044. 784-97.e4. [DOI] [PubMed] [Google Scholar]
- 30.Jew B., Alvarez M., Rahmani E., et al. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat Commun. 2020;11(1):1971. doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emont M.P., Jacobs C., Essene A.L., et al. A single-cell atlas of human and mouse white adipose tissue. Nature. 2022 doi: 10.1038/s41586-022-04518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ecker J., Scherer M., Schmitz G., Liebisch G. A rapid GC-MS method for quantification of positional and geometric isomers of fatty acid methyl esters. J Chromatogr B Anal Technol Biomed Life Sci. 2012;897:98–104. doi: 10.1016/j.jchromb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 33.Lepage G., Roy C.C. Direct transesterification of all classes of lipids in a one-step reaction. J Lipid Res. 1986;27(1):114–120. [PubMed] [Google Scholar]
- 34.Weidlich D., Honecker J., Gmach O., et al. Measuring large lipid droplet sizes by probing restricted lipid diffusion effects with diffusion-weighted MRS at 3T. Magn Reson Med. 2019;81(6):3427–3439. doi: 10.1002/mrm.27651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Commission on Radiological Protection . Pergamon Press; New York: 1975. ICRP Publication 23, Reference Man: Anatomical, Physiological, and Metabolic Characteristics. [Google Scholar]
- 36.Thomas L.W. The chemical composition of adipose tissue of man and mice. Q J Exp Physiol Cogn Med Sci. 1962;47:179–188. doi: 10.1113/expphysiol.1962.sp001589. [DOI] [PubMed] [Google Scholar]
- 37.Gmach O., Bertsch A., Bilke-Krause C., Kulozik U. Impact of oil type and pH value on oil-in-water emulsions stabilized by egg yolk granules. Colloids Surf A. 2019;581 [Google Scholar]
- 38.Guilmineau F., Kulozik U. Influence of a thermal treatment on the functionality of hen's egg yolk in mayonnaise. J Food Eng. 2007;78:648–654. [Google Scholar]
- 39.Ruschke S, Weidlich D, Wu M, Hoch A, Karampinos DC. Single-voxel short-TR multi-TI multi-TE (SHORTIE) STEAM for water–fat magnetic resonance spectroscopy. Proceedings International Society for Magnetic Resonance in Medicine, Montréal, QC, Canada 2019. p. 27(4230).
- 40.Bydder M., Hamilton G., Yokoo T., Sirlin C.B. Optimal phased-array combination for spectroscopy. Magn Reson Imaging. 2008;26(6):847–850. doi: 10.1016/j.mri.2008.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis J.E., Gay D.M., Welsch R.E. Algorithm 573: NL2SOL-an adaptive nonlinear least-squares algorithm [E4] ACM Trans Math Softw. 1981;7(3):369–383. [Google Scholar]
- 42.Hamilton G., Yokoo T., Bydder M., et al. In vivo characterization of the liver fat ¹H MR spectrum. NMR Biomed. 2011;24(7):784–790. doi: 10.1002/nbm.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 44.Anderson C.M., Stahl A. SLC27 fatty acid transport proteins. Mol Aspects Med. 2013;34(2-3):516–528. doi: 10.1016/j.mam.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Q., Ortegon A.M., Tsang B., Doege H., Feingold K.R., Stahl A. FATP1 is an insulin-sensitive fatty acid transporter involved in diet-induced obesity. Mol Cell Biol. 2006;26(9):3455–3467. doi: 10.1128/MCB.26.9.3455-3467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammarstedt A., Graham T.E., Kahn B.B. Adipose tissue dysregulation and reduced insulin sensitivity in non-obese individuals with enlarged abdominal adipose cells. Diabetol Metab Syndr. 2012;4(1):42. doi: 10.1186/1758-5996-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poitou C., Viguerie N., Cancello R., et al. Serum amyloid a: production by human white adipocyte and regulation by obesity and nutrition. Diabetologia. 2005;48(3):519–528. doi: 10.1007/s00125-004-1654-6. [DOI] [PubMed] [Google Scholar]
- 48.Jernas M., Palming J., Sjoholm K., et al. Separation of human adipocytes by size: hypertrophic fat cells display distinct gene expression. FASEB J. 2006;20(9):1540–1542. doi: 10.1096/fj.05-5678fje. [DOI] [PubMed] [Google Scholar]
- 49.Joint FAO/WHO Codex Alimentarius Commission . World Health Organization; 1999. Codex Alimentarius: Standard for Named Vegetable Oils: Food and Agriculture Organization of the United Nations. [Google Scholar]
- 50.Castro J.P., Grune T., Speckmann B. The two faces of reactive oxygen species (ROS) in adipocyte function and dysfunction. Biol Chem. 2016;397(8):709–724. doi: 10.1515/hsz-2015-0305. [DOI] [PubMed] [Google Scholar]
- 51.Fried S.K., Lee M.J., Karastergiou K. Shaping fat distribution: new insights into the molecular determinants of depot- and sex-dependent adipose biology. Obesity. 2015;23(7):1345–1352. doi: 10.1002/oby.21133. (Silver Spring, Md) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosen Evan D., Spiegelman Bruce M. What we talk about when we talk about fat. Cell. 2014;156(1):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schleinitz D., Krause K., Wohland T., et al. Identification of distinct transcriptome signatures of human adipose tissue from fifteen depots. Eur J Hum Genet. 2020;28(12):1714–1725. doi: 10.1038/s41431-020-0681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nedergaard J., Cannon B. UCP1 mRNA does not produce heat. Biochim Biophys Acta (BBA) - Mol Cell Biol Lipids. 2013;1831(5):943–949. doi: 10.1016/j.bbalip.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 55.Vijay J., Gauthier M.F., Biswell R.L., et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat Metab. 2020;2(1):97–109. doi: 10.1038/s42255-019-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H., Ghosh S., Perrard X.D., et al. T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation. 2007;115(8):1029–1038. doi: 10.1161/CIRCULATIONAHA.106.638379. [DOI] [PubMed] [Google Scholar]
- 57.Keophiphath M., Rouault C., Divoux A., Clément K., Lacasa D. CCL5 promotes macrophage recruitment and survival in human adipose tissue. Arterioscler Thromb Vasc Biol. 2010;30(1):39–45. doi: 10.1161/ATVBAHA.109.197442. [DOI] [PubMed] [Google Scholar]
- 58.Arner P., Rydén M. Human white adipose tissue: a highly dynamic metabolic organ. J Intern Med. 2021 doi: 10.1111/joim.13435. [DOI] [PubMed] [Google Scholar]
- 59.Gustafson B., Hammarstedt A., Hedjazifar S., Smith U. Restricted adipogenesis in hypertrophic obesity: the role of WISP2, WNT, and BMP4. Diabetes. 2013;62(9):2997–3004. doi: 10.2337/db13-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vishvanath L., Gupta R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J Clin Invest. 2019;129(10):4022–4031. doi: 10.1172/JCI129191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weidlich D., Zamskiy M., Maeder M., Ruschke S., Marburg S., Karampinos D.C. Reduction of vibration-induced signal loss by matching mechanical vibrational states: Application in high b-value diffusion-weighted MRS. Magn Reson Med. 2020;84(1):39–51. doi: 10.1002/mrm.28128. [DOI] [PubMed] [Google Scholar]
- 62.Morozov D., Quirk J.D., Beeman S.C. Toward noninvasive quantification of adipose tissue oxygenation with MRI. Int J Obes. 2020;44(8):1776–1783. doi: 10.1038/s41366-020-0567-x. (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.