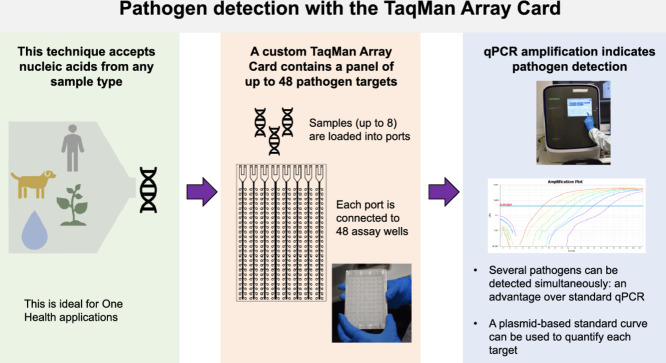
Abstract
Quantitative polymerase chain reaction (qPCR) is a gold standard method for the detection and quantification of pathogenic organisms. Standard qPCR is inexpensive, sensitive and highly specific to the pathogen of interest. While qPCR assays can be multiplexed to allow the detection of multiple organisms in one reaction, it is prohibitively labour intensive to screen large numbers of samples for several pathogens at the same time. The TaqMan Array Card (TAC) is a cost-effective and accurate technique that expands the number of assays that can be simultaneously performed on a sample, with no increase in set-up time and only small reductions in sensitivity. This approach is highly beneficial in settings where there is a need to monitor a large panel of pathogens. We illustrate the application of TAC to the monitoring of gastrointestinal pathogens, which span viral, bacterial, protist and helminth taxa. This protocol outlines the laboratory set-up of a TaqMan Array Card, and some recommended data processing steps to aid in accurate interpretation of the results. A video protocol is additionally provided to assist in the use of the technique.
-
•
The TAC is designed primarily for gene expression assays, but has recently been utilised in several studies for pathogen detection in human clinical samples.
-
•
We expand the use of TAC for pathogen detection across human, animal and environmental sample types, and have developed a protocol and guidelines for the processing and interpretation of results that circumvents issues with the automated outputs.
-
•
This technique is applicable to pathogen or organism detection in any context, if quality nucleic acid extracts can be obtained from the sample type of interest.
Keywords: qPCR, Enteropathogen, One health, Surveillance
Graphical abstract
Specification table
| Subject Area: | Immunology and Microbiology |
| More specific subject area: | Pathogen detection with qPCR arrays |
| Method name: | Customising a TaqMan Array Card for pathogen detection across multiple sample types |
| Name and reference of original method: |
Development of TAC for pathogen detection: Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, et al. Application of TaqMan Low-Density Arrays for Simultaneous Detection of Multiple Respiratory Pathogens. J Clin Microbiol. 2011;49: 2175–2182. doi:10.1128/JCM.02270-10 Original enteropathogen TAC assays and optimisation: Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, et al. A Laboratory-Developed TaqMan Array Card for Simultaneous Detection of 19 Enteropathogens. J Clin Microbiol. 2013;51: 472–480. doi:10.1128/JCM.02658-12 Liu J, Gratz J, Amour C, Nshama R, Walongo T, Maro A, et al. Optimization of Quantitative PCR Methods for Enteropathogen Detection. PLOS ONE. 2016;11: e0158199. doi:10.1371/journal.pone.0158199 |
| Resource availability: |
|
Method details
Background
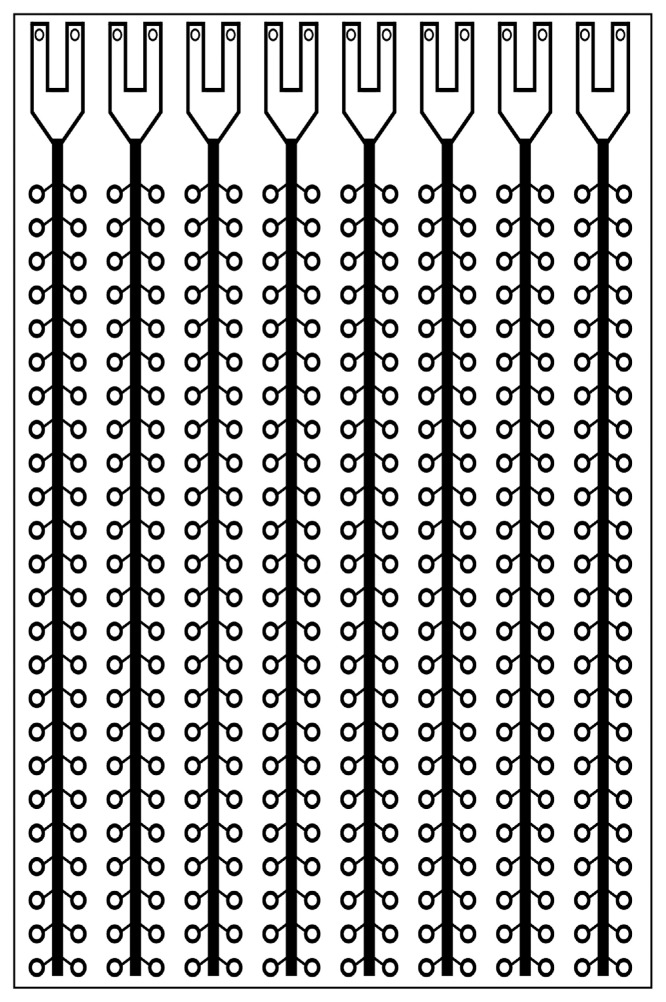
The TaqMan Array Card (TAC), previously known as the TaqMan Low Density Array, is a 384-well microfluidic card that is designed to streamline the process of applying a panel of different quantitative polymerase chain reaction (qPCR) assays to a set of samples. The cards, originally designed to measure mammalian gene expression, have been adapted for pathogen detection [1]. A card (Fig. 1) contains 8 sample loading ports and 48 reaction wells connected to each port, allowing 1-8 samples and 12-384 gene targets to be analysed per card (e.g. 8 samples with 12 assays in quadruplicate, or 4 samples with 32 assays in triplicate, or 8 samples with 48 non-replicated assays, etc.). The card uses TaqMan probe chemistry with the primers and probe for each target manufactured onto the card in lyophilized form when the cards are made to order. To run the card, the user only needs to load combined nucleic acid sample and master mix reagents into the ports on the card, centrifuge it to resuspend the reactions throughout the card, and perform the analysis on a real-time qPCR instrument with an array card block (e.g. a ViiA 7 or QuantStudio 7 real-time PCR system). The card can detect both DNA and RNA targets with the use of a one-step RT-qPCR master mix (which contains both reverse transcriptase and DNA polymerase).
Fig. 1.
Schematic of a TaqMan Array Card. Each of the eight sample loading ports (top) are connected to 48 individual reaction wells with an approximate volume of 1.5 µl. The cards are made to order with lyophilized primers and probes applied to the wells during manufacturing.
TAC is approximately tenfold less sensitive than equivalent individual qPCR assays [2], [3], [4]. However, for applications where there is a need to screen samples for a substantial number of pathogens, this is outweighed by the ability to run 47 custom assays on 8 samples simultaneously. Additionally, the target throughput can be increased if wells are multiplexed. TAC has previously been utilized to detect a range of pathogens involved in gastrointestinal disease [5,6], respiratory illness [4,7], febrile illness [8], meningitis [9,10] and sepsis [11] as well as arrays of drug resistance markers in tuberculosis [12,13], malaria [14], and E. coli [15]. In the context of enteric pathogen detection, multiple viruses, bacteria, protists and helminths contribute to the burden of gastrointestinal disease. Monitoring an array of individual pathogens provides more detailed information on pathogen abundance and transmission than focusing on a few pathogens or relying on faecal indicator organisms to assess environmental contamination [3,16]. TAC enables high throughput for both target numbers and sample numbers, is rapid and straightforward to set up, and is cost-effective per pathogen.
In this protocol, we outline how to use TAC semi-quantitatively to detect pathogens by 1) designing a custom card layout, 2) generating a standard curve, 3) preparing a TAC in the laboratory, and 4) processing the raw data for downstream analysis applications. We focus on enteropathogen detection in a One Health (human, animal and environmental) surveillance context and provide representative results from a custom enteropathogen TAC with DNA extracted from human faecal, animal scat, wastewater, and soil samples. However, the same approach may be used for any panel of gene targets and any sample type from which sufficient nucleic acid can be extracted.
Protocol
Designing a custom card and creating an experiment template
TaqMan Array Cards have traditionally been used for human, mouse and rat gene expression assays. However, a custom card with the user's desired panel of assays can be made to order, which enables researchers to tailor a card design to their targets of interest. The most common card layout is 47 × 8, which allows eight samples to be assayed for 47 custom targets and one compulsory manufacturing control per card. In this protocol, we use the 47 × 8 layout cards and a QuantStudio 7 Flex Instrument with Array Card Block (SKU 4485696).
-
1.
Compile a list of the desired custom qPCR primer and probe sets; for the 47 × 8 card, 47 custom assays can be added to the card in singleplex. Existing assays that use a TaqMan probe with FAM (fluorophore) and MGB (quencher) and have been validated by standard qPCR under TAC reaction conditions are best suited to the card.
NOTE: Each well on the card contains final concentrations of 900 nM for each primer, and 250 nM of probe [17,18].
-
2.
Select the manufacturing control to be included on the card (human GAPDH, or 18S rRNA). The universal 18S rRNA control is suitable for analyses on most sample types.
-
3.
Contact your local ThermoFisher representative to order the custom cards, providing them with the list of custom primer and probe sequences. The cards are manufactured in the USA. Further details of the custom enteropathogen TAC used in this protocol can be found in Lappan et al. (2021) [3]. The primer and probe sequences from each custom assay on this card are reproduced in Supplementary material 1.
NOTE: The catalogue number for the 47 × 8 layout is SKU 4342253.
-
4.Download the QuantStudio software and create an experiment template file.
-
4.1.Select “New Experiment” and in Experiment Properties select your instrument type, “Array Card”, “Relative Standard Curve” “TaqMan Reagents” and “Standard”.
-
4.2.Move to the “Define” window. Select File > Import Plate Setup and import the file provided with your card order (SDS_[production number].txt). This will import the assay IDs and their positions on the plate. Ensure the correct Reporter and Quencher are selected (FAM and NFQ-MGB respectively).
-
4.3.In “Assign”, double check these positions are correct based on the card map provided with your card order.
-
4.4.In “Run Method”, enter the cycling conditions as in Fig. 2. The cycling conditions are as follows: 45°C for 20 minutes, 95°C for 10 minutes, then 45 cycles of 95°C for 15 seconds and 60°C for 1 minute.
-
4.5.Move to the “Export” window. Select the columns of data to export when an experiment is run (in this protocol, we use: Results tab only: Omit, Sample Name, Target Name, CT and Comments).
-
4.6.Go to File > Save as Template to save these settings as an experiment template.
-
4.1.
Fig. 2.
Cycling conditions as seen in the QuantStudio PCR software. The cycling conditions for a TaqMan Array Card, described in Liu et al. (2013)[17], as they should appear in the software. The reverse transcription and initial denaturing steps occur in the “Hold Stage”, and the PCR cycles are specified in the PCR stage. Data capture occurs only at the end of each cycle.
Generating a standard curve
While the TaqMan Array Card is best suited to presence/absence detection, a standard curve may be generated to estimate target abundance in a sample. The most ideal standard curve would contain equal copies of purified genomic DNA or RNA from each pathogen on the card. As this can be impractical, we use a modification of the method proposed by Kodani and Winchell (2012) [19] whereby all primer and probe sequences are synthesized as a plasmid insert. For simplicity, all targets (DNA or RNA) are included in the insert and the plasmid DNA is used directly. This additionally serves as a positive control for all assays on the card.
-
1.Design the plasmid inserts. An example can be found in the supplementary material of Liu et al. (2013) [17]. See also the original plasmid design method [19]. The plasmid insert sequences used for the custom card in this protocol are presented in Supplementary material 1.
-
1.1.The insert sequence consists of a concatenated sequence containing a) the forward primer, b) probe (or reverse complement of the probe, depending on the strand the assay is designed to) and c) reverse complement of the reverse primer, for each assay. For example: Assay 1 forward – Assay 1 probe – Assay 1 reverse – Assay 2 forward – Assay 2 probe – Assay 2 reverse etc.
-
1.2.If the primers contain degenerate nucleotides, use the sequence present in the reference genome.
-
1.3.Limit the insert to approximately 1 kb, designing multiple plasmids as necessary to cover all card targets.
-
1.1.
-
2.
Order the plasmids from a plasmid synthesis company.
-
3.
Resuspend the plasmids as directed by your plasmid synthesis company.
-
4.
Quantify the stock plasmids to calculate the number of copies per microlitre using the total size of the plasmid (e.g. http://scienceprimer.com/copy-number-calculator-for-realtime-pcr)
-
5.
Combine each of your plasmids at equal copy numbers into one solution with a final concentration in the millions for each plasmid (e.g. 7 × 106 copies per microlitre of each plasmid). To do this, you may need to dilute the stock solutions first and then account for the dilution of each plasmid upon mixing all plasmids.
-
6.
Serially dilute this solution tenfold with PCR-grade nuclease-free water to create a seven-point standard curve (from millions of copies per microlitre down to 1-10 copies per microlitre of each plasmid).
-
7.
Run these seven standard solutions plus a no-template control (using the same aliquot of water used to dilute the plasmids) on a TaqMan Array Card following the protocol below. Each card is one replicate of the standard curve. We recommend dedicating three cards to produce a standard curve (triplicate), and generating a new triplicate standard curve for each new batch of cards. Data demonstrating the reproducibility of standard curve replicates for the custom card in this protocol can be found in Supplementary material 2.
NOTE: We recommend creating multiple aliquots of plasmid stocks to reduce the likelihood of contamination.
Laboratory set up of a TaqMan Array Card
This protocol applies to standard curve cards and the analysis of samples. Please see Supplementary material 3 for a video demonstration of this protocol. A no-template control should be run regularly (e.g. every ten cards) to monitor contamination. The TaqMan Array Card should also be set up within a biosafety cabinet to reduce contamination. Further detail on this procedure can be found in the Applied Biosystems TaqMan Array Micro Fluidic Cards User Guide (https://tools.thermofisher.com/content/sfs/manuals/cms_062836.pdf). The reaction mixture and cycling conditions follow those described by Liu et al. (2013) [17].
We recommend pre-aliquoting the master mix reagents for single use. In this protocol, we use the AgPath-ID One-Step RT-PCR Reagents (ThermoFisher, catalogue number 4387391). This 1000-reaction kit supplies enough reagents to run 27 array cards if aliquoted for single use (one card) as follows: 475 µl buffer, 40 µl enzyme and 60 µl nuclease-free water.
-
1.
Wipe the biosafety cabinet thoroughly with 80% ethanol. Place 1000 µl, 200 µl and 20 µl tip boxes, the bottle of nuclease-free water, a 1.5 ml tube rack and nine 1.5 ml microfuge tubes inside the cabinet. Run the UV sterilization procedure for 30 minutes.
-
2.
Take a TaqMan Array Card out of the fridge and allow it to sit at room temperature inside the packaging. The card must have been at room temperature for at least 15 minutes before it is used.
-
3.
Place an aliquot of AgPath buffer (475 µl), AgPath enzyme (40 µl) and nuclease-free water (60 µl) on ice. Single-use aliquots (enough to run one card) are recommended to avoid contamination of reagents.
-
4.
Wipe a P1000, P200 and P20 pipette with 80% ethanol, DNA Away (or 10% bleach) and RNase Away (if the card includes RNA targets).
NOTE: RNase Away may be used in place of DNA Away/bleach.
-
5.When the UV procedure is complete, wipe the biosafety cabinet with RNase Away (for RNA targets) and make the master mix in a 1.5 ml tube on ice:
-
5.1.Combine 450 µl AgPath buffer, 54 µl nuclease-free water and 36 µl AgPath enzyme in the tube.
-
5.2Mix by pipetting gently up and down and discard any remaining volume from the single-use aliquots.
-
5.1.
-
6.
Transfer 60 µl of master mix into each of the remaining eight 1.5 ml tubes and keep them on ice.
-
7.
If the DNA/RNA samples require dilution, add nuclease-free water to the tubes that require it, followed by the sample and mix well.
NOTE: Dilution of concentrated samples ensures that the sample port is not overloaded with DNA, and may also be required if samples contain PCR inhibitors. Generally, use up to 1 µg of total DNA per port, though this may be exceeded if low concentrations of targets are expected (e.g. enteropathogens in non-clinical samples). The user will need to consider their sample type and balance the requirement for dilution with the potential for low concentration targets to be diluted below detectable levels.
-
8.
Open the TaqMan Array Card. In the event it is opened but not immediately going to be used, ensure it is kept in the dark. Take care not to touch the area where the sample ports are.
-
9.Load 100 µl of each sample into the larger hole of the corresponding port on the card (1-8).
-
9.1.Keep the pipette tip at an angle and take care not to puncture the foil and the bottom of the port.
-
9.2.Load the sample slowly, and do not push the pipette to the second stop to avoid adding an air bubble to the port (alternatively, reverse pipetting may be used).
-
9.3.If there is spillage around the port, aspirate it and return it to the port.
-
9.1.
-
10.
Centrifuge the card:
NOTE: A centrifuge compatible with the array cards is required. See the Applied Biosystems TaqMan Array Micro Fluidic Cards User Guide (https://tools.thermofisher.com/content/sfs/manuals/cms_062836.pdf) for more information. In this protocol, we use a Heraeus Megafuge 40 with array card buckets (ThermoFisher, catalogue number 75004503) using bucket type 3618 and rotor type TX-750.
-
10.1.
Place the card, foil side inwards, in the Array Card holder inside the centrifuge bucket. Put it in the outermost slot.
-
10.2.
Place another card (e.g. a previous calibration card) as a balance in the opposite bucket.
-
10.3
Centrifuge for 1 minute at 1200 rpm (331 x g) with ramp speeds set to 9.
-
10.4
Check that the channels are filling properly (the port is not completely drained and not still full, see Fig. 3A) before repeating the spin for another minute at 1200 rpm. Do not combine these centrifugation steps.
Fig. 3.
Loading the TaqMan Array Card. Representative images of (A) a correctly filled card after centrifugation (the ports are no longer full, but not completely drained) and (B) the preferred direction from which to cut off the ports: from port 8 to port 1. Left-handed individuals should also cut in this direction (pictured). This cut should be made as close to the plastic of the card as possible, leaving no overhang.
NOTE: It is rare for a channel to fill incorrectly. If this occurs, a third centrifugation step can be performed but not exceeded. If the port remains full, the channel may be blocked. If the port is completely empty, the channel may be punctured. If this is the case, disregard the results for any samples that have not filled correctly in the card and repeat the sample on another card.
-
11.
Seal the card by placing it foil side up into the sealer while the carriage is down (closest to you), pressing it in so the pins hold the card. Push the carriage to the top in one swift motion, then remove the card before sliding the carriage back down.
-
12.
Check that the foil is not damaged before trimming off the ports with scissors. Ensure the ports have been cut clearly and there is no overhang. Cut from port 8 whether left or right handed, to avoid a jagged edge (Fig. 3B).
-
13.
Check that the Array Card block and holder are in the QuantStudio 7. Turn on the machine and use the Eject button to open the side tray.
-
14.
Place the card flat in the tray foil side down, with the A1 well in the top left corner.
-
15.
Close the tray and use the template file created previously to begin the run.
-
16.
When the run is complete, transfer the .eds file off the QuantStudio 7 and dispose of the used array card in the biohazard PC2 waste bin.
Inspecting the amplification curves
The QuantStudio software stores the fluorescence data and applies threshold values, generating an .eds file for each experiment. However, as each assay on the card must run under the same conditions the amplification curves may be of variable quality, and these thresholds must be inspected along with quality control flags to check the integrity and accuracy of the data prior to downstream analysis. The Multicomponent Plot is useful for observing the true fluorescence in the well, and the Amplification Plot is useful for diagnosing issues with the automatic threshold.
-
1.
In “Analysis Settings” > “Flag Settings”, select “Revert to Default Settings” to restore all flags (these are removed when the cycling conditions are changed from the default).
-
2.Check the “Analysis” > “QC Summary” for any wells that have been flagged and may have issues. Generally:
-
2.1.BADROX, NOISE, SPIKE and PRFDROP indicate an issue with the well: consider removing results with these flags after inspecting the Multicomponent Plot.
-
2.2.NOAMP is useful for flagging negative results (in “Flag Settings”, we use a NOAMP value of 1.5 to distinguish between positive and negative wells).
-
2.1.
-
3.
Select all negative results in the Well Table and right click > “Omit” them. This will remove their Cq values if they have any and recalculate thresholds for the remaining amplification curves. Click “Analyse” to update.
-
4.Using the “Group by” function on the well table, group results by target. Check the calculated Cq values against the Multicomponent Plot for all targets with positive results, as the software may place an inappropriate threshold for targets where one or more wells has produced a spurious or poor amplification curve.
-
4.1.If the well has been assigned a Cq value but a fluorescence signal is absent from the Multicomponent Plot, right click and “Omit” the well. Click “Analyse” to update the automatic thresholds.
-
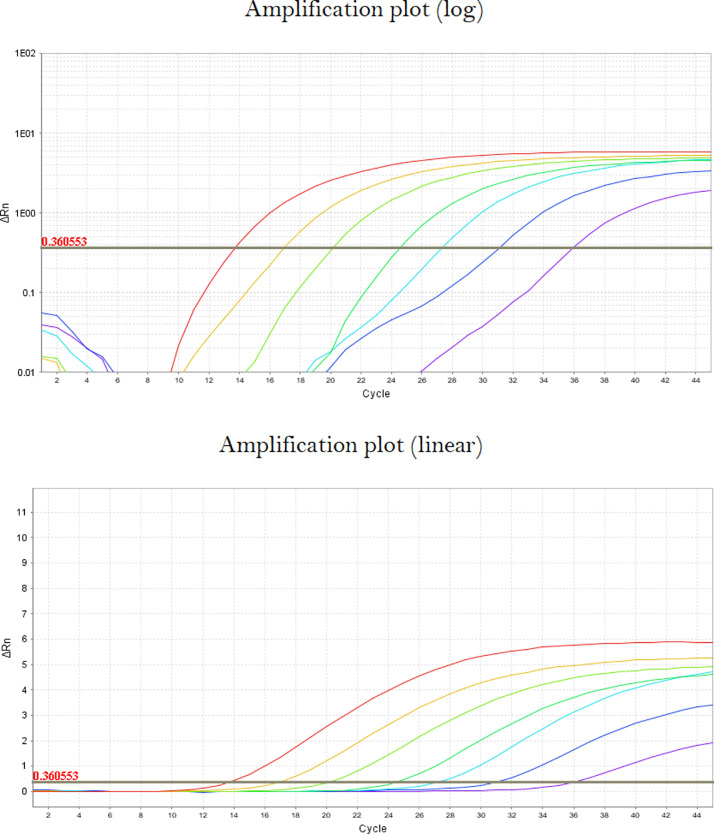
4.2.Check the Cq values for all positive wells. If the Cq value does not correspond to the point at which the fluorescence line begins to rise (this often occurs with poorer amplification curves), we recommend adjusting the threshold for the target. Viewing the Amplification Plot, untick “Auto” for the threshold line to drag it manually. Check the log and linear plots for all wells corresponding to the target until it is appropriately placed (Fig. 4).
-
4.3.Click “Analyse” to update all Cq values.
-
4.4.Repeat as necessary for all targets.
-
4.5.In the “Export” window, export the desired columns by selecting/deselecting as desired. Ensure that “Skip Omitted Wells” is unticked to export negative results.
-
4.1.
Fig. 4.
Appropriately placed Cq threshold. As in standard (plate) qPCR, the threshold line used to determine the Cq value for an amplification curve should be placed approximately in the middle of the exponential phase of the curve (log view). Ideally, it is placed just above the point at which the signal rises significantly above the baseline (linear view).
Calculation of gene copies
These calculations may be done manually or via a script (e.g. in R). See Supplementary material 4 for an example R script.
-
1.
Export the Cq values generated by the plasmid standard curve. Calculate a standard curve equation for each target using all standard curve replicates (e.g. using a linear model in R) to define the relationship between Cq value and gene copies per microlitre.
NOTE: Outlier points on the standard curve may be removed if necessary.
-
2.
Consider labelling any results with a Cq value above 40 as “negative” results, and any between the lower limit of quantification/limit of detection and Cq 40 as “inconclusive”. We define the lower limit of quantification as the lowest point on the standard curve at which all standard curve replicates were detected, and recommend distinguishing strong positives (with concentrations at or above this limit) from weak or inconclusive results (below this limit).
NOTE: These thresholds are at the user's discretion, and may depend on the sample type, project needs (e.g. seeking clear positives only, or capturing low levels of targets) and the behaviour of no template controls.
-
3.
Use the standard curve equation for the appropriate target to convert the Cq values from samples analysed on TAC to gene copies per microlitre. Ensure that any sample dilution performed for TAC is taken into account. This may be further adjusted to copies per gram of sample or equivalent if applicable.
Method validation
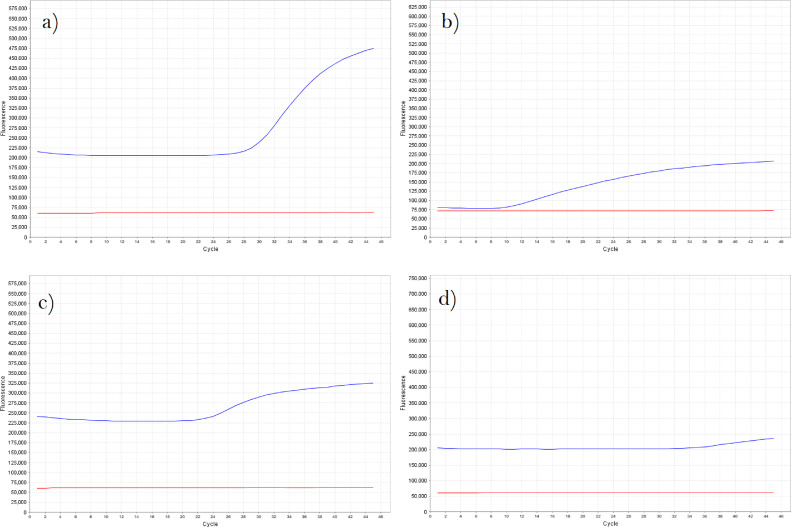
Fig. 5 contains examples of amplification curves viewed as a Multicomponent Plot (unadjusted fluorescence values). An ideal amplification curve is similar to Fig. 5A: a sigmoid curve, where the software will likely choose an appropriate threshold. Some less ideal curves that may require manual threshold adjustment may have a more linear increase in fluorescence (Fig. 5B), have a smaller fluorescence increase (Fig. 5C) or be ambiguously positive due to a late Cq, low fluorescence or a linear increase (Fig. 5D). In these cases, the threshold should be manually adjusted to best fit all curves for a given target. Highly ambiguous results where it is difficult to determine the Cq value can be considered inconclusive or counted as negative results in the downstream analysis, depending on project needs.
Fig. 5.
Multicomponent plots for various amplification curve shapes. Optimal assays will produce a tall sigmoid curve (A), for which the QuantStudio software's default threshold value will often be appropriate. Some assays may produce more linear amplification curves (B), a smaller increase in fluorescence (C) or ambiguous curves (D). This may be a result of the assay's performance under the universal cycling conditions on the card, or may occur in certain sample types prone to PCR inhibition. For poorer curves, the appropriate Cq value may be difficult to determine.
To demonstrate the utility across a variety of sample types, we applied a custom gastrointestinal pathogen TAC to two sets of samples. Table 1 shows human clinical stool samples deemed positive or negative for a variety of pathogens via multiplexed-tandem PCR (MT-PCR, using the AusDiagnostics Faecal Pathogens M panel), and the comparative TAC results. TAC successfully and correctly identified the pathogen present in 19 of 20 samples. The C. difficile result not detected by TAC was a low concentration (Cq = 36.28) MT-PCR result. TAC additionally detected Campylobacter pan and Giardia in some samples where they were not detected by MT-PCR. These discrepancies may represent false positive or false negative results in either technique that occur due to low target concentration, off-target amplification, or variation in the coverage of particular species or strains by the different PCR assays.
Table 1.
Enteropathogen detection in clinically positive and negative samples by a custom enteropathogen TAC. Human stool samples from a previous study [20] were clinically determined positive or negative for enteropathogens using the AusDiagnostics Faecal Pathogen M panel. The 16S rRNA and 18S rRNA controls on the TAC were positive for all samples. Pathogens congruent with the AusDiagnostics result are bolded.
| Sample | AusDiagnostics result | TaqMan Array Card result |
|---|---|---|
| 1 | No pathogen | No pathogen |
| 2 | Sapovirus | Sapovirus, Giardia, Campylobacter pan |
| 3 | No pathogen | No pathogen |
| 4 | Campylobacter | Campylobacter jejuni/coli, Campylobacter pan |
| 5 | No pathogen | No pathogen |
| 6 | No pathogen | No pathogen |
| 7 | No pathogen | No pathogen |
| 8 | Campylobacter, Cryptosporidium | Campylobacter (pan only), Cryptosporidium |
| 9 | No pathogen | No pathogen |
| 10 | Salmonella | Salmonella enterica, Campylobacter pan |
| 11 | Shigella | Shigella/EIEC (ipaH), Shigella flexneri periplasmic protein, Campylobacter pan |
| 12 | Norovirus GII | Norovirus GII, Giardia, Campylobacter pan |
| 13 | Clostridioides difficile | No pathogen |
| 14 | No pathogen | No pathogen |
| 15 | No pathogen | No pathogen |
| 16 | Norovirus GI | Norovirus GI |
| 17 | No pathogen | No pathogen |
| 18 | STEC (Shiga toxin 1) | STEC (Shiga toxin 1) |
| 19 | Giardia | Giardia |
| 20 | No pathogen | Campylobacter pan |
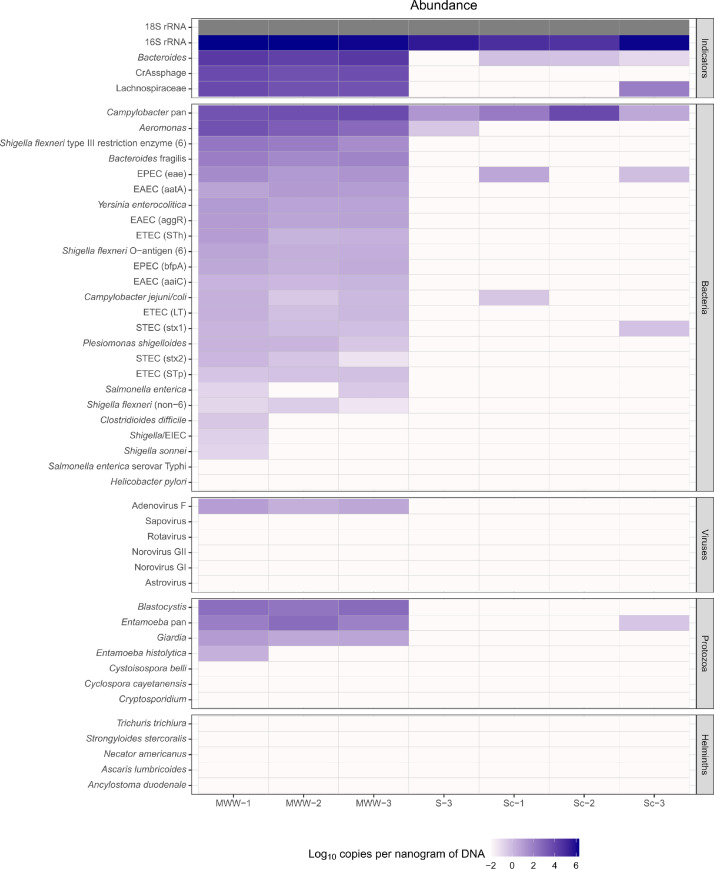
TAC may also be used to detect pathogens across multiple sample types. Fig. 6 illustrates the range of pathogens detected via TAC in samples from wastewater, soil, and animal (dog) scats. In wastewater samples, overall bacterial load and all faecal indicators were detected at high concentrations, with a range of bacterial and protist enteropathogens also present. Minimal pathogens were found in the urban park soil and dog scats, though Campylobacter was common.
Fig. 6.
Estimated gene copies per nanogram of DNA across a custom enteropathogen TAC panel. TAC was run using the protocol described on three wastewater samples (MWW-1, -2 and -3), a soil sample from an urban park area (S-3) and three domestic dog scat samples (Sc-1, -2 and -3) from Melbourne, Australia.
Discussion
The TaqMan Array Card provides an efficient technique for the simultaneous detection of typically 47 custom targets across 8 samples in an assay that takes approximately 1 hour of lab preparation time and 2.5 hours of PCR runtime. The number of targets per card can be expanded if fewer samples are run, or if wells are multiplexed through the use of two different fluorophores. Once established, the laboratory protocol involves relatively few steps, with the more challenging steps lying in the interpretation of suboptimal curves and the quantification of gene copies.
While the use of a plasmid standard curve enables an estimation of gene copies in a sample, this quantification approach has limitations. TAC is therefore ideally suited to presence/absence data generation, and should be considered “semi-quantitative”. The first limitation, shared with conventional qPCR, is that for multi-copy genes (especially ribosomal RNA genes), the number of gene copies per genome may be unknown and vary greatly, impacting quantification. Further to this, the reverse transcriptase step in the TAC protocol ensures that transcript copies and rRNA copies in addition to genomic copies can be amplified. This is beneficial for increased sensitivity, but again introduces challenges in gene copy quantification, though treatment with DNase may assist in estimating the contribution of RNA copies. Finally, the efficiency of primer binding and amplification from a synthesized plasmid insert is likely to differ from that of the samples, where amplification from a large genomic DNA template occurs in the presence of large proportions of non-target DNA, potentially in the presence of inhibitory substances. Collectively, these will heavily impact the accurate calculation of target gene copies and are not easily resolved. If highly accurate quantification is required for certain pathogens of interest, TAC may be used to screen for positive samples after which individual qPCR reactions can be run for more accurate measurement of gene copies.
TAC is most beneficial in studies where the number of different pathogen assays exceeds that which can practicably be undertaken by standard qPCR. Generally, TAC is less sensitive than equivalent individual qPCR reactions [2], [3], [4], so is ideal when prioritizing breadth (many pathogens) over depth (highly accurate and sensitive detection of a few pathogens). Typically, we observe the second-lowest concentration of the plasmid standard to be consistently detectable on TAC (∼72 copies per microlitre of DNA) and the lowest concentration (∼7 copies per microlitre of DNA) is detectable 90% of the time, with some targets more consistent than others. Conversely, standard qPCR prioritizes depth over breadth with higher accuracy and sensitivity [3]. Another consideration is that, while TAC is cost-effective per sample per pathogen, the discrete cost of the custom cards is high (approx. $550 (USD) per card). However, it may be lower than the cost of moderately deep metagenomic sequencing (approx. $300-400 per sample).
For pathogen detection, TAC has largely been used for enteropathogens. Multicentre studies on childhood diarrhoea have successfully employed TAC to quantify pathogens and determine diarrhoeal aetiologies [5,6]. We recently evaluated TAC for enteropathogen detection on human stool, soil, water and animal scat samples and found it to be comparable to standard qPCR, providing a substantial benefit in surveillance contexts where multiple pathogens and pathogen reservoirs need to be monitored to understand transmission [3]. As there are no steps in the TAC protocol that are specific to enteropathogens or particular sample types, TAC is a useful technique for the detection of any set of pathogens (in clinical, animal or environmental surveillance settings) or other target organisms in any sample type, as long as good quality DNA extracts can be obtained and TaqMan assays exist or can be designed to the target genes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was funded by the Wellcome Trust Our Planet Our Health grant 205222/Z/16/Z, a National Health and Medical Research Council (NHMRC) EL2 Fellowship (APP1178715; salary for CG) and NHMRC Equipment Grant (awarded to S.L.C.). We thank Eric Houpt, Darwin Operario, James Platts-Mills, Karin Leder and Stephen Luby for helpful discussions, David Rayner for technical support and advice, and Sarah Thomas and Julie Bines for providing rotavirus RNA for quality control purposes.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.mex.2022.101707.
Appendix. Supplementary materials
Supplementary material 2: Results from three replicates of a standard curve (serial dilutions of the combined plasmid standards). Typically, the plasmid standard curve on TAC is highly consistent between replicates and the amplification efficiency for each target is within the acceptable range.
Supplementary material 3: Video protocol covering the TaqMan Array Card technique, including recommended laboratory and data analysis steps.
Supplementary material 4: An example R script that can be used to generate linear models from a TAC standard curve, and calculate gene copies per microlitre of DNA sample from the curve.
References
- 1.Heaney J., Rolfe K., Gleadall N.S., Greatorex J.S., Curran M.D. In: Methods in Microbiology. Sails A., Tang Y.-W., editors. Academic Press; 2015. Chapter 6 - low-density TaqMan® array cards for the detection of pathogens; pp. 199–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harvey J.J., Chester S., Burke S.A., Ansbro M., Aden T., Gose R., et al. Comparative analytical evaluation of the respiratory TaqMan Array Card with real-time PCR and commercial multi-pathogen assays. J. Virol. Methods. 2016;228:151–157. doi: 10.1016/j.jviromet.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lappan R., Henry R., Chown S.L., Luby S.P., Higginson E.E., Bata L., et al. Monitoring of diverse enteric pathogens across environmental and host reservoirs with TaqMan array cards and standard qPCR: a methodological comparison study. Lancet Planet Health. 2021;5:e297–e308. doi: 10.1016/S2542-5196(21)00051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodani M., Yang G., Conklin L.M., Travis T.C., Whitney C.G., Anderson L.J., et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J. Clin. Microbiol. 2011;49:2175–2182. doi: 10.1128/JCM.02270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogawski E.T., Liu J., Platts-Mills J.A., Kabir F., Lertsethtakarn P., Siguas M., et al. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob. Health. 2018;6:e1319–e1328. doi: 10.1016/S2214-109X(18)30351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Platts-Mills J.A., Juma J., Kabir F., Nkeze J., Okoi C., et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet. 2016;388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steensels D., Reynders M., Descheemaeker P., Curran M.D., Jacobs F., Denis O., et al. Clinical evaluation of a multi-parameter customized respiratory TaqMan® array card compared to conventional methods in immunocompromised patients. J. Clin. Virol. 2015;72:36–41. doi: 10.1016/j.jcv.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., Ochieng C., Wiersma S., Ströher U., Towner J.S., Whitmer S., et al. Development of a TaqMan array card for acute-febrile-illness outbreak investigation and surveillance of emerging pathogens, including Ebola Virus. J. Clin. Microbiol. 2016;54:49–58. doi: 10.1128/JCM.02257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao C., Wang X., Zhang C., Liu B., Jing H., Ming L., et al. Development of a TaqMan Array card to target 21 purulent meningitis-related pathogens. BMC Infect. Dis. 2019;19:289. doi: 10.1186/s12879-019-3856-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwambana-Adams B.A., Liu J., Okoi C., Mwenda J.M., Mohammed N.I., Tsolenyanu E., et al. Etiology of pediatric meningitis in West Africa using molecular methods in the era of conjugate vaccines against Pneumococcus, Meningococcus, and Haemophilus influenzae Type b. Am. J. Trop. Med. Hyg. 2020;103:696–703. doi: 10.4269/ajtmh.19-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore C.C., Jacob S.T., Banura P., Zhang J., Stroup S., Boulware D.R., et al. Etiology of sepsis in Uganda using a quantitative polymerase chain reaction-based TaqMan array card. Clin. Infect. Dis. 2019;68:266–272. doi: 10.1093/cid/ciy472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banu S., Pholwat S., Foongladda S., Chinli R., Boonlert D., Ferdous S.S., et al. Performance of TaqMan array card to detect TB drug resistance on direct specimens. PLOS One. 2017;12 doi: 10.1371/journal.pone.0177167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foongladda S., Banu S., Pholwat S., Gratz J., O-Thong S., Nakkerd N., et al. Comparison of TaqMan® Array Card and MYCOTBTM with conventional phenotypic susceptibility testing in MDR-TB. Int. J. Tuberc. Lung Dis. 2016;20:1105–1112. doi: 10.5588/ijtld.15.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pholwat S., Liu J., Stroup S., Jacob S.T., Banura P., Moore C.C., et al. The malaria TaqMan array card includes 87 assays for Plasmodium falciparum drug resistance, Identification of species, and genotyping in a single reaction. Antimicrob. Agents Chemother. 2017:61. doi: 10.1128/AAC.00110-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pholwat S., Liu J., Taniuchi M., Chinli R., Pongpan T., Thaipisutikul I., et al. Genotypic antimicrobial resistance assays for use on E. coli isolates and stool specimens. PLOS One. 2019;14 doi: 10.1371/journal.pone.0216747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korajkic A., McMinn B.R., Harwood V.J. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health. 2018;15 doi: 10.3390/ijerph15122842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Gratz J., Amour C., Kibiki G., Becker S., Janaki L., et al. A laboratory-developed TaqMan array card for simultaneous detection of 19 enteropathogens. J. Clin. Microbiol. 2013;51:472–480. doi: 10.1128/JCM.02658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Gratz J., Amour C., Nshama R., Walongo T., Maro A., et al. Optimization of quantitative PCR methods for enteropathogen detection. PLOS One. 2016;11 doi: 10.1371/journal.pone.0158199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kodani M., Winchell J.M. Engineered combined-positive-control template for real-time reverse transcription-PCR in multiple-pathogen-detection assays. J. Clin. Microbiol. 2012;50:1057–1060. doi: 10.1128/JCM.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson D.A., Chow E.P.F., Lee D., Maddaford K., Sait M., Easton M., et al. Risk factors for asymptomatic enteric pathogen detection among men who have sex with men. Open Forum Infect. Dis. 2019;6:ofz326. doi: 10.1093/ofid/ofz326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 2: Results from three replicates of a standard curve (serial dilutions of the combined plasmid standards). Typically, the plasmid standard curve on TAC is highly consistent between replicates and the amplification efficiency for each target is within the acceptable range.
Supplementary material 3: Video protocol covering the TaqMan Array Card technique, including recommended laboratory and data analysis steps.
Supplementary material 4: An example R script that can be used to generate linear models from a TAC standard curve, and calculate gene copies per microlitre of DNA sample from the curve.