Summary
Background
N6-methyladenosine (m6A) is the most common and abundant mRNA modification and it plays crucial roles in many biological processes. However, as a key RNA demethylase, alkylation repair homolog protein 5 (ALKBH5) has not been well studied in human osteosarcoma. The present study sought to explore ALKBH5-mediated m6A modification and the underlying mechanisms in human osteosarcoma.
Methods
The expression of ALKBH5 and its correlation with clinicopathological features were examined by bioinformatics analysis and tissue microarrays. Cellular proliferation was detected by CCK8 assays. Cell cycle and apoptosis were analyzed by TUNEL and Flow cytometry assay. Finally, investigation of the regulatory mechanism of ALKBH5 in human osteosarcoma was performed by MeRIP assay, RNA-sequencing, dual luciferase reporter assay, RNA pull-down and RNA stability assay. Tumor xenograft models were established for in vivo experiments.
Findings
Our data showed that low expression of ALKBH5 was associated with worse overall survival for osteosarcoma patients. Reducing m6A mRNA levels in human osteosarcoma cells through ALKBH5 up-regulation lead to cell proliferation inhibition, cell apoptosis and cycle arrest. We identified SOCS3, a negative regulator of STAT3, as a downstream target of ALKBH5-mediated m6A modification. And the m6A modified SOCS3 mRNA was recognized by YTHDF2, which promotes the decay of SOCS3. Mechanistically, our data revealed that ALKBH5 inactivated STAT3 pathway by increasing SOCS3 expression via an m6A-YTHDF2-dependent manner.
Interpretation
M6A methylation is rising as a pathway affecting tumorigenicity and tumor progression. Our findings illuminate the clinical significance of ALKBH5-mediated m6A modification in human osteosarcoma and the regulatory mechanisms underlying tumor proliferation and growth, suggesting that ALKBH5 is a potential biomarker for treatment in human osteosarcoma.
Funding
This work was supported by and Science and Technology foundation of Hubei, China (Grant No.2017CFB762); the Tongji hospital foundation (Grant No.2201103013); and the National Natural Science Foudation of China (No.82002849).
Keywords: Osteosarcoma, m6A, ALKBH5, YTHDF2, STAT3, SOCS3
Research in context.
Evidence before this study
Many studies have revealed that ALKBH5-mediated m6A modification plays a vital role in tumor progression. The effects of m6A mRNA methylation and its underlying mechanisms in human osteosarcoma are controversial. It is reported that ALKBH5-mediated m6A demethylation suppressed osteosarcoma progression through regulation of YAP. In another study, ALKBH5 interacted with PVT1 and increased its stability, which contributes to osteosarcoma tumorigenesis. Which suggested the significance of ALKBH5-mediated m6A on progression of osteosarcoma.
Added value of this study
The current study combines LC-MS quantification of m6A with RNA sequencing which show that reduced m6A mRNA methylation mediated by ALKBH5 could suppress cell growth by regulating STAT3 signal pathway. We identified SOCS3, a negative regulator of STAT3, as a downstream target of ALKBH5-mediated m6A modification in further study. ALKBH5 could be a potential biomarker for treatment in human osteosarcoma.
Implications of all the available evidence
This findings advise that regulation of STAT3 signal activity through ALKBH5-mediated m6A methylation could be a universal growth regulatory mechanism that affects diverse other tumors. In this study the methylated SOCS3 was read by YTHDF2 alone .It should be noted that YTHDF2-mediated mRNA degradation might play a significant role in tumors with high m6A methylation levels.
Alt-text: Unlabelled box
Introduction
N6-methyladenosine (m6A) is the most common and abundant mRNA modification and it plays crucial roles in many biological processes.1,2 This modification is regulated by different kinds of regulators, including ‘writer’, ‘eraser’ and ‘reader’ proteins.3 RNA m6A is enriched near stop codon and 3′ untranslated terminal region (UTR) and translated near 5′ UTR in a cap-independent manner, thereby affecting RNA transcription, processing, translation and metabolism.4 In line with these roles, dysregulation of m6A methylation influences various biological processes in mammals.5, 6, 7, 8 Growing evidence suggests that m6A methylation is rising as a pathway affecting tumorigenicity and tumor progression.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 m6A mRNA methylation affects the growth and proliferation of stem cells and tumor cells.5,10, 11, 12, 13, 14,21,22 However, the underlying pathways and mechanisms of m6A modification in tumor have yet to be thoroughly illuminated.
Osteosarcoma is the most common primary malignant bone tumor. Although the improved survival rate of osteosarcoma have been noted in recent years, the cure rate of patients of osteosar coma is still very low because of the rapid progression of tumor. Currently, the effects of m6A mRNA methylation and its underlying mechanisms in human osteosarcoma are controversial. Herein, we investigate this question in human osteosarcoma, which may reveal new directions for osteosarcoma treatment.
To investigate the underlying mechanisms of m6A modification in osteosarcoma, we focused on ALKBH5, an m6A eraser, which has been shown to play an essential role in many tumors.13,23 ALKBH5 was found to be expressed at a low level in some cancer tissues, and depletion of ALKBH5 was markedly associated with poorer prognosis in some kinds of tumors.23, 24, 25, 26 It was found that expression of ALKBH5 was positively associated with overall survival of osteosarcoma patients using tissue microarrays assay (Figure 1). ALKBH5 may exert a tumor-suppressor function in osteosarcoma, which needs further experimental verification.
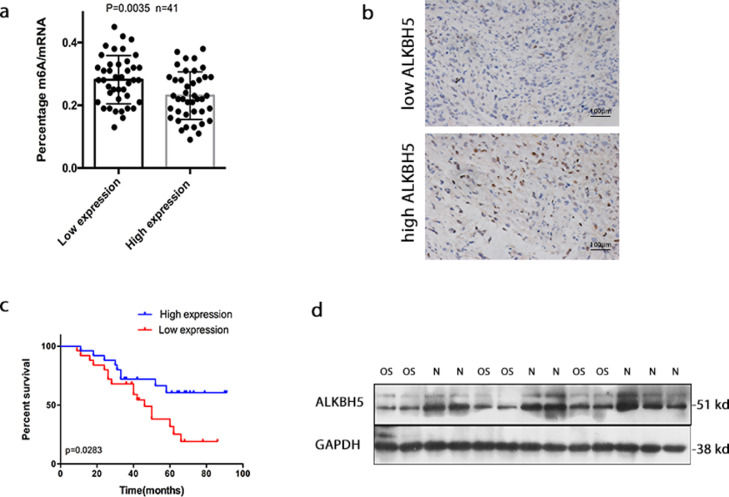
Figure 1.
ALKBH5 expression in osteosarcoma and its relationship to patient survival. a. High ALKBH5 tumor tissues exhibited lower total m6A methylation levels compared to low ALKBH5 group (t-test p = 0.0035, n = 41 per group). b. Representative images showing low or high expression of ALKBH5 by IHC staining. c. Kaplan-Meier survival analysis showed that the overall survival time of patients with low ALKBH5 expression was significantly shorter than that of patients with high ALKBH5 expression (log-rank p = 0.0283,n = 41 per group). d. Representative western blot showed the expression of ALKBH5 was markedly decreased in osteosarcoma (OS) compared with the normal bone (N). The data represent the mean± S.D. *P < 0.05 or **P < 0.01 indicates a significant difference between the indicated groups.
In this study, decreasing m6A methylation level in human osteosarcoma cells through ALKBH5 overexpression could inhibit cell proliferation, promote cell apoptosis and cycle arrest in vitro and in vivo. We demonstrate that ALKBH5 overexpression affect tumor progression by inactivating STAT3 signal pathway. We identify SOCS3, a negative regulator of STAT3,27 as a downstream target of ALKBH5-mediated m6A modification. And the m6A modified SOCS3 mRNA was recognized by YTHDF2, which promotes the decay of SOCS3. Mechanistically, our data reveal that ALKBH5 inactivate STAT3 pathway by increasing SOCS3 expression via an m6A-YTHDF2-dependent manner. Overall, these results suggest that ALKBH5 is a potential biomarker for treatment in human osteosarcoma.
Materials and methods
Clinical specimens
Eighty-two tumor tissues of osteosarcoma with complete follow-up were collected under the protocols approved by the ethics committee of Tongji Hospital (Ethics number TJ-IRB20210939). None of the patients received antitumor treatment before surgery. Informed consents (written in the light of the ethical guidelines) were obtained from all of the patients. The clinical characteristics of these patients were displayed in Table 1. Fresh tissues were stored in liquid nitrogen before RNA extraction. Clinical and histopathologic information was recorded through a retrospective review of patient records.
Table 1.
The relationship between ALKBH5 expression and clinicopathological variables of osteosarcoma.
| Items | ALKBH5 |
P-value |
|---|---|---|
| Low High | ||
| All cases Age ≥18 <18 Gender Male Female Anatomical location Limb bone Axial bone Grade of tumor Low High |
41 41 15 18 26 23 25 28 16 13 29 27 12 14 11 27 30 14 |
0.4993 0.4884 0.6350 0.0004 |
Cell culture and reagents
The human osteosarcoma cell lines U2OS (RRID:CVCL_0042) and KHOS (RRID:CVCL_2544) cells were purchased from the American Type Culture Collection (ATCC). KHOS and U2OS cells were cultured with RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum (Gibco). Cells lines were cultured at 37 °C with 5% CO2. 3-deazaadenosine (DAA) (CAS No.1338466-77-5)were purchased from Sigma Chemical Co. (St. Louis, MO,USA). These following antibodies were used in the experiments:anti-ALKBH5(#80283S),anti-STAT3(#12640S), anti-p-STAT3(#9145S), anti-CyclinD1(#2978S), anti-c-Myc (#18583S),anti-bcl-2(#4223S) and anti-GAPDH (#5174S) were from Cell Signalling Technology (Beverly, MA, USA). Anti-YTHDF2(ab220163), and anti-SOCS3(ab236519) was from Abcam (Cambridge, MA, USA). Orthogonal strategies were used for validating antibody specificity. The cell lines have been sending out for STR authentication and mycoplasma testing. The cell identification report and antibody product datasheets have been provided in the supplemental data.
Three primary OS cell cultures, OS1, OS2, OS3, were patient derived from biopsies obtained from Tongji Hospital in accordance with the protocol approved by the ethics committee of Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology (Ethics number TJ-IRB20210939). Informed consents were obtained from all of the patients. Tumor anatomic location, tumor type and personal details of the patients of the derived OS primary cell lines (OS-1, OS-2 and OS-3) are listed in Table 2. Tumor tissues were mechanically dissociated with surgical scissor into 1∼2mm3 pieces. Each tumor tissue was washed with Hanks solution (Gibco) three times in the plate, then digested with 0.25% trypsin (Invitrogen) at 37 °C for 30 min. Then complete medium was used to neutralize trypsin after the digestive liquid was removed. Primary tumor cells were cultured in plate with 37 °C and 5% CO2 in a professional complete culture system. And the media was changed every 2 days. Once cells reached 70–80% confluency they were lifted using 0.25% trypsin and frozen stock vials for each of the cell cultures were stored in liquid nitrogen.
Table 2.
Clinical features of the three human OS tumors from which primary cells derived.
| Name | Gender | Race | Age | Location | Histological characterization |
|---|---|---|---|---|---|
| OS-1 OS-2 OS-3 |
Male Female Male |
Asian Asian Asian |
14 16 21 |
Humerus Distal femur Distal femur |
High-grade osteosarcoma High-grade osteosarcoma with chondroid differentiation High-grade osteosarcoma |
Transfection
ShSOCS3, shYTHDF2, siALKBH5 and scrambled negative control were synthesized in GenePharma (Suzhou, China). The sequences are listed in Supplementary Table S1. RNAs were transfected into tumor cells using Lipofectamine3000 (Invitrogen). ALKBH5 stably expressed osteosarcoma cells were infected with the lentivirus and selected with puromycin (1 μg/ml) for 4 weeks.
CCK-8 assay
Cells were plated in 96-well plates at a density of 5000 cells in 100 μL medium per well 1 day before the experiment. The cell viability was examined by CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan)according to the manufacturer's instruction.
Western blot analysis
Equal amounts of proteins collected from different kinds of cell lysates were loaded on 10–15% SDS-PAGE gels using a NuPAGE system (Invitrogen) and then transferred onto PVDF membranes as previously described.28
Flow cytometry experiments
Cells for cell cycle analysis were fixed in 70% ethanol, digested with RNase A, and labeled with propidium iodide (PI). Apoptotic cells were analyzed with Annexin V/FITC kit (BD Biosciences, San Jose, CA, USA) according to the manufacturers instructions and analyzed by flow cytometry after compound treatment as previously described.28
Immunohistochemistry and tunel assay
IHC staining was performed as previously described.29 Paraffin sections were reacted with rabbit polyclonal anti-ALKBH5, anti-STAT3, anti-SOCS3 and anti-YTHDF2 antibodies (1:100 dilution). Sections stained with non-immune rabbit serum (1:200 dilution) in phosphate-buffered saline (PBS) instead of primary antibody served as negative controls. Cells displaying positive staining were counted in at least 12 representative fields and the mean percentage of positive cells was calculated. Immunostaining was assessed by two independent pathologists blinded to clinical characteristics and outcomes. TUNEL (terminal deoxynucleotidyl transferase-mediated nick end labeling) assay was performed on cells as described earlier.29
Quantitative RT-PCR (qRT-PCR)
The total RNA was extracted by Trizol reagent (Invitrogen). The reverse transcription was performed as described previously.5 QRT-PCR Primers were purchased from RiboBio, which are listed in Supplementary Table S2. GAPDH were used as endogenous controls.
RNA-sequencing
The total RNA was extracted using TRIzol reagent (15596018, Invitrogen), and the RNA sample quantification, qualification, library preparation and subsequent RNA-sequencing were conducted by Novogene Co., LTD (Beijing, China). Differential expression analysis of the two conditions was performed using the edgeR R package (3.12.1). Corrected P-values of 0.05 and absolute fold-changes of 2 were set as the threshold for significantly differential expression.30
LC-MS/MS quantification of m6A in poly (A)-mRNA
These assays were conducted as described previously. Total RNA was extracted using TRIzol reagent (Invitrogen, CA, USA), and purified using a Dynabeads mRNA DIRECT kit and RiboMinus Eukaryote Kit (Ambion, CA, USA) following the manufacturer's instruction. The sample was diluted to a total volume of 90μl and filtered (0.22 μm pore size, Millipore). In total, 10μL of the solution was injected into LC-MS/MS (Agilent Technologies, CA, USA). Quantification was carried out by comparison with a standard curve obtained from pure nucleoside standards run with the same batch of samples. The ratio of m6A to A was calculated based on the calibrated concentrations.5
MeRIP-qPCR
M6A modifications of individual genes were determined using MeRIP-qPCR assay, which was performed as described earlier.15
RNA pull-down assay
RNA pull-down assays were performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (20,164, Thermo Scientific). Up to 50 pmol of biotinylated RNAs was mixed with 2 mg of protein lysates and 50 μl of streptavidin beads. After incubation and three washes, the streptavidin beads were boiled and used for the immunoblotting assay.15
RNA stability assays
Stable cells were incubated with actinomycin D (HY-17,559, MedChemExpress)for 0 h, 3 h or 6 h and then followed by RNA extraction. And the half-life of SOCS3 mRNA was analyzed by quantitative RT-PCR as previously described.31
Luciferase reporter and mutation assay
SOCS3-MUTs mutants were conducted by Mut Express Ⅱ Fast Mutagenesis Kit (Vazyme). Osteosarcoma cells were seeded into the 24-well plate and then were transfected with 250 ng 3′ UTR luciferase reporter plasmid (Promega). The luciferase activity was tested by Dual-Glo Luciferase Assaw kit (Promega) as previously described.31
Generation of xenografts
Twenty-four Six-week-old BALB/c female athymic nude mice (Vitalriver, Beijing, China) were randomly divided into two groups. Four nude mice were fed in each cage. Tow group nude mice were subcutaneously injected in the right flank with osteosarcoma cells (2 × 106 in 100 ul PBS) at the same time. The experimental group (Lv-ALKBH5,n = 12) and control group (Lv-NC,n = 12) were fed separately (A group of 4 nude mice were repeated three times). All animal care and handling procedures were performed in accordance with the National Institutes of Health guide for the care and use of Laboratory animals, which was approved by the Laboratory Animal Welfare & Ethics Committee of Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology (Ethics number TJH-201903021) .The volume of xenografts was measured every five days (tumor volume = (length × width2)/2). The mice were sacrificed 30 days later. The experiments were conducted between March and June 2020. The operation was performed on a sterile laboratory. Tumor samples were processed for IHC.
Statistical analysis
The SPSS19.0 software package and GraphPad Prism 7 (GraphPad Software, La Jolla, CA,USA) were used to perform all statistical analyses. Log-rank test was used for Kaplan-Meier survival analysis. Data are expressed as the mean±SD of at least 3 independent experiments, and statistical evaluation was performed using one-way analysis of variance (ANOVA) or Student′s t-tests. Values of p < 0.05 or p < 0.01 were considered statistically significant.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The authors had full access to all the data in the study and accept responsibility to submit for publication.
Results
ALKBH5 expression in osteosarcoma and its relationship to patient survival
Previous studies have indicated that ALKBH5 was dysregulated in various tumors.25,26,32 Our data validated a negative correlation between ALKBH5 and overall survival in human OS (osteosarcoma). OS group was divided into high expression group (n = 41) and low expression group (n = 41) according to median of relative expression of ALKBH5 mRNA. None of the patients received antitumor treatment before surgery. As shown in Figure 1a, total RNA was extracted and m6A levels were determined as a percentage of all adenosine residues in RNA, high ALKBH5 tumor tissues exhibited lower total m6A methylation levels compared to low ALKBH5 group. The ALKBH5 expression levels were further assessed by immunohistochemistry (IHC) staining. Representative high or low expression of ALKBH5 staining images were shown in Figure 1b. We examined the correlation between ALKBH5 expression and osteosarcoma patient prognosis. Kaplan-Meier survival analysis showed that the overall survival time of patients with low ALKBH5 expression was significantly shorter than that of patients with high ALKBH5 expression (Figure 1c). Furthermore, western bolt analysis was performed on 15 samples taken from the normal bone and osteosarcoma tissues. The expression of ALKBH5 was detected in both tissues, but was markedly decreased in osteosarcoma compared with the normal bone (Figure 1d).
Reduced m6A methylation-mediated by ALKBH5 suppresses osteosarcoma cell growth and promotes cell apoptosis
We next examined whether the reduced m6A methylation observed in the human osteosarcoma tissue affects functions associated with tumor progression in human osteosarcoma cells. To explore the effects of ALKBH5 mediated m6A methylation in U2OS and KHOS cell lines, we used lentiviral transfection to upregulate ALKBH5, and ALKBH5 overexpression was confirmed by western blot (Figure 2a).
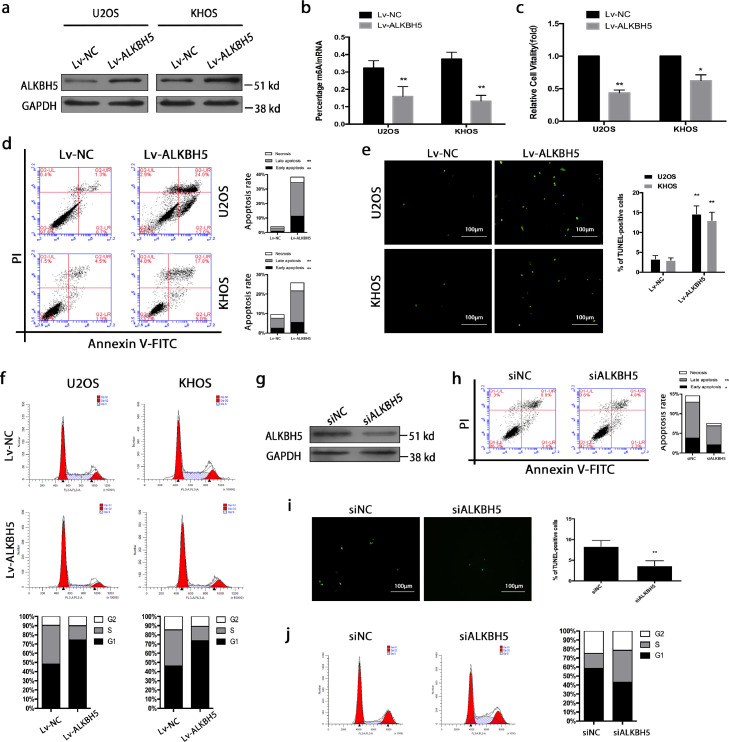
Figure 2.
Reduced m6A methylation-mediated by ALKBH5 suppresses osteosarcoma cell growth and promotes cell apoptosis. a. ALKBH5 overexpression was confirmed by western blot in U2OS and KHOS cell lines. b. ALKBH5 up-regulation reduced m6A mRNA methylation. c. Cell viability was assessed by CKK-8 assay. d. The percentage of apoptosis was determined by FCM analysis. e. TUNEL staining was detected from osteosarcoma cells with transfection of Lv-ALKBH5, positive cells were labeled with TUNEL (green) (magnification × 400) . f. Cell cycle analysis was performed using FCM, which revealed osteosarcoma cells were mostly arrested in the G0/G1 phase. g. SiRNA was used to down-regulate ALKBH5, and ALKBH5 down-regulation was confirmed by western blot. h,i. Flow cytometry (h)and TUNEL (i) assay were carried out to confirm the reduction of apoptosis. j. Cell cycle assay showed that there was an increase in the number of dividing tumor cells following the knockdown of ALKBH5. The data represent the mean± S.D. of three independent experiments. ANOVA or t-tests was used for statistical analysis. *P < 0.05 or **P < 0.01 indicates a significant difference between the indicated groups.
As expected, the ALKBH5 up-regulated cells exhibited reduced m6A mRNA methylation (Figure 2b). Functionally, overexpression of ALKBH5 suppressed cell proliferation, induced cell cycle arrest and promoted apoptosis. Decreased cell viability was observed by a CKK-8 assay (Figure 2c). Apoptosis assays were carried out at 48 h post-transfection. Substantial apoptosis was observed by flow cytometry (FCM) in these two cell lines (Figure 2d). TUNEL staining was performed to confirm the induction of apoptosis (Figure 2e). Our analysis of cell cycle assay revealed that osteosarcoma cells were mostly arrested in the G0/G1 phase, implying that there was a reduction in the number of dividing tumor cells following the overexpression of ALKBH5 (Figure 2f). To further confirm these findings, the above experiments were performed in three primary cells derived from osteosarcoma. As expected, overexpression of ALKBH5 leads to direct growth inhibition of primary cells through G0/G1 arrest and apoptosis (Figure S1).
To better understand the role of ALKBH5 in osteosarcoma, we also performed knockdown experiments in KHOS cell lines. SiRNA was used to down-regulate ALKBH5, and ALKBH5 down-regulation was confirmed by western blot (Figures 2g and S2a). Flow cytometry (Figure 2h) and TUNEL (Figure 2i) assay were carried out to confirm the reduction of apoptosis caused by ALKBH5 down-regulation (siALKBH5-1). Cell cycle assay (Figure 2j) implied that there was an increase in the number of dividing tumor cells following the knockdown of ALKBH5 (siALKBH5-1). Functional experiments of different siALKBH5 (siALKBH5-2) were performed to exclude the off-target effect of siRNA sequences (Figure S2b,c).
RNA sequencing identifies transcripts with altered methylation in osteosarcoma
We performed RNA sequencing (RNA-seq) of ALKBH5 overexpression KHOS cells which showed altered expression of transcripts relative to conrtols, both upregulated (n = 41) and downregulated (n = 114) (Figure 3a). Gene Set Enrichment Analysis (GSEA) demonstrated those differentially expressed genes (DEGs) have a strong relationship with cell proliferation, cell cycle and apoptosis. We also found the STAT3 signalling pathway was significantly altered by reduced m6A methylation in the osteosarcoma cell lines (Figure 3b,c). Because the STAT3 signalling pathway plays a vital role in cell survival and growth and is generally activated in various tumors,33,34 we supposed that reduced m6A methylation might suppress tumor growth through inactivation of STAT3 pathway.
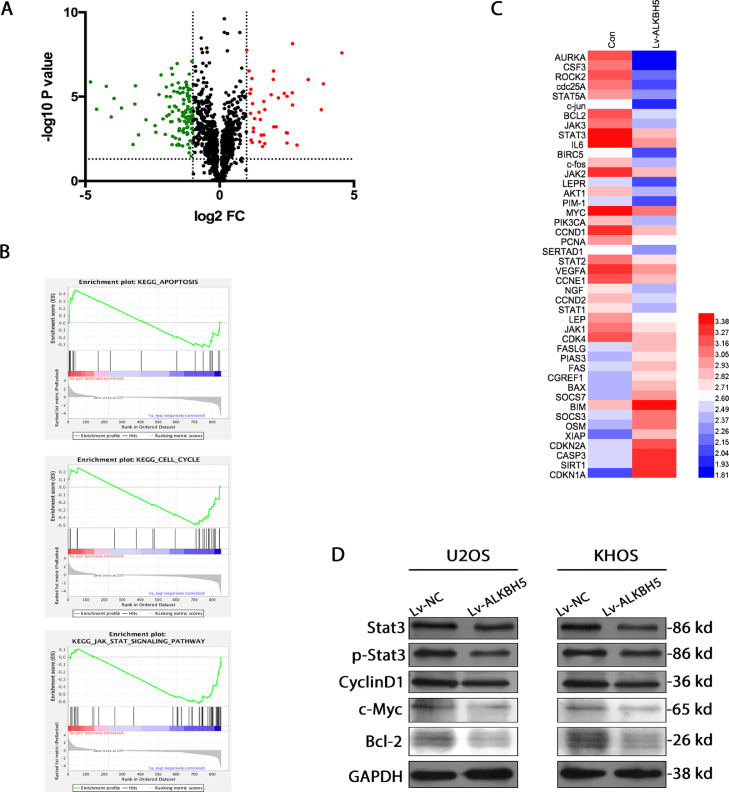
Figure 3.
RNA sequencing identifies transcripts with altered methylation in osteosarcoma. a. Volcano plots for ALKBH5 overexpression KHOS cells versus controls. Significantly upregulated (red) and downregulated (green) transcripts are shown (|logFC| > 1, P < 0.001, FDR < 0.01). b. Gene set enrichment analysis (GSEA) validated enrichment of cell cycle, apoptosis, and tumor-related pathway. c. A heatmap showing changes in representative cell proliferation, cell cycle and apoptosis related genes and pathways, including STAT3. d. By western blot, it was found that downstream targets of STAT3 (CyclinD1,c-Myc and bcl-2)showed decreased activation in the cells relative to control.
We next determined if reduced m6A methylation in osteosarcoma cells affects STAT3 signalling by investigating the phosphorylation status of STAT3.The ALKBH5 overexpression cell lines showed decreased phosphorylation levels of STAT3 when compared with the relevant control cell lines (Figure 3d). To figure out whether these changes in STAT3 phosphorylation stimulate STAT3 signalling, we assessed the expression levels of downstream targets of STAT3. CyclinD1,c-Myc and bcl-2 showed decreased activation in the cells relative to control. These results suggest that reducing m6A methylation inactivates the STAT3 pathway.
m6A methylation regulates the expression of regulator of STAT3 activation
Interestingly, there was no distinct effect on the STAT3 m6A modification after ALKBH5 overexpression (Figure 4a),suggesting that STAT3 may not be a direct target of ALKBH5 in osteosarcoma. To determine the mechanisms underlying inhibition of STAT3 upon reduced m6A methylation, we examined SOCS3, a negative regulator of STAT3.
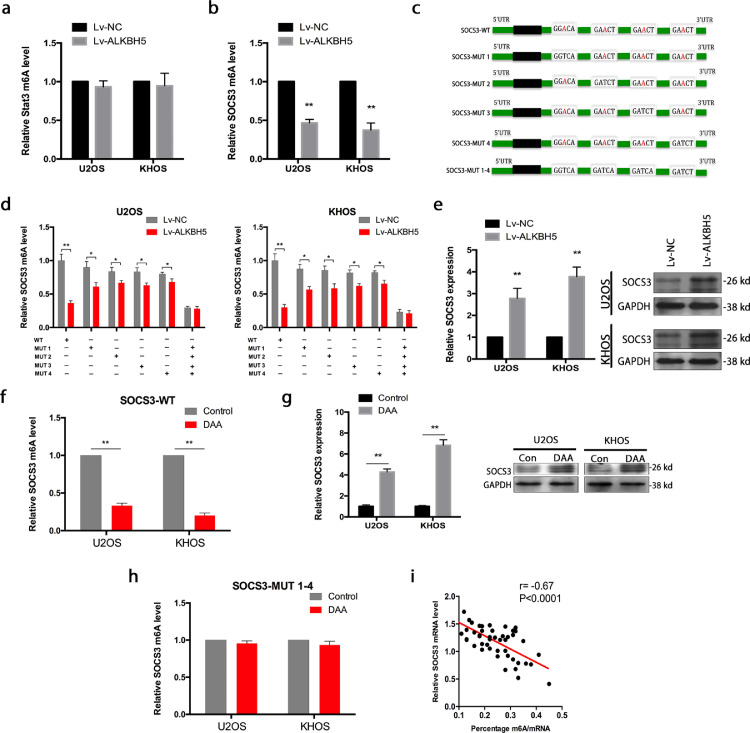
Figure 4.
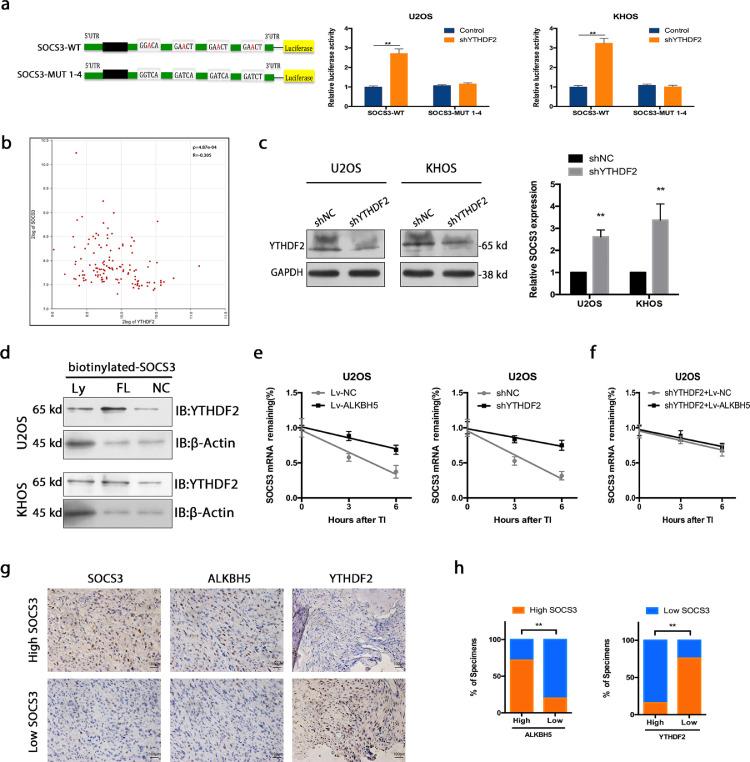
m6A methylation regulates the expression of regulators of STAT3 activation. a. There was no distinct effect on the STAT3 m6A modification after ALKBH5 overexpression. b. SOCS3 m6A methylation decreased after ALKBH5 overexpression. c. Mutations at the four putative m6A sites in SOCS3 (A to T) were generated. d. ALKBH5 decreased the m6A modifications of SOCS3-WTand SOCS3-MUTs (which contains only one potential m6A site) in osteosarcoma cells. And the decrease degree of m6A modification was suppressed in SOCS3-MUTs compared to SOCS3-WT.While the m6A modification of SOCS3-MUT1–4 (which contains all four potential m6A sites mutation) was not decreased in osteosarcoma cells with overexpression of ALKBH5. e. SOCS3 expression was elevated when ALKBH5 was overexpressed both in U2OS and KHOS cells. f. SOCS3 m6A modification level of osteosarcoma cells was decreased when treated with DAA. g. SOCS3 expression was augmented strongly when treated with DAA. h. The decrease of m6A modification caused by DAA was not detected in SOCS3-MUT1–4 cells. i. SOCS3 expression negatively correlated with m6A methylation in osteosarcoma tissues. The data represent the mean± S.D. of three independent experiments. ANOVA or t-tests was used for statistical analysis. *P<0.05 or **P<0.01 indicates a significant difference between the indicated groups.
We overexpressed ALKBH5 in U2OS and KHOS cells. As expected, the SOCS3 m6A modification decreased after ALKBH5 overexpression (Figure 4b). Through analysis of published m6A-seq data set (GSE37005),we found that SOCS3 mRNA 3′ UTRs have highly enriched and specific m6A peaks and identified four m6A sites of SOCS3-3′ UTR in RMBase v2.0 (rna.sysu.edu.cn)(Figure S3). We performed mutations at the four putative m6A sites in SOCS3 (A to T), which indicated as SOCS3-MUTs: SOCS3-MUT1, SOCS3-MUT2, SOCS3-MUT3, SOCS3-MUT4 (which contains only one potential m6A site) and SOCS3-MUT1-4 (which contains all four potential m6A sites) (Figure 4c). We then tested the m6A modification levels in SOCS3-WT and SOCS3-MUTs by using MeRIP-qPCR.ALKBH5 decreased the m6A modifications of SOCS3-WT, SOCS3-MUT1, SOCS3-MUT2, SOCS3-MUT3 and SOCS3-MUT4 in osteosarcoma cells compared to the control vector of ALKBH5. And yet, the decrease degree of m6A modification was suppressed in SOCS3-MUTs (which contains only one potential m6A site) compared to SOCS3-WT because of the presence of m6A site mutation. Importantly, the m6A modification of SOCS3-MUT1-4 (which contains all four potential m6A sites mutation) was not decreased in U2OS and KHOS cells with overexpression of ALKBH5 (Figure 4d).
Based on RT-qPCR, SOCS3 expression was elevated when ALKBH5 was overexpressed both in U2OS and KHOS cells (Figure 4e). In addition, we showed the SOCS3 m6A modification level of osteosarcoma cells was decreased when treated with 3-deazaadenosine (DAA), a global methylation inhibitor (Figure 4f). Meanwhile, the SOCS3 expression was augmented strongly (Figure 4g). But this decrease of m6A modification caused by DAA was not detected in SOCS3-MUT1–4 cells (Figure 4h). We also detected a linear correlation between m6A methylation and SOCS3 expression in osteosarcoma tissues (Figure 4i). DAA is a nonselective drug, which can globally alter methylation of transcripts. This methylation inhibitor was used here to verify the influence of ALKBH5-mediated m6A on SOCS3 mRNA stability.
Overexpression of ALKBH5 enhances SOCS3 mRNA stability via an m6A-YTHDF2-dependent mechanism
Next, the effects of m6A-mediated regulation on SOCS3 were examined. Epigenetic m6A modification is recognized largely by reader proteins, such as YTHDF1, YTHDF2, or YTHDF33. YTHDF1 and YTHDF3 function in translation regulation, while YTHDF2 mainly increases the decay of m6A methylated transcripts.35 Another point view is that three YTHDF proteins all work together to mediate methylated mRNA degradation.36
Since reduction of m6A methylation seemed to increase the expression of SOCS3, we speculated that SOCS3 transcripts are targets of YTHDF2, which increases the decay of m6A-modified transcripts.17,31 To test whether YTHDF2 directly binds the m6A modification sites of SOCS3 mRNA, dual-luciferase reporter was performed. As shown in Figure 5a, knockdown of YTHDF2 increased luciferase activity of reporter carrying wild-type 3′ UTR fragment of SOCS3, while these changes were abolished when the m6A sites were mutated. In bioinformatics analysis of osteosarcoma (http://hgserver1.amc.nl), a negative correlation between YTHDF2 and SOCS3 was observed (Figure 5b). In line with the above hypothesis, inhibition of YTHDF2 in osteosarcoma cells improved the expression of SOCS3 to a similar degree to overexpression of ALKBH5 (Figure 5c). The YTHDF2 knock down efficiency was confirmed by western blot (Figures 5c and S4). We also knocked down three YTHDF paralogs (YTHDF1, YTHDF2, YTHDF3) to figure out whether three YTHDF proteins work together to enhance the degradation of m6A-modified SOCS3. Interestingly, a significantly increase in SOCS3 expression is seen with triple knockdown. However, comparing with downregulation of YTHDF2 alone, a small increase is seen when these three are down-regulated together. In this study, triple knockdown did not show a significantly larger effect compared to knockdown of YTHDF2 alone (Figure S5a). Meanwhile, YTHDF1 and YTHDF3 had no significant correlation with SOCS3 in bioinformatics analysis of osteosarcoma (http://hgserver1.amc.nl) (Figure S5b).
Figure 5.
Overexpression of ALKBH5 enhances SOCS3 mRNA stability via an m6A-YTHDF2-dependent mechanism. a. Dual-luciferase reporter showed that knockdown of YTHDF2 increased luciferase activity of reporter carrying wild-type 3′ UTR fragment of SOCS3, while these changes were abolished when the m6A sites were mutated. b. Bioinformatics analysis of osteosarcoma showed a negative correlation between YTHDF2 and SOCS3 (http://hgserver1.amc.nl)(n = 88). c. inhibition of YTHDF2 in osteosarcoma cells improved the expression of SOCS3 to a similar degree to overexpression of ALKBH5. d. Streptavidin RNA pull-down assay demonstrated that YTHDF2 bound the SOCS3 full-length transcripts in osteosarcoma cells. e. The SOCS3 mRNA half-lives were significantly increased upon ALKBH5 overexpression and YTHDF2 inhibition in osteosarcoma cells. f. There was no significant difference in SOCS3 mRNA half-lives between ALKBH5 overexpression and control cells with YTHDF2 deficiency. g. Representative images showing high or low expression of SOCS3,ALKBH5 and YTHDF2 in osteosarcoma tissues. h. Correlation between SOCS3 and ALKBH5 or YTHDF2 in osteosarcoma microarray specimens. The data represent the mean± S.D. of three independent experiments. ANOVA or t-tests was used for statistical analysis. **P<0.01 indicates a significant difference between the indicated groups.
Streptavidin RNA pull-down assay was performed to demonstrate that YTHDF2 bound the SOCS3 full-length transcripts in osteosarcoma cells (Figure 5d). Additionally, we examined the RNA decay rate in ALKBH5 overexpressed or YTHDF2 inhibited osteosarcoma cells and the comparable control cells. The SOCS3 mRNA half-lives were significantly increased upon ALKBH5 overexpression and YTHDF2 inhibition in osteosarcoma cells (Figure 5e). To confirm the involvement of m6A reader YTHDF2 in regulation of SOCS3 by ALKBH5, we modulated ALKBH5 expression in cells with YTHDF2 deficiency and examined the SOCS3 RNA decay rate (Figure 5f).
We made further exploration of the clinical correlation between ALKBH5, YTHDF2, and SOCS3 in our study. IHC assay of SOCS3, ALKBH5 and YTHDF2 was performed using the human osteosarcoma tissue microarray. Markedly, SOCS3 expression positively correlated with ALKBH5. However, YTHDF2 expression negatively correlated with SOCS3 in human osteosarcoma tissues (Figure 5g,h).
Our study indicated that the methylated SOCS3 transcripts were directly identified by YTHDF2, which promotes the degradation of methylated SOCS3 mRNA in human osteosarcoma cells via an ALKBH5-m6A-YTHDF2-dependent mechanism.
Inhibition of STAT3 signal mediates the effects of reduced m6A methylation on osteosarcoma cell proliferation and apoptosis
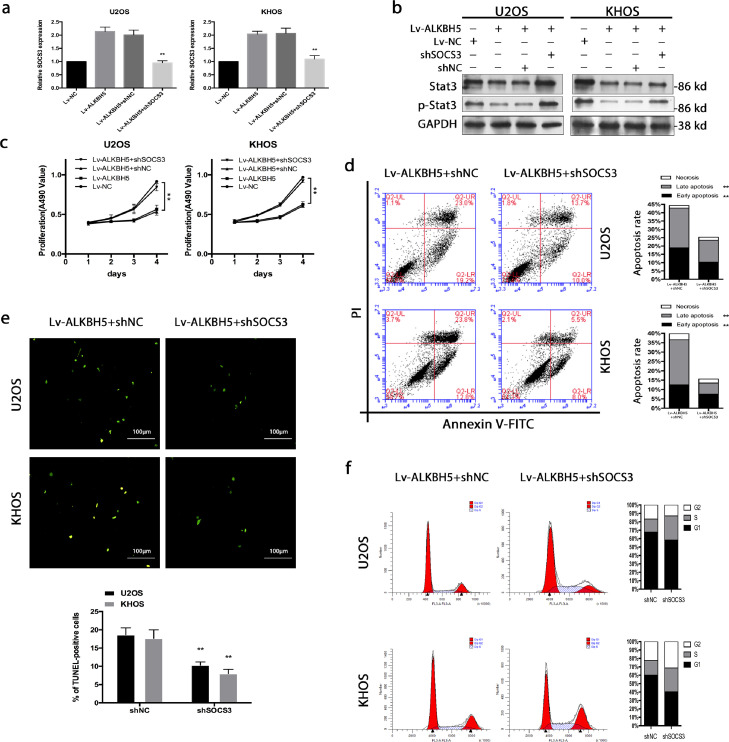
To figure out if inactivation of STAT3 underlies the suppressed proliferation, increased apoptosis and cycle arrest observed upon decreasing m6A methylation in osteosarcoma cells, we attempted to rescue these phenotype by knockdown of SOCS3. The SOCS3 knock down efficiency was confirmed by western blot (Figure S6a).As expected, down-regulation of SOCS3 increased the expression of p-STAT3 in ALKBH5 overexpression cells (Figure 6a,b). Functionally, SOCS3 knockdown (shSOCS3-1) rescued the proliferation inhibition (Figure 6c), apoptosis elevation (Figure 6d,e) and cycle arrest promotion (Figure 6f) caused by up-regulation of ALKBH5. To exclude the off-target effect of shSOCS3, another shSOCS3 sequence (shSOCS3-3) was used to validate functional experiments in flow cytometry assay (Figure S6b) and cell cycle assay (Figure S6c).
Figure 6.
Inhibition of STAT3 signalling mediates the effects of reduced m6A methylation on osteosarcoma cell proliferation and apoptosis. a. The efficiency of SOCS3 down-regulation was validated by qRT-PCR. b. Down-regulation of SOCS3 increased the expression of p-STAT3 in ALKBH5 overexpression cells. c. SOCS3 knockdown rescued the proliferation inhibition. d and e Elevation of cell apoptosis was reversed by SOCS3 knockdown. f. SOCS3 knockdown rescued the cycle arrest promotion caused by up-regulation of ALKBH5. The data represent the mean± S.D. of three independent experiments. ANOVA or t-tests was used for statistical analysis. *P<0.05 or **P<0.01 indicates a significant difference between the indicated groups.
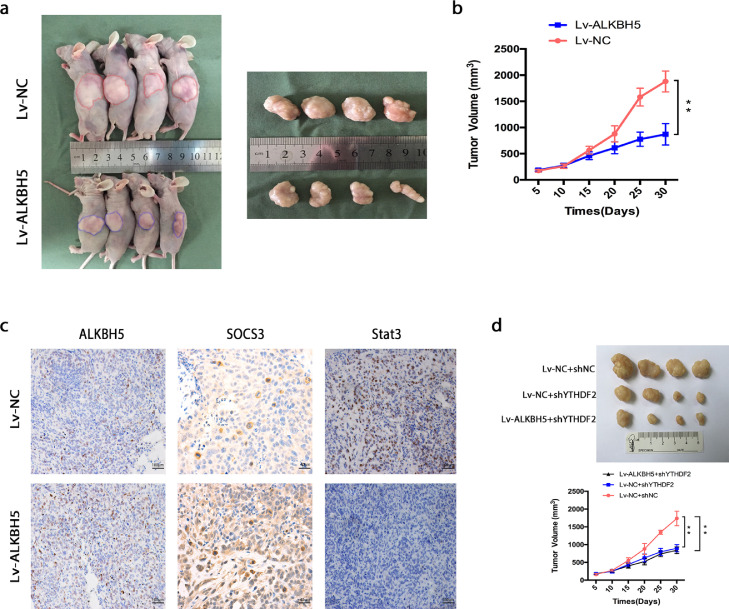
Overexpression of ALKBH5 suppresses human osteosarcoma growth in vivo
To further investigate the function of ALKBH5 in human osteosarcoma, we performed a subcutaneous transplantation assay in nude mice to test whether ALKBH5 exert a crucial influence on osteosarcoma tumorigenicity. As expected, the growth of tumor was effectively inhibited, when implanted with U2OS cells stably expressing ALKBH5 compared to the control group (Figure 7a,b). Moreover, the tumor samples were analyzed by IHC staining, which showed increased staining of SOCS3 and reduced staining of STAT3 expression in the ALKBH5 overexpressed group (Figure 7c). To confirm the involvement of m6A reader YTHDF2 in regulation of SOCS3 in vivo, tumor xenograft models with YTHDF2 down-regulated U2OS cells were constructed. As expected, the tumor growth was effectively inhibited in YTHDF2 down-regulated group. However, ALKBH5 had no significant effect on tumor growth in YTHDF2 deficiency group (Figure 7d). In general, our data suggest that ALKBH5 acts as a tumor suppressor via an m6A-YTHDF2-dependent manner in human osteosarcoma.
Figure 7.
Overexpression of ALKBH5 suppresses human osteosarcoma growth in vivo. a. Overexpression of ALKBH5 effectively inhibited human osteosarcoma subcutaneous tumor growth in nude mice (n = 12 per group). b. The size of tumor was monitored every 5 days. c. Representative images of IHC staining, showing ALKBH5, SOCS3, and STAT3 in two subcutaneous tumor models. d. The tumor growth was effectively inhibited in YTHDF2 down-regulated group. However, ALKBH5 had no significant effect on tumor growth in YTHDF2 deficiency group (n = 4 per group). The data represent the mean± S.D. of three independent experiments. ANOVA test was used for statistical analysis. *P<0.05 or **P<0.01 indicates a significant difference between the indicated groups.
In addition, the expression of differentiation-related genes (COLL1,ALP, OSTEONECTIN and OPG) in osteosarcoma cell lines derived tumor xenograft models were examined (Figure S7). The relative expression of selected genes in subcutaneous tumor models measured using RT-qPCR. The levels of relative gene expression are presented as fold changes compared to the levels detected in control group. Expression of COLL1,OSTEONECTIN and OPG increased slightly. While ALP decreased slightly. However, there were no statistically significant differences in the expression of these differentiation-related genes. Mineralization in osteosarcoma cell lines derived tumor xenograft models was detected by Nanoscale observation (Figure S8).These suggest that osteosarcoma cells maintained tumor cell characteristics throughout the experiment, but did not differentiate.
Discussion
It is a proven fact that m6A mRNA methylation affects the growth and proliferation of tumor cells.21,22 However, the functions of m6A mRNA methylation and underlying mechanisms in human osteosarcoma remain largely obscure. In the present study, we revealed that m6A mRNA methylation modulates the STAT3 pathway to affect cell proliferation and growth in human osteosarcoma. The STAT3 signal pathway plays significant roles in diverse biological processes, and abnormal activation of STAT3 signalling conduces to various tumors.37,38 In our previous study, STAT3 functions as an oncogene in chondrosarcoma.28
In a precious study on osteosarcoma, METTL3-based m6A methylation promotes tumor cell progression by regulation of LEF1.39 However, METTL3 expression level was not significantly correlated with prognosis in our preliminary experiment. Many studies have revealed that ALKBH5-mediated m6A modification plays a vital role in tumor progression.13,23 It is reported that ALKBH5-mediated m6A demethylation suppressed osteosarcoma progression through regulation of YAP.40 In another study, ALKBH5 interacted with PVT1 and increased its stability, which contributes to osteosarcoma tumorigenesis.41 According to our study, reducing m6A methylation levels in human osteosarcoma cells through ALKBH5 up-regulation lead to cell proliferation inhibition, cell apoptosis and cycle arrest. To make the effect of ALKBH5 more visible, a very high level of overexpression is used. What is important is that we have demonstrated this finding in primary osteosarcoma cells. RNA sequencing characterization of ALKBH5 overexpressed cell lines showed that reduced m6A mRNA methylation could suppress cell growth by regulating STAT3 signal pathway. Interestingly, there was no distinct effect on the STAT3 m6A modification after ALKBH5 overexpression, suggesting that STAT3 may not be a direct target of ALKBH5 in osteosarcoma. We identified SOCS3, a negative regulator of STAT3, as a downstream target of ALKBH5-mediated m6A modification in further study.
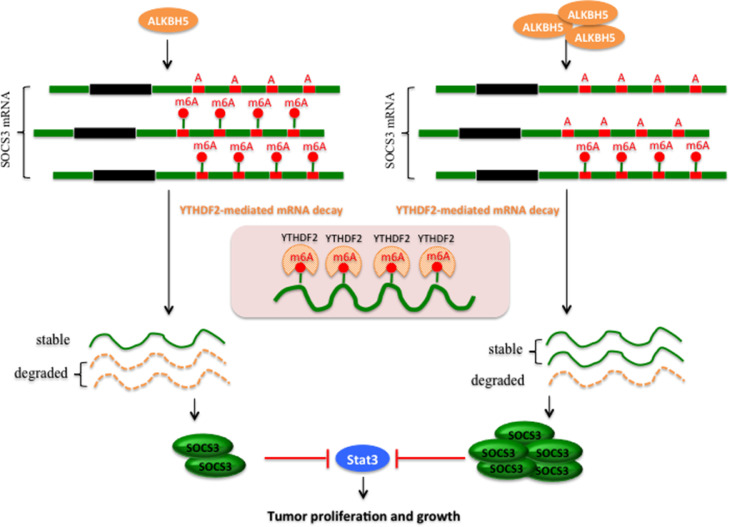
M6A readers have been reported to affect the translation, stability or splicing of target mRNAs.31,42 Epigenetic m6A modification is recognized largely by reader proteins, such as YTHDF1, YTHDF2, or YTHDF33. YTHDF1 and YTHDF3 function in translation regulation, while YTHDF2 mainly increases the decay of m6A methylated transcripts.35 A new point view is that three YTHDF proteins all work together to mediate methylated mRNA degradation.36 Another study shows that METTL3 regulates STAT3 pathway by mediating the expression SOCS3 in an m6A-YTHDF1/YTHDF2-dependent manner.43 Here, we coincidentally demonstrated the methylated SOCS3 was read by YTHDF2 alone, which increases the decay of m6A-methylated transcripts. These results revealed that ALKBH5 inactivated STAT3 pathway by increasing SOCS3 expression via an m6A-YTHDF2-dependent mechanism (Figure 8). It should be noted that YTHDF2-mediated mRNA degradation might play a significant role in tumors with high m6A methylation levels.
Figure 8.
Proposed working model summarizing the mechanism of this study. m6A demethylase ALKBH5 decreases the m6A modification of SOCS3 mRNA, leading to inhibiting YTHDF2-dependent degradation of SOCS3, resulting in up-regulation of SOCS3 expression, thereby inactivates STAT3 pathway and suppresses the tumor proliferation and growth.
A variety of signalling pathways mediated by m6A are associated with tumor prognosis.44 M6A can affect the expression of various of transcripts to regulate the STAT3 signalling pathway, which leads to methylation mediated by different m6A-related enzymes can activate or inhibit the STAT3 signalling pathway in different tumors. Decreased STAT3 activation is possibly one of the major factors of declined proliferation in cells with decreased m6A methylation, as up-regulation of STAT3 is sufficient to reverse the influence on cell growth. Because STAT3 is known to play an important role in regulation of cell proliferation and survival in multiple tumors,28 these discoveries may be suitable beyond osteosarcoma to other malignant tumors mediated by aberrant activated STAT3 signal pathway. Some other tumors could achieve rapid growth via m6A-mediated abnormal STAT3 activation. Nonetheless, we cannot exclude the possible involvement of other signal pathways that could be regulated directly or indirectly by fluctuation in m6A methylation. Our findings advise that regulation of STAT3 signal activity through ALKBH5-mediated m6A methylation could be a universal growth regulatory mechanism that affects diverse other tumors. Although our results reveal that reduced m6A methylation inhibits tumorigenesis in osteosarcoma, decreased m6A methylation could promote cell proliferation and growth in some other tumors.45, 46, 47 A growing number of studies indicated varied mechanisms underlying the effect of m6A methylation in different tumors.48, 49, 50 And further investigations will be necessary.
Contributors
Zechuan Yang and Zhuo Cai are co-first authors. Zhengqian Luo and Xing Bao conceived and designed the study. Xing Bao and Zechuan Yang performed the experiments. Caihong Yang are responsible for grouping experimental animals. Xing Bao analyzed and interpreted the data. Xing Bao wrote the manuscript. Xing Bao, Zhengqian Luo and Zechuan Yang have verified the underlying data. All authors read and approved the final manuscript.
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgments
We thank Dr Tingting Ren for technical assistance.This work was supported by and Science and Technology Foundation of Hubei, China (Grant No. 2017CFB762); the Tongji Hospital Foundation (Grant No. 2201103013); and the National Natural Science Foudation of China (No. 82002849).
Data sharing statement
The datasets generated and/or analysed during the current study are available in the GEO database (https://www.ncbi.nlm.nih.gov/gds/?term=); Correlation between YTHDF1,2,3 and SOCS3 are available in R2 database (http://hgserver1.amc.nl); m6A peaks and sites are available in RMBase v2.0 (rna.sysu.edu.cn).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104019.
Contributor Information
Zhengqiang Luo, Email: luozhengqianghust@163.com.
Xing Bao, Email: baoxing0301@163.com.
Appendix. Supplementary materials
References
- 1.Liu N., Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 2.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Xiao J., Bai J., et al. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol Cancer. 2019;14:137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol Cancer. 2019;29:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Eckert M.A., Harada B.T., et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geula S., Moshitch-Moshkovitz S., Dominissini D., et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 7.Visvanathan A., Patil V., Arora A., et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B.S., Wang X., Beadell A.V., et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S., Sun C., Li J., et al. Roles of RNA methylation by means of N6-methyladenosine (m6A) in human cancers. Cancer Lett. 2017;408:112–120. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C., Samanta D., Lu H., et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:2047–2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu X., Peng W.X., Zhou H., et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27(6):1782–1794. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M., Wong C.M. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020;19:44. doi: 10.1186/s12943-020-01172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S., Zhao B.S., Zhou A., et al. mA demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Q., Shi H., Ye P., et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T., Hu P.S., Zuo Z., et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Y., Ma P., Liu Y., Li W., Shu Y. Multiple functions of m6A RNA methylation in cancer. J Hematol Oncol. 2018;11:48. doi: 10.1186/s13045-018-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M., Wei L., Law C.T., et al. RNA N6-methyladenosine methyltransferase METTL3 promotes liver cancer progression through YTHDF2 dependent post-transcriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 18.Vu L.P., Pickering B.F., Cheng Y., et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng H., Huang H., Wu H., et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su R., Dong L., Li C., et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer K.D., Saletore Y., Zumbo P., et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 23.Cho S.H., Ha M., Cho Y.H., et al. ALKBH5 gene is a novel biomarker that predicts the prognosis of pancreatic cancer: a retrospective multicohort study. Ann Hepatobiliary Pancreat Surg. 2018;22:305–309. doi: 10.14701/ahbps.2018.22.4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok C.T., Marshall A.D., Rasko J.E., Wong J.J. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:39. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang B., Yang Y., Kang M., et al. m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19:3. doi: 10.1186/s12943-019-1128-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Deng X., Su R., Feng X., Wei M., Chen J. Role of N6-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan K., Lei Y., Chen H.N., et al. HBV-induced ROS accumulation promotes hepatocarcinogenesis through Snail-mediated epigenetic silencing of SOCS3. Cell Death Differ. 2016;23:616–627. doi: 10.1038/cdd.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao X., Ren T., Huang Y., et al. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017;8:e2605.27. doi: 10.1038/cddis.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu K., Ren T., Huang Y., et al. Apatinib promotes autophagy and apoptosis through VEGFR2/STAT3/BCL-2 signaling in osteosarcoma. Cell Death Dis. 2017;8:e3015. doi: 10.1038/cddis.2017.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Y., Guo W., Ren T., et al. Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis. Cancer Lett. 2019;440:116–125. doi: 10.1016/j.canlet.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Zhong L., Liao D., Zhang M., et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W., Qi X., Liu L., Ma S., Liu J., Wu J. Epigenetic Regulation of m6A modifications in human cancer. Mol Ther Nucleic Acids. 2019;19:405–412. doi: 10.1016/j.omtn.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin L., Deangelis S., Foust E., et al. A novel small molecule inhibits STAT3 phosphorylation and DNA binding activity and exhibits potent growth suppressive activity in human cancer cells. Mol Cancer. 2010;9:217. doi: 10.1186/1476-4598-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Shen Y., Wang S., Shen Q., Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117–128. doi: 10.1016/j.canlet.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin D., Guo J., Wu Y., et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Zaccara S., Jaffrey S.R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell. 2020;181(7):1582–1595. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuo D., Shogren K.L., Zang J., et al. Inhibition of STAT3 blocks protein synthesis and tumor metastasis in osteosarcoma cells. J Exp Clin Cancer Res. 2018;37:244. doi: 10.1186/s13046-018-0914-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B., Liu Q., Pan S., et al. The HOTAIR/miR-214/ST6GAL1 crosstalk modulates colorectal cancer procession through mediating sialylated c-met via JAK2/STAT3 cascade. J Exp Clin Cancer Res. 2019;38:455. doi: 10.1186/s13046-019-1468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao W., Chen J., Jia L., Ma J., Song D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019;516(3):719–725. doi: 10.1016/j.bbrc.2019.06.128. [DOI] [PubMed] [Google Scholar]
- 40.Yuan Y., Yan G., He M., et al. ALKBH5 suppresses tumor progression via an m6A-dependent epigenetic silencing of pre-miR-181b-1/YAP signaling axis in osteosarcoma. Cell Death Dis. 2021;12(1):60. doi: 10.1038/s41419-020-03315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S., Zhou L., Wang Y. ALKBH5-mediated m6A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. doi: 10.1186/s12935-020-1105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorci M., Ianniello Z., Cruciani S., et al. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018;9:796. doi: 10.1038/s41419-018-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu R., Liu Y., Zhao Y., et al. m6A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 2019;10(3):171. doi: 10.1038/s41419-019-1417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng C., Huang W., Li Y., Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13(1):117. doi: 10.1186/s13045-020-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo X., Chen Z., Gao W., et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L., Wang J., Sun G., et al. m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol Cancer. 2019;18:188. doi: 10.1186/s12943-019-1119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang S., Wei J., Cui Y.H., et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan Q., Liu P.Y., Haase J., Bell J.L., Hüttelmaier S., Liu T. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 49.He L., Li H., Wu A., Peng Y., Shu G., Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng M., Sheng L., Gao Q., et al. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38:3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.