Summary
Background
Emerging evidence highlights that targeting the gut microbiota could be an interesting approach to improve alcohol liver disease due to its important plasticity. This study aimed to evaluate the effects of inulin supplementation on liver parameters in alcohol use disorder (AUD) patients (whole sample) and in a subpopulation with early alcohol-associated liver disease (eALD).
Methods
Fifty AUD patients, hospitalized for a 3-week detoxification program, were enrolled in a randomized, double-blind, placebo-controlled study and assigned to prebiotic (inulin) versus placebo for 17 days. Liver damage, microbial translocation, inflammatory markers and 16S rDNA sequencing were measured at the beginning (T1) and at the end of the study (T2).
Findings
Compared to placebo, AST (β = 8.55, 95% CI [2.33:14.77]), ALT (β = 6.01, 95% CI [2.02:10.00]) and IL-18 (β = 113.86, 95% CI [23.02:204.71]) were statistically significantly higher in the inulin group in the whole sample at T2. In the eALD subgroup, inulin supplementation leads to specific changes in the gut microbiota, including an increase in Bifidobacterium and a decrease of Bacteroides. Despite those changes, AST (β = 14.63, 95% CI [0.91:28.35]) and ALT (β = 10.40, 95% CI [1.93:18.88]) at T2 were higher in the inulin group compared to placebo. Treatment was well tolerated without important adverse events or side effects.
Interpretation
This pilot study shows that 17 days of inulin supplementation versus placebo, even though it induces specific changes in the gut microbiota, did not alleviate liver damage in AUD patients. Further studies with a larger sample size and duration of supplementation with adequate monitoring of liver parameters are needed to confirm these results. Gut2Brain study: https://clinicaltrials.gov/ct2/show/NCT03803709
Funding
Fédération Wallonie-Bruxelles, FRS-FNRS, Fondation Saint-Luc.
Keywords: Alcohol use disorder, Prebiotics, Inulin, Alcohol-associated liver disease, Gut microbiota, Inflammation
Research in context.
Evidence before the study
Alcohol use disorder (AUD) patients displayed concomitant liver and gut microbiota alterations. Recent studies have demonstrated the potential role of the gut-liver axis in alcohol liver disease progression. Inulin is considered a prebiotic and promotes the growth of beneficial bacteria especially Bifidobacteria. Modulating the gut microbiota could be an interesting approach to improve inflammatory status and liver damage in AUD.
Added value of the study
In this pilot interventional randomized placebo-controlled study we demonstrate that 17 days of inulin supplementation in AUD patients has no beneficial effect on liver, microbial translocation or inflammatory markers over alcohol abstinence alone even in patients with early alcohol-associated liver disease (eALD). Although inulin administration does modulate the microbiota inducing specific changes in its composition, AUD patients supplemented with inulin had higher level of AST, ALT and IL-18 after 17 days of supplementation compared to placebo. In particular, in AUD patients presenting an eALD, inulin further decreased microbial diversity of an already disturbed microbiota at baseline.
Implications of all the available evidence
Inulin does modulate the gut microbiota in a similar way independently of the severity of liver disease. However, this prebiotic intervention did not add any additional benefit of alcohol abstinence alone regarding improvement of liver function or inflammatory markers. Further studies with a larger sample size and duration of supplementation with accurate monitoring of liver parameters are needed to confirm these results.
Alt-text: Unlabelled box
Introduction
Alcohol-associated liver disease (ALD) is one of the leading causes of chronic liver disease worldwide. Although most patients with an alcohol use disorder (AUD) present with steatosis, only 10–20% develop progressive forms of liver disease and its related complications. Currently, the pathophysiological mechanisms implicated in liver disease progression are not completely understood and there is no drug approved for treatment of ALD.1
Recent reports have highlighted the potential role of the gut-liver axis in ALD progression. Gut barrier dysfunction together with alterations in the composition of the intestinal microbiota as well as elevated systemic microbial translocation have been associated with ALD progression.2
Murine models of chronic ethanol exposure have been used to discover new potential therapeutic targets at the frontier of the gut-liver axis in ALD.3 Manipulations designed to restore gut barrier function, thus preventing microbial translocation, or alleviating dysbiosis all improved liver disease in animals.4, 5, 6, 7, 8 However, these data cannot necessarily be extrapolated to human pathology for several reasons. Animals have a natural aversion to alcohol,9,10 a 5 times faster ethanol metabolism,11 and profound differences in their immune system12 and their microbiota13 compared to humans. Animals do only develop mild forms of ALD upon chronic alcohol feeding and do not resume the liver-damage pattern observed in humans.14 Clinical studies targeting the gut microbiota in AUD patients are scarce and generally focused on patients with severe alcoholic hepatitis and decompensated cirrhosis.15 Little is known about the impact of a gut microbiota modifying strategy in AUD patients on earlier non-cirrhotic disease stages of ALD. One potential way of modulating the gut microbiota in those patients might be the use of prebiotics. Prebiotics are defined as ‘substrates that are selectively used by host microorganisms conferring a health benefit’16 meaning that they promote the growth of some specific bacteria. Of particular interest, inulin-type fructan (ITF) is known to favour Bifidobacterium and F. prausnitzii two well-recognized beneficial bacteria which are decreased in AUD patients.17, 18, 19 It has been shown that Bifidobacteria negatively correlate with pro-inflammatory cytokines and improve intestinal health in humans.20,21 F.prausnitzii, a butyrate producer, exhibits anti-inflammatory properties both in vitro and in vivo studies.22,23 ITF supplementation and subsequent microbial modulation, have been shown to exert beneficial effects on gut barrier function, reduce microbial translocation and thus attenuate systemic inflammation.15,18,24,25 We therefore designed a randomized, double-blind, placebo-controlled study to modulate the gut microbiota of AUD patients using a 3-week inulin supplementation. Since the liver is closely connected to the gut via the portal vein, we hypothesized that restoring the microbial balance with subsequent improvement of the gut barrier function could exert a beneficial effect on ALD development.
The principal aim of this sub-study was to investigate the effect of inulin supplementation on liver parameters and systemic inflammation in AUD subjects. The secondary objective was to study the effect of supplementation in a subgroup of patients with early alcohol-associated liver disease (eALD).
Methods
Study design and ethics
The Gut2Brain study was a randomized, double blind, placebo-controlled trial. Each subject was randomly assigned to daily intake of prebiotic inulin (Fibruline®; Inulin group) or maltodextrin (Placebo group). To reduce potential gastrointestinal side effects, the dose of inulin or maltodextrin increased gradually from 4 to 16 g per day during the 17 days of treatment (4 g from day 3 to day 4; 8 g from day 5 to day 14 and 16 g from day 15 to day 19 of the detoxification program; Supplementary Figure 1). We have previously shown that 16 g of inulin per day was well tolerated and had a bifidogenic effect in obese patients.18,26,27
The trial protocol was published on protocols.io (dx.doi.org/10.17504/protocols.io.bvs2n6ge).
This study was approved by the ethics committee of the hospital (Nb: 190616V1). All participants signed informed consent prior to inclusion and the trial was registered in the clinicaltrials.gov registry (ClinicalTrials.gov identifier: NCT03803709). Due to an administrative problem beyond our control, a slight delay between enrolment of study participants and registration on the website had occurred. During this period, the trial was conducted as if the trial had been registered, and only six patients were enrolled in that first phase (October-December 2018). In this study, we focused on the outcomes of the Gut2Brain study related to liver and inflammation (primary outcome see28).
Patients
Fifty AUD patients undergoing elective alcohol withdrawal were recruited between October 2018 and December 2019. They followed a 3-week highly standardized alcohol-detoxification and rehabilitation program (Supplementary Figure 1) in an academic hospital (Brussels, Belgium). This program includes 1 week in the hospital, followed by 1 week at home and another week in the hospital (Supplementary Figure 1).
AUD patients were diagnosed by a psychiatrist according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition.
Eighteen to 65 years old male or female subjects who were actively drinking until at least 48 h prior to admission were included in the study. The following exclusion criteria applied: presence of another addiction (except tobacco), inflammatory bowel disease, chronic inflammatory diseases (such as rheumatoid arthritis), cancer, obesity (BMI ≥30 kg/m2), diabetes, bariatric surgery, or severe cognitive impairment (Mini Mental State Examination (MMSE) < 24). Patients with known cirrhosis or significant hepatic fibrosis (≥F2) detected by Fibroscan (>7.6 kPa) immediately after admission were also excluded from the study. Patients who have been taking antibiotics, probiotics or prebiotics during the 3 months prior to enrolment or who had regularly used non-steroidal anti-inflammatory drugs or glucocorticoids during the month prior to enrolment were excluded.
Randomization and masking
The randomization was performed using the method of randomly permuted blocks via the website www.randomization.com, by a researcher not clinically involved to ensure the double blind. The treatment allocation sequence was conserved in a sealed envelope kept in a separate location, inaccessible to the investigators, and was only made available at the time of database lock. Neither the patient nor the investigators knew which boxes contained inulin or placebo until unblinding at the end of the study.
Examinations and sample collections
A Fibroscan® (Echosense, Paris, France) combined with the controlled attenuation parameter (CAP) was performed at admission and repeated after 19 days. Fasting blood samples were drawn and stool samples collected twice on day 2 (T1) and 19 (T2) after the admission. Blood samples were centrifuged at 1000g for 15 min at 4°C. Plasma, serum and stool samples were stored at -80 °C until use.
Inflammatory markers
Plasma concentrations of inflammatory markers (IL-18, MCP-1, IFN-γ, IL-8, IL-10, TNF-a, IL-6) and Fibroblast growth factor 21 (FGF-21) were determined using the Meso Scale Discovery (MSD) U-PLEX assay (Rockville, MD, USA) following the manufacturer's instructions.
Microbial translocation and biomarkers of liver cell damage
Microbial translocation was determined in plasma using Lipopolysaccharide Binding Protein (LBP), soluble CD14 (sCD14) (Human LBP duoset ELISA and Human CD14 Quantikine ELISA kit sCD14, Biotechne Ltd, Abingdon, United Kingdom) and Peptidoglycan Recognition Proteins (Human PGRPs ELISA kit, Thermofisher, Merelbeke, Belgium). Liver damage was assessed by measuring serum Keratin 18 (K18) (K18-M65 ELISA kit; TECOmedical AG, Sissach, Switzerland). All assays were performed in duplicate following the manufacturer's instructions. Routine biochemical analyses were performed by the clinical biochemistry laboratory of the hospital.
Classification of patients according to severity of liver disease
Currently, non-invasive testing allows for a good estimation of steatosis and fibrosis. Controlled attenuation parameter measurements > 250 dB/m is a generally accepted cut-off for steatosis29,30 and liver elasticity measurements by Fibroscan reliably predict fibrosis stage.31 In addition, a novel biomarker, keratin 18 (K18), correlates with histological changes32 and has the potential to distinguish severe forms of ALD from less severe ones.33,34 Our own studies also showed that K18-M65 can be used to discriminate patients with simple steatosis from those that have progressed to early stages of alcohol-associated liver disease or steato-fibrosis.2 Validation of those findings in a cohort of 200 patients identified a level of K18-M65> 270 U/L as a cut-off with excellent sensitivity and specificity (own unpublished data). Therefore, AUD patients were classified according to the severity of liver disease using these clinical biomarkers. Briefly, patients with a normal or increased CAP values (> 250 dB/m)29,30 but normal transaminases, normal bilirubin and K18-M65 < 270 U/L were considered as having minimal liver disease (minimal liver involvement or simple steatosis). Patients with CAP values >250 dB/m AND K18-M65 >270 U/L and/or AST/ALT >40 U/L were considered as having progressed to early alcohol-associated liver disease (eALD). In addition to CAP values, the presence of steatosis was further confirmed by Doppler ultrasound examination performed on the second day after admission. According to the predefined inclusion/exclusion criteria, patients with liver stiffness values > 7.6 kPa on Fibroscan at admission (significant fibrosis) were excluded from the study.
Patients’ care and monitoring
Patients are taken care of by a multidisciplinary team consisting of a gastroenterologist, a psychiatrist, a psychologist, a dietician, a social assistant, and a dedicated nursing team. At admission, a complete medication and past medical history was taken, and a complete physical examination was performed including collection of basic demographic data such as age, gender, weight, and height. The amount of alcohol consumed the week before hospitalization was evaluated with the time line follow-back approach as previously described.35,36 During hospitalization, patients underwent daily clinical monitoring by the nursing and medical team following the standardized scheme of the unit. All adverse events considered as not related to alcohol withdrawal were recorded. Psychological support was provided throughout the study.
Potential gastro-intestinal side effects of inulin were assessed by a questionnaire commonly used to evaluate the symptoms of irritable bowel syndrome.37 This questionnaire assessed the presence of abdominal pain and bloating, satisfaction about intestinal transit and whether gastrointestinal symptoms affected daily life by using visual analogic scale from 0 to 100. Patients were also asked about the frequency of stools and completed the Bristol Stool Form Scale (BSFS)38 allowing to identify stool types ranging from the hardest (type 1) to the softest (type 7). Evaluation was performed at admission, during the second week of treatment and at the end of the study.
Dietary intake was evaluated by a trained dietician at admission of week 1 and week 3. During the week at home, dietary changes were monitored using a food diary. Detailed results of the dietary aspects are published elsewhere.28,39 Dietary advice was provided “on demand” at the end of study. All patients received standard hospital diet during hospitalization.
16S rRNA sequencing and data analysis
Stool samples were collected at Day 2 (T1) and at the end of the intervention (Day 19 – T2). They were collected and stored immediately at -20 °C and then transferred to -80 °C within 5 to 10 h. Genomic DNA was extracted from the faeces using a QIAamp DNA Stool Mini Kit (Qiagen, Germany), including a bead‐beating step and following the protocol Q.40 The composition of the gut microbiota was analysed by Illumina sequencing of the 16S rRNA gene and qPCR of 16S rDNA was used to quantify the abundance of total bacteria and Bifidobacterium spp. (see supplementary Material-Microbiota analysis).
Statistical analysis
Data were presented as mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). The efficacy analysis has been conducted in the modified intention to treat (mITT) population including all the randomized patients that completed the study. According to data distribution, baseline parameters were compared using T-test or Mann–Whitney U test. Linear regression models adjusted for gender, the quantity of ethanol consumed during the second week of the program and the baseline measurement of the outcome were performed to analyse the difference between placebo and inulin groups at T2.
For the gut microbiota analysis only the phyla, families, and genera with an average relative abundance superior to 0.1% were analysed. We used a Mann–Whitney U test in R to compare the relative abundance between groups and the within group analyses were evaluated using a Wilcoxon paired test.
Spearman partial correlations (adjusted for gender and the quantity of ethanol consumed during the second week of the program) were performed to assess the relationships between biological outcomes and microbial data. A pvalue < .05 was considered as statistically significant. Statistical analyses were performed using SAS version 9.4 and Graphpad Prism 8.0. As this was a pilot study with a small sample size, the multiplicity of outcomes was not taken into account.
Sample size
Sample size was estimated using G*Power based on the bifidogenic effect of inulin.18 Therefore, we estimated that a total sample size of 50 participants, with a 20% drop out during the study and 20 patients in each group completing the study provides 80% power to observe an effect size of 0.34 (target difference, SD: 4.25) for the relative abundance of Bifidobacterium genus using a power calculation test with a 0.05 two-sided significance level.
Role of funding source
The funders had no role in collection, analysis, and interpretation of the data or in the writing of this publication.
Results
Study population
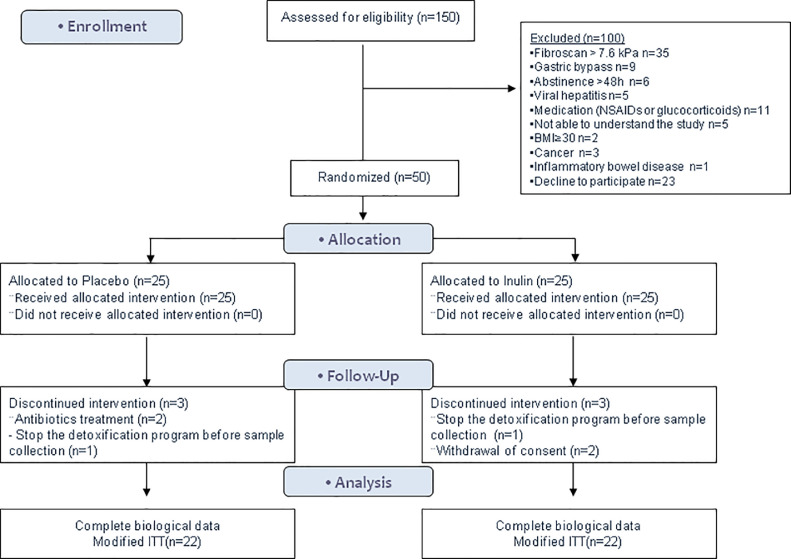
To reach the final sample, 150 patients were screened, 77 did not meet the inclusion criteria and 23 refused to participate. Finally, 50 AUD patients were randomized. Among those, 44 patients had complete biological data and composed the modified intention to treat (ITT) population (Figure 1). Compliance with the nutritional supplementation was 96% in the placebo group and 98% in the inulin group. The sociodemographic characteristics and biological parameters at baseline are presented in Tables 1 and 2. There was a women predominance in the inulin group (20% in placebo vs 48% in the inulin group, Chi-Square test p = 0.11) and they had one additional criterion in the DSM-5 classification compared to the placebo group (7.9 ± 1.9 in placebo vs 9.3 ± 1.4 in inulin group, Mann Whitney U test p = 0.02). During the second week of the detoxification program, among the patients who completed the study, 7 patients relapsed in the placebo group vs 10 in the inulin group (32% vs 45% respectively, Fisher's test p = 0.53). Patients who relapsed in placebo group consumed 53 g/d alcohol on average vs 74 g/d in inulin group (Mann Whitney U test, p = 0.47).
Figure 1.
CONSORT flow diagram of the gut2Brain study.
Table 1.
Baseline characteristics of participants in the modified ITT population.
| Placebo n = 22 | Inulin n = 22 | |
|---|---|---|
| Sociodemographic characteristics | ||
| Age (y) | 45.8 [41.0; 56.3] | 47.8 [38.9; 55.9] |
| Gender, n (%) | ||
| Male | 17 (77.3) | 11 (50.0) |
| Female | 5 (22.7) | 11 (50.0) |
| Marital status, n (%) | ||
| Couple/ married | 10 (45.4) | 7 (32.0) |
| Single | 8 (36.4) | 12 (52.0) |
| Separated/divorced | 4 (18.2) | 3 (16.0) |
| Educational level, n (%) | ||
| Primary | 2 (9.1) | 2 (9.1) |
| Secondary | 9 (40.9) | 6 (27.3) |
| University | 11 (50.0) | 14 (63.6) |
| Clinical examination | ||
| Weight (kg) | 71.2 ± 10.2 | 73.4 ± 14.7 |
| BMI (kg/m2) | 23.3 ± 3.5 | 24.4 ± 3.1 |
| MMSE score | 28.7 ± 1.2 | 27.7 ± 2.9 |
| Smoking, n (%) | 18 (81.8) | 16 (72.7) |
| Alcohol history | ||
| DSM-5 AUD score | 7.9 ± 1.9 | 9.3 ± 1.4 |
| Age of loss of control (years) | 29.5 [25.0; 36.0] | 32.0 [22.0; 40.0] |
| Number of alcohol withdrawal cures | 2.5 ± 2.4 | 2.0 ± 2.1 |
| Duration of drinking habit (years) | 16.4 [5.7; 21.2] | 14.9 [5.3; 22.4] |
| Alcohol consumption (g/day) | 130.0 [85.6; 178.5] | 143.3 [96.0; 180.0] |
Data are described as Mean ± standard deviation or numbers (n) with (%). Age, age of loss of control, duration of drinking habits and alcohol consumption are described as median [Q1; Q3].
AUD, Alcohol use disorders; Alcohol Use Disorders Test; BMI, Body mass index; DSM-5, Diagnostic and Statistical Manual of Mental Disorders fifth edition; MMSE, Mini Mental State Examination
Table 2.
Liver related and biochemical markers at baseline.
| Placebo n = 22 | Inulin n = 22 | |
|---|---|---|
| Liver related markers | ||
| AST, IU/L | 32.0 [25.0; 76.0] | 36.0 [22.0; 70.0] |
| ALT, IU/L | 32.5 [22.0; 62.0] | 32.5 [19.0; 78.0] |
| yGT, IU/L | 71.5 [52.0; 185.0] | 63.0 [47.0; 131.0] |
| Liver elasticity, kPa | 5.5 [4.5; 6.0] | 5.7 [4.6; 6.3] |
| CAP, dB/m | 267.0 [200.0; 312.0] | 307.0 [234.0; 333.0] |
| K18-M-65, pg/ml | 215.0 [164.6; 322.9] | 278.7 [162.3; 417.5] |
| FGF-21, pg/ml | 2073.0 [1633.0;3915.0] | 2632.4 [2048.0; 4769.1] |
| Biochemical markers | ||
| Bilirubin, mg/dL | 0.40 [0.30; 0.60] | 0.40 [0.30; 0.50] |
| Albumin, g/L | 46.0 [44.0; 48.0] | 47.0 [46.0; 48.0] |
| Creatinine mg/dl | 0.78 [0.67; 0.84] | 0.76 [0.66; 0.86] |
| Bacterial translocation | ||
| LBP, ng/ml | 18.6 [18.0; 20.1] | 19.1 [18.1; 19.7] |
| sCD14, ng/ml | 1601.4 [1470.0; 1845.6] | 1739.2 [1516.0; 2081.3] |
| PGRP, ng/ml | 44.9 [35.7; 57.1] | 49.5 [37.2; 74.9] |
| Inflammation | ||
| IL-18, pg/ml | 462.8 [368.4; 656.6] | 496.3 [370.6; 679.1] |
| IL-8, pg/ml | 5.3 [4.0; 7.5] | 5.4 [4.9; 8.5] |
| MCP-1, pg/ml | 249.2 [215.5; 335.5] | 240.4 [221.0; 285.6] |
| TNFa, pg/ml | 2.1 [1.9; 3.1] | 3.0 [2.0; 4.2] |
| IL-6, pg/ml | 4.2 [2.3; 5.8] | 4.3 [3.0; 6.9] |
| IL-10, pg/ml | 1.5 [0.6; 2.1] | 1.7 [1.2; 2.6] |
Data are described as median [Q1; Q3].
AST, Aspartate transaminase; ALT, Alanine transaminase; CAP, controlled attenuation parameter; K18-M65, Serum Keratin 18; sCD14, soluble CD14.
We first studied the effect of inulin supplementation compared to the placebo group in the whole sample (n = 50).
Then, we stratified the population according to the severity of the liver disease (minimal liver disease (LD) vs early ALD) to study the effect of inulin supplementation in patients with more severe liver disease.
Inulin supplementation did not improve liver damage, microbial translocation, and inflammatory markers over abstinence alone
The univariate analysis highlighted that inulin supplementation did not have a beneficial effect on liver enzymes, microbial translocation, and inflammatory markers (Table 3). Since an imbalance between the groups was initially observed between gender (50% of woman in inulin group vs 23% in placebo group), we adjusted the analysis for this parameter as well as for the amount of ethanol consumed during the intermediate week and the baseline parameter. Indeed, these parameters are known to influence the recovery of liver and inflammatory parameters.41,42 Linear regression analysis revealed that the levels of AST and ALT at T2 were higher in the inulin group than in the placebo group (Linear regression models β = 8.55, 95% CI [2.33; 14.77], p = 0.008 and β = 6.01, 95% CI [2.02; 10.00] p = 0.004 respectively; Table 3). Five patients (22.7%) in the inulin group had an AST value above the upper limit of normal, while all patients in the placebo group returned to a normal value at T2 (Fisher test, p = 0.047). CAP values and FGF-21 were not significantly different between the placebo group and the inulin group (Table 3).
Table 3.
Effect of inulin supplementation on liver function, bacterial translocation, and inflammatory markers in the global sample.
| Placebo n = 22 | Inulin n = 22 | Adjusted difference at T2 (inulin vs placebo) β [95% CI] | P1 | |
|---|---|---|---|---|
| T2 | T2 | |||
| Liver related markers (normal range) | ||||
| AST (<40 IU/L) | 24.9 ± 7.4 | 31.6 ± 17.7 | 8.55 [2.33; 14.77] | 0.008 |
| ALT (<40 IU/L) | 12.2 ± 6.8 | 16.6 ± 12.8 | 6.01 [2.02; 10.00] | 0.004 |
| CAP (< 250 dB/m) | 226.4 ± 39.5 | 243.9 ± 57.3 | 4.76 [-23.98; 33.51] | 0.73 |
| K18-M-65 (<270 pg/ml) | 221.1 ± 109.7 | 306.9 ± 204.9 | 74.48 [-1.89; 150.84] | 0.06 |
| FGF-21, pg/ml | 2240.2 ± 1409.9 | 2590.3 ± 1515.7 | -164.70 [-867.38; 537.97] | 0.64 |
| Bacterial translocation | ||||
| LBP, ng/ml | 18.7 ± 1.6 | 19.1 ± 1.7 | 0.46 [-0.30; 1.22] | 0.23 |
| sCD14, ng/ml | 1406.8 ± 282.2 | 1647.8 ± 448.7 | 94.71 [-92.61; 282.04] | 0.31 |
| PGRP, ng/ml | 50.2 ± 18.5 | 58.9 ± 24.7 | 2.39 [-7.61; 12.40] | 0.63 |
| Inflammation | ||||
| IL-18, pg/ml | 474.4 ± 208.4 | 545.3 ± 239.5 | 102.85 [15.46; 190.25] | 0.02 |
| IL-8, pg/ml | 5.3 ± 2.2 | 5.5 ± 2.2 | 0.22 [-1.09; 1.53] | 0.73 |
| MCP-1, pg/ml | 249.1 ± 81.8 | 240.0 ± 87.5 | -2.71 [-47.15; 41.7] | 0.90 |
| TNFα, pg/ml | 2.5 ± 1.2 | 3.1 ± 1.5 | 0.20 [-0.41; 0.80] | 0.51 |
| IL-6, pg/ml | 4.7 ± 2.7 | 4.7 ± 2.9 | -0.31 [-1.90; 1.27] | 0.69 |
| IL-10, pg/ml | 1.8 ± 1.1 | 1.9 ± 1.0 | -0.03 [-0.59; 0.53] | 0.92 |
Linear regression model adjusted for gender, the baseline measurement of the outcome and the quantity of ethanol consumed during the second week of the program
AST, Aspartate transaminase; ALT, Alanine transaminase; CAP, controlled attenuation parameter; FGF-21, Fibroblast growth factor 21; K18-M65, Serum keratin 18; sCD14, soluble CD14.
No difference was observed neither for microbial translocation nor for inflammatory markers between groups except for IL-18. The adjusted linear regression analysis showed a statistically significant higher level of IL-18 in inulin group compared to placebo group at T2 (Linear regression model β = 102.85, 95% CI [1.46; 190.25], p = 0.02; Table 3).
Taken together, our results suggest that 17 days of inulin supplementation has no beneficial effect on liver, bacterial translocation, and inflammatory markers in our population of AUD patients during a period of 3 weeks of abstinence.
The impact of inulin supplementation in AUD patients with early alcohol-associated liver disease (eALD)
We have previously shown that the gut microbiota of patients with more severe liver disease was particularly impaired.2 Therefore, we wanted to study the effect of inulin supplementation in a subgroup of patients with eALD. For this exploratory analysis, we stratified the study population into patients with minimal LD and eALD according to clinical parameters. As by definition, patients with eALD were characterized by elevated AST, ALT, CAP, and serum K18-M65 compared to minimal LD patients. In addition, we found that among the upregulated markers, two of them, FGF-21, and IL-8, also distinguished minimal LD from eALD with statistically significantly higher levels in the latter one (Supplementary Figure 2).
Inulin supplementation induced different changes in gut microbiota composition at the phylum, family and genus level in minimal LD and eALD groups
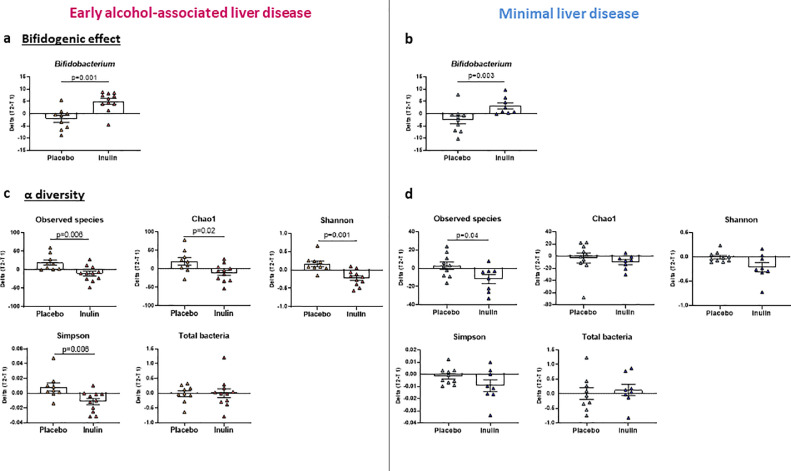
Microbiota analysis at baseline revealed that the eALD group displayed a decreased richness since the Chao-1 index was statistically significantly decreased compared to minimal LD patients (Supplementary Figure 3a). A trend was also observed towards a lower number of species observed and a lower Shannon index (p = 0.09 and p = 0.10 respectively; Supplementary Figure 3a). Total bacteria measured by qPCR, was not different between the groups (Supplementary Figure 3b). The LEfSE analysis showed that Firmicutes and especially Clostridia were higher in minimal LD group while Proteobacteria were statistically significantly higher in eALD patients (Supplementary Figure 3c and d). At the genus level, the relative abundance of NK4A214 group and UCG-005 from Oscillospiraceae family, Ruminococcus and Fusicatenibacter were statistically significantly higher in minimal LD group while Flavonifractor and Veillonella were statistically significantly increased in eALD (Supplementary Figure 3d). The effects of inulin supplementation on the gut microbiota composition have been already published elsewhere, showing namely an increase in Bifidobacterium as a “signature” of inulin intake.28 We compared the change from baseline of Bifidobacterium between placebo and the inulin group. Bifidobacterium increased statistically significantly in the inulin group compared to placebo in eALD as well as minimal LD group (Figure 2a and b). The bifidogenic effect of inulin was therefore confirmed in both groups. Inulin supplementation induced a statistically significant decrease of all α diversity indexes compared to placebo in the eALD group (Figure 2c) while only the number of observed species was decreased in the minimal LD group (Figure 2d). In the eALD group, analysis of phylum and family levels of bacteria revealed minor changes in those who received placebo while an increase of Bifidobacteriaceae (Actinobacteriota) and a decrease of Bacteroidaceae (Bacteroidota) was found in patients supplemented with inulin (Supplementary Figure 4a). In the minimal LD group, inulin supplementation has no effect at phylum level, but similar changes were observed at family level with an additional increase of Veillonellaceae family (Supplementary Figure 4b). At the genus level, prebiotic treatment also increased Dialister whereas Bacteroides, Ruminococcus torques and Dorea decreased in the eALD group (Supplementary Table 1). In the placebo group, Fusicatenibacter, Oscillospiraceae UCG-002, Monoglobus, Alistipes and Lachnospiraceae ND3007 increased statistically significantly between T1 and T2 while Oscillibacter, Flavonifractor, Colidextribacter and Sutterella decreased (Supplementary Table 1).
Figure 2.
Effect of inulin supplementation on gut microbiota composition in minimal liver disease and early ALD patients.
Early alcohol-associated liver disease (eALD): n = 9 and n = 11 in placebo and inulin groups respectively. Minimal liver disease: n = 10 and n = 8 in placebo and inulin groups respectively (a, b) Change in relative abundance of Bifidobacterium between T1 and T2. (c, d) Changes in alpha-diversity indexes: Number of observed species, Chao-1, Shannon, Simpson and total bacteria measured by qPCR. Mann-Whitney U tests were performed to compare placebo and inulin groups. *p < 0.05, **p < 0.01,*** p < 0.001.Data are expressed as mean ± SEM.
The genera modified between T1 and T2 in minimal LD group are presented in Supplementary Table 2. Bifidobacterium and Veillonella increased statistically significantly after 17 days of inulin supplementation. In contrast, Lachnoclostridium, Ruminococcus torques group, Dorea, Tizzerella, Oscillibacter, Colidextribacter, Erysipelotrichaceae UCG-003 and Bacteroides decreased statistically significantly (Supplementary Table 2). In the placebo group, Lachnospiraceae NK4A136 group increased while Blautia, Eubacterium eligens group and Acidaminococcus decreased after 17 days (Supplementary Table 2).
Globally, inulin supplementation exacerbated the decrease in α-diversity but did not impact the bacteria shown to be already altered at baseline in eALD patients.
Inulin supplementation did not improve liver damage, microbial translocation, and inflammatory markers in the subgroup of patient suffering from early alcohol-associated liver disease
We compared liver enzymes, microbial translocation and inflammatory markers between inulin and placebo at T2 and found no difference (Table 4). However, the adjusted linear regression models revealed that AST and ALT at T2 remained higher in the inulin group compared to placebo (Linear regression models β = 14.63, 95% CI [0.91; 28.35], p = 0.038 and β = 10.40, 95% CI [1.93; 18.88] p = 0.02 respectively; Table 4). No statistically significant differences were observed between placebo and inulin for the other markers after adjustment.
Table 4.
Effect of inulin supplementation on liver function, bacterial translocation, and inflammatory markers in patients with early alcohol-associated liver disease.
| Placebo n = 9 | Inulin n = 13 | Adjusted difference at T2 (inulin vs placebo) β [95% CI] | p1 | |
|---|---|---|---|---|
| T2 | T2 | |||
| Liver related markers (normal range) | ||||
| AST (<40 IU/L) | 27.7 ± ± 7.5 | 37.4 ± 20.6 | 14.63 [0.91; 28.35] | 0.04 |
| ALT (<40 IU/L) | 16.3 ± 9.3 | 21.8 ± 13.7 | 10.40 [1.93; 18.88] | 0.02 |
| CAP (< 250 dB/m) | 240.6 ± 37.4 | 256.1 ± 58.1 | 0.20 [-43.54; 43.93] | 0.99 |
| K18-M-65 | 265.0 ± 140.2 | 390.5 ± 217.8 | 123.41 [-30.93; 277.76] | 0.11 |
| FGF-21, pg/ml | 2589.6 ± 1903.5 | 2457.2 ± 1218.2 | -258.20 [-1428.38; 911.98] | 0.65 |
| Bacterial translocation | ||||
| LBP, ng/ml | 19.1 ± 1.4 | 18.7 ± 1.2 | -0.25 [-1.31; 0.82] | 0.63 |
| sCD14, ng/ml | 1293.1 ± 271.6 | 1639.8 ± 423.2 | 210.04 [-88.54; 508.61] | 0.16 |
| PGRP, ng/ml | 43.0 ± 7.7 | 60.8 ± 23.2 | 3.01 [-10.85; 16.86] | 0.65 |
| Inflammation | ||||
| IL-18, pg/ml | 457.9 ± 179.5 | 503.9 ± 222.8 | 99.97 [-15.84; 215.78] | 0.09 |
| IL-8, pg/ml | 6.2 ± 2.6 | 5.2 ± 2.0 | -0.39 [-2.24; 1.47] | 0.66 |
| MCP-1, pg/ml | 282.5 ± 95.0 | 210.3 ± 49.3 | -45.67 [-102.64; 11.29] | 0.11 |
| TNFα, pg/ml | 2.4 ± 1.3 | 2.6 ± 0.8 | 0.06 [-0.60; 0.72] | 0.85 |
| IL-6, pg/ml | 5.3 ± 2.9 | 4.3 ± 3.1 | -1.12 [-3.19; 0.94] | 0.27 |
| IL-10, pg/ml | 2.0 ± 1.3 | 1.6 ± 0.8 | 0.23 [-0.43; 0.88] | 0.47 |
Linear regression model adjusted for gender, the baseline measurement of the outcome and the quantity of ethanol consumed during the second week of the program
AST, Aspartate transaminase; ALT, Alanine transaminase; CAP, controlled attenuation parameter; K18-M65, Serum cytokeratin 18; sCD14, soluble CD14.
Overall, our data do not support a beneficial effect of inulin supplementation compared with placebo, even in the sub-group of patients with more severe liver disease at baseline.
Link between liver alterations, inflammatory markers, and microbiota in the subgroup of patient with eALD
To investigate whether changes in liver or inflammatory markers after treatment could be related to certain bacterial genera, we performed partial correlations adjusted for gender and the amount of alcohol consumed during the second week in the eALD group. We found moderate negative correlations between the variations of Shannon and Simpson indexes, the variation of Fusicatenibacter and the variation of AST level (r = -0.49, p = 0.03; r = -0.47, p = 0.04 and r = -0.46, p = 0.048 respectively; Supplementary Figure 4). The variation of Shannon and Simpson indexes, Oscillospiraceae UCG-002, and Alistipes were negatively associated with the variation of the translocation marker sCD14 level (r = -0.49, p = 0.03; r = -0.50, p = 0.027; r = -0.55, p = 0.01 and r = -0.59, p = 0.007 respectively) while Sutterella was positively associated (r = 0.46, p = 0.048). A strong positive correlation was observed between Oscillibacter and IL-10 while Colidextribacter were moderately correlated with IL-10 (r = 0.60, p = 0.007; r = 0.48, p = 0.04 respectively). No correlation between alpha diversity indexes or bacterial genera and IL-18 levels was observed (supplementary Figure 4).
Tolerability and safety
Tolerability and safety endpoints are described in Table 5. No difference was observed for abdominal pain, bloating, stool frequency, the total score of gastrointestinal tolerance or the creatinine level between placebo and inulin group at the end of the study. Only the BSFS score was statistically significantly higher in inulin group compared to placebo (3.2 ± 1.5 vs 4.5 ± 3.3 in placebo and inulin group, respectively). However, a BSFS score of 4.5 is considered as a normal score.38 One patient complained of diarrhoea in the inulin group which has been considered as possibly related to the compound. Three patients reported being constipated in the placebo group. One patient developed an allergic contact dermatitis that was not related to the intervention. No severe adverse events have been reported during the study.
Table 5.
Tolerability and safety assessments
| Placebo n=22 |
Inulin n=22 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Study entry | End of the study | Study entry | End of the study | |||||
| Abdominal pain, n (%) | 8 | (34.8) | 2 | (11.1) | 13 | (59.1) | 7 | (31.8) |
| Bloating, n (%) | 8 | (34.8) | 6 | (33.3) | 14 | (63.6) | 10 | (45.5) |
| Stool frequency, mean ± SD | 1.7 ± 1.2 | 1.5 ± 0.9 | 1.5 ± 0.9 | 1.9 ± 1.4 | ||||
| Stool consistency, mean ± SD | 4.7 ± 1.6 | 3.2 ± 1.5 | 5.2 ± 1.9 | 4.5 ± 3.3 | ||||
| Global score of GI symptoms, mean ± SD | 106.6 ± 80.5 | 53.2 ± 62.1 | 152.5 ± 78.7 | 70.7 ± 74.4 | ||||
| Diarrhoea, n (%) | 0 | (0.00) | 0 | (0.00) | 0 | (0.00) | 1 | (4.5) |
| Constipation, n (%) | 0 | (0.00) | 3 | (13.6) | 0 | (0.00) | 0 | (0.00) |
| Creatinine (mg/dl) | 0.77 ± 0.15 | 0.89 ± 0.16 | 0.75 ± 0.12 | 0.92 ± 0.16 | ||||
| Allergic contact dermatitis, n (%) | 0 | (0.00) | 0 | (0.00) | 0 | (0.00) | 1 | (4.5) |
| Serious adverse events, n (%) | 0 | (0.00) | 0 | (0.00) | 0 | (0.00) | 0 | (0.00) |
Discussion
This work aimed at studying the effect of inulin supplementation on the gut-liver axis in AUD patients during alcohol withdrawal. We hypothesized that inulin, through restoration of gut dysbiosis, could improve liver damage in AUD patients especially in a subgroup of patients with eALD defined by clinical parameters. We demonstrated that 17 days of inulin supplementation did not elicit additional benefit over abstinence alone on liver disease and inflammatory parameters in AUD patients. Surprisingly, we observed even a less pronounced reduction of AST, ALT and IL-18 in patients supplemented with inulin compared to placebo. Even after stratifying patients according to the severity of liver disease, we did not find a benefit of inulin in the subgroup of patient suffering from more severe ALD at baseline. AST and ALT remained statistically significantly higher in inulin group than in placebo group at the end of the intervention after adjustment for gender and the quantity of ethanol consumed during the second week of the detoxification program.
Our findings that inulin does not improve liver function or inflammatory markers in the specific context of AUD is in opposite to animal studies where some benefits have been observed43, 44, 45, 46 after alcohol feeding. In obese or diabetic patients inulin supplementation also decreased circulating cytokines and ameliorated liver function tests as did oligofructose supplementation, a short chain ITF, in patients with non-alcoholic steatohepatitis.27,47, 48, 49
The less pronounced decrease of ALT and AST in our patients occurred in parallel to higher IL-18 levels in inulin supplemented subjects than in placebo subjects at T2. This is in line with the proposal that IL-18 could play a specific role in liver injury.50,51 One might postulate that some specific inulin-related changes in the gut microbiota composition could be responsible for this intriguing result. However, we did not find any correlation between the changes in IL-18 levels and changes in genera abundance. Alternatively, we did not assess changes in metabolites from the gut microbiota or bile acids that potentially could explain the changes observed in IL-18. Some studies have revealed that Bifidobacterium can produce ethanol through the fermentation of fibres, which is susceptible to partially counteract the beneficial effect of inulin.52,53 It would therefore be of interest to ensure that inulin does not promote the growth of ethanol-producing bacteria, which could explain the smaller decrease not only in liver enzymes but also in IL-18. This effect might be especially relevant in an AUD population.
Our previous study showed that patients with eALD had a particularly altered microbiota.2 We here observed that inulin supplementation reduced microbial richness compared to placebo in these patients in line with other studies showing a decrease in microbial diversity with inulin supplementation.54,55 Decreased microbial diversity has often been associated with a pathological context in the literature56,57 including ALD.2,58 In our study, this decrease in diversity was negatively correlated with the decrease AST and sCD14 levels in eALD patients and as such might rather be an unfavourable outcome. Early ALD patients are characterized by an overrepresentation of the phylum Proteobacteria compared to minimal LD patient. Proteobacteria has been shown to be increased in patients with liver cirrhosis59,60 and in non-alcoholic steatohepatitis patients with advanced fibrosis and cirrhosis.61 Alcohol use has also been associated with an increase in Proteobacteria considered to be responsible for endotoxemia and hepatic inflammation.62, 63, 64 At least in our context, inulin was not able to counteract this potentially negative impact.
At the genus level, inulin increased the abundance of Bifidobacterium and Dialister while it decreased Bacteroides, Ruminococcus torques group, Dorea and Eubacterium ruminantium group. Most of these changes have already been observed in a different context19,65 but their significance in AUD and ALD is not clear. Addolorato et al showed that Bacteroides was enriched in AUD patients and that this is associated with circulating LPS.58 By contrast, a preclinical study in mice fed a Lieber De Carli diet found that liver alterations due to alcohol are associated with a low level of Bacteroides.66 Bifidobacteria, a core genus in human gut microbiota, are generally associated with good health outcome including improvement of gut barrier function and reduced liver alterations.20,21,67 Our group previously reported that AUD patients had a decreased abundance of Bifidobacteria compared to healthy individuals17 but another study showed an increased faecal abundance of Bifidobacterium in AUD patients with cirrhosis.68 Inulin supplementation in dysbiotic TLR5-KO mice induced hepatic alterations via the increase of butyrate69 indicating a possible negative impact that results from butyrate production via inulin fermentation. It is conceivable that potential beneficial effects of inulin only apply to particular pathological contexts70,71 and that promoting Bifidobacteria, in the context of AUD patients might not be beneficial.
Multiple alterations of the intestinal barrier have been associated with chronic alcohol consumption.2,7,8 It is possible that inulin is not the right compound to restore dysbiosis or that a prebiotic approach alone is not sufficiently powerful to achieve this objective. One small randomized, placebo-controlled study72 and one small open-label study73 used FMT to modulate the microbiota in patients with advanced forms of ALD (cirrhosis and severe alcoholic hepatitis, respectively). They showed some beneficial biological and behavioural effects in the FMT groups indicating that a more “radical” change of the microbiota in patients with advanced ALD might be needed to observe improvements of biological and behavioural parameters. Our study took a different approach looking for benefits of microbiota modulation using a prebiotic in patients with less severe forms of liver disease who followed an in-hospital detoxification program. Given these profound differences in study design, a direct comparison of the results of the studies is not possible. It is also possible that the duration of the supplementation and the dose of inulin were not sufficient to induce significant changes in biological parameters. Indeed, to avoid side effects, the dose of inulin was progressively increased from 4 to 16g/d during the study, thus patients were supplemented with 16g only for 5 days. In addition, we recruited a highly selected population already on abstinence therapy. As abstinence alone already has a strong impact on liver and inflammatory parameters,2,74 this could mask an additional positive effect of inulin. Finally, we have chosen maltodextrin as a placebo. Although a study in mice showed that maltodextrin could lead to a decrease in the gut barrier function,75 no significant changes were observed in microbial translocation markers neither in the microbial composition.
The principal strength of our pilot study is the blinded-randomized-placebo controlled design. However, some limitations do apply. Since gender is known to influence the biological parameters including liver damage,41,42 a higher proportion of females in the inulin group could have impacted the results. Although relapse rates were similar between both groups of treatment, we cannot exclude that this parameter could have influenced liver recovery. To limit the consequences of these potential biases, we adjusted the linear regression models for both variables. The sample size for the subgroup analyses was small which could have impacted the results.
Overall, our pilot study shows that modulation of the microbiota can be obtained via inulin in AUD patients. However, 17 days of inulin supplementation versus placebo did not have a beneficial effect on liver recovery after short-term alcohol withdrawal. Further studies with a larger sample size and longer duration of supplementation are needed to confirm the results. Future trials targeting the gut microbiota in AUD subjects, especially those where a decrease in bacterial diversity could be anticipated, should be planned and performed carefully with a particular attention given to liver alterations.
Contributors
Conceptualization & design: PS, SL, NMD, PdT, AMN.
Data curation: CA, SL, PS, NMD, PdT, AMN.
Formal analysis: CA, SL, LM, PS.
Funding acquisition: NMD, PdT, PS.
Investigation: CA, SL, PS.
Methodology: SL, NMD, PdT, PS, AMN.
Project administration: AMN, PS, SL, NMD, PdT.
Resources: NMD, PdT, PS.
Software: CA, SL, PS.
Supervision: PS, PdT,NMD, SL.
Validation: PS, SL, NMD, PdT, AMN.
Writing original draft: CA, LM, PS
Writing review & editing: CA, LM, PS, SL, NMD, PdT, AMN.
All authors read and approved the final manuscript.
Data sharing statement
The study protocol is available on protocol.io (dx.doi.org/10.17504/protocols.io.bvs2n6ge). The statistical analysis plan is provided in supplementary data files. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. The accession number for the raw data generated with the 16S rRNA gene sequencing reported in this paper is BioProject PRJNA745947 (SRA) and data are available through the following link https://www.ncbi.nlm.nih.gov/bioproject/PRJNA745947/.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
We are grateful to the study participants for their participation in the study. We thank Alejandra Ruiz Moreno, as well as the nurses of the Unité Intégrée d'Hépatologie of Saint-Luc Hospital for their technical help. We also thank Isabelle Blave and Bouazza Es Saadi for their excellent technical and experimental assistance in this study. We thank Cosucra for kindly providing the inulin. We thank Ana Beloqui for her help with the MSD cytokines assays. Finally, we are grateful to the INRAE MIGALE bioinformatics facility (MIGALE, INRAE, 2020, Migale bioinformatics Facility, doi: 10.15454/1.5572390655343293E12) for providing help and computing and storage resources.
This study was supported by the Fédération Wallonie-Bruxelles (Action de Recherche Concertée ARC18-23/092). SL is supported by ARC. PS is supported by grants from Fond National de Recherche Scientifique Belgium (FRS-FNRS J.0146.17 and T.0217.18). NMD is a recipient of grants from the Fonds de la Recherche Scientifique (FRS-FNRS, convention PDR T.0068.19 and convention PINTMULTI R.8013.19 (NEURON, call 2019)). PdT is a recipient of other grant support from the Fondation Saint-Luc.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104033.
Appendix. Supplementary materials
References
- 1.Global status report on alcohol and health 2018. Geneva: World Health Organisation; 2018. https://www.who.int/publications/i/item/9789241565639. Accessed 22 September 2021.
- 2.Maccioni L., Gao B., Leclercq S., et al. Intestinal permeability, microbial translocation, changes in duodenal and fecal microbiota, and their associations with alcoholic liver disease progression in humans. Gut Microbes. 2020;12 doi: 10.1080/19490976.2020.1782157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starkel P., Leclercq S., de Timary P., Schnabl B. Intestinal dysbiosis and permeability: the yin and yang in alcohol dependence and alcoholic liver disease. Clin Sci. 2018;132:199. doi: 10.1042/CS20171055. (Colch) [DOI] [PubMed] [Google Scholar]
- 4.Chen P., Stärkel P., Turner J.R., Ho S.B., Schnabl B. Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology. 2015;61:883–894. doi: 10.1002/hep.27489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrikx T., Duan Y., Wang Y., et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68:1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P., Torralba M., Tan J., et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148:203–214. doi: 10.1053/j.gastro.2014.09.014. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartmann P., Chen P., Wang H.J., et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58:108–119. doi: 10.1002/hep.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Fouts D.E., Stärkel P., et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19:227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkin R.J.W., Lalor P.F., Parker R., Newsome P.N. Murine models of acute alcoholic hepatitis and their relevance to human disease. Am J Pathol. 2016;186:748–760. doi: 10.1016/j.ajpath.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Tabakoff B., Hoffman P.L. Animal models in alcohol research. Alcohol Res Health. 2000;24:77–84. [PMC free article] [PubMed] [Google Scholar]
- 11.Cederbaum A.I. Alcohol metabolism. Clin Liver Dis. 2012;16:667–685. doi: 10.1016/j.cld.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javier M., Christopher C.W.H. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172 doi: 10.4049/jimmunol.172.5.2731. (Baltimore, Md : 1950) [DOI] [PubMed] [Google Scholar]
- 13.Nguyen T.L.A., Vieira-Silva S., Liston A., Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao B., Xu M.J., Bertola A., Wang H., Zhou Z., Liangpunsakul S. Animal models of alcoholic liver disease: pathogenesis and clinical relevance. Gene Expr. 2017;17:173–186. doi: 10.3727/105221617X695519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leclercq S., de Timary P., Stärkel P. Targeting the gut microbiota to treat alcoholic liver diseases: evidence and promises. Acta Gastroenterol Belg. 2020;83:616–621. [PubMed] [Google Scholar]
- 16.Gibson G.R., Hutkins R., Sanders M.E., et al. Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq S., Matamoros S., Cani P.D., et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci USA. 2014;111:E4485–E4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewulf E.M., Cani P.D., Claus S.P., et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type Fructans in obese women. Gut. 2013;62:1112–1121. doi: 10.1136/gutjnl-2012-303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bastard Q., Chapelet G., Javaudin F., Lepelletier D., Batard E., Montassier E. The effects of inulin on gut microbial composition: a systematic review of evidence from human studies. Eur J Clin Microbiol Infect Dis. 2020;39:403–413. doi: 10.1007/s10096-019-03721-w. [DOI] [PubMed] [Google Scholar]
- 20.Ouwehand A.C., Bergsma N., Parhiala R., et al. Bifidobacterium microbiota and parameters of immune function in elderly subjects. FEMS Immunol Med Microbiol. 2008;53:18–25. doi: 10.1111/j.1574-695X.2008.00392.x. [DOI] [PubMed] [Google Scholar]
- 21.Whorwell P.J., Altringer L., Morel J., et al. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol. 2006;101:1581–1590. doi: 10.1111/j.1572-0241.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 22.Sokol H., Pigneur B., Watterlot L., et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miquel S., Martín R., Rossi O., et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Kellow N.J., Coughlan M.T., Reid C.M. Metabolic benefits of dietary prebiotics in human subjects: a systematic review of randomised controlled trials. Br J Nutr. 2014;111:1147–1161. doi: 10.1017/S0007114513003607. [DOI] [PubMed] [Google Scholar]
- 25.Stachowska E., Portincasa P., Jamioł-Milc D., Maciejewska-Markiewicz D., Skonieczna-Żydecka K. The relationship between prebiotic supplementation and anthropometric and biochemical parameters in patients with NAFLD-A systematic review and meta-analysis of randomized controlled trials. Nutrients. 2020;12:E3460. doi: 10.3390/nu12113460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leyrolle Q., Cserjesi R., D G H Mulders M., et al. Prebiotic effect on mood in obese patients is determined by the initial gut microbiota composition: a randomized, controlled trial. Brain Behav Immun. 2021;94:289–298. doi: 10.1016/j.bbi.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Hiel S., Gianfrancesco M.A., Rodriguez J., et al. Link between gut microbiota and health outcomes in inulin -treated obese patients: lessons from the Food4Gut multicenter randomized placebo-controlled trial. Clin Nutr. 2020 doi: 10.1016/j.clnu.2020.04.005. S0261561420301606. [DOI] [PubMed] [Google Scholar]
- 28.Amadieu C., Coste V., Neyrinck A.M., et al. Restoring an adequate dietary fiber intake by inulin supplementation: a pilot study showing an impact on gut microbiota and sociability in alcohol use disorder patients. Gut Microbes. 2022;14 doi: 10.1080/19490976.2021.2007042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Lédinghen V., Wong G.L.H., Vergniol J., et al. Controlled attenuation parameter for the diagnosis of steatosis in non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2016;31:848–855. doi: 10.1111/jgh.13219. [DOI] [PubMed] [Google Scholar]
- 30.Thiele M., Rausch V., Fluhr G., et al. Controlled attenuation parameter and alcoholic hepatic steatosis: diagnostic accuracy and role of alcohol detoxification. J Hepatol. 2018;68:1025–1032. doi: 10.1016/j.jhep.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 31.Salavrakos M., Piessevaux H., Komuta M., Lanthier N., Stärkel P. Fibroscan reliably rules out advanced liver fibrosis and significant portal hypertension in alcoholic patients. J Clin Gastroenterol. 2019;53:772–778. doi: 10.1097/MCG.0000000000001119. [DOI] [PubMed] [Google Scholar]
- 32.Atkinson S.R., Grove J.I., Liebig S., et al. In severe alcoholic hepatitis, serum keratin-18 fragments are diagnostic, prognostic, and theragnostic biomarkers. Am J Gastroenterol. 2020;115:1857–1868. doi: 10.14309/ajg.0000000000000912. [DOI] [PubMed] [Google Scholar]
- 33.Vatsalya V., Cave M.C., Kong M., et al. Keratin 18 is a diagnostic and prognostic factor for acute alcoholic hepatitis. Clin Gastroenterol Hepatol. 2020;18:2046–2054. doi: 10.1016/j.cgh.2019.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClain C.J., Vatsalya V., Mitchell M.C. Keratin-18: diagnostic, prognostic, and theragnostic for alcohol-associated hepatitis. Am J Gastroenterol. 2021;116:77–79. doi: 10.14309/ajg.0000000000001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maisto S.A., Sobell L.C., Cooper A.M., Sobell M.B. Comparison of two techniques to obtain retrospective reports of drinking behavior from alcohol abusers. Addict Behav. 1982;7:33–38. doi: 10.1016/0306-4603(82)90022-3. [DOI] [PubMed] [Google Scholar]
- 36.de Timary P., Cani P.D., Duchemin J., et al. The loss of metabolic control on alcohol drinking in heavy drinking alcohol-dependent subjects. PLoS One. 2012;7:e38682. doi: 10.1371/journal.pone.0038682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francis C.Y., Morris J., Whorwell P.J. The irritable bowel severity scoring system: a simple method of monitoring irritable bowel syndrome and its progress. Aliment Pharmacol Ther. 1997;11:395–402. doi: 10.1046/j.1365-2036.1997.142318000.x. [DOI] [PubMed] [Google Scholar]
- 38.Blake M.R., Raker J.M., Whelan K. Validity and reliability of the Bristol stool form scale in healthy adults and patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2016;44:693–703. doi: 10.1111/apt.13746. [DOI] [PubMed] [Google Scholar]
- 39.Amadieu C., Leclercq S., Coste V.., et al. Dietary fiber deficiency as a component of malnutrition associated with psychological alterations in alcohol use disorder. Clin Nutr. 2021;40:2673–2682. doi: 10.1016/j.clnu.2021.03.029. [DOI] [PubMed] [Google Scholar]
- 40.Costea P.I., Zeller G., Sunagawa S., et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 41.Guy J., Peters M.G. Liver disease in women: the influence of gender on epidemiology, natural history, and patient outcomes. Gastroenterol Hepatol. 2013;9:633. [PMC free article] [PubMed] [Google Scholar]
- 42.Buzzetti E., Parikh P.M., Gerussi A., Tsochatzis E. Gender differences in liver disease and the drug-dose gender gap. Pharmacol Res. 2017;120:97–108. doi: 10.1016/j.phrs.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Cani P.D., Possemiers S., Van de Wiele T., et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasuda K., Dawson H.D., Wasmuth E.V., et al. Supplemental dietary inulin influences expression of iron and inflammation related genes in young pigs. J Nutr. 2009;139:2018–2023. doi: 10.3945/jn.109.110528. [DOI] [PubMed] [Google Scholar]
- 45.Yang X., He F., Zhang Y., et al. Inulin ameliorates alcoholic liver disease via suppressing LPS-TLR4-Mψ axis and modulating gut microbiota in mice. Alcohol Clin Exp Res. 2019;43:411–424. doi: 10.1111/acer.13950. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z., Zhang X., Zhu L., et al. Inulin alleviates inflammation of alcoholic liver disease via SCFAs-inducing suppression of M1 and facilitation of M2 macrophages in mice. Int Immunopharmacol. 2020;78 doi: 10.1016/j.intimp.2019.106062. [DOI] [PubMed] [Google Scholar]
- 47.Farhangi M.A., Javid A.Z., Dehghan P. The effect of enriched chicory inulin on liver enzymes, calcium homeostasis and hematological parameters in patients with type 2 diabetes mellitus: a randomized placebo-controlled trial. Prim Care Diabetes. 2016;10:265–271. doi: 10.1016/j.pcd.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Dehghan P., Gargari B.P., Jafar-Abadi M.A., Aliasgharzadeh A. Inulin controls inflammation and metabolic endotoxemia in women with type 2 diabetes mellitus: a randomized-controlled clinical trial. Int J Food Sci Nutr. 2014;65:117–123. doi: 10.3109/09637486.2013.836738. [DOI] [PubMed] [Google Scholar]
- 49.Daubioul C.A., Horsmans Y., Lambert P., Danse E., Delzenne N.M. Effects of oligofructose on glucose and lipid metabolism in patients with nonalcoholic steatohepatitis: results of a pilot study. Eur J Clin Nutr. 2005;59:723–726. doi: 10.1038/sj.ejcn.1602127. [DOI] [PubMed] [Google Scholar]
- 50.Flisiak-Jackiewicz M., Bobrus-Chociej A., Tarasów E., Wojtkowska M., Białokoz-Kalinowska I., Lebensztejn D.M. Predictive role of interleukin-18 in liver steatosis in obese children. Can J Gastroenterol Hepatol. 2018;2018 doi: 10.1155/2018/3870454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachmann M., Pfeilschifter J., Mühl H. A prominent role of interleukin-18 in acetaminophen-induced liver injury advocates its blockage for therapy of hepatic necroinflammation. Front Immunol. 2018;9:161. doi: 10.3389/fimmu.2018.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Meulen R., Makras L., Verbrugghe K., Adriany T., De Vuyst L. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl Environ Microbiol. 2006;72:1006–1012. doi: 10.1128/AEM.72.2.1006-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van der Meulen R., Adriany T., Verbrugghe K., De Vuyst L. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD + through its growth-associated production. Appl Environ Microbiol. 2006;72:5204–5210. doi: 10.1128/AEM.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reimer R.A., Willis H.J., Tunnicliffe J.M., Park H., Madsen K.L., Soto-Vaca A. Inulin-type fructans and whey protein both modulate appetite but only fructans alter gut microbiota in adults with overweight/obesity: a randomized controlled trial. Mol Nutr Food Res. 2017;61 doi: 10.1002/mnfr.201700484. [DOI] [PubMed] [Google Scholar]
- 55.Vandeputte D., Falony G., Vieira-Silva S., et al. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut. 2017;66:1968–1974. doi: 10.1136/gutjnl-2016-313271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng F., Li Y., Zhao J. The gut microbiome of healthy long-living people. Aging. 2019;11:289–290. doi: 10.18632/aging.101771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosca A., Leclerc M., Hugot J.P. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Addolorato G., Ponziani F.R., et al. Gut microbiota compositional and functional fingerprint in patients with alcohol use disorder and alcohol-associated liver disease. Liver Int. 2020;40:878–888. doi: 10.1111/liv.14383. [DOI] [PubMed] [Google Scholar]
- 59.Qin N., Yang F., Li A., et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y., Yang F., Lu H., et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 61.de Faria Ghetti F., Oliveira D.G., de Oliveira J.M., de Castro Ferreira L., DE C., Moreira A.P.B. Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur J Nutr. 2018;57:861–876. doi: 10.1007/s00394-017-1524-x. [DOI] [PubMed] [Google Scholar]
- 62.Fukui H. Role of gut dysbiosis in liver diseases: what have we learned so far? Diseases. 2019;7:58. doi: 10.3390/diseases7040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Philips C.A., Augustine P., Yerol P.K., et al. Modulating the intestinal microbiota: therapeutic opportunities in liver disease. J Clin Transl Hepatol. 2020;8:87–99. doi: 10.14218/JCTH.2019.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mutlu E.A., Gillevet P.M., Rangwala H., et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302:G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Healey G., Murphy R., Butts C., Brough L., Whelan K., Coad J. Habitual dietary fibre intake influences gut microbiota response to an inulin-type fructan prebiotic: a randomised, double-blind, placebo-controlled, cross-over, human intervention study. Br J Nutr. 2018;119:176–189. doi: 10.1017/S0007114517003440. [DOI] [PubMed] [Google Scholar]
- 66.Ferrere G., Wrzosek L., Cailleux F., et al. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J Hepatol. 2017;66:806–815. doi: 10.1016/j.jhep.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 67.O'Callaghan A., van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dubinkina V.B., Tyakht A.V., Odintsova V.Y., et al. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome. 2017;5:141. doi: 10.1186/s40168-017-0359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh V., Yeoh B.S., Chassaing B., et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell. 2018;175:679–694. doi: 10.1016/j.cell.2018.09.004. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weitkunat K., Schumann S., Petzke K.J., Blaut M., Loh G., Klaus S. Effects of dietary inulin on bacterial growth, short-chain fatty acid production and hepatic lipid metabolism in gnotobiotic mice. J Nutr Biochem. 2015;26:929–937. doi: 10.1016/j.jnutbio.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 71.Li L.L., Wang Y.T., Zhu L.M., Liu Z.Y., Ye C.Q., Qin S. Inulin with different degrees of polymerization protects against diet-induced endotoxemia and inflammation in association with gut microbiota regulation in mice. Sci Rep. 2020;10:978. doi: 10.1038/s41598-020-58048-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bajaj J.S., Gavis E.A., Fagan A., et al. A randomized clinical trial of fecal microbiota transplant for alcohol use disorder. Hepatology. 2021;73:1688–1700. doi: 10.1002/hep.31496. [DOI] [PubMed] [Google Scholar]
- 73.Philips C.A., Pande A., Shasthry S.M., et al. Healthy donor fecal microbiota transplantation in steroid-ineligible severe alcoholic hepatitis: a pilot study. Clin Gastroenterol Hepatol. 2017;15:600–602. doi: 10.1016/j.cgh.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 74.Leclercq S., De Saeger C., Delzenne N., de Timary P., Stärkel P. Role of inflammatory pathways, blood mononuclear cells, and gut-derived bacterial products in alcohol dependence. Biol Psychiatry. 2014;76:725–733. doi: 10.1016/j.biopsych.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 75.Nickerson K.P., Homer C.R., Kessler S.P., et al. The dietary polysaccharide maltodextrin promotes salmonella survival and mucosal colonization in mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.