Abstract
1,3-Butadiene (BD) exposure is known to cause numerous adverse health effects, including cancer, in animals and humans. BD is metabolized to reactive epoxide intermediates, which are genotoxic, but it is not well know what other effects of BD has on cellular metabolism. We examined the effects of exposure to BD on the mouse lung metabolome in the genetically heterogeneous collaborative cross outbred mouse model. Mice were exposed to 3 concentrations of BD for 10 days (2, 20, and 200 ppm), and lung tissues were analyzed using high-resolution mass spectrometry-based metabolomics. As compared to controls (0 ppm BD), BD had extensive effects on lung metabolism at all concentrations of exposure, including the lowest concentration of 2ppm, as reflected by reprogramming of multiple metabolic pathways. Metabolites participating in glycolysis and the tricarboxylic acid cycle were elevated, with 6 out of 10 metabolites demonstrating a 2 to 8-fold increase, including the oncometabolite fumarate. Fatty acid levels, sphingosine, and sphinganine were decreased (2 to 8-fold), and fatty acyl-CoAs were significantly increased (16 to 31-fold), suggesting adjustments in lipid metabolism. Furthermore, metabolites involved in basic amino acid metabolism, steroid hormone metabolism, and nucleic acid metabolism were significantly altered. Overall, these changes mirror the metabolic alterations found in lung cancer cells, suggesting that very low doses of BD induce metabolic adaptations that may prevent or promote adverse health effects such as tumor formation.
Introduction
1,3-butadiene (BD) is a ubiquitous occupational and environmental pollutant that is carcinogenic in exposed animals and humans. BD exposure occurs through inhalation, with the major sources of exposure being emissions from manufacturing of BD synthetic rubber, plastics, and resins, as well as inhalation of cigarette smoke, vehicle exhaust, and other products of incomplete combustion[1]. The major nonlethal effects of acute exposure include irritation of skin, eye and respiratory tract, and adverse effects on the central nervous system[2]. Chronic and accidental occupational exposures to BD have been shown to result in some neurotoxicity[3], reduced lung function[4], cardiovascular disease (CVD), [5–7], and cancer[8–10]. In mice, the lung is highly susceptible to cancer development with BD exposure, while in humans, lymphopoietic cancers have been reported in exposed industry workers, and are the basis of BD classification as a human carcinogen[8–10].
BD toxicity and carcinogenicity result from its metabolism to reactive epoxide intermediates which form DNA adducts. In the lung, BD is actively metabolized to its epoxide intermediates, which is the site of first contact of BD after inhalation[11]. The epoxide intermediates are deactivated by GST enzymes, and BD exposure leads to a significant depletion of GSH in lung tissues [12, 13]. BD activation, deactivation, and glutathione depletion have been shown to occur most efficiently in the mouse lung as compared to other species, suggesting a direct effect of BD on mouse lung metabolism[14–16].
There is huge amount of data on the metabolism of BD to reactive epoxides and aldehydes and the accompanying depletion of GSH, but to our knowledge there is no report on BD effects on general metabolism [17]. To address this limitation, we applied high-resolution mass spectrometry-based metabolomics (HRM) methods, which are capable of simultaneously measuring thousands of small molecule metabolites in tissues, cells, or biofluids. The levels of these metabolites reflect changes in metabolic activity and biochemical processes associated with environmental or dietary exposures, disease states, or other health status measures. Thus, the purpose of this study was to use untargeted HRM methodology to assess metabolic variation associated with BD exposure in mouse lung, which is a tissue active in BD metabolism and prone to development of BD-induced cancer. Collaborative Cross mice were exposed to 0, 2, 20, or 200 ppm BD for 10 days, and the lung tissue was analyzed using HRM. The lowest exposure groups are highly relevant to current OSHA exposure limit of 1 ppm, while the higher exposure groups are similar to acute temporary occupational exposures of 10 ppm and higher in butadiene manufacturing plants [18–20]. In Rats, exposure to 62.5 ppm BD is carcinogenic[17]. The resulting metabolomics data was analyzed for shifts in metabolic pathways possibly associated with adverse effects of BD in the lung.
Methods
Animal treatment
Female mice (6–8 weeks old) from 60 Collaborative Cross strains were obtained from the University of North Carolina Systems Genetics Core (Chapel Hill, NC)31. Animals were fed a diet of PicoLab® Rodent Diet 20–5053 (Lab Diet; further details available at lab.diet.com) composed of 20% protein, 33.9% starch, 10.6% fat, 4.7% crude fiber, and 6.1% ash, with essential vitamins and minerals and water ad libitum. Animals were housed on a 12-h light-dark cycle and allowed to acclimate to the room for at least one week prior to exposures. Female animals were used because previous studies did not show any sex difference in mice regardining formation of N-terminal valine adducts, and it also allowed comparisons with previously studies in female mice[21, 22] The first goal was to simulate a genetically diverse human population. We used one CC mouse per strain form 60 strains to build four genetically diverse populations, all carrying an identical a gene pool. The collaborative Cross populations, containing one female mouse from each strain (n=60 for each group), were exposed to air (0 ppm BD), 2, 20 or 200 ppm BD by inhalation in whole body exposure chambers for 5 days per week, for 2 weeks for a total of 10-days of exposure[23]. Mice were euthanized immediately after the last exposures, blood was obtained by cardiac puncture, and tissues (lung, liver, spleen) were harvested and flash-frozen. The Institutional Animal Care and Use Committee of the University of Texas Medical Branch Galveston Texas (Protocol number 0808053B) approved all experiments. All experiments were carried out in accordance with established guiding principles for animal research. All BD inhalation studies were performed at the Inhalation Toxicology Core facility at the University of Texas Medical Branch (UTMB) with strict compliance approval of the UTMB Environmental Health and Safety and Chemical Safety committees.
High Resolution Mass Spectrometry-based Metabolomics
Samples were prepared and analyzed as previously described[24, 25]. In brief, 100 μl of water, 200 μl of acetonitrile, and 5 μl of an internal standard mixture containing 14 stable isotope standards were added to each lung sample. Samples were sonicated and incubated on ice for 30 minutes, followed by centrifugation at 16,100 x g at 4°C for 10 minutes. Extracted sample supernatants were transferred to autosampler vials for LC-MS analysis. Triplicates of each sample were loaded onto both C18 (Higgins C18 stainless steel column,2.1 × 50mm) and HILIC (Waters XBridge BEH Amide XP HILIC column, 2.1 × 50mm) liquid chromatography columns and then subjected to negative (C18) or positive (HILIC) electrospray ionization and high-resolution mass spectrometry (Thermo Orbitrap Fusion Tribrid). Mass spectral data were collected over a 5-minute run period at a resolution of 120,000 and mass-to-charge (m/z) scan range of 85–1275. Each batch of 40 samples included six replicates of a pooled human plasma reference (Qstd3) for which over 200 metabolites have confirmed identification[26].
Raw data were extracted and aligned using xMSanalyzer[27] with apLCMS[28]. Unique features were defined by accurate mass m/z, retention time, and ion intensity. Sample feature intensities were normalized relative to the overall mean sample intensity, in order to compensate for any differences in tissue mass. Filtering of the features was performed to include those features with a median coefficient of variation between technical replicates less than or equal to 30% and with a Pearson correlation coefficient greater than 0.7.
Measurement of protein adducts
BD-derived protein adducts were quantified as previously described with minor modifications [21, 29, 30]. In brief, globin was isolated from red blood cells according to Mowrer et al.[31]. Globin (1 mg) was dissolved in 1.5 mL of 0.1 M NH4HCO3 and 2 pmol of 11-mer internal standard [13C5]HB-Val, [13C5]pyr-Val, [13C5]THB-Val peptides were added. Samples were digested for 18 hours at 37 °C with 100 μL of trypsin-biotin-agarose suspension. Each sample was transferred to a centrifugal filter device and filters were rinsed with water for a final volume of 380–430 μL. Samples were brought to dryness in a speed vac and stored at −80 °C until analyzed. Peptide standards were accurately quantified as described by our group previously[32].
Tandem Mass Spectrometry Analysis. For quantitation, samples were re-dissolved in 50 μl of 15 mM ammonium formate-0.1% formic acid. The quantitative analysis of the adducted N-terminal Val (1–7) peptides by LC-MS/MS was performed with a UPLC (Agilent 1290 Infinity LC) coupled to a triple quad mass analyzer (Agilent 2490). An Agilent HPLC Poroshell 120 C18 column (2.7μm 4.6 × 150 mm) was operated with a linear gradient of 5% 15 mM ammonium formate/0.1% formic acid for 1 min to 40% acetonitrile (ACN) in 12 min, at a flow rate of 0.4 mL/min. Between injections, the column was washed at 90% ACN for 5 min before re-equilibration for 8 min. The retention times of analytes were determined with synthesized peptide standards. The peptides were detected in multiple reaction monitoring (MRM) mode, monitoring the transition of the doubly charged precursor ions to the a1-and y6-fragments; HB-Val m/z 409.2 to 141.1, [13C5]HB-Val m/z 409.2 to 141.1,pyr-Val m/z 417.2 to 158.1, [13C5]pyr-Val m/z 419.2 to 162.1, THB-Val 426.2 to 176.1 and [13C5]THB-Val 428.2 to 180.1. The MS conditions were as follows: column temperature 60°C, fragmentor 380 V, sheath gas temperature 400°C, sheath gas flow 12 L/min, gas temperature 350°C, gas flow 15 L/min, nebulizer 40 psi. Sample injection volume was 20 μl. Amounts of adducts were calculated based on the corresponding ion transition peak area and the standard curve. Standard curves were constructed daily and repeatedly showed linear response from 2 to 500 fmol/injection.
Data analysis
Data was preprocessed by filtering features to be included in the analysis if they were present in at least 20% of all samples, and at least 80% of samples within a group. After filtering, the data was normalized by log2 transformation and quantile normalization. One-way ANOVA and partial least squares discriminant analysis (PLS-DA) were performed to identify discriminatory features using thresholds of P<0.05 and variable importance for projection (VIP) ≥ 1.5. Multiple testing correction to avoid type I error (false positives) was performed by false discovery rate (FDR) adjustment using the Benjamini-Hochberg method. Metabolites were identified and annotated by comparing with an in-house library of confirmed metabolites and by using the R package xMSannotator [33]. Identity scores provided are based on Schymanski et al [34].
Discriminatory features that met cutoffs of P<0.05 and VIP≥ 1.5 were analyzed by mummichog (v2.0.6), which performs pathway enrichment analysis using permutation testing with differentially expressed features. For the features selected in the pathway analysis, a Wilcoxon p-value and the fold-change were calculated to compare the controls to the 2 ppm BD-exposed mice. The fold-change indicates whether the metabolite was increased (positive) or decreased (negative) in BD-exposed mice relative to controls.
xMWAS analysis
The levels of protein adducts were correlated with metabolomic feature intensities using xMWAS, an R package that provides integration of multiple data sets for network visualization and cluster analysis[35]. Correlated features were analyzed using mummichog (v2.0.6) to look for enriched pathways associated with the levels of protein adducts.
Results
Butadiene exposure resulted in numerous effects on lung metabolism.
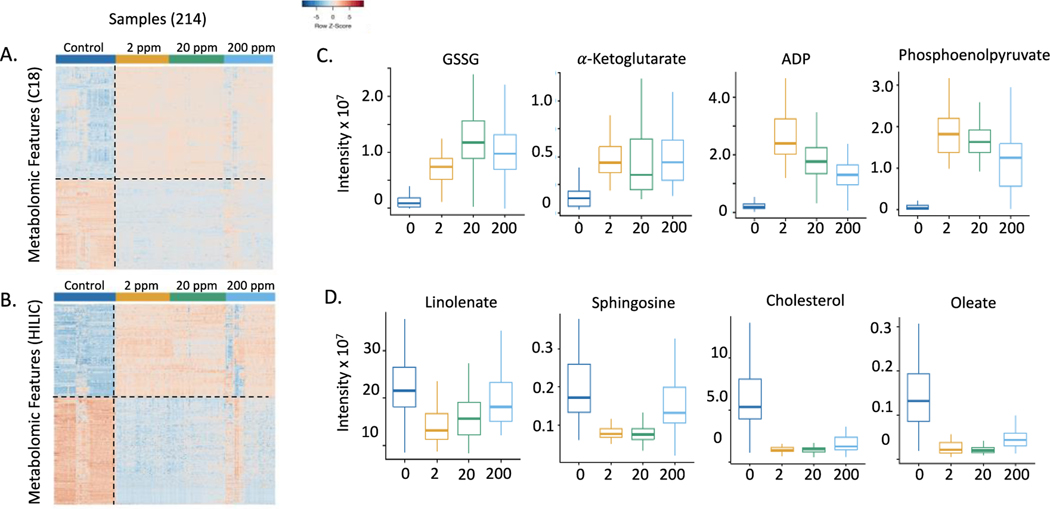
Analysis of the metabolomics data by one-way ANOVA showed that, in data from both platforms, more than half the detected signals were changed even with FDR at 0.05. For the C18/ESI- data, 6021 out of 9,248 features (65%) were differentially expressed between control and BD-exposed mice, and in the HILIC/ESI+ data, 10,401 out of 12,865 features (81%) were differentially expressed (shown in supplemental data). Because the number of differentially expressed metabolites was so large, we performed PLS-DA to select metabolites contributing most to separation. PLS-DA reduces the dimensions of the data by maximizing the covariance of metabolite intensity based on exposure group. Analysis by PLS-DA resulted in 1,370 differentially expressed features in the C18/ESI- data with a VIP ≥1.5. Of these features, similar numbers were increased (771) and decreased (599) in the BD-exposed mice relative to the control. PLS-DA analysis of the HILIC/ESI+ data yielded 1807 differentially expressed features having a VIP ≥ 1.5, with similar numbers increased (835) and decreased (972) in the BD-exposed mice relative to the controls. One-way hierarchical cluster analysis of the top discriminatory features showed that the control mice clustered together, and the BD-exposed mice clustered together (Figure 1A and 1B). Some selected examples of metabolites changing with BD exposure concentration are provided in boxplots, which demonstrate variations in the intensities at the different exposure levels of BD (Figure 1C and 1D). The overall results show that the metabolic effects were present at all levels of BD exposure. Thus, without any consideration of pathways affected, BD exposure resulted in numerous effects on lung metabolism at the lowest exposure level (2 ppm BD) studied.
Figure 1.
One-way hierarchical clustering analysis (HCA) of discriminatory metabolites selected by PLS-DA analysis of control and BD-exposed mice. A. For the C18/ESI- method, there were 771 features that increased and 599 features that decreased in the BD-exposed mice relative to the controls. B. For the HILIC/ESI+ method, there were 835 features that increased and 972 features that decreased in BD-exposed mice relative to the controls. C. Metabolic changes representative of the patterns seen for differentially expressed features in control mice and mice exposed to 2 ppm, 20 ppm, and 200 ppm butadiene. Metabolic effects were seen at the lowest level of butadiene exposure. In panels C and D, metabolites were significantly different in exposure groups vs. controls with p<0.001
Pathways altered by butadiene
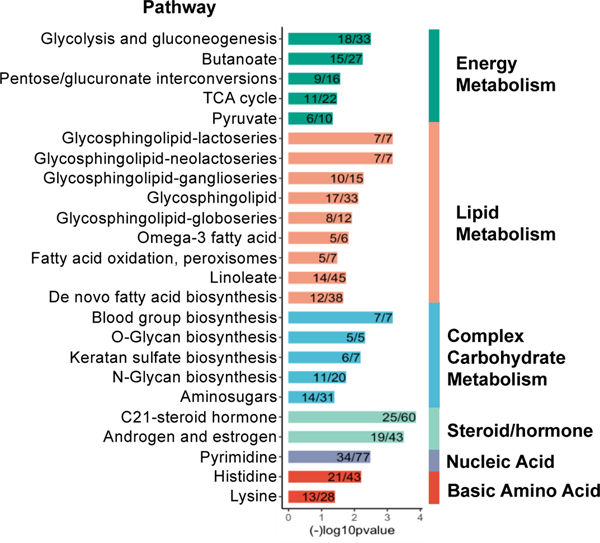
Pathway enrichment analysis was performed to identify potential metabolic pathways that were altered by BD. We performed the analysis separately for each method (C18/ESI− and HILIC/ESI+) using the respective discriminatory features with a VIP ≥ 1.5 selected by PLS-DA. The analysis showed BD exposure was associated with alterations of multiple pathways related to the metabolism of energy, lipids, complex/structural carbohydrates, steroid-hormones, pyrimidines, and basic amino acids (Figure 2). Selected differentially expressed features from these pathways are shown in Table 1.
Figure 2.
Pathways altered in butadiene exposed mice compared to control mice. Pathway analysis was performed using mummichog 2.0.6 with features identified by PLS-DA with a VIP ≥ 1.
Table 1:
Selected features involved in pathways enriched in butadiene exposed mice
| Category | Pathway(s) | Metabolitea | Wilcoxon Pb | Fold-changec | Identity Score |
|---|---|---|---|---|---|
| Energy metabolism | TCA cycle | citrate/isocitrate | 6.21E-16 | 3.0 | 1 |
| α-ketoglutarate | 2.15E-18 | 3.8 | 1 | ||
| fumarate | 2.80E-16 | 2.0 | 1 | ||
| malate | 2.56E-15 | 2.1 | 1 | ||
| Multiple pathways | glutamate | 1.02E-16 | 2.3 | 1 | |
| Glycolysis | fructose 1,6-bisphosphate | 4.48E-16 | 2.1 | 1 | |
| glyceraldehyde −3-phosphate/DHAP | 6.84E-08 | 1.6 | 1 | ||
| 1,3-bisphosphoglycerate | 4.35E-15 | 5.9 | 3 | ||
| 2- or 3- phosphoglycerate | 1.43E-09 | 2.3 | 1 | ||
| phosphoenolpyruvate | 6.66E-14 | 4.9 | 1 | ||
| Multiple pathways | pyruvate | 1.66E-14 | 1.7 | 1 | |
| Pentose glucuronate interconversion | xylose; ribose | 5.43E-05 | 1.5 | 1 | |
| ribose phosphate/ribulose phosphate | 2.03E-13 | 2.6 | 1 | ||
| Energy (multiple) | UMP | 2.75E-16 | 4.1 | 1 | |
| ADP | 1.56E-18 | 9.4 | 1 | ||
| GDP | 1.34E-16 | 5.9 | 1 | ||
| CoA | 1.66E-14 | 4.3 | 1 | ||
| Lipid metabolism | Glycosphingolipid (multiple) | galactosylceramidesulfate | 1.43E-11 | 2.6 | 4 |
| phosphoethanolamine | 2.99E-18 | 3.8 | 1 | ||
| sphingosine | 1.41E-15 | −2.4 | 1 | ||
| sphinganine | 2.99E-04 | −1.9 | 1 | ||
| Omega-3 fatty acid | stearidonic acid | 2.24E-18 | −5.3 | 3 | |
| 9(S)-HPOT | 2.17E-13 | −2.3 | 3 | ||
| Linoleate | linolenic acid | 9.88E-13 | −2.6 | 1 | |
| Fatty acid | oleic acid | 3.18E-17 | −5.5 | 1 | |
| palmitic acid | 8.99E-19 | −8.3 | 1 | ||
| stearic acid | 2.08E-05 | −1.8 | 1 | ||
| arachidonic acid | 3.29E-16 | −5.9 | 1 | ||
| docosahexaenoic acid | 3.18E-17 | −5.8 | 1 | ||
| eicosapentaenoic acid | 1.59E-12 | −4.7 | 1 | ||
| Fatty acid oxidation, peroxisomes | palmitoyl-CoAd | 5.37E-23 | 31.4 | 1 | |
| stearoyl-CoAd | 3.94E-22 | 16.4 | 3 | ||
| Steroid hormone metabolism | C21-steroid hormone; Androgen/estrogen | estradiol | 3.52E-16 | −3.7 | 1 |
| testosterone | 7.92E-06 | −1.5 | 3 | ||
| androsterone | 7.92E-06 | −1.5 | 3 | ||
| C21-steroid hormone | hydroxycholesterole | 1.10E-10 | −3.5 | 1 | |
| cholesterole | 1.09E-18 | −5.1 | 1 | ||
| Complex carbohydrate metabolism | Multiple pathways | CMP-N-Acetylneuraminate | 1.42E-18 | 4.9 | 3 |
| UDP-glucose | 3.69E-18 | 4.1 | 3 | ||
| N-Glycan biosynthesis | GDP-fucose | 1.61E-16 | 3.2 | 3 | |
| Nucleic acid metabolism | Pyrimidine | cytosine | 1.22E-07 | −1.9 | 1 |
| dihydrouracil | 6.99E-13 | −2.1 | 4 | ||
| dihydrothymine | 8.57E-07 | −5.3 | 4 | ||
| Basic amino acid metabolism | Histidine | histidine | 8.7E-03 | 1.2 | 1 |
| imidazole acetaldehyde | 8.62E-09 | −1.7 | 4 | ||
| methylimidazole acetaldehyde | 1.35E-11 | −2.1 | 4 | ||
| acetyl-lysine | 6.00E-05 | −1.1 | 4 | ||
| Lysine | dimethyllysine | 1.22E-15 | 4.7 | 4 | |
| trimethyllysine | 1.61E-17 | 2.8 | 1 |
All adducts are [M+H]+ or [M-H]− unless noted
2ppm vs Control, FDR adjusted
Fold change was determined in 2ppm BD-exposed vs Control mice;
positive: higher in exposed mice
negative: lower in exposed mice
[M-2H]2−
[M-H20+H]+
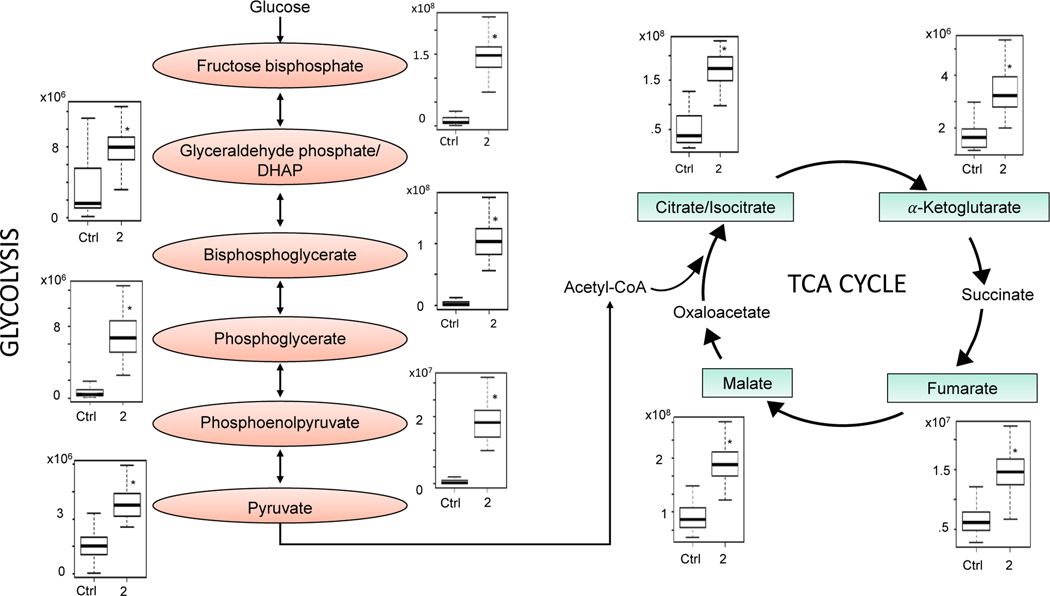
For the category of energy metabolism, the glycolysis and the citric acid cycle pathways were observed to be perturbed. Key intermediates in these pathways are listed in Table 1, and the changes in these intermediates after exposure to 2 ppm BD are illustrated in Figure 3. Statistically significant increases were observed in the glycolytic metabolites fructose-1,6-bisphosphate (2-fold), glyceraldehyde phosphate (2-fold), phosphoglycerate (2-fold), bisphosphoglycerate (6-fold), phosphoenolpyruvate (5-fold), and pyruvate (2-fold). Similarly, increases were observed in key features of the TCA cycle, including citrate (isocitrate) (3-fold), malate (2-fold), fumarate (2-fold), and α-ketoglutarate (4-fold).
Figure 3.
Key metabolites in glycolysis and the TCA cycle that were significantly changed with low level butadiene inhalation exposure. The raw intensity for each metabolite is plotted for control mice and for mice exposed to 2 ppm butadiene;*P < 1 × 10−8.
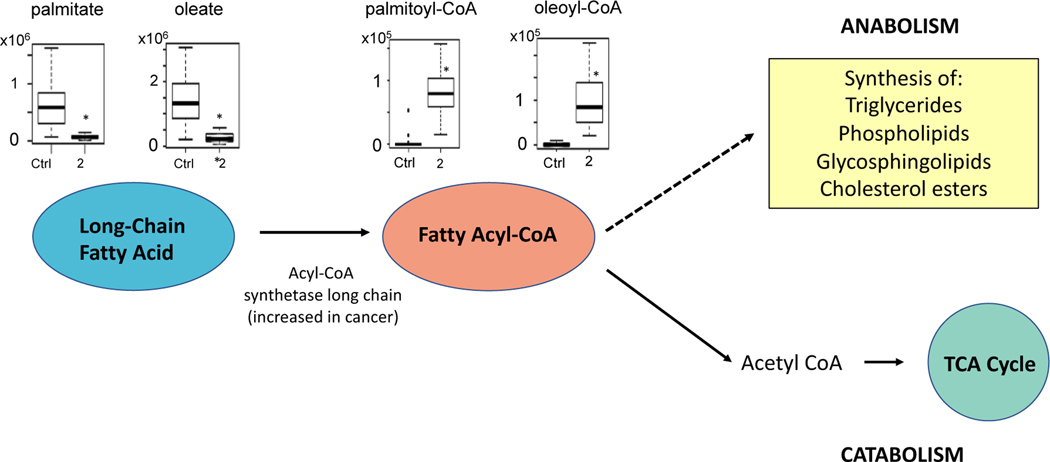
Pathway analysis also indicated there were changes in fatty acid and lipid metabolism in BD-exposed mice. Exposure to 2 ppm BD resulted in multiple fatty acid levels being decreased (Table 1). Figure 4 shows a statistically significant reduction of two of these fatty acids, palmitate (8-fold) and oleate (5.5-fold), and the increase in the corresponding fatty acyl-CoAs: palmitoyl-CoA (31-fold) and oleoyl-CoA (16-fold). Changes were observed in several metabolites related to glycosphingolipid metabolism, including a decrease in backbone moieties sphinganine (2-fold) and sphingosine (2-fold) and an increase phosphoethanolamine (4-fold) and in metabolites involved in glycosylation. Metabolites involved in sterol and steroid hormone metabolism were also decreased (Table 1).
Figure 4.
Schematic of pathways related to lipid metabolism that were altered with butadiene exposure. Representative metabolites palmitate, oleate, palmitoyl-CoA, and oleoyl-CoA are plotted for the control and 2 ppm BD-exposed mice. The observed decreases in long-chain fatty acids and increases in fatty acyl-CoAs could be related to an upregulation of acyl-CoA synthetase long chain (ACSL), which has been previously shown to be upregulated in lung cancer. Metabolites in both the anabolic and catabolic pathways were differentially expressed in BD-exposed mice (see Table 1); *P<1×10−8
Changes were noted in pyrimidine, histidine, and lysine pathways (Table 1). Metabolites related to pyrimidine metabolism were decreased (2–5-fold). In the histidine pathway, metabolites related to the degradation of histamine were mainly decreased. Finally, in the lysine pathway, metabolites related to lysine degradation were increased (5–6-fold).
Variability in metabolite response to butadiene exposure
Of the 1370 metabolites that were classified as differentially expressed by PLS in the C18/ESI- data, there were 374 metabolites with a CV below the 50th percentile in the control group that maintained a CV below the 50th percentile for all exposed groups, suggesting a stable response and little genetic contribution to the response for these metabolites. There were 197 metabolites with a CV below the 50th percentile in the control group where the CV increased above the 50th percentile in all treated groups, suggesting large amount of variability in the response to butadiene exposure, which may be due to genetic variation. Of the metabolites that had a CV of greater than 50th percentile at the onset, the after treatment strain variability is reduced in 78 metabolites suggesting a common response after variable base line. There were 306 metabolites where the CV was above the 50th percentile in all groups, suggesting strain variability throughout. Similar variability was observed in the HILIC/ESI+ data. Further analysis will be needed to determine specific genetic variations that contribute to the varied response of certain metabolites.
Butadiene protein adducts
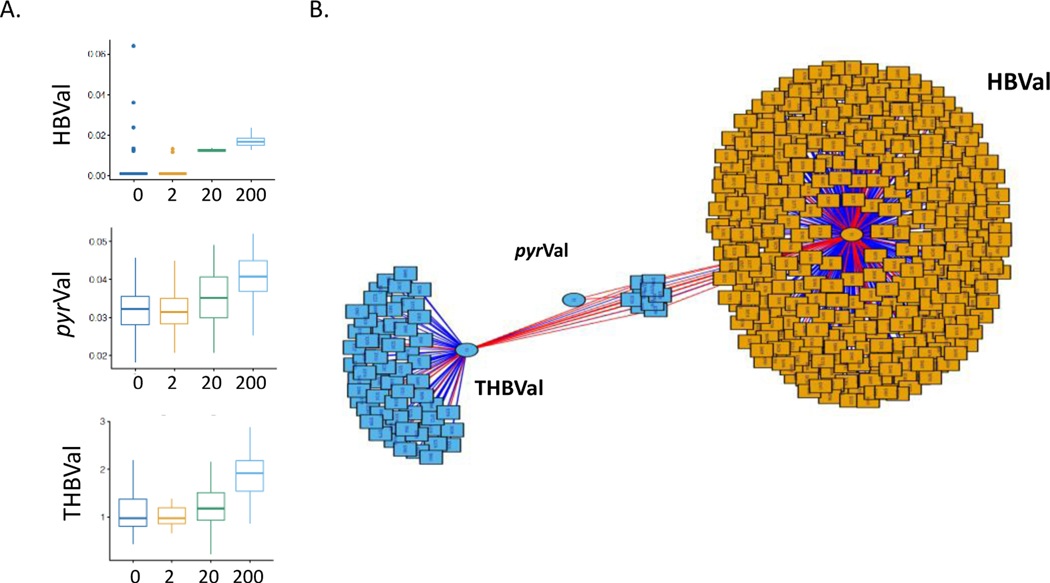
Protein adducts formed from the reactive epoxide metabolites of BD and hemoglobin were measured after exposure to each dose of BD (Figure 5A). The levels of these protein adducts were correlated with metabolomics data using xMWAS. Results of the correlation analysis showed two clusters of features that were primarily correlated with HBVal and/or THBVal (Figure 5B).
Figure 5.
A. The protein adducts formed from butadiene metabolites were measured in blood. B. xMWAS was used to look for correlations between the butadiene protein adducts and the metabolomic features with a minimum correlation of 0.4. Red lines represent positive correlations and blue lines represent negative correlations.
Discussion
In this study, we used high resolution metabolomics to examine the effects of BD exposure on mouse lung metabolism. BD is a known carcinogen that is metabolized to form genotoxic epoxide intermediates, but it is unknown whether it has additional direct or indirect metabolic effects that prevent or promote adverse effects. We found that BD exposure induces extensive and bidirectional changes in the mouse lung metabolome. Multiple metabolic pathways are disturbed, including pathways associated with energy, lipid, carbohydrate, amino acid and nucleic acid metabolism. The observed re-programming of many of these pathways are reflective of metabolic changes that are observed in lung models and lung tumor studies.
We observed increases in multiple metabolites in the bioenergetic pathways of glycolysis and the TCA cycle suggesting that these pathways are upregulated with BD exposure. Both pathways are important for generating energy required for cell proliferation and biosynthesis, and contribute to the redox balance in cells. Previous studies show that glycolysis and the TCA cycle are more active in lung tumor tissue in humans and mice[36–39]. In a study of lung cancer patients infused with 13C glucose, the downstream metabolites lactate, succinate, and citrate were enriched with 13C in lung tumors, suggesting that glycolysis and the TCA cycle had increased activity in the tumors[40]. Similarly, stable isotope studies in mice showed that glucose contribution to the TCA cycle is increased in lung tumors relative to normal lung tissue and that glucose carbon contribution to the TCA cycle is required for tumor formation[41]. Our data from mice exposed to BD for two weeks suggests that there is increased flux through glycolysis and the TCA cycle in lung tissue before tumors are present, possibly influencing subsequent tumor formation in the lung.
The buildup of TCA cycle intermediates has previously been shown to affect cellular metabolic functions related to cancer progression [42, 43]. We found that fumarate, malate, citrate/isocitrate, and α-ketoglutarate were all increased in BD exposed mouse lung. Fumarate has been deemed an oncometabolite and acts by inducing pseudohypoxia and affecting epigenetic regulation of gene expression[44]. Accumulation of fumarate in cells has been found to promote metastasis by inhibiting Tet-mediated demethylation of an antimetastatic miRNA cluster[45]. The TCA cycle intermediate α-ketoglutarate is the precursor to 2-hydroxyglutarate, which is an oncometabolite that triggers changes in DNA and histone methylation[38]. Citrate has been shown to play a role in both cancer suppression and cancer promotion[43]. Thus, there are many mechanisms by which BD modulates the TCA cycle in the lung, but further studies will be required to reveal specific downstream effects related to promotion or prevention of tumorigenesis.
Our results showed that BD exposure resulted in various alterations in lipid metabolism in the mouse lung. For example, fatty acid levels were decreased, which is consistent with other studies that have found serum fatty acids to be decreased in lung cancer [36, 46]. Fatty acyl-CoAs were increased with BD exposure. This suggests a shift towards the formation of fatty acyl-CoAs from fatty acids, which is the common first step that occurs prior to fatty acid catabolism or synthesis of complex lipids. This could result from an upregulation of acyl-CoA synthetase long chain (ACSL), which has been found to be upregulated in lung cancer cells in mice and humans [47]. Fatty acyl-CoAs can be catabolized through the fatty acid oxidation process to yield acetyl-CoA for entry into the TCA cycle, which could partially explain the increased activity of the TCA cycle that we observed. Fatty acyl-CoAs are also be used in the synthesis of complex lipids. We observed changes in metabolites involved in the synthesis of complex lipids, but the specific pathway responses could not be discerned, because these metabolites are involved in multiple interconnected pathways. Sphingosine was decreased, phosphoethanolamine was increased, and metabolites related to glycosylation were increased, suggesting there were disturbances in sphingolipid metabolism. This is consistent with the finding that sphingolipid metabolism is disturbed in lung cancer[48]. Deviations that occur in the synthesis or modification of any of these complex lipids can contribute to changes in lung cell surface and cellular signaling, which are disruptions that have been linked to lung cancer progression[49, 50].
Hemoglobin adducts of the reactive epoxides of BD were measured in exposed mice and were found to increase with the concentration of BD exposure, although there was little change at the 2 ppm concentration. Metabolomics analysis revealed that for many differentially expressed metabolites, the metabolite levels were altered significantly at all concentrations of BD exposure, including the lowest concentration of 2 ppm. Pyr-Val and THB-Val were readily detected and background levels were similar to the ones found in previous studies[22, 30]. The fact that the protein adduct levels in the 2 ppm exposure group were not increased over background found in the air control, yet there were significant changes in the lung metabolome at low exposure concentration, suggests that the metabolic response to BD may not be entirely dependent on formation of reactive epoxides. The fact the BD exposure alters expression of over 6,000 metabolic features in the lungs of mice exposed to 2 ppm BD, suggests that BD, has a significant effect on general metabolism of the lung in addition to the knonwn genotoxic mechanisms, has a significant effect on general metabolism of the lung.
The metabolic adaptations discussed above represent (i) normal tissue response to BD- exposure and defense or (ii) may in part contribute to tumor development later on. However, at this time one cannot distinguish mechanisms of normal tissue defense from pro-tumorigenic re-programming. Thus, further studies on low concentrations of inhaled BD are needed to clarify the metabolism-altering mechanisms that may ultimately prevent or contribute to the development of adverse effecs in lung such as lung cancer.
In summary, high-resolution mass spectrometry-based metabolomics analyses show that BD exposure causes considerable reprogramming of the mouse lung metabolome. The observed alterations in metabolites involved in glycolysis, the TCA cycle, and lipid metabolism in BD-exposed mouse lung are consistent with the metabolic reprogramming found in lung tumors[38–40, 50]. This provides evidence that along with its known genotoxic mechanisms, BD exposure induced many metabolic changes that may in part prevent or contribute to tumorigenesis. Importantly, the apparent sensitivity of metabolic response to ultra-low BD exposures could be utilized to identify individuals at risk for BD exposure.
Supplementary Material
Acknowledgements
The authors are grateful to Nicholas Hallberg, Spotswood Miller, Anita Reno, Walter Spear and Barbara Rolls for assistance with the animal exposures and harvest. Support has been provided in part by the National Institutes of Health (R21ES023046, R21ES019684, R21ES031824, P30ES019776, UL1TR000039, T32ES007254), the Health Effects Institute (HEI4907-RFPA09–5/12–1), the Arkansas Bioscience Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000, The Sealy Center for Environmental Health and Medicine (SCEHM), and The Brown Foundation.
Footnotes
Supplemental material
Supplemental Tables 1 and 2 contain the one-way anova results of the log2-transformed/quantile-normalized metabolomics data from mouse lung (HILIC/ESI+, C18/ESI-, respectively) of mice exposed to butadiene. Mean values for control and each exposure group (2ppm, 20ppm, and 200ppm) are included, as well as the FDR-adjusted p-value for all groups and the post-hoc p-values for comparisons between each group.
Supplemental Tables 3 and 4 contain the PLS-DA results of the same data(HILIC/ESI+, C18/ESI-, respectively). Mean values for each group as well as the variable importance of projection(VIP) score are included.
References
- 1.Humans, I.W.G.o.t.E.o.C.R.t., IARC monographs on the evaluation of carcinogenic risks to humans. Volume 97. 1,3-butadiene, ethylene oxide and vinyl halides (vinyl fluoride, vinyl chloride and vinyl bromide). IARC Monogr Eval Carcinog Risks Hum, 2008. 97: p. 3–471. [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson R, Health Hazards encountered in the manufacture of synthetic rubber. JAMA 1944. 124: p. 701–703. [Google Scholar]
- 3.Khalil M, Abudiab M, and Ahmed AE, Clinical evaluation of 1,3-butadiene neurotoxicity in humans. Toxicol Ind Health, 2007. 23(3): p. 141–6. [DOI] [PubMed] [Google Scholar]
- 4.Sadeghi-Yarandi M, Golbabaei F, and Karimi A, Evaluation of pulmonary function and respiratory symptoms among workers exposed to 1,3-Butadiene in a petrochemical industry in Iran. Arch Environ Occup Health, 2020. 75(8): p. 483–490. [DOI] [PubMed] [Google Scholar]
- 5.Lin CY, et al. , The association between urinary levels of 1,3-butadiene metabolites, cardiovascular risk factors, microparticles, and oxidative stress products in adolescents and young adults. J Hazard Mater, 2020. 396: p. 122745. [DOI] [PubMed] [Google Scholar]
- 6.Penn A. and Snyder CA, 1,3-Butadiene exposure and cardiovascular disease. Mutat Res, 2007. 621(1–2): p. 42–9. [DOI] [PubMed] [Google Scholar]
- 7.Pulliero A, et al. , Environmental carcinogens and mutational pathways in atherosclerosis. Int J Hyg Environ Health, 2015. 218(3): p. 293–312. [DOI] [PubMed] [Google Scholar]
- 8.Owen PE, et al. , Inhalation toxicity studies with 1,3-butadiene. 3. Two year toxicity/carcinogenicity study in rats. Am Ind Hyg Assoc J, 1987. 48(5): p. 407–13. [DOI] [PubMed] [Google Scholar]
- 9.Melnick RL, Shackelford CC, and Huff J, Carcinogenicity of 1,3-butadiene. Environ Health Perspect, 1993. 100: p. 227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graff JJ, et al. , Chemical exposures in the synthetic rubber industry and lymphohematopoietic cancer mortality. J Occup Environ Med, 2005. 47(9): p. 916–32. [DOI] [PubMed] [Google Scholar]
- 11.Seaton MJ, Plopper CG, and Bond JA, 1,3-Butadiene metabolism by lung airways isolated from mice and rats. Toxicology, 1996. 113(1–3): p. 314–7. [DOI] [PubMed] [Google Scholar]
- 12.Degner A, et al. , Interindividual Differences in DNA Adduct Formation and Detoxification of 1,3-Butadiene-Derived Epoxide in Human HapMap Cell Lines. Chem Res Toxicol, 2020. 33(7): p. 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boysen G, et al. , Effects of GSTT1 Genotype on the Detoxification of 1,3-Butadiene Derived Diepoxide and Formation of Promutagenic DNA-DNA Cross-Links in Human Hapmap Cell Lines. Chem Res Toxicol, 2021. 34(1): p. 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himmelstein MW, Asgharian B, and Bond JA, High concentrations of butadiene epoxides in livers and lungs of mice compared to rats exposed to 1,3-butadiene. Toxicol Appl Pharmacol, 1995. 132(2): p. 281–8. [DOI] [PubMed] [Google Scholar]
- 15.Bond JA, et al. , Metabolism of butadiene by mice, rats, and humans: a comparison of physiologically based toxicokinetic model predictions and experimental data. Toxicology, 1996. 113(1–3): p. 48–54. [DOI] [PubMed] [Google Scholar]
- 16.Filser JG, et al. , Metabolism of 1,3-butadiene to toxicologically relevant metabolites in single-exposed mice and rats. Chem Biol Interact, 2007. 166(1–3): p. 93–103. [DOI] [PubMed] [Google Scholar]
- 17.Himmelstein MW, et al. , Toxicology and epidemiology of 1,3-butadiene. Crit Rev Toxicol, 1997. 27(1): p. 1–108. [DOI] [PubMed] [Google Scholar]
- 18.Ahmadkhaniha R, Ghoochani M, and Rastkari N, Application of biological monitoring for exposure assessment of 1.3 Butadiene. J Environ Health Sci Eng, 2020. 18(2): p. 1265–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albertini RJ, et al. , Biomarkers in Czech workers exposed to 1,3-butadiene: a transitional epidemiologic study. Res Rep Health Eff Inst, 2003(116): p. 1–141; discussion 143–62. [PubMed] [Google Scholar]
- 20.Fustinoni S, et al. , Biological monitoring in occupational exposure to low levels of 1,3-butadiene. Toxicol Lett, 2004. 149(1–3): p. 353–60. [DOI] [PubMed] [Google Scholar]
- 21.Boysen G, et al. , Formation of 1,2:3,4-diepoxybutane-specific hemoglobin adducts in 1,3-butadiene exposed workers. Toxicol Sci, 2012. 125(1): p. 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgieva NI, et al. , Exposure-response of 1,2:3,4-diepoxybutane-specific N-terminal valine adducts in mice and rats after inhalation exposure to 1,3-butadiene. Toxicol Sci, 2010. 115(2): p. 322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartman JH, et al. , 1,3-Butadiene-induced mitochondrial dysfunction is correlated with mitochondrial CYP2E1 activity in Collaborative Cross mice. Toxicology, 2017. 378: p. 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soltow QA, et al. , High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics, 2013. 9(1 Suppl): p. S132–S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandler JD, et al. , Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am J Physiol Regul Integr Comp Physiol, 2016. 311(5): p. R906–R916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu KH, et al. , Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal Chem, 2020. 92(13): p. 8836–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uppal K, et al. , xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics, 2013. 14: p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu T, et al. , Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. J Proteome Res, 2013. 12(3): p. 1419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boysen G, et al. , Analysis of diepoxide-specific cyclic N-terminal globin adducts in mice and rats after inhalation exposure to 1,3-butadiene. Cancer Res, 2004. 64(23): p. 8517–20. [DOI] [PubMed] [Google Scholar]
- 30.Boysen G, et al. , A simplified method for detection of N-terminal valine adducts in patients receiving treosulfan. Rapid Commun Mass Spectrom, 2019. 33(21): p. 1635–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mowrer J, et al. , Modified Edman degradation applied to hemoglobin for monitoring occupational exposure to alkylating agents. Toxicological & Environmental Chemistry, 1986. 11(3): p. 215–231. [Google Scholar]
- 32.Bordeerat NK, et al. , Accurate quantitation of standard peptides used for quantitative proteomics. PROTEOMICS, 2009. 9(15): p. 3939–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uppal K, Walker DI, and Jones DP, xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal Chem, 2017. 89(2): p. 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schymanski EL, et al. , Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol, 2014. 48(4): p. 2097–8. [DOI] [PubMed] [Google Scholar]
- 35.Uppal K, et al. , xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics, 2018. 34(4): p. 701–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mu Y, et al. , Serum Metabolomics Study of Nonsmoking Female Patients with Non-Small Cell Lung Cancer Using Gas Chromatography-Mass Spectrometry. J Proteome Res, 2019. 18(5): p. 2175–2184. [DOI] [PubMed] [Google Scholar]
- 37.Hensley CT, et al. , Metabolic Heterogeneity in Human Lung Tumors. Cell, 2016. 164(4): p. 681–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanhove K, et al. , The Metabolic Landscape of Lung Cancer: New Insights in a Disturbed Glucose Metabolism. Front Oncol, 2019. 9: p. 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noreldeen HAA, Liu X, and Xu G, Metabolomics of lung cancer: Analytical platforms and their applications. J Sep Sci, 2020. 43(1): p. 120–133. [DOI] [PubMed] [Google Scholar]
- 40.Fan TW, et al. , Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol Cancer, 2009. 8: p. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidson SM, et al. , Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab, 2016. 23(3): p. 517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Meulenaere V, et al. , In vivo selection of the MDA-MB-231br/eGFP cancer cell line to obtain a clinically relevant rat model for triple negative breast cancer brain metastasis. PLoS One, 2020. 15(12): p. e0243156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eniafe J. and Jiang S, The functional roles of TCA cycle metabolites in cancer. Oncogene, 2021. 40(19): p. 3351–3363. [DOI] [PubMed] [Google Scholar]
- 44.O’Flaherty L, et al. , Dysregulation of hypoxia pathways in fumarate hydratase-deficient cells is independent of defective mitochondrial metabolism. Hum Mol Genet, 2010. 19(19): p. 3844–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sciacovelli M. and Frezza C, Oncometabolites: Unconventional triggers of oncogenic signalling cascades. Free Radic Biol Med, 2016. 100: p. 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. , Serum unsaturated free Fatty acids: potential biomarkers for early detection and disease progression monitoring of non-small cell lung cancer. J Cancer, 2014. 5(8): p. 706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Padanad MS, et al. , Fatty Acid Oxidation Mediated by Acyl-CoA Synthetase Long Chain 3 Is Required for Mutant KRAS Lung Tumorigenesis. Cell Rep, 2016. 16(6): p. 1614–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, et al. , Biomarker identification and pathway analysis by serum metabolomics of lung cancer. Biomed Res Int, 2015. 2015: p. 183624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo X, et al. , The implications of signaling lipids in cancer metastasis. Exp Mol Med, 2018. 50(9): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merino Salvador M, et al. , Lipid metabolism and lung cancer. Crit Rev Oncol Hematol, 2017. 112: p. 31–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.