Abstract
In the past two decades, adeno-associated virus (AAV) vector manufacturing has made remarkable advancements to meet large-scale production demands for preclinical and clinical trials. In addition, AAV vectors have been extensively studied for their safety and efficacy. In particular, the presence of empty AAV capsids and particles containing “inaccurate” vector genomes in preparations has been a subject of concern. Several methods exist to separate empty capsids from full particles; but thus far, no single technique can produce vectors that are free of empty or partial (non-unit length) capsids. Unfortunately, the exact genome compositions of full, intermediate, and empty capsids remain largely unknown. In this work, we used AAV-genome population sequencing to explore the compositions of DNase-resistant, encapsidated vector genomes produced by two common production pipelines: plasmid transfection in human embryonic kidney cells (pTx/HEK293) and baculovirus expression vectors in Spodoptera frugiperda insect cells (rBV/Sf9). Intriguingly, our results show that vectors originating from the same construct design that were manufactured by the rBV/Sf9 system produced a higher degree of truncated and unresolved species than those generated by pTx/HEK293 production. We also demonstrate that empty particles purified by cesium chloride gradient ultracentrifugation are not truly empty but are instead packaged with genomes composed of a single truncated and/or unresolved inverted terminal repeat (ITR). Our data suggest that the frequency of these “mutated” ITRs correlates with the abundance of inaccurate genomes in all fractions. These surprising findings shed new light on vector efficacy, safety, and how clinical vectors should be quantified and evaluated.
Keywords: adeno-associated virus, gene therapy, HEK293, Sf9, vector heterogeneity
Introduction
Adeno-associated virus (AAV) vectors have established themselves as ideal vehicles for delivering therapeutic transgenes. To date, more than 140 recombinant (r)AAV-based clinical trials to treat a wide range of diseases have been carried out. Of these, 51 trials have met efficacy endpoints,1 and three have been approved for commercialization.1–5 The rAAV genome, which can be engineered to carry an array of transgene cassette designs, requires inverted terminal repeats (ITRs) that need to be present at both ends of the DNA strand. Most of what is known about the function of ITRs, which remain the last viral elements in rAAVs, is based on the ITR of AAV serotype 2 (AAV2).
The ITR of AAV2 is 145 nt in length. The first 125 nts fold into a T-shaped hairpin structure consisting of two internal inverted repeat sequences called the B-B′ and C-C′ arms and a main stem called the A-A′ region. The rest of the ITR forms the single-stranded D sequence. Embedded within the A-A′ sequence is the Rep-binding element (RBE). The RBE serves as the binding site for Rep68/78, which nicks the terminal resolution site (TRS) to initiate replication of the self-priming ITR structure. This terminates DNA synthesis. The 3′-end of the ITR then folds in on itself to begin a round of replication. The process is the repeated in what is termed rolling-hairpin replication.6 Rep also binds a sequence called RBE′, which is located at the tip of the cross arms.
The wild-type ITR has two formal configurations called “flip” and “flop,”7 where the B-B′ or C-C′ palindromes are, respectively, close to the genome ends. Consequentially, rAAVs that harbor wild-type ITRs will also exhibit both orientations. For example, in production formats that employ the widely used plasmid transfection method into human embryonic kidney cells (pTx/HEK293), where the vector genome-bearing plasmid (cis plasmid) harbors ITRs in the flop configuration at both the left and right ends (Lflop/Rflop), the progeny genomes will have near equal distributions of the four flip and flop confirmations, Lflip/Rflip: Lflip/Rflop: Lflop/Rflip: Lflop/Rflop.8
This equal distribution of configurations results from rolling hairpin replication,6 whereby the B-B′ and C-C′ arms swap positions following each resolution event. As a result, the 3′-ITR of the template genome becomes the new 5′-ITR of the replicated genome. Interestingly, it has been demonstrated that vector genomes that carry a mutation in one of its ITRs can be repaired in cis, via replication, using the opposing ITR as the template.9,10
The biology of ITRs makes them indispensable with contemporary vector designs and under current production platforms. Mutations within the ITR have been shown to compromise packaging yields.11–13 The ITRs also imbue AAV vectors with the ability to persist in nondividing cells. After vectors enter the cell and traffic into the nucleus, the genome is uncoated. Using the host DNA polymerase machinery, the self-primed ITRs drive the conversion of the single-strand genome into the double-stranded species, also known as second-strand synthesis. The genome is then converted into stable episomal forms through intra- or intermolecular recombination, also via the ITRs.
Despite the successes for rAAVs in the clinic, manufacturing of these biotherapies is still met by many challenges. The production of AAV vectors requires the expression of the rep and cap genes (usually provided in trans) and a helper virus, such as adenovirus (Ad).14 Alternatively, the expression of the essential Ad helper genes (e.g., E1a, E2a/b, E4, VARNA), also provided in trans, is sufficient to carry out rAAV genome replication and packaging. The pTx/HEK293 platform is a popular approach for research-grade rAAV.15 Unfortunately, this method is not very scalable for meeting most clinical needs.15 To overcome this limitation, other rAAV production methods have been pursued.15–17
Recombinant baculovirus vectors (rBVs) that are derived from Autographa californica multiple nucleopolyhedrovirus (AcMNPV) can be used to deliver rAAV production components into invertebrate cell lines for large-batch clinical vector production.17 In current systems, the rep and cap cassettes are combined into a single baculovirus, whereas a second baculovirus harbors the vector genome.18 The two rBVs are used to transduce stable Sf9 cell lines to produce rAAVs.18,19 These platforms have been coined rBV/Sf9 production systems. Today, pTx/HEK293 and rBV/Sf9 are the two dominant systems for rAAV production. There is growing interest in rBV/Sf9 platforms, owing to their advantages over pTx/HEK293 production.
The rBV/Sf9 platform does not directly require plasmid transfections for rAAV production, lowering the cost of raw materials and making it easier to scale-up. Additionally, there is no possible risk associated with the replication of contaminating human agents with rBV/Sf9 platforms.20 The prevalence of residual DNAs in vector preparations has been a subject of interest for many years. Direct head-to-head comparisons of pTx/HEK293 and rBV/Sf9 production methods have been reported21,22; however, very little has been defined for vector heterogeneity generated by the two strategies.
Recent advancements in sequencing technology, namely with next-generation sequencing (NGS) methods, have been used with great success to characterize and quantify the abundance of residual DNA, reverse packaged genomes, and vector heterogeneity in preparations.23–27 Short-read fragment sequencing has been used to interrogate the abundances of DNA contaminants and the presence of plasmid- or production-associated mutations, whereas long-read sequencing has been able to characterize heterogeneity of vector genomes, namely the presence of truncated and chimeric genomes.26,28,29
Among long-read sequencing approaches, AAV-genome population sequencing (AAV-GPseq),26 a vector sequencing method based on single molecule real-time sequencing (SMRT), has the capacity to sequence vectors from ITR to ITR without the need for bioinformatic reconstruction of the full genome.26,28 Because of this unique feature, the outcomes of vector genomes can be interrogated at a resolution that can reveal whether they are faithfully replicated and packaged.
In this work, we sought to compare identically designed vectors that were generated by either pTx/HEK293 or rBV/Sf9 platforms to determine whether genome heterogeneity between the two production methods can be inherently different. We found using AAV-GPseq that truncated genomes from pTx/HEK293 and rBV/Sf9 production were indeed dissimilar. Our findings suggest that the differences were attributed to mutated and unresolved ITRs, which were more ubiquitous among rBV/Sf9-produced vectors.
We were able to validate this observation by revealing that genomes from both full and partial fractions of rBV/Sf9 vectors purified by cesium chloride (CsCl) gradient ultracentrifugation had a higher degree of unresolved and truncated genomes than pTx/HEK293 vectors. Curiously, upon sequencing empty fractions, we found that particles can be packaged with short reads that map to ITRs, potentially raising concerns regarding innate immune recognition. The implications of their presence on vector biology/performance for in vivo gene delivery are unknown. Nevertheless, our findings may lead to further study in vector design and capsid purification methods to improve AAV vector production.
Materials and Methods
Plasmid construct and baculovirus generation
The pFB-GFP vector plasmid, which was used for pTx/HEK293 production (see Vector production) and served as the donor plasmid for the generation of the bacmids, contains the human cytomegalovirus (hCMV) promoter, the enhanced green fluorescence protein (EGFP) reporter gene, and the 3′-untranslated region (UTR) of the human HBB gene. The cassette is flanked by two flop-oriented AAV2 ITRs derived from the pSub-201 plasmid.30 The ITR-CMV-EGFP and Rep2Cap8 rBVs were generated by Tn7 transposition using the Invitrogen™ Bac-to-Bac™ Baculovirus Expression System (Thermo Fisher Scientific, Waltham, MA), as described previously.31 The integrality of the rAAV plasmids, bacmids, and rBV genomes were verified by Sanger sequencing.
Vector production
For vector production by pTx/HEK293, HEK293 cells were cotransfected with the pDP8 helper and pFB-GFP vector plasmids, as described previously.32 For rBV/Sf9 production, Sf9 cells (Invitrogen/Thermo Fisher Scientific) were seeded at a density of 1E6 cells per milliliter in a 400-mL spinner flask or in a 2-L bioreactor (Sartorius) and grown at 27°C in Sf-900 III SFM. Cells were coinfected with rBV-ITR-CMV-EGFP and rBV-Rep2Cap8 at a multiplicity of infection of 0.05 plaque-forming units/baculovirus. Four days after infection, chemical cell lysis and baculovirus inactivation were performed with 0.5% Triton X-100 (Sigma–Aldrich, St. Louis, MO) incubation for 2.5 h at 27°C under agitation.
The lysate was then treated for an additional 2 h at 37°C with Benzonase (5 U/mL; Merck, Darmstadt, Germany). The crude bulks were clarified by centrifugation for 5 min at 1,000 g and then purified by double CsCl gradient ultracentrifugation, as described previously,32 or by immune affinity chromatography with a single POROS CaptureSelect AAV8 column (Thermo Fisher Scientific) and formulated in Dulbecco's phosphate-buffered saline (Lonza) containing 0.001% Poloxamer 188 (Merck Millipore).
Molecular quantification and analytical ultracentrifugation of AAV vectors
The AAV vector genome copy number was determined by real-time polymerase chain reaction (PCR) using primers targeting the ITR sequences.33 Total particle titers were determined by the AAV8 titration enzyme-linked immunosorbent assay (ELISA) kit (Progen) following recommended procedures. To assess viral protein purity, 1E10 of total particles was subjected to sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) and silver staining with the PlusOne Silver Stain Kit (GE Healthcare Life Sciences).
Empty-to-full particle ratios were calculated with quantitative polymerase chain reaction (qPCR) and ELISA titers, or with analytical ultracentrifugation (AUC) (Grenoble, FR). Briefly, AUC was performed with 100–500 μL of purified vector. Sedimentation velocity experiments were performed at 20,000 rpm and 20°C, on a Beckman XLI analytical ultracentrifuge using an AN-60 or AN-50 Ti Rotor (Beckman Coulter, Brea) and double-sector cells with optical path lengths of 12 or 1.5 mm, equipped with sapphire windows (Nanolytics, Potsdam, DE). Sedimentation data were processed with REDATE34 and then analyzed, considering solvent density and viscosity of 1.005 g/mL and 1.017 cp, respectively; and particle partial specific volume of 0.73 mL/g, with SEDFIT, for sedimentation coefficient distribution (c(s)) analysis.35
The resulting data were exported to GUSSI for integration and figures.36 Capsid-type classification was performed with the following criteria for empty capsids, 60–65S and A260/J ≈ 0.5; for full capsids, 90–100S and A260/J ≈ 2.5; and for high-molecular-weight species representing those that are attributed to particles containing oversized genomes, or duplexed and aggregated capsids, 100–120S. S is the sedimentation coefficient, A260 is the 260 nm absorbance signal, and J is the fringe shift signal. The fringe shift signal was used to determine the percentage of the different types of capsids.
Vector DNA extraction and agarose gel electrophoresis
Extractions of vector DNA from 8E11 to 1E13 genome copies (or particles) were performed by phenol:chloroform and ethanol precipitation, as described previously.28 Samples were then heated in annealing buffer (25 mM NaCl, 10 mM Tris-HCl [pH 8.5], 0.5 mM EDTA [pH 8]) at 95 °C for 5 min and then cooled to 25 °C (1 min for every −1°C) on a thermocycler (Eppendorf Mastercycler). Vector DNAs were subjected to standard agarose or alkaline gel electrophoresis stained with ethidium bromide.37
Illumina sequencing and single-stranded DNA virus sequencing
Single-stranded DNA virus sequencing (SSV-Seq) was performed, as described previously.38 Illumina sequencing was conducted on the HiSeq2500 system at GenoBiRD (Nantes, France).
SMRT sequencing and AAV-GPseq
Extracted vector DNAs were spiked with lambda phage DNA (λDNA) digested with BstEII (NEB, Ipswich) and used as a normalizer for size loading bias.26 Vectors separated by CsCl density gradient centrifugation were spiked with different amounts of λDNA. Full fractions (pTx/HEK293 and rBV/Sf9) and partial fraction of the rBV/Sf9 DNAs were spiked with 10% λDNA. Since the partial fraction of pTx/HEK293 and empty capsids (pTx/HEK293 and rBV/Sf9) of DNAs were quantified to be less than 100 ng total (under the requirements of input DNA needed for SMRT sequencing), they were mixed with 500 ng of λDNA per library to serve as both carrier DNA and fragments for size normalization as described.
Libraries for vector DNA along with spike-ins were constructed using the Express Template Prep Kit 2.0 (End-Repair/A-tailing) (PN 100-938-900) and ligated to indexed SMRTbell adapters with the Barcoded Overhang Adapter Kit (PN 101-628-400/500). Libraries were pooled and purified using 1.8 × AMPure beads. Sequencing was performed on a Sequel II instrument following standard procedures defined by the manufacturer and the UMMS Deep Sequencing Core: Pacific Biosciences Core Enterprise.
To ensure accurate interlibrary comparisons and to maximize experimental conditions in a cost-effective manner, data reported in this study were executed on two flow cells: vectors purified by immunoaffinity column (Supplementary Table S1) were ran on one flow cell, whereas data related to full, partial, and empty fractions of pTx/HEK293- and rBV/Sf9-produced vectors were ran on a separate flow cell.
Data analysis
Subreads from SMRT sequencing were first preprocessed by running recalladapters (version 9.0.0) with the following options: –disableAdapterCorrection and –minSnr = 2.0. This preprocessing step is needed when working with subreads generated from SMRT-Link 7 (and up), which artificially cleaves long palindromic reads. The subsequent reads were then processed by the circular consensus sequencing (CCS) tool from SMRT Link version 8.0.0.79519 with the following options: –min-snr = 3.75, –min-passes = 2, and –by-strand. Downstream analyses were carried out by using custom workflows on Galaxy.39
The consensus read fastq files generated from CCS were demultiplexed into six libraries. The libraries were mapped to the vector and λDNA references using Burrows-Wheeler aligner-maximal exact match (BWA-MEM).40 The total number of mapped reads, their lengths, and the relative read abundances were calculated for each library to adjust for size bias.28 Read alignments were displayed on Integrative Genomics Viewer tool version 2.6. × and higher with soft-clipping on.41
The procedure for categorizing and quantifying ITR variants was carried out using custom workflows using USEARCH and publicly available tools provided on Galaxy. Reads from all libraries were mapped to the pFB-GFP vector reference from ITR-to-ITR or to the AAV2 ITR reference using BWA-MEM. Mapped reads to both references were selected, and the ITR sequences were extracted from these mapped reads by selecting sequences that flank the D region. ITRs were analyzed for their length distribution, and three major peaks were identified.
For each identified peak, the population was extracted and unique ITR lengths were selected to cluster reads with 95% similarity using USEARCH v.10 with the command -cluster_fast and using the parameter, -id 0.95.42 Clusters with five or more members were selected, and the consensus sequences were defined as the ITR configuration.43 Through this method, 13 ITR configurations were identified. The left and right (5′ and 3′, respectively) ITRs were separated and mapped to the defined ITR configurations (as reference genomes) to separate into categories.
Data availability
The data sets generated and/or analyzed in the current study have been deposited in the National Center for Biotechnology Information Sequence Read Archive: PRJNA794197.
Results
Vector genomes packaged by pTx/HEK293 and rBV/Sf9 platforms are grossly different
To investigate the differences between packaged genomes from pTx/HEK293 and rBV/Sf9 platforms, we opted to use a simple rAAV genome consisting of AAV2 ITRs and an EGFP reporter gene, whose expression is driven by the hCMV promoter, the second intron of the β-globin gene (HBB), and the 3′-UTR, also from the HBB gene (Fig. 1). This 3.3-kb vector construct was then flanked by Tn7 elements and served as the final cis-plasmid construct. Therefore, the same plasmid preparation used for pTx/HEK293 production can also be used to generate the bacmid for recombinant baculovirus production with the Bac-to-Bac transposition system.31 Vectors were then produced using serotype 8 (AAV8) under standard conditions and procedures for the respective platforms.18,19,44
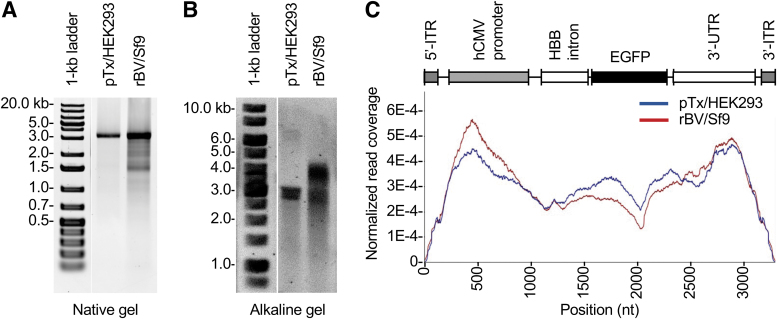
Figure 1.
Encapsidated DNA evaluation of pTx/HEK293- and rBV/Sf9-produced AAV vectors. (A) Native agarose gel of rAAV DNA. The 3.3-kb band represents the full-length ITR-to-ITR vector genomes. Additional bands represent heterogeneous species. (B) Alkaline agarose gel of rAAV DNAs. (C) Profiling vector genomes by SSV-Seq. The reference genome is displayed above the individual coverage traces of the pTx/HEK293-produced vector (blue) and the rBV/Sf9-produced vector (red). Reads are normalized to sequencing read counts from a vector plasmid construct prepared by restriction enzyme digestion to liberate a double-strand fragment spanning from the 5′-ITR to the 3′-ITR. Coverage of the ITRs is reduced compared with the rest of the genome as a result of the ITRs strong secondary structure, differences in flip and flop configurations, and PCR bias across GC-rich sequences and homopolymers.46 ITR, inverted terminal repeat; PCR, polymerase chain reaction; rAAV, recombinant adeno-associated virus; rBV, recombinant baculovirus vector; SSV-Seq, single-stranded DNA virus sequencing.
Both vectors were then subjected to immunoaffinity column purification. To assess the quality of the vectors, preparations were subjected to CsCl gradient ultracentrifugation to observe the distribution of particle compositions (Supplementary Fig. S1A). We found that the pTx/HEK293-produced vector exhibited two well-defined bands attributed to empty and full particles. In contrast, the rBV/Sf9-produced vector revealed a distinct band associated with empty particles, but an ill-defined band for the full particle fraction. The full particle fraction displayed a large gradient range, suggesting the presence of heterogeneously packaged capsids. Based on these results, we predicted that the rBV/Sf9-produced vector also harbors a population of oversized genomes.
To quantify the abundances of empty versus full particles, the two vectors were subjected to AUC analysis (Supplementary Table S1 and Supplementary Fig. S1B). The pTx/HEK293-produced vector showed a major sedimentation coefficient (S) peak at 100S, which corresponds to “full” capsids, and a nearly negligible peak at 60S (9.1%), which is the range that empty particles are found. In contrast, the rBV/Sf9-produced vector batch had a more pronounced empty fraction peak (41.1%).
Notably, the 100S peak exhibited a broad range and a shoulder peak at 80S, suggesting the presence of partial genome species. To better assess this observation, vectors were subjected to DNA purification and subsequent agarose gel analyses. Based on native gel analysis (Fig. 1A), we observed that the genomes of the pTx/HEK293-produced vector were predominantly a single species, migrating at the full-length size of 3.3 kb. In contrast, the rBV/Sf9-produced vector genomes exhibited a high degree of heterogeneity, displaying multiple species from 1.5 to 3 kb in size.
As demonstrated previously,45 we predicted that genome heterogeneity was in part attributed to the formation of truncated self-complementary genomes. To determine the nature of these species, vector DNAs were subjected to alkaline gel electrophoresis (Fig. 1B). As expected, the pTx/HEK293-produced vector yielded a single band with an approximate length of 3 kb. However, the rBV/Sf9-produced vector produced two major visible bands: one at ∼3 kb and one near 4 kb, the latter of which is greater than the 3.3 kb ITR-to-ITR design. Additionally, the rBV/Sf9-produced vector displayed more smearing, suggesting that this vector had a higher degree of heterogeneity.
We sought to determine whether NGS analysis could reveal any differences between the two vectors. We selected to employ SSV-Seq,38 a method that utilizes Illumina short-read sequencing to capture vector genome representation across a reference. This analysis revealed that normalized reads between pTx/HEK293- and rBV/Sf9-produced vectors were similarly distributed across the entire 3.3 kb genome (Fig. 1C). We note that due to the strong secondary structure of the ITR and differences in flip and flop configurations, coverage of the ITRs was reduced compared with the rest of the genome. Furthermore, the sequencing coverage was uneven across the genome as a result of PCR bias across GC-rich sequences and homopolymers.46
Although both vectors showed near equal representation across the reference, the sequencing analysis using 100-base paired-end Illumina reads did not reveal why the rBV/Sf9-produced vector showed a higher degree of heterogeneity than the pTx/HEK293-produced vector (Fig. 1A, B).
SMRT sequencing reveals that pTx/HEK293 and rBV/Sf9 platforms produce different proportions of heterogeneous vectors
Since short-read sequencing was not able to reveal clear differences between vector genomes generated by pTx/HEK293 and rBV/Sf9, we opted to employ SMRT sequencing and AAV-GPseq to profile the composition of packaged genomes.28 Libraries were prepared as previously described28 and sequenced on a single SMRT cell. Produced subreads were processed as described in the Materials and Methods section to generate CCS reads. The pTx/HEK293-produced vector reads were mapped to the vector plasmid sequence (Fig. 2A), whereas the reads of the rBV/Sf9-produced vector were mapped to the vector sequence flanked by the Tn7 elements (Fig. 2B).
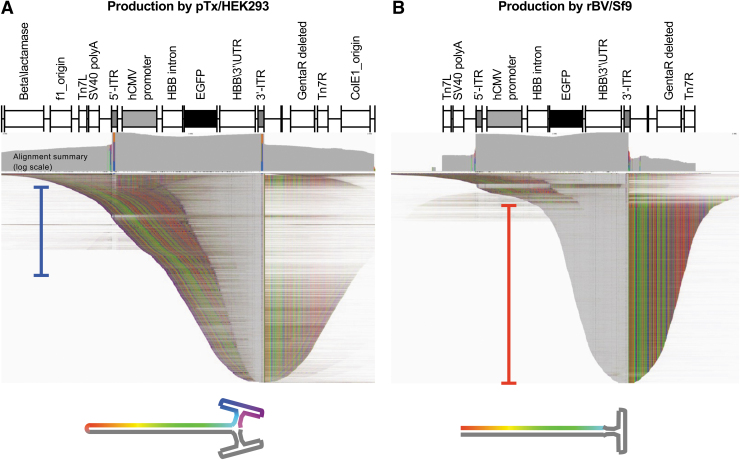
Figure 2.
Production platform-related differences in vector genomes. (A) IGV display of pTx/HEK293-produced vector reads aligned to the trans plasmid reference. (B) rBV/HEK293-produced vector reads were aligned to a reference spanning from the left and right Tn7 elements. Alignment summaries are shown above in log scale. Reads are shown with 100-read down sampling with soft-clipped bases shown to highlight reads of unique length and composition. The portion of reads matching the reference are in gray, mismatches are shown as colored bases. Deletions appear as speckles in this squished display. Structures depicting truncated double-ITR species (blue-bracketed reads) and truncated single-ITR species (red-bracketed reads) are shown below each plot. IGV, Integrative Genomics Viewer.
Notably, the vector plasmid constructs contain ITRs in the flop configuration; and thus, the references also contain ITRs in the flop orientation. Alignments of the reads revealed that they predominantly mapped to the reference between the 5′-ITR and 3′-ITR elements, consistent with SSV-Seq analysis. Some reads did map to regions beyond the ITRs and into the plasmid backbone. These sequences likely represent reverse-packaged genomes.26 Interestingly, when the aligned reads of the vectors were displayed to highlight reads of unique length and composition by showing soft-clipped bases and with a 100-read down sampling, the major truncated forms were revealed to be different between the vectors.
The truncated species produced from the pTx/HEK293 platform exhibited mapping behaviors that reflect self-complementary structures that contained two ITRs, one at the 5′ end and one at the 3′ end, as described previously for other vector designs.26,45 The “looped” end of the genome resides within the body of the vector genome and originates from template-switching events occurring throughout the vector genome (Fig. 2A, blue-bracketed population in the panel).45
In contrast, the rBV/Sf9-produced vectors yielded truncated self-complementary genomes with open ends terminating within the body of vector genome (Fig. 2B, red-bracketed population in the panel). These genomes also showed an unresolved 3′-ITR, forming reads consisting of a single ITR anchored at the center of the read. We therefore hypothesized that rBV/Sf9-produced vectors have a higher degree of unresolved ITRs, which corresponded to vector genomes with a self-complementary single ITR configuration.
We sought to determine whether the mapped vector reads correspond to what was represented by agarose gels (Fig. 1A, B). The lengths of each mapped read were determined and graphed (Supplementary Fig. S2). As expected, the mapped pTx/HEK293-produced vector reads showed a single prominent peak at 3.3 kb. Interestingly, the trace for length distribution of vectors produced by the rBV/Sf9 platform showed a large range of different sizes, including full-length-sized genomes (Supplementary Fig. S2). This result suggested that the vector exhibited a high degree of heterogeneity.
While the length distribution of the pTx/HEK239-produced vector conformed to the agarose gel results, the lengths of the rBV/Sf9-produced vector reads did not reflect the two major populations observed by alkaline gel (Fig. 1B). Nonetheless, both agarose gel analyses and SMRT sequencing support the notion that the heterogeneity of the rBV/Sf9-produced vector was considerably higher.
The presence of truncated genomes in the rBV/Sf9-produced vectors correlates with unresolved mutant ITRs
To better illustrate the differences in unresolved genomes between pTx/HEK293- and rBV/Sf9-produced vectors, we generated a reference sequence consisting of three genome units linked together by ITRs (Fig. 3). This reference configuration allows the ability to capture species that have unresolved 5′-ITRs (left) or 3′-ITRs (right). The SMRT reads of the vectors from the two production systems were then mapped to this “trimer” reference. As predicted, the pTx/HEK293-produced vector reads predominantly mapped between the boundaries of the left and right ITRs (Fig. 3A). In contrast, the majority of the rBV/Sf9 vector reads spanned across two genome units with the center of the read anchored at the ITRs (Fig. 3B).
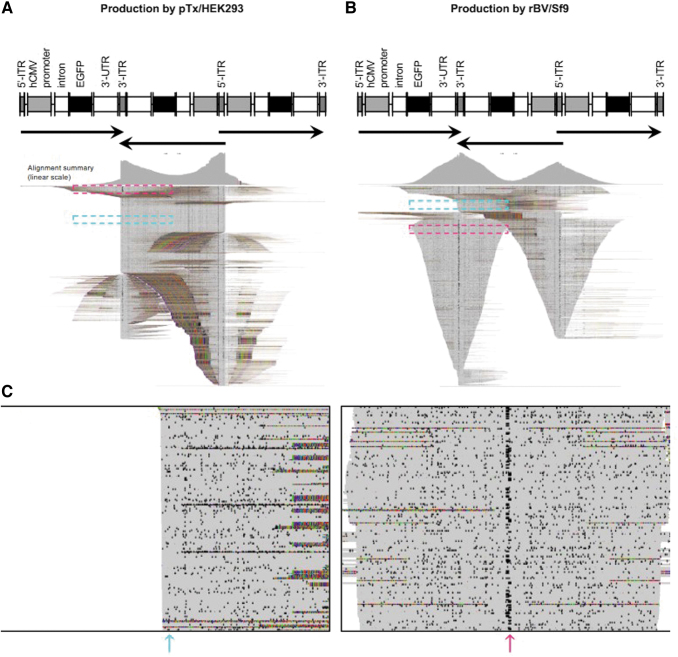
Figure 3.
Vector reads aligned to an unresolved trimer reference reveal differential vector heterogeneity. (A, B) Displays of the pTx/HEK293-produced (A) and rBV/Sf9-produced (B) vector reads aligned to a reference that represents a predicted three-unit genome linked together by unresolved ITRs to capture species that have unresolved 5′-ITRs or 3′-ITRs. Alignment summaries are shown. Arrows indicate transgene direction. Dashed cyan and magenta boxes show populations of resolved and unresolved 3′-ITRs, respectively. (C) Zoom-in of cyan and magenta boxes from (B), respectively, representing reads with resolved (left) and unresolved 3-ITRs (right). Read matches (gray), mismatches (colored), and deletions (speckles) are shown in squished display. Cyan and magenta arrows show read regions with low and high degrees of deletions at the ITR, respectively.
Interestingly, some of the truncated genomes from the rBV/Sf9-produced vector were similar in structure as those of the pTx/HEK293-produced genomes, where the reads terminate at resolved ITRs (Fig. 3B, dashed cyan box, and Fig. 3C, left panel). Upon closer inspection of the ITR-spanning sequences, we discovered that the truncated genomes with unresolved ITRs had a high frequency of deletions (Fig. 3B, dashed magenta box, and Fig. 3C, right panel), whereas the reads terminating at resolved ITRs had visibly fewer deletions (Fig. 3C, left panel at position of cyan arrow). The high frequency of nonrandom deletions within the ITRs among unresolved genomes suggests that there is a correlation between mutations within the ITRs and failure of these genomes to resolve.
Inspection of the truncated genomes from pTx/HEK293-produced vectors that also span two genome units also revealed deletions within the ITR sequence; however, these populations are less abundant (Fig. 3A, dashed magenta box). These findings suggest that mutated unresolved genomes are not a specific hallmark of either production methods but are more abundantly found in the rBV/Sf9-produced vector.
The genome compositions between full, partial, and empty capsid fractions of pTx/HEK293- and rBV/Sf9-produced vectors are distinct
We next aimed to determine whether the differences in genome heterogeneity found among the immunoaffinity column-purified vectors might be caused by a higher abundance of partial genomes generated by the rBV/Sf9 platform. Typically, these partial genomes can be removed by methods such as CsCl density gradient centrifugation, whereby vectors are subjected to high-speed ultracentrifugation in a CsCl gradient. Full particle fractions can therefore be enriched and isolated by density to obtain more than 90% full capsids. We therefore hypothesized that the partial fractions are enriched in these truncated unresolved species; and thus, differences between the production platforms can be interrogated directly.
To investigate the compositions of the partial fraction vectors, we used the pTx/HEK293 and rBV/Sf9 platforms for production as before. The vectors were then subjected to two rounds of CsCl gradient ultracentrifugation to separate the full+oversized (high density), partial (medium density), and empty particle (low density) fractions (Supplementary Fig. S3). As predicted, we found that the composition of vector particle bands from the first centrifugation was different between the two preparations (Supplementary Fig. S3A). Specifically, the rBV/Sf9-produced vector showed a less well-defined full particle band.
Following the second centrifugation, we observed among the high-density fractions, which consist of full and oversized particles (blue boxes), that the pTx/HEK293-produced vector demonstrated a clear delineated band, whereas the rBV/Sf9-produced vector showed a poorly defined high-density population.
These observations suggest that the pTx/HEK293 preparation exhibits a uniform genome structure, whereas the rBV/Sf9 preparation exhibits a high diversity of vector genome sizes. Based on the distribution of bands following density gradient separation, three fractions were collected for pTx/HEK293- and rBV/Sf9-produced vectors: low density, medium density, and high density (Supplementary Table S1 and Supplementary Fig. S3B). Vector titers for the three fractions were determined by qPCR and ELISA (Supplementary Table S1).
We first analyzed the composition of the partial fractions by AUC analysis (Supplementary Table S1 and Supplementary Fig. S3C). The partial fractions of the pTx/HEK293-produced vector showed 52% empty particles, 43.9% partial particles, and no full particles (Supplementary Table S1), whereas the partial fractions of the rBV/Sf9-produced vector showed 1.1% empty particles, 92.3% partial particles, and 4.7% full particles. Based on these results, we were cautious of drawing direct comparisons between the partial fractions of the vectors since the composition of the partial genomes among the pTx/HEK293-produced vectors was half composed of empty particles.
Finally, to ensure that the quality of the produced capsids met expected standards, the fractions were subjected to PAGE, followed by silver staining (Supplementary Fig. S3D). The high- and low-density fractions of the pTx/HEK293- and rBV/Sf9-produced vectors displayed the approximate and expected 1:1:10 ratio of VP1:VP2:VP3 capsids.47 Notably, the rBV/Sf9-produced vectors showed degradation bands of VP1/2. These bands are suggested to be a product of baculoviral cathepsin cleavage of AAV8 capsids.48,49 Silver staining of capsids for the partial fractions was similar to full and empty fractions (unpublished observations).
SMRT sequencing reads from the full and partial fractions of rBV/Sf9-producted vectors share similar genome compositions
The full, partial, and empty fractions of the pTx/HEK293- and rBV/Sf9-produced vectors were subjected to SMRT sequencing as before. When mapped to an ITR-to-ITR reference, we observed that reads from the full fraction of the pTx/HEK293-produced vector were a mixture of full-length and truncated genomes (Fig. 4A). When reads were plotted by their relative abundances as a function of length (Supplementary Fig. S4A), the expected 3.3 kb full-length peak was identified. Interestingly, this read peak only accounted for 9.14% of reads spanning a range of 0.5–5 kb. The remaining reads were truncated genomes with various sizes under 3.3 kb in length.
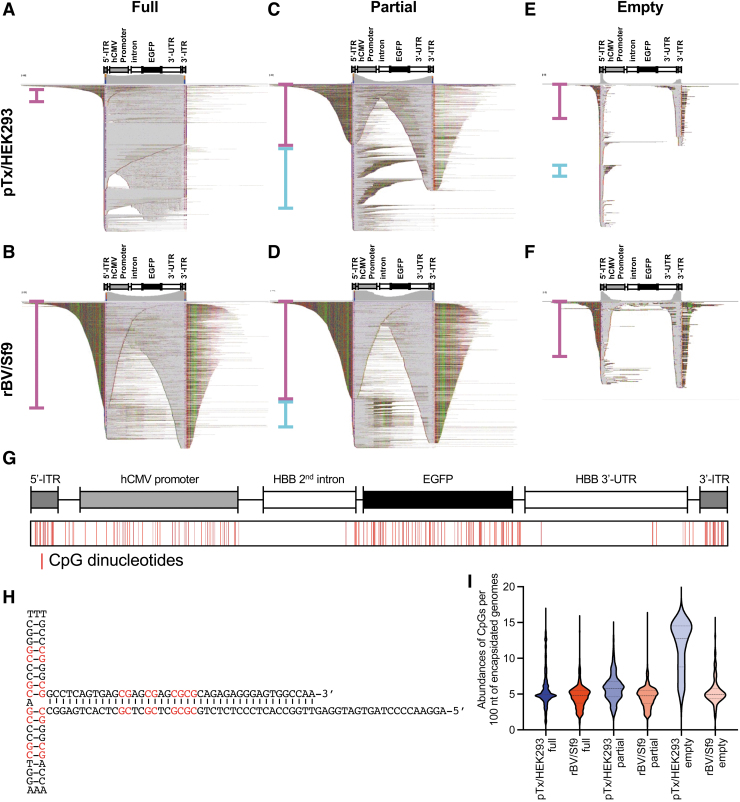
Figure 4.
Differential vector heterogeneity between density gradient fractions. (A–F) Sequencing reads obtained from full (A, B), partial (C, D), and empty fractions (E, F) from pTx/HEK293-produced vectors (A, C, E) and rBV/Sf9-produced vectors (B, D, F). 5′-sorted alignments reveal variable abundances of single-ITR (magenta brackets) and double-ITR (cyan brackets) self-complementary genomes. (G) Diagram depicting the position of CpGs throughout the reference genome. (H) Structure of the AAV2 ITR highlighting the 16 CpGs (red). (I) Violin plot showing the abundances of CpGs per 100 nt of sequence among pTx/HEK293- (blue) and rBV/Sf9-produced (red) vector fractions. Dashed lines represent the data median, and dotted lines indicate upper and lower quartiles.
This observation was predicted since full capsid fractions from CsCl gradient purifications have been demonstrated previously to contain truncated genomes.26,28 However, the large percentage of reads under 1 kb in length was unexpected and may reflect strong bias in the representation of the smaller genomes. Interestingly, the full fraction of the rBV/Sf9-produced vectors was predominantly partial/truncated self-complementary, unresolved genomes (Fig. 4B). When plotted by length, the reads were largely 0.5–1.5 kb in length (Supplementary Fig. S4A). As mentioned above, the alkaline gels of the rBV/Sf9-produced vector purified by immunoaffinity column produced a high degree of oversized genomes with a single-strand length of ∼4 kb (Fig. 1A, B).
Since these larger molecular weight bands are presumed to be unresolved genomes that form partial self-complementary configurations (Supplementary Fig. S4B), they may fail to adapter with high efficiency during the library construction process. In addition, the polishing of fragment ends (DNA damage/end repair) that is standard for SMRT sequencing library preparation may possibly cleave the single-stranded DNA at variable positions to form self-complementary single-ITR configurations (Supplementary Fig. S4B); but this possibility remains unsubstantiated in this study. Although the read outcomes reflect this possibility, it is not clear whether this is strictly due to the library preparation steps or related to another phenomenon.
As predicted, the partial fraction of the pTx/HEK293-produced vector exhibited a reduction of full-length reads (Fig. 4C), accounting for 1.47% of all reads spanning a range of 0.5–5 kb (Supplementary Fig. S4A). We also found that truncated genomes are enriched in this population. Interestingly, among the truncated forms, we observed multiple single-ITR self-complementary forms as well as double-ITR forms (Fig. 4C, magenta and cyan brackets, respectively). Curiously, the mapping behavior of reads related to the partial fraction of rBV/Sf9-produced vectors was similar to those of the full fraction (Fig. 4B, D). This observation suggests that both full and partial genomes of the rBV/Sf9-produced vector were populated by unresolved ITRs.
Empty particles are not entirely empty but packaged with short DNA fragments that contain ITRs inherently abundant with CpG motifs
In our pursuit to understand the types of vector genomes that were packaged into full and partial genome fractions, we also had an opportunity to profile the contents of empty particle fractions acquired from the pTx/HEK293- and rBV/Sf9-produced vectors (Fig. 4E, F). It has been demonstrated previously that empty particles may contain small fragments of genomic material,50 but the contents were unknown. We found that the empty fractions for both vector preparations indeed contained short DNA fragments. When mapped to the vector reference, the reads predominantly spanned the ITRs.
Interestingly, the majority of these short reads from pTx/HEK293-produced vectors are smaller than 500 bp, similar in nature to snapback genomes.51 In contrast, the short fragments in the rBV/Sf9-produced vector are smaller than 1 kb (Supplementary Fig. S4A) and predominantly have unresolved self-complementary configurations. Nevertheless, read coverage of these short fragments was also enriched at the ITRs. These results were indeed surprising since they not only demonstrate that empty capsids are not in fact empty, but also suggest that the packaging of short fragment DNA is not a passive event. The packaging of short fragments is instead nonrandom and favors ITR-bearing DNAs.8
The finding that empty capsids are packaged with short ITR-containing DNAs is not only striking but also concerning. The presence of unmethylated CpG dinucleotides in vectors has recently been viewed as a priority concern since these motifs have been known to trigger the innate immune response via toll-like receptor 9 (TLR9) activation.52,53 These observations have in turn raised questions regarding the safety and durability of rAAVs that contain CpG motifs in their design, and whether gene therapy vectors that contain a high abundance of CpGs could lead to immunological responses that may coincide with adverse host responses or T cell responses to capsid or transgene products.54,55
There have been several efforts to reduce the abundance of CpG dinucleotides in vector designs, as well as approaches to mitigate the host cell response to unmethylated CpGs.56–58 In fact, each ITR harbors 16 CpG dinucleotides (Fig. 4G, H) and can therefore represent an enrichment of sequences that may compromise the safety of vector preparations that are abundant with empty capsids. To probe this, the relative abundances of CpG motifs found in the vector genome reads from full, partial, and empty fractions from both pTx/HEK293 and rBV/Sf9 production schemes were determined (Fig. 4I).
These analyses revealed that reads from the empty fractions for the pTx/HEK239 vector showed an increase in the median CpG content over its full fraction by ∼2.5-fold. All other fractions had approximately five CpG motifs per 100 nts. This finding suggested that although empty capsids package DNAs that prominently package ITRs, the abundance of CpG motifs on a per fragment basis is not drastically enriched in empty fractions.
Wild-type ITRs are abundant in HEK293-produced vectors, while unresolved and mutant ITRs are ubiquitous in rBV/Sf9-produced vectors
As noted, we observed a correlation between mutated ITRs and unresolved genomes. We therefore aimed to address the hypothesis that mutated ITRs in pTx/HEK293- and rBV/Sf9-produced vectors correlate with vector heterogeneity and the generation of partial and empty particles. The ITR sequences from each of the particle fractions from pTx/HEK293- and rBV/Sf9-produced vectors were extracted from the reads. The left ITRs (5′-ITRs) and right ITRs (3′-ITRs) were also partitioned for separate evaluation.
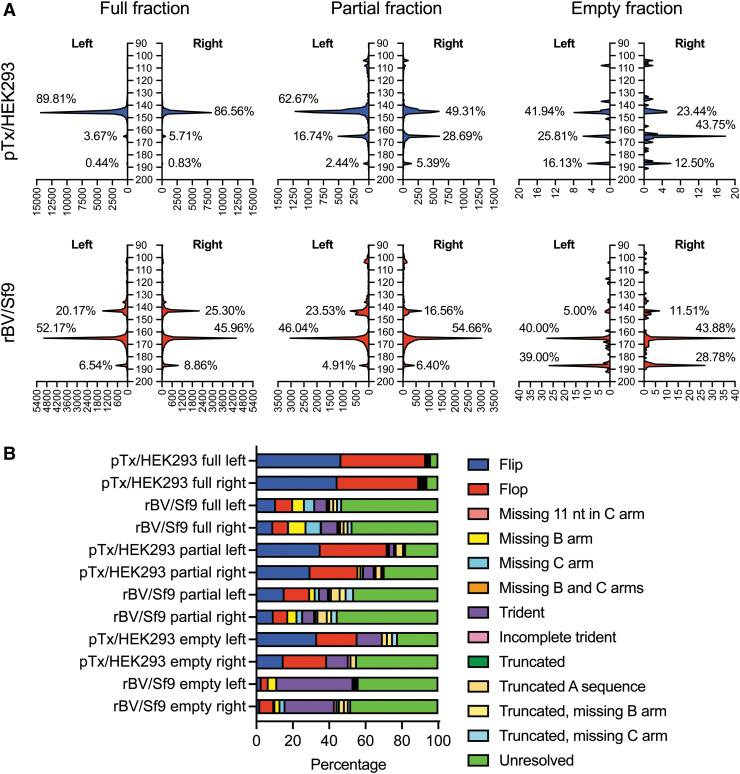
The diversity of ITRs was first profiled by length distribution (Fig. 5A). When comparing the full fractions of pTx/HEK293- and rBV/Sf9-produced vectors, we observed that the former predominantly harbored ITRs with lengths of ∼145 nt at both the left and right ends of the genome (89.81% and 86.56%, respectively). Notably, the wild-type AAV2 ITR is 145 nt in length, thus suggesting that the ITRs of the pTx/HEK293-produced vector have primarily intact ITRs. In contrast, the major population at both the left and right ITRs of the rBV/Sf9-produced vector has peak lengths of 165 nt (52.17% and 45.96%, respectively).
Figure 5.
Differences in ITR heterogeneity between capsid fractions and production methods. (A) Distribution of ITR lengths among full, partial, and empty capsid fractions. Traces of left and right ITRs are shown for pTx/HEK293-produced vectors (top) and rBV/Sf9-produced vectors (bottom). The percentages of ITRs are calculated as the ITR counts within each peak over all counts integrated across sequences with lengths of 90–200 nt. (B) Thirteen ITR species were identified among all capsid fractions of the two production methods. Their abundances in samples are expressed as a percentage of all ITR counts properly categorized into the designated types.
Additionally, the ∼145-nt peak composed of only 20.17% and 25.3% of the total left and right ITR populations, respectively. Notably, an ITR that is unresolved and retains a D element at both the 5′ and 3′ ends of the ITR is 165 nt in length. This result was somewhat expected since we observed that the majority of reads from the full fraction had unresolved genome configurations (Fig. 4B).
We also detected a small fraction of unresolved genomes in the pTx/HEK293-produced vector at both the left and right ITRs (3.67% and 5.71%, respectively). Interestingly, we also observed a third minor peak at 187 nt at both the left and right ITRs of the rBV/Sf9-produced full vectors (Fig. 5A). These larger ITRs exist at relatively low percentages (6.54%, left ITR; and 8.86%, right ITR) and are near absent in the pTx/HEK293-produced full vector (<1%).
Among the pTx/HEK293-produced partial fraction vectors, we observed an increase in the abundance of the 165-nt species at the left and right ITRs (16.74% and 28.69%, respectively). This observation coincides with the increase in unresolved species found in this fraction (Fig. 4C). In addition, an increase in the 187-nt species were also detected in the partial fraction (Fig. 5B), suggesting that pTx/HEK293-produced vectors can also harbor mutated ITRs.
Interestingly, the partial fraction of the rBV/Sf9-produced vector contained a similar distribution of variable-length ITRs as the full fraction. There was a slight decrease in 165-nt left ITRs (52.17–46.04%), but a 9% increase among right ITRs. Since this study only utilized one vector preparation from each production scheme, these differences cannot be utilized to establish significance. As with the full fraction, the partial fraction of the rBV/Sf9-produced vector also contained 187-nt species at both the left and right ITRs.
When analyzing the ITR compositions of empty fractions for the vectors produced by either the pTx/HEK293 and rBV/Sf9 platforms, an overall decrease in the 145-nt species was observed at both left and right ITRs (Fig. 5A). At the same time, we observed an increase in the percentages of 165- and 187-nt species for both vectors. For example, the empty fraction of the rBV/Sf9-produced vector is composed of 39% and 28.76% 187-nt species at the left and right ITRs, respectively. Altogether, these data demonstrate that partial and empty fractions have a high degree of ITRs with variable lengths. Shorter ITRs (<145 nt) are also present in partial and empty fractions but persist at low amounts (Fig. 5A).
These new findings support the hypothesis that genomes that fail to properly resolve can lead to genome heterogeneity.
The distribution of ITR lengths revealed common ITR configurations in each capsid fraction for pTx/HEK293- and rBV/Sf9-produced vectors. We therefore aimed to determine the structural compositions of these variable ITRs from each production method and gradient fraction. As mentioned before, wild-type AAVs and standard rAAVs contain ITRs that take on either flip or flop orientations, and these two configurations are equally distributed if replication of the vector genome goes unperturbed. However, we have revealed that rBV/Sf9 vectors have a high degree of variable ITRs (Fig. 5A), even in full particle fractions. Using clustering analysis, we were able to identify at least 13 common and unique configurations across all the groups (Fig. 5B).
Full fractions of the pTx/HEK293-produced vector harbored ITRs that were predominantly flip or flop. These configurations were equally distributed at both the left and right ITRs, as expected of wild-type structures. Importantly, the full fraction of the rBV/Sf9-produced vector showed a low percentage of flip and flop (10.75% and 9.4%, respectively, at the left ITR; and 9.23% and 8.61% at the right ITR). As predicted by the large abundance of 165-nt ITRs (Fig. 5A), these vector genomes had ∼50% unresolved ITRs (Fig. 5B).
Strikingly, the full fraction of the rBV/Sf9-produced vector harbored ITRs with missing B or C arms (6.5% and 5.6%, respectively, at the left ITR; and 9.6% and 8.37% at the right ITR) (Fig. 5B) or had trident configurations where either the B or C arm was duplicated (6.81% at the left ITR and 8.99% at the right ITR). Additional truncated configurations were also identified, but these made up ∼2% of all classified ITR configurations, or less.
The ITRs of the partial fraction for pTx/HEK293-produced vectors also exhibited a mixture of configurations. At the left ITR, 35.27% were in the flip orientation and 36.81% were in the flop orientation; at the right ITR, 29.71% were in the flip orientation and 26.27% were in the flop orientation. Although the percentages of flip and flop were comparatively lower than what was observed among the full fractions, they still were detected at equal abundances. This finding suggests that the partial fraction pTx/HEK293 vector genomes with intact ITRs were still able to replicate accurately during production. For both the left and right ITRs, the third most abundant configuration was unresolved ITRs (17.5% and 29.5%, respectively). Interestingly, the partial fraction also contained a small percentage of trident-shaped ITRs, suggesting that this configuration is not exclusive to the rBV/Sf9-produced vector.
As expected, the partial fraction of the rBV/Sf9-produced vector revealed a similar distribution of unresolved genomes as the full fraction, where ∼50% of the ITRs detected were the 165-nt unresolved species.
There was a smaller percentage of ITRs with missing B and C arms, and a slight increase in truncated ITRs that carry missing B and C arms. But as stated above, because these data cover just a single vector, the observed differences may not be substantial. Interestingly, the empty capsid fractions of both vectors showed a considerable increase in the percentage of trident-shaped ITRs compared with other fractions. In the case of the rBV/Sf9 empty fraction, 41.9% and 27% of the left and right ITRs were respectively trident shaped. The remaining ITRs were predominantly unresolved (Fig. 5B).
Altogether, our observations suggest that there is a strong correlation between ITR integrity and vector heterogeneity.
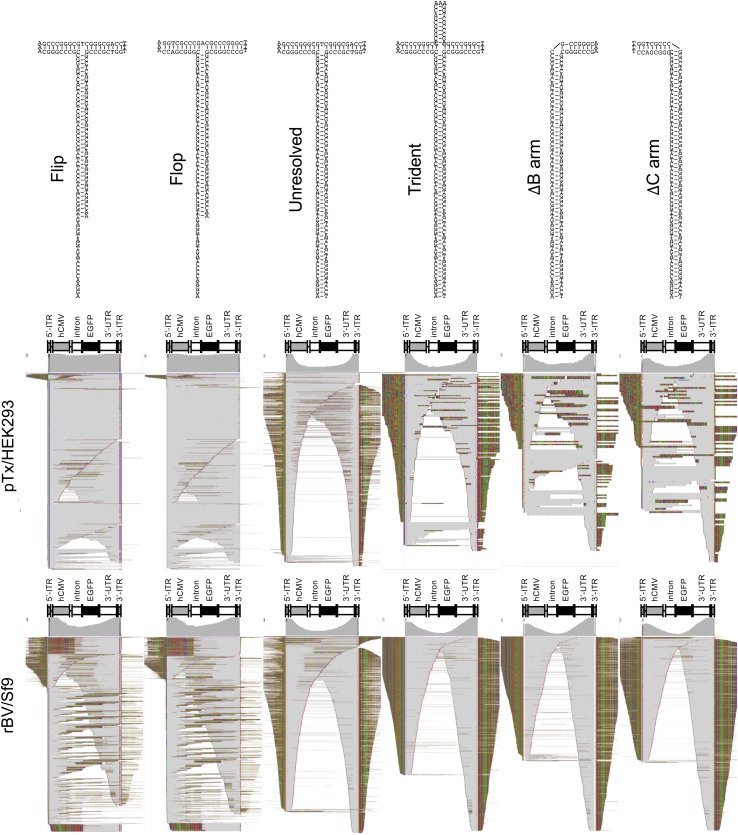
Trident-shaped, B arm-deleted, and C arm-deleted ITRs are poorly resolved
We next aimed to address whether specific ITR configurations can impact resolution. Working with the full capsid fractions of both pTx/HEK293- and rBV/Sf9-produced vectors, we segregated reads into six categories defined by the most prominent ITR configurations identified: flip, flop, unresolved, trident, deleted B arm, and deleted C arm ITRs. These reads were then mapped back to the reference to observe their mapping behaviors (Fig. 6). As expected, the flip and flop configurations of pTx/HEK293 vectors led to the highest degree of ITR-to-ITR spanning genomes.
Figure 6.
Differential genome heterogeneity among vectors that harbor different ITR species. Sequencing reads that harbor the five ITR species: flip, flop, unresolved, trident-shaped, B arm-deleted, or C arm-deleted configurations were extracted from the full capsid fractions of the pTx/HEK293-produced vectors (top) and rBV/Sf9-produced vectors (bottom). Reads were remapped to the vector genome reference. Aligned reads are displayed with soft-clipped bases shown. Alignment summary tracks are displayed above each plot. The portion of reads that perfectly align with the reference are in gray; mismatched bases are colored; deletions are shown as black dashes and appear as speckles throughout the squished display.
We did observe a few reads that appeared to contain unresolved genomes, but these may be due to misclustered reads. Interestingly, rBV/Sf9-produced vector genomes bearing flip and flop configurations exhibited resolved ITRs, yet the majority of these genomes were also truncated, demonstrating that despite isolating reads harboring resolved ITRs, full-length genomes were still not well enriched. As before, multiple reads were found to have mapping behaviors indicative of unresolved genomes, but these are likely due to misclustering from the workflow and remain in the minority of the reads. As expected, reads that contained unresolved ITRs revealed genomes with mapping behaviors that indicated unresolved self-complementary genomes (Fig. 6).
We next probed the mapping behaviors of the genomes harboring trident-shaped ITRs found in both pTx/HEK293- and rBV/Sf9-produced vectors (Fig. 6). Surprisingly, despite having intact B and C arms, and an unchanged TRS sequence, the majority of reads from both vectors demonstrated mapping behaviors that were indicative of unresolved self-complementary genomes. This finding suggests that ITR resolution not only relies on the presence of sequence elements for recognition of Rep and subsequent nicking of the strand on the TRS, but also that secondary structure might be critical for resolution as well. However, ITRs with B or C arm deletions had a higher degree of resolved ITRs than the trident-shaped counterparts among the pTx/HEK293-produced vectors. Interestingly, the rBV/Sf9-produced vector failed to demonstrate a strong capacity to resolve B or C arm-deleted ITRs.
These data seem to suggest that the rBV/Sf9-production system has a lower tolerance for resolving ITRs with deletions. Alternatively, rBV/Sf9 systems may be less efficient at ITR resolution.
Discussion
Our collective work has shown that not all rAAV genomes within a preparation are made the same.26,28,29,45 Previously, we demonstrated that vector heterogeneity can be influenced by the vector design. Now, we show that production platforms can also impact genome structural differences. Using AAV-GPseq, we discovered that compared with pTx/HEK293-produced vectors, rBV/Sf9-produced vectors yielded a high degree of unresolved genomes, which directly correlated with the generation of truncated, self-complementary single ITR species. These species were also present in the pTx/HEK293-produced vector but were much more infrequent.
Due to the highly recombinogenic nature of AcMNPV replication and the inherent generation of defective interfering (DI) baculoviral particles, rBV genomes are intrinsically instable throughout its amplification.59,60 Large deletions occur frequently in the AcMNPV genome.61 Baculovirus replication is based on the recombination of DNA fragments by either the annealing of homologous single strands or through strand invasion.62 The inherent nature of baculovirus replication may favor ITR mutation/deletion forms. Therefore, a standing question following this investigation is whether mutations and/or deletions happen during rBV propagation or during rAAV rescue and replication.
Nevertheless, Sanger sequencing of the bacmid and rBV used to produce the vector did not reveal any clear ITR mutations (unpublished observations). Furthermore, we found that the vectors investigated in our study had at least 13 unique ITR species shared among both vectors. We concluded that these forms were either inherent to the plasmid or rBV, or the mechanisms that drive their synthesis are inherent to both platforms.
With this investigation, we also demonstrated that empty capsids are not in fact empty but can be packaged with short DNA fragments. Although our report is not the first to discover this phenomenon,50 we were able to formally characterize these DNA fragments by sequencing analysis. Surprisingly, these sequences were chiefly ITR-bearing reads, opening new questions for AAV biology and vectorology. The capacity for rAAVs to trigger the innate immune response through TLR9-mediated recognition of unmethylated CpG dinucleotides52,53 raises an interesting quagmire for production schemes that produce high proportions of empty capsids. The fact that ITRs have 16 CpG motifs within its inverted repeat structure suggests that vector preparations with a high percentage of empty capsids are not only less effective but can also trigger TLR9 activation.
Additionally, our findings that empty capsids can contain DNA may implicate an inherent issue with how vectors titers are quantified. Since all contemporary rAAV vectors designed for clinical application contain AAV2 ITRs, many qPCR- or Droplet Digital (dd)PCR-based quantification methods utilize sequences proximal to ITRs as a standardized means for determining vector titers. Since partial and empty fractions tend to harbor genomes that are enriched with sequences that are most proximal to ITRs, the functional titers based on these “ITR-probes” are likely less accurate than probe/primer sets designed against sequences set more central to the vector transgene. Nonetheless, the removal of empty and partial capsids seems imperative to determining accurate titers, as they are essential for establishing dosing for clinical vectors.
We showed that mutated ITRs were more frequent in partial and empty particles, shedding light on the possibility that ITR integrity can influence vector heterogeneity, and may point toward an unexplored path for reducing the abundance of empty capsids and improving vector production. Unfortunately, the mechanisms underpinning why and how heterogeneous ITRs are packaged are not fully understood. One potential confounding element in this investigation is that the vectors were produced outside the context of natural AAV helper viruses. The pTx/HEK293 platform utilizes a helper plasmid and complementary cell line that expresses all the essential Ad genes. In contrast, the baculovirus vector seems to contribute helper function on its own.63
Requirements for additional auxiliary genes during rAAV genome replication in Sf9 cells has not been fully investigated. Insects are natural hosts for densoviruses and autonomous parvoviruses with ssDNA genomes that are also flanked by ITRs. Both AAV and insect densoviruses such as Junonia coenia densovirus belong to the Parvoviridae family and demonstrate similar replication mechanisms.64 Therefore, an invertebrate host such as Spodoptera frugiperda should be a suitable surrogate for AAV vector DNA replication.17 Nevertheless, the maintenance of intact ITRs by different parvoviruses and their penchant to produce DI particles need to be reconsidered.
The relatively high frequency of genomes bearing trident-shaped ITRs in partial and empty capsid fractions is quite intriguing. In essence, they carry all the necessary motifs required for resolution: the Rep binding site (RBS), the RBS′ sequence at the tips of the B or C arms, and the D sequence. Our findings suggest that ITR resolution also depends on the secondary structure of the ITR, and not just the sequence. In addition, the observation shows that terminal resolution is not a prerequisite to packaging, which has long been proven as such with the generation of scAAVs via mutation of the ITR at the TRS.65,66 Our data suggest that other mutant ITR forms can also be used to drive unresolved scAAV configurations, opening the door to designing gene therapy vectors that depart from native ITR sequences to further improve vector performance.
In conclusion, despite the differences observed in this investigation between pTx/HEK293 and rBV/Sf9 production methods, we have shown that heterogeneity of ITRs is correlated with the diversity of vector genomes independent of the production method. It is universally accepted that ITR stability is critical for vector production, owing to the multiple purposes that the ITR serves.8,67 We have now molecular evidence for how ITR diversity may directly influence genome composition. One remaining question regarding the presence of mutant ITRs in preparations is how they impact transgene expression and stability in the cell. ITRs have shown multiple postentry roles (e.g., second strand synthesis, transcription, episome/concatemer formation, integration).8
Therefore, additional work is needed to determine the extent to which ITR heterogeneity impacts vector safety and efficacy. Hybrid ITR vector designs have shown increased directional intermolecular recombination,68 thus implicating the importance of the ITR structure in forming episomal species. Perhaps high ITR heterogeneity can compromise therapeutic persistence. If the observations shown in our report also hold true for other vector designs, further exploration into platform-based differences in ITR heterogeneity and their impacts is indeed warranted.
Supplementary Material
Acknowledgments
We would like to thank the UMass Chan Medical School Deep Sequencing core (Maria Zapp, Daniella Wilmot, and Ellie Kittler) for their help with sample preparations and SMRT sequencing. We are most grateful to the Genomics and Bioinformatics Core Facility of Nantes (GenoBiRD, Biogenouest) for its technical support concerning Illumina sequencing. AUC was performed at the platforms of the Grenoble Instruct-ERIC center (ISBG; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB). We thank Aline Le Roy and Christine Ebel from IBS and ISBG (Grenoble, FR) for AUC analyses.
Authors' Contributions
N.T.T., M.P.-B., and P.W.L.T. designed, conducted, and interpreted the bioinformatics analysis. P.W.L.T., M.P.-B., E.A., and G.G. conceived and directed the project. M.P.-B. designed the rAAV vectors. N.T.T. and P.W.L.T. developed the AAV-GPseq pipelines and performed the analyses. E.L. and S.S. performed SSV-Seq experimental and bioinformatics analyses. C.R., E.D., and V.B. supervised the production of AAV vectors and corresponding quality controls. O.A. and E.D. provided funding and led the INSERM UMR 1089 laboratory and vector core, respectively. S.N. prepared vector DNAs for sequencing. K.W. provided the recalladaptor script. N.T.T., G.G., M.P.-B., and P.W.L.T. wrote and finalized the article.
Author Disclosure
G.G. is a scientific cofounder of Voyager Therapeutics and Aspa Therapeutics and holds equity in these companies. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics, Aspa Therapeutics, and other biopharmaceutical companies. The remaining authors declare no competing interests. M.P.-B. and E.A. are inventors of patents related to AAV gene therapy licensed to biopharma companies. K.W. is a full-time employee of Pacific Biosciences, a company commercializing SMRT sequencing technologies.
Funding Information
G.G. is supported by grants from the UMass Chan Medical School (an internal grant) and by the National Institutes of Health (R01NS076991-01, P01HL131471-05, R01AI121135, UG3HL147367-01, R01HL097088, R01HL152723-02, U19AI149646-01, and UH3HL147367-04). This research was also supported by the Région Pays de la Loire, the University of Nantes, the Centre Hospitalier Universitaire (CHU) of Nantes and INSERM. The Grenoble Partnership for Structural Biology (PSB) is supported by The French Infrastructure for Integrated Structural Biology (ANR-10-INBS-0005-02) and Labex GRAL (Grenoble Alliance pour la biologie structurale et cellulaire intégrées), which is financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003).
Supplementary Material
References
- 1. Kuzmin DA, Shutova MV, Johnston NR, et al. The clinical landscape for AAV gene therapies. Nat Rev Drug Discov 2021;20:173–174. [DOI] [PubMed] [Google Scholar]
- 2. Keeler AM, Flotte TR. Recombinant adeno-associated virus gene therapy in light of luxturna (and zolgensma and glybera): where are we, and how did we get here? Annu Rev Virol 2019;6:601–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodrigues GA, Shalaev E, Karami TK, et al. Pharmaceutical development of AAV-based gene therapy products for the eye. Pharm Res 2018;36:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mendell JR, Al-Zaidy S, Shell R, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 2017;377:1713–1722. [DOI] [PubMed] [Google Scholar]
- 5. Yla-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European Union. Mol Ther 2012;20:1831–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cotmore SF, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res 1987;33:91–174. [DOI] [PubMed] [Google Scholar]
- 7. Lusby E, Fife KH, Berns KI. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J Virol 1980;34:402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berns KI. The unusual properties of the AAV inverted terminal repeat. Hum Gene Ther 2020;31:518–523. [DOI] [PubMed] [Google Scholar]
- 9. Samulski RJ, Berns KI, Tan M, et al. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci U S A 1982;79:2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Samulski RJ, Srivastava A, Berns KI, et al. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 1983;33:135–143. [DOI] [PubMed] [Google Scholar]
- 11. Zhou Q, Tian W, Liu C, et al. Deletion of the B-B′ and C-C′ regions of inverted terminal repeats reduces rAAV productivity but increases transgene expression. Sci Rep 2017;7:5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Savy A, Dickx Y, Nauwynck L, et al. Impact of inverted terminal repeat integrity on rAAV8 production using the Baculovirus/Sf9 cells system. Hum Gene Ther Methods 2017;28:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilmott P, Lisowski L, Alexander IE, et al. A user's guide to the inverted terminal repeats of adeno-associated virus. Hum Gene Ther Methods 2019;30:206–213. [DOI] [PubMed] [Google Scholar]
- 14. Bulcha JT, Wang Y, Ma H, et al. Viral vector platforms within the gene therapy landscape. Signal Transduct Target Ther 2021;6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clement N, Grieger JC. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol Ther Methods Clin Dev 2016;3:16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grieger JC, Soltys SM, Samulski RJ. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol Ther 2016;24:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther 2002;13:1935–1943. [DOI] [PubMed] [Google Scholar]
- 18. Smith RH, Levy JR, Kotin RM. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther 2009;17:1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mietzsch M, Grasse S, Zurawski C, et al. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1–12 vectors for gene therapy. Hum Gene Ther 2014;25:212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurasawa JH, Park A, Sowers CR, et al. Chemically defined, high-density insect cell-based expression system for scalable AAV vector production. Mol Ther Methods Clin Dev 2020;19:330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kondratov O, Marsic D, Crosson SM, et al. Direct head-to-head evaluation of recombinant adeno-associated viral vectors manufactured in human versus insect cells. Mol Ther 2017;25:2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Penaud-Budloo M, Francois A, Clement N, et al. Pharmacology of recombinant adeno-associated virus production. Mol Ther Methods Clin Dev 2018;8:166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lecomte E, Tournaire B, Cogne B, et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol Ther Nucleic Acids 2015;4:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maynard LH, Smith O, Tilmans NP, et al. Fast-Seq: a simple method for rapid and inexpensive validation of packaged single-stranded adeno-associated viral genomes in academic settings. Hum Gene Ther Methods 2019;30:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guerin K, Rego M, Bourges D, et al. A novel next-generation sequencing and analysis platform to assess the identity of recombinant adeno-associated viral preparations from viral DNA extracts. Hum Gene Ther 2020;31:664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tai PWL, Xie J, Fong K, et al. Adeno-associated virus genome population sequencing achieves full vector genome resolution and reveals human-vector chimeras. Mol Ther Methods Clin Dev 2018;9:130–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Radukic MT, Brandt D, Haak M, et al. Nanopore sequencing of native adeno-associated virus (AAV) single-stranded DNA using a transposase-based rapid protocol. NAR Genom Bioinform 2020;2:lqaa074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tran NT, Heiner C, Weber K, et al. AAV-genome population sequencing of vectors packaging CRISPR components reveals design-influenced heterogeneity. Mol Ther Methods Clin Dev 2020;18:639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ibraheim R, Tai PWL, Mir A, et al. Self-inactivating, all-in-one AAV vectors for precision Cas9 genome editing via homology-directed repair in vivo. Nat Commun 2021;12:6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Samulski RJ, Chang LS, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol 1987;61:3096–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Penaud-Budloo M, Lecomte E, Guy-Duche A, et al. Accurate identification and quantification of DNA species by next-generation sequencing in adeno-associated viral vectors produced in insect cells. Hum Gene Ther Methods 2017;28:148–162. [DOI] [PubMed] [Google Scholar]
- 32. Ayuso E, Blouin V, Lock M, et al. Manufacturing and characterization of a recombinant adeno-associated virus type 8 reference standard material. Hum Gene Ther 2014;25:977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. D'Costa S, Blouin V, Broucque F, et al. Practical utilization of recombinant AAV vector reference standards: focus on vector genomes titration by free ITR qPCR. Mol Ther Methods Clin Dev 2016;5:16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao H, Ghirlando R, Piszczek G, et al. Recorded scan times can limit the accuracy of sedimentation coefficients in analytical ultracentrifugation. Anal Biochem 2013;437:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 2000;78:1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brautigam CA. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol 2015;562:109–133. [DOI] [PubMed] [Google Scholar]
- 37. Joseph S, Russell D. Molecular cloning: a laboratory manual. In: Alkaline Agarose Gel Electrophoresis. New York: Cold Spring Harbor Laboratory Press, 2012:636–666. [Google Scholar]
- 38. Lecomte E, Leger A, Penaud-Budloo M, et al. Single-stranded DNA virus sequencing (SSV-Seq) for characterization of residual DNA and AAV vector genomes. Methods Mol Biol 2019;1950:85–106. [DOI] [PubMed] [Google Scholar]
- 39. Afgan E, Sloggett C, Goonasekera N, et al. Genomics virtual laboratory: a practical bioinformatics workbench for the cloud. PLoS One 2015;10:e0140829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. Oxford: Oxford University Press, 2013. [Google Scholar]
- 41. Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 43. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003;31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao G, Sena-Esteves M. Introducing genes into mammalian cells: viral vectors. Mol Clon Lab Manual 2012;2:1209–1313. [DOI] [PubMed] [Google Scholar]
- 45. Xie J, Mao Q, Tai PWL, et al. Short DNA hairpins compromise recombinant adeno-associated virus genome homogeneity. Mol Ther 2017;25:1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lecomte E, Saleun S, Bolteau M, et al. The SSV-Seq 2.0 PCR-free method improves the sequencing of adeno-associated viral vector genomes containing GC-rich regions and homopolymers. Biotechnol J 2021;16:e2000016. [DOI] [PubMed] [Google Scholar]
- 47. Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 2019;18:358–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Galibert L, Savy A, Dickx Y, et al. Origins of truncated supplementary capsid proteins in rAAV8 vectors produced with the baculovirus system. PLoS One 2018;13:e0207414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cecchini S, Virag T, Kotin RM. Reproducible high yields of recombinant adeno-associated virus produced using invertebrate cells in 0.02- to 200-liter cultures. Hum Gene Ther 2011;22:1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Levy HC, Bowman VD, Govindasamy L, et al. Heparin binding induces conformational changes in Adeno-associated virus serotype 2. J Struct Biol 2009;165:146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang J, Yu X, Guo P, et al. Satellite subgenomic particles are key regulators of adeno-associated virus life cycle. Viruses 2021;13:1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wright JF. Codon modification and PAMPs in clinical AAV vectors: the tortoise or the hare? Mol Ther 2020;28:701–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wright JF. Quantification of CpG motifs in rAAV genomes: avoiding the toll. Mol Ther 2020;28:1756–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiang Z, Kurupati RK, Li Y, et al. The effect of CpG sequences on capsid-specific CD8(+) T cell responses to AAV vector gene transfer. Mol Ther 2020;28:771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bertolini TB, Shirley JL, Zolotukhin I, et al. Effect of CpG depletion of vector genome on CD8(+) T cell responses in AAV gene therapy. Front Immunol 2021;12:672449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Faust SM, Bell P, Cutler BJ, et al. CpG-depleted adeno-associated virus vectors evade immune detection. J Clin Invest 2013;123:2994–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pan X, Yue Y, Boftsi M, et al. Rational engineering of a functional CpG-free ITR for AAV gene therapy. Gene Ther 2021. Oct 6 [Epub ahead of print]; DOI: 10.1038/s41434-021-00296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chan YK, Wang SK, Chu CJ, et al. Engineering adeno-associated viral vectors to evade innate immune and inflammatory responses. Sci Transl Med 2021;13:eabd3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jacob A, Brun L, Jimenez Gil P, et al. Homologous recombination offers advantages over transposition-based systems to generate recombinant baculovirus for adeno-associated viral vector production. Biotechnol J 2021;16:e2000014. [DOI] [PubMed] [Google Scholar]
- 60. Pijlman GP, van den Born E, Martens DE, et al. Autographa californica baculoviruses with large genomic deletions are rapidly generated in infected insect cells. Virology 2001;283:132–138. [DOI] [PubMed] [Google Scholar]
- 61. Giri L, Feiss MG, Bonning BC, et al. Production of baculovirus defective interfering particles during serial passage is delayed by removing transposon target sites in fp25k. J Gen Virol 2012;93:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Okano K, Vanarsdall AL, Mikhailov VS, et al. Conserved molecular systems of the Baculoviridae. Virology 2006;344:77–87. [DOI] [PubMed] [Google Scholar]
- 63. Kotin RM. Large-scale recombinant adeno-associated virus production. Hum Mol Genet 2011;20:R2–R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kotin RM, Snyder RO. Manufacturing clinical grade recombinant adeno-associated virus using invertebrate cell lines. Hum Gene Ther 2017;28:350–360. [DOI] [PubMed] [Google Scholar]
- 65. McCarty DM, Fu H, Monahan PE, et al. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther 2003;10:2112–2118. [DOI] [PubMed] [Google Scholar]
- 66. Wang Z, Ma HI, Li J, et al. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther 2003;10:2105–2111. [DOI] [PubMed] [Google Scholar]
- 67. Maurer AC, Weitzman MD. Adeno-associated virus genome interactions important for vector production and transduction. Hum Gene Ther 2020;31:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yan Z, Zak R, Zhang Y, et al. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J Virol 2005;79:364–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and/or analyzed in the current study have been deposited in the National Center for Biotechnology Information Sequence Read Archive: PRJNA794197.