Abstract
Background
MICAL-like protein 2 (MICAL-L2), a member of the molecules interacting with CasL (MICAL) family of proteins, is strongly associated with the malignancy of multiple types of cancer. However, the role of MICAL-L2 in colon adenocarcinoma (COAD) has not been well characterized.
Methods
In this study, we analyzed the role of MICAL-L2 in COAD using datasets available from public databases. The mRNA and protein expression of MICAL-L2 was investigated using TCGA, UALCAN, and independent immunohistochemical assays. Overall survival (OS) and disease-specific survival (DSS) of COAD patients were assessed based on the MICAL-L2 expression level using the Kaplan–Meier method. Univariate and multivariate analysis was employed to determine whether MICAL-L2 could serve as an independent prognostic indicator of OS. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set enrichment analysis (GSEA) were further utilized to explore the possible cellular mechanism underlying the role of MICAL-L2 in COAD. In addition, the correlation between MICAL-L2 expression and immune cell infiltration levels was investigated via single-sample gene set enrichment analysis (ssGSEA).
Results
Data from TCGA, HPA, and UALCAN datasets indicated that MICAL-L2 expression was significantly higher in COAD tissue than in adjacent normal tissues, and this was confirmed by immunohistochemical assays. Kaplan–Meier survival analysis revealed that patients with MICAL-L2 had shorter OS and DSS. Furthermore, multivariate Cox analysis indicated that MICAL-L2 was an independent risk factor for OS in COAD patients. ROC analysis confirmed the diagnostic value of MICAL-L2, and a prognostic nomogram involving age, M stage, and MICAL-L2 expression was constructed for OS. Functional enrichment analyses revealed that transport-related activity was closely associated with the role of MICAL-L2 in COAD. Regarding immune infiltration levels, MICAL-L2 was found to be positively associated with CD56bright NK cells.
Conclusions
Our results suggested that MICAL-L2 is a promising biomarker for determining prognosis and correlated with immune infiltration levels in COAD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-022-09614-0.
Keywords: MICAL-L2, Overall survival, Prognosis, COAD
Background
Colon cancer is a commonly diagnosed malignant tumor of the digestive tract and a leading cause of cancer-related death worldwide [1]. It usually affects adults at 40–50 years of age and occurs more often in males than females. The etiology of colon cancer is mainly associated with a high-fat diet, colonic polyps, genetic make-up, and chronic inflammation [2]. Patients in the early stage of colon cancer may not present obvious clinical symptoms. As the tumor volume increases, patients may display abdominal distension and dyspepsia, and may even be able to feel a lump/mass in the abdomen. Although the 5-year survival rate of patients in the early stages of colon cancer can be higher than 90%, that of patients diagnosed at an advanced stage is lower than 20% [3–5]. These observations underline the need to further unravel the mechanisms underlying colon cancer progression and identify novel therapeutic targets for the treatment of this disease.
Molecules interacting with CasL (MICALs) represent an evolutionarily conserved family of proteins with roles in the regulation of cytoske leton dynamics [6]. MICAL-like protein 2 (MICAL-L2), a member of the MICAL family, has three conserved domains, namely, a calponin homology (CH) domain; a Lin11, Isl-1, and Mec-3 (LIM) domain; and a C-terminal coiled-coil (CC) domain [7]. The CH and LIM domains link MICAL-L2 to the actin cytoskeleton, while the CC domain is required for interaction with Rab GTPases. MICAL-L2 exerts its multiple biological functions primarily via processes involving cargo transportation. For example, by binding to Rab13, MICAL-L2 triggers the transportation of glucose transporter-4 (GLUT4) and mediates GLUT4-containing vesicle localization and fusion with the muscle cell membrane [8]. Rab13 and MICAL-L2 also act together in the transfer of actinin-4 from the cell body to the tips of neurites [9]. It has been well documented that MICAL-L2 is highly expressed and promotes cell migration and invasion in multiple types of cancer, including gastric cancer, ovarian cancer, and breast cancer [10–12]. In ovarian cancer cells, the silencing of MICAL-L2 was shown to inhibit canonical Wnt/β-catenin signaling and induce mesenchymal–epithelial transition [11]. We have previously shown that MICAL-L2 facilitates the proliferation of lung cancer cells via the de-ubiquitination of c-Myc, which blocks its degradation [13]. Recently, another MICAL family member, MICAL1, which shares sequence similarity with MICAL-L2 [14], was found to play a key role in the migration and growth of colorectal cancer cells by suppressing the ERG1/β-catenin signaling pathway [15]. However, the role of MICAL-L2 in the prognosis and possible pathogenesis of colon cancer has not been fully elucidated.
Colon adenocarcinoma (COAD) is one the most common type of colon cancer. In this study, several informatics tools were used to evaluate the expression profile and the prognostic significance of MICAL-L2 in COAD. Moreover, the correlation between MICAL-L2 expression and immune infiltration, and the putative mechanisms underlying the role of MICAL-L2 in COAD, were also investigated. This is the first comprehensive study of the association between MICAL-L2 expression and its clinical characteristics in COAD and our findings may contribute to our understanding of MICAL-L2-related processes in this cancer.
Methods
Ethics statement
All immunohistochemical assays with human tumor specimens were conducted according to the institutional guidelines of Jiangsu Province.
MICAL-L2 mRNA expression and analysis of prognosis
The mRNA expression of MICAL-L2 in COAD and the corresponding clinical information data were downloaded from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/) [16]. MICAL-L2 mRNA expression and its association with overall survival (OS) and disease-specific survival (DSS) of patients with COAD were also analyzed using the TCGA–COAD dataset. The expression of MICAL-L2 was assessed in 456 COAD and 41 adjacent normal tissue samples from the TCGA database. According to the median values of mRNA expression, patients with COAD were divided into high and low expression groups. Data were collected and analyzed using R3.6.3 software [17].
MICAL-L2 protein expression analysis
The Human Protein Atlas (HPA) database (https://www.proteinatlas.org) and the University of Alabama Cancer Database (UALCAN) (http://ualcan.path.uab.edu/index.html) were used to compare MICAL-L2 protein expression between normal and COAD tissues.
Immunohistochemistry
Immunohistochemistry was performed as previously described [18]. COAD tissue microarrays were purchased from Outdo Biotech (Shanghai, China). Thirty paired COAD and paracancerous tissue samples were used for MICAL-L2 immunohistochemical assays. After dewaxing and hydration, the microarray was incubated with 3% H2O2 for 30 min, subjected to antigen retrieval with citric acid at 95 °C for 20 min, blocked for 2 h at room temperature, incubated with primary antibody against MICAL-L2 at 4 °C overnight, and then with a species-matched secondary antibody for 2 h at room temperature. DAB staining was employed to detect the expression of MICAL-L2, with hematoxylin serving as the counterstain. Images were captured using an Olympus BX51 microscope. The immunoreactivity score (IRS) was obtained by multiplying the percentage of stained cells by the staining intensity scores of MICAL-L2, as previously described [19, 20].
Enrichment analysis for MICAL-L2 function
An ordered list of genes was generated based on the correlation between all genes and MICAL-L2 expression. Enriched pathways were determined using Gene Ontology (GO) [21, 22], KEGG [23–25], and GSEA [26, 27]. In the KEGG analysis, genes were determined to be differentially expressed based on a log2 fold-change of > 1.0 and an adjusted P-value < 0.05 (www.kegg.jp/kegg/kegg1.html), as previously reported [28]. “GSEA is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states” [26, 27]. In this study, the predefined gene set was obtained from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). STRING (https://cn.string-db.org/) and Cytoscape were used to predict and display the protein-protein interaction network of MICAL-L2 co-expressed genes.
Immune cell infiltration analysis using single-sample GSEA
Immune infiltration analysis of COAD tissue was performed using single-sample GSEA (ssGSEA) [27, 29]. The infiltration levels of 24 immune cell types were quantified from gene expression profiles, as previously described [30]. In addition, Spearman’s correlation was used to investigate the association between MICAL-L2 expression and immune cell infiltration.
Statistical analysis
SPSS 22.0 software was used for statistical analysis. The chi-square test was used to analyze and compare the clinical and pathological conditions of the two groups. The Kaplan–Meier method was used to evaluate the survival of patients and the log rank test was used to test the significance. A Cox proportional hazards regression model was used to identify significant and independent prognostic factors for COAD patients. Finally, R language was used to draw a nomogram and build a prediction model. P < 0.05 indicates significance (two-tailed).
Results
MICAL-L2 is highly expressed in COAD samples
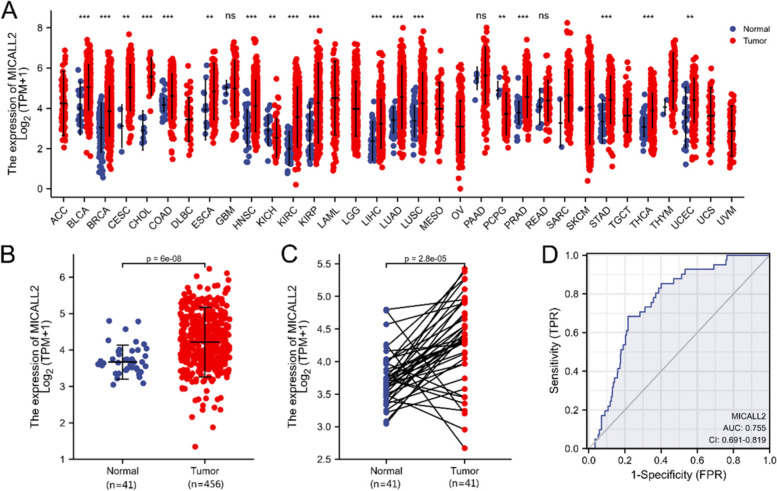
Data mining in TCGA database showed that the mRNA expression of MICAL-L2 was elevated in most types of cancer (Fig. 1A). Focusing on COAD, a common histological subtype of colon cancer, we then examined the expression of MICAL-L2 in 456 COAD samples and 41 adjacent normal tissue samples from TCGA. We found that the mRNA expression of MICAL-L2 was significantly upregulated in COAD tissues compared with that in adjacent normal tissues (P < 0.001) (Fig. 1B). Similarly, in 41 paired cancerous and adjacent normal tissues, MICAL-L2 mRNA expression was also markedly higher in the cancer samples (P < 0.001) (Fig. 1C). Receiver operating characteristic (ROC) curve analysis was also applied to evaluate the diagnostic value of MICAL-L2 expression levels in COAD, and the area under the curve (AUC) was found to be 0.755 (95% CI = 0.691–0.819) (Fig. 1D).
Fig. 1.
mRNA expression data for MICAL-L2 in colon adenocarcinoma (COAD) obtained from The Cancer Genome Atlas (TCGA) database. A MICAL-L2 expression levels in different human tumor types according to TCGA data. B The differential expression of MICAL-L2 between COAD tissues and adjacent normal tissues. C The differential expression of MICAL-L2 between COAD tissues and paired adjacent normal tissues. D The receiver operating characteristic (ROC) curve for MICAL-L2 shows promising discrimination power between normal and COAD samples. **: P < 0.01, ***: P < 0.001
Analysis of the HPA and UALCAN data showed that the protein expression level of MICAL-L2 was higher in COAD tissues than in normal adjacent tissues (Fig. 2A, B). MICAL-L2 protein levels were also analyzed in a tissue microarray containing COAD and paracancerous tissues. Although some signal was lost during sample preparation, the immunohistochemical analysis nevertheless showed that MICAL-L2 protein levels were significantly higher in COAD tissues than in paracancerous normal tissues (Fig. 2C). Combined, these results indicated that MICAL-L2 is highly expressed in COAD at both the mRNA and protein levels.
Fig. 2.
The protein expression of MICAL-L2 in colon adenocarcinoma (COAD) tissues. A Differential protein expression of MICAL-L2 between COAD and adjacent normal tissues. B Representative images of MICAL-L2 expression in COAD. C Representative images of MICAL-L2 staining in COAD tissues. A scatterplot showing correlations between protein levels in COAD or paracancerous tissue as determined by immunoreactivity scores (IRS). ***: P < 0.001
Correlation between MICAL-L2 expression and clinicopathological features
The characteristics of 454 patients with COAD, including gene expression and clinical data, were collected from TCGA database. The patients were divided into high and low MICAL-L2 expression groups based on the mean value of MICAL-L2 expression (Table 1), following which putative correlations between MICAL-L2 expression and clinical characteristics were evaluated using logistic regression analysis. The results showed that MICAL-L2 mRNA expression was significantly associated with lymphatic invasion and primary therapy outcome (progressive disease [PD] + stable disease [SD] + partial response [PR] vs complete response [CR]) (Table 2).
Table 1.
Association between MICAL-L2 expression and clinicopathologic features in the validation cohort
| Characteristic | levels | Low expression of MICALL2 | High expression of MICALL2 | p |
|---|---|---|---|---|
| n | 227 | 227 | ||
| T stage, n (%) | T1 | 4 (0.9%) | 7 (1.5%) | 0.389 |
| T2 | 43 (9.5%) | 34 (7.5%) | ||
| T3 | 155 (34.2%) | 154 (34%) | ||
| T4 | 24 (5.3%) | 32 (7.1%) | ||
| N stage, n (%) | N0 | 140 (30.8%) | 127 (28%) | 0.301 |
| N1 | 52 (11.5%) | 53 (11.7%) | ||
| N2 | 35 (7.7%) | 47 (10.4%) | ||
| M stage, n (%) | M0 | 164 (41.3%) | 169 (42.6%) | 0.666 |
| M1 | 34 (8.6%) | 30 (7.6%) | ||
| Pathologic stage, n (%) | Stage I | 41 (9.3%) | 34 (7.7%) | 0.185 |
| Stage II | 94 (21.2%) | 82 (18.5%) | ||
| Stage III | 54 (12.2%) | 74 (16.7%) | ||
| Stage IV | 34 (7.7%) | 30 (6.8%) | ||
| Primary therapy outcome, n (%) | PD | 10 (4.3%) | 15 (6.4%) | 0.214 |
| SD | 1 (0.4%) | 3 (1.3%) | ||
| PR | 4 (1.7%) | 8 (3.4%) | ||
| CR | 105 (44.7%) | 89 (37.9%) | ||
| Gender, n (%) | Female | 104 (22.9%) | 110 (24.2%) | 0.638 |
| Male | 123 (27.1%) | 117 (25.8%) | ||
| Age, n (%) | <=65 | 100 (22%) | 88 (19.4%) | 0.295 |
| > 65 | 127 (28%) | 139 (30.6%) | ||
| Lymphatic invasion, n (%) | NO | 137 (33.3%) | 111 (27%) | 0.048 |
| YES | 73 (17.8%) | 90 (21.9%) |
Table 2.
Association between MICAL-L2 expression and clinicopathologic characteristics (logistic regression analysis)
| Characteristics | Total(N) | Odds Ratio (OR) | P value |
|---|---|---|---|
| T stage (T3&T4 vs. T1&T2) | 453 | 1.191 (0.748–1.904) | 0.462 |
| N stage (N1&N2 vs. N0) | 454 | 1.267 (0.872–1.845) | 0.215 |
| M stage (M1 vs. M0) | 397 | 0.856 (0.499–1.463) | 0.570 |
| Age (> 65 vs. <=65) | 454 | 1.244 (0.856–1.810) | 0.253 |
| Gender (Female vs. Male) | 454 | 1.112 (0.769–1.609) | 0.573 |
| Pathologic stage (Stage III&Stage IV vs. Stage I&Stage II) | 443 | 1.375 (0.944–2.008) | 0.098 |
| Lymphatic invasion (YES vs. NO) | 411 | 1.522 (1.024–2.268) | 0.038 |
| Perineural invasion (YES vs. NO) | 179 | 1.150 (0.585–2.255) | 0.684 |
| BMI (> = 25 vs. < 25) | 232 | 0.714 (0.412–1.232) | 0.226 |
| Primary therapy outcome (PD&SD&PR vs. CR) | 235 | 2.045 (1.032–4.181) | 0.044 |
| Colon polyps present (YES vs. NO) | 225 | 1.073 (0.617–1.860) | 0.803 |
Prognostic value of MICAL-L2 in COAD patients
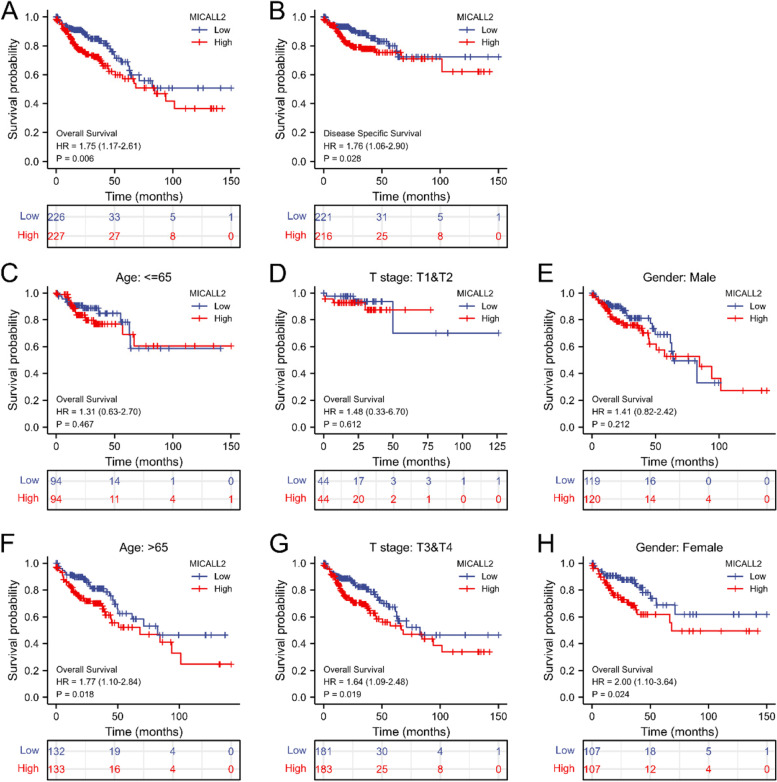
We next determined the prognostic value of MICAL-L2 in COAD. For this, we evaluated the relationship between MICAL-L2 expression and clinical follow-up data using Kaplan–Meier analysis. Significance was assessed using the log rank test. The results showed that high MICAL-L2 expression was negatively correlated with OS (n = 453, P = 0.006; Fig. 3A) and DSS (n = 437, P = 0.028; Fig. 3B), indicating that MICAL-L2 expression levels were significantly associated with the prognosis of COAD patients. High MICAL-L2 expression was associated with poor OS in COAD patients who were over the age of 65, had stage T3 and T4 disease, or were female (Fig. 3C–H).
Fig. 3.
Prognostic value of MICAL-L2 expression for clinical outcomes in colon adenocarcinoma (COAD) patients. A&B Kaplan–Meier analysis of overall survival (OS) (A) and disease-specific survival (DSS) (B) in patients with COAD. C–H Kaplan–Meier analysis of subgroup OS in patients with COAD. (C) Age ≤ 65, (D) stages T1 and T2, (E) male, (F) age > 65, (G) stages T3 and T4, and (H) female
To further identify the risk factors associated with OS in patients with COAD, univariate and multivariate analyses were performed using TCGA–COAD dataset. Univariate analysis showed that T stage, N stage, M stage, age, lymphatic invasion, and MICAL-L2 expression were the factors influencing OS. The multivariate analysis showed that MICAL-L2 expression (P = 0.032), age, and M stage were independent risk factors for OS (Table 3 & Fig. 4). Combined, these data suggested that MICAL-L2 may serve as a biomarker for the prediction of OS among COAD patients.
Table 3.
Univariate and multivariate Cox proportional hazards analysis of the correlation between overall survival (OS) and MICAL-L2 expression levels
| Characteristics | Total(N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| T stage (T3&T4 vs T1&T2) | 452 | 2.962 (1.372–6.395) | 0.006 | 2.759 (0.842–9.047) | 0.094 |
| N stage (N1&N2 vs N0) | 453 | 2.519 (1.691–3.752) | < 0.001 | 1.346 (0.744–2.434) | 0.326 |
| M stage (M1 vs M0) | 396 | 4.327 (2.763–6.776) | < 0.001 | 3.179 (1.785–5.664) | < 0.001 |
| Age (> 65 vs < =65) | 453 | 1.649 (1.077–2.526) | 0.021 | 2.372 (1.400–4.018) | 0.001 |
| Gender (Male vs FeMale) | 453 | 1.118 (0.756–1.654) | 0.575 | ||
| History of colon polyps (YES vs NO) | 385 | 0.783 (0.466–1.313) | 0.354 | ||
| Lymphatic invasion (YES vs NO) | 410 | 2.315 (1.520–3.525) | < 0.001 | 1.436 (0.848–2.432) | 0.178 |
| MICALL2 (High vs Low) | 453 | 1.751 (1.174–2.612) | 0.006 | 1.703 (1.046–2.773) | 0.032 |
Fig. 4.
Forest plot of the multivariate Cox regression analysis for overall survival (OS) in colon adenocarcinoma (COAD) patients
Based on multivariate Cox regression analysis for OS, a nomogram was generated for internal validation. Prediction models were constructed for 1-, 3-, and 5-year OS in patients with COAD (Fig. 5A) while calibration plots to validate the efficiency of the nomograms for predicting OS were also generated. As shown in Fig. 5B, the bias-corrected line in the calibration plot was close to the ideal curve, indicative of an intimate relationship between the observed and predicted values.
Fig. 5.
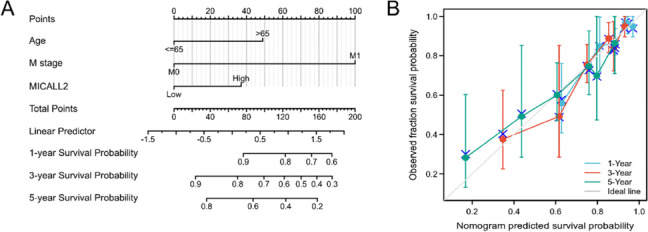
Construction and validation of a nomogram to show the relationship between MICAL-L2 and other clinical factors with survival probability. A A nomogram for predicting the probability of 1-, 3-, and 5-year overall survival (OS) in colon adenocarcinoma (COAD) patients. B Calibration plots validating the efficiency of the nomograms for OS
Function enrichment analysis of MICAL-L2 in COAD
As we found that COAD patients with high levels of MICAL-L2 expression have worse OS and DSS than those with low MICAL-L2 expression, we explored the possible underlying cellular mechanism through KEGG and GSEA. As shown in Fig. 6A, 434 differentially expressed genes (DEGs) (|logFC| > 1, adjusted P-value < 0.05) were identified between the high and low MICAL-L2 expression groups, including 338 that were upregulated and 96 that were downregulated. The 10 genes showing the greatest positive or negative correlation with MICAL-L2 expression are shown in Fig. 6B. A network of potential co-expressed genes of MICAL-L2 in COAD are shown in Fig. S1.
Fig. 6.
Function enrichment analysis of MICAL-L2 in colon adenocarcinoma (COAD) using Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set enrichment analysis (GSEA). A Volcano plot of differentially expressed genes (DEGs) between the high and low MICAL-L2 expression groups. B Heat map showing the top 10 DEGs between the high and low MICAL-L2 expression groups. C GO and KEGG analysis of the DEGs. D Enrichment plots from GSEA. Hallmark_kras_signaling_DN (left) and hallmark_apical_junction (right)
The identified DEGs were submitted to GO term and KEGG pathway enrichment analysis. The following biological processes were found to be significantly affected: Chylomicron assembly, triglyceride-rich lipoprotein particle remodeling, and regulation of sensory perception of pain. The most enriched cellular component terms were apical plasma membrane, apical part of cell, and chylomicron. For molecular function, the most enriched terms were passive transmembrane transporter activity, channel activity, substrate-specific channel activity. The most enriched KEGG terms were cholesterol metabolism, neuroactive ligand–receptor interaction, and bile secretion (Fig. 6C). The GSEA results indicated that the co-expressed genes were mainly associated with the hallmark_kras_signaling_DN and hallmark_apical_junction pathways (Fig. 6D). We will further explore these pathways in future studies to better understand the function of MICAL-L2 in COAD.
Correlation between immune cell infiltration and MICAL-L2 expression levels in TCGA
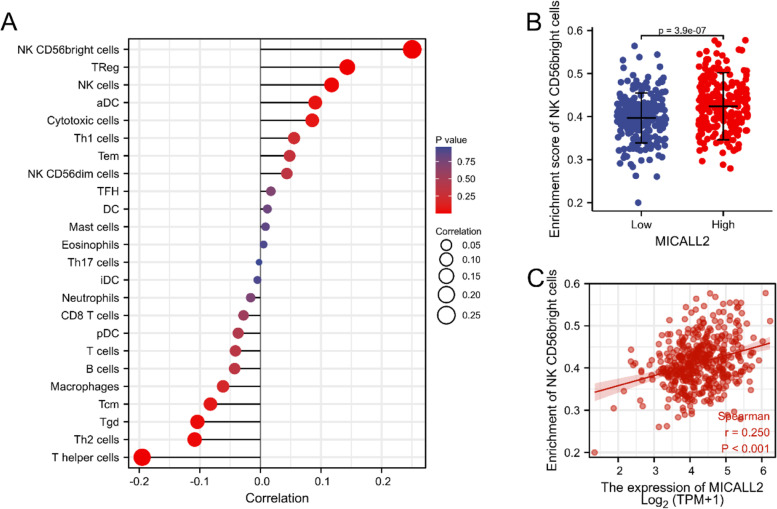
The correlation between MICAL-L2 expression and immune infiltrate abundance in COAD was evaluated by ssGSEA using Spearman’s correlation tests (Fig. 7A). As shown in Fig. 7A, CD56bright natural killer (NK) cells, regulatory T cells (Tregs), and NK cells were all positively correlated with MICAL-L2 expression, whereas the opposite was seen for T-helper (Th) cells, gamma delta T (Tgd) cells, and Th2 cells. We further evaluated the infiltration levels of CD56bright NK cells, which displayed the greatest positive correlation with MICAL-L2 expression. The results showed that MICAL-L2 was significantly and positively correlated with the infiltration levels of CD56bright NK cells (P < 0.01, Fig. 7B, C).
Fig. 7.
Analysis of the association between MICAL-L2 expression and immune cell infiltration in colon adenocarcinoma (COAD). A Association between MICAL-L2 expression and 24 immune infiltrating cells. B Enrichment of CD56bright NK cells between high and low MICAL-L2 expression groups. C Correlation between MICAL-L2 and CD56bright NK cells. aDCs, activated DCs; DCs, dendritic cells; iDCs, immature DCs; NK, natural killer; pDCs, plasmacytoid DCs; Tcm, T central memory; Tem, T effector memory; Tfh, T follicular helper; Tgd, T gamma delta; Th, T helper cells; Th1, type 1 Th cells; Th2, type 2 Th cells; Th17, type 17 Th cells; Treg, regulatory T cell
Discussion
While there is only one MICAL-encoding gene in Drosophila, vertebrate genomes express genes encoding three MICAL (MICAL1–3) and two MICAL-like (MICAL-L1, MICAL-L2) isoforms. The disruption of MICAL1 activity was shown to impair cytoskeleton organization and breast tumor growth in an orthotopic model [31]. Additionally, high MICAL2 expression has been associated with lymphatic metastasis and shorter OS in lung cancer patients [32]. The three MICAL isoforms (MICAL1–3) contain a FAD domain and exhibit flavoprotein monooxygenase catalytic activity. Of note, MICAL1 exerts its effect on proliferation via reactive oxygen species (ROS)-sensitive PI3K/AKT/ERK signaling in breast cancer cells [33]. Similarly, MICAL2-induced ROS generation has also been reported to enhance the migratory potential of gastric cancer cells [34]. MICAL-L2 lacks the FAD domain and cannot generate ROS [35], and although several studies have suggested that MICAL-L2 may positively influence cancer progression [10, 13, 36], whether and how MICAL-L2 may be involved in this process remains unclear.
MICAL-L2 has been shown to be significantly upregulated in ovarian cancer tissues in a FIGO stage-dependent manner and has also been associated with histologic subgroups of ovarian cancer [11]. Consistent with these observations, our results revealed that, compared with adjacent normal tissues, MICAL-L2 expression was significantly upregulated in COAD tissues at both the mRNA and protein levels. ROC curve analysis also confirmed the diagnostic value of MICAL-L2. These findings strongly suggested that MICAL-L2 may play an oncogenic role in COAD. Accordingly, we assessed the prognostic value of MICAL-L2 in COAD using Kaplan–Meier survival analysis and found that patients with high MICAL-L2 expression have shorter OS and DSS compared with those with low MICAL-L2 expression. Univariate and multivariate analysis further revealed that high MICAL-L2 expression was an independent risk factor for OS in individuals with COAD. Collectively, these results indicated that MICAL-L2 may predict the prognosis of COAD and may represent a promising therapeutic target for the treatment of this cancer.
Abundant evidence supports that MICAL-L2 serves as a regulator of actin cytoskeleton organization, affecting processes such as cell vesicle trafficking and cellular morphology, among many other cytological behaviors [37–39]. For example, the interaction between MICAL-L2 and Rab8 and Rab13 was found to coordinate tight junction and adherens junction assembly [40, 41]. As expected, GO, KEGG, and GSEA indicated that MICAL-L2 localized to the apical part of the cell and regulated multiple types of transportation events. MICAL-L2 has been reported to mediate the endocytic recycling of occludin [42], while we have also recently shown that MICAL-L2 promotes the migration of gastric cancer cells via inhibiting EGFR transportation and degradation [10]. Although these findings demonstrated that MICAL-L2 might play a critical role in COAD progression through its transport-related activity, how MICAL-L2 precisely regulates cellular trafficking and then promotes COAD progression requires further exploration.
Over the past few years, the treatments used for COAD have transited from traditional chemical remedies to the use of targeted or immunotherapeutic drugs [43, 44]. In this study, ssGSEA was used to explore the association between MICAL-L2 expression and immune cell infiltration in COAD. Among the immune cell subpopulations, CD56bright NK cells showed the most enrichment in the high-MICAL-L2 expression group. NK cells are effective at killing tumors, and are commonly divided into CD56bright and CD56dim subtypes. Until recently, CD56bright NK cells were thought to exhibit potent antitumor activity [45, 46]. However, it has since been shown that CD56bright NK cells, which are enriched in human non-small-cell lung cancer infiltrate, display an impaired capability to kill tumor cells [47]. Further investigation, especially involving reciprocal activating crosstalk between immune cells and MICAL-L2, is necessary to delineate the regulatory immune mechanisms associated with MICAL-L2.
In conclusion, the results obtained in this study provide promising clues for a new mechanistic connection between MICAL-L2 expression and prognosis in COAD patients. Our findings indicated that MICAL-L2 may serve as an independent prognostic factor for patients with COAD, and further suggested that MICAL-L2 regulates cellular trafficking and promotes immune cell infiltration in COAD. However, how MICAL-L2 precisely regulates COAD progression remains to be characterized.
Supplementary Information
Additional file 1: Figure S1. Network of co-expressed genes of MICAL-L2.
Acknowledgements
Not applicable.
Abbreviations
- AUC
Area Under Curve
- CI
Confidence Interval
- CH
Calponin homology
- CC
C-terminal coiled-coil
- COAD
Colon Adenocarcinoma
- DEGs
Differentially expressed genes
- DSS
Disease Specific Survival
- GSEA
Gene Set Enrichment Analysis
- HPA
Human Protein Atlas Database
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- HR
Hazard Ratio
- LIM
Lin11, Isl-1 and Mec-3
- IRS
Immunoreactivity score
- MICALs
Molecules Interacting With CasLs
- NCBI
National Center for Biotechnology Information
- NES
Normalized Enrichment Score
- NK
Natural killer
- OR
Odds Ratio
- OS
Overall Survival
- ROC
Receiver Operating Characteristic
- ROS
Reactive Oxygen Species
- ssGSEA
Single-sample Gene Set Enrichment Analysis
- Tregs
Regulatory T Cells
- TNM
Tumor Node Metastasis
- TCGA
The Cancer Genome Atlas
- UALCAN
University of Alabama Cancer Database
Authors’ contributions
JD designed the study. YY, FY performed the statistical analysis. TX performed the immunohistochemistry assays. JD, QW,YZ drafted the manuscript. JD supervised the experimental work. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82073226), “Shilv” Plan founded by School of Basic Medical Sciences of NJMU.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Declarations
Ethics approval and consent to participate
All methods were carried out in accordance with the Declaration of Helsinki. No ethics approval was required for this work. All utilized public data sets were generated by others who obtained ethical approval.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yixing Yang and Fengwen Ye contributed equally to this work.
Contributor Information
Yixing Yang, Email: njmuyangyixing@163.com.
Fengwen Ye, Email: yefengwen1998@163.com.
Tianxiang Xia, Email: 310654177@qq.com.
Qianwen Wang, Email: sandyqianwenwang@163.com.
Yujie Zhang, Email: zeater87@126.com.
Jun Du, Email: dujun@njmu.edu.cn.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Rubin DC, Shaker A, Levin MS. Chronic intestinal inflammation: inflammatory bowel disease and colitis-associated colon cancer. Front Immunol. 2012;3:107. doi: 10.3389/fimmu.2012.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akinkuotu AC, Maduekwe UN, Hayes-Jordan A. Surgical outcomes and survival rates of colon cancer in children and young adults. Am J Surg. 2021;221(4):718–724. doi: 10.1016/j.amjsurg.2021.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Araghi M, Arnold M, Rutherford MJ, Guren MG, Cabasag CJ, Bardot A, Ferlay J, Tervonen H, Shack L, Woods RR, et al. Colon and rectal cancer survival in seven high-income countries 2010-2014: variation by age and stage at diagnosis (the ICBP SURVMARK-2 project) Gut. 2021;70(1):114–126. doi: 10.1136/gutjnl-2020-320625. [DOI] [PubMed] [Google Scholar]
- 5.Qaderi SM, Dickman PW, de Wilt JHW, Verhoeven RHA. Conditional survival and cure of patients with Colon or rectal Cancer: a population-based study. J Natl Compr Cancer Netw. 2020;18(9):1230–1237. doi: 10.6004/jnccn.2020.7568. [DOI] [PubMed] [Google Scholar]
- 6.Yoon J, Terman JR. MICAL redox enzymes and actin remodeling: new links to classical tumorigenic and cancer pathways. Mol Cell Oncol. 2018;5(1):e1384881. doi: 10.1080/23723556.2017.1384881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giridharan SS, Caplan S. MICAL-family proteins: complex regulators of the actin cytoskeleton. Antioxid Redox Signal. 2014;20(13):2059–2073. doi: 10.1089/ars.2013.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Jaldin-Fincati J, Liu Z, Bilan PJ, Klip A. A complex of Rab13 with MICAL-L2 and alpha-actinin-4 is essential for insulin-dependent GLUT4 exocytosis. Mol Biol Cell. 2016;27(1):75–89. doi: 10.1091/mbc.E15-05-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakane A, Honda K, Sasaki T. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol Cell Biol. 2010;30(4):1077–1087. doi: 10.1128/MCB.01067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min P, Zhao S, Liu L, Zhang Y, Ma Y, Zhao X, Wang Y, Song Y, Zhu C, Jiang H, et al. MICAL-L2 potentiates Cdc42-dependent EGFR stability and promotes gastric cancer cell migration. J Cell Mol Med. 2019;23(6):4475–4488. doi: 10.1111/jcmm.14353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu LY, Zhang WM, Yang XM, Cui L, Li J, Zhang YL, Wang YH, Ao JP, Ma MZ, Lu H, et al. Silencing of MICAL-L2 suppresses malignancy of ovarian cancer by inducing mesenchymal-epithelial transition. Cancer Lett. 2015;363(1):71–82. doi: 10.1016/j.canlet.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Ioannou MS, Bell ES, Girard M, Chaineau M, Hamlin JN, Daubaras M, Monast A, Park M, Hodgson L, McPherson PS. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J Cell Biol. 2015;208(5):629–648. doi: 10.1083/jcb.201407068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min P, Zhang L, Wang Y, Qi C, Song Y, Bibi M, Zhang Y, Ma Y, Zhao X, Yu M, et al. MICAL-L2 is essential for c-Myc Deubiquitination and stability in non-small cell lung Cancer cells. Front Cell Dev Biol. 2020;8:575903. doi: 10.3389/fcell.2020.575903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanoni MA. Structure-function studies of MICAL, the unusual multidomain flavoenzyme involved in actin cytoskeleton dynamics. Arch Biochem Biophys. 2017;632:118–141. doi: 10.1016/j.abb.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Gu H, Li Y, Cui X, Cao H, Hou Z, Ti Y, Liu D, Gao J, Wang Y, Wen P. MICAL1 inhibits colorectal cancer cell migration and proliferation by regulating the EGR1/beta-catenin signaling pathway. Biochem Pharmacol. 2022;195:114870. doi: 10.1016/j.bcp.2021.114870. [DOI] [PubMed] [Google Scholar]
- 16.Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 17.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi C, Min P, Wang Q, Wang Y, Song Y, Zhang Y, Bibi M, Du J. MICAL2 contributes to gastric Cancer cell proliferation by promoting YAP Dephosphorylation and nuclear translocation. Oxidative Med Cell Longev. 2021;2021:9955717. doi: 10.1155/2021/9955717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W, Gu L, Li X, Zheng J, Zhang Y, Duan B, Cui J, Dong J, Du J. CD24 associates with EGFR and supports EGF/EGFR signaling via RhoA in gastric cancer cells. J Transl Med. 2016;14:32. doi: 10.1186/s12967-016-0787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumitru CA, Bankfalvi A, Gu X, Zeidler R, Brandau S, Lang S. AHNAK and inflammatory markers predict poor survival in laryngeal carcinoma. Plos One. 2013;8(2):e56420. doi: 10.1371/journal.pone.0056420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gene Ontology C. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Xia T, Qi C, Du J, Ye C. High expression of S100A2 predicts poor prognosis in patients with endometrial carcinoma. BMC Cancer. 2022;22(1):77. doi: 10.1186/s12885-022-09180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan Y, Nie K, Zheng J, Jiang X, Huang Y, Zheng Z, Wen Y, Li P. High endothelin receptor type a expression as an independent prognostic biomarker and correlated with immune infiltrates in stomach adenocarcinoma. Cancer Manag Res. 2021;13:5013–5026. doi: 10.2147/CMAR.S313078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGarry DJ, Armstrong G, Castino G, Mason S, Clark W, Shaw R, McGarry L, Blyth K, Olson MF. MICAL1 regulates actin cytoskeleton organization, directional cell migration and the growth of human breast cancer cells as orthotopic xenograft tumours. Cancer Lett. 2021;519:226–236. doi: 10.1016/j.canlet.2021.07.039. [DOI] [PubMed] [Google Scholar]
- 32.Zhou W, Liu Y, Gao Y, Cheng Y, Chang R, Li X, Zhou Y, Wang S, Liang L, Duan C, et al. MICAL2 is a novel nucleocytoplasmic shuttling protein promoting cancer invasion and growth of lung adenocarcinoma. Cancer Lett. 2020;483:75–86. doi: 10.1016/j.canlet.2020.04.019. [DOI] [PubMed] [Google Scholar]
- 33.Deng W, Wang Y, Zhao S, Zhang Y, Chen Y, Zhao X, Liu L, Sun S, Zhang L, Ye B, et al. MICAL1 facilitates breast cancer cell proliferation via ROS-sensitive ERK/cyclin D pathway. J Cell Mol Med. 2018;22(6):3108–3118. doi: 10.1111/jcmm.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Min P, Qi C, Zhao S, Yu M, Zhang Y, Du J. MICAL2 facilitates gastric Cancer cell migration via MRTF-A-mediated CDC42 activation. Front Mol Biosci. 2021;8:568868. doi: 10.3389/fmolb.2021.568868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giridharan SS, Rohn JL, Naslavsky N, Caplan S. Differential regulation of actin microfilaments by human MICAL proteins. J Cell Sci. 2012;125(Pt 3):614–624. doi: 10.1242/jcs.089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakane A, Yoshizawa S, Yokota H, Sasaki T. Dancing styles of collective cell migration: image-based computational analysis of JRAB/MICAL-L2. Front Cell Dev Biol. 2018;6:4. doi: 10.3389/fcell.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyake K, Sakane A, Tsuchiya Y, Sagawa I, Tomida Y, Kasahara J, Imoto I, Watanabe S, Higo D, Mizuguchi K, et al. Actin cytoskeletal reorganization function of JRAB/MICAL-L2 is fine-tuned by intramolecular interaction between first LIM zinc finger and C-terminal coiled-coil domains. Sci Rep. 2019;9(1):12794. doi: 10.1038/s41598-019-49232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakane A, Abdallah AA, Nakano K, Honda K, Ikeda W, Nishikawa Y, Matsumoto M, Matsushita N, Kitamura T, Sasaki T. Rab13 small G protein and junctional Rab13-binding protein (JRAB) orchestrate actin cytoskeletal organization during epithelial junctional development. J Biol Chem. 2012;287(51):42455–42468. doi: 10.1074/jbc.M112.383653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahajeng J, Giridharan SS, Cai B, Naslavsky N, Caplan S. Important relationships between Rab and MICAL proteins in endocytic trafficking. World J Biol Chem. 2010;1(8):254–264. doi: 10.4331/wjbc.v1.i8.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamura R, Nishimura N, Nakatsuji H, Arase S, Sasaki T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell. 2008;19(3):971–983. doi: 10.1091/mbc.e07-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakatsuji H, Nishimura N, Yamamura R, Kanayama HO, Sasaki T. Involvement of actinin-4 in the recruitment of JRAB/MICAL-L2 to cell-cell junctions and the formation of functional tight junctions. Mol Cell Biol. 2008;28(10):3324–3335. doi: 10.1128/MCB.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terai T, Nishimura N, Kanda I, Yasui N, Sasaki T. JRAB/MICAL-L2 is a junctional Rab13-binding protein mediating the endocytic recycling of occludin. Mol Biol Cell. 2006;17(5):2465–2475. doi: 10.1091/mbc.e05-09-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andre T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal Cancer. N Engl J Med. 2020;383(23):2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 44.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner JA, Rosario M, Romee R, Berrien-Elliott MM, Schneider SE, Leong JW, Sullivan RP, Jewell BA, Becker-Hapak M, Schappe T, et al. CD56bright NK cells exhibit potent antitumor responses following IL-15 priming. J Clin Invest. 2017;127(11):4042–4058. doi: 10.1172/JCI90387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang S, Han J, Baek JS, Tak E, Song GW, Lee SG, et al. Cytotoxicity of human hepatic Intrasinusoidal CD56(bright) natural killer cells against hepatocellular carcinoma cells. Int J Mol Sci. 2019;20(7):1564. doi: 10.3390/ijms20071564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrega P, Morandi B, Costa R, Frumento G, Forte G, Altavilla G, Ratto GB, Mingari MC, Moretta L, Ferlazzo G. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(−) cells and display an impaired capability to kill tumor cells. Cancer. 2008;112(4):863–875. doi: 10.1002/cncr.23239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Network of co-expressed genes of MICAL-L2.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.