Abstract
Aging is associated with declines in a host of cognitive functions, including attentional control, inhibitory control, episodic memory, processing speed, and executive functioning. Theoretical models attribute the age-related decline in cognitive functioning to deficits in goal maintenance and attentional inhibition. Despite these well-documented declines in executive control resources, older adults endorse fewer episodes of mind-wandering when assessed using task-embedded thought probes. Furthermore, previous work on the neural basis of mind-wandering has mostly focused on young adults with studies predominantly focusing on the activity and connectivity of a select few canonical networks. However, whole-brain functional networks associated with mind-wandering in aging have not yet been characterized. In this study, using response time variability—the trial-to-trial fluctuations in behavioral responses—as an indirect marker of mind-wandering or an “out-of-the-zone” attentional state representing suboptimal behavioral performance, we show that brain-based predictive models of response time variability can be derived from whole-brain task functional connectivity. In contrast, models derived from resting-state functional connectivity alone did not predict individual response time variability. Finally, we show that despite successful within-sample prediction of response time variability, our models did not generalize to predict response time variability in independent cohorts of older adults with resting-state connectivity. Overall, our findings provide evidence for the utility of task-based functional connectivity in predicting individual response time variability in aging. Future research is needed to derive more robust and generalizable models.
Keywords: Functional connectivity, Aging, Mind-wandering, Response time variability, Connectome-based modeling, Functional magnetic resonance imaging, Machine learning
1. Introduction
Humans spend a considerable amount of time engaged in thoughts involving spontaneous shifts of attention away from their external environment to their inner thoughts (Kane et al., 2007; Killingsworth and Gilbert, 2010). This phenomenon, colloquially known as mind-wandering, has been studied in the literature under various titles, including task-unrelated thoughts, off-task thoughts, self-generated thoughts, zoning-out, and tuning-out (Esterman et al., 2013; Seli et al., 2018). Mind-wandering has important consequences for exogenous processing, with previous studies implicating off-task thinking in performance on tasks of reading comprehension (Unsworth and McMillan, 2013), working memory (Kane et al., 2007), episodic memory (Maillet and Rajah, 2016; Risko et al., 2012), as well as activities of daily living, like driving performance (Galéra et al., 2012; He et al., 2011). Given the ubiquitous downstream effects of these task-unrelated thoughts on cognitive performance, there has been an increasing interest in examining the developmental evolution of mind-wandering in the context of cognitive aging. Older adults show notable declines on tasks of cognitive functioning, including on tasks assessing attentional control, inhibitory control, episodic memory, processing speed, and executive functioning (Craik and Salthouse, 2011; Glisky, 2007; Park et al., 2002; Tucker-Drob et al., 2009). These ubiquitous declines in controlled processing have largely been attributed to deficits in goal maintenance and attentional inhibition (Hasher and Zacks, 1988; Braver and West, 2008), thus suggesting that mind-wandering, involving the inability to suppress internally-focused thoughts, may be amplified with increasing age.
However, contrary to the anticipated positive relationship between mind-wandering and increasing age, there is unequivocal support for mind-wandering, as assessed through quasi-randomly presented thought probes during cognitive tasks, to decrease with increasing age (see Maillet and Schacter, 2016 for a review). Older adults endorse fewer episodes of off-task thinking when probed during experimental tasks (Fountain-Zaragoza et al., 2018; Jackson and Balota, 2012; McVay et al., 2013), in everyday life as assessed using experience sampling techniques (Maillet et al., 2018), and on trait measures of mind-wandering (Seli et al., 2017). Additionally, individuals with Alzheimer’s disease endorse even fewer episodes of mind-wandering (Gyurkovics et al., 2018), evincing support for the further decline in off-task thinking with advancing age and deteriorating cognitive resources. This perplexing developmental trajectory of mind-wandering has catapulted several explanations for these age-related declines in mind-wandering. Some theoretical models propose an attenuation of age-related reductions in mind-wandering endorsement after controlling for older adults’ increased motivation and interest of these laboratory-based tasks (Jackson and Balota, 2012; Krawietz et al., 2012; Shake et al., 2016). Other models posit declining executive control abilities to constrain resources directed to internally-focused thoughts, thereby decreasing off-task thinking during demanding cognitive tasks (Smallwood and Schooler, 2006). Age-related declines in mind-wandering have also been attributed to older adults’ need to perform well on lab-based cognitive tasks (Soubelet and Salthouse, 2011) and poorer meta-cognition (Maillet and Schacter, 2016), thus questioning the veridicality of thought probes in measuring mind-wandering with increasing age.
Although there is some evidence that these endorsements of mind-wandering may indeed reflect older adults’ mind-wandering propensity (Frank et al., 2015), there has also been an increasing interest in identifying and developing more objective indicators of mind-wandering. Response time variability, or the trial-to-trial variance in reaction time, is one such identified marker. Increased individual variability in reaction time has been associated with self-reports of mind-wandering episodes (Bastian and Sackur, 2013; Henríquez et al., 2016; Jubera-García et al., 2020; Kucyi et al., 2016; Maillet et al., 2020) as well as other lapses in attention (Cheyne et al., 2009; Schooler et al., 2014). Using the metronome response task, Seli et al. (2013) reported higher variability in response times preceding off-task thought probes compared with variability prior to on-task thought reports. Moreover, there is evidence from neuroimaging studies supporting the involvement of the frontoparietal, dorsal attention, and ventral attention networks during off-task processing (Christoff et al., 2016; Esterman et al., 2013; Golchert et al., 2017; Hasenkamp et al., 2012; Kucyi, 2018; Smallwood et al., 2016; Turnbull et al., 2019; Yamashita et al., 2021), suggesting that mind-wandering is a controlled, regulatory process requiring goal-directed neural processing. Indeed, mindfulness meditation training, which involves the cultivation of sustained attention, has been shown to notably reduce both self-reported and behaviorally measured mind wandering, like response time variability (Mrazek et al., 2012; Whitmoyer et al., 2020). Thus, with the re-direction of executive control resources to off-task thinking, less consistent (more variable) responding could be an important online behavioral correlate of mind-wandering.
Additionally, behavioral response variability has been employed to characterize distinct states of attentional fluctuation (Esterman et al., 2014, 2013; Yamashita et al., 2021). Consistent responding denotes an optimal “in-the-zone” state, whereas high response time variability is reflective of an “out-of-the-zone” state (Esterman et al., 2013; Esterman and Rothlein, 2019). Functional magnetic resonance imaging (fMRI) studies comparing optimal to suboptimal states highlight the contribution of activity in the default-mode network (Esterman et al., 2013; Fortenbaugh et al., 2018; Kucyi et al., 2016), including nodes in the precuneus, the anterior cingulate cortex, and the temporoparietal junction (Christoff et al., 2009; Mason et al., 2007; Wang et al., 2009) in supporting consistent responding. In contrast, activity in the dorsal attention network, known traditionally to subserve externally focused attention, was present during erratic responding (Esterman et al., 2013; Kucyi et al., 2017).
Altogether, response time variability is an important metric that has been characterized to reflect fluctuations in attentional states and may be an important marker of mind-wandering, especially in older adults. Although, the vast majority of findings from fMRI investigations of response time variability and thought probes as markers of mind-wandering to date have largely been based on data from young adults, there have been a handful of studies examining neural correlates of mind-wandering in older adults. Mind-wandering episodes have been linked with reduced communication between temporal and prefrontal regions of the default mode network (Martinon et al., 2019) and a reduced engagement of the medial and lateral prefrontal cortex as well as the left superior temporal gyrus (Maillet et al., 2019) in older adults. Other work has shown that mind-wandering frequency in older adults is associated with changes in connectivity within the default mode network. Specifically, these include an increased connectivity between regions of the lateral and medial temporal lobes and a decreased connectivity between the temporal pole and the dorsomedial prefrontal cortex (O’Callaghan et al., 2015). One common limitation is that previous fMRI studies were guided by a priori assumptions that involved focusing on specific brain regions or networks. Additionally, most previous studies typically employed classical univariate analysis based on general linear model or group-level inferences. One prominent drawback of these methods is the lack of proper characterization of brain function at the individual level (Dubois and Adolphs, 2016).
More recently, network neuroscience methods have been complimented by advances in machine learning, allowing us to gain unprecedented insight into the neural mechanisms underlying cognitive functioning. One such approach is connectome-based predictive modeling (CPM; Shen et al., 2017). CPM is a novel whole-brain, data-driven technique that allows for the derivation of brain-based predictive models from individualized functional connectivity patterns. Moreover, CPM allows for the identification of whole-brain functional connections that are associated with target cognitive function. Using whole brain, task-based, or resting state functional connectivity, several recent studies have demonstrated the utility of the CPM technique in identifying individual differences in brain functional architecture. These have allowed for the construction of brain-based models capable of predicting fluid intelligence (Finn et al., 2015), processing speed (Gao et al., 2020), attention (Rosenberg et al., 2016), reading ability (Jangraw et al., 2018), working memory (Avery et al., 2020; Manglani et al., 2021), loneliness (Feng et al., 2019), mind-wandering (Kucyi et al., 2021), or even diseased states such as Alzheimer’s disease (Lin et al., 2018) and attention deficit hyperactivity disorder (ADHD; Barron et al., 2020). A recent fMRI study demonstrated the utility of CPM to build generalizable models of mind-wandering, as measured using an experience sampling method, in healthy young adults and adults with ADHD (Kucyi et al., 2021).
Here, leveraging the CPM framework, we examined whether response time variability can be predicted from whole-brain functional connectivity in a cross-sectional sample of cognitively normal older adults. Response time variability was quantified using the reaction time coefficient of variation (RT_CV) calculated as the standard deviation of reaction time/mean reaction time. Using data from publicly available datasets, we first investigated whether functional connectivity during an intrascanner Go/NoGo task and resting-state fMRI could predict response time variability. We also explored the utility of our model in predicting individual response time variability in two independent cohorts of healthy older adults. We hypothesized that task-based, whole-brain models of functional connectivity would explain significant individual differences in response time variability in older adults with nodes of the default mode network, dorsal attention network, and the ventral attention network showing the highest contributions.
2. Materials and methods
2.1. Datasets overview
We analyzed functional MRI and behavioral data in 407 cognitively normal older adults between the ages of 65–85 years from three independent cohorts. These datasets were from the publicly available Human Connectome Project in Aging (Bookheimer et al., 2019; Harms et al., 2018) (HCP-Aging) and Cambridge Centre for Ageing Neuroscience (Cam-CAN; Shafto et al., 2014; Taylor et al., 2017) databases. The third dataset, Studying Cognitive and Neural Aging (SCAN; Fountain-Zaragoza et al., 2021), was acquired by our group here at The Ohio State University, Columbus, OH. In the present study, the HCP-Aging was used as the primary dataset for model derivation and internal validation. Cam-CAN and SCAN served as the validation datasets used to assess the generalizability of the derived models.
2.2. Participants
2.2.1. HCP-Aging
HCP-Aging is an ongoing multisite study designed to acquire normative neuroimaging and behavioral data for examining changes in brain organization during typical aging. The dataset used in the current study was drawn from the first release (Lifespan HCP Release 1.0) and comprised of 689 cognitively healthy older adults (36–105 years). Participants were excluded from the HCP-Aging study if they had been diagnosed and treated for major neuropsychiatric and neurological disorders. Those with impaired cognitive abilities were also excluded. In the current study, we began by restricting our analysis to participants who are unrelated, aged 65–85 years, right-handed, and with fMRI and behavioral data available (n = 189). We then excluded participants with outlier performance (see Datasets description: Cognitive tasks) during cognitive task (n = 8). Of the remaining participants, those with excessive head motion during fMRI (n = 35, described below in Head Motion Control) and incidental finding (n = 1) were also excluded. Data for 145 participants (73 women, mean age = 73.8 years, SD = 5.8) met inclusion criteria and were analyzed further. All HCP-Aging participating sites obtained Institutional Review Board approval. Participants gave written informed consent at study enrollment.
2.2.2. Cam-CAN
Cam-CAN is a cross-sectional, population-based study established with the purpose of acquiring cognitive and multimodal neuroimaging data to investigate the neural mechanisms underlying successful cognitive aging. This dataset has been described elsewhere (Shafto et al., 2014; Taylor et al., 2017). We analyzed data from 168 right-handed cognitively normal adults aged 65 to 85 years. We excluded participants with missing cognitive data (n = 8). We also excluded an additional 58 participants due to excessive head motion during fMRI (see Head Motion Control). Two additional participants were excluded due to error in generating their connectivity matrices. These exclusions resulted in a final sample that consisted of 100 individuals (40 women, mean age = 73.12 years, SD = 5.8). The Cam-CAN study was approved by the Cambridgeshire 2 Research Ethics Committee. Each participant gave written informed consent before participating in the study.
2.2.3. SCAN
SCAN was a cross-sectional study designed to characterize the neuromarker of sustained attention in healthy aging. We recruited 50, healthy, right-handed older adults between the ages of 65 and 85 years from the greater Columbus, Ohio area. All participants had normal or corrected-to-normal visual acuity, no contraindication to MR environment, and reported no history of learning disability, psychiatric disorder, neurological disorder or terminal illness. Participants were not using medication that could significantly alter brain activity. No participant scored less than 26 on the Montreal Cognitive Assessment (Nasreddine et al., 2005) (MoCA), indicating no evidence of mild cognitive impairment or dementia. Participants attended two experimental sessions separated by 7–14 days. Of the 50 participants recruited, we excluded participants due to incidental findings during MRI (n = 2) and MRI data acquisition error (n = 1). The resulting analytic sample comprised of 47 participants (25 women, mean age = 70.2, SD = 4.5). The study was approved by The Ohio State University Institutional Review Board. Each participant gave written informed consent at study enrollment and received monetary compensation for their time.
2.3. Datasets description: cognitive tasks
The target dependent variable for prediction was the reaction time coefficient of variability (RT_CV). Individual rate of mind-wandering can be indirectly quantified using the RT_CV, which is calculated as the standard deviation of reaction time (RT) divided by mean RT (RT standard deviation/RT mean). Higher RT_CV score in the context of reaction time data is considered to be reflective of higher rate of mind-wandering or a suboptimal attentional state (Bastian and Sackur, 2013; Hu et al., 2012).
2.3.1. HCP-Aging
Participants performed a Go/NoGo task known as the Conditioned Approach Response Inhibition Task (CARIT; Somerville et al., 2018) inside the MRI. During task performance, participants were instructed to rapidly press a button (“Go”) in response to seeing all shapes except for a circle and a square (“NoGo”). Go stimuli were six different previously unseen shapes including hexagon, octagon, parallelogram, pentagon, trapezoid, and plaque. Each stimulus was presented for 600 ms, followed by a jittered inter-trial interval of fixation, ranging from 1 s to 4.5 s. The allowable stimulus response time was 800 ms (including 600 ms during stimulus presentation and the first 200 ms of fixation). Data was acquired over a single 4-minute task run consisting of 68 Go and 24 NoGo trials. We obtained the correct responses on the Go trials (Hits) for each participant and identified participants with outlier behavior as those with less than 50% Hits rate (n = 8). Individual reaction time data on correct Go trials was used to compute a RT_CV score for each participant.
2.3.2. Cam-CAN
Participants performed a Choice Reaction Time outside of the scanner. During the task, participants viewed an onscreen image of a hand with four blank circles above each finger. Responses were recorded with a 4-button response box on which the participants were asked to place four fingers in their right hand on each of the four buttons. During each trial, any of the four blank circles above each finger could turn black, and participants must press the corresponding button as quickly as possible. For example, when the index finger circle turns black on the image, participants press with their index finger as quickly as possible. The circle turns blank again after each response. The allowable stimulus response time was 3 s. Pseudo-random inter-trial intervals with the following details were used: minimum 1.8 s, mean 3.7 s, median 3.9 s, and maximum 6.8 s. Participants completed a total of 67 trials. The reaction time from stimulus onset to button press was averaged across all four fingers for all correct trials. The CamCAN dataset provides already calculated RT_CV scores.
2.3.3. SCAN
Participants performed a Go/NoGo task outside of the scanner. Task and imaging data acquisition were conducted on separate days. In this task, participants pressed a key in response to frequently presented non-targets (or Go trials) and withheld their responses on NoGo trials that were preceded by an auditory tone. Each trial began with a fixation cross for 750 ms, followed by the stimulus presented for 750 ms with a maximum response window of 1500 ms. The task consisted of 6 blocks with each block consisting of 54 Go trials, 6 NoGo trials, and 3 self-report mind-wandering probes presented pseudo-randomly after 15–20 trials (data not analyzed here). The entire task lasted approximately 35 min. We computed RT_CV score for each participant using reaction time data on correct Go trials.
2.4. Datasets description: MRI data
2.4.1. HCP-Aging
Neuroimaging data was acquired with a Siemens 3 Tesla Prisma system with a 32-channel head coil. Anatomical T1-weighted images were acquired via a multiecho magnetization-prepared gradient-echo sequence (MPRAGE; repetition time (TR) = 2500 ms; echo time (TE) = 1.8 ms; voxel size = 0.8 × 0.8 × 0.8 mm; 208 sagittal slices; flip angle = 8°). Task and resting-state fMRI images were acquired using a 2D multiband gradient-recalled echo echo-planar imaging (EPI) sequence (TR = 800 ms, TE = 37 ms; 72 axial slices; voxel size = 2.0 × 2.0 × 2.0 mm; flip angle = 52°; multiband factor = 8). A total of 300 volumes were acquired during a single run of task-fMRI that lasted 4 min 11 s. Two sessions of eyes-open resting state fMRI (REST1 and REST2) were performed in a single day or across two days depending on site-specific constraints. Each resting state session consisted of two, separate 6 min 5 s runs, with opposite phase-encoding direction (i.e. REST1 Anterior-Posterior, REST1 Posterior-Anterior, REST2 Anterior-Posterior and REST2 Posterior-Anterior, hereafter collectively referred to as four resting-state scans). A total of 488 volumes were acquired for each of the four resting-state scans. Additionally, a pair of opposite phase-encoding spin-echo fieldmaps (anterior-to-posterior and posterior-to-anterior; TR = 8000 ms, TE = 66 ms, flip angle = 90°) were acquired separately for task and resting-state and were used to correct functional images for signal distortion.
2.4.2. Cam-CAN
Structural and functional MRI data were acquired at the Medical Research Council Cognition and Brain Science Unit in Cambridge UK using a 3 Tesla Siemens TIM Trio scanner with a 32-channel head-coil. 3D T1-weighted structural images were acquired using a T1-weighted sequence using MPRAGE sequence with the following protocol parameters: TR = 2250 ms; TE = 2.98 ms; voxel size = 1.0 × 1.0 × 1.0 mm; 192 sagittal slices, flip angle = 9°, GRAPPA acceleration factor = 2. Eyes-closed resting-state fMRI data was acquired using a gradient-echo EPI sequence with the following parameters: TR = 1970 ms, TE = 30 ms; 32 axial slices; voxel size = 3.0 × 3.0 × 3.7 mm; flip angle = 78°; acquisition time=8 min 40 s, total number of volumes = 261). Phase-encoded gradient echo fieldmaps (TR = 400 ms, TE = 5.19/7.65 ms, flip angle = 60°) were acquired and used for distortion correction.
2.4.3. SCAN
MRI data acquisition was performed at the Center for Cognitive and Behavioral Brain Imaging at The Ohio State University, using a 3 Telsa Siemens Magnetom Prisma MRI scanner with a 32-channel head coil. Structural T1-weighted images were acquired using a MPRAGE sequence (TR = 1900 ms; TE=4.44 ms; voxel size = 1.0 × 1.0 × 1.0 mm; 176 sagittal slices, flip angle=12°). Eyes-open resting-state images were acquired using a whole-brain multiband EPI sequence (TR = 1000 ms, TE = 28 ms; 45 axial slices; voxel size = 3.0 × 3.0 × 3.0 mm; flip angle = 50°; multiband factor = 3, total number of volumes = 480). Fieldmaps were also acquired (TR = 512 ms, TE = 5.19/7.65 ms, flip angle = 60°) to correct the EPI images for signal distortion.
2.5. Preprocessing of MRI data
MRI NIfTI files were first organized according to the Brain Imaging Data Structure (BIDS) format (Gorgolewski et al., 2016) and validated with the BIDS validator v.1.5.6 (https://bids-standard.github.io/bids-validator/). Preprocessing of structural and functional MRI data was similar in all three datasets and was performed using fMRIprep v1.5.0rc1, a Nipype based tool (Esteban et al., 2019). Each T1-weighted image was corrected for bias field with N4BiasFieldCorrection v2.1.0 (Tustison et al., 2010). The T1w was then skull stripped using antsBrainExtraction.sh v2.1.0. Next, brain-extracted T1-weighted image was normalized to MNI space through nonlinear registration with antsRegistration tool (Avants et al., 2008). Brain tissue segmentation of cerebrospinal fluid (CSF), white matter, and gray matter was performed on the brain-extracted T1-weighted using FAST (Zhang et al., 2001). Functional data preprocessing involved the following steps: functional MRI data was corrected for susceptibility distortions using 3dQwarp (Cox and Hyde, 1997), co-registered to corresponding T1w configured with boundary-based registration with nine degrees-of-freedom (Greve and Fischl, 2009) and motion corrected using FSL MCFLIRT v5.0.9 (Jenkinson et al., 2012). Slice-timing correction was not performed on the HCP or the SCAN datasets. This step is considered to be unnecessary for datasets acquired using multiband pulse sequences with a short Time-to-Repetition as all slices in a volume are acquired much closer together (Glasser et al., 2013; Smith et al., 2013). However, due to the relatively long Time-to-Repetition used for the CamCAN data acquisition, we additionally performed slice-time correction (Sladky et al., 2011) in this dataset using 3dTshift from AFNI (Cox and Hyde, 1997). Motion correcting transformations, functional-to-T1w transformation, and T1w-to-MNI-template warp were concatenated and applied in a single step with antsApplyTransforms (ANTs v2.1.0) using Lanczos interpolation. Physiological noise regressors were extracted based on CompCor procedure (Behzadi et al., 2007). Several confounding timeseries, including framewise displacement (FD) and global signal (Power et al., 2014), were calculated based on the functional data using the implementation of Nipype and used during nuisance regression (see Nuisance removal and filtering). Many internal operations of fMRIPrep use Nilearn (Abraham et al., 2014), principally within the blood-oxygen-level-dependent (BOLD)-processing workflow. Additional details of the pipeline can be found here: https://fmriprep.readthedocs.io/en/latest/workflows.html. Following implementation of this pipeline, we obtained preprocessed functional data for each participant in their native BOLD space.
2.6. Nuisance removal and filtering
Preprocessed BOLD images were denoised using the signal.clean function within Nilearn (Abraham et al., 2014), which allowed for removal of several sources of spurious variance orthogonal to the temporal filters (Lindquist et al., 2019). For HCP-Aging, functional volumes acquired during the 8-s countdown period of task-fMRI were discarded. The first 10 volumes of the resting-state data in all three datasets were also excluded to minimize magnetization equilibrium effects. Next, we created confound files for each scan for each participant with the following regressors: six rigid body motion parameters, six temporal derivatives and their squares (Friston et al., 1996), mean white matter, mean CSF, single timepoint regressor for outlier timeframes – defined in the current study as volumes with FD value ≥ 0.9 mm (Lemieux et al., 2007; Siegel et al., 2014), and mean global signal as prior work has shown that global signal regression strengthens the association between functional connectivity and behavior (Li et al., 2019). These confounds were regressed out of each preprocessed data. Temporal filtering was performed with a high-pass filter of 0.01 Hz to remove the effects of slow fluctuating noise such as scanner drift. Finally, spatial smoothing of functional data was performed using a Gaussian-smoothing kernel of 6-mm full width half maximum (FWHM).
2.7. Whole-brain parcellation and functional network construction
We next constructed whole brain functional connectivity matrices for each participant in the HCP-Aging dataset. Functional network nodes were defined based on the Shen 268-node functional parcellation scheme covering the cortical, subcortical, and cerebellar regions (Shen et al., 2013). First, we obtained subject and scan specific atlases by transforming the Shen atlas from its original standard space to each subject’s native functional MRI space. The derived subject and scan specific atlases were used to parcellate the brain into 268 non-overlapping regions. Functional connectivity was defined as the Fisher z-transformed Pearson correlation coefficients between the mean timeseries of all pairs of the parcellated regions (i.e., nodes). As prior research has provided empirical support for task-based model performance to decrease following regression of task-evoked activations (Greene et al., 2020), our task-based functional connectivity matrices were constructed based on raw timeseries with no regression of task-evoked activations. Ultimately, we obtained a symmetric matrix representing the task fMRI functional connectivity (“task-FC”) for each subject. We repeated the above steps for each of the resting state scans, resulting in four resting state functional connectivity (“rest-FC”) matrices for each subject. Rest-FC from all four runs were then averaged to derive a single rest-FC per participant. The above-described procedures resulted in a task-FC and rest-FC for each participant in HCP-Aging dataset and were used in constructing predictive models independently. Finally, we used the above-described pipeline to construct a rest-FC matrix for each participant in the Cam-CAN and SCAN datasets.
2.8. Connectome-based predictive modeling
We adopted CPM with a leave-one-out cross validation approach in n = 145 participants to determine whether whole-brain functional connectivity could predict individual mind-wandering. CPM analysis procedures involve three main stages including features extraction, model building, and prediction. During feature extraction, for each participant, we correlated each of the 35,778 edges (or functional connections between nodes) in the connectivity matrix with their observed RT_CV score using Spearman’s partial correlation with each participant’s mean FD value entered as a covariate. The resulting correlation coefficients, across participants, were thresholded at p < .01 (Beaty et al., 2018; Rosenberg et al., 2016; Shen et al., 2017). We then extracted sets of surviving edges that are positively (i.e., positive r values) or negatively (i.e., negative r values) associated with RT_CV scores. The weights of the positive edges and negative edges were averaged to obtain a positive-feature network and negative-feature network, respectively. In the current study, we defined the positive-feature network as the high response time variability network and the negative-feature network as the low response time variability network. The high response time variability network represents functional edges that are stronger in people with high trial-to-trial variability in response time, whereas the low response time variability network includes edges that are stronger in those with low variability in response time. A combined response time variability network was also computed as the difference between strengths in the high and low response time variability networks.
For model building, we used robust regression to fit single-subject summary network strength values from the high, low, and combined networks with RT_CV scores separately to build three predictive models namely the high response time variability, low response time variability and the combined models. At the prediction stage, we used the regression coefficients obtained in the trained models to predict RT_CV for the left-out participant. The entire process was repeated iteratively 145 times until each participant had served as the test participant. Predictive power was assessed via the Spearman’s rank correlation between observed and predicted RT_CV scores. This approach has been widely used in CPM and machine learning literature.
Statistical significance of prediction accuracy was assessed with permutation testing (Dosenbach et al., 2010). This involved repeating the entire CPM analysis 1000 times during which observed RT_CV scores were randomly shuffled across participants while preserving the structure of each functional connectivity matrix. For instance, functional connectivity matrix for subject 001 was paired with participant 002′s RT_CV score. The resulting nonparametric p-value from this null distribution, calculated as p = (1 + number of null prediction correlation values ≥ true prediction correlation value)/1001, allowed to determine whether model performance was significantly better than expected by chance. The entire CPM analysis was performed in MATLAB (The MathWorks, Inc.; version r2019b).
2.9. Anatomical distribution of predictive edges
To gain insight into the spatial distribution of relevant predictive edges (or connections) identified in the feature extraction stage of CPM, we defined consensus whole-brain network masks containing the edges that appeared in each iteration of leave-one-out cross validation across all subjects (Rosenberg et al., 2016). Focusing on the models derived from task-FC, we constructed two separate masks to include edges that correlate positively (high response time variability network) or negatively (low response time variability network) with RT_CV. These masks were used to assess model generalizability between task and resting-state brain states and across datasets.
Next, to visualize the edges in each respective mask, all 268 nodes from the Shen atlas were grouped according to nine canonical brain networks that comprised seven networks from the Yeo’s 7-network cortical parcellation scheme (Yeo et al., 2011) and two networks from the Shen parcellation scheme (Greene et al., 2018; Noble et al., 2017; Shen et al., 2013). Specifically, each node in the cortical region of the Shen atlas was assigned to the networks in the Yeo atlas based on their Dice similarity (Dice, 1945). As the Yeo atlas lacks full brain coverage, the original network assignment of the nodes in the subcortical and cerebellar networks of the Shen atlas was retained. The final nine networks included the visual, somatomotor, dorsal attention, ventral attention, limbic, frontoparietal, default mode, subcortical, and cerebellar networks. We next computed the number of connections between and within each pair of canonical networks. Crucially, we normalized the edge counts within each network and between each possible pair of networks to account for network sizes (Greene et al., 2018). This allowed us to explicitly characterize the contribution of each network and subsequently determine the networks that were overrepresented in our predictive models. Edges were visualized using BrainNet viewer toolbox (Xia et al., 2013) and Chord (https://pypi.org/project/chord/) in Python 3.
2.10. Cross-condition and external validation of predictive models
Trained models and consensus masks were applied to calculate network summary strength for each subject in the testing set. Network strength scores were then fitted with observed RT_CV scores to obtain predicted RT_CV scores. Performance was evaluated as the Spearman correlation between predicted and observed RT_CV scores with head motion added as a covariate. For cross-condition prediction analysis (Greene et al., 2018; Jiang et al., 2020), models generated from task-FC in n − 1 subjects were applied to the rest-FC matrix in the left out participant to predict trial-to-trial response time variability. In the cross-datasets’ application, models generated from task-FC were applied to resting state functional connectivity data in the CamCAN and SCAN datasets, separately.
2.11. Head motion control
Head motion during fMRI was quantified using mean FD. Following data preprocessing, participants (or runs in the case of HCP-Aging resting state scans) with mean FD values greater than 0.15 mm were excluded from further analyses. We also examined potential associations between RT_CV and head motion. Crucially, RT_CV was significantly associated with mean FD during task-fMRI (r = .22, p = .008), but not during resting-state fMRI (average FD across four runs: r = .15, p = .079). There was a significant association between RT_CV and head motion during resting-state in the CamCAN dataset (r = 0.23, p = .019). Although there was no significant association between head motion and RT_CV in the SCAN dataset (r = .02, p = .903), we chose to control for possible effects of head motion in all our analyses by including mean FD as a covariate. Specifically, for model derivation analysis, we included mean FD as a covariate at the edge selection stage of CPM as described above. For model application analysis, we calculated the Spearman’s partial correlation between predicted and observed RT_CV scores while controlling for mean FD.
2.12. Control analysis
We performed validation analyses to assess the potential influence of several variables on our main results. (1) Covariates: We repeated CPM analysis by entering age, sex, and study sites independently as a covariate in addition to mean FD at the edge selection stage of CPM analysis. (2) Functional parcellation scheme: To validate our results with an alternative functional atlas and to ensure full coverage of the cortical, subcortical, and cerebellar regions, we used an atlas (McNabb et al., 2018) obtained by merging the cortical and subcortical regions of the human Brainnetome atlas (Fan et al., 2016) with a probabilistic atlas of the human cerebellum (Diedrichsen et al., 2009). We subsequently derived a whole-brain atlas with 272 regions and reran the entire CPM analysis using the 272 × 272 matrices constructed with this atlas. (3) K-fold: Recent work suggests that the leave-one-out cross validation approach may lead to unstable and biased estimates (Varoquaux et al., 2017). To test if our main results are robust to the choice of cross-validation method used, we repeated the CPM analysis procedures described above using a repeated k-fold cross-validation approach (k = 2, 5, 10). For example, in the 2-fold (i.e., split-half) cross-validation, data was randomly split into 2 approximately equal subsets (i.e., 72 and 73), with one subset used as training set while the other was held out as the testing set. The 2-fold cross-validation was then repeated 1000 times to allow random assignment of participants into folds to avoid bias in fold assignment. In line with a previous CPM study (Goldfarb et al., 2020), we calculated model predictive power as the average Spearman’s correlation across all 1000 iterations. A null distribution predicting randomly shuffled RT_CV scores was also performed 1000 times. The correlation values from the models predicting true RT_CV scores were compared to those from the null distribution to compute effect sizes.
2.13. Statistical analyses
Tests of normality were performed to determine the distribution of RT_CV scores in each cohort. Pearson correlation was used to determine if there was a relationship between RT_CV scores and other variables such as head motion, age, sex, and study sites. These statistical tests were performed using Pingouin v0.3.8 (https://pingouin-stats.org/; Vallat, 2018) in Python 3. A p < .05 was considered significant and p-values are noted as reported by the statistical package used.
2.14. Correction for multiple comparisons
Multiple comparisons were corrected for where necessary by using the false discovery rate (FDR). Because the models (i.e., high, low, and combined) derived using the CPM approach are not independent, we opted to adjust only the permuted p values from the task-FC combined and rest-FC combined models by controlling the FDR at 5% using the Benjamin-Hochberg procedure (Benjamini and Hochberg, 1995). Note that the combined model consolidates information from both the high and low RT_CV models within each of the two respective brain states (i.e., task and rest). FDR correction was performed using the p.adjust function in R. A FDR corrected p < .05 was set as the significant threshold for multiple comparisons.
3. Results
3.1. Predicting mind-wandering from whole-brain functional connectivity
Using the leave-one-subject-out cross-validation approach, we found that connectome-based models trained solely on whole-brain functional connectivity during task fMRI, but not resting state fMRI, can predict response time variability in healthy older adults. Specifically, using task-FC, the CPM based on high response time variability model (ρ = .25, pperm = .028, Fig. 1A), low response time variability model (ρ = .22, pperm = .043, Fig. 1A), and overall combined model (ρ = 0.25, pperm = .021, pFDR = .042, Fig. 1A) successfully predicted individual response time variability. We observed that the task-FC trained model utilized 268 functional connections, with 134 functional connections each in the high and low response time variability networks. However, we did not see significant associations between observed RT_CV scores and predicted RT_CV scores using resting-state CPM models for the high response time variability model (ρ = .11, pperm = .202, Fig. 1B), the low response time variability model (ρ = .08, pperm = .283, 1B), or the combined model (ρ = .09, pperm = .214, pFDR = .214, Fig. 1B).
Fig. 1.
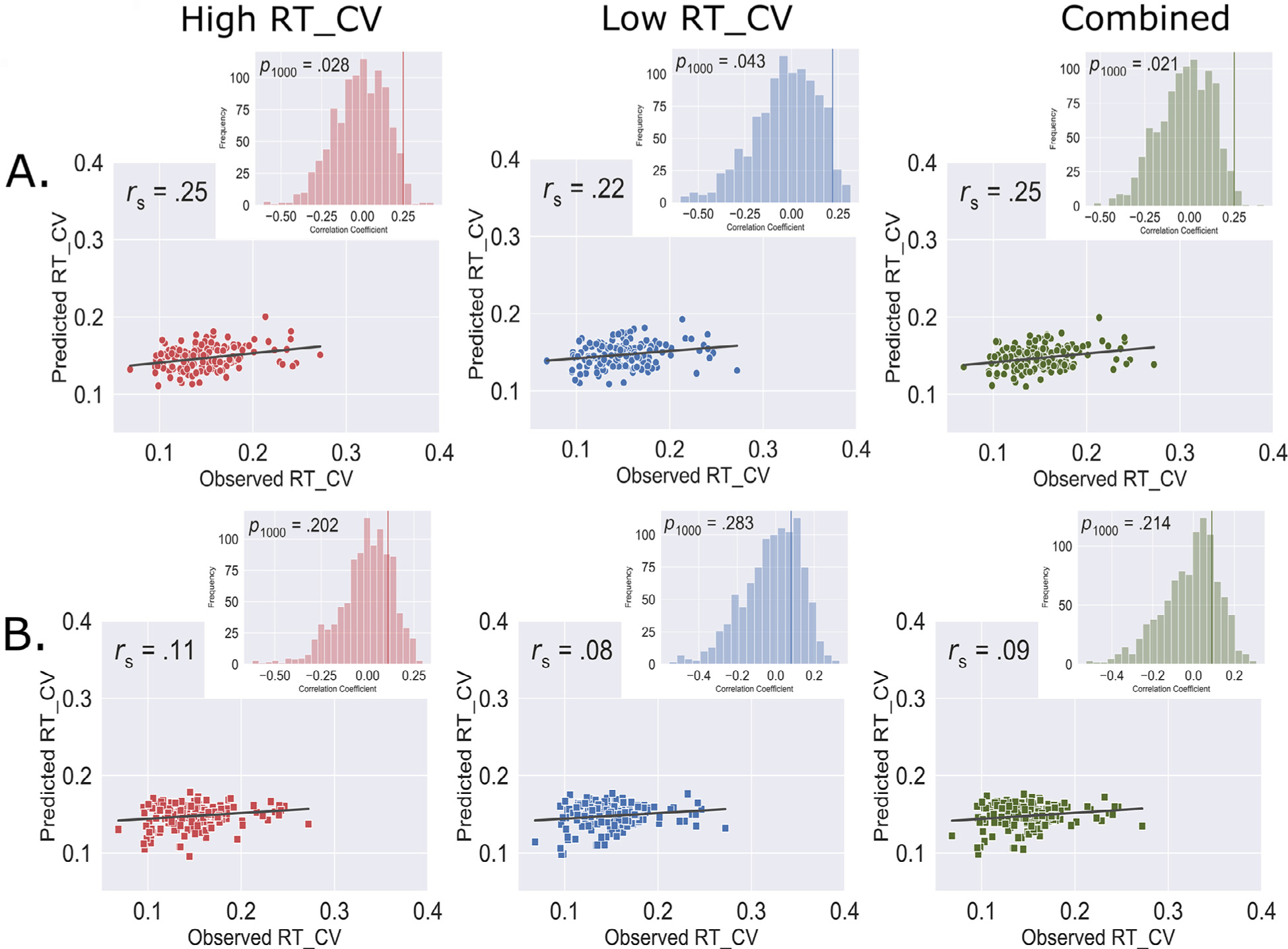
Predictive model performance showing cross-validated prediction of individual variability in response time from whole-brain A) Task functional connectivity, and B) Resting-state functional connectivity. Scatterplot represents Spearman correlation between the observed RT_CV and the predicted RT_CV scores. Histogram shows distribution of correlation values obtained by randomly shuffling connectivity matrices and respective RT_CV scores 1000 times to determine model significance. RT_CV: Reaction time coefficient of variation; rs: Spearman correlation; p1000: p value obtained after 1000 iterations permutation.
3.2. Assessing cross-condition model generalizability
After establishing that response time variability can be predicted from individual functional connectivity during task fMRI, we next sought to investigate whether the task-FC derived models generalize across brain state in the same cohort. To prevent circularity, we applied models trained on task-FC matrices in n – 1 participants (i.e., 144 participants) to predict individual response time variability from rest-FC matrix in the left-out participant. We repeated this process until each participant has been held out as the test participant. This analysis revealed that task-FC trained models did not significantly predict response time variability from rest-FC data for high response time variability (ρ = .10, p = .392), low response time variability (ρ = .15, p = .078), and combined (ρ = .15, p = .072) models after covarying out mean FD during resting-state fMRI (Fig. 2).
Fig. 2.
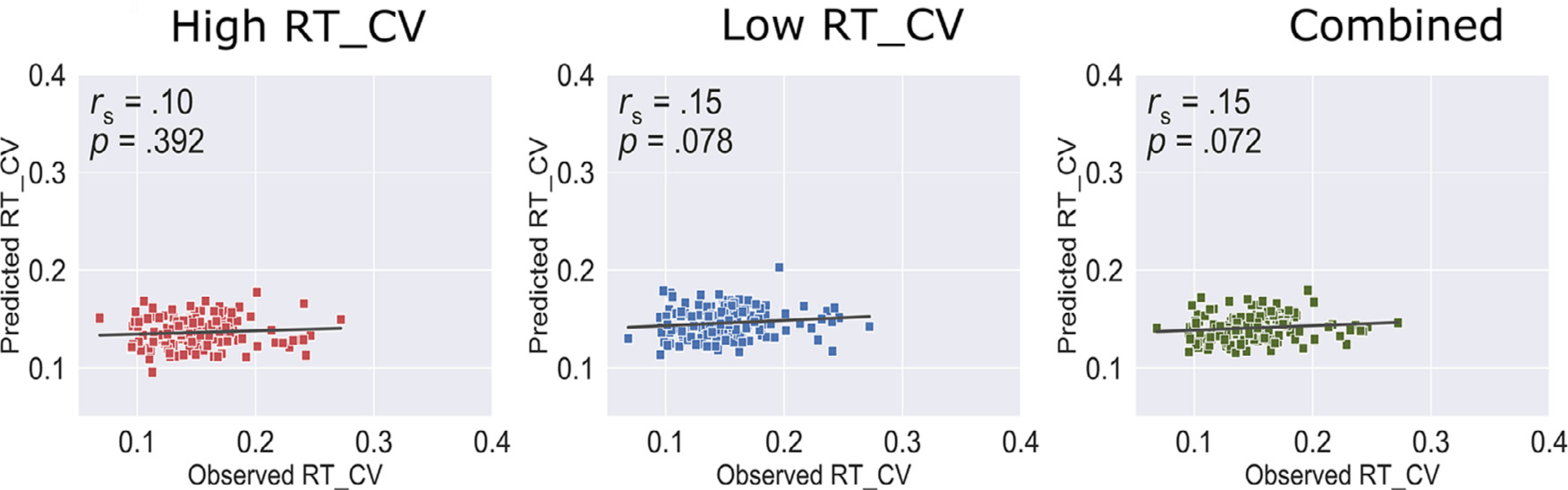
Cross-condition model performance. Models trained on task fMRI functional connectivity were applied to resting-state functional connectivity in the same sample to predict response time variability. Head motion was added as a covariate. RT_CV: Reaction time coefficient of variation, rs: Spearman correlation.
3.3. Neuroanatomy of predictive edges
Having shown that task-FC models successfully predict response time variability, we next explored the spatial distribution of brain connections that contributed most to predictive power. Because slightly different edges were selected in each leave-one-out cross validation, we restricted our analysis to those that were repeatedly selected across all iterations. Consistent with previous CPM studies, these edges were widely distributed across the brain (Fig. 3A and B).
Fig. 3.
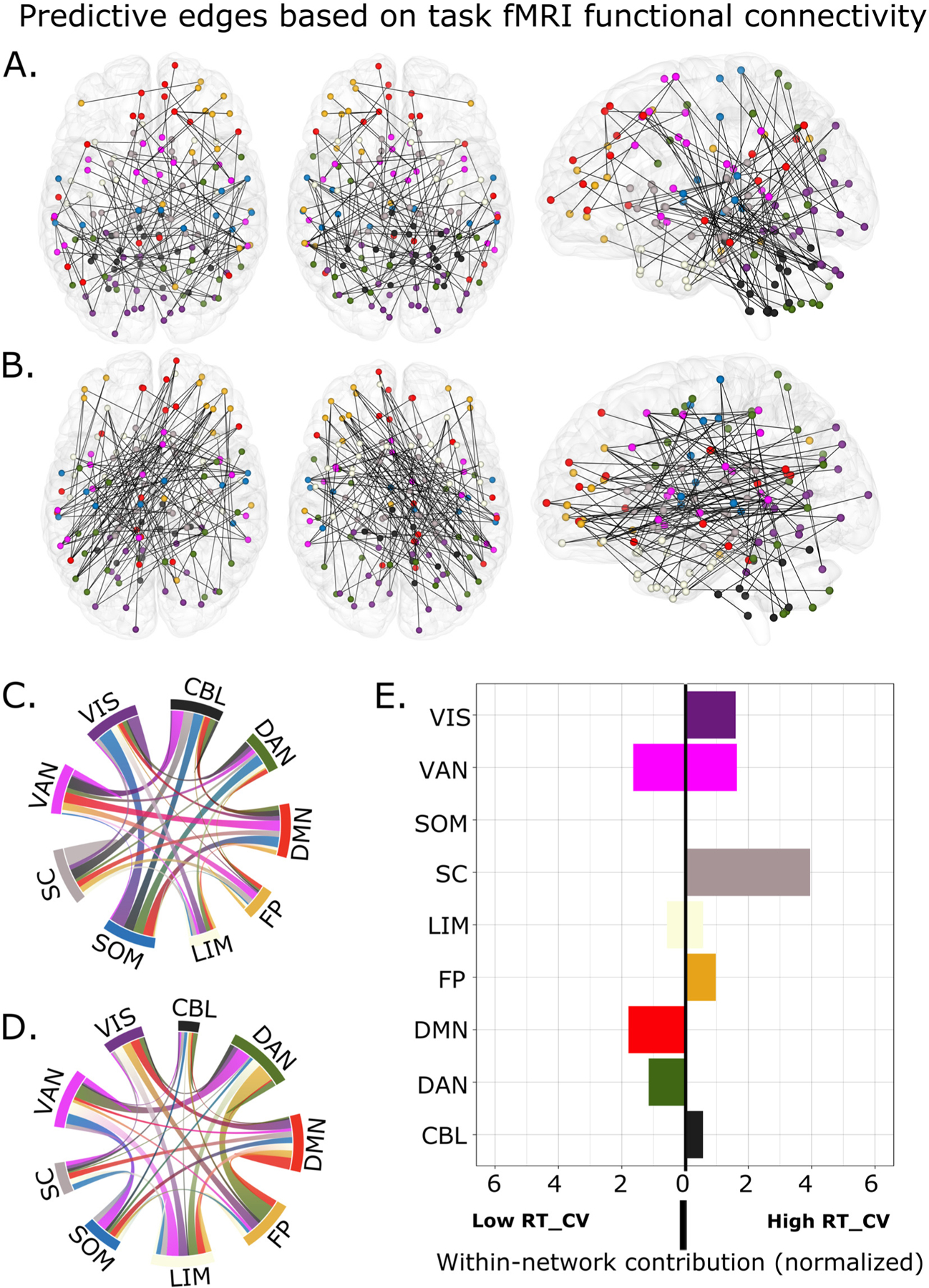
Anatomical distribution of predictive edges from models trained on task functional connectivity. A) Predictive edges (n = 134) in the high RT_CV model. B) Predictive edges (n = 134) in the low RT_CV model. Each node in the brain-based figure is colored based on nine canonical networks. To aid visualization, predictive edges in C) high and D) low RT_CV models were further grouped according to their canonical network and visualized using chord plots. E) Normalized contribution of edges within each network in the low RT_CV and high RT_CV models. CBL: cerebellar network; DAN: dorsal attention network, DMN: default mode network, FP: frontoparietal network, LIM: limbic network; SOM: somatomotor network; SC: subcortical, VAN: ventral attention network; VIS: visual attention network; RT_CV: Reaction time coefficient of variation.
We began by summarizing all connections by canonical networks (Shen et al., 2013; Thomas Yeo et al., 2011) (Fig. 3C and D). We next calculated within-network and between-network connections normalized by network size to allow for network-level visualization. In the high response time variability network, we identified the overrepresented networks as those with a value >1 (Fig. 4A and B, left panel). These overrepresented networks include the somatomotor, ventral attention, subcortical, cerebellar, and the default mode networks. Furthermore, we found that in the high response time variability model, within-network connectivity of the subcortical network was the most utilized relative to connectivity within other brain networks (Fig. 3E). Additionally, we observed an overrepresentation of some network pairs that included the somatomotor—visual, somatomotor—dorsal attention, cerebellar—ventral attention, somatomotor—default mode, cerebellar—somatomotor, frontoparietal—ventral attention, and default mode—ventral attention networks (Fig. 4A).
Fig. 4.
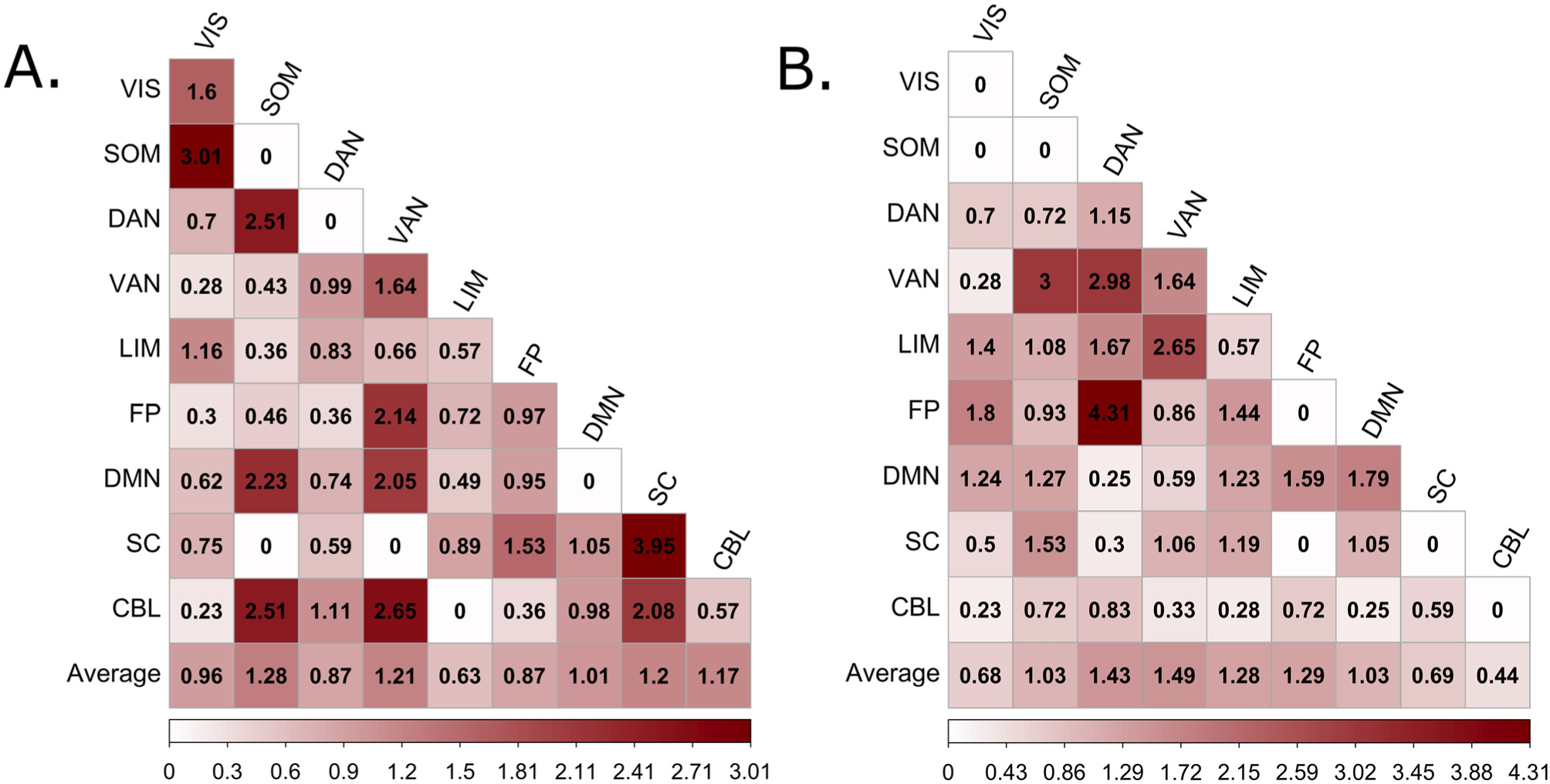
Relative contribution of each canonical network to model performance. The predictive edge counts were normalized between and within each possible pair of networks to account for network sizes (see Anatomical Distribution of Predictive Edges). Overrepresented networks were identified as those with values > 1. Representation of each network in the A) High RT_CV, and B) Low RT_CV. CBL: cerebellar network; DAN: dorsal attention network, DMN: default mode network, FP: frontoparietal network, LIM: limbic network; SOM: somatomotor network; SC: subcortical, VAN: ventral attention network; VIS: visual attention network; RT_CV: Reaction time coefficient of variation.
In the same vein, we assessed the spatial distribution of connections in the low response time variability network i.e., those functional connections that were stronger during low trial-to-trial variability in response time. The frontoparietal, dorsal attention, limbic, and ventral attention networks were the most overrepresented networks in the low response time variability network. Moreover, connectivity between the dorsal attention—frontoparietal, dorsal attention—ventral attention, ventral attention—somatomotor, visual—frontoparietal, limbic—visual, limbic—ventral attention, and the limbic—dorsal attention networks contributed the most to the low response time variability network (Fig. 4B). Furthermore, we found an overrepresentation of intra-connectivity of the default mode, dorsal attention, and ventral attention networks (Fig. 3E), indicating that connections within these networks exhibited negative correlations with RT_CV.
3.4. Assessing model generalizability in independent datasets
Having established a robust within-sample prediction for task-FC derived models, we next aimed to investigate whether those models derived in HCP-Aging cohort would generalize to predict individual response time variability in two independent samples of n = 100 (Cam-CAN data) and n = 47 (SCAN data) healthy older adults. Resting-state data is available in majority of publicly available datasets thus allowing for assessment of model generalizations in large samples. To this end, we obtained individual RT_CV scores and analyzed resting-state fMRI data in both cohorts using data analysis pipelines identical to those used in the HCP-Aging dataset, and subsequently constructed individual resting state functional connectivity matrix. We next applied our task-FC derived response time variability models to the rest-FC and RT_CV scores in both datasets, separately. We found that despite the successful in-sample application within the HCP-Aging cohort, our models did not generalize to predict response time variability in either the CamCAN cohort (high response time variability model: ρ = .07, p = .48; low response time variability model: ρ = −.11, p = .29; combined model: ρ = −.06, p = .59, Fig. 5A) or in the SCAN cohort (high response time variability model: ρ = .01, p = .96; low response time variability model: ρ = .16, p = .27; combined model: ρ = .11, p = .48, Fig. 5B).
Fig. 5.
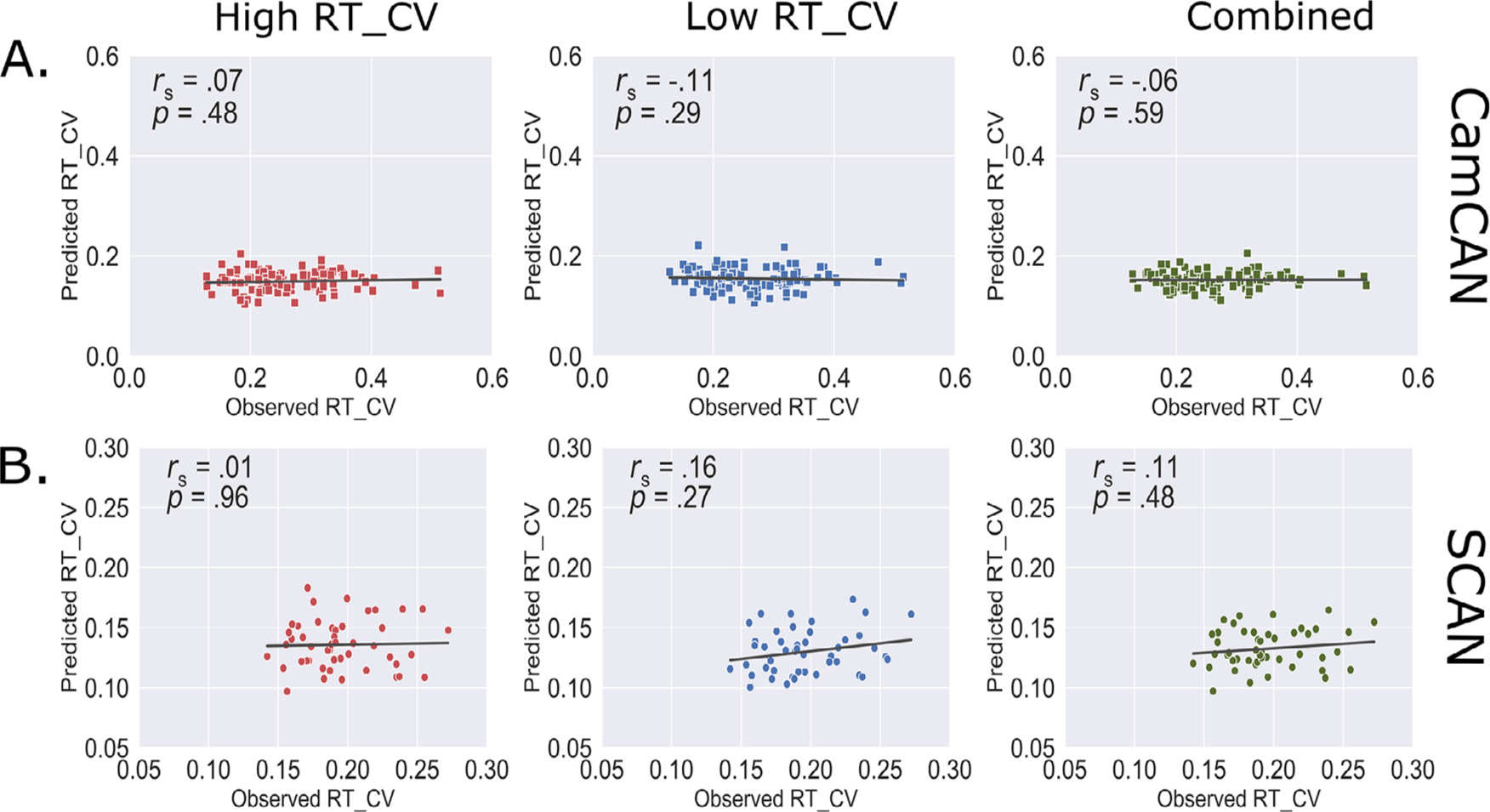
Summary of model performance in two independent datasets. Model trained on task functional connectivity in the HCP-Aging cohort was applied to predict individual response time variability in the A) CamCAN and B) SCAN cohorts. RT_CV: Reaction time coefficient of variation.
3.5. Influence of known confounding variables on model performance
Lastly, we performed several control analyses to examine if the models that significantly predicted response time variability (i.e., task-FC derived models) in the HCP-Aging dataset are robust to known confounding variables such as age, sex, study sites, choice of cross-validation method, and choice of parcellation scheme. To this end, we included head motion jointly with each of the variables as covariates at the edge selection stage of CPM LOOCV analysis. Focusing on the combined model (i.e., model built by subtracting the network strength of the low RT_CV model from the network strength of the high RT_CV model), we found robust, but relatively low model performance when controlling for the potential effect of sex and age (marginally significant). Predictions remained largely unchanged after controlling for the potential effect of study sites (Supplementary Table 1). In line with a recent CPM study (Goldfarb et al., 2020), we determined the robustness of model performance in the repeated k-fold (k = 2, 5, and 10) cross-validation analysis using Cohen’s d effect size. Crucially, we found that the combined models robustly predict true RT_CV scores better than null values as demonstrated by large-to-moderate effect size (Supplementary Table 2). Finally, we found that model performance did not generalize to an alternative functional parcellation scheme (Supplementary Fig. 1).
4. Discussion
In this study, we developed a predictive model of response time variability in healthy older adults, using whole-brain functional connectivity, and tested the model’s ability to predict response time variability in two independent healthy elderly cohorts. First, we were able to demonstrate that individual response time variability, assessed using RT_CV, can be reliably predicted from task-based, whole-brain functional connectivity, but not from resting-state connectivity. The task-based predictive model was robust to the effect of sex, study sites, cross-validation method, and age (marginally significant); however, the model was not robust to an alternative functional parcellation scheme. Second, our analysis provided support for the differential involvement of key canonical networks, namely the somatomotor network, dorsal attention network, ventral attention network, visual network, frontoparietal network and the default mode network in high and low response time variability networks. Finally, despite within-sample prediction using task connectivity, our models did not generalize to predict individual response time variability using resting-state fMRI data either within the derivation sample or in the two independent healthy elderly cohorts. We discuss our results in detail below.
This work, to our knowledge, is the first study to utilize a machine learning approach and whole-brain functional connectivity to predict individual response time variability as an avenue to study brain correlates of mind-wandering in healthy older adults. Our results are consistent with the emerging literature demonstrating that machine learning approaches, particularly the CPM framework, can be utilized to predict individual cognitive outcomes from functional connectivity (Avery et al., 2020; Feng et al., 2019; Finn et al., 2015; Gao et al., 2020; Jangraw et al., 2018; Kucyi et al., 2021; Rosenberg et al., 2016). The finding that rest-FC variance solely did not predict RT_CV is consistent with prior CPM work demonstrating that predictive utility is suboptimal when brain-based models of individual cognitive measures are trained on resting-state functional connectivity (Greene et al., 2018; Jiang et al., 2020; Tomasi and Volkow, 2020). It also suggests that task functional connectivity captures more connections relevant for the prediction of response time variability than resting-state functional connectivity.
Our supplementary analyses tested for the robustness of our predictive models against key confounds known to impact brain-behavior relationships. Our results suggested that the task-based combined model, providing an estimate of the fit between observed scores and the relative strengths of the high vs. low RT_CV networks, was robust after controlling for variance associated with age, sex, and study sites. The effect sizes from repeated k-fold cross-validation analysis also revealed that, despite the relatively low mean correlations, the combined models robustly predict RT_CV better than null models as demonstrated by the moderate-to-large effect size differences. In contrast, however, we observed a significant drop in the fit between observed and predicted scores from the combined model using an alternate functional parcellation scheme. The choice of brain parcellation scheme in machine learning pipelines has recently been garnering attention with variations of parcellation schemes demonstrating a sizeable impact on the reliability and prediction accuracy of machine learning algorithms (Dadi et al., 2019). Regions of interest (ROIs), which serve as nodes to extract timeseries data, can be defined based on coordinates from existing literature; these functional parcellations were the basis of the Shen atlas and McNabb atlas employed in the current study. ROIs can additionally be defined based on anatomy, like the AAL (Tzourio-Mazoyer et al., 2002) or using data-driven approaches, such as k-means clustering or Independent Component Analysis (Beckmann and Smith, 2004). Dadi et al. (2019), comparing these approaches, found that connectomes built using functional parcellations outperformed those built using anatomical atlases with 150 brain parcellations being an optimal dimensionality for prediction accuracy. Thus, coarser parcellation resolutions may improve performance and yield more reliable biomarkers for network analyses (Abraham et al., 2017) or age prediction tasks (Liem et al., 2017). In our study, although the two parcellation schemes used were functionally defined, the number of brain regions was approximately 270. It is thus likely that the varied and specialized functional parcellations across the two atlases contributed to our lack of generalization across the two brain parcellation schemes. Future studies could benefit from adopting a more optimal approach that involves averaging across models trained on connectivity matrices from different parcellation schemes since determining the optimal atlas among the possible alternatives may not be feasible (Khosla et al., 2019).
Consistent with previous studies using the CPM framework, we demonstrate that the critical edges in our response time variability models were widespread across multiple brain networks. We begin by discussing the key findings in the high response time variability model. Emerging evidence suggests that along with the default mode network, mind-wandering is supported by other brain networks such as the frontoparietal and the dorsal attention networks (Christoff et al., 2009; Golchert et al., 2017; Hasenkamp et al., 2012; Smallwood et al., 2012). In line with these studies, we demonstrate that the high response time variability model, potentially indexing off-task thinking or fluctuations in attentional state, utilized large-scale networks that included the somatomotor, ventral attention, cerebellar, visual, and default mode networks. Specifically, our findings revealed that relative to the other brain networks, connectivity within the visual, ventral, and subcortical networks were the most represented in the high response time variability model. Our finding that the within-network connectivity in the visual network contributed substantially to the high response time variability model may be related to a previous finding that implicated certain networks, including the visual network in a brain state that associates with suboptimal task performance and increased reaction time variability (Yamashita et al., 2021). Another recent study suggests that the visual network is sensitive to perturbations from mind-wandering (Zuberer et al., 2021). Likewise, previous studies have highlighted the importance of the nodes within the subcortical network in greater reaction time variability (Bellgrove et al., 2004) and mind-wandering (Christoff et al., 2016). Furthermore, we established that although the connectivity within the somatomotor network was not utilized for high reaction time variability prediction, this network had the highest overall representation when considering its interactions with other networks. This finding suggests that its role in predicting high reaction time variability may be more reflected in its interaction with other networks. Additionally, we observed that the inter-connectivity of the default mode network with the somatomotor and ventral attention networks were highly overrepresented. This pattern of result is consistent with previous research highlighting that the default mode network couples with other prominent brain networks during mind-wandering (Godwin et al., 2017).
The low response time variability model, characterizing functional connections that were stronger during consistent responding, had high representation of nodes from the frontoparietal, dorsal attention, limbic, and the ventral attention networks. We also found that the strongest inter-network representation was between the frontoparietal and dorsal attention networks, with both networks represented in the top networks utilized by the low response time variability model when considering their mean overall contributions. Broadly, these findings are not surprising given existing evidence demonstrating that the frontoparietal network couples with the dorsal attention network in support of externally focused cognition (Spreng et al., 2010), which may reflect reduced variability in response time. Moreover, the high intra-network connectivity of the dorsal attention network within the low response time variability model may be related to evidence suggesting that the dorsal attention network directs attention externally to the task at hand (Corbetta et al., 2008), which may help explain why the connectivity within this network is only represented in the low response time variability model. Additionally, we found comparable contribution of the ventral attention network to the high and low response time variability models, which leads us to speculate that the functional contribution of this network to response time variability is not homogenous. Future research is needed to elucidate the potential functional heterogeneity within this brain network during response time variability.
The finding that our models did not generalize to predict response time variability from resting-state connectivity in two independent datasets of healthy, older adults is particularly striking given that previous studies have demonstrated the utility of the CPM framework to build generalizable models of various cognitive outcomes in novel datasets (Avery et al., 2020; Beaty et al., 2018; Gao et al., 2019; Jiang et al., 2018; Kucyi et al., 2021; Rosenberg et al., 2020). For example, a recent fMRI study in younger adults operationalized mind-wandering as stimulus-independent, task-unrelated thoughts (SITUT) and showed that the CPM methodology can be used to build generalizable models of mind-wandering in healthy young adults and ADHD adults (Kucyi et al., 2021). A major difference between the current findings and those from Kucyi et al. (2021) is that many of the intra-network and inter-network connectivity that contributed strongly to the SITUT models were not utilized to the same extent by our models. Nonetheless, we observed some similarities between the top network pairs utilized by the high response time variability model in the current study and the high SITUT model (i.e., the model with positive correlation with SITUT) from Kucyi et al. (2021). These include connectivity between the frontoparietal—ventral attention, ventral attention—default mode, subcortical—frontoparietal, and somatomotor—dorsal attention networks. Despite both models having some top network pairs in common, the network pairs with the greatest contribution to each model were different. Relatedly, the low response time variability and low SITUT models utilized different top network for prediction. On the one hand, these largely discrepant observations align with prior report of independent neural correlates of response time variability and self-reports of mind-wandering (Kucyi et al., 2016). On the other hand, the similarities observed between the high response time variability and high SITUT models may suggest an association between increased response time variability and increased self-reports of mind-wandering episodes in older adults (Bastian and Sackur, 2013; Henríquez et al., 2016; Jubera-García et al., 2020; Kucyi et al., 2016; Maillet et al., 2020).
The inability of our model to generalize to independent cohorts may be related to inherent methodological differences, such as the variation between task and resting-state connectivity, between the derivation and validation datasets. Although there is evidence that functional connectivity at rest reflects reliable fingerprints of individual differences (Finn et al., 2015; Miranda-Dominguez et al., 2014), it is plausible that resting-state connectivity in both derivation and validation datasets did not sufficiently capture variance in intrinsic connectivity necessary for predicting response time variability. Another possible explanation is the effect of varying degree of difficulty across the tasks performed by participants in the derivation and both validation datasets. According to the executive-resource hypothesis, task-related and task-unrelated thoughts compete for limited executive resources, implying that when the primary task is difficult, there is reduced or no resources available for mind-wandering to occur, and consequently result in a decrease in response time variability. This pattern is reversed when the primary task is easy as mind-wandering tends to utilize unused executive resources available (Smallwood and Schooler, 2006). Empirical support for this hypothesis comes from prior studies that showed relationship between task difficulty and mind-wandering (Baird et al., 2012; Konishi et al., 2015; Smallwood et al., 2011; Thomson et al., 2013). Although all three datasets employed in the current study included variants of Go/No-Go tasks, it is plausible that the three tasks taxed executive control resources differentially. As such, our models derived based on the HCP data, did not fully capture nuances in response time variability patterns.
Although the current study provides novel contribution to the understanding of brain networks implicated in response time variability in healthy aging, several limitations should be noted. First, the task duration in the derivation dataset was relatively shorter than those used in the majority of previous behavioral studies that assessed mind-wandering objectively with RT_CV during Go/NoGo task performance (Carriere et al., 2010; Hu et al., 2012). A longer task time could allow the capture of more robust response time variability during ongoing task. Second, although we controlled for the potential effect of known confounds such as sex, age, head motion, and study sites on model performance, we did not control for potential effect of individual differences in cortical thickness. Prior work has shown that task-unrelated thought associates with individual differences in cortical thickness of the medial prefrontal and anterior cingulate cortices (Bernhardt et al., 2014). Third, our models were derived based on an objective marker that has shown promise as an indirect measure of mind-wandering. Future studies that combine self-report, behavioral measures, and neurocognitive measures (Martinon et al., 2019; Smallwood and Schooler, 2015) may allow for the development of a more robust and generalizable models of mind-wandering in aging. Fourth, the finding that our model performance generalizes poorly to an alternative atlas suggests an influence of the choice of parcellation scheme. There is a clear and urgent need to better understand the impact of parcellation pipelines on prediction accuracy in older adults and clinical populations. Finally, considering the inability of our model to generalize to independent datasets of older adults, future studies leveraging a design that addresses these limitations could potentially result in an improved predictive model that generalizes to predict response time variability and mind-wandering in multiple validation datasets.
Despite these limitations, the present study demonstrates that within a population of healthy older adults, response time variability—an indirect marker of mind-wandering—can be reliably predicted from whole-brain task-based functional connectivity. However, resting-state functional connectivity alone did not explain the significant variance. Our results also provide support for evidence that the default mode network, along with other networks such as frontoparietal, dorsal attention, visual, somatomotor and ventral attention, contributes to individual variability in response time. Collectively, these findings provide new insights into the neural mechanisms underlying an indirect marker of mind-wandering and further underscore the importance of individual differences and a whole-brain approach in fMRI studies of mind-wandering. Future research with task-based fMRI datasets of RT_CV is necessary to further assess the generalizability of our model.
Supplementary Material
Acknowledgments
This work was supported by the National Institute on Aging of the National Institutes of Health (R01AG054427 awarded to RSP). Some of the data and/or research tools used in the preparation of this manuscript were obtained from the National Institute of Mental Health (NIMH) Data Archive (NDA). NDA is a collaborative informatics system created by the National Institutes of Health to provide a national resource to support and accelerate research in mental health. Dataset identifier(s): NIMH Data Archive Collection ID: #1155; NIMH Data Archive Digital Object Identifier: 10.15154/1521345. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or of the Submitters submitting original data to NDA. Additional data used in the preparation of this article was provided by the Cambridge Centre for Ageing and Neuroscience (CamCAN). CamCAN funding was provided by the United Kingdom Biotechnology and Biological Sciences Research Council (grant number BB/H008217/1), together with support from the United Kingdom Medical Research Council and University of Cambridge, United Kingdom. Finally, we would like to thank the Ohio Supercomputer Center for providing valuable computational resources used for data preprocessing.
Footnotes
Declaration of Competing Interest
The authors declare no competing financial interests.
Credit authorship contribution statement
Oyetunde Gbadeyan: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Visualization. James Teng: Formal analysis, Writing – review & editing. Ruchika Shaurya Prakash: Conceptualization, Methodology, Writing – review & editing, Supervision, Funding acquisition.
Code and data availability
The code used to run the CPM analysis is available at: https://github.com/YaleMRRC/CPM. HCP-Aging brain imaging and behavioral data are available at: https://www.humanconnectome.org/study/hcp-lifespan-aging/data-releases. Likewise, brain imaging and behavioral data from the Cam-CAN project are available at: https://camcan-archive.mrc-cbu.cam.ac.uk/dataaccess/. Behavioral and brain imaging data from the SCAN cohort will be made available upon request by qualified investigators and approval from the institution.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2022.118890.
References
- Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, Varoquaux G, 2017. Deriving reproducible biomarkers from multi-site resting-state data: an Autism-based example. Neuroimage 147, 736–745. [DOI] [PubMed] [Google Scholar]
- Abraham A, Pedregosa F, Eickenberg M, Gervais P, Mueller A, Kossaifi J, Gramfort A, Thirion B, Varoquaux G, 2014. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform 8, 14. doi: 10.3389/fninf.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan Cheyne J, Solman GJF, Carriere JSA, Smilek D, 2009. Anatomy of an error: a bidirectional state model of task engagement/disengagement and attention-related errors. Cognition 111, 98–113. doi: 10.1016/j.cognition.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal 12, 26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery EW, Yoo K, Rosenberg MD, Greene AS, Gao S, Na DL, Scheinost D, Constable TR, Chun MM, 2020. Distributed patterns of functional connectivity predict working memory performance in novel healthy and memory-impaired individuals. J. Cognit. Neurosci 32, 241–255. doi: 10.1162/jocn_a_01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird B, Smallwood J, Mrazek MD, Kam JWY, Franklin MS, Schooler JW, 2012. Inspired by distraction: mind wandering facilitates creative incubation. Psychol. Sci 23, 1117–1122. doi: 10.1177/0956797612446024. [DOI] [PubMed] [Google Scholar]
- Barron DS, Gao S, Dadashkarimi J, Greene AS, Spann MN, Noble S, Lake EMR, Krystal JH, Constable RT, Scheinost D, 2020. Transdiagnostic, connectome-based prediction of memory constructs across psychiatric disorders. Cereb. Cortex doi: 10.1093/cercor/bhaa371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Sackur J, 2013. Mind wandering at the fingertips: automatic parsing of subjective states based on response time variability. Front. Psychol.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty RE, Kenett YN, Christensen AP, Rosenberg MD, Benedek M, Chen Q, Fink A, Qiu J, Kwapil TR, Kane MJ, Silvia PJ, 2018. Robust prediction of individual creative ability from brain functional connectivity. Proc. Natl. Acad. Sci 115, 1087–1092. doi: 10.1073/pnas.1713532115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM, 2004. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging 23, 137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT, 2007. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage 37, 90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellgrove MA, Hester R, Garavan H, 2004. The functional neuroanatomical correlates of response variability: evidence from a response inhibition task. Neuropsychologia 42, 1910–1916. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. [Google Scholar]
- Bernhardt BC, Smallwood J, Tusche A, Ruby FJM, Engen HG, Steinbeis N, Singer T, 2014. Medial prefrontal and anterior cingulate cortical thickness predicts shared individual differences in self-generated thought and temporal discounting. Neuroimage 90, 290–297. doi: 10.1016/j.neuroimage.2013.12.040. [DOI] [PubMed] [Google Scholar]
- Bookheimer SY, Salat DH, Terpstra M, Ances BM, Barch DM, Buckner RL, Burgess GC, Curtiss SW, Diaz-Santos M, Elam JS, Fischl B, Greve DN, Hagy HA, Harms MP, Hatch OM, Hedden T, Hodge C, Japardi KC, Kuhn TP, Ly TK, Smith SM, Somerville LH, Uğurbil K, van der Kouwe A, Van Essen D, Woods RP, Yacoub E, 2019. The lifespan human connectome project in aging: an overview. Neuroimage 185, 335–348. doi: 10.1016/j.neuroimage.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braver TS, West R, 2008. Working memory, executive control, and aging.
- Carriere JSA, Cheyne JA, Solman GJF, Smilek D, 2010. Age trends for failures of sustained attention. Psychol. Aging 25, 569–574. doi: 10.1037/a0019363. [DOI] [PubMed] [Google Scholar]
- Christoff K, Gordon AM, Smallwood J, Smith R, Schooler JW, 2009. Experience sampling during fMRI reveals default network and executive system contributions to mind wandering. Proc. Natl. Acad. Sci. U.S.A 106, 8719–8724. doi: 10.1073/pnas.0900234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoff K, Irving ZC, Fox KCR, Spreng RN, Andrews-Hanna JR, 2016. Mind-wandering as spontaneous thought: a dynamic framework. Nat. Rev. Neurosci 17, 718–731. doi: 10.1038/nrn.2016.113. [DOI] [PubMed] [Google Scholar]
- Corbetta M, Patel G, Shulman GL, 2008. The reorienting system of the human brain: from environment to theory of mind. Neuron 58, 306–324. doi: 10.1016/j.neuron.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, Hyde JS, 1997. Software tools for analysis and visualization of fMRI data. NMR Biomed. 10, 171–178 doi:. [DOI] [PubMed] [Google Scholar]
- Craik FIM, Salthouse TA, 2011. The Handbook of Aging and Cognition. Psychology press. [Google Scholar]
- Dadi K, Rahim M, Abraham A, Chyzhyk D, Milham M, Thirion B, Varoquaux G, 2019. Benchmarking functional connectome-based predictive models for resting-state fMRI. Neuroimage 192, 115–134. doi: 10.1016/j.neuroimage.2019.02.062. [DOI] [PubMed] [Google Scholar]
- Dice LR, 1945. Measures of the amount of ecologic association between species. Ecology 26, 297–302. [Google Scholar]
- Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N, 2009. A probabilistic MR atlas of the human cerebellum. Neuroimage 46, 39–46. doi: 10.1016/j.neuroimage.2009.01.045. [DOI] [PubMed] [Google Scholar]
- Dosenbach NUF, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR, Barch DM, Petersen SE, Schlaggar BL, 2010. Prediction of individual brain maturity using fMRI. Science (80-.) 329, 1358. doi: 10.1126/science.1194144, LP–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J, Adolphs R, 2016. Building a science of individual differences from fMRI. Trends Cognit. Sci 20, 425–443. doi: 10.1016/j.tics.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, Kent JD, Goncalves M, DuPre E, Snyder M, Oya H, Ghosh SS, Wright J, Durnez J, Poldrack RA, Gorgolewski KJ, 2019. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116. doi: 10.1038/s41592-018-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterman M, Noonan SK, Rosenberg M, Degutis J, 2013. In the zone or zoning out? Tracking behavioral and neural fluctuations during sustained attention. Cereb. Cortex 23, 2712–2723. doi: 10.1093/cercor/bhs261. [DOI] [PubMed] [Google Scholar]
- Esterman M, Rosenberg MD, Noonan SK, 2014. Intrinsic fluctuations in sustained attention and distractor processing. J. Neurosci 34, 1724. doi: 10.1523/JNEUROSCI.2658-13.2014, LP –1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterman M, Rothlein D, 2019. Models of sustained attention. Curr. Opin. Psychol 29, 174–180. [DOI] [PubMed] [Google Scholar]
- Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, Yang Z, Chu C, Xie S, Laird AR, Fox PT, Eickhoff SB, Yu C, Jiang T, 2016. The human brainnetome atlas: a new brain atlas based on connectional architecture. Cereb. Cortex 26, 3508–3526. doi: 10.1093/cercor/bhw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Wang L, Li T, Xu P, 2019. Connectome-based individualized prediction of loneliness. Soc. Cognit. Affect. Neurosci 14, 353–365. doi: 10.1093/scan/nsz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT, 2015. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci 18, 1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortenbaugh FC, Rothlein D, McGlinchey R, DeGutis J, Esterman M, 2018. Tracking behavioral and neural fluctuations during sustained attention: a robust replication and extension. Neuroimage 171, 148–164. doi: 10.1016/j.neuroimage.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain-Zaragoza S, Manglani HR, Rosenberg MD, Andridge R, Prakash RS, 2021. Defining a connectome-based predictive model of attentional control in aging. bioRxiv doi: 10.1101/2021.02.02.429232, 2021.02.02.429232. [DOI] [Google Scholar]
- Fountain-Zaragoza S, Puccetti NA, Whitmoyer P, Prakash RS, 2018. Aging and attentional control: examining the roles of mind-wandering propensity and dispositional mindfulness. J. Int. Neuropsychol. Soc 24, 876–888. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Nara B, Zavagnin M, Touron DR, Kane MJ, 2015. Validating older adults’ reports of less mind-wandering: an examination of eye movements and dispositional influences. Psychol. Aging 30, 266–278. doi: 10.1037/pag0000031. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RSJ, Turner R, 1996. Movement-related effects in fMRI time-series. Magn. Reson. Med 35, 346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Galéra C, Orriols L, M’Bailara K, Laborey M, Contrand B, Ribéreau-Gayon R, Masson F, Bakiri S, Gabaude C, Fort A, Maury B, Lemercier C, Cours M, Bouvard MP, Lagarde E, 2012. Mind wandering and driving: responsibility case-control. BMJ 345, 1–7. doi: 10.1136/bmj.e8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wong CHY, Huang H, Shao R, Huang R, Chan CCH, Lee TMC, 2020. Connectome-based models can predict processing speed in older adults. Neuroimage 223, 117290. doi: 10.1016/j.neuroimage.2020.117290. [DOI] [PubMed] [Google Scholar]
- Gao S, Greene AS, Constable RT, Scheinost D, 2019. Combining multiple connectomes improves predictive modeling of phenotypic measures. Neuroimage 201, 116038. doi: 10.1016/j.neuroimage.2019.116038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, 2013. The minimal preprocessing pipelines for the human connectome project. Neuroimage 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisky EL, 2007. Changes in cognitive function in human aging. Brain Aging 3–20. [PubMed] [Google Scholar]
- Godwin CA, Hunter MA, Bezdek MA, Lieberman G, Elkin-Frankston S, Romero VL, Witkiewitz K, Clark VP, Schumacher EH, 2017. Functional connectivity within and between intrinsic brain networks correlates with trait mind wandering. Neuropsychologia 103, 140–153. doi: 10.1016/j.neuropsychologia.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Golchert J, Smallwood J, Jefferies E, Seli P, Huntenburg JM, Liem F, Lauckner ME, Oligschläger S, Bernhardt BC, Villringer A, Margulies DS, 2017. Individual variation in intentionality in the mind-wandering state is reflected in the integration of the default-mode, fronto-parietal, and limbic networks. Neuroimage 146, 226–235. doi: 10.1016/j.neuroimage.2016.11.025. [DOI] [PubMed] [Google Scholar]
- Goldfarb EV, Rosenberg MD, Seo D, Constable RT, Sinha R, 2020. Hippocampal seed connectome-based modeling predicts the feeling of stress. Nat. Commun 11, 1–10. doi: 10.1038/s41467-020-16492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, Flandin G, Ghosh SS, Glatard T, Halchenko YO, Handwerker DA, Hanke M, Keator D, Li X, Michael Z, Maumet C, Nichols BN, Nichols TE, Pellman J, Poline JB, Rokem A, Schaefer G, Sochat V, Triplett W, Turner JA, Varoquaux G, Poldrack RA, 2016. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 1–9. doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AS, Gao S, Nobel S, Scheinost D, Constable RT, 2020. How Tasks Change Whole-Brain Functional Organization to Reveal Brain-Phenotype Relationships. Cell reports 8, 25–32. doi: 10.1016/j.celrep.2020.108066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AS, Gao S, Scheinost D, Constable RT, 2018. Task-induced brain state manipulation improves prediction of individual traits. Nat. Commun 9. doi: 10.1038/s41467-018-04920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Fischl B, 2009. Accurate and robust brain image alignment using boundary-based registration. Neuroimage 48, 63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics M, Balota DA, Jackson JD, 2018. Mind-wandering in healthy aging and early stage Alzheimer’s disease. Neuropsychology 32, 89–101. doi: 10.1037/neu0000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MP, Somerville LH, Ances BM, Andersson J, Barch DM, Bastiani M, Bookheimer SY, Brown TB, Buckner RL, Burgess GC, Coalson TS, Chappell MA, Dapretto M, Douaud G, Fischl B, Glasser MF, Greve DN, Hodge C, Jamison KW, Jbabdi S, Kandala S, Li X, Mair RW, Mangia S, Marcus D, Mascali D, Moeller S, Nichols TE, Robinson EC, Salat DH, Smith SM, Sotiropoulos SN, Terpstra M, Thomas KM, Tisdall MD, Ugurbil K, van der Kouwe A, Woods RP, Zöllei L, Van Essen DC, Yacoub E, 2018. Extending the human connectome project across ages: imaging protocols for the lifespan development and aging projects. Neuroimage 183, 972–984. doi: 10.1016/j.neuroimage.2018.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenkamp W, Wilson-Mendenhall CD, Duncan E, Barsalou LW, 2012. Mind wandering and attention during focused meditation: a fine-grained temporal analysis of fluctuating cognitive states. Neuroimage 59, 750–760. doi: 10.1016/j.neuroimage.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Hasher L, Zacks RT, 1988. Working memory, comprehension, and aging: A review and a new view. Psychology of learning and motivation 22, 193–225. doi: 10.1016/S0079-7421(08)60041-9. [DOI] [Google Scholar]
- He J, Becic E, Lee YC, McCarley JS, 2011. Mind wandering behind the wheel: performance and oculomotor correlates. Hum. Factors 53, 13–21. doi: 10.1177/0018720810391530. [DOI] [PubMed] [Google Scholar]
- Henríquez RA, Chica AB, Billeke P, Bartolomeo P, 2016. Fluctuating minds: spontaneous psychophysical variability during mind-wandering. PLoS One 11, e0147174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, He S, Xu B, 2012. Different efficiencies of attentional orienting in different wandering minds. Conscious. Cognit 21, 139–148. doi: 10.1016/j.concog.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Jackson JD, Balota DA, 2012. Mind-wandering in younger and older adults: converging evidence from the sustained attention to response task and reading for comprehension. Psychol. Aging 27, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangraw DC, Gonzalez-Castillo J, Handwerker DA, Ghane M, Rosenberg MD, Panwar P, Bandettini PA, 2018. A functional connectivity-based neuromarker of sustained attention generalizes to predict recall in a reading task. Neuroimage 166, 99–109. doi: 10.1016/j.neuroimage.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, 2012. FSL. Neuroimage 62, 782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jiang R, Calhoun VD, Zuo N, Lin D, Li J, Fan L, Qi S, Sun H, Fu Z, Song M, Jiang T, Sui J, 2018. Connectome-based individualized prediction of temperament trait scores. Neuroimage 183, 366–374. doi: 10.1016/j.neuroimage.2018.08.038. [DOI] [PubMed] [Google Scholar]
- Jiang R, Zuo N, Ford JM, Qi S, Zhi D, Zhuo C, Xu Y, Fu Z, Bustillo J, Turner JA, Calhoun VD, Sui J, 2020. Task-induced brain connectivity promotes the detection of individual differences in brain-behavior relationships. Neuroimage 207, 116370. doi: 10.1016/j.neuroimage.2019.116370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubera-García E, Gevers W, Van Opstal F, 2020. Influence of content and intensity of thought on behavioral and pupil changes during active mind-wandering, off-focus, and on-task states. Atten. Percept. Psychophys 82, 1125–1135. doi: 10.3758/s13414-019-01865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MJ, Brown LH, McVay JC, Silvia PJ, Myin-Germeys I, Kwapil TR, 2007. For whom the mind wanders, and when. Psychol. Sci 18, 614–621. doi: 10.1111/j.1467-9280.2007.01948.x. [DOI] [PubMed] [Google Scholar]
- Khosla M, Jamison K, Kuceyeski A, Sabuncu MR, 2019. Ensemble learning with 3D convolutional neural networks for functional connectome-based prediction. Neuroimage 199, 651–662. doi: 10.1016/j.neuroimage.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killingsworth MA, Gilbert DT, 2010. A wandering mind is an unhappy mind. Science (80-.) 330, 932. 10.1126/science.1192439 [DOI] [PubMed] [Google Scholar]
- Konishi M, McLaren DG, Engen H, Smallwood J, 2015. Shaped by the past: the default mode network supports cognition that is independent of immediate perceptual input. PLoS One 10, e0132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawietz SA, Tamplin AK, Radvansky GA, 2012. Aging and mind wandering during text comprehension. Psychol. Aging 27, 951. [DOI] [PubMed] [Google Scholar]
- Kucyi A, 2018. Just a thought: how mind-wandering is represented in dynamic brain connectivity. Neuroimage 180, 505–514. doi: 10.1016/j.neuroimage.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Esterman M, Capella J, Green A, Uchida M, Biederman J, Gabrieli JDE, Valera EM, Whitfield-Gabrieli S, 2021. Prediction of stimulus-independent and task-unrelated thought from functional brain networks. Nat. Commun 12, 1793. doi: 10.1038/s41467-021-22027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucyi A, Esterman M, Riley CS, Valera EM, 2016. Spontaneous default network activity reflects behavioral variability independent of mind-wandering. Proc. Natl. Acad. Sci. U. S. A 113, 13899–13904. doi: 10.1073/pnas.1611743113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucyi A, Hove MJ, Esterman M, Hutchison RM, Valera EM, 2017. Dynamic brain network correlates of spontaneous fluctuations in attention. Cereb. Cortex 27, 1831–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux L, Salek-Haddadi A, Lund TE, Laufs H, Carmichael D, 2007. Modelling large motion events in fMRI studies of patients with epilepsy. Magn. Reson. Imaging 25, 894–901. doi: 10.1016/j.mri.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Li J, Kong R, Liégeois R, Orban C, Tan Y, Sun N, Holmes AJ, Sabuncu MR, Ge T, Yeo BTT, 2019. Global signal regression strengthens association between resting-state functional connectivity and behavior. Neuroimage 196, 126–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem F, Varoquaux G, Kynast J, Beyer F, Masouleh SK, Huntenburg JM, Lampe L, Rahim M, Abraham A, Craddock RC, 2017. Predicting brain-age from multimodal imaging data captures cognitive impairment. Neuroimage 148, 179–188. [DOI] [PubMed] [Google Scholar]
- Lin Q, Rosenberg MD, Yoo K, Hsu TW, O’Connell TP, Chun MM, 2018. Resting-state functional connectivity predicts cognitive impairment related to Alzheimer’s disease. Front. Aging Neurosci 10, 1–10. doi: 10.3389/fnagi.2018.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist MA, Geuter S, Wager TD, Caffo BS, 2019. Modular preprocessing pipelines can reintroduce artifacts into fMRI data. Hum. Brain Mapp 40, 2358–2376. doi: 10.1002/hbm.24528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet D, Beaty RE, Adnan A, Fox KCR, Turner GR, Spreng RN, 2019. Aging and the wandering brain: age-related differences in the neural correlates of stimulus-independent thoughts. PLoS One 14, e0223981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet D, Beaty RE, Jordano ML, Touron DR, Adnan A, Silvia PJ, Kwapil TR, Turner GR, Spreng RN, Kane MJ, 2018. Age-related differences in mind-wandering in daily life. Psychol. Aging 33, 643–653. doi: 10.1037/pag0000260. [DOI] [PubMed] [Google Scholar]
- Maillet D, Rajah MN, 2016. Assessing the neural correlates of task-unrelated thoughts during episodic encoding and their association with subsequent memory in young and older adults. J. Cognit. Neurosci 28, 826–841. doi: 10.1162/jocn_a_00935. [DOI] [PubMed] [Google Scholar]
- Maillet D, Schacter DL, 2016. From mind wandering to involuntary retrieval: age-related differences in spontaneous cognitive processes. Neuropsychologia 80, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet D, Yu L, Hasher L, Grady CL, 2020. Age-related differences in the impact of mind-wandering and visual distraction on performance in a go/no-go task. Psychol. Aging 35, 627–638. doi: 10.1037/pag0000409. [DOI] [PubMed] [Google Scholar]
- Manglani HR, Fountain-Zaragoza S, Shankar A, Nicholas JA, Prakash RS, 2021. Employing Connectome-Based Models to Predict Working Memory in Multiple Sclerosis. Brain Connect. doi: 10.1089/brain.2021.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon LM, Smallwood J, McGann D, Hamilton C, Riby LM, 2019. The disentanglement of the neural and experiential complexity of self-generated thoughts: a users guide to combining experience sampling with neuroimaging data. Neuroimage 192, 15–25. doi: 10.1016/j.neuroimage.2019.02.034. [DOI] [PubMed] [Google Scholar]
- Mason MF, Norton MI, Horn J.D.Van, Wegner DM, Grafton ST, Macrae CN, Mason MF, Norton MI, Horn JDV, Wegner DM, Grafton ST, Macrae CN, 2007. Wandering minds: stimulus-independent thought. Science (80-.) 315, 393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb CB, Tait RJ, McIlwain ME, Anderson VM, Suckling J, Kydd RR, Russell BR, 2018. Functional network dysconnectivity as a biomarker of treatment resistance in schizophrenia. Schizophr. Res 195, 160–167. doi: 10.1016/j.schres.2017.10.015. [DOI] [PubMed] [Google Scholar]
- McVay JC, Meier ME, Touron DR, Kane MJ, 2013. Aging ebbs the flow of thought: adult age differences in mind wandering, executive control, and self-evaluation. Acta Psychol. (Amst). 142, 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Mills BD, Carpenter SD, Grant KA, Kroenke CD, Nigg JT, Fair DA, 2014. Connectotyping: model based fingerprinting of the functional connectome. PLoS One 9, e111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek MD, Smallwood J, Schooler JW, 2012. Mindfulness and mind-wandering: finding convergence through opposing constructs. Emotion 12, 442. [DOI] [PubMed] [Google Scholar]
- Nasreddine ZS, Phillips NA, Bédirian V, Charbonneau S, Whitehead V, Collin I, Cummings JL, Chertkow H, 2005. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J. Am. Geriatr. Soc 53, 695–699. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- Noble S, Spann MN, Tokoglu F, Shen X, Constable RT, Scheinost D, 2017. Influences on the test-retest reliability of functional connectivity MRI and its relationship with behavioral utility. Cereb. Cortex 27, 5415–5429. doi: 10.1093/cercor/bhx230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan C, Shine JM, Lewis SJG, Andrews-Hanna JR, Irish M, 2015. Shaped by our thoughts - a new task to assess spontaneous cognition and its associated neural correlates in the default network. Brain Cognit. 93, 1–10. doi: 10.1016/j.bandc.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Park DC, Lautenschlager G, Hedden T, Davidson NS, Smith AD, Smith PK, 2002. Models of visuospatial and verbal memory across the adult life span. Psychol. Aging 17, 299. [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE, 2014. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 84, 320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risko EF, Anderson N, Sarwal A, Engelhardt M, Kingstone A, 2012. Everyday attention: variation in mind wandering and memory in a lecture. Appl. Cognit. Psychol 26, 234–242. doi: 10.1002/acp.1814. [DOI] [Google Scholar]
- Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen X, Constable RT, Chun MM, 2016. A neuromarker of sustained attention from whole-brain functional connectivity. Nat. Neurosci 19, 165–171. doi: 10.1038/nn.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Scheinost D, Greene AS, Avery EW, Kwon YH, Finn ES, Ramani R, Qiu M, Todd Constable R, Chun MM, 2020. Functional connectivity predicts changes in attention observed across minutes, days, and months. Proc. Natl. Acad. Sci. U. S. A 117, 3797–3807. doi: 10.1073/pnas.1912226117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooler JW, Mrazek MD, Franklin MS, Baird B, Mooneyham BW, Zedelius C, Broadway JM, 2014. The middle way: finding the balance between mindfulness and mind-wandering. Psychol. Learn. Motiv 60, 1–33. [Google Scholar]
- Seli P, Cheyne JA, Smilek D, 2013. Wandering minds and wavering rhythms: linking mind wandering and behavioral variability. J. Exp. Psychol. Hum. Percept. Perform 39, 1. [DOI] [PubMed] [Google Scholar]
- Seli P, Kane MJ, Smallwood J, Schacter DL, Maillet D, Schooler JW, Smilek D, 2018. Mind-wandering as a natural kind: a family-resemblances view. Trends Cognit. Sci 22, 479–490. doi: 10.1016/j.tics.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seli P, Maillet D, Smilek D, Oakman JM, Schacter DL, 2017. Cognitive aging and the distinction between intentional and unintentional mind wandering. Psychol. Aging 32, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafto MA, Tyler LK, Dixon M, Taylor JR, Rowe JB, Cusack R, Calder AJ, Marslen-Wilson WD, Duncan J, Dalgleish T, Henson RN, Brayne C, Matthews FE, Cam-CAN, 2014. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) study protocol: a cross-sectional, lifespan, multidisciplinary examination of healthy cognitive ageing. BMC Neurol 14, 204. doi: 10.1186/s12883-014-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shake MC, Shulley LJ, Soto-Freita AM, 2016. Effects of individual differences and situational features on age differences in mindless reading. J. Gerontol. Ser. B Psychol. Sci. Soc. Sci 71, 808–820. [DOI] [PubMed] [Google Scholar]
- Shen X, Finn ES, Scheinost D, Rosenberg MD, Chun MM, Papademetris X, Constable RT, 2017. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc 12, 506–518. doi: 10.1038/nprot.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Tokoglu F, Papademetris X, Constable RT, 2013. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage 82, 403–415. doi: 10.1016/j.neuroimage.2013.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Power JD, Dubis JW, Vogel AC, Church JA, Schlaggar BL, Petersen SE, 2014. Statistical improvements in functional magnetic resonance imaging analyses produced by censoring high-motion data points. Human brain mapping 35, 1981–1996. doi: 10.1002/hbm.22307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladky R, Friston KJ, Tröstl J, Cunnington R, Moser E, Windischberger C, 2011. Slice-timing effects and their correction in functional MRI. Neuroimage 58, 588–594. doi: 10.1016/j.neuroimage.2011.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood J, Brown K, Baird B, Schooler JW, 2012. Cooperation between the default mode network and the frontal-parietal network in the production of an internal train of thought. Brain Res 1428, 60–70. doi: 10.1016/j.brainres.2011.03.072. [DOI] [PubMed] [Google Scholar]
- Smallwood J, Brown KS, Tipper C, Giesbrecht B, Franklin MS, Mrazek MD, Carlson JM, Schooler JW, 2011. Pupillometric evidence for the decoupling of attention from perceptual input during offline thought. PLoS One 6, e18298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood J, Karapanagiotidis T, Ruby F, Medea B, de Caso I, Konishi M, Wang HT, Hallam G, Margulies DS, Jefferies E, 2016. Representing representation: integration between the temporal lobe and the posterior cingulate influences the content and form of spontaneous thought. PLoS One 11, e0152272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood J, Schooler JW, 2015. The science of mind wandering: empirically navigating the stream of consciousness. Annu. Rev. Psychol 66, 487–518. doi: 10.1146/annurev-psych-010814-015331. [DOI] [PubMed] [Google Scholar]
- Smallwood J, Schooler JW, 2006. The restless mind. Psychol. Bull doi: 10.1037/0033-2909.132.6.946. [DOI] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Andersson J, Auerbach EJ, Bijsterbosch J, Douaud G, Duff E, Feinberg DA, Griffanti L, Harms MP, 2013. Resting-state fMRI in the human connectome project. Neuroimage 80, 144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville LH, Bookheimer SY, Buckner RL, Burgess GC, Curtiss SW, Dapretto M, Elam JS, Gaffrey MS, Harms MP, Hodge C, Kandala S, Kastman EK, Nichols TE, Schlaggar BL, Smith SM, Thomas KM, Yacoub E, Van Essen DC, Barch DM, 2018. The lifespan human connectome project in development: a large-scale study of brain connectivity development in 5–21 year olds. Neuroimage 183, 456–468. doi: 10.1016/j.neuroimage.2018.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubelet A, Salthouse TA, 2011. Influence of social desirability on age differences in self-reports of mood and personality. J. Pers 79, 741–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng RN, Stevens WD, Chamberlain JP, Gilmore AW, Schacter DL, 2010. Default network activity, coupled with the frontoparietal control network, supports goal-directed cognition. Neuroimage 53, 303–317. doi: 10.1016/j.neuroimage.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JR, Williams N, Cusack R, Auer T, Shafto MA, Dixon M, Tyler LK, Cam-CAN, Henson RN, 2017. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) data repository: structural and functional MRI, MEG, and cognitive data from a cross-sectional adult lifespan sample. Neuroimage 144, 262–269. doi: 10.1016/j.neuroimage.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fisch B, Liu H, Buckner RL, 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol 106, 1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D, Besner D, Smilek D, 2013. In pursuit of off-task thought: mind wandering-performance trade-offs while reading aloud and color naming. Front. Psychol 4, 360. doi: 10.3389/fpsyg.2013.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND, 2020. Network connectivity predicts language processing in healthy adults. Hum. Brain Mapp 41, 3696–3708. doi: 10.1002/hbm.25042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Drob EM, Johnson KE, Jones RN, 2009. The cognitive reserve hypothesis: a longitudinal examination of age-associated declines in reasoning and processing speed. Dev. Psychol 45, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull A, Wang HT, Schooler JW, Jefferies E, Margulies DS, Smallwood J, 2019. The ebb and flow of attention: between-subject variation in intrinsic connectivity and cognition associated with the dynamics of ongoing experience. Neuroimage 185, 286–299. doi: 10.1016/j.neuroimage.2018.09.069. [DOI] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: improved N3 Bias correction. IEEE Trans. Med. Imaging 29, 1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M, 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Unsworth N, McMillan BD, 2013. Mind wandering and reading comprehension: examining the roles of working memory capacity, interest, motivation, and topic experience. J. Exp. Psychol. Learn. Mem. Cognit 39, 832–842. doi: 10.1037/a0029669. [DOI] [PubMed] [Google Scholar]
- Vallat R, 2018. Pingouin: statistics in Python. J. Open Source Softw. 3, 1026. [Google Scholar]
- Varoquaux G, Raamana PR, Engemann DA, Hoyos-Idrobo A, Schwartz Y, Thirion B, 2017. Assessing and tuning brain decoders: cross-validation, caveats, and guidelines. Neuroimage 145, 166–179. doi: 10.1016/j.neuroimage.2016.10.038. [DOI] [PubMed] [Google Scholar]
- Wang K, Yu C, Xu Lijuan, Qin W, Li K, Xu Lin, Jiang T, 2009. Offline memory reprocessing: involvement of the brain’s default network in spontaneous thought processes. PLoS One 4. doi: 10.1371/journal.pone.0004867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmoyer P, Fountain-Zaragoza S, Andridge R, Bredemeier K, Londeree A, Kaye L, Prakash RS, 2020. Mindfulness training and attentional control in older adults: a randomized controlled trial. Mindfulness 11, 203–218. [Google Scholar]
- Xia M, Wang J, He Y, 2013. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS One 8, e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Rothlein D, Kucyi A, Valera EM, Esterman M, 2021. Brain state-based detection of attentional fluctuations and their modulation. Neuroimage 236, 118072. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S, 2001. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zuberer A, Kucyi A, Yamashita A, Wu CM, Walter M, Valera EM, Esterman M, 2021. Integration and segregation across large-scale intrinsic brain networks as a marker of sustained attention and task-unrelated thought. Neuroimage 229, 117610. doi: 10.1016/j.neuroimage.2020.117610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


