Summary:
Resident memory T cells (Trms) predominantly reside within tissue and are critical for providing rapid protection against invasive viruses and bacteria. Given that tissues are heavily impacted and shaped by the microbiota, it stands to reason that Trms are also influenced by the microbiota that inhabits barrier sites. The influence of the microbiota is largely mediated by microbial production of metabolites which are crucial to the immune response to both viral infection and cancerous tumors. In addition to the effects of metabolites, antigens derived from the microbiota can activate T cell responses. While microbiota-specific T cells may assist in tissue repair, control of infection and anti-tumor immunity, the actual ‘memory’ potential of these cells remains unclear. Here, we hypothesize that memory responses to antigens from the microbiota must be ‘licensed’ by inflammatory signals activated by invasion of the host by microorganisms.
Keywords: Microbiota, Tissue resident memory T cells, CD4 T cells, CD8 T cells, Short Chain Fatty Acids
INTRODUCTION
Immune memory, the ability of B and T cells to make antigen-specific recall responses to counteract reinfection, is the cardinal attribute of the adaptive immune response. The evolution of adaptive immune cells with rearranged, variable antigen receptors that can respond to an almost unlimited number of antigens, coincides with the acquisition of a complex intestinal microbiome. It has been theorized that the purpose of the immune memory is not only to protect the host against re-infection, but also to ‘remember’ benign interactions with the microbiome to prevent continuous and potentially damaging responses1. Accordingly, the development, maintenance, and function of memory lymphocytes is likely responsive to signals derived from the microbiota.
Memory T cells can be broadly divided into three main categories Central (Tcm), Effector (Tem) Memory cells that circulate through secondary lymphoid tissue and Tissue Resident memory (Trm)2,3 that permanently reside in tissue. Tcm cells largely reside in secondary lymphoid tissue and are defined by the expression of surface proteins (CD62L, CCR7) and transcription factors (KLF2) that mediate that program. In contrast, Tem cells lack the expression of these surface molecules and traffic back and forth between the secondary lymphoid tissue and the peripheral blood/tissues. Tem and Tcm can interconvert in response to re-activation or the contraction of an immune response. In contrast, Trms reside permanently in the tissues and do not require reconstitution from the secondary lymphoid tissues, though this can occur under specific circumstances, such as re-infection4,3,5. Trms are important for protective immunity as they reside directly within the barrier tissue sites most commonly accessed by invasive microorganisms and can act immediately to prevent or limit infection6–8. While tissue resident memory has been most thoroughly studied in CD8+ T cells, it is also a property of CD4+ T cells and perhaps even B cells9–11. In this review, we will focus on T cells.
Barrier surfaces are inhabited by diverse populations of bacteria, fungi, protists and viruses that together compose the microbiota12. Metazoans have evolved in concert with a microbiota and as such the microbiota modulates multiple aspects of host physiology. For example, anaerobic intestinal bacteria are necessary for the digestion of complex carbohydrates and metabolites produced by that digestion are an important carbon source for the colonic epithelium. Microbiome-derived products and metabolites are also critical for the development and function of the immune system13. Environmental factors, most notably the diet, directly impact the composition of the microbiome, which in turn influences both the local immune response in adjacent host tissues and systemic immunity14,15. The microbiota affects both the innate and adaptive arms of the immune system and as such, is an important modifier of immunological memory.
Tissue resident memory T cells (Trms) are shaped by the tissue in which they reside. Since the microbiota is a dominant factor in shaping immunity at barrier surfaces, it follows that it also affects the Trm cells. The microbiota varies at different barrier tissue sites (small intestine, colon, mouth, skin, etc)16 and can differ enormously according to the condition of the tissue (infection, wounding, antibiotics, etc)17–20. Thus, Trms will be subject to microbiota-derived cues that differ according to the tissue and also local environmental conditions. In addition, the microbiome also drives the development of microbiota-specific T cell responses that are shaped by the type of microbe carrying the antigenic peptide, tissue location, and environmental context. Here we will review how the microbiota shapes CD8+ and CD4+ Trm development, survival, and function.
SECTION 1. THE MICROBIOTA AND MICROBIOTA-DERIVED METABOLITES IN TRM DEVELOPMENT
The development of Trms (and all other memory T cell subsets) begins when naïve T cells are activated in the lymph nodes by dendritic cells carrying antigens from the surrounding tissue. Signals imparted on dendritic cells by the tissue from which they are derived are critical to the differentiation of naïve T cells to effector T cells and ultimately, tissue-resident memory T cells. CD8+ Trms predominantly develop from ‘memory precursor’ CD8+ T cells, though there is also a route for Trms to develop from KLRG1+ ‘terminal effectors’21,22. Development of Trms is driven by inflammatory cytokines, such a IFNα and IL-12 and is directed by expression of transcription factors such as Blimp-1, Hobit and Ahr23. Upon activation, CD69 is upregulated transiently in T cells, but this signal is maintained in Trm cells24. CD69 expression antagonizes S1PR1 surface expression, thereby reducing T cell egress signals and results in tissue residency. In addition, CD8+ Trms that reside at epithelial sites such as the skin, but not the intestine upregulate adhesion molecules (CD103), chemokine receptors (CXCR3, CXCR6) and downregulate lymphoid homing receptors (CCR7), aiding in their ability to remain tissue-resident22,25. Once in the tissue, Trms continue to receive signals from their local environment, including cytokines (TGFβ)26–29 that are important for tissue retention and survival (IL-7 and IL-15)22,30,31. As we will discuss below, the microbiota can shape all of these aspects of Trm development and function at various tissue sites (Figure 1).
Figure 1 – Tissue resident memory signals regulated by the microbiota.
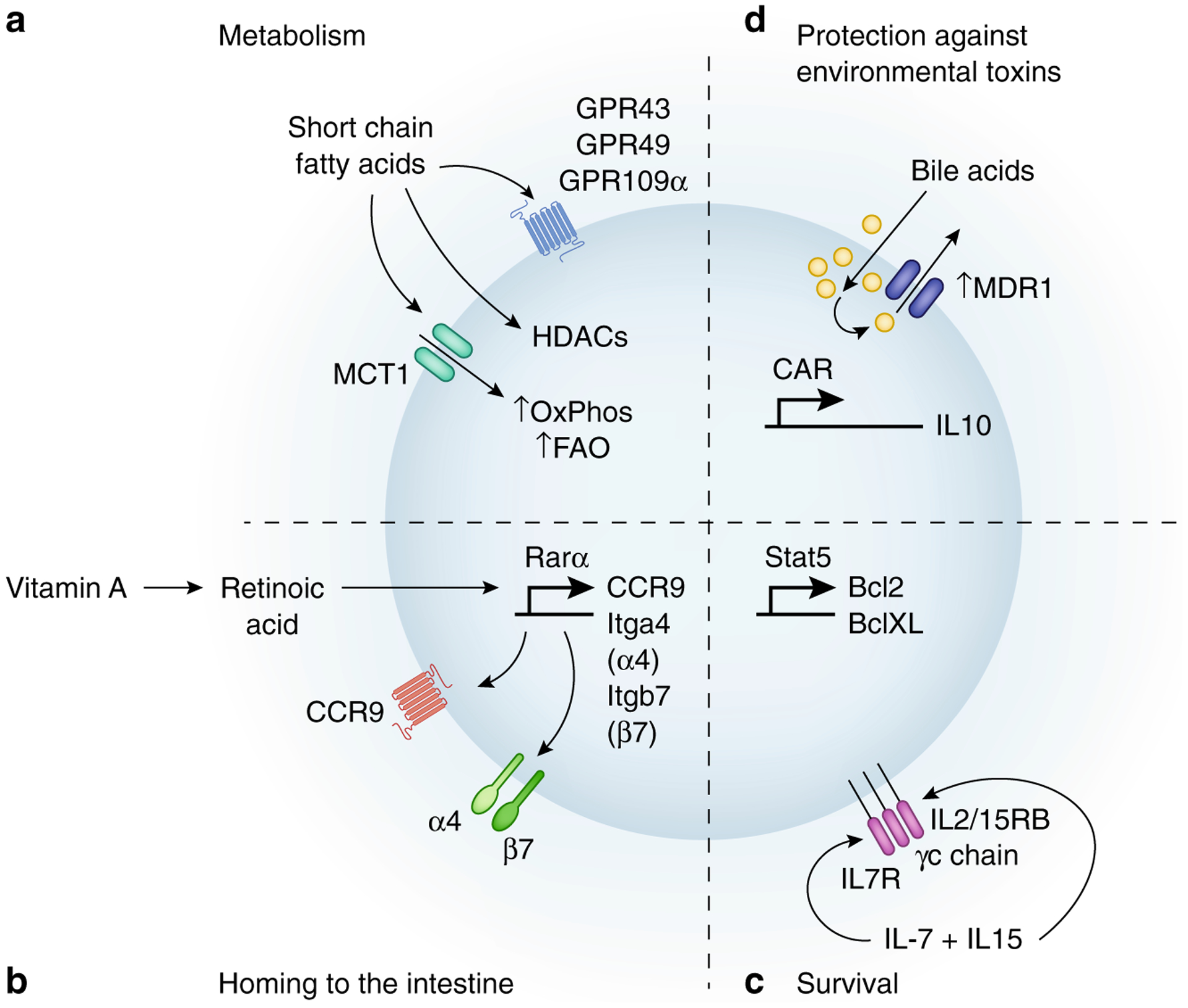
Trms reside in tissues and are directed by signals in their local environment. The microbiota is one of the dominant factors controlling the barrier tissue environment and microbiota-derived signals may be important Trm survival. A) Microbiota derived SCFAs direct T cell metabolism toward Oxidative Phosphorylation (OxPhos) and Fatty Acid Oxidation (FAO) that benefit long-term survival. B) Microbiota controls the production of Retinoic Acid from Vitamin A, which via activation of the transcription factor Rarα activates expression of the chemokine receptor CCR9 and the integrin α4β7 which guide T cells back to the intestine. C) Microbiota-induced IFNγ and dendritic cell activation can increase IL-7 and IL-15 production, which activates anti-apoptotic genes (Bcl-2, Bcl-XL). D) MDR1 and CAR protect T cells against bile acid induced toxicity allowing for survival in the small intestine and decrease inflammation via increasing IL-10 and decreasing TNFα.
Beyond any specific effects on Trms, the microbiota is important for the overall function and development of effector and memory T cells. Germ-free mice, that are propagated entirely without any microbiota, are predisposed to ‘Type 2’ adaptive immune responses that are characterized by CD4+ T cells and NKT cells that produce IL-4, IL-5 and IL-1332,33. Antiviral CD8+ T cell responses are also affected in microbiota-depleted mice. Antibiotic depletion of the microbiota in mice leads to a reduction in the accumulation of virus-specific effector CD8+ T cells34,35. This reduction was dependent primarily on the absence of Gram positive bacteria which produce various TLR ligands and lead ultimately to the upregulation of IL-1β by APCs. Indeed, T cell numbers and viral protection could be restored by replicating the impact of the microbiome through direct TLR stimulation34,36,37. CD8+ T cells from microbiota-depleted mice are not only reduced in number, but also display a dysfunctional phenotype, characterized by an increase in inhibitory receptors (PD-1, 2B4, CD160, and LAG3) and a reduction in cytokine production, especially IFNγ and TNFα38. Microbiota depletion leads both to more severe disease in response to the initial infection and also results in reduced virus-specific serum IgM and IgG and a reduction in viral specific CD8+ memory T cells, showing that the microbiota is required for optimal viral control35,39. This is a complex issue however, as depletion of the microbiota can reduce ‘colonization resistance’ to oral Listeria monocytogenes infection and thus increase the resultant intestinal infection and production of Trms40. Therefore, the absence of specific microorganisms within the microbiota, can affect T cell responses to infection. An example of this property are comparisons between wild and pet store mice and specific pathogen free (SPF) that are used for most experiments to control for environmental variables. However, SPF mice are not always reliable representations of humans and one primary difference is reduced diversity and increased stability of the microbiome41. Laboratory mice are not devoid of a microbiota, but because they are continuously propagated in cages and do not freely interact with their environment and other animals, their microbiomes are generally less diverse and subject to significant ‘cage effects’, where lineages of mice can differ in their microbiome because of ‘drift’ that is driven by their provenance and not any underlying genotype/phenotype42. This overall lack of microbial diversity results in an altered immune system compared to mice propagated in the wild or in pet stores and most importantly, humans. Laboratory mice show a deficit in CD8+ Trms compared to ‘wild’ mice or humans and possess a surplus of naïve CD8+ T cells43. In addition, laboratory mice show a reduction in CD8+ T cell infiltration into mucosal barrier sites, such as the reproductive tract or salivary gland, both of which are readily infiltrated in pet-store mice and humans43. Fascinatingly, these immune defects can be restored by co-housing laboratory mice with ‘wild’ mice. The exact microorganisms that drive the shifts between SPF mice co-housed with pet store mice or mice carrying a wild microbiome have not comprehensively described, but host infecting viruses that would not typically considered part of the microbiome likely have a dominant effect43. However, the bacterial microbiota of wild mice also differs substantially from SPF mice and likely is also an important modifier of the T cell response41,44. ‘Wilding’ of laboratory mice has also revealed an important role for fungi in inducing Th17 immune cells44–47. Together these results demonstrate that the microbiota can directly impact long-term T cell responses at mucosal barriers and that modification of the composition of the microbiota may be a mechanism to improve T cell responses and augment vaccines.
Microbial derived or modified metabolites as modulators of Trms.
The metabolites produced by the microbial digestion of food can impact the immune system within the gut and beyond. For example, short chain fatty acids (SCFAs; acetate, propionate or butyrate), which are primarily derived from the breakdown of dietary fiber by the gut microbiome, contribute to and stabilize the gut-resident regulatory T cells and reduce inflammatory innate immune cells48–52. In addition to regulatory T cells, SCFAs have also been associated with increased CD8+ memory T cell survival and function. SCFAs have many cellular targets and it is likely that they affect memory T cell development and homeostasis indirectly via effects on innate immune cells and Treg cells, which regulate memory cell differentiation through cytokine expression51,53. CD4 and CD8 T cells also be directly affected by SCFAs as they express multiple SCFA receptors (GPR41, GPR43 and GPR109a) and transporters (MCT1). For example, acetate, can be directly taken up by CD8+ T cells and boost glycolytic activity through an increase in acetyl-coA, which supports immediate CD8+ effector functions after infection (such as Listeria) and thus an increased CD8+ memory T cell pool54. High fiber diets are associated with an increase in Bacteroidetes a reduction in Firmicutes, an increase in SCFA production, especially butyrate and reduced severity in a mouse influenza infection model52. Increases in SCFAs mediated by the high fiber diet supported an increase in IL-10 and IL-4 production in the lung, and importantly, a high fiber diet led to an increase in virus-specific CD8+ T cells and a dampening of inflammatory monocytes52,54. With a high fiber diet, flu-specific CD8+ T cells cells not only produce more cytokine (IFNγ, TNFα) during primary infection but are also more metabolically robust through elevated fatty acid oxidation52. SCFAs also promote the development of long-term memory. For example, butyrate, increased the uptake of fatty acids and oxidative phosphorylation in CD8+ memory T cells55, which supports memory cell survival56. SCFAs also promotes IL-10 production, which is critical to the development of memory CD8 T cells57,58. Beyond SCFAs, other metabolites that are modified by the microbiota are also important for tissue-resident T cell function. Retinoic acid (RA) is a metabolite of Vitamin A whose production by intestinal epithelial cells is regulated by spore-forming Clostridia59. Conversely, the specific bacteria within the microbiota (Segmented filamentous bacteria, Bacillus cereus, Bifidobacteria bifidum) can produce RA from Vitamin A, independent of host effects175. The presence of RA is critical for robust intestinal T cell responses (and thus intestinal Trms) to infection because it is necessary for the development of CD11b+CD103+ DCs in the intestine and for full activation of T cells via the TCR60,61. Also, RA is necessary for the expression of the chemokine receptor CCR9 and the integrin α4β7, two surface proteins necessary for the traffic of T cells and therefore Trm development, in the small intestine and colon62,63. Bile acids, which are produced by the liver to aid in the digestion of fat, are also modified by the microbiota and have been shown to modulate intestinal T cells. For example, microbiota modified bile acids are critical for the development of a subset of FOXP3+ Tregs that co-express the transcription factor RORγT64. Additionally, bile acids can directly modulate effector/memory T cell homeostasis in the intestine. Activated CD4+ T cells in the intestine upregulate the expression of the nuclear bile acid receptor CAR and as a result the Multi drug resistance transporter (MDR1, Abcb1a)65,66. Expression of CAR/MDR1 by CD4+ T cells is important to prevent bile acid toxicity, but also for the expression of IL-10 and the regulation of TNFα production, which together reduces intestinal inflammation65,66. Finally, the microbiota can be important for the production of the cytokines that mediate TRM survival: IL-7 and IL-15. Experiments using IL-7 reporter mice have revealed that high levels of IL-7 are expressed in the intestine67. Intestinal IL-7 expression was induced by microbiota-dependent induction of IFNγ which might explain why anti-viral T cell responses are so subdued in mice where the microbiota has been depleted by antibiotics67,68. The microbiota-dependent expression of IL-7 might be particularly important for intestinal Trms since their survival has been shown to be independent of signals from IL-1569. IL-15 expression could also be affected by the microbiota. For instance, IL-15 and an altered composition of the microbiota have both been associated Celiac disease and Inflammatory Bowel Disease (IBD)70,71. Indeed, NOD2, increases IL-15 expression in the intestine through sensing of the microbiota and single nucleotide polymorphisms in the Nod2 gene have been associated with IBD72. Altogether, these studies suggest that microbial metabolites have both direct and indirect impacts on Trm cell differentiation, function and survival in tissues.
The microbiota shapes the T cell response to cancer.
Activating the immune system to eradicate cancerous cells has revolutionized cancer therapy. Most immunotherapy approaches rely on activating or re-activating T cells to kill tumor cells. While still a nascent area of research, tumor-resident Trm cells are an important target of immunotherapy approaches, and accordingly, the composition of the microbiota is important for therapeutic success73–79. Indeed, select gut bacterial species have been associated with better prognosis in melanoma patients and specifically, a better response to immunotherapies like anti-PD1antibodies80–83. While the exact mechanism by which these bacteria provide a benefit is still unclear, increased tumor infiltration by CD8+ T cells and a reduction of Tregs within the tumor microenvironment has been associated with specific members of the microbiota. One potential mechanism by which the microbiota may benefit tumor immunotherapy is via the provision of metabolites. For example, a collection of eleven human members of the microbiota have been demonstrated to improve anti-tumor immunity, likely by shifting the metabolism of the microbiota84. Similarly, inosine derived from intestinal populations of Bifidobacteria, Lactobacillus and Olsenella has been associated to increased numbers CD8+IFNγ+ T cells and control over tumor growth85.
T cell responses raised against antigens-derived from microbes, including bacteria within the microbiota, can also contribute to anti-tumor immunity. While tumor-specific T cells are undoubtedly critical to anti-tumor immunotherapy, a large proportion of tumor infiltrating CD8+ T cells are not specific to the tumor, but rather are ‘bystander’ T cells that can be specific to microbes86,87. For example, CD8+ T cells activated by repeated bacterial and viral infections that traffic to tumors can contribute to anti-tumor immunity88. Virus-specific T cells can also be ‘boosted’ for better anti-tumor immunity through direct tumoral injection of viral peptides89. Enhanced anti-tumor immunity after a ‘peptide boost’ is associated with activation of dendritic cells and NK cells, leading to increased recruitment of T cells. This response can unleash an effective immune response against the tumor after anti-PDL1 in otherwise resistant tumors and suggests that T cells specific to viruses can be leveraged to enhance the anti-tumor immunity89.
Antigen-driven interactions with less infectious microorganisms that are found within the microbiota might also contribute to the antitumor immunity. The ‘wild’ mouse microbiome is associated with improved immune responses to colorectal cancer44. This response can be transferred to laboratory mice through a gavage of the ‘wild’ microbiome and is associated with an increase in the relative abundance of Proteobacteria (Helicobacter spp.), a decrease in Firmicutes and reduced colonic inflammation44. In concert with these findings, our group has shown that colonization of SPF mice with Helicobacter hepaticus is sufficient to provide long-term control over colorectal cancer. Protection provided by H. hepaticus colonization required the development of Helicobacter-specific T Follicular Helper T cells (TFH) and Tertiary Lymphoid Structures (TLS) in and around the tumor. Together the increase in microbiota-driven TFH and TLS led to greater invasion of the tumor with cytotoxic T cells and NK cells90. However, not all strains of Helicobacter spp. may have this affect as other configurations of the microbiota containing large amounts of this taxa have been associated with failure of the CD8+ anti-tumor response91. In addition, this property is not strictly limited to Helicobacter, since a member of the Bacteroides phylum Odoribacter splanchnicus, also controls colorectal cancer via the induction of Th17 T cells92. Together these studies support the notion that leveraging the microbiota and microbe-specific memory T cells could be a useful strategy to improve the efficacy of anti-tumor imunotherapy. In the future, it will be important to define the members of the microbiota that are best associated with improved treatment outcomes and to define the distinct microbiota compositions necessary to augment immunotherapy against different tumor types.
In addition to the effects of relatively distant microbiotas of barrier tissue sites, the tumor itself can harbor its own microbiome. Many cancer types, including breast, bone, pancreas, ovarian, lung, melanoma, and colorectal cancer harbor ‘tumor microbiotas’, which are typically dominated by anaerobic bacteria93–95. Though how exactly anaerobic bacteria traffic through the body to tumors is unclear, once there anaerobic bacteria can thrive in the hypoxic environment at the center of many tumors and can impact tumor growth93–95. The type of tumor and location has a great impact on the bacteria present, with breast cancer harboring the most diverse microbiome, consisting of S. infantis, L. iners, and F. nucleatum96,97. In contrast, melanoma is far more restricted in microbiota diversity and is predominantly composed of P. marcusii and S. aureus96. However, other factors such as lifestyle can impact the tumor microbiome as well. In lung cancer, smokers show an increase in Acetobacteraceae, Rhodospirillales, Roseomonas, and Alcaligenes taxa compared to patients who have never smoked96,98. Perhaps most interestingly, melanoma patients that respond favorably to treatment with anti-PD1 show a distinct tumor microbiome, with an increase in Clostridia, Mycobacteria, and Novosphingoblum and a somewhat surprising reduction in Bifidobacteria given that this taxon has been associated with positively associated with tumor immunotherapy success in mouse models96. It is important to mention that the immune microbiota can also contribute to carcinogenesis and tumor growth, such as in liver cancer, pancreatic cancer and colon cancer99–103. In colon cancer, bacteria can contribute directly to tumorigenesis via damaging the DNA (pks+E. coli) and also by inducing inflammatory STAT3 activating cytokines from T cells that can aid tumor growth and suppress anti-tumor immune responses (Fusobacteria, enterotoxigenic Bacteroides fragilis)104–106. It will be of great interest to learn further about the potential bacterial functionality, characteristics, or metabolites of tumor resident bacteria that either drive carcinogenesis or increase response to immunotherapies in hopes of developing novel microbial-based therapeutics.
SECTION 2. MICROBIOTA-SPECIFIC T CELL RESPONSES
There is more diversity in the microbiota between different barrier tissue sites within an individual than there is diversity between different individuals at the same site16. To put more simply, the average person’s intestinal microbiota looks more like their neighbor’s intestinal microbiota than it does their own skin or mouth microbiota. This is evidence that various microbiotas are not random assemblages of whatever happens to land on a particular barrier site, but instead that the host is shaping which microorganisms can inhabit a given barrier tissue and the bacteria are also colonizing according to the environmental conditions. Within tissue-specific microbial communities there is also heterogeneity. For example, bacterial communities within the mammalian intestine often contain five or more different phyla and dozens of individual isolates within those phyla, each of which possess diverse genomes, structures and behavioral strategies to survive host colonization. The various members of the microbiota compete for resources within a constantly changing environment and the composition of the microbiota is not fixed. In response to microbiota heterogeneity, anti-microbiota T cell responses are tailored according to the tissue environment. This phenomenon is best demonstrated by comparing the immune responses of the adult skin, intestine and mouth.
Intestine-Resident Microbiota-specific T cells
The intestine contains the densest and most rich microbiota and the number of bacteria and their diversity generally increases from the duodenum to the rectum. Given this diversity, it is perhaps unsurprising that there is substantial diversity in the T cell responses induced in the intestine. CD4+ T cells specific to bacterial members of the microbiota have been found to differentiate into Th1, Th17, T Follicular Helper (TFH) and regulatory T cell (Treg) states107. In line with their diverse differentiation states, microbiota-specific T cells have been shown to perform a variety of important functions in the intestine, including limiting bacterial mucus colonization via the induction of anti-microbial peptides and the increase of IgA production and secretion108,109. Which of the various T cell differentiation states is induced in microbiota-specific T cells depends upon both the biology of the inducing bacteria and the specific site along the intestine where the microbe resides. One property that is shared by all of the intestinal bacteria that have been demonstrated to induce ‘spontaneous’ immune responses is attachment or interaction with the intestinal epithelium/mucus layer110–118. For example, Segmented Filamentous Bacteria (SFB), Akkermansia muciniphilia, Mucispirillum schaedleri and H. hepaticus either directly adhere to the intestinal epithelium or live within the inner mucus layer, while Bacteroides thetaiotaomicron secretes outer membrane vesicles that can be absorbed by the host118. In contrast, bacteria that lack the ability to invade the mucus and epithelial surface require a breakdown of the intestinal barrier for recognition by host T cells112,119. Despite the shared property of epithelial association, the immune responses induced by each of these organisms at steady state is very different (Figure 2), where SFB induces predominantly Th17 responses but other bacterial taxa (A. muciniphilia, H. hepaticus, M. schaedleri and the ‘CBir’ Lachnospiraceae isolate) induce varying mixtures of TFH and Tregs. Undoubtedly, biological attributes of each of these bacteria are partially responsible for the differences in the induction of T cell response, but where along the intestine that a particular bacteria colonizes is also important. Each part of the intestine drains to a different LN within the mesenteric lymph node (LN) string and each node has different predisposition to various T cell states120,121. In a broad sense, mucus resident/epithelium adherent bacteria, such as SFB in the small intestine drive responses that are dominated by Th17 T cells, with a minor population of Th1 cells111,122. Conversely bacteria that inhabit a similar mucosal niche in the colon (H. hepaticus, A. muciniphilia etc.) induce responses that are mixed between Treg and TFH115,116,123. Within the small intestine, there are also differences between bacteria that live in the proximal versus the distal small intestine. The LN that drains the duodenum is more skewed towards the induction of Tregs while in contrast, the ileal draining LN, is more skewed to the induction of Th17 T cells120. The reason for this is that presumably the duodenum-draining LN is more important for inducing tolerance to food, while the ileum, which is more anaerobic and less acidic, is home to more mucus-resident bacteria which must be contained to prevent inflammation. These differences also extend beyond the bacterial microbiome to protists where Tritrichomonas spp. colonization of the colon induces Th1 immune responses, whereas the same or similar protists colonizing the small intestine induce ILC2 activation and a more type 2 skewed response124–126.
Figure 2 – Various members of the microbiota induce heterogeneous T cell responses.
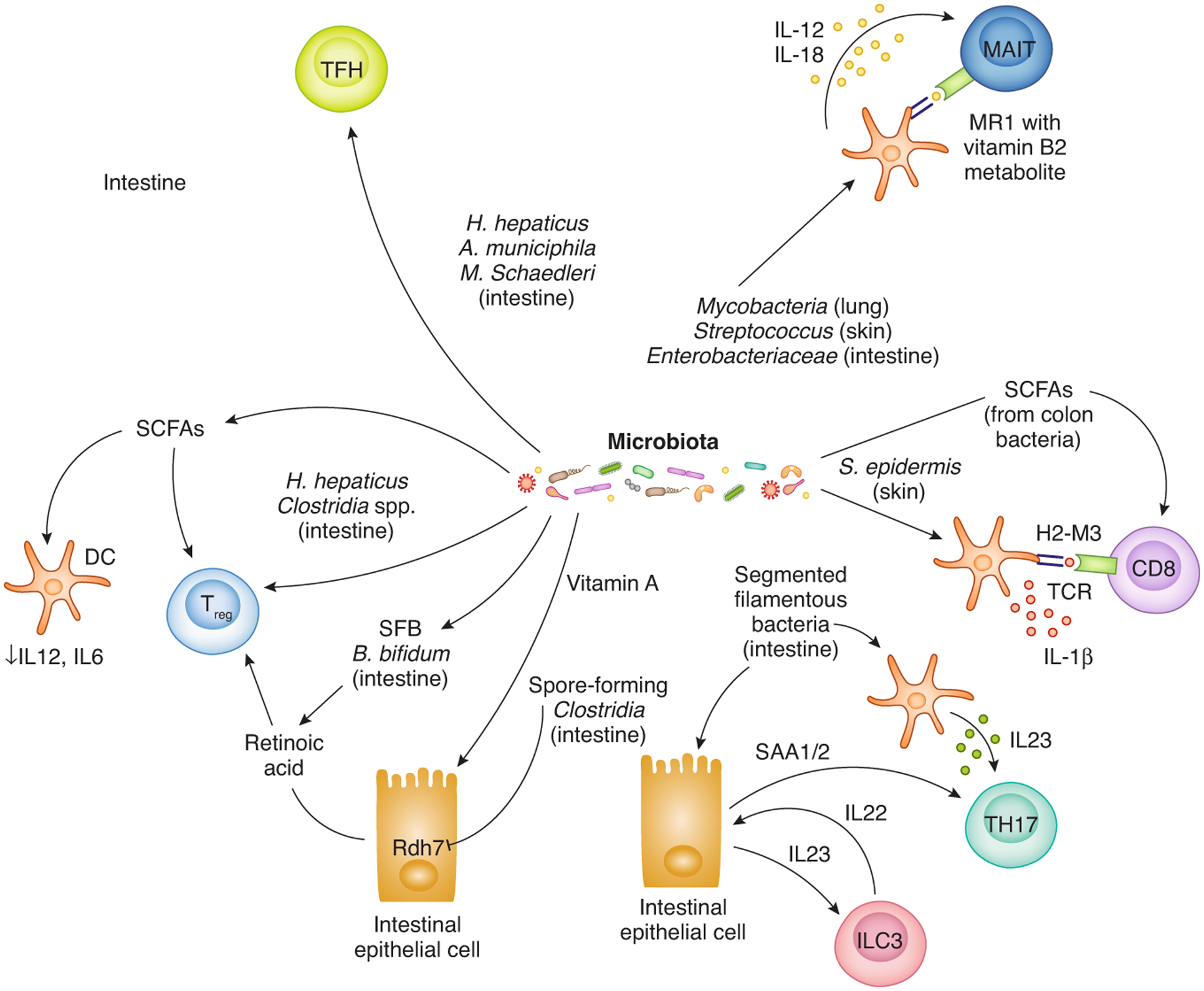
At different barrier tissue sites antigens-derived from the microbiota can induce microbiota-specific T cell responses. The differentiation state of microbiota-specific T cells differs according to the nature of the host/microbe interaction, the tissue environment and the type and magnitude of the innate immune response that is induced. Microbiota-derived metabolites also regulate effector/memory T cells directly via receptors on T cells and indirectly via modulation cytokine expression by antigen-presenting and epithelial cells. While microbiota-specific MHC class II restricted CD4 T cells (Treg, TFH, TH17) are the best understood, CD8 T cells and MAIT cells specific to microbiota-derived peptides and Vitamin B2 metabolites expressed on non-classical MHC class I molecules (H2-M3, MR1) have also been described. Where each bacteria predominantly resides on the host is described in parentheses.
In the intestine the majority of the anti-microbiota T cell responses that have been described are CD4+ Helper T cells. It is possible that the relevant intestinal conditions/microorganisms where robust anti-microbiota CD8+ T cell responses are evident have not been identified. Alternatively, MHC class I restricted CD8+ T cell responses may compose a relatively minor population because benign members of the microbiota rarely invade the host cytoplasm or induce apoptosis and are thus poor initiators of MHC class I cross-presentation by antigen presenting cells. There are effects of the intestinal microbiota on systemic CD8 T cell differentiation and function and though rare, CD8+ T cells that are cross-reactive to microbiome-derived peptides have been observed84,127. There is also evidence of CD8 T cells that are specific to bacteriophages carried in microbiota-derived bacteria128. Intestinal resident CD8 T cell populations that possess either inflammatory (TNFα) or regulatory (IL-26) transcriptional modules are associated with Ulcerative colitis, so anti-microbiota CD8 T cell responses are potentially very important129 and more research in this area is needed.
Microbiota-Specific T cells outside the intestine
In the skin, the microbiome-specific T cell population is comprised of both CD8+ and CD4+ T cells. Colonization of the skin of rodents with Staphylococcus epidermidis induces CD4+ Tregs, CD4+ Th1 cells and IL-17-expressing CD4+ and CD8+ T cells130,131 (Figure 2). The antigens that drive the CD8+ T cell response to S. epidermidis have been identified and are formylated peptides presented upon H2-M3 MHC class I like molecules132. Unlike CD8+ T cells raised in response to viral infections, microbiota-specific T cells in the skin do not express gene modules associated with cytolytic killing of infected cells and instead express functions associated with tissue repair and alarmin expression, which can help close wounds and control fungal infections130,133,134. These dual functions are reflective of the primary function of the skin, to act as a barrier for the internal organs to the environment. One interesting aspect of skin-resident microbiota-specific CD8+ T cells is the co-expression of transcription factors (GATA-3 and RORγT) and gene modules (Th2 and Th17) that define distinct developmental lineages of Innate Lymphoid cells133. Similar to the intestine, the microbiome of the skin varies from site to site (sebaceous, dry, and damp) and it will be interesting to determine whether the T cell response in the skin also varies according to these changes135.
The female reproductive tract (FRT) is also colonized by a microbiome, but here in stark contrast to the intestine, diversity is not associated with health and many women are colonized with only one or two isolates of Lactobacilli136. To our knowledge it has not been shown whether the vaginal microbiota induces antigen-specific T cells, but Lactobacilli tend to be relatively immunologically inert or even immunoregulatory, so T cell responses may be relatively muted in the FRT137,138.
In contrast to the epithelial layer of the intestine, the mouth is dominated by CD4+ T cells rather than CD8+ T cells. In the tongue, TCRαβ+ CD4+ IL-17A+ T cells are responsive to the microbiome and can provide antigen-independent protection against subsequent Candida infection139. Whether these cells are specific to antigens derived from the mouth microbiota is not clear, but an interesting hypothesis is that these tongue-resident CD4+ T cells are induced by the microbiome to provide heterologous protection against infection. In contrast, the TCRαβ+ CD4+ IL-17A+ T cells residing in the gingiva develop independently of the microbiota and instead respond to the act of mastication140. It is surprising to see such diversity in T cell responses at physically connected oral surfaces and it will be exciting to learn the mechanisms by which these cells develop and traffic to their respective sites.
Development and differentiation of microbiota-specific T cells
The anti-microbiota T cell response is not just shaped by where the bacteria colonize but also when the bacteria colonize during development. While still a very contentious area of research141, the process of microbiota colonization and therefore anti-microbiota T cell responses may begin in utero, where some studies have shown that fetuses have a small population of bacteria on their barrier surfaces, particularly the intestine142,143. In accord, in human fetuses, the intestine is home to a relatively robust population of effector/memory T cells some of which are phenotypically similar to TRMs and specific to bacteria present in the ‘fetal microbiota’142,144. However, caution should be taken in interpreting the paired discoveries of the fetal microbiota and T cell memory development, since the re-derivation of germ-free animals would argue against the presence of viable bacteria in the final stages of pregnancy. Further, the long-term implications of the population of the intestine with a microbiota and TRMs prior to delivery are unclear since they will soon be overwhelmed by colonization of the microbiota post birth. Independent of the possibility of a fetal microbiota, maternal infection can pre-dispose infants to increased CD4+ IL-17+ T cell responses in the intestine after delivery and these T cells protect against intestinal infection, so there is evidence of in utero effects on T cell immunity post-delivery145. The colonization of infant intestines by the microbiota post-delivery is a regulated process whereby both maternal (Human Milk Oligosaccharides and Immunoglobulin A from breast milk) and host inputs combine to protect the infant from damaging inflammatory micoorganisms and shape the microbiome to support health146. In mice prior to weaning, the intestinal resident CD4+ T cell response is subdued with the majority of intestinal and mesenteric lymph node CD4+ T cells being unactivated147. Both IgA from breast milk and IgG transferred via the placenta are important for the maintenance of naïve T cells in the neonatal mesenteric lymph nodes and thus the maternal antibody response is critical in shaping Trms in the early stages of immune development post-birth147,148.
In the intestine, Tregs can be generally subdivided based upon their expression of the transcription factor RORγT and the surface protein Neuropilin-1 (Nrp1), where microbiota-specific Tregs are RORγT+Nrp1−, self-antigen specific Tregs are RORγT−Nrp1+ and food antigen-specific Tregs are negative for both markers149–151. A wide variety of bacteria appear to be capable of inducing RORγT+ Tregs150,152, and the induction and accumulation of RORγT+Tregs begins at weaning in response to the expansion of the microbiota diversity that occurs at this time153. Importantly, if RORγT+Nrp1− Tregs are not induced at the time of weaning they cannot be restored by subsequent bacterial colonization, which makes the host more susceptible to autoinflammatory disorders such as IBD and allergy153. The frequency of RORγT+Nrp1− Tregs is controlled maternally through the provision of IgA in the breast milk through multiple generations154. Similarly, in the skin, early colonization of mouse pups predominantly induces Tregs which can assist long-term in preventing exacerbation of skin inflammation by the skin microbiota131. Taken together, there seems to be a predisposition of early life anti-microbiota T cell responses toward regulation and tolerance that is mediated by a combination of linked antibody and Treg responses that limit access and activation against microbiota-derived antigens experienced early in life. Given the substantial shifts in the composition of the microbiota that occurs during this time, such responses make intuitive sense as a mechanism to prevent constant autoinflammation. It will be interesting to determine if such ‘openness’ is punished by microorganisms that use this window to gain a foothold when otherwise they would induce a protective inflammatory immune response. One such example of this phenomena could be pediatric malnutrition, which allows for increased colonization by inflammatory isolates of Proteobacteria155,156 and in mouse models leads to both considerable immune cell infiltration and pathology and as a result the induction of RORγT+Nrp1− Tregs that inhibit effector CD4+ T cell responses in the small intestine157.
Do microbiota-specific T cells form memory?
Putting all these findings together, it is clear that a subset of bacteria, protists and fungi at multiple barrier tissue sites induce T cell responses. What has been less clear is whether these T cells are forming true memory T cell responses, defined by the ability to survive for long periods of time without cognate antigen restimulation while maintaining the ability to reconstitute secondary responses to provide rapid protection. Most microbiota-specific T cell responses that have been described to date are CD4+ T cells, which even when responding to bacterial or viral infection do not possess the vast memory potential of CD8+ T cells158,159, so even if CD4+ microbiota-specific T cells do survive to memory the expectation is that they would diminish over time. Indeed long-lived microbiota-specific CD4+ T cells that dwindle over time have been demonstrated under conditions where infection and physical breakdown of the intestine leads to systemic translocation of the microbiota from barrier surfaces, though in these experiments the antigen is likely continuously present within the intestinal microbiota119. In support of the concept that access to antigen could be important microbiota-specific CD4+ T cells, such cells are significantly increased in patients with Inflammatory Bowel Disease compared to healthy controls whose intestinal barriers are presumably more consistently intact160. Conversely, experiments in mice show that colitogenic CD4+ T cells can survive after transfer into lymphopenic germ-free mice and drive colitis after subsequent colonization with a microbial flora161. In contrast to the increase in CD4+ T cell responses, patients with Crohn’s Disease (CD) or Ulcerative Colitis (UC) present with lower numbers of CD8+ Trms129,162. Trms isolated from CD or UC patients expressed a number of regulatory markers including IL-10, IL-26 and CD39, suggesting that they may aid in maintaining homeostasis and reducing inflammation, and that in diseases such as Crohn’s or Ulcerative Colitis, an overall reduction in Trms contributes to inflammatory phenotypes, perhaps by allowing systemic immune responses against the microbiota129,163.
Outside of their effects on inflammatory disease, it has been hypothesized that microbiota-specific Trm cells might act as a heterologous defense system, where infection could lead to their re-activation and induction of a generic immune response119. Heterologous protection has been demonstrated in principle in the intestine where γδ T cells responding to infection with the Gram positive bacteria Listeria monocytogenes infection can provide protection against Gram negative Salmonella infections164. Additionally, microbiota-specific T cells in the skin offer protection against subsequent skin infection with Candida130. Thus, while ‘antigen-free’ survival of microbiota-specific T cells remains an open question, it is clear that long-lived microbiota-specific T cells contribute to both immune-mediated protection and inflammatory disease.
It makes intuitive sense that T cell responses against benign microorganisms that do not invade the host would be short-lived, since if they do not invade or secrete toxins that damage the host’s cells, they are not generally a life-threatening problem. Thus, it might be a benefit to the host to make T cell responses to the microbiota ‘as required’ to maintain plasticity and responsiveness in the face of constantly changing non-invasive microbiota. For example, microbiota-specific IgA producing B cells turnover in response to a changing microbiota, with new specificities against the dominant colonizers replacing the old but, in contrast, IgA+ plasma cells induced against enterotoxins show long-life165,166. The mechanism of the replacement of microbiota-specific clones is not clear, but it has been postulated that there is a ceiling on the niche available in the intestine for IgA producing B cells and because they are much more numerous newly activated B cells end up dominating these niches165. Interestingly no such ceiling has been detected for CD8+ Trms either in the secondary lymphoid tissues or the barrier sites such as the intestine167,168. Whether the intestine and skin can accommodate all microbiota-specific T cell clones or if there is competition and turnover in these populations remains unknown. As a corollary to the hypothesis that memory against benign, non invasive members of the microbiota would be short-lived, memory functionality amongst T cells at barrier surfaces would then be confined to T cells that have been ‘licensed’ by cytokine signals derived from the immune response to invasive microorganisms (IFNγ, IL-6, IL-12 and IL-18 etc.). Amongst microbiota-specific T cells activated at steady state antigen-free memory has only demonstrated in Mucosa-Associated Invariant T cells (MAIT) cells that inhabit barrier sites such as the skin and intestine169. MAIT cells are activated by riboflavin metabolites presented in the context of the of the MHC class I molecule MR1170,171. In support of the ‘licensing’ hypothesis the riboflavin metabolites that activate MAIT cells are more commonly produced by more invasive ‘pathogenic’ bacteria, such as Enterobacteriaceae that will be associated with the induction of inflammatory cytokines169,172,173. ‘Licensing’ is also evident in the large differences in the transcriptome and metabolism of Th17 T cells responding to either the microbiota (SFB) or a toxin-producing pathogen (Citrobacter rodentium)174. Taken together, our knowledge of microbiota-specific T cell responses is in its infancy because we have been restricted to studying a limited number of bacteria one at a time and lack a comprehensive view of the system. Hopefully, single cell RNA sequencing combined with TCR specificity screening techniques might allow us to capture a more complete view of the anti-microbiota T cell immune response and how it is shaped over time. Further, we hope that a better understanding of the ‘licensing signals’ required for the formation of microbiota-specific memory T cells might allow us to limit these responses in the context of diseases of autoinflammation and activate them to form a population of heterologous Trm protection.
Conclusion
Tissue resident memory T cells reside long-term in host tissues and contribute to protection of that tissue from re-infection. Tissue resident memory T cells are shaped by the tissue that they inhabit in way that is critical to their long-term survival and function. Barrier tissues are the most important sites patrolled by Trms and these tissues are colonized by tissue-specific microbiomes that are integral to the biology of the host tissue including the population of said tissue with Trms. Learning how to harness both microbiota-derived metabolites and antigens to shape the development of Trms is a potentially important area for future research as we have seen already that shifting the composition of the microbiota can be used to augment anti-tumor immunity. In the future, identification of the specific microorganisms and their metabolites that are capable of modulating Trms could lead to breakthroughs in the development of mucosal vaccines, tumor immunotherapy regimens, and treatments for chronic autoinflammatory disorders.
Acknowledgements:
We apologize that not all papers in this expanding field could be discussed and referenced. AEO is supported by a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship (2360-19 to A.O.D.). This work was supported by grants from the NIH (R21AI142051, R21CA249074; TWH) and a Burroughs Wellcome Foundation Investigator in the Pathogenesis of Infectious Disease award (TWH). We would like to thank the members of the Hand lab for helpful discussions.
Footnotes
Conflict of interest statement: The authors declare no conflicts related to this paper.
Bibliography
- 1.McFall-Ngai M Adaptive immunity: care for the community. Nature 445, 153 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Schenkel JM & Masopust D Tissue-resident memory T cells. Immunity 41, 886–897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gebhardt T, Palendira U, Tscharke DC & Bedoui S Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunol. Rev 283, 54–76 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Schenkel JM, Fraser KA & Masopust D Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J. Immunol 192, 2961–2964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen QP, Deng TZ, Witherden DA & Goldrath AW Origins of CD4+ circulating and tissue-resident memory T-cells. Immunology 157, 3–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amezcua Vesely MC et al. Effector TH17 Cells Give Rise to Long-Lived TRM Cells that Are Essential for an Immediate Response against Bacterial Infection. Cell 178, 1176–1188.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin H & Iwasaki A A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature 491, 463–467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schenkel JM et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisel NM et al. Comprehensive analyses of B-cell compartments across the human body reveal novel subsets and a gut-resident memory phenotype. Blood 136, 2774–2785 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allie SR et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol 20, 97–108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allie SR & Randall TD Resident memory B cells. Viral Immunol 33, 282–293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK & Knight R Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burr AHP, Bhattacharjee A & Hand TW Nutritional modulation of the microbiome and immune response. J. Immunol 205, 1479–1487 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wastyk HC et al. Gut-microbiota-targeted diets modulate human immune status. Cell 184, 4137–4153.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hand TW, Vujkovic-Cvijin I, Ridaura VK & Belkaid Y Linking the microbiota, chronic disease, and the immune system. Trends Endocrinol. Metab 27, 831–843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costello EK et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molloy MJ et al. Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 14, 318–328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalan LR et al. Strain- and Species-Level Variation in the Microbiome of Diabetic Wounds Is Associated with Clinical Outcomes and Therapeutic Efficacy. Cell Host Microbe 25, 641–655.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupp C et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Dethlefsen L, Huse S, Sogin ML & Relman DA The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6, e280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herndler-Brandstetter D et al. KLRG1+ Effector CD8+ T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity 48, 716–729.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackay LK et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol 14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Behr FM, Chuwonpad A, Stark R & van Gisbergen KPJM Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells. Front. Immunol 9, 1770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankovich AJ, Shiow LR & Cyster JG CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J. Biol. Chem 285, 22328–22337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dijkgraaf FE, Kok L & Schumacher TNM Formation of Tissue-Resident CD8+ T-Cell Memory. Cold Spring Harb. Perspect. Biol 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammed J et al. Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-β. Nat. Immunol 17, 414–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirai T et al. Competition for Active TGFβ Cytokine Allows for Selective Retention of Antigen-Specific Tissue-Resident Memory T Cells in the Epidermal Niche. Immunity 54, 84–98.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N & Bevan MJ Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity 39, 687–696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nath AP et al. Comparative analysis reveals a role for TGF-β in shaping the residency-related transcriptional signature in tissue-resident memory CD8+ T cells. PLoS One 14, e0210495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackay LK et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Adachi T et al. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med 21, 1272–1279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazmanian SK, Liu CH, Tzianabos AO & Kasper DL An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Olszak T et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ichinohe T et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 108, 5354–5359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abt MC et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw MH, Kamada N, Kim Y-G & Núñez G Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med 209, 251–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshi N et al. MyD88 signalling in colonic mononuclear phagocytes drives colitis in IL-10-deficient mice. Nat. Commun 3, 1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke TB et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med 16, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka K, Sawamura S, Satoh T, Kobayashi K & Noda S Role of the indigenous microbiota in maintaining the virus-specific CD8 memory T cells in the lung of mice infected with murine cytomegalovirus. J. Immunol 178, 5209–5216 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Becattini S et al. Enhancing mucosal immunity by transient microbiota depletion. Nat. Commun 11, 4475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosshart SP et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ubeda C et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med 209, 1445–1456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beura LK et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosshart SP et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–1028.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J-D et al. Rewilding nod2 and atg16l1 mutant mice uncovers genetic and environmental contributions to microbial responses and immune cell composition. Cell Host Microbe 27, 830–840.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abolins S et al. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat. Commun 8, 14811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reese TA et al. Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe 19, 713–719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arpaia N et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith PM et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furusawa Y et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Maslowski KM et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trompette A et al. Dietary Fiber Confers Protection against Flu by Shaping Ly6c-Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity 48, 992–1005.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Kaech SM & Cui W Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balmer ML et al. Memory CD8(+) T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44, 1312–1324 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Bachem A et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8+ T Cells. Immunity 51, 285–297.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Pearce EL, Poffenberger MC, Chang C-H & Jones RG Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laidlaw BJ et al. Production of IL-10 by CD4(+) regulatory T cells during the resolution of infection promotes the maturation of memory CD8(+) T cells. Nat. Immunol 16, 871–879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui W, Liu Y, Weinstein JS, Craft J & Kaech SM An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35, 792–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grizotte-Lake M et al. Commensals Suppress Intestinal Epithelial Cell Retinoic Acid Synthesis to Regulate Interleukin-22 Activity and Prevent Microbial Dysbiosis. Immunity 49, 1103–1115.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klebanoff CA et al. Retinoic acid controls the homeostasis of pre-cDC-derived splenic and intestinal dendritic cells. J. Exp. Med 210, 1961–1976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall JA et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity 34, 435–447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwata M et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21, 527–538 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Hammerschmidt SI et al. Stromal mesenteric lymph node cells are essential for the generation of gut-homing T cells in vivo. J. Exp. Med 205, 2483–2490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song X et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen ML et al. CAR directs T cell adaptation to bile acids in the small intestine. Nature 593, 147–151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao W et al. The xenobiotic transporter mdr1 enforces T cell homeostasis in the presence of intestinal bile acids. Immunity 47, 1182–1196.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shalapour S et al. Commensal microflora and interferon-gamma promote steady-state interleukin-7 production in vivo. Eur. J. Immunol 40, 2391–2400 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Oshima S et al. Interferon regulatory factor 1 (IRF-1) and IRF-2 distinctively up-regulate gene expression and production of interleukin-7 in human intestinal epithelial cells. Mol. Cell. Biol 24, 6298–6310 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schenkel JM et al. IL-15-Independent Maintenance of Tissue-Resident and Boosted Effector Memory CD8 T Cells. J. Immunol 196, 3920–3926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caminero A, Meisel M, Jabri B & Verdu EF Mechanisms by which gut microorganisms influence food sensitivities. Nat. Rev. Gastroenterol. Hepatol 16, 7–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gevers D et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang W et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J. Exp. Med 210, 2465–2476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nizard M et al. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat. Commun 8, 15221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milner JJ et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malik BT et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci. Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Enamorado M et al. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat. Commun 8, 16073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savas P et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med 24, 986–993 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Duhen T et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun 9, 2724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ganesan A-P et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat. Immunol 18, 940–950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McQuade JL, Daniel CR, Helmink BA & Wargo JA Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol 20, e77–e91 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Gopalakrishnan V et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Routy B et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Matson V et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanoue T et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Mager LF et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489 (2020). [DOI] [PubMed] [Google Scholar]
- 86.Scheper W et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med 25, 89–94 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Simoni Y et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Danahy DB, Berton RR & Badovinac VP Cutting Edge: Antitumor Immunity by Pathogen-Specific CD8 T Cells in the Absence of Cognate Antigen Recognition. J. Immunol 204, 1431–1435 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rosato PC et al. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat. Commun 10, 567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Overacre-Delgoffe AE et al. Microbiome-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity (2021). In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu AI et al. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep 31, 107471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xing C et al. Microbiota regulate innate immune signaling and protective immunity against cancer. Cell Host Microbe (2021). doi: 10.1016/j.chom.2021.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bullman S et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dang LH, Bettegowda C, Huso DL, Kinzler KW & Vogelstein B Combination bacteriolytic therapy for the treatment of experimental tumors. Proc. Natl. Acad. Sci. USA 98, 15155–15160 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yazawa K, Fujimori M, Amano J, Kano Y & Taniguchi S Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther 7, 269–274 (2000). [DOI] [PubMed] [Google Scholar]
- 96.Nejman D et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xuan C et al. Microbial dysbiosis is associated with human breast cancer. PLoS One 9, e83744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peters BA et al. The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol. Biomarkers Prev 28, 731–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma C et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miyazaki K et al. Colonic epithelial cells express specific ligands for mucosal macrophage immunosuppressive receptors siglec-7 and -9. J. Immunol 188, 4690–4700 (2012). [DOI] [PubMed] [Google Scholar]
- 101.Sun J & Kato I Gut microbiota, inflammation and colorectal cancer. Genes Dis 3, 130–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zeng H, Lazarova DL & Bordonaro M Mechanisms linking dietary fiber, gut microbiota and colon cancer prevention. World J Gastrointest Oncol 6, 41–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Farrell JJ et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arthur JC et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dejea CM et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kostic AD et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Honda K & Littman DR The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Kumar P et al. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity 44, 659–671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bos NA, Jiang HQ & Cebra JJ T cell control of the gut IgA response against commensal bacteria. Gut 48, 762–764 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Atarashi K et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ivanov II et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chiaranunt P, Tometich JT, Ji J & Hand TW T cell proliferation and colitis are initiated by defined intestinal microbes. J. Immunol 201, 243–250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Herp S et al. Mucispirillum schaedleri Antagonizes Salmonella Virulence to Protect Mice against Colitis. Cell Host Microbe 25, 681–694.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 114.Melo-Gonzalez F et al. Antigen-presenting ILC3 regulate T cell-dependent IgA responses to colonic mucosal bacteria. J. Exp. Med 216, 728–742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chai JN et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci. Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu M et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wegorzewska MM et al. Diet modulates colonic T cell responses by regulating the expression of a Bacteroides thetaiotaomicron antigen. Sci. Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hickey CA et al. Colitogenic Bacteroides thetaiotaomicron Antigens Access Host Immune Cells in a Sulfatase-Dependent Manner via Outer Membrane Vesicles. Cell Host Microbe 17, 672–680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hand TW et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Esterházy D et al. Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 569, 126–130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Houston SA et al. The lymph nodes draining the small intestine and colon are anatomically separate and immunologically distinct. Mucosal Immunol 9, 468–478 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Gaboriau-Routhiau V et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). [DOI] [PubMed] [Google Scholar]
- 123.Ansaldo E et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Howitt MR et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chudnovskiy A et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 167, 444–456.e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Escalante NK et al. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J. Exp. Med 213, 2841–2850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hebbandi Nanjundappa R et al. A Gut Microbial Mimic that Hijacks Diabetogenic Autoreactivity to Suppress Colitis. Cell 171, 655–667.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 128.Fluckiger A et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 369, 936–942 (2020). [DOI] [PubMed] [Google Scholar]
- 129.Corridoni D et al. Single-cell atlas of colonic CD8+ T cells in ulcerative colitis. Nat. Med 26, 1480–1490 (2020). [DOI] [PubMed] [Google Scholar]
- 130.Naik S et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scharschmidt TC et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity 43, 1011–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Linehan JL et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell 172, 784–796.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Harrison OJ et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nosbaum A et al. Cutting edge: regulatory T cells facilitate cutaneous wound healing. J. Immunol 196, 2010–2014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grice EA et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gajer P et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med 4, 132ra52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Devkota S et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mu Q, Tavella VJ & Luo XM Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol 9, 757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Verma AH et al. Oral epithelial cells orchestrate innate type 17 responses to Candida albicans through the virulence factor candidalysin. Sci. Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dutzan N et al. On-going Mechanical Damage from Mastication Drives Homeostatic Th17 Cell Responses at the Oral Barrier. Immunity 46, 133–147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Perez-Muñoz ME, Arrieta M-C, Ramer-Tait AE & Walter J A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5, 48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mishra A et al. Microbial exposure during early human development primes fetal immune cells. Cell 184, 3394–3409.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rackaityte E et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med 26, 599–607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stras SF et al. Maturation of the human intestinal immune system occurs early in fetal development. Dev. Cell 51, 357–373.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 145.Lim AI et al. Prenatal maternal infection promotes tissue-specific immunity and inflammation in offspring. Science 373, (2021). [DOI] [PubMed] [Google Scholar]
- 146.Gopalakrishna KP & Hand TW Influence of maternal milk on the neonatal intestinal microbiome. Nutrients 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Torow N et al. Active suppression of intestinal CD4(+)TCRαβ(+) T-lymphocyte maturation during the postnatal period. Nat. Commun 6, 7725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koch MA et al. Maternal igg and iga antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim KS et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351, 858–863 (2016). [DOI] [PubMed] [Google Scholar]
- 150.Sefik E et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ohnmacht C et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349, 989–993 (2015). [DOI] [PubMed] [Google Scholar]
- 152.Geva-Zatorsky N et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Al Nabhani Z et al. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 154.Ramanan D et al. An immunologic mode of multigenerational transmission governs a gut treg setpoint. Cell 181, 1276–1290.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huus KE et al. Immunoglobulin recognition of fecal bacteria in stunted and non-stunted children: findings from the Afribiota study. Microbiome 8, 113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kau AL et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med 7, 276ra24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bhattacharjee A et al. Environmental enteric dysfunction induces regulatory T cells that inhibit local CD4+ T cell responses and impair oral vaccine efficacy. Immunity 54, 1745–1757.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pepper M et al. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat. Immunol 11, 83–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Murali-Krishna K et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8, 177–187 (1998). [DOI] [PubMed] [Google Scholar]
- 160.Hegazy AN et al. Circulating and Tissue-Resident CD4+ T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology 153, 1320–1337.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nemoto Y et al. Long-lived colitogenic CD4+ memory T cells residing outside the intestine participate in the perpetuation of chronic colitis. J. Immunol 183, 5059–5068 (2009). [DOI] [PubMed] [Google Scholar]
- 162.Smillie CS et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Noble A et al. Deficient resident memory T cell and CD8 T cell response to commensals in inflammatory bowel disease. J. Crohns Colitis 14, 525–537 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Khairallah C et al. A blend of broadly-reactive and pathogen-selected Vγ4 Vδ1 T cell receptors confer broad bacterial reactivity of resident memory γδ T cells. Mucosal Immunol (2021). doi: 10.1038/s41385-021-00447-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hapfelmeier S et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bemark M et al. Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization. Nat. Commun 7, 12698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wijeyesinghe S et al. Expansible residence decentralizes immune homeostasis. Nature 592, 457–462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vezys V et al. Memory CD8 T-cell compartment grows in size with immunological experience. Nature 457, 196–199 (2009). [DOI] [PubMed] [Google Scholar]
- 169.Constantinides MG et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kjer-Nielsen L et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). [DOI] [PubMed] [Google Scholar]
- 171.Treiner E et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003). [DOI] [PubMed] [Google Scholar]
- 172.Nel I, Bertrand L, Toubal A & Lehuen A MAIT cells, guardians of skin and mucosa? Mucosal Immunol 14, 803–814 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lupp C et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204 (2007). [DOI] [PubMed] [Google Scholar]
- 174.Omenetti S et al. The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells. Immunity 51, 77–89.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Woo et al. Commensal segmented filamentous bacteria-derived retinoic acid primes host defense to intestinal infection. Cell Host Microbe 10.1016/j.chom.2021.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]


