SUMMARY
Lung progenitor cells are crucial for regeneration following injury, yet it is unclear whether lung progenitor cells can be functionally engrafted after transplantation. We transplanted organoid cells derived from alveolar type II (AT2) cells enriched by SCA1-negative status (SNO) or multipotent SCA1-positive progenitor cells (SPO) into injured mouse lungs. Transplanted SNO cells are retained in the alveolar regions, whereas SPO cells incorporate into airway and alveolar regions. Single-cell transcriptomics demonstrate that transplanted SNO cells are comparable to native AT2 cells. Transplanted SPO cells exhibit transcriptional hallmarks of alveolar and airway cells, as well as transitional cell states identified in disease. Transplanted cells proliferate after re-injury of recipient mice and retain organoid-forming capacity. Thus, lung epithelial organoid cells exhibit progenitor cell functions after reintroduction to the lung. This study reveals methods to interrogate lung progenitor cell potential and model transitional cell states relevant to pathogenic features of lung disease in vivo.
Graphical Abstract
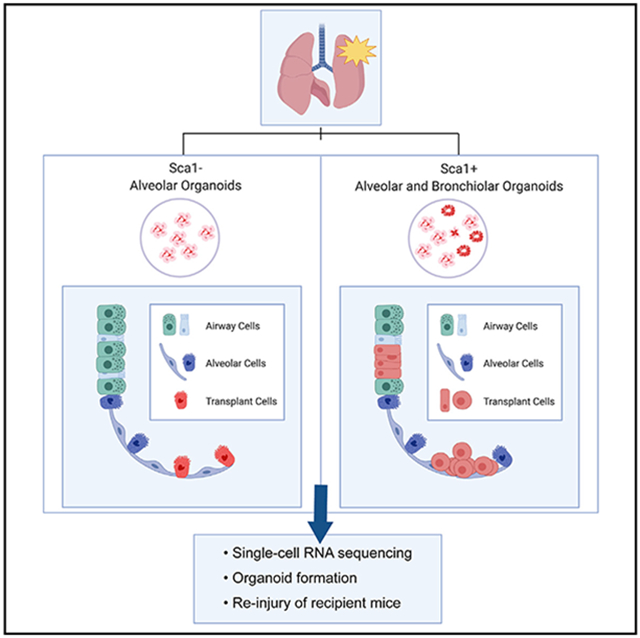
In brief
Upon return to the lung via transplantation, lung epithelial progenitor cells from organoid cultures restore their native transcriptional state and retain progenitor cell function. Louie et al. show future cell-based therapies, reveal methods to interrogate lung progenitor cell potential, and model transitional cell states relevant to lung disease in vivo.
INTRODUCTION
Lung diseases are among the leading causes of morbidity and mortality worldwide (Soriano et al., 2020). Transplantation is currently the only option for patients with different chronic, end-stage lung diseases, but there is a shortage of donor lungs, making this option limited. Recent discoveries in lung biology (Barkauskas et al., 2013; Hong et al., 2001; Kathiriya et al., 2020; Kim et al., 2005; Rawlins et al., 2009; Rock et al., 2009; Vaughan et al., 2015) have identified progenitor cells that could potentially treat various pulmonary diseases. For example, AT2 progenitor cells and alveolar type I (AT1) cells in the distal alveolar region, the cells responsible for surfactant production and gas exchange, respectively, may be beneficial for treating idiopathic pulmonary fibrosis. Club and goblet cells, secretory epithelial cells in the bronchiolar region, may be beneficial for treating chronic obstructive pulmonary disease. The complex cellular composition along the proximal to distal axis of the lung requires a deeper understanding of cell transplantation potential before cell-based therapies can become a reality. Transplantation is the gold-standard assessment of hematopoietic stem cell function in vivo and for treatment of a number of hematological disorders (Rowe et al., 2016); similarly, transplanted lung progenitor cells could be used to develop cellular therapy and as tools to characterize progenitor cell potential in vivo.
Animal models have been used to study several aspects of lung cell transplantation, including the use of different types of lung cells, pre-conditioning requirements, administration routes, and functional capabilities (Basil et al., 2020). Studies have shown that cells from different lung lineages, including multipotent embryonic progenitors, bulk embryonic lung tissue, cultured distal airway stem cells, and alveolar progenitor cells, can be transplanted into mouse lungs (Cao et al., 2017; Kathiriya et al., 2020; Milman Krentsis et al., 2018; Nichane et al., 2017; Rosen et al., 2015; Vaughan et al., 2015; Weiner et al., 2019; Zuo et al., 2015). Retention of various fluorescent donor cell types into recipient lungs by immunostaining has been reported. These studies suggest that pre-conditioning of the recipient mouse lungs by one or a combination of injuries, including bleomycin, naphthalene, influenza, and irradiation, may be required for donor cell retention. A side-by-side comparison of the transcriptional identities and functional capacity of endogenous lung progenitor cells, cells maintained in organoid cultures, and their transplanted counterparts has not been reported.
We compared the transcriptional profiles of distal and proximal lung epithelial progenitor cells from cultured organoid cells, native epithelial cells, and transplanted cells to illustrate the in vivo potential of the organoid cells. Transplanted cells from donor organoid cells regained their native transcriptional identity and maintained proliferative potential in vivo. Furthermore, organoid cells underwent transitional cell states after transplantation. These approaches are informative for developing robust assays for assessment of lung progenitor cell function in vivo and cell-based therapies for respiratory diseases.
RESULTS
Lung organoid cells are retained in distal lung compartments after transplantation
Based on previous studies showing that pre-conditioning of the lung was necessary for donor cell retention (Rosen et al., 2015), we utilized bleomycin injury to develop an in vivo transplantation assay using lung organoid cells. Immunodeficient recipient mice were given a single dose of bleomycin 1 day before cell transplantation (Figure 1A). Donor lung epithelial cells from β-actin DsRed mice were isolated by fluorescence-activated cell sorting (FACS) using a gating strategy to isolate non-endothelial, non-hematopoietic epithelial cells (CD31− CD45− EpCAM+), from which the surface antigen SCA1 was used to distinguish two different types of epithelial progenitors as previously established: AT2 cells enriched in the fraction negative for SCA1 (hereafter SCA1−) and multipotent progenitor cells enriched in the SCA1-expressing fraction (hereafter SCA1+; Kim et al., 2005; Lee et al., 2014, 2017; Figure S1A). Lung epithelial populations were cultured in our previously described 3D organoid co-culture system to yield alveolar and bronchiolar organoids (Lee et al., 2014). Organoid typology was based on immunofluorescence (IF) for lung epithelial cell markers, such as surfactant protein C (SPC) for alveolar organoids and club cell secretory protein (CCSP) for bronchiolar organoids. As expected, SCA1+ lung epithelial cells gave rise to both alveolar and bronchiolar organoids, whereas SCA1− lung epithelial cells gave rise to alveolar organoids (Figure S1B; Lee et al., 2014, 2017). After 21 days of culture, dissociated cells from organoids derived from SCA1− cells (hereafter SNO) or SCA1+ cells (hereafter SPO) were delivered orthotopically (Figure 1A). Recipients of SNO cells are referred to as SNO-transplanted mice or SNO recipients, and recipients of SPO cells are referred to as SPO-transplanted mice or SPO recipients. Cells retained in the lungs of recipient mice will be referred to as transplanted cells.
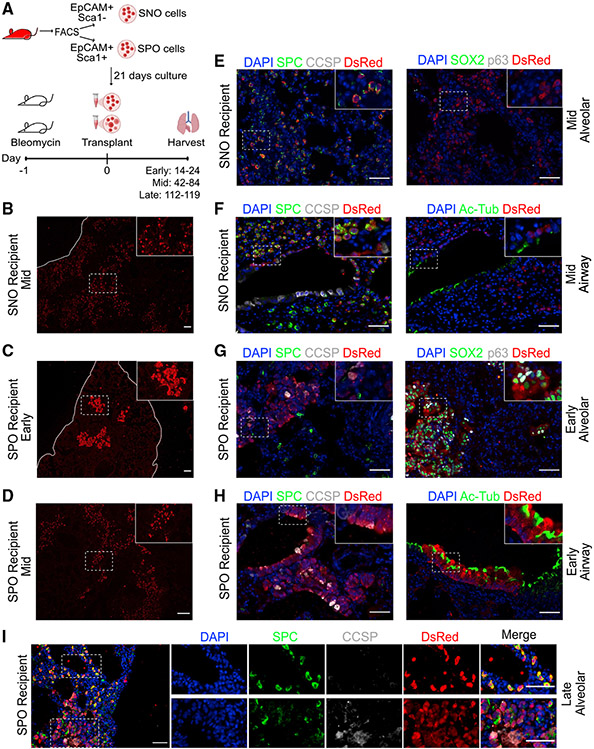
Figure 1. Lung organoid cells are retained in distinct lung compartments after transplantation.
(A) Experimental strategy for transplantation of lung organoid cells in pre-conditioned mouse lungs.
(B) Representative picture of IF staining on mouse lung transplanted with SNO cells and harvested at a middle time point. Red, DsRed. Scale bar represents 100 μm.
(C) Representative pictures of IF staining on mouse lungs transplanted with SPO cells and harvested at an early time point. Red, DsRed. Scale bar represents 100 μm.
(D) Representative pictures of IF staining on mouse lungs transplanted with SPO cells and harvested at a middle time point. Red, DsRed. Scale bar represents 100 μm.
(E and F) Representative pictures of IF staining on mouse lungs transplanted with SNO cells and harvested at a middle time point. Images show cells retained in the (E) distal alveolar region or (F) airway region. (E and F) Left: blue, DAPI; green, SPC; gray, CCSP; red, DsRed. (E) Right: blue, DAPI; green, SOX2; gray, p63; red, DsRed. (F) Right: blue, DAPI; green, Ac-Tub; red, DsRed. Scale bars represent 50 μm.
(G and H) Representative pictures of IF staining on mouse lungs transplanted with SPO cells and analyzed at an early time point. Images show cells retained in the (G) distal alveolar region or (H) airway region. (G and H) Left: blue, DAPI; green, SPC; gray, CCSP; red, DsRed. (G) Right: blue, DAPI; green, SOX2; gray, p63; red, DsRed.
(H) Right: blue, DAPI; green, Ac-Tub; red, DsRed. Scale bars represent 50 μm.
(I) Representative pictures of IF staining on mouse lungs transplanted with SPO cells and harvested at a late time point. Top inset depicts areas that express the alveolar marker SPC. Bottom inset depicts areas that express the airway marker CCSP but also express SPC. Blue, DAPI; green, SPC; gray, CCSP; red, DsRed. Scale bars represent 50 μm.
See also Figure S1.
IF showed that both SNO and SPO cells were retained in the recipient mouse lungs. Recipient mouse lungs were analyzed at different time points to assess potential of transplanted cells; recipients were initially analyzed at an early time point, 14–24 days after delivery, and then at a middle time point, 42–84 days after delivery. Flow cytometry using a gating strategy to identify DsRed+ cells demonstrated donor cell retention (Figure S1C). IF revealed that SNO cells were detected as DsRed+ cells found throughout the distal alveolar region at early (Figure S1D) and middle time points (Figure 1B) after transplantation. SPO cells were also detected in the lungs, but the pattern of cell retention differed from that of SNO cells; at early time points after transplantation, DsRed+ SPO-transplanted cells were found in airways or in large clusters of cells in the alveolar space (Figure 1C). At middle time points after transplantation, DsRed+ SPO-transplanted cells were still detected in airways and the alveolar clusters appeared less dense (Figure 1D). This prompted us to examine a late time point after SPO transplantation (112–119 days after delivery), at which we observed transplanted SPO cells in a scattered pattern in the alveolar space, more similar to the SNO recipients (Figure 1I).
IF of canonical lung epithelial cell markers was used to confirm the cell type potential of transplanted cells. In SNO-transplanted mice analyzed at early (Figure S1H) and middle (Figure 1E) time points, DsRed+ cells expressed the AT2 cell marker SPC. DsRed+ SNO-transplanted cells rarely expressed HOPX (Figure S1E), an AT1 cell marker. Furthermore, the DsRed+ HOPX+ cells were cuboidal with large, rounded nuclei, suggesting that they may not be bona fide AT1 cells. There was no significant difference in the percentage of SPC+ or HOPX+ cells in transplanted versus non-transplanted areas of SNO-recipient mice (Figures S1F and S1G). In addition, DsRed+ SNO-transplanted cells did not express airway cell markers, such as CCSP, SOX2, and p63 (Figures 1E and S1H). Airways of SNO-transplanted mice occasionally had DsRed+ cells, yet these DsRed+ cells did not express airway markers and were positive for SPC (Figures 1F and S1I). In contrast, at early time points, the DsRed+ SPO cells in alveolar clusters expressed airway cell markers, such as SOX2, p63, and CCSP, and did not express alveolar markers, such as SPC (Figures 1C-1G). DsRed+ cells were also detected in SPO-transplanted airways; these transplanted cells expressed airway markers, such as SOX2, p63, CCSP, and acetylated tubulin (Ac-Tub) (Figures 1H and S1J). SPO-transplanted cells showed a change in marker expression through the time points examined. Middle and late time point SPO lungs had alveolar areas with transplanted cells that expressed AT2 cell markers, such as SPC (Figures 1I and S1K). These results suggested that transplanted SNO cells contributed to AT2 cells, whereas transplanted SPO cells either had limited capacity or required more time to establish alveolar and bronchiolar cell types in vivo.
Single-cell RNA sequencing reveals that SNO-transplanted cells are transcriptionally similar to native alveolar epithelial cells
We next investigated the transcriptional profiles of the transplanted cells to further understand their identity and progenitor cell capacity. We used a single-cell RNA sequencing (scRNA-seq) approach to define the transcriptome of DsRed+ transplanted cells from SNO-transplanted mice, native SCA1-epithelial cells from the same SNO-transplanted mice, and epithelial cells from SNO (Figure 2A). The 10× Genomics scRNA-seq platform was used to examine gene expression, and data were analyzed using the SCANPY toolkit (Wolf et al., 2018). Transplanted epithelial cells were isolated from SNO-transplanted mice at a middle time point, 84 days post-transplant, using the gating strategy CD31− CD45− DsRed+ EpCAM+ for donor cell detection. Native SCA1-epithelial cells from the same SNO-transplanted mice were isolated as CD31− CD45− DsRed+ EpCAM+ SCA1− cells. Lastly, epithelial organoid cells were isolated from SNO after 21 days in co-culture using the gating strategy DsRed+ EpCAM+. Cells from all three groups, consisting of transplanted cells, native cells, and organoid cells, were captured and analyzed in parallel (Figures 2B and S2A-S2C).
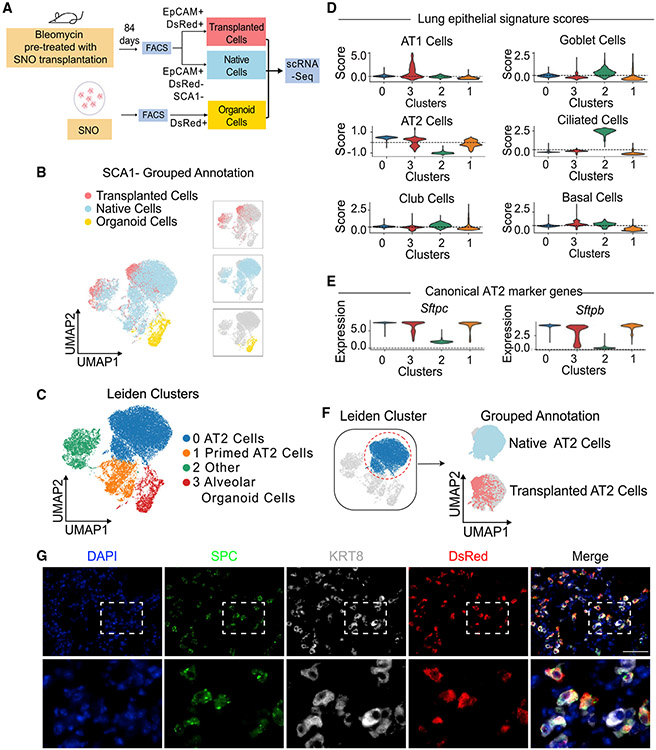
Figure 2. Single-cell RNA sequencing reveals that SNO-transplanted cells are transcriptionally similar to native alveolar epithelial cells.
(A) Experimental strategy. SNO cells, native EpCAM+ SCA1− cells, and transplanted SNO cells from bleomycin pre-treated mice were collected for scRNA-seq analysis (image created with Biorender.com).
(B and C) Clustering of transcriptomes using UMAP. Cells are colored based on (B) grouped annotation and (C) annotated Leiden clusters.
(D) Violin plots showing Z scores of indicated signatures in Leiden clusters.
(E) Violin plots showing expression levels of Sftpc and Sftpb in Leiden clusters.
(F) Cluster 0 AT2 cells were subset and colored by Leiden (left) and grouped annotation (right).
(G) Representative pictures of IF on mouse lungs transplanted with SNO cells. Lung is shown from RagKO recipient middle time point. Blue, DAPI; green, SPC; gray, KRT8; red, DsRed. Scale bar represents 50 μm.
Transplanted cells and native cells were transcriptionally similar, being present in the same clusters, whereas organoid cells were transcriptionally distinct. Using the Leiden algorithm and differential expression analysis, we identified four major clusters containing more than 200 cells for further downstream analysis, clusters 0–3 (Figures 2B and 2C; Table S2; McInnes et al., 2020; Traag et al., 2019). Clusters 0, 1, and 2 were composed of a mixture of both transplanted cells and native cells, whereas cluster 3 was almost exclusively organoid cells (Figures 2C and S2C). Cluster 3 included AT1-like and AT2-like cells, based on cell signature scores calculated using marker genes derived from the Panglao database (Table S1), as expected for alveolar organoids (Figure 2D; Franzén et al., 2019). Using RNA velocity to predict the cell state directionality in scRNA-seq datasets, we identified a transcriptional transition from AT2 to AT1 cells in our SNO cells (Figure S2E; La Manno et al., 2018). This result is in line with studies showing that AT2 cells can differentiate into AT1 cells in vivo and in organoid cultures (Adamson and Bowden, 1975; Evans et al., 1975; Barkauskas et al., 2013; Basil et al., 2020; Choi et al., 2020). In addition, AT1 and AT2 signatures in the organoid cluster correlated with expression of AT1 cell marker genes, Hopx and Ager, or AT2 cell marker genes, Lyz2 and Sftpc, respectively, with relatively little to no expression of club or ciliated cell marker genes (Figures S2E and S2F). These results were consistent with our previous studies that suggested organoids derived from SCA1-negative cells contain alveolar epithelial cell types most closely resembling AT2 cells and AT1 cells; day 21 SNO cells used for transplantation were composed of these alveolar cell types.
Next, we used differential expression analysis to identify cluster-specific gene expression patterns. We calculated gene signatures of common lung epithelial cells, including AT1, AT2, club, goblet, ciliated, and basal cells, using marker genes derived from the Panglao database (Franzén et al., 2019; Figure 2D; Table S1). Based on a high AT2 signature score and top marker genes, including the canonical AT2 cell markers Lyz2, Sftpc, Sftpb, and Etv5, we annotated cluster 0, which consisted of both native and transplanted cells, as AT2 cells (Figures 2D, 2E, S2D, and S2H; Table S2). Cluster 1, which also included both native and transplanted cells, is defined by a high signature score for AT2 cells, while expressing lower levels of Lyz2, Sftpc, Sftpb, and Etv5 relative to cluster 0 (Figures 2D, 2E, S2D, and S2I). Based on the signature score, expression profile, and comparison to the canonical AT2 cells in cluster 0, cluster 1 was annotated as the primed AT2 cell state. Cluster 2 consisted of both native and transplanted cells and contains a mixture of canonical marker gene expression and signature scores; hence, we annotated cluster 2 as “other” (Figures 2D, S2G, and S2H). When we focused on cluster 0, the canonical AT2 cells, we found that both the grouped annotation projected on a uniform manifold approximation and projection (UMAP) and hierarchical clustering revealed similar expression profiles of the transplanted and native cells, suggesting that the two populations are transcriptionally similar (Figures 2F and S2H). We then performed differential expression and Gene Ontology analysis to better understand the differences between SNOs and AT2 cells of native and transplanted origin. When directly comparing the AT2 cell cluster (cluster 0) with the SNO cluster, after AT1 cells were removed (cluster 3; Figure 2C), we observed an enrichment for cholesterol, peptide, and macromolecule synthesis pathways in addition to pathways important for translation in the AT2 cell cluster. In contrast, the SNO cluster was enriched for inflammation, hypoxia response, and metabolic pathways, indicating that, in organoids, progenitor cells activate pathways normally stimulated during lung injury (Table S3). Although transcriptionally distinct compared with in vivo derived native and transplanted cells of cluster 0, the SNO cells retained their alveolar epithelial lung lineage identity (Figure 2D).
Additional differential expression analysis comparing the native AT2 cells, transplanted cells, and SNO cells using their group annotation showed that, while SNO cells and native AT2s have transcriptionally distinct states, SNO cells re-established a native transcriptional state when reintroduced to their native environment (Figures S2J and S2K; Table S2). Genes reactivated by transplanted cells included Cited1, a positive regulator of SMAD transcription. SMAD/BMP signaling has been shown to be important for AT2 cell proliferation and differentiation (Chung et al., 2018; Lee et al., 2014). In addition, Ctgf, which is highly expressed in SNO cultures and has been linked to AT2 cell proliferation in the normal lung (Pan et al., 2001), is suppressed by SNOs following transplantation, likely reflecting the reduced proliferation requirements of AT2 cells in vivo relative to SNO cultures. To determine key remaining differences between native AT2 cells and transplanted cells, we performed Gene Ontology analysis using genes differentially expressed by native AT2 compared with transplanted cells Table S2. Transplanted cells were enriched for genes associated with energy production and the neutrophil immune response, while native AT2 cells were enriched for translation pathways and macromolecule synthesis pathways, potentially connected to surfactant production (Figure S2K; Table S4).
Although AT1 cells from transplanted cells were not detected in SNO recipients, we uncovered transcriptional similarity of transplanted cells and progenitor cell states linked to AT1 cell fate potential. As noted above, cluster 1 was annotated as primed AT2 cells based on shared marker gene expression; some transplanted SNO cells were primed AT2 cells. KRT8-positive cell states indicative of AT2 cell to AT1 cell differentiation were recently described in a bleomycin model of fibrosis (Choi et al., 2020) and in human lung fibrosis (Strunz et al., 2020). IF of transplanted lungs revealed the presence of DsRed+-transplanted cells that co-expressed SPC and KRT8 (Figure 2G). Thus, transplanted cells have progenitor cell states linked to AT1 cell fate potential.
Single-cell RNA sequencing captures transitional progenitor cell states following transplantation of SPO cells
We next examined the transcriptional states of transplanted SPO cells to understand their potential in vivo. We performed scRNA-seq of FACS-isolated native SCA1+ epithelial cells from uninjured wild-type mice (CD31− CD45− EpCAM+ SCA1+; Kim et al., 2005; Lee et al., 2014, 2017), DsRed+ epithelial cells from SPO-transplanted mice, and epithelial cells from SPO cultures (Figure 3A). Native SCA1+ epithelial cells from uninjured wild-type mice were isolated using a previously described gating strategy. DsRed+ epithelial transplanted cells were isolated from SPO recipients at a middle time point 70 days post-transplant using the gating strategy for quantification of donor cell retention (CD31− CD45− DsRed+ EpCAM+). Finally, epithelial cells were isolated from SPO after 21 days in co-culture using the gating strategy DsRed+ EpCAM+. We categorized the cells into three groups: transplanted SPO cells, native SCA1+ cells, and organoid cells (SPOs) and visualized them by a UMAP (Figure 3B). Using the Leiden algorithm, we identified seven major clusters, clusters 0–6 (Figure 3C). No clusters were composed of cells from a single sample (Figures S3A and S3B). The grouped annotation indicated that some transplanted, native, and organoid cells clustered closely and may share transcriptional similarities, as in clusters 2 and 4. The majority of cluster 3 consisted of transplanted cells, while clusters 1 and 5 primarily included organoid cells (Figure S3C).
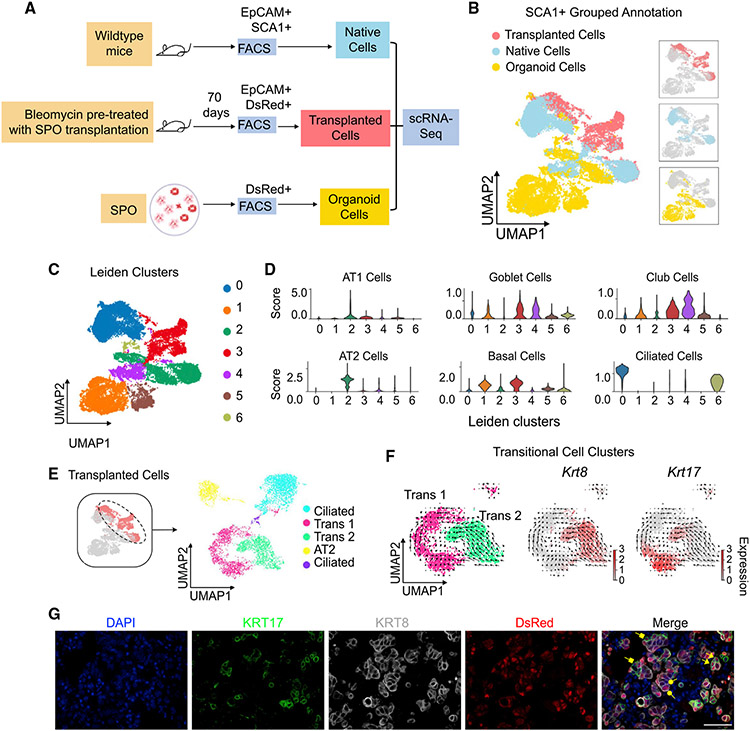
Figure 3. Single-cell RNA sequencing captures transitional progenitor cell states following transplantation of SPO cells.
(A) Experimental strategy. SPO cells, native EpCAM+ SCA1+ cells from uninjured mice, and transplanted SPO cells from bleomycin pre-treated mice were collected for scRNA-seq analysis (image created with Biorender.com).
(B and C) Clustering of transcriptomes using UMAP. Cells are colored based on (B) grouped annotation and (C) Leiden clusters.
(D) Violin plots showing Z scores of indicated signatures in Leiden clusters.
(E) (Left) Cells from the transplanted groups were colored on the UMAP. Transplanted cells are subset and re-analyzed, and newly determined Leiden clusters are shown on the right. Leiden clusters are annotated based on cell signature scores.
(F) RNA velocity analysis of “transitional 1” and “transitional 2” cells from subset analysis of transplanted cells. Cells are colored based on annotated Leiden clusters on the left. Krt8 and Krt17 expression are visualized on the right.
(G) Representative pictures of IF staining on mouse lungs transplanted with SPO cells and harvested at middle time point. Arrowheads, donor cells that dually express KRT8 and KRT17; square vectors, donor cells that express KRT17; round vectors, donor cells that express KRT8. Blue, DAPI; green, KRT17; gray, KRT8; red, DsRed. Scale bar represents 50 μm.
We next annotated clusters using differential expression analysis and cell signature scores for common lung epithelial cells, including AT1, AT2, club, goblet, ciliated, and basal cells, using marker genes derived from the Panglao database (Franzén et al., 2019; Figures 3D, S3D, and S3E; Table S2). We identified cluster 0 as ciliated cells with top marker genes, such as Fam183b and Tmem212 (known effector genes of the ciliated cell marker Foxj1), and a high ciliated cell signature score. Cluster 1 expressed the basal cell marker Krt5 while also expressing a high basal cell signature score. Cluster 2 has high levels of AT2 marker genes Sftpc and Cd74 and a high AT2 cell signature score. From our analysis of common lung epithelial cell signature scores, cluster 3, which primarily included transplanted cells, expressed high scores for AT1, club, goblet, and basal cells. Cluster 4 has high expression of club cell marker genes Scgb1a1 and Scgb3a2 while also expressing a high club-cell signature score. Cluster 6, which included organoid cells and a subset of transplanted cells, had elevated levels of goblet-, basal-, and ciliated-cell signature scores.
To better understand the cell types present in the day 21 SPO prior to transplantation, we examined cell-type signatures specifically in the organoid cells. To do this, we subset the SPO cluster, recomputed the UMAP, and checked epithelial marker gene expression. We observed a range of gene expression patterns in SPO cells suggesting that they exhibit multiple transcriptional states (Figure S3H). Our previous work demonstrated that SCA1+ lung epithelial cells can form alveolar, bronchiolar, and bronchioalveolar organoids (Lee et al., 2014). Our scRNA-seq analysis of SPO cultures revealed the presence of distinct AT2, club, and ciliated cell signatures. Interestingly, subsets of cells with high basal, neuroendocrine, AT1, and goblet cell signatures were detected. AT2 stem cell markers Axin2 and Tm4sf1 were also detected, but cells positive for these markers did not overlap. The majority of SPO cells expressed Ly6a, the gene that encodes SCA1, with the exception of cells with high AT1 and AT2 cell canonical gene expression. A large percentage of cells also contained high expression of Itga6 and Itgb4, the subunits for α6β4 integrin previously identified as markers for alveolar progenitors (Chapman et al., 2011), and overlapped with SPOs that highly expressed Ly6a. Overall, these data showed day 21 SPO cells used for transplantation were a mixture of cells with alveolar and airway cell states.
To characterize the transcriptional identity of the transplanted SPO cells, we subset and re-clustered the transplanted cells. Whereas some transplanted cells were transcriptionally similar to native and organoid cells based on overlapping grouped annotations on the UMAP, other transplanted cells, like those in cluster 3, appeared transcriptionally distinct from all other groups. Using the Leiden algorithm, five clusters were identified in the transplanted cell data (Figure 3E). We annotated each cluster using differential expression gene analysis and cell signature scores (Figure S3F). Of the transplanted cells, we identified clusters with ciliated cell and AT2 cell gene expression. The remaining two clusters had features of several cell types, including club, goblet, AT1, and basal cells. While we were not able to definitively annotate the latter two clusters using cell signature scores, we termed these “transitional” cells on the basis that they exhibit features of multiple common lung epithelial cell types. Using differential expression gene analysis, we discovered that Krt17 and Krt8 can be used to identify each of the two transitional transplanted cell clusters, which we termed “transitional 1” and “transitional 2,” respectively. Transitional 1 cells expressed Krt17 and basal cell markers Trp63 and Krt5, yet these cells also shared AT1-, club-, goblet-, and basal-cell signatures (Figure S3F). Transitional 2 cells were Krt8 expressing and expressed some AT2 markers, such as Adh6a and Aldh1a3, while also identifying with AT1, club, goblet, and basal cell signatures. Recent studies have identified both KRT8- and KRT17-expressing epithelial cells in the distal lung either in a pathological state or during alveolar regeneration (Choi et al., 2020; Strunz et al., 2020). Transitional 1 and 2 cells also show heightened correlation with several signaling pathways known to be involved with pathologic and regenerative states, such as Wnt, Notch, and transforming growth factor β (TGF-β) signaling based on hallmark and Gene Ontology signature scores (Figure S3G; Table S1; Fernandez and Eickelberg, 2012; Guseh et al., 2009; Königshoff and Eickelberg, 2010; Pardo-Saganta et al., 2015; Tammela et al., 2017; Zacharias et al., 2018; Zepp and Morrisey, 2019).
We used RNA velocity to identify cell-state transitions in the transplanted cells we identified with scRNA-seq (La Manno et al., 2018). Several studies have identified KRT8-expressing transitional AT2 cells (Choi et al., 2020; Strunz et al., 2020), supporting the idea that, in our study, KRT8-expressing cells are transitional AT2 cells, giving rise to KRT17-expressing cells in a regenerative state. Although RNA velocity cannot definitively deduce progenitor cell activity and differentiation, the results revealed a possible transition from Krt8-expressing transitional 2 cells to Krt17-expressing transitional 1 cells based on gene expression dynamics (Figure 3F). IF confirmed the presence of KRT8-expressing, KRT17-expressing, and dual KRT8- and KRT17-expressing transplanted cells in the distal lung of SPO-transplanted mice (Figure 3G).
Transplanted cells retain progenitor cell potential in vivo and in vitro
To evaluate the functional abilities of transplanted SNO or SPO cells, we first tested whether the transplanted cells retained their progenitor cell function in culture. We isolated DsRed+ transplanted cells from the SNO or SPO recipients by FACS and then cultured them in our organoid conditions (Figure 4A). DsRed+ cells from SNO-transplanted mice only produced alveolar organoids, and DsRed+ cells from SPO-transplanted mice produced both alveolar and bronchiolar organoids (Figure 4A). Thus, both SPO and SNO cells retain their progenitor cell function and lineage potential when transplanted in vivo and in repeated organoid cultures.
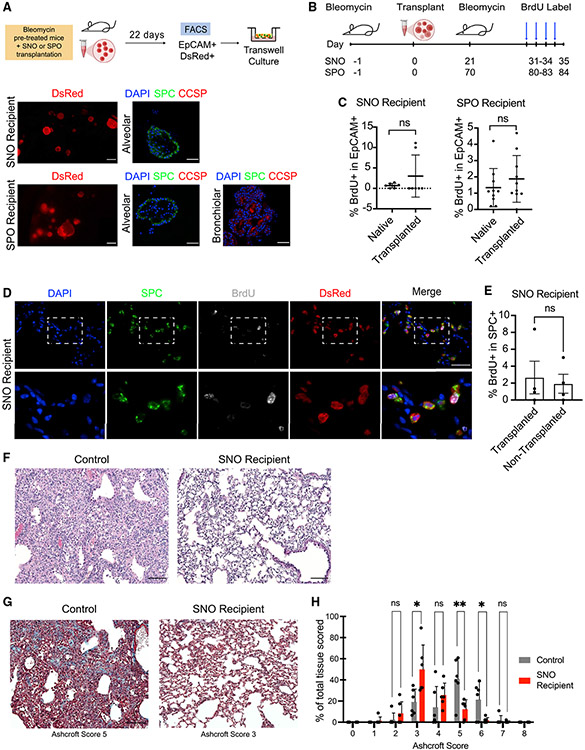
Figure 4. Transplanted cells retain progenitor cell potential in vivo and in vitro.
(A) Top row: experimental strategy to isolate DsRed+ transplanted cells from transplanted mice followed by in vitro culture in a 3D organoid system. Lower left column: representative IF pictures of in-well 3D organoids cultured from DsRed+ cells isolated from transplanted mice are shown. Scale bars represent 200 μm. Lower middle and right columns: representative pictures of IF staining on organoids cultured from transplanted cells are shown. Right: blue, DAPI; green, SPC; red, CCSP. Scale bars represent 50 μm.
(B) Experimental strategy to induce a second injury following transplantation and to observe donor cell proliferation capacity.
(C) Flow cytometry quantification of epithelial (EpCAM+) cells by native or transplanted cell origin that incorporate BrdU in SNO- and SPO-recipient mice. p value was determined using the Student’s t test. Not significant (ns), p ≥ 0.05. Data are represented as mean ± SEM.
(D) Representative pictures of IF staining of SNO transplant lungs that received a second injury and BrdU incorporation. An engrafted area is shown. Blue, DAPI; green, SPC; gray, BrdU; red, DsRed. Scale bar represents 50 μm.
(E) IF quantification of transplanted and non-transplanted areas of SNO-recipient mice: BrdU/SPC double-positive cells normalized to total SPC+ cells. Quantification was performed on three fields of transplanted or non-transplanted regions of recipient mouse lung. p value was determined by Student’s t test. ns, p ≥ 0.05; n = 4 per group. Data are represented as mean ± SEM.
(F) Representative H&E images of control pre-injured lung and pre-injured SNO-transplanted lung at a middle time point after bleomycin. Scale bar represents 200 μm.
(G) Representative images of Masson’s trichrome staining of control pre-injured lung (no transplant) and pre-injured SNO-transplanted lung at a late time point after bleomycin. Ashcroft scores are indicated. Scale bar represents 200 μm.
(H) Comparison of Ashcroft score in RagKO SNO-recipient mice and control mice. p value was determined using the Student’s t test. *p < 0.05; **p < 0.01; n = 6 per group. Data are represented as mean ± SEM.
See also Figure S4.
We next asked whether transplanted cells retained progenitor cell function in vivo by examining their ability to proliferate after a subsequent injury. SNO- and SPO-recipient mice were administered a second injury of bleomycin after transplantation at the corresponding time points when we observed contribution of transplanted cells to SPC-expressing cells (21 days later for SNO and 70 days later for SPO; Figures 4B and S1H-S1K). Ten days after the second injury, mice were administered bromodeoxyuridine (BrdU) to label all proliferative cells for 4 consecutive days. IF confirmed the presence of proliferative cells in the distal lungs of transplanted mice that received a second dose of bleomycin (Figures S4A and S4B). Flow cytometry analysis for BrdU+ cells within the EpCAM+ lung epithelial cell population of transplanted mice revealed that there were proliferative native and transplanted cells (Figure 4C). In both SNO- and SPO-transplanted mice, native and transplanted cells were equally capable of proliferating in response to a second bleomycin injury (Figure 4C). Furthermore, IF confirmed the presence of BrdU+-transplanted cells in the distal alveolar region of SNO- and SPO-transplanted mice (Figures 4D and S4C). The abundance of BrdU+ AT2 cells was comparable between transplanted and non-transplanted areas of SNO-recipient lungs (Figure 4E). HOPX was detected in SNO-transplanted cells in recipient mice that were given a second bleomycin injury (Figure S4D). Notably, DsRed+ HOPX+ cells were cuboidal with rounded nuclei rather than the flat morphology of AT1 cells, similar to the DsRed+ HOPX+ cells in single-injury mice (Figure S1E).
Finally, we asked whether transplanted cells had a beneficial impact on the lungs of recipient mice. Given that SNO-transplanted cells resembled native alveolar epithelial cells in transcriptomic analyses (Figure 2) while SPO-transplanted cells resembled a mixture of native and transitional cell types (Figure 3), we focused on single-injury SNO recipients. Histological analysis of SNO-transplanted mouse lungs suggested relatively healthy lungs post-transplant based on lung structure by hematoxylin &eosin (H&E) staining, cell morphology by IF, and expression of AT2 cell markers by IF (Figures 1 and 4F). We used the Ashcroft scale to evaluate immune cell infiltrate and fibrosis caused by bleomycin treatment. Bleomycin pre-conditioned SNO recipients from early and middle time points were compared with injury- and time-point-matched mice that did not receive transplanted cells (Figures S4E and S4F; Hübner et al., 2008). Transplanted mouse lungs had a distribution of lower Ashcroft scores (Figures 4G and 4H). SNO-recipient mice had significantly fewer fields with Ashcroft scores 5 or 6 as compared with controls, and SNO recipients had significantly more fields with a score of 3 (Figures 4G and 4H). These data suggested transplanted cells improved some aspects of the bleomycin injury response in vivo.
DISCUSSION
Our study demonstrates that lung organoid cells retain progenitor cell function after transplantation. Previous studies have utilized different techniques for pre-conditioning along with different sources of cells, making direct comparisons between studies challenging. Together, our work provides a system using organoid cultures, in vivo transplantation assays, and scRNA-seq to interrogate the functional capacity of lung progenitor cells.
Our data suggest that lung alveolar epithelial progenitor cells in organoid conditions maintain their identity and progenitor cell potential that can be restored after they are returned to the lung environment. When SNO cells were transplanted, the transcriptional signatures of the transplanted cells correlated highly with native AT2 cells. Some key differences between transplanted and native AT2 cells remain. For example, transplanted cells are enriched for expression of neutrophil-activating pathways, including genes such as Lgals3 (galectin-3), which can regulate epithelial proliferation and neutrophil activation (Pilette et al., 2007; Table S4). These pathways have been linked to the rejection of transplanted tissue and organs (Scozzi et al., 2017). Methods to suppress neutrophil activation in response to SNO or SPO transplantation may result in higher rates of engraftment and an improved ability for transplanted cells to avoid immune clearance and transition into a native state. SCA1− cells from uninjured mice and transplanted mice only gave rise to alveolar organoids. Thus, they are limited to an alveolar lineage and retain their identity upon retention in recipient lungs. Importantly, transplanted SNO cells proliferated in response to repeated alveolar injury.
Despite the presence of AT1 cells in SNO cultures, they were not retained or produced in recipient mice at the time points analyzed. However, the presence of DsRed+ DATP-like KRT8-expressing cells and DsRed+ cells with HOPX staining in SNO-recipient mice suggests that some transplanted cells may be in a transitional state towards AT1 cell fate. Others have described an increase in HOPX expression in the alveolar epithelium during injury-induced alveolar regeneration, such as after bleomycin instillation and pneumonectomy (Ota et al., 2018; Wang et al., 2018). Weiner et al. (2019) showed that freshly sorted AT2 cells can differentiate into AT1 cells upon transplantation, yet this was not observed with cultured AT2 cells in the same study. It may be possible to define conditions that allow the SNO cells to further differentiate into AT1 cells in vivo. Despite the lack of contribution to AT1 cells, transplanted SNO cells had a beneficial effect on recipient mouse lung response to injury seen by Ashcroft analysis. Although we ruled out an effect on the proliferative response of native AT2 cells, it will be of great interest to further explore non-cell-autonomous effects of the transplanted organoid cells to explain this observation.
Similar to alveolar progenitors, organoids derived from multi-potent lung progenitor cells retained their potential in vivo. SCA1+ cells generated both bronchiolar and alveolar organoids, and this potential was reflected in our transplantation studies. The transcriptional signatures of transplanted SPO cells aligned with a variety of epithelial cells, including AT2, ciliated, basal, club, and goblet cells. At the middle time point we analyzed by scRNA-seq, transplanted SPO cells included distinct clusters with transcriptional hallmarks of AT2 cells and ciliated cells. Although transplanted SPO cells did not contain a transcriptionally discrete club cell cluster, we observed intermediate cell states that we hypothesized to transition towards AT2, AT1, goblet, or club cell fates. We additionally observed transplanted SPO cells contribute to alveolar cells by scRNA-seq at middle time points and by IF at late time points. These data suggest that transplanted SPO cells may have broad lineage potential and different timelines for differentiation in vivo, and it may be possible to define time frames or altered conditions that allow SPO cells to differentiate into club cells and other bronchiolar cell types.
Our findings demonstrate the utility of bleomycin injury to study donor cell contributions to airway epithelia in addition to alveolar epithelia. Although traditionally used to model alveolar injury, our results and others’ indicate that bleomycin injures other cell types. We observe injuries of the proximal and distal lung, as evidenced by loss of airway and alveolar cells. Others have also described changes in CCSP expression following bleomycin administration (Daly et al., 1998). Consistent with these findings, we show that SPO cells are retained in both distal alveolar and airway regions. scRNA-seq and IF results demonstrated that SPO-transplanted cells contributed to ciliated cells in the bleomycin-injured airway. In the bleomycin-injured alveolar niche, engrafted SNO cells had AT2 cell features and retained proliferative potential post-transplantation. Thus, the transplantation assay can be applied to model cell-based therapy for lung diseases with different candidate donor cells.
We find evidence that cellular programs resembling multiple lineages can be modeled in transplantation and further suggest that specific microenvironmental cues can cause epithelial cells to switch between pathologic and healthy states. A subset of transplanted SNO cells were in a primed AT2 transcriptional state, and we identified DsRed+ KRT8+ cells in SNO-recipient lungs reminiscent of transitional progenitors in fibrotic disease states (Choi et al., 2020; Strunz et al., 2020) not present in culture. We observed similar transitional cells in SPO transplantation, which express a spectrum of features belonging to alveolar, secretory, and airway epithelial cells, raising the possibility that a common transitional phenotype can give rise to multiple cell lineages in disease repair. These transitional SPO-transplanted cells formed patches that resemble atypical adenomatous hyperplasia, a precursor lesion of lung adenocarcinoma (Mori et al., 2001), which resolved at later time points. Several pathways were upregulated in these transitional cell types, including the TGF-β and Wnt pathways, both of which have been described to regulate cell differentiation and tumorigenesis (Chen et al., 2007; Eser and Jänne, 2018; Nabhan et al., 2018; Tammela et al., 2017; Zacharias et al., 2018). Future studies using in vivo transplantation could have important implications for understanding how cues from the environment can drive cellular programs, such as signaling that distinguishes a resolution cell fate from a hyperplastic fate. Such studies would be important to understanding several pulmonary diseases, including lung adenocarcinoma and pulmonary fibrosis, among others.
Limitations of the study
Reactivation of native AT2 transcriptional profiles by transplanted SNOs was partial; hence, additional work is required to identify ways to promote transplanted SNOs to fully reactivate native transcriptional profiles. A mechanistic understanding of how SNOs can suppress and reactivate native AT2-gene-expression programs remains unknown. In addition, the ability of SNOs to contribute to transplanted lungs after a secondary injury using lineage tracing and a larger cohort of mice would strengthen our conclusions. Further analysis of the complexity of SPO cultures, the engraftment abilities of this heterogeneous population, and the ability of populations of cells in the SPO cultures to contribute to lungs post-transplantation requires additional work. Future studies are also required to determine how SNO cells benefit recipient mouse lung injury response; the long-term functions of transplanted SNOs, such as their ability to produce surfactant proteins; and whether pulmonary functions are altered in breathing after transplantation. Lastly, future work will interrogate the ability of SNOs and SPOs to engraft into the lungs after different types of injury and the physiological effects of lung progenitor cell transplantation through pulmonary function testing.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Carla F. Kim (carla.kim@childrens.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All raw and processed scRNA-Seq data were deposited to the NCBI Gene Expression Omnibus (GEO) and Sequencing Read Archive (SRA) under the accession code GEO: GSE190565. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code for the scRNA-Seq analysis are available on Github as Jupyter notebooks: https://github.com/alm8517/Lung_organoid_transplantation.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL DETAILS
For organoid generation, 8–12 weeks old DsRed.T3 mice of both sexes were used. This strain was purchased from the Jackson Laboratory. Mice were maintained in virus-free conditions and were housed in groups. All mouse experiments were approved by the BCH Animal Care and Use Committee, accredited by AAALAC, and were performed in accordance with relevant institutional and national guidelines and regulations.
For injury and transplantation assays, 8–12 weeks old Rag1 KO and Nude mice of both sexes were used. Rag1 KO mice are available from The Jackson Laboratory and Nude mice are available from ENVIGO. Mice were maintained in virus-free conditions and were housed in groups. Littermates were randomly assigned to experimental groups. All mouse experiments were approved by the BCH Animal Care and Use Committee, accredited by AAALAC, and were performed in accordance with relevant institutional and national guidelines and regulations.
METHOD DETAILS
Lung preparation and flow cytometry
Lungs were prepared and cells were flow sorted as previously described (Lee et al., 2014). Briefly, mice were anesthetized with avertin overdose. Lungs were perfused with cold PBS followed by intratracheal instillation of 2 mL dispase (Corning). Lungs were placed on ice, minced, and incubated in 0.0025% DNase (Sigma-Aldrich) and 100 mg/mL collagenase/dispase (Roche) in PBS for 45 min at 37°C. Cells were then sequentially filtered through 100- and 40-μm cell strainers (Falcon) and centrifuged at 1000 rpm for 5 min at 4°C. Cells were resuspended in red blood cell lysis buffer (0.15 M NH4Cl, 10mM KHCO3,0.1 mM EDTA) for 90 s at room temperature, followed by addition of DMEM (Gibco) and FBS. Cells were centrifuged at 1000 rpm for 5 min at 4°C and then resuspended in PBS/10% FBS for further staining. The following antibodies were used: anti-CD31 APC, anti-CD45 APC, anti-Ly-6A/E (SCA1) APC/Cy7 (all Thermo Fisher Scientific), anti-CD326 (EpCAM) PE/Cy7 (Biolegend) (all 1:100). DAPI (Sigma-Aldrich) was used to eliminate dead cells. Single stain controls and fluorescence minus one (FMO) controls were included for each experiment. FACS was performed on a FACSAria II and analysis was done on FlowJo (BD).
Lung organoid culture
Lung organoids were established as previously described (Lee et al., 2014). Briefly, freshly sorted cells from DsRed.T3 mice were resuspended in 3D media (DMEM/F12 (Gibco), 10% FBS, penicillin/streptomycin, 1mM HEPES, and insulin/transferrin/selenium (Corning) at a concentration of 5,000 live cells per well (trypan blue negative). As supporting cells, a mix of neonatal stromal cells was isolated as described previously (Lee et al., 2014). The stromal cells were resuspended in growth factor reduced Matrigel (Corning) at a concentration of 50,000 cells per well. Equal volumes of cells in 3D media and stromal cells in growth factor reduced Matrigel were mixed and pipetted into a Transwell (Corning). Plates were incubated for 20 min at 37°C and 5% CO2 until Matrigel solidified. Lastly, 3D media was added to the lower chamber and media was changed every other day.
Preparing single cell suspensions of organoid cultures
At day 21 of organoid culture, 100 μL dispase (Corning) was added to the top chamber of the transwell and incubated for 1 h at 37°C and 5% CO2. Following dissociation of Matrigel, the wells were washed with PBS and organoids were collected. Organoids were centrifuged at 300 × g for 5 min and then resuspended in 0.25% Trypsin-EDTA (Invitrogen) at 37°C and 5% CO2 for 10 min to obtain a single cell suspension. Trypsin was quenched by addition of PBS/10% FBS.
Bleomycin administration
8-12 weeks old Rag1 KO and Athymic Nude mice were anesthetized with avertin until mice were unresponsive to toe pinch. The mice were positioned on an intratracheal intubation stand and 1.5U/kg of bleomycin (Sigma-Aldrich) was delivered intratracheally through a 22G catheter.
Transplantation assay
8-12 weeks old Rag1 KO or Athymic Nude mice were given 1.5U/kg bleomycin intratracheally one day before transplantation. The following day, single cell suspensions from organoid cultures were prepared as described in the section “preparing single cell suspensions of organoid cultures.” Mice were anesthetized with avertin until they were unresponsive to toe pinch. Organoid cells were resuspended in 40 μL of PBS/10% FBS and were administered into the lungs of the mice intratracheally through a 22G catheter. Mice were sacrificed and the left lung lobe was used for histological analysis, while the right lung lobes were used for flow cytometry analysis. For histology analysis, the left lung lobe was fixed by injection with 10% neutral-buffered formalin. For flow cytometry analysis, the lungs were prepared as described in the section “lung preparation and flow cytometry.” Flow cytometry analysis of recipient mice lungs was performed on a LSRII and analysis was done on FlowJo (BD). Early time point transplant mice were transplanted with 0.1–1.0 million cells and sacrificed 14–24 days after transplantation (SNO, n = 14; SPO, n = 7). Middle time point transplant mice were transplanted with 0.1–1 million cells and sacrificed 42–84 days after transplantation (SNO, n = 14; SPO, n = 8). The late time point transplant mice were transplanted with 0.2–0.3 million cells and sacrificed 112–119 days after transplantation (SPO, n = 7).
Reinjury mouse model and BrdU incorporation
Transplantation assays were performed on Athymic Nude mice as described in the section “Transplantation Assay.” 21 days following transplantation of SNO cells (n = 7), mice were administered another dose of bleomycin at 1.5U/kg intratracheally. 10 days following the subsequent bleomycin administration, 50mg/kg of bromodeoxyuridine (BrdU) (Sigma-Aldrich) was administered intraperitoneally once daily for 4 days before mice were sacrificed.
Mice were sacrificed and the left lung lobe was used for histological analysis, while the right lung lobes were used for flow cytometry analysis. Detection of BrdU by flow cytometry was performed with a BrdU Flow kit (BD). For histology analysis, the left lung lobe was fixed by injection with 10% neutral-buffered formalin. For flow cytometry analysis, the lungs were prepared as described in the section “lung preparation and flow cytometry.” Flow cytometry analysis of recipient mice lungs was performed on a LSRII and analysis was done on FlowJo (BD).
Histology and immunofluorescence
Mouse lung tissues were perfused, inflated, and fixed with 10% neutral-buffered formalin overnight at room temperature. After rinsing with 70% ethanol, paraffin sections were used for histology and IF analysis. Organoid cultures were fixed with10% neutral-buffered formalin overnight at room temperature. After rinsing with 70% ethanol, the organoid culture containing Matrigel plug was immobilized with Histogel (Thermo Scientific) for paraffin embedding. Paraffin sections were used for histology and IF analysis. Sectioned lung tissues or organoid slides underwent deparaffinization by incubation with xylene and rehydration in 100%, 95%, and 70% ethanol successively. Slides were stained with hematoxylin and eosin (H&E) or processed for IF staining. For IF, antigen was retrieved by incubating the slides in citric acid buffer (pH 6) at 95°C for 20 min. Slides were washed with PBS/0.2% Triton X-(PBS-T) and blocked with 10% normal donkey serum for 1 h at room temperature. Primary antibodies were incubated overnight at 4°C at the indicated dilutions: goat anti-SPC (1:100, Santa Cruz Biotechnology), rabbit anti-SPC (1:500, Abcam), mouse anti-RFP (1:100, Invitrogen), rabbit anti-CC10 also known as rabbit anti-CCSP (1:100, Santa Cruz Biotechnology), rat anti-SOX2 (1:100, eBioscience), rabbit anti-p63 (1:100, Cell Signaling Technology), rat anti-cytokeratin-8 (1:300, DSHB), rabbit anti-keratin 17 (1:100, Cell Signaling Technology), rat anti-BrdU (1:100, Abcam), rabbit anti-acetyl-α-tubulin (1:100, Cell Signaling Technology) and mouse anti-Hop (1:200, Santa Cruz Biotechnology). Slides were incubated with Alexa Fluor-coupled secondary antibodies for 1 h at room temperature (all Invitrogen, 1:200, see Key resources table). Slides were mounted using Prolong Gold with DAPI (Invitrogen). For all images shown, either RagKO or nude mice were used and transplanted with the number of organoid cells described in the section “Transplantation assay.” Early, middle, and late time points used for IF are also described in the section “Transplantation assay.” Quantification as seen in Figures S1G and 4E were completed by acquiring 3 fields each of transplanted and non-transplanted areas of SNO recipient mice and analyzing each field with ImageJ. All images used in these analyses were scaled equivalently to subtract background staining, which was determined with negative controls for primary antibodies.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-CD45 APC [30-F11, BD] | Thermo Fisher Scientific | Cat# BDB559864; RRID:AB_398672 |
| Rat monoclonal anti-CD31 APC [MEC 13.3, BD] | Thermo Fisher Scientific | Cat# BDB551262; RRID:AB_398497 |
| Rat monoclonal anti-CD326 (Ep-CAM) PE/Cy7 [G8.8] | Biolegend | Cat# 118216; RRID:AB_1236471 |
| Rat monoclonal anti-Ly-6A/E (Sca1) APC/Cy7 [D7] | Thermo Fisher Scientific | Cat# 560654; RRID:AB_1727552 |
| Rabbit monoclonal anti-SP-C [EPR19839] | Abcam | Cat# ab211326 |
| Goat polyclonal anti-SPC (M-20) | Santa Cruz Biotechnology | Cat# sc-7706; RRID:AB_2185507 |
| Mouse monoclonal anti-RFP (RF5R) | Invitrogen | Cat# MA5-15257; RRID:AB_10999796 |
| Rabbit polyclonal anti-CC10 (FL-96) | Santa Cruz Biotechnology | Cat# sc-25555; RRID:AB_2269914 |
| Rat monoclonal anti-Sox2 (Btjce) | eBioscience | Cat# 14-9811-82; RRID:AB_11219471 |
| Rabbit monoclonal anti-p63-α (D2K8X) | Cell Signaling Technology | Cat# 13109S; RRID:AB_2637091 |
| Rat monoclonal anti-cytokeratin-8 | Developmental Studies Hybridoma Bank | Cat# TROMA-I; RRID:AB_531826 |
| Rabbit monoclonal anti-Keratin 17 (D73C7) | Cell Signaling Technology | Cat# 4543; RRID:AB_2133014 |
| Rat monoclonal anti-Brdu [BU1/75 (ICR1)] | Abcam | Cat# ab6326; RRID:AB_305426 |
| Rabbit monoclonal anti-Acetyl-α-Tubulin (Lys40) (D20G3) | Cell Signaling Technology | Cat# 5335; RRID:AB_10544694 |
| Mouse monoclonal anti-HopX antibody (E-1) | Santa Cruz Biotechnology | Cat# 398703; RRID:AB_2687966 |
| Donkey anti-rat Alexa Fluor 488 | Invitrogen | Cat# A-21208; RRID:AB_141709 |
| Donkey anti-rat Alexa Fluor 594 | Invitrogen | Cat# A-21209; RRID:AB_2535795 |
| Donkey anti-goat Alexa Fluor 488 | Invitrogen | Cat# A-11055; RRID:AB_2534102 |
| Donkey anti-rabbit Alexa Fluor 488 | Invitrogen | Cat# A-21206; RRID:AB_2535792 |
| Donkey anti-rabbit Alexa Fluor 594 | Invitrogen | Cat# A-21207; RRID:AB_141637 |
| Donkey anti-mouse Alexa Fluor 647 | Invitrogen | Cat# A-31571; RRID:AB_162542 |
| Chemicals, peptides, and recombinant proteins | ||
| Growth Factor Reduced Matrigel | Corning | Cat# 356231 |
| Bleomycin Sulfate | Sigma-Aldrich | Cat# B2434 |
| Dispase | Corning | Cat# 354235 |
| Collagenase/Dispase | Roche | Cat# 10269638001 |
| DNase | Sigma-Aldrich | Cat# D4527 |
| 0.25% Trypsin-EDTA | Invitrogen | Cat# 25200-056 |
| ITS | Corning | Cat# 25-800-CR |
| BrdU | Sigma-Aldrich | Cat# B9285 |
| Critical commercial assays | ||
| FITC BrdU Flow Kit | BD | Cat# 559619 |
| Chromium Single Cell 3′ Library & Gel Bead Kit v3, 16 reactions | 10x Genomics | Cat# 1000075 |
| Chromium Chip B Single Cell Kit, 48 reactions | 10x Genomics | Cat# PN-10000153 |
| Chromium i7 Sample Index Plate - 96 reactions | 10x Genomics | Cat# 220103 |
| Deposited data | ||
| Single cell RNA-seq raw data | This paper | GEO: GSE190565 |
| Single cell RNA-seq features/matrix/barcode files | This paper | GEO: GSE190565 |
| Jupyter notebooks for single cell RNA-Seq analysis | This paper | https://github.com/alm8517/Lung_organoid_transplantation |
| Experimental models: Organisms/strains | ||
| Hsd:Athymic Nude-Foxn1nu (Nude) | ENVIGO | Cat# 6903F |
| B6.129S7-Rag1tm1Mom/J (Rag1 KO) | The Jackson Laboratory | Cat# 02216 |
| B6.Cg-Tg(CAG-DsRed*MST)1Nagy/J (DsRed.T3) | The Jackson Laboratory | Cat# 6051 |
| Software and algorithms | ||
| Image J | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | Becton, Dickinson & Company | https://www.flowjo.com/ |
| Scanpy 1.4.6 | Wolf et al. (2018) Genome Biology | https://github.com/theislab/scanpy |
| Velocyto 0.17.17 | La Manno et al. (2018) Nature | https://github.com/velocyto-team/velocyto.py |
| scVelo 0.2.1 | Theis lab | https://github.com/theislab/scvelo |
| CellRanger 3.1.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Markov Affinity-based Graph Imputation of Cells (MAGIC) 1.5.5 | van Dijk et al. (2018) Cell | https://github.com/KrishnaswamyLab/MAGIC |
| Scanorama 1.6 | Hie et al. (2019) Nature Biotechnology | https://github.com/brianhie/scanorama |
| Other | ||
| ProLongGold Antifade Mountant with DAPI | Invitrogen | Cat# P36935 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Transwells | Corning | Cat# 3470 |
| SPRIselect Reagent | Beckman Coulter | Cat# NC0406407 |
| Qubit dsDNA HS Assay Kit | Invitrogen | Cat# Q32851 |
| DMEM/F12 | Gibco | Cat# 11330-057 |
| DMEM | Gibco | Cat# 11995-065 |
| HEPES | Invitrogen | Cat# 15630-080 |
| Penicillin-Streptomycin | Gibco | Cat# 15140-122 |
| Histogel | Thermo Scientific | Cat# HG4000012 |
Ashcroft scoring
Fibrosis scoring was performed as described in Hübner et al., (2008). Briefly, one paraffin section per mouse was stained with Masson’s Trichrome. Slides were de-identified and assessed by a researcher who was blinded to the groups. Forty-five images at 10x magnification were scored per mouse and the average score for each mouse was used to calculate the mean Ashcroft score for each group.
Generation and analysis of scRNA-Seq data
Mouse and organoid cell preparation
For single-cell RNA sequencing experiments, transplanted mice are 8–12 weeks old RagKO mice that were given 1.5U/kg bleomycin intratracheally one day before transplantation. Mice were transplanted with 0.4 million SNO cells (n = 3) or 0.35 million SPO cells (n = 2) the following day. 84 and 70 days after transplantation, SNO and SPO recipient mice were sacrificed, respectively. For SNO recipient mice, an EpCAM + DsRed+ (transplanted cells) and an EpCAM + DsRed− SCA1− (native cells) population were FACS sorted followed by downstream preparation for sequencing. For SPO recipient mice, an EpCAM + DsRed+ (transplanted cells) population was FACS sorted followed by downstream preparation for sequencing. Wildtype mice for SCA1+ (Figure 3) native cell analysis did not receive bleomycin injury or transplanted cells (n = 3). Wildtype mice were FACS sorted for an EpCAM + SCA1+ (native cells) population followed by downstream preparation for sequencing.
At 21 days of culture, organoids were dissociated as described in the section “preparing single cell suspensions of organoid cultures.” Dissociated cells were FACS sorted for DsRed + cells for both SNO (n = 2) and SPO (n = 3) cells followed by downstream preparation for sequencing.
Library preparation
ScRNA-Seq was performed using the 10X Genomics platform (10X Genomics, Pleasanton, CA). FACS sorted cells from either mice or organoid cultures were encapsulated with a 10X Genomics Chromium Controller Instrument using the Chromium Single Cell B Chip Kit. Encapsulation, reverse transcription, cDNA amplification, and library preparation reagents are from the Chromium Single Cell Library & Gel Bead Kit v3. Libraries were indexed using index sequences from a Chromium i7 Sample Index Plate (10X Genomics). Briefly, single cells were resuspended in PF10 at a concentration of 1000 cells mL−1. The protocol was performed as per 10X Genomics protocols without modification (chromium single cell 3 reagent kits user guide v3 chemistry). Nucleic acid size selection steps were performed using SPRIselect reagent (Beckman Coulter). Total cDNA and cDNA quality following amplification and cleanup was determined using a QubitTM dsDNA HS assay kit and the Agilent TapeStation High Sensitivity D5000 ScreenTape System. Library quality pre-sequencing was determined using an Agilent TapeStation and QPCR prior to sequencing. TapeStation analysis and library QPCR was performed by the Biopolymers Facility at Harvard Medical School. Libraries were sequenced using an Illumina NextSeq500 using paired-end sequencing with single indexing (Read 1 = 26 cycles, Index (i7) = 8 cycles, and Read 2 = 98 cycles). Reads were aligned to the mm10 reference genome and count matrices were generated using CellRanger Count 3.1.0 (10X Genomics).
Bioinformatics for SCA1-cell scRNA-Seq data
Count matrices for SCA1-samples were read into the Python single cell analysis environment SCANPY (v 1.4.6) (Wolf et al., 2018). In brief, cells with >20% mitochondrial content and outlier read counts were removed. The data was normalized, logarithmized, and the significant number of principal components determined using in-built Scanpy functions. Data was denoised using Markov Affinity-based Graph Imputation (v 1.5.5) using the following settings (Gene to return = all_genes, knn = 5, t = 5, n_pca = 20) (van Dijk et al., 2018). Cells with high expression of the stromal marker Vim and Leiden clusters with fewer than 200 cells were removed before downstream analysis.
Bioinformatics for SCA1+ cell scRNA-Seq data
Count matrices for SCA1+ samples were read into the Python single cell analysis environment SCANPY (v 1.4.6) (Wolf et al., 2018). In brief, cells with >20% mitochondrial content and outlier read counts were removed. The data was normalized, logarithmized, and the significant number of principal components determined using in-built Scanpy functions. Because scRNA-Seq libraries for native SCA1+ cells were prepared at a different time than transplanted SCA1+ cells (DsRed+) and SCA1+ organoid libraries, we performed batch correction using Scanorama (v 1.6) (Hie et al., 2019) prior to data denoising using Markov Affinity-based Graph Imputation (v 1.5.5) (van Dijk et al., 2018). Cells with high expression of the stromal marker Vim were removed before downstream analysis.
RNA velocity
Velocyto (0.17.17) was run on the CellRanger Count output files using the run10X shortcut and the mm10 genome annotation file provided with the CellRanger pipeline. Loom files generated by Velocyto for each sample were concatenated into an anndata object. To visualize velocity on the original UMAP embedding a new anndata was created by merging the velocity and original anndata objects using the utils.merge() function in scVelo (0.2.1). Velocity was calculated using the merged anndata object and in-built velocity functions.
Data visualization
Data was visualized using in-built Scanpy plotting functions, Seaborn (v 0.10.1) (https://seaborn.pydata.org/), and Matplotlib (v 3.0.2) (Hunter, 2007).
Genes used for signature score calculation
Murine AT2 marker genes are from the PanglaoDB (Franzén et al., 2019). Primed AT2 genes are from Choi et al., (2020). Hallmark gene lists are from the MSigDB (Liberzon et al., 2011, 2015; Subramanian et al., 2005). Gene Ontology gene lists are from the Gene Ontology Database (Ashburner et al., 2000).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical testing was performed using GraphPad Prism. The tests used to determine statistical significance are quoted in the appropriate figure legends. Data in graphs are shown as mean ± SEM. p values are indicated in the figures and p values <0.05 were considered significant. The researcher was blinded to the experimental groups for Ashcroft scoring in Figures 4F and 4G.
Supplementary Material
Highlights.
Alveolar organoid cells engraft into the alveolar space
Transplanted lung alveolar cells have transcriptional signature of native AT2 cells
Transplanted alveolar cells proliferate and retain organoidforming capacity
Organoid cells undergo changes to transitional cell states upon transplantation
ACKNOWLEDGMENTS
We thank members of the Kim Lab, D. Kotton, members of the Kotton Lab, E. Morrisey, and W. Zacharias for helpful discussion and feedback. We thank the flow cytometry core facility at Boston Children’s Hospital (BCH), the single-cell core facility at Harvard Medical School (HMS), the HMS biopolymers facility, the DFHCC rodent histopathology facility, and the BCH Pathology Department histology lab. Graphical abstract was created with BioRender.com. This work was supported in part by the Hope Funds for Cancer Research Postdoctoral Fellowship (S.M.L.), a Damon Runyon Cancer Research Foundation postdoctoral fellowship (no. DRG:2368-19) and a Postdoctoral Enrichment Program Award from the Burroughs Wellcome Fund (no. 1019903; A.L.M.), F31 HL159919 (I.G.W.), R01 HL090136, R01 HL132266, R01 HL125821, U01 HL100402, RFA-HL-09-004, R35HL150876, the Cystic Fibrosis Foundation Award KIM19P0, LONGFONDS ∣ Accelerate, project BREATH, Gilda and Alfred Slifka, Gail and Adam Slifka, the Cystic Fibrosis/Multiple Sclerosis Fund Foundation Inc., and the Harvard Stem Cell Institute (C.F.K.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110662.
DECLARATION OF INTERESTS
C.F.K. had a sponsored research agreement with Celgene/BMS Corporation. S.M.L., E.A., and C.F.K. have filed a patent application (PCT/US2020/042961) related to this work. C.F.K. and all other authors declare no additional competing interests.
INCLUSION AND DIVERSITY
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper received support from a program designed to increase minority representation in science. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper self-identifies as living with a disability.
REFERENCES
- Adamson IY, and Bowden DH (1975). Derivation of type 1 epithelium from type 2 cells in the developing rat lung. Lab. Investig. J. Tech. Methods Pathol 32, 736–745. [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BLM (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basil MC, Katzen J, Engler AE, Guo M, Herriges MJ, Kathiriya JJ, Windmueller R, Ysasi AB, Zacharias WJ, Chapman HA, et al. (2020). The cellular and physiological basis for lung repair and regeneration: past, present, and future. Cell Stem Cell 26, 482–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Ye T, Sun Y, Ji G, Shido K, Chen Y, Luo L, Na F, Li X, Huang Z, et al. (2017). Targeting the vascular and perivascular niches as a regenerative therapy for lung and liver fibrosis. Sci. Transl. Med 9, eaai8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, and Vu TH (2011). Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J. Clin. Invest 121,2855–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Desai TJ, Qian J, Niederreither K, Lu J, and Cardoso WV (2007). Inhibition of Tgf signaling by endogenous retinoic acid is essential for primary lung bud induction. Development 134, 2969–2979. [DOI] [PubMed] [Google Scholar]
- Choi J, Park J-E, Tsagkogeorga G, Yanagita M, Koo B-K, Han N, and Lee J-H (2020). Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, and Hogan BLM (2018). Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development 145, dev163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly HE, Baecher-Allan CM, Paxhia AT, Ryan RM, Barth RK, and Finkelstein JN (1998). Cell-specific gene expression reveals changes in epithelial cell populations after bleomycin treatment. Lab. Investig. J. Tech. Methods Pathol 78, 393–400. [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. (2018). Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser PÖ, and Jänne PA (2018). TGFβ pathway inhibition in the treatment of non-small cell lung cancer. Pharmacol. Ther 184, 112–130. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Cabral LJ, Stephens RJ, and Freeman G(1975).Transformation of alveolar Type 2 cells to Type 1 cells following exposure to NO2. Exp. Mol. Pathol 22, 142–150. [DOI] [PubMed] [Google Scholar]
- Fernandez IE, and Eickelberg O (2012). The impact of TGF-β on lung fibrosis: from targeting to biomarkers. Proc. Am. Thorac. Soc 9, 111–116. [DOI] [PubMed] [Google Scholar]
- Franzén O, Gan L-M, and Björkegren JLM (2019). PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, and Rajagopal J (2009). Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 136, 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hie B, Bryson B, and Berger B (2019). Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol 37, 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, and Stripp BR (2001). Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am. J. Respir. Cell Mol. Biol 24, 671–681. [DOI] [PubMed] [Google Scholar]
- Hübner R-H, Gitter W, El Mokhtari NE, Mathiak M, Both M, Bolte H, Freitag-Wolf S, and Bewig B (2008). Standardized quantification of pulmonary fibrosis in histological samples. BioTechniques 44, 507–511, 514-517. [DOI] [PubMed] [Google Scholar]
- Hunter JD (2007). Matplotlib: a 2D graphics environment. Comput. Sci. Eng 9, 90–95. [Google Scholar]
- Kathiriya JJ, Brumwell AN, Jackson JR, Tang X, and Chapman HA (2020). Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell 26, 346–358.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CFB, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, and Jacks T (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835. [DOI] [PubMed] [Google Scholar]
- Königshoff M, and Eickelberg O (2010). WNT signaling in lung disease: a failure or a regeneration signal? Am. J. Respir. Cell Mol. Biol 42, 21–31. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, Wagers AJ, Tseng Y-H, Ryeom S, and Kim CF (2014). Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, et al. (2017). Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170, 1149–1163.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinforma. Oxf. Engl 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclnnes L, Healy J, and Melville J (2020). UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at arXiv. 10.48550/arXiv.1802.03426. [DOI] [Google Scholar]
- Milman Krentsis I, Rosen C, Shezen E, Aronovich A, Nathanson B, Bachar-Lustig E, Berkman N, Assayag M, Shakhar G, Feferman T, et al. (2018). Lung injury repair by transplantation of adult lung cells following preconditioning of recipient mice. Stem Cells Transl. Med 7, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Rao SK, Popper HH, Cagle PT, and Fraire AE (2001). Atypical adenomatous hyperplasia of the lung: a probable forerunner in the development of adenocarcinoma of the lung. Mod. Pathol 14, 72–84. [DOI] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, and Desai TJ (2018). Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichane M, Javed A, Sivakamasundari V, Ganesan M, Ang LT, Kraus P, Lufkin T, Loh KM, and Lim B (2017). Isolation and 3D expansion of multipotent Sox9+ mouse lung progenitors. Nat. Methods 14, 1205–1212. [DOI] [PubMed] [Google Scholar]
- Ota C, Ng-Blichfeldt J-P, Korfei M, Alsafadi HN, Lehmann M, Skronska-Wasek W, M De Santis M, Guenther A, Wagner DE, and Königshoff M (2018). Dynamic expression of HOPX in alveolar epithelial cells reflects injury and repair during the progression of pulmonary fibrosis. Sci. Rep 8, 12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan LH, Yamauchi K, Uzuki M, Nakanishi T, Takigawa M, Inoue H, and Sawai T (2001). Type II alveolar epithelial cells and interstitial fibroblasts express connective tissue growth factor in IPF. Eur. Respir. J 17, 1220–1227. [DOI] [PubMed] [Google Scholar]
- Pardo-Saganta A, Law BM, Tata PR, Villoria J, Saez B, Mou H, Zhao R, and Rajagopal J (2015). Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell 16, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilette C, Colinet B, Kiss R, André S, Kaltner H, Gabius H-J, Delos M, Vaerman J-P, Decramer M, and Sibille Y (2007). Increased galectin-3 expression and intra-epithelial neutrophils in small airways in severe COPD. Eur. Respir. J 29, 914–922. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, and Hogan BLM (2009). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U S A 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C, Shezen E, Aronovich A, Klionsky YZ, Yaakov Y, Assayag M, Biton IE, Tal O, Shakhar G, Ben-Hur H, et al. (2015). Preconditioning allows engraftment of mouse and human embryonic lung cells, enabling lung repair in mice. Nat. Med 21, 869–879. [DOI] [PubMed] [Google Scholar]
- Rowe RG, Mandelbaum J, Zon LI, and Daley GQ (2016). Engineering hematopoietic stem cells: lessons from development. Cell Stem Cell 18, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzi D, Ibrahim M, Menna C, Krupnick AS, Kreisel D, and Gelman AE (2017). The role of neutrophils in transplanted organs. Am. J. Transpl 17, 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano JB, Kendrick PJ, Paulson KR, Gupta V, Abrams EM, Adedoyin RA, Adhikari TB, Advani SM, Agrawal A, Ahmadian E, et al. (2020). Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med 8, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, Tsidiridis G, Lange M, Mattner LF, Yee M, et al. (2020). Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun 11, 3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K, Park Y, Azimi R, Kerper NR, Wesselhoeft RA, et al. (2017).A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep 9, 5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. (2015). Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang Z, Huang H, Li J, Wang Z, Yu Y, Zhang C, Li J, Dai H, Wang F, et al. (2018). Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc. Natl. Acad. Sci. U S A 115, 2407–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AI, Jackson SR, Zhao G, Quansah KK, Farshchian JN, Neupauer KM, Littauer EQ, Paris AJ, Liberti DC, Scott Worthen G, et al. (2019). Mesenchyme-free expansion and transplantation of adult alveolar pro-genitor cells: steps toward cell-based regenerative therapies. Npj Regen. Med 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp JA, and Morrisey EE (2019). Cellular crosstalk in the development and regeneration of the respiratory system. Nat. Rev. Mol. Cell Biol 20, 551–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew A-A, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. (2015). p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517, 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed scRNA-Seq data were deposited to the NCBI Gene Expression Omnibus (GEO) and Sequencing Read Archive (SRA) under the accession code GEO: GSE190565. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code for the scRNA-Seq analysis are available on Github as Jupyter notebooks: https://github.com/alm8517/Lung_organoid_transplantation.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.