Abstract
Histone nonenzymatic covalent modifications (NECMs) have recently emerged as an understudied class of posttranslational modifications that regulate chromatin structure and function. These NECMs alter the surface topology of histone proteins, their interactions with DNA and chromatin regulators, as well as compete for modification sites with enzymatic posttranslational modifications. NECM formation depends on the chemical compatibility between a reactive molecule and its target site, in addition to their relative stoichiometries. Here we survey the chemical reactions and conditions that govern the addition of NECMs onto histones as a manual to guide the identification of new physiologically relevant chemical adducts. Characterizing NECMs on chromatin is critical to attain a comprehensive understanding of this new chapter of the so-called “histone code”.
Keywords: Epigenetics, Histone, Nucleosome, Non-enzymatic covalent modifications, Post-translational modification, Acylation, Glycation, Lipidation, Antioxidant
Introduction
Posttranslational modifications (PTMs) on histones regulate gene expression by organizing chromatin into conformations ranging from closed and transcriptionally silent heterochromatin to open and transcriptionally active euchromatin [1]. In 2000, Strahl and Allis proposed a model which they defined as ‘the histone code’, where ‘writer’ enzymes are responsible for installing a wide range of site-specific modifications, and ‘eraser’ enzymes for removing them [2]. In this model, PTMs can alter chromatin structure directly, by modulating the strength of interactions between histones and DNA through charge or steric effects, or indirectly, by recruiting ‘reader’ proteins which translate permutations of PTMs to a transcriptional output [3].
In recent years, it has become clear that nonenzymatic covalent modifications (NECMs) are prevalent on histones and represent a novel family of PTMs which deviate from this established paradigm of the histone code. In contrast to canonical PTMs, NECMs are formed when reactive molecules within the nucleus spontaneously form covalent bonds to reactive moieties on proteins. Some proteins are particularly susceptible to NECMs, such as the propensity of mitochondrial proteins to react with lipid peroxidation products because of the higher concentration of reactive oxygen species in these organelles [4]. Histones are uniquely prone to accumulating NECMS because of their high percentage of basic, nucleophilic residues and extremely long half-lives [5]. As such, rather than being enzymatically guided and installed on a specific residue, NECM formation is governed by the intrinsic reactivity of the small molecule toward amino acids, its local concentration in the nucleus, and the accessibility of these histone residues [6].
While the chemical reactions that occur in histone NECM formation are conceptually simple, conclusively identifying these adducts in a biological context, resolving the mechanisms for their formation and defining their physiological function have proven challenging. To address the first of these issues, powerful tools, including discovery-based mass spectrometry (MS) methods and chemical probes, have been developed to detect and track NECMs in the complex environment of the cell [7-9]. However, proving a newly discovered histone modification is a bona fide NECM is nontrivial; absence of a known endogenous writer enzyme is not enough, as it is possible that one has simply not yet been discovered. Demonstration of a spontaneous reaction with free histones or reconstituted nucleosomes in vitro provides more compelling evidence but is still insufficient as this may not be physiologically relevant. Moreover, some PTMs can arise both enzymatically and nonenzymatically, and the former may dominate in the cellular context. Finally, as with PTMs, conclusively linking an NECM with an epigenetic effect is arduous. Here, we seek to highlight recently identified hallmarks of histone NECMs and challenges facing their identification. We emphasize the chemical reactions governing these adducts, which may guide identification of new small molecules with the potential to form NECMs.
General properties of histone NECMs
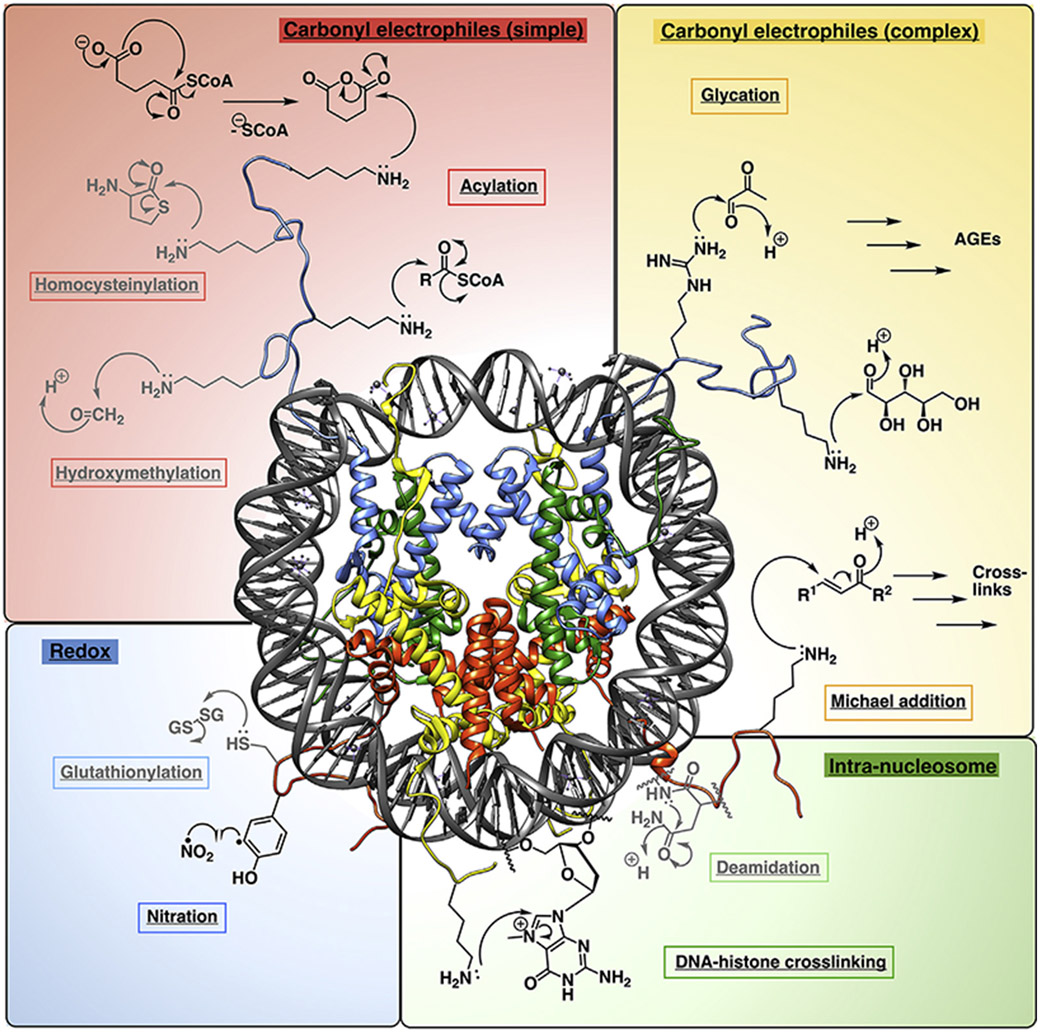
Of the twenty canonical amino acids, nucleophilic side chains display the greatest intrinsic reactivity toward small molecules. These include residues bearing amino (lysine, arginine, and histidine), hydroxyl (serine, threonine, and tyrosine), or thiol groups (cysteine); the latter of which is also susceptible to redox reactions [10]. Therefore, reactive electrophiles form the bulk of known histone NECMs (Figure 1). In principle, histones may also be subject to radical species present in the nucleus; however, in practice, owing to efficient cellular defense mechanisms, there are much fewer examples of these NECMs than what might be expected.
Figure 1. Representative NECM-forming reactions on histones.
Clockwise from top left: reactions with single carbonyl electrophiles including, from top to bottom: acetyl-coenzyme A (coA) derivatives via cyclic intermediates (top) or direct displacement of coA (bottom, R = variable functional group) (acylations), homocysteine (homocysteinylation), and formaldehyde (hydroxymethylation). Top right: reactions with carbonyl electrophiles at multiple carbonyls or distal positions, which give rise to complex mixtures of products. Electrophiles depicted include methylglyoxal (MGO, top), ribose (bottom) (glycation), which both rearrange to form advanced glycation end-products (AGEs) and α,β-unsaturated carbonyls (Michael addition, R1 and R2 = variable functional groups), giving rise to crosslinked products. Bottom right: Intranucleosomal reactions. Electrophiles depicted include asparagine (deamidation) and damaged DNA bases (DNA-histone crosslinking). Bottom left: redox reactions. Species depicted include oxidized glutathione (glutathionylation) and nitrate radical (nitration). Reactions depicted with lysine as the nucleophile may also occur with arginines, and vice-versa. Reactions shown in grey are not discussed in depth in this review.
NECM adducts are formed on accessible, reactive side chains and thus tend to have a broad and variable distribution. This is in contrast to enzymatic PTMs, which have well-defined sites of distribution across the histone, with most concentrated at the N-terminal tails which protrude from the nucleosome core particle and serve as the primary site of interaction with readers. In particular, NECMs accumulate specifically on histones because of their longevity; while they occur more readily on the same exposed lysine- and arginine-rich N-terminal tails as enzymatic PTMs, they have also been observed in their globular and C-terminal domains [11,12].
Examples of histone NECM-forming small molecules
Carbonyl electrophiles
Acyl-coAs
Many cellular metabolic pathways generate reactive carbonyl species as either intermediates or by-products. These readily react with nucleophilic side chains and nucleic acids and comprise a major class of NECM-forming reactive electrophiles. While some species such as formaldehyde participate in simple nucleophilic addition reactions, many contain a good leaving group and are capable of addition-elimination reactions to acylate histone residues. The most common and biologically relevant example of this is the acyl-coenzyme As (acyl-coAs) (Figure 2), where -coA can be readily displaced by spontaneous nucleophilic attack. However, an important caveat to note is that many acylations also occur enzymatically via promiscuous lysine acyltransferases (KATs) such as p300, making it nontrivial to distinguish true NECMs [12].
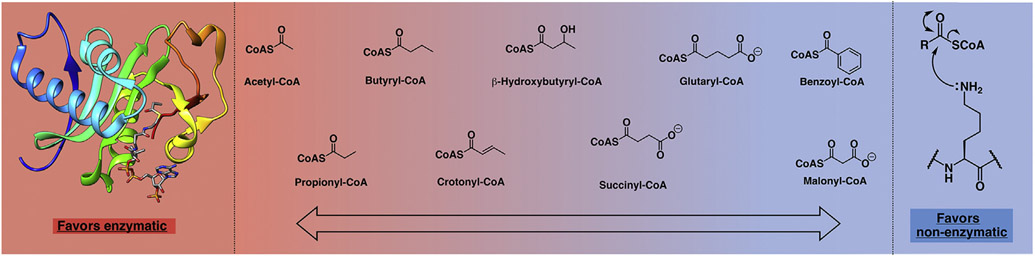
Figure 2. Spectrum depicting the propensity of selected acetyl-coA derivatives to acylate histones ranging from enzymatic (left) to nonenzymatic (right) mechanisms.
Human acyltransferase Gcn5 in complex with propionyl-coA (PDB ID 5H84) is shown as a representative example of an acyltransferase enzyme, and reaction with histone lysine is shown as a representative nucleophile. Substrates, arranged from left to right in order of decreasing amenability to enzymatic reaction and/or increasing propensity for nonenzymatic reaction are acetyl-coA, propionyl-coA, butyryl-coA, crotonyl-coA, β-hydroxybutyryl-coA, succinyl-coA, glutaryl-coA, benzoyl-coA (no evidence for enzymatic acylation), and malonyl-coA (no evidence for enzymatic acylation). Adapted from the study by Xie et al. [11].
Recent analysis has determined that the balance between an acyl-coA’s intrinsic reactivity and how easily it is processed by KATs determines how predominant its corresponding acylation NECM is (Figure 2) [12]. In general, KATs can more easily use the smaller, linear, and charge-neutral derivatives (acetyl-coA, propionyl-coA, and butyryl-coA). In contrast, acidic (malonyl-coA, succinyl-coA, and glutaryl-coA) and bulkier derivatives (β-hydroxybutyryl-coA, benzoyl-coA) are less compatible with KAT active sites and are more likely to be outcompeted by their nonenzymatic counterparts. In addition, some longer derivatives (e.g. succinyl-coA and glutaryl-coA) can access a more reactive 5-membered ring intermediate; for these, nonenzymatic acylation is particularly rapid. Indeed, some modifications which only occur nonenzymatically have been observed to outcompete enzymatic PTMs; for example, benzoylation at H4K8 corresponded to a decrease in H4K8Ac levels compared with untreated controls [13].
Acidic substrates-malonyl-coA
As a negatively charged electrophile, malonyl-coA is a challenging substrate for KATs. Indeed, enzymatic malonylation has not yet been identified, leaving open the possibility that this exists as a pure NECM. Interestingly, histone malonylation has been detected via MS-based methods in yeast under normal conditions on core histones at H2BK116, H3K56, and H2AK119 [11]. The latter modification is particularly significant, as the first example of a nonacetyl, and presumably nonenzymatic acylation to demonstrate crosstalk with an enzymatic phosphorylation PTM. Ishiguro et al. demonstrated that H2AK119mal blocks the Bub1 kinase from interacting with the proximal H2AS121 (H2AT120 in humans), preventing its phosphorylation [14]. This prevents the subsequent recognition of H2AS121p by the Shugoshin proteins, responsible for ensuring sister chromatid cohesion during meiosis, and ultimately leads to chromosome segregation defects.
Bulky substrates
In 2018, Huang et al. reported nonenzymatic histone lysine modification in vivo by benzoyl-coA, which is produced by metabolic processing of the common food additive sodium benzoate at physiologically relevant concentrations [13]. Distributed across common acetylation and crotonylation sites, ChIP-seq analysis identified Kbz as an activating mark; its hydrophobicity and bulkiness are speculated to force chromatin into a more open conformation and induce the transcription of the embedded genes [6]. Moreover, recent studies have identified histone lysine isobutyrylation (Kibu) as a novel PTM [15]. While several acyltransferase enzymes have been confirmed as writers for this mark, structural similarities to other acyl-coAs known to form NECMs, such as β-hydroxybutyryl-coA, suggest isobutyryl-coA may also react nonenzymatically [16].
Reaction via cyclic intermediates-succinyl-coA and glutaryl-coA
Both succinyl- and glutaryl-coA are formed during the TCA and amino acid synthesis cycles. As 4- and 5-C acidic acetyl-coA derivatives, they are also challenging substrates for KATs and able to access cyclic 5- and 6-membered intermediates, respectively, via intramolecular general base catalysis [17,18]. However, an enzymatic writer does exist for these marks at some sites; Gcn5 (KAT2A) has been shown to install H3K79succ [19] and H4K91Glu [20] as part of the Spt-Ada-Gcn5 acetyltransferase complex [21]. Despite the identification of Gcn5 sites, several additional modification sites have been detected via MS-based characterization [11] for which the mode of attachment has not yet been demonstrated. These modifications reverse the charge of histone lysines from positive to negative, leading to pronounced chromatin decompaction; perhaps for this reason, machine learning predictions of novel succinylation and glutarylation sites favor modification proximal to other positive residues [22-26]. Interestingly, desuccinylation at H3K122 by SIRT7 in response to DNA damage was shown to lead to chromatin condensation in a key step in double-strand break repair, suggesting a key role for a potentially nonenzymatic succinylation mark in maintaining genome stability [27,28].
Alternative leaving groups-lactoyl-GSH
Glutathione has recently emerged as another good leaving group for nonenzymatic acylations. Inspired by the discovery of nonenzymatic acetylation of lysines by acetyl-GSH [29], Gaffney et al. demonstrated that several H4 lysines can similarly undergo lactoylation (Kla) in vitro by lactoylGSH, a by-product of the glycolysis pathway [30]. In parallel, Zhang et al. showed that histone lysine lactoylation induces epigenetic changes in M1 macrophages in response to increasing anaerobic glycolytic flux, which in turn promotes homeostasis [31].
Complex reaction products
Glycation
The acyl-CoA and acyl-GSH derivatives discussed so far bear only a single reactive carbonyl, meaning modification is only possible through one site. In contrast, species containing multiple sites for nucleophilic attack, such as dicarbonyls or α,β unsaturated carbonyls, give rise to a complex mixture of products. A salient example of NECMs with multiple carbonyls is glycation, which has recently emerged as a histone NECM with enormous potential for understanding diseases where the glycolysis pathway is perturbed, such as cancer and diabetes (Figure 3a).
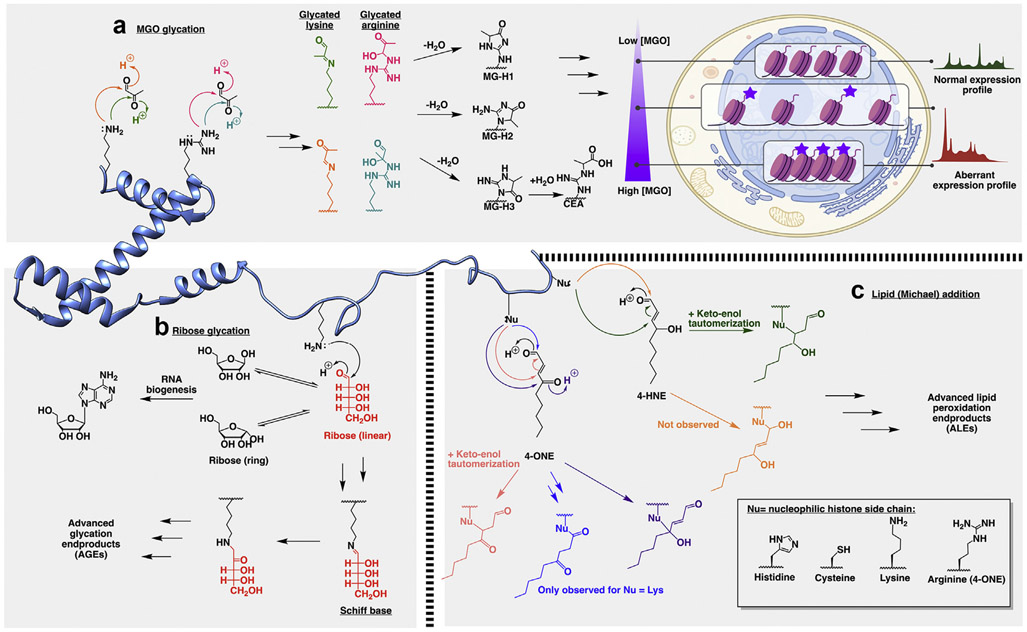
Figure 3. Selected reactions of histones with substrates bearing multiple electrophilic sites.
(a) Reaction of lysine (left) or arginine (right) with methylglyoxal (MGO) results in a mixture of reaction products. The two arginine derivatives (right, pink and teal) can then undergo further dehydration and hydride shift to form methylglyoxal derivatives MG-H1, MG-H2, and MG-H3, which can undergo further rearrangement to form the AGE CEA. The glycated lysine and arginine derivatives can further rearrange to form an array of AGEs including inter- and intra-histone crosslinks. At low concentrations, glycation (represented by a purple star) induces chromatin decompaction, while at high concentrations, it results in crosslinks and condensed chromatin. In both cases, the epigenetic landscape is altered. (b) Ribose, present in high concentrations in the nucleus due to its role in nucleic acid biosynthesis, rapidly interconverts between two anomers. The linear intermediate (Fischer projection) can react with lysines, followed by Schiff base formation and subsequent rearrangement. (c) Modification of nucleophilic amino acids (Nu; histidine, cysteine, lysine, or arginine) with lipid peroxidation products 4-ONE (left) or 4-HNE (right) gives rise to a range of adducts, depending on the modification site. 4-ONE can undergo Michael addition (salmon) or nucleophilic addition to the carbonyls at the 1 position (blue-product only observed for lysine) or the 4 position (purple). 4-HNE undergoes Michael addition (green). While nucleophilic addition at the carbonyl is chemically plausible (orange), it has not yet been observed. The lipid adducts can undergo further rearrangement to give advanced lipid peroxidation end-products (ALEs). Reactions adapted from the study by Maksimovic et al. [38].
Glycation by MGO
Two major sources of histone glycation identified thus far in vivo are the 1,2 dicarbonyl species glyoxal (GO) and methylglyoxal (MGO). GO is formed via a combination of retroaldol condensation and autooxidation pathways [32], while MGO arises from the fragmentation of glyceraldehyde-3-phosphate and dihydroxyacetone phosphate [33]. Modification by both species has been identified as particularly prevalent in cells under metabolic stress [33]. As dicarbonyls, GO and MGO present two possible sites for nucleophilic attack by lysines and arginines, and the resulting permutations lead to complex mixtures of products (Figure 3a). At low concentrations, these NECMs neutralize the charge interactions between DNA and histones yielding a more open, permissive chromatin structure. However, at high concentrations, these adducts rearrange and react further to form histone-histone and histone-DNA crosslinks, resulting in a chromatin fiber that is constrained, inaccessible, and silenced [34,35]. Moreover, MGO glycation has also recently been demonstrated to induce epigenetic dysregulation by competing with essential regulatory PTMs such as acetylation, as well as H3K4me3, H3R8me2, and H3K9me3 [34,36].
Glycation by intact sugars; ribose
In theory, all reducing sugars can exist as aldoses and are capable of glycating histones via Maillard chemistry. In practice, ribose has stood out as particularly reactive and biologically significant (Figure 3b) [37]. This is thought to arise from the high concentrations of ribose and its derivatives in ribonucleotide biosynthetic pathways. Moreover, ribose spends more time in its labile linear form than other sugars, as its five-membered furanose ring form is sterically frustrated because of a steric clash between equiplanar hydroxyl groups. A recent study using an azidoribose probe to track glycation in cellulo has shown that ribose glycation (RiboLys) occurs on histones and competes with some enzymatic PTMs such as lysine acetylation and H3K9 methylation, but not H3K36 methylation [38]. This observation suggests a differential distribution of RiboLys rather than simple global competition with all enzymatic lysine PTMs.
Michael addition: 4-HNE and 4-ONE
Lipid metabolism by-products such as α,β unsaturated carbonyls are another source of complex reaction products, as nucleophilic attack can occur directly at the carbonyl or at the β-carbon (Figure 3c) [39]. Two examples which have drawn much attention in recent years in the context of chromatin are 4-hydroxynonenal (4-HNE) and 4-oxo-2-nonenal (4-ONE). 4-HNE is one of the most abundant and cytotoxic products of lipid peroxidation, a result of oxidative stress in the cell. In 2004, it was shown to modify histones in vitro, subsequently disrupting nucleosome structure and stability [40]. Similar to glycation, it has been suggested that the 4-HNE adduct neutralizes DNA-histone charge interactions, as well as disrupts chromatin compaction with its steric bulk. Physiological roles for the modification have since been identified in age-related degradation of oocytes [41] and osteoarthritis [42]. Intriguingly, when the sites of modification were analyzed via MS/MS, it was found to occur primarily via Michael addition on histidines, a less common residue for enzymatic PTMs. In particular, modifications at H2BH50, H4H76, H2AH83, and H2BH110 were detected, which lie in the globular domain center and C-terminal end, part of the DNA-binding helix-turn-helix motif [43]. Similarly, adduction by 4-ONE has been shown to prevent nucleosome assembly [44]. Its known sites of modification include lysines H3K23 and H3K37, suggesting it competes with enzymatic PTMs such as methylation.
Cellular methods for regulating NECMs
‘Chemical warfare’
Left unchecked, the perturbation of chromatin structure by histone NECMs would pose a threat to cell survival as they accumulate over time and can disrupt normal gene expression.
Therefore, various mechanisms have evolved to prevent this by sequestering reactive small molecules before they can interact with histones, or by enzymatically erasing the NECM once formed. The former strategy is particularly effective for preventing damage due to oxidative stress. The latter provides a mechanism for the cell to regulate levels of NECMs on histones and has been comprehensively explored in other reviews [45].
Radical reactive oxygen species (ROS) and reactive nitrogen species rapidly modify any biomolecules they encounter. This generates several electrophilic species which can then go on to react with histone nucleophiles; in addition to the lipids mentioned previously, modified DNA bases are a ready source of DNA-histone crosslinks [46]. While they might also be expected to react with histones, surprisingly few examples of radical histone NECMs have been discovered; a notable exception is tyrosine nitration generated by the peroxynitrite radical, which has been identified as an immunogenic modification on H2B [47]. One reason for the relative scarcity of direct radical modifications on histones is the presence of small-molecule scavengers which are able to quench and sequester these reactive species before they are able to reach the chromatin.
Antioxidants
Many of the small molecules produced within the cell have the capacity to act as radical scavengers. Melatonin and vitamin D are two endogenous antioxidants found in the nucleus which are of particular interest, as additional epigenetic effects have been associated with them. Melatonin has been shown to protect against ROS-induced genotoxicity by sequestering these reactive species [48] and was also found to stimulate HATs such as p300 to promote histone acetylation, modulating the epigenetic landscape [49,50]. Vitamin D acts as an ROS sensor, with its oxidized form 1,25(OH)D shown to bind vitamin-D responsive elements in the genome and stimulate further expression of antioxidant genes [51,52].
Dietary antioxidants, such as ascorbate and vitamin E, are also key for neutralizing radicals. Ascorbate in particular is more effective than glutathione at reducing primary ROS and reactive nitrogen species such as , •NO2, and and is therefore thought to provide a major mechanism for removing these species in vivo [53]. Similar to melatonin, it also has a direct epigenetic reprogramming function, as it is able to induce the removal of 5-methyl-C on DNA as well as histone lysine methylation by stimulating ten-eleven translocation [54] and JmjC [55] demethylases, respectively. Carnosine, a β-Ala-His dipeptide, also traps ROS via reaction at the histidine [56] and, similar to vitamin D, activates expression of antioxidant genes via the Nrf2 pathway [57,58]. In addition, it is known to sequester dicarbonyls such as 4-ONE and GO/MGO, preventing NECM formation by these species [59,60].
Conclusions and future directions
Histone NECMs are emerging as a novel family of modifications forming a new key component of epigenetic regulation and the histone code. Many new NECMs have been characterized in recent years, and we anticipate this trend will continue as methods for their detection and study become increasingly sophisticated. Here, we have aimed to highlight some of their key identifying features and complicating factors which make verifying true NECMs a major technical challenge. In general, the distribution of a modification nearer the globular domain, intrinsic reactivity toward nucleophilic residues or in radical reactions, lack of an enzymatic writer, and high concentration of the reactive substrate in the nucleus may be indicators that a PTM may also be an NECM. However, it is important to also account for the mechanisms by which the cell fights NECM addition to histones. We anticipate future research into NECM formation, control, and dysregulation will reduce key gaps in our understanding of the epigenetic regulation of transcription. As NECMs provide a direct link between the metabolic and epigenetic states of the cell, we further expect this knowledge will prove a powerful tool in understanding and treating a range of applicable diseases.
Acknowledgements
The authors thank Dr. Rasmus Pihl for his assistance and insight while editing this review. Work in the David lab is supported by the Josie Robertson Foundation, the Pershing Square Sohn Cancer Research Alliance, the NIH (CCSG core grant P30 CA008748, MSK SPORE P50 CA192937, R21 DA044767, and R35 GM138386), the Parker Institute for Cancer Immunotherapy (PICI), and the Anna Fuller Trust. In addition, the David lab is supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center. Elements of Figures created with BioRender.com.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Bannister AJ, Kouzarides T: Regulation of chromatin by histone modifications 2011, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD: The language of covalent histone modifications. Nature 2000, 403:41–45. [DOI] [PubMed] [Google Scholar]
- 3.Rothbart SB, Strahl BD: Interpreting the language of histone and DNA modifications. Biochim Biophys Acta - Gene Regul Mech 2014, 1839:627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson EJ, Katunga LA, Willis MS: Mitochondria as a source and target of lipid peroxidation products in healthy and diseased heart. Clin Exp Pharmacol Physiol 2012, 39:179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmel R, Fiedler D: Features and regulation of non-enzymatic post-translational modifications. Nat Chem Biol 2018, 14: 244–252. [DOI] [PubMed] [Google Scholar]
- 6.Chan JC, Maze I: Nothing is yet set in (Hi)stone: novel post-translational modifications regulating chromatin function. Trends Biochem Sci 2020, 45:829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X, Zhao Q, Yang T, Fung YME, Li XD: A chemical probe for lysine malonylation. Angew Chemie Int Ed 2013, 52:4883–4886. [DOI] [PubMed] [Google Scholar]
- 8.Bos J, Muir TW: A chemical probe for protein crotonylation. J Am Chem Soc 2018, 140:4757–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Q, Maksimovic I, Upad A, Guber D, David Y: Synthesis of an alkynyl methylglyoxal probe to investigate nonenzymatic histone glycation. J Org Chem 2020, 85:1691–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dadová J, Galan SR, Davis BG: Synthesis of modified proteins via functionalization of dehydroalanine. Curr Opin Chem Biol 2018, 46:71–81. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y: Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics 2012, 11:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.**. Simithy J, Sidoli S, Yuan Z-F, Coradin M, Bhanu NV, Marchione DM, Klein BJ, Bazilevsky GA, McCullough CE, Magin RS, et al. : Characterization of histone acylations links chromatin modifications with metabolism. Nat Commun 2017, 8:1141. Using MS based assays, the authors systematically characterize the reactivity of a series of acyl-coAs with histones via spontaneous chemical reaction and via enzymatic acyl transfer. Comparing these two measurements, the authors were able to categorise the different acyl-coAs according to their propensity to acylate histones via enzymatic vs non-enzymatic means. This paper represents the first comprehensive attempt to deconvolute the contribution of non-enzymatic reactions to histone acylation PTMs.
- 13.*. Huang H, Zhang D, Wang Y, Perez-Neut M, Han Z, Zheng YG, Hao Q, Zhao Y: Lysine benzoylation is a histone mark regulated by SIRT2. Nat Commun 2018, 9:3374. The authors identify a novel histone acylation NECM from an environmental source, map its distribution using proteomics, analysed the effect on the genome using ChIP-seq and RNA-seq experiments, and identified its eraser enzyme SIRT2. This represents discovery and thorough characterization of a new NECM.
- 14.*. Ishiguro T, Tanabe K, Kobayashi Y, Mizumoto S, Kanai M, Kawashima SA: Malonylation of histone H2A at lysine 119 inhibits Bub1-dependent H2A phosphorylation and chromosomal localization of shugoshin proteins. Sci Rep 2018, 8:7671. In order to study the effects of malonylation on histones, the authors constructed a range of K-> E and K->D mutants in yeast histones at known modification sites to mimic the negative charge of the malonyl group. This was then followed by detailed studies on the identified H2AK119mal peptide. This paper represents the first example of a non-enzymatic acylation demonstrating crosstalk with an enzymatic phosphorylation PTM, and elucidates a biological role for this malonylation mark.
- 15.Zhu Z, Han Z, Halabelian L, Yang X, Ding J, Zhang N, Ngo L, Song J, Zeng H, He M, et al. : Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res 2020, 10.1093/nar/gkaa1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y, et al. : Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol Cell 2016, 62:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sreedhar A, Wiese EK, Hitosugi T: Enzymatic and metabolic regulation of lysine succinylation.Genes Dis 2020, 7:166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bender ML: Mechanisms of catalysis of nucleophilic reactions of carboxylic acid derivatives.Chem Rev 1960, 60:53–113. [Google Scholar]
- 19.Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y, Tan L, Yang P, Lee J-H, Li X, et al. : KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 2017, 552:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao X, Liu Z, Zhang W, Gladysz K, Fung YME, Tian G, Xiong Y, Wong JWH, Yuen KwY, Li XD: Glutarylation of histone H4 lysine 91 regulates chromatin dynamics. Mo/ Ce// 2019, 76:660–675.e9. [DOI] [PubMed] [Google Scholar]
- 21.Espinola-Lopez JM, Tan S: The Ada2/Ada3/Gcn5/Sgf29 histone acetyltransferase module. Biochim Biophys Acta - Gene Regul Mech 2020, 1864:194629, 10.1016/j.bbagrm.2020.194629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AL-barakati HJ, Saigo H, Newman RH, Kc DB, RF-GlutarySite: A random forest based predictor for glutarylation sites.Mol Omi 2019, 15:189–204. [DOI] [PubMed] [Google Scholar]
- 23.Dou L,Li X, Zhang L, Xiang H, Xu L: iGlu_AdaBoost: identification of lysine glutarylation using the AdaBoost classifier. J Proteome Res 2020, 10.1021/acs.jproteome.0c00314. [DOI] [PubMed] [Google Scholar]
- 24.Arafat ME, Ahmad MW, Shovan SM, Dehzangi A, Dipta SR, Hasan MA, Taherzadeh G, Shatabda S, Sharma A: Accurately predicting glutarylation sites using sequential Bi-Peptide-Based evolutionary features. Genes 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang KY, Hsu JBK, Lee TY: Characterization and identification of lysine succinylation sites based on deep learning method.Sci Rep 2019, 9:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, Jia C, Li F, Song J:Inspector: a lysine succinylation predictor based on edited nearest-neighbor undersampling and adaptive synthetic oversampling. Anal Biochem 2020, 593:113592. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L, et al. : SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun 2016, 7:12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bi S, Liu Z, Wu Z, Wang Z, Liu X, Wang S, Ren J, Yao Y, Zhang W, Song M, et al. : SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell 2020, 11:483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James AM, Hoogewijs K, Logan A, Hall AR, Ding S, Fearnley IM, Murphy MP: Non-enzymatic N-acetylation of lysine residues by AcetylCoA often occurs via a proximal S-acetylated thiol intermediate sensitive to glyoxalase II. Cell Rep 2017, 18:2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.*. Gaffney DO, Jennings EQ, Anderson CC, Marentette JO, Shi T, Schou Oxvig A-M, Streeter MD, Johannsen M, Spiegel DA, Chapman E, et al. : Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem Biol 2020, 27:206–213.e6. The authors discover a new class of histone acylating substrates, acyl-GSH derivatives, in demonstrating non-enzymatic histone lactoylation.
- 31.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al. : Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange JN, Wood KD, Knight J, Assimos DG, Holmes RP: Glyoxal formation and its role in endogenous oxalate synthesis. Adv Urol 2012, 2012:819202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allaman I, Bélanger M, Magistretti PJ: Methylglyoxal, the dark side of glycolysis. Front Neurosci 2015, 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.**. Zheng Q, Omans ND, Leicher R, Osunsade A, Agustinus AS, Finkin-Groner E, D’Ambrosio H, Liu B, Chandarlapaty S, Liu S, et al. : Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat Commun 2019, 10. Using Western blot- based methods, the authors identified MGO glycation on histone proteins. Combining this data with ATAC-seq and MNAse- based chromatin accessibility assays, and chromatin compaction assays, they formulated a model for the biphasic effect of MGO glycation on the genome. They also identified DJ-1 as a histone deglycase, and found an increased sensitivity to MGO glycation in cancer cell lines. This represents systematic characterization of a new histone NECM with clinical relevance in disease.
- 35.Zheng Q, Prescott NA, Maksimovic I, David Y: (De)Toxifying the epigenetic code. Chem Res Toxicol 2019, 32:796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galligan JJ, Wepy JA, Streeter MD, Kingsley PJ, Mitchener MM, Wauchope OR, Beavers WN, Rose KL, Wang T, Spiegel DA, et al. : Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc Natl Acad Sci U S A 2018, 115:9228–9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talasz H, Wasserer S, Puschendorf B: Nonenzymatic glycation of histones in vitro and in vivo. J Cell Biochem 2002, 85:24–34. [PubMed] [Google Scholar]
- 38.*. Maksimovic I, Zheng Q, Trujillo MN, Galligan JJ, David Y: Anazidoribose probe to track ketoamine adducts in histone ribose glycation. J Am Chem Soc 2020, 142:9999–10007. The authors use an azidoribose probe to follow ribose glycation in vivo, and identify crosstalk with regulatory enzymatic PTMs. This represents the development and use of a bespoke chemical tool to study an NECM.
- 39.Doorn JA, Petersen DR: Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol 2002, 15:1445–1450. [DOI] [PubMed] [Google Scholar]
- 40.Drake J, Petroze R, Castegna A, Ding Q, Keller JN, Markesbery WR, Lovell MA, Butterfield DA: 4-Hydroxynonenal oxidatively modifies histones: implications for Alzheimer’s disease. Neurosci Lett 2004, 356:155–158. [DOI] [PubMed] [Google Scholar]
- 41.Mihalas BP, De luliis GN, Redgrove KA, McLaughlin EA, Nixon B: The lipid peroxidation product 4-hydroxynonenal contributes to oxidative stress-mediated deterioration of the ageing oocyte. Sci Rep 2017, 7:6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.*. Geib T, lacob C, Jribi R, Fernandes J, Benderdour M, Sleno L: Identification of 4-hydroxynonenal-modified proteins in human osteoarthritic chondrocytes. J Proteomics 2020:104024, 10.1016/j.jprot.2020.104024. The authors identify a role for non-enzymatic histone modification on histidine, a less commonly adducted residue.
- 43.Brennan RG, Matthews BW: The helix-turn-helix DNA binding motif. J Biol Chem 1989, 264:1903–1906. [PubMed] [Google Scholar]
- 44.Galligan JJ, Rose KL, Beavers WN, Hill S, Tallman KA, Tansey WP, Marnett LJ: Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J Am Chem Soc 2014, 136:11864–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Q, Maksimovic I, Upad A, David Y: Non-enzymatic covalent modifications: a new link between metabolism and epigenetics. Protein Cell 2020, 11:401–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang K, Park D, Tretyakova NY, Greenberg MM: Histone tails decrease N7-methyl-2′-deoxyguanosine depurination and yield DNA–protein cross-links in nucleosome core particles and cells. Proc Natl Acad Sci U S A 2018, 115:E11212–E11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan MA, Alam K, Hassan SM, Rizvi MMA: Nitration of H2B histone elicits an immune response in experimental animals. Autoimmunity 2017, 50:232–240. [DOI] [PubMed] [Google Scholar]
- 48.Bernardini L, Barbosa E, Chaãro MF, Goethel G, Muller D, Bau C, Steffens NA, Santos Stein C, Moresco RN, Garcia SC, et al. : Oxidative damage, inflammation, genotoxic effect, and global DNA methylation caused by inhalation of formaldehyde and the purpose of melatonin. Toxicol Res (Camb) 2020, 9:778–779, 10.1093/toxres/tfaa079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keshavarzi S, Salehi M, Farifteh-Nobijari F, Hosseini T, Hosseini S, Ghazifard A, Novin MG, Fallah-Omrani V, Nourozian M, Hosseini A: Melatonin modifies histone acetylation during in vitro maturation of mouse oocytes. Cell J 2018, 20:244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Chen X, Zhou W, Ji S, Li X, Li G, Liu G, Wang F, Hao A: Effect of melatonin on neuronal differentiation requires CBP/p300-mediated acetylation of histone H3 lysine 14. Neuroscience 2017, 364:45–59. [DOI] [PubMed] [Google Scholar]
- 51.Wimalawansa SJ: Vitamin D deficiency: effects on oxidative stress, epigenetics, gene regulation, and aging. Biology (Basel) 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer MB, Pike JW: Mechanistic homeostasis of vitamin D metabolism in the kidney through reciprocal modulation of Cyp27b1 and Cyp24a1 expression. J Steroid Biochem Mol Biol 2020, 196:105500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhitkovich A: Nuclear and cytoplasmic functions of vitamin C. Chem Res Toxicol 2020, 33:2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hore TA, von Meyenn F, Ravichandran M, Bachman M, Ficz G, Oxley D, Santos F, Balasubramanian S, Jurkowski TP, Reik W: Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc Natl Acad Sci Unit States Am 2016, 113. 12202 LP – 12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ebata KT, Mesh K, Liu S, Bilenky M, Fekete A, Acker MG, Hirst M, Garcia BA, Ramalho-Santos M: Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b. Epigenet Chromatin 2017, 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ihara H, Kakihana Y, Yamakage A, Kai K, Shibata T, Nishida M, Kenichi Y, Uchida K: 2-Oxo-histidine-containing dipeptides are functional oxidation products. J Biol Chem 2019, 294:1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alsheblak MM, Elsherbiny NM, El-Karef A, El-Shishtawy MM: Protective effects of L-carnosine on CCl4 -induced hepatic injury in rats. Eur Cytokine Netw 2016, 27:6–15. [DOI] [PubMed] [Google Scholar]
- 58.Zhao K, Li Y, Wang Z, Han N, Wang Y: Carnosine protects mouse podocytes from high glucose induced apoptosis through PI3K/AKT and Nrf2 Pathways. BioMed Res Int 2019, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tatsuno F, Lee SH, Oe T: Imidazole dipeptides can quench toxic 4-oxo-2( E )-nonenal: molecular mechanism and mass spectrometric characterization of the reaction products. J Pept Sci 2018, 24, e3097. [DOI] [PubMed] [Google Scholar]
- 60.Zhao J, Posa DK, Kumar V, Hoetker D, Kumar A, Ganesan S, Riggs DW, Bhatnagar A, Wempe MF, Baba SP: Carnosine protects cardiac myocytes against lipid peroxidation products. Amino Acids 2019, 51:123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]