Abstract
Introduction:
Receptor tyrosine kinase (RTK) fusions in colorectal cancers (CRC) are rare but potentially therapeutically relevant. We describe clinical, molecular and pathologic attributes of RTK fusion-associated CRC.
Methods:
We identified all cases with RTK fusions in colorectal cancer patients seen at Dana-Farber Cancer Institute who underwent OncoPanel testing between 2013–2018. Clinical, histological and molecular features were extracted from the patient charts and molecular testing results.
Results:
We identified 12 driver oncogenic fusions in various RTKs. These fusions occurred exclusively in BRAF and RAS wild-type tumors and were enriched in right-sided and mismatch repair-deficient (MMR-D) CRC. All of the MMR-D CRCs with RTK fusions were found in tumors with acquired MMR-D due to MLH1 promoter hypermethylation and one was associated with a sessile serrated polyp. Molecular profiles of MMR-D CRC with RTK fusions largely resembled BRAF V600E-mutated MMR-D CRC rather than those secondary to Lynch syndrome. We describe two patients with fusion-associated microsatellite stable (MSS) CRC patients who derived clinical benefit from therapeautic targeting of their translocation. The first harbored an ALK-CAD fusion and received sequential crizotinib and alectinib therapy for a total of 7.5 months until developing an ALK L1196Q gatekeeper mutation. The second patient, whose tumor contained a ROS1-GOPC fusion, continues to benefit from entrectinib after 9 months of therapy.
Conclusions:
RTK fusions in CRC are a rare but important disease subgroup that occur in RAS and BRAF wild-type tumors. Despite enrichment in acquired MMR-D tumors, RTK fusions also occur in microsatellite-stable CRC and provide an important therapeutic target.
Keywords: colorectal cancer, translocation, fusion, targeted therapy
Introduction
In recent years, there has been an expanding list of molecularly targeted treatment options for patients with metastatic colorectal cancer (CRC). These options target EGFR (1), BRAF V600E mutation (2) and ERBB2 (HER2) amplification (3). In addition, CRC patients with mismatch repair-deficient (MMR-D) tumors can have a prolonged response to PD1-directed immunotherapy (4). Although these targeted therapies constitute a major advance, limited options are available for patients with metastatic CRC, particularly mismatch repair proficient tumors that are resistant to PD-1 directed immunotherapy.
Chromosomal translocations are powerful oncogenic drivers, and inhibitors of ALK, ROS1, ABL, NTRK1, FGFR2, FGFR3 and RET are currently FDA approved (5–7). In addition to these FDA-approved therapies, there are additional promising fusion-targeting agents in the experimental phases of development. For example, Schram et al. have reported two patients with tumors bearing a Neuregulin 1 (NRG1) fusion who achieved a partial response to the MCLA-128 bispecific antibody directed against HER2 and HER3 (8).
Oncogenic gene fusions have been reported to occur in approximately 0.9–1.8% of CRCs (9–14). Most targetable fusions involve receptor tyrosine kinases (RTKs), which lead to the constitutive activation of MAP kinase and related mitogenic pathways (7). Recent reports have suggested that these fusions occur more often in right-sided colon adenocarcinomas and are particularly abundant in MMR-D cancers (9,10,15).
Our objective in the current study was to assess the molecular landscape of CRCs harboring RTK fusions. Our data confirm the enrichment of RTK fusions in right-sided MMR-D CRCs and demonstrate that these cases exhibit molecular features that are largely similar to those of MMR-D CRC with BRAF V600E mutations. The similarity of fusion-associated MMR-D CRC cases to BRAF V600E MMR-D related cases might reflect common pathways of development with origins in sessile serrated polyps and/or the acquisition of microsatellite instability via MLH1 hypermethylation in both groups (16). Since MSS colorectal cancers have more limited therapeutic options than MMR-D CRCs, we also investigated the clinical importance of RTK fusions in these tumors and observed two patients with MSS CRC who benefited from therapeutic targeting of their RTK translocations.
Materials and Methods
Clinical data review:
Patient epidemiologic and clinical information was abstracted by chart review as part of an institutional review board (IRB) approved protocol.
OncoPanel testing and computational analysis:
Targeted next-generation sequencing was performed with the OncoPanel assay (17) in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory. OncoPanel testing was part of an IRB approved protocol; patients provided informed written consent before molecular testing.
In this study, two versions of OncoPanel were used:
POPv2 (302 genes) and POPv3 (447 genes) (17,18). Briefly, between 50 and 200 ng of tumor DNA was prepared, hybridized to custom RNA bait sets (Agilent SureSelect™, San Diego, CA) and sequenced on the Illumina HiSeq 2500 platform with 2×100 paired-end reads. Sequence reads were aligned to the reference sequence b37 edition from the Human Genome Reference Consortium by using bwa and further processed with Picard (version 1.90, http://broadinstitute.github.io/picard/) to remove duplicates and with Genome Analysis Toolkit (GATK, version 1.6–5-g557da77) to perform localized realignment around indel sites (19). Single nucleotide variants were called with MuTect v1.1.4 (20), insertions and deletions were called with GATK Indelocator, and variants were annotated with Oncotator (21). Copy number variants and structural variants, including possible gene fusions, were called with the internally developed algorithms RobustCNV and BreaKmer (22), followed by manual review by a molecular pathologist. To filter out potential germline variants, the standard pipeline removed SNPs present at >0.1% in Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/, accessed May 30, 2013), present in either dbSNP or in an in-house panel of normals, but rescued those present two or more times in the COSMIC database (23). For this study, variants were further filtered by removing those present at >0.1% in the gnomAD v.2.1.1 database or annotated as benign or likely benign in the ClinVar database (24). The following alterations were considered likely to be oncogenic: 1) missense variants annotated as oncogenic or likely to be oncogenic in OncoKB (25) and 2) loss-of-function mutations in genes classified by OncoKB as tumor suppressor genes.
Cases were defined as MSS or MMR-D, on the basis of results of either immunohistochemistry (IHC) for mismatch repair proteins or microsatellite repeat expansion by PCR or by OncoPanel(26). Cases were identified to be MMR-D through OncoPanel as recently described (27).
Comparative analysis of various types of mutational signatures was performed using a two-step approach as previously described (28). First, de novo signature extraction was performed across the entire cohort with the SomaticSignatures package in R (29). To account for the inherent heuristic quality of this non-negative matrix factorization approach, we repeated this analysis 100 times and identified signatures with a Pearson correlation coefficient >0.6, as compared with the 30 established COSMIC signatures v2 (https://cancer.sanger.ac.uk/cosmic/signatures_v2) (30). We then used the DeconstructSigs package in R to estimate the contribution of identified signatures by using a regression model (31). Signature contributions were compared among the three groups with a Kruskal-Wallis test.
Statistical analysis:
Enrichment of genomic alterations was assessed with the Fisher’s exact test, and two-tailed p-values are reported. The Benjamini-Hochberg procedure was applied to correct for multiple comparisons. The medians of continuous variables were compared with the Mann-Whitney rank-sum test, for which two-tailed p-values are reported. Multiple testing correction was performed with the Bonferroni method.
RNA-based next generation sequencing assay for fusion confirmation:
The Solid Tumor FusionPlex (Archer) assay uses target enrichment followed by paired-end RNA-sequencing on tumor-derived RNA for the identification of fusion transcripts in 53 genes (32). All genes identified as putative fusions with OncoPanel analysis were included in this gene panel. RNA was isolated from macro-dissected formalin-fixed paraffin-embedded tumor tissue slides with the Maxwell RSC RNA FFPE Kit (Promega, USA). Data from this assay were analyzed with proprietary software from Archer Dx.
Pan-TRK IHC for NTRK fusion confirmation:
IHC was performed on 4-μm-thick formalin-fixed paraffin-embedded whole-tissue sections after antigen retrieval with 1 mM EDTA (pH 8.0; Thermo Fisher Scientific, Cheshire, UK) in a pressure cooker, by using a rabbit anti-pan-TRK monoclonal antibody (1:100 dilution; clone EPR17341; Abcam, Cambridge, MA) and the Novolink Polymer Detection System (Leica, Buffalo Grove, IL). The intensity of immunoreactivity (weak, moderate or strong) and the pattern of staining (cytoplasmic, nuclear or both) were recorded.
Targeting ALK-CAD and ROS1-GOPC fusion:
The patients with CRC bearing ALK-CAD and ROS1-GOPC fusions were treated with crizotinib and entrectinib, respectively, as part of clinical trials (crizotinib: ClinicalTrials.gov number, NCT00585195; entrectinib: ClinicalTrials.gov number, NCT02568267). Radiological assessment of the responses in both clinical trials, as well as that of the patient treated with off-label alectinib, was reviewed by an attending radiologist according to RECISTv1.1. The clinical trials were conducted according to the principles of the Declaration of Helsinki and the International Conference on Harmonization of Good Clinical Practice guidelines. Both patients provided signed IRB-approved consent forms before enrollment.
Pathologic analysis of RTK fusion-associated colorectal carcinomas:
Hematoxylin and eosin staining was performed on formalin-fixed paraffin-embedded tissue sections. Adenocarcinomas were classified according to the 2019 (5th edition) WHO Classification of Digestive System Tumors as adenocarcinoma not otherwise specified (NOS), mucinous adenocarcinoma (>50% of the lesion with pools of extracellular mucin containing tumor cells), signet-ring-cell adenocarcinoma (>50% of the tumor cells with prominent intracytoplasmic mucin, typically with displacement of the nucleus) or medullary carcinoma (sheets of tumor cells with vesicular nuclei, prominent nucleoli and abundant eosinophilic cytoplasm, with prominent infiltration by lymphocytes and neutrophils) (33). Adenocarcinomas were designated as having mucinous, signet-ring-cell and/or medullary “features” when such patterns were identified but amounted to <50% of the lesions.
Results:
Identification of RTK fusions in colorectal cancer using a targeted next generation sequencing assay
We analyzed patients with CRC seen at the Dana-Farber Cancer Institute who underwent OncoPanel testing between the years 2013 and 2018 for the presence of RTK translocations (18). We screened 1559 patients with CRC and identified 22 patients with RTK fusions. We considered fusions observed in OncoPanel to represent true positives if: (1) if they were detected by an orthogonal method such as IHC or fluorescence in situ hybridization (FISH) or an RNA-based fusion detection assay (Archer Dx) (32) or (2) if the translocation had a previously described fusion partner and breakpoint. Using these predefined criteria to reduce false positives, we selected 12 of the 22 patients with an RTK fusion for further analysis. All four NTRK1 fusions were validated for the presence of the TRK protein by IHC (Suppl. Fig. 1) (34). The CAD-ALK fusion was confirmed by using a breakapart FISH assay (Fig. 3B). RNA-based next generation sequencing was used to confirm the presence of an RTK fusion in six separate tumors (Table 1). The results of this assay confirmed RNA expression of the fusion protein in four of the six fusions identified in OncoPanel analysis. The only two cases not validated by the RNA-based assay were NTRK-LMNA fusions. However, these cases were both confirmed orthogonally by IHC (Suppl. Fig. 1).
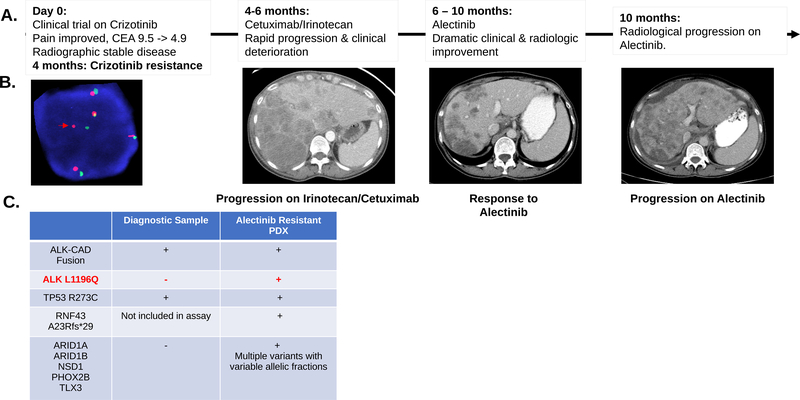
Figure 3.
A. Clinical and radiographic correlates of patient treatment history and response. The patient experienced a dramatic albeit short-lived response on alectinib. B. Break-apart FISH probes demonstrated the continued presence of ALK fusion in the alectinib-resistant tumor-derived xenograft. Red and green signals hybridize to the 5’ and 3’ ends of the ALK gene. In the case of an intact ALK gene the two signals map close together, often appearing as a yellow, or fused, signal. Rearranged alleles are scored when at least a two-signal diameter spacing is observed between the red and green ALK signals (Red arrow). In addition to ALK rearrangement the FISH assay shows evidence of ALK gene polysomy as evidenced by 5 copies of ALK gene in the representative image. C. Comparison of SNVs detected in diagnostic and alectinib-resistant samples for this index case. Genetic testing showed a shared TP53 mutation and the continued detection of ALK-CAD fusion. An acquired ALK L1196Q gatekeeper mutation was detected in the alectinib-resistant sample along with several other mutations at low allelic frequency (Suppl. Table 3). RNF43 frameshift (fs) mutation was detected in the alectinib-resistant sample. This gene was not part of the earlier version of the assay performed on the diagnostic sample.
Table 1.
RTK fusions: Fusion partners and additional details for each of the 12 RTK fusion cases identified in our cohort
| Fusion oncogene | Fusion partner | Known fusion partner | Validation method | Validation result | Mismatch Repair Status | Method of detection MSS/MSI status | MLH1 promoter hypermethylation (Y/N) | Site of primary tumor | Stage at diagnosis | Histology |
|---|---|---|---|---|---|---|---|---|---|---|
| RET | NCOA4 | Yes | - | - | Deficient | IHC and PCR | Yes | Ascending | I | Signet-ring-cell |
| RET | NCOA4 | Yes | - | - | Deficient | IHC and PCR | Yes | Descending | II | NA |
| NTRK1 | TPR | Yes | IHC and Archer Dx | Confirm | Deficient | IHC | Yes | Ascending | IV | Mucinous features |
| NTRK1 | LMNA | Yes | IHC | Confirm | Deficient | IHC | Yes | Ascending | II | Mucinous |
| NTRK1 | LMNA | Yes | IHC and Archer Dx | Confirm | Deficient | IHC | Yes | Ascending | II | Medullary |
| NTRK1 | LMNA | Yes | IHC | Confirm | Deficient | IHC and PCR | Yes | Cecum | III | Medullary features |
| ALK | CAD | Yes | FISH | Confirm | Proficient | IHC and PCR | NA | Cecum | IV | Poorly differentiated |
| ALK | EML4 | Yes | - | - | Deficient | IHC | Yes | Ascending | IV | Medullary and signet-ring-cell features |
| BRAF | ARMC10 | Yes | - | - | Deficient | IHC | Yes | Ascending | III | Poorly differentiated |
| FGFR2 | TACC2 | Yes | Archer Dx | Confirm | Proficient | IHC and PCR | NA | Rectosigmoid | IV | NA |
| ROS1 | GOPC | Yes | - | - | Proficient | IHC and PCR | NA | Rectosigmoid | IV | Mucinous features |
| NRG1 | KIF13B | Yes | Archer Dx | Confirm | Proficient | IHC | NA | Unknown | IV | NA |
FISH: Fluorescence in situ hybridization. IHC: Immunohistochemistry. - : Tissue not available for further testing.
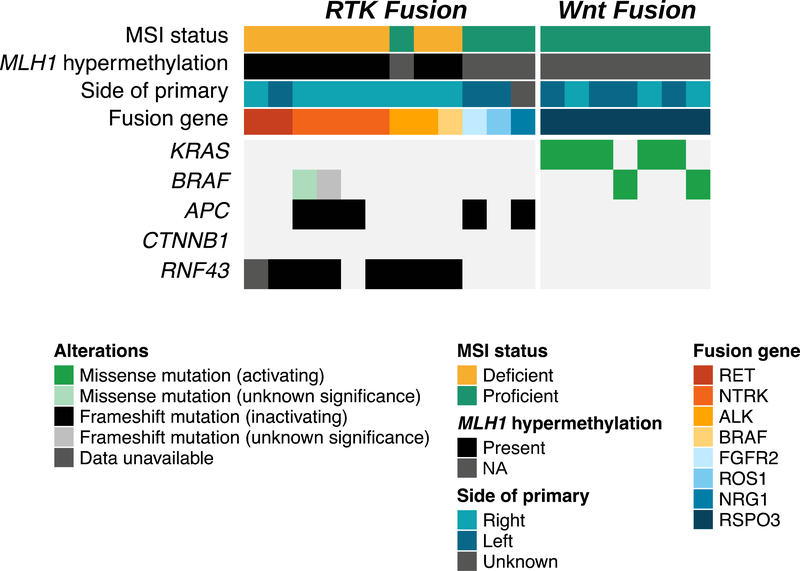
In addition to identifying fusions involving RTKs (n = 12/1559, 0.7%), we found seven patients (n = 7/1559, 0.4%) with PTPRK-RSPO3 fusions, which are known to activate the Wnt pathway. General epidemiological details are listed in Table 2. Similar to the results in prior studies, RSPO3 fusions occurred in MSS CRCs that did not bear other genomic alterations known to cause aberrant Wnt activation, such as APC loss, inactivating RNF43 alterations or CTNNB1 activating mutations (Fig. 1) (35,36). No patient with a Wnt-fusion had an RTK rearrangement. Given the immediate therapeutic relevance and higher prevalence of RTK fusions, we focus on these alterations for the remainder of the article.
Table 2.
Epidemiologic details of the fusion-associated CRC cases identified in our cohort.
| Characteristic | RTK fusions (n = 12) | Wnt fusions (n = 7) |
|---|---|---|
| Median age, yr (range) | 63.5 (50–76) | 51 (28–68) |
| Sex | ||
| Female | 9 (75%) | 2 (29%) |
| Male | 3 (25%) | 7 (71%) |
| Smoking history | ||
| Current or former smoking | 6 (50%) | 4 (57%) |
| Tumor stage | ||
| I-II | 4 (33%) | 0 |
| III | 2 (17%) | 5 (71%) |
| IV | 6 (50%) | 2 (29%) |
| Side of primary | ||
| Right colon | 8 (67%) | 4 (57%) |
| Left colon and rectum | 3 (25%) | 3 (43%) |
| Microsatellite status | ||
| High | 8 (67%) | 0 (0%) |
| Stable | 4 (33%) | 7 (100%) |
Figure 1.
Co-mutation plot for RTK and Wnt fusions identified in our study. All cases of RTK fusions are BRAF and KRAS wild-type, whereas all cases of Wnt fusions are wild-type for mutations in canonical Wnt pathway genes including APC, CTNNB1 and RNF43.
Molecular landscape of RTK fusions:
All CRC cases with RTK fusions were KRAS and NRAS wild-type. In addition, tumors bearing these RTK fusions did not have co-occurring BRAF p.V600E mutations or ERBB2 amplification or activating mutation (Fig. 1). Two patients with NTRK1 fusions had a BRAF missense mutation (p.S333G, exon 8) and a BRAF frameshift (fs) mutation (p.P403Lfs*8, exon 10). Neither of these BRAF mutations has been previously reported in the COSMIC database (23); they are likely to be passenger mutations that arose because of a high tumor mutational burden, given that both tumors were MMR-D (described further below).
Most (8 of 12; 67%) of the CRC tumors with RTK fusions were also MMR-D. In addition, 8 of the 12 patients (67%) whose tumors contained RTK fusions had a right-sided CRC, and seven of the eight right-sided tumors (88%) were MMR-D (Fig. 1). This high frequency of MMR-D tumors contrasts with the findings for the entire CRC patient cohort, in which MMR-D CRC cases were observed in only 8.8% of patients. This enrichment in MMR-D tumors in the RTK fusion population compared with the entire CRC cohort was statistically significant (67% vs. 8.8%, chi-square statistic 46, p < 0.0001). Importantly, all patients with MMR-D tumors were found to have MLH1 promoter hypermethylation, thus indicating an acquired rather than an inherited deficit in mismatch repair. The remaining 4 of 12 (33%) RTK fusions were detected in MSS CRCs. We detected one fusion each in ALK, ROS1, FGFR2 and NRG1 in this cohort of patients (MSS CRC with RTK fusion). Two of four MSS tumors with RTK fusions originated from left-sided CRC, whereas the one case with NRG1 fusion had metastatic disease and an indeterminate primary tumor location (Fig. 1, Table 1).
All patients with NTRK1 fusions (n = 4) and RET fusions (n = 2) had MMR-D tumors. Interestingly, two patients in our cohort had an ALK rearrangement: one with MSS CRC and the other with a MMR-D ALK-translocated tumor. Both ALK-rearranged tumors were right-sided primary and bore no inactivating alterations of APC. Instead, both ALK-translocated tumors bore biallelic RNF43 inactivating mutations (Suppl. Fig. 2A); however, they significantly differed in their copy number profiles. The MSS ALK-translocated CRC contained extensive copy number changes, whereas the MMR-D ALK-translocated case had a relatively silent copy number profile; both findings were consistent with the classic copy number profiles associated with these microsatellite subtypes (Suppl. Fig. 2B) (37).
Next we examined the stage distribution of RTK fusions on the basis of the tumor microsatellite status. Whereas there was a trend toward MMR-D patients presenting with early stage disease, this finding was not statistically significant. Two of eight patients with MMR-D CRC had stage IV disease (one NTRK1 fusion and one ALK fusion), compared with three of four patients with MSS CRC (chi-square statistic 2.74, p = 0.09). However, a significant difference was observed in the number of patients who had stage IV disease at any time point during their disease course. The one patient with an RTK fusion-associated, localized MSS CRC developed metastatic disease shortly after finishing adjuvant treatment. None of the six MMR-D patients with localized disease developed metastatic lesions. Hence, four of four patients (100%) with RTK fusion-associated MSS CRC developed or were initially diagnosed with metastatic cancer, as compared with only two of eight patients (25%) with RTK fusion-associated MMR-D CRC (chi-square statistic 6, p = 0.014; Suppl. Table 1). This finding is particularly relevant when considering the therapeautic relevance of these fusions (4). In our cohort, none of MMR-D CRC patients with RTK fusion underwent therapeautic targeting of their translocation. Instead, both patients with stage IV MMR-D CRC were treated with anti-PD-1 monotherapy and were having ongoing responses at the time of their last clinic visits. In contrast to this, two patients (ALK-CAD and ROS1-GOPC) with MSS CRC received agents targeting their respective fusions (described in detail below). The remaining two patients with MSS CRC (FGFR2-TACC and NRG1-KIF13B) visited only to obtain a second opinion at our institution and did not have further follow up.
Comparative molecular analysis of RTK fusion+ MMR-D CRC with BRAF V600E+ MMR-D and Lynch syndrome+ CRC
We were struck by the finding that all of the MMR-D CRCs with RTK fusions were found in tumors with acquired MMR-D due to MLH1 promoter hypermethylation. Typically, MLH1 promoter hypermethylation is associated with BRAF V600E mutations (38,39). Therefore, we hypothesized that the RTK fusion MMR-D tumors might represent a unique biologic subtype of acquired MMR-D CRC. To examine this hypothesis, we performed a comparative analysis of the molecular features of these RTK fusion MMR-D tumors to better understand their pathogenesis.
To identify appropriate controls for this comparison, we queried 124 MMR-D CRC cases with available OncoPanel testing in our institutional database and categorized the instances into groups, as described below, according to the likely etiology of the MMR-D, by using available germline genetic testing, somatic mutation data from OncoPanel and MLH1 promoter hypermethylation analysis. MMR-D CRC was divided into three categories with the above approach: MLH1 promoter hypermethylated (n = 42), Lynch syndrome (n = 29) and somatic mismatch repair gene mutations (n = 9) (Suppl. Table 2). Cases with MLH1 hypermethylation were further divided on the basis of their oncogenic drivers: (1) BRAF V600E mutation, (2) fusion associated or (3) Not otherwise specificied (NOS) for cases in which no clear oncogenic driver was identified. We chose the BRAF V600E mutation (n = 41) and Lynch syndrome (n = 26), for which raw mutation data calls were available, as the most common forms of MMR-D CRC, then proceeded with comparative molecular analysis of fusion-associated MMR-D cancers. For MSS CRC comparators, we used four cases with RTK-fusion+MSS CRC and seven cases with RSPO3-fusion+MSS CRC. Compared with MSS CRCs, the three subtypes of MMR-D CRCs showed characteristics known to be associated with MMR-D cancers, such as genomes with few copy number changes (Suppl. Fig. 3A) and significantly elevated tumor mutational burden (TMB) and homopolymer indels (HPIs) (Fig. 2A and Suppl. Fig. 3B) (37,40).
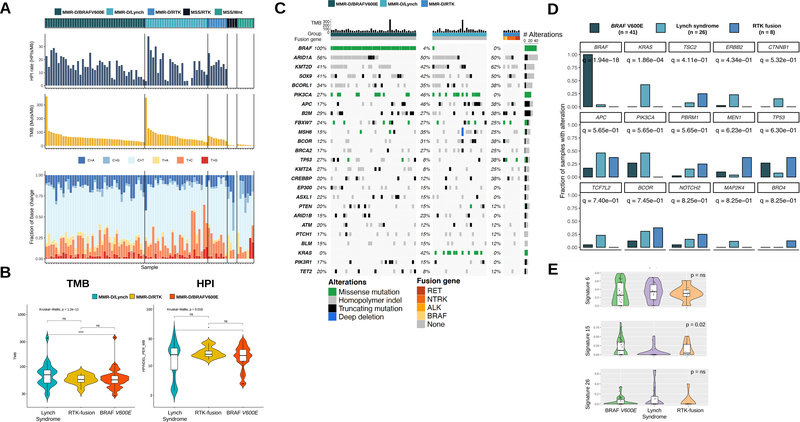
Figure 2.
A. Tumor mutational burden (TMB), homopolymer indel (HPI) frequency and base change spectrum in our identified groups: MMR-D: BRAF V600E mutated, Lynch syndrome and RTK fusions. MSS: RTK fusions and RSPO fusion. All three MMR-D subgroups have a higher TMB and HPI than MSS tumors with RTK or RSPO fusions. B. Lynch syndrome tumors demonstrate a higher TMB and HPI than MMR-D BRAF V600E tumors. Median TMB and HPI levels of MMR-D RTK fusion tumors are closer to MMR-D BRAF V600E tumors (text) but not significantly (ns) different from Lynch syndrome tumors, probably because of low case numbers. C. Co-mutation plot for the pre-defined groups. D. Varying frequencies of various oncogenes among the three MMR-D cancer subgroups. Q values for each oncogene are listed beneath their identifiers. Oncogenic mutations in KRAS, ERBB2 and CTNNB1 show relative predominance in Lynch syndrome. E. Lynch syndrome-associated MMR-D cancers demonstrate a lower median contribution from signature 15 than do BRAF V600E and RTK fusion-associated MMR-D cancers.
After determining that the RTK fusion-associated MMR-D CRCs displayed the common molecular features of MMR-D tumors, we performed a detailed comparison of the three subtypes of MMR-D tumors. Interestingly, MMR-D tumors from patients with Lynch syndrome had a significantly higher TMB and HPI rate than did MMR-D tumors with the BRAF V600E mutation (Fig. 2B) (Lynch syndrome vs. MMR-d BRAF V600E median TMB 69.6 vs. 56.3, p = 1.2e-11 by Kruskal-Wallis test), a result possibly reflecting a longer duration of microsatellite instability. The TMB of the RTK fusion group (median TMB 55.6) was numerically similar to that observed in the BRAF V600E mutation cohort (median TMB 56.3), although the difference between the RTK fusion cohort and Lynch cohort was not statistically significant, probably because of the low sample numbers in the former cohort.
Next, we compared the rates of various oncogenic mutations among the three MMR-D cohorts (Fig. 2C–D). Tumors from patients with Lynch syndrome were significantly enriched in activating KRAS (q = 1.86e-04, p = 3.21e-05) mutations, and a trend toward enrichment in ERBB2 activating mutations was seen (q = 0.43, p = 0.014). When evaluating mutational changes in the Wnt pathway, we observed that all three groups collectively had mutations in APC (BRAF V600E cohort 17% vs. Lynch syndrome cohort 46% vs. RTK fusion cohort 37%) and RNF43 (BRAF V600E cohort 90% vs. Lynch syndrome cohort 62% vs. RTK fusion cohort 75%). We observed activating beta-catenin (CTNNB1) mutations only in patients with Lynch syndrome but not in other cohorts; however, the case numbers were too low to allow us to draw any definitive conclusions.
In further analysis, we compared the DNA mutational signatures among the groups. All three groups displayed enrichment in signatures 6, 15 and 26, which are characteristic of MMR-deficiency (Fig. 2E) (30). Our analysis showed that patients with Lynch syndrome had lower mutational signature contributions of signature 15 than patients with BRAF V600E- and RTK fusion-positive cases (Kruskal-Wallis, p = 0.02; Fig. 2E). None of the other identified signatures were statistically significantly different in any of the three groups.
Pathology of RTK fusions
Pathologic analysis of the available cases of RTK fusion-associated CRC showed varied histologies including mucinous, medullary and signet-ring-cell features (Suppl. Fig. 4A, 4B). In agreement with the known pathological features of MMR-D CRC, none of the cancers had conventional moderately differentiated morphology (41).
Colon cancer develops via two predominant pathways: the more common adenoma-carcinoma sequence and the sessile serrated polyp pathway (16,42). Most right-sided BRAF V600E mutated CRCs emerge from the sessile serrated polyp pathway. Given the observed molecular similarities between RTK fusion-associated and BRAF V600E-associated MMR-D CRC, we wondered whether this finding might reflect a shared origin from sessile serrated polyps. Ascertaining this uniformly is technically challenging because most colectomy resections do not show evidence of precursor lesions, probably because of overgrowth of the primary tumor. However, in one case with a RET-NCOA4 fusion, we were able to identify a tumor-adjacent 1.5-cm sessile serrated polyp with dysplasia.
Therapeutic targeting of MSS CRC with RTK fusions:
Case 1:
A 53-year-old woman with metastatic right-sided colon cancer presented for evaluation after experiencing progression on FLOX (bolus 5-fluorouracil, leucovorin, and oxaliplatin)/bevacizumab and irinotecan/bevacizumab. OncoPanel testing performed on her diagnostic colonoscopic biopsy detected an ALK-CAD fusion (Suppl. Fig. 2B). No co-occurring KRAS, BRAF or ERBB2 alterations were identified. On the basis of this finding, the patient was enrolled in a clinical trial with single agent crizotinib at a dosage of 250 mg twice daily (ClinicalTrials.gov number, NCT00585195). During the crizotinib trial, her abdominal pain resolved, and her CEA decreased from 9.5 to 4.8. Radiologically, her CT scan after two cycles showed stable disease. She remained on crizotinib for 16 weeks, after which her disease progressed radiographically. She subsequently progressed rapidly through irinotecan/cetuximab over the next 6 weeks (Fig. 3A). After the irinotecan/cetuximab, she began to deteriorate rapidly (Eastern Cooperative Oncology Group (ECOG) performance status 3) and was started on off-label alectinib 600 mg twice daily (a highly potent and selective second generation oral ALK inhibitor) (43). Within days after initiation of alectinib, her abdominal pain and large volume ascites both resolved. In agreement with the significant improvement in her performance status, CT scans after 8 weeks of therapy revealed a 36% decrease, according to RECIST 1.1 criteria, in her tumor burden (Fig. 3A).
Unfortunately, this response was short lived, and she experienced dramatic radiological progression after 15 weeks on alectinib (Fig. 3A). Alectinib was discontinued, and she underwent a large volume paracentesis for symptom management. A portion of the malignant ascites was collected as part of an institutional research protocol and was used to derive a mouse patient-derived xenograft line (PDX). She ultimately died 1 week after the cessation of alectinib.
The patient’s short-lived responses to both crizotinib and alectinib raised questions regarding the potential mechanisms of resistance to ALK-directed therapy. Using FISH, we confirmed the presence of ALK fusion in the PDX line derived from the malignant ascites at the time of alectinib resistance (Fig. 3B). OncoPanel testing on the resistant PDX again confirmed the presence of ALK-CAD fusion and a prior TP53 mutation (Fig. 3C), as well as acquisition of the ALK p.L1196Q gatekeeper mutation in the kinase domain of the ALK gene. A repeat query of the raw data from the original OncoPanel showed ~300× coverage of this region in ALK with no evidence of the ALK p.L1196Q mutation in the diagnostic sample. The ALK p.L1196Q gatekeeper mutation has been described as a resistance mutation to both crizotinib and alectinib (44). We also noted multiple other mutations (Fig. 3C; Suppl. Table 3) at varying but mostly low allelic frequencies, which were not detected in the original OncoPanel test results, thus suggesting sub-clonal divergence of her tumor over time.
Case 2:
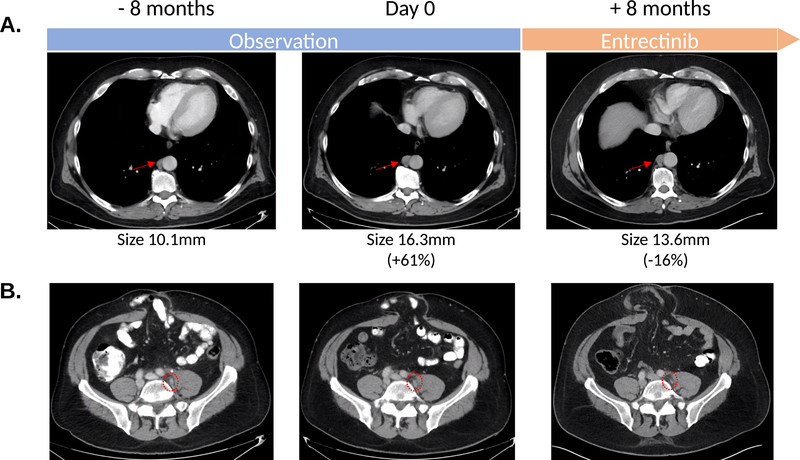
A 48-year-old man who presented with severe anemia was found to have a mismatch repair proficient, locally advanced rectosigmoid adenocarcinoma. He underwent a low anterior resection of his cancer, and pathological evaluation revealed a T4aN2bM0 rectosigmoid adenocarcinoma. After the low anterior resection, he was treated with adjuvant FOLFOX (bolus and infusional 5-fluorouracil, leucovorin and oxaliplatin) and 5-FU chemoradiation. Two and half years after chemoradiation, imaging studies showed enlarging retroperitoneal nodes, and these lymph nodes were surgically removed. Pathology confirmed recurrent colorectal adenocarcinoma. One year after the surgical removal of the retroperitoneal lymph nodes, the patient was again found to have enlarging, FDG avid retroperitoneal lymph nodes. In considering his therapeutic options, we noted that molecular analysis of both his primary rectal cancer and recurrent retroperitoneal lymph node revealed that the cancer was KRAS/NRAS/BRAF wild-type and bore a ROS1-GOPC fusion. He was enrolled in a phase 2 clinical trial of entrectinib (NCT02568267). He has tolerated entrectinib well and has had a 16% reduction in his tumor burden (Fig. 4). He continues on entrectinib after 9 months of therapy.
Figure 4.
A. Radiographic trajectory of patient with ROS1-GOPC fusion showing consistent growth in retrocrural (A) and left internal iliac lymph nodes (B) between April and December 2019 whilst the patient was being closely observed. Entrectinib was initiated in early January 2020. Scans 8 months into therapy in August 2020 show stable disease with slight reduction in size of adenopathy. Measurements of index retrocrural nodes used for RECIST1.1 measurements are noted. The left internal iliac node is too small for measurements but was FDG avid on a prior PET CT.
Discussion:
There is an urgent clinical need to identify new targets and therapies for patients with CRC. In this study, we describe clinical, molecular and pathological features of patients with RTK fusion-associated CRC. Although RTK fusion-associated CRCs are uncommon, this population of patients nonetheless has additional potential therapeutic options beyond those of most CRC patients. We collated the molecular features of patients with RTK fusion-associated CRC, as detected with a targeted DNA sequencing panel (17,18).
We observed a striking enrichment in RTK fusions in MMR-D and right-sided CRC. Interestingly, the fusions involving MMR-D cancers in our cohort occurred only in cases of acquired MMR-D CRCs, as evidenced by the presence of MLH1 promoter hypermethylation. Our finding that fusions were restricted to sporadic MMR-D tumors is in agreement with the results of recent studies (9,10,15); however, the exact reasons leading to this condition remain unclear and prompt important biologic questions regarding the oncogenesis of these tumors. A detailed molecular analysis of various subtypes of MMR-D CRC (Lynch vs. BRAF V600E+ vs. RTK) revealed several molecular features unique to Lynch syndrome, including enrichment in specific oncogenic mutations such as KRAS and ERRB2 and an overall lower contribution of mutational signature 15. The Lynch syndrome cohort also had a significantly higher mutational burden and HPI rates than the BRAF V600E mutation cohort. Data from multiple groups have shown that signatures 6, 15 and 26 are all characteristic of microsatellite instability (30). In our dataset, signature 15 had a lower relative contribution in Lynch syndrome CRC patients compared to CRC patients whose tumor harbored a BRAF V600E mutation or RTK fusion. Given the small samples size of our dataset, this observation will need validation in larger cohorts. There is no clear mechanistic data to explain the lower signature 15 contribution in Lynch syndrome CRC. Both signatures 6 and 15 are driven by large numbers of substitutions and short indels at nucleotide repeats. However, signature 15 favors C>T at GpCpN trinucleotides whereas signature 6 occurs at NpCpG (30). The hypermethylation that occurs in acquired MMR-D tumors is one possible explanation for what drives the phenotypic difference in signature 15. Another possibility is that CRCs arising in patients with Lynch syndrome, by virtue of its inherited nature, have a much longer duration of microsatellite instability driving the difference in mutational signature. The latter might also explain the higher mutational burden seen in Lynch syndrome compared to those with acquired MMR-D CRC.
Although our series included small patient numbers, thus preventing rigorous statistical comparisons between subtypes, MMR-D RTK fusion CRC cases appear to be molecularly more similar to BRAF V600E-mutated MMR-D cases than Lynch syndrome cases. Interestingly, both these groups show MLH1 hypermethylation, thus suggesting the possibility of shared pathways of tumorigenesis. Comparison of global methylation profiles of MMR-D RTK fusion CRC with BRAF V600E-mutated MMR-D CRC cases will be useful to understand important shared and divergent methylation patterns between these two disease subgroups. BRAF V600E mutation-associated tumors arise in sessile serrated polyps, and the BRAF V600E mutation is thought to be an important initial oncogenic event in these polyps (16,45). Although assessing all colectomy specimens in our cohort for evidence of precursor lesions was technically challenging, we nonetheless were able to demonstrate that one case with a RET fusion arose from a sessile serrated polyp. We hypothesize that most RTK fusion-associated tumors, like CRC with BRAF V600E mutation, also originate in sessile serrated polyps and RTK fusions might represent an early oncogenic driver event. In support of this model, recent work in lung cancer has shown that common oncogenic fusions such as EML4-ALK occur very early in oncogenesis (46). Further investigation of fusion associated CRCs using whole genome sequencing on early precursor sessile serrated polyps, dysplastic polyps and associated tumors will shed light on the sequence of oncogenic hits and underlying mechanisms involved in the genesis of these tumors.
Approximately one-third of RTK fusions occurred in MSS CRC in our patient cohort. Because MMR-D early stage CRCs have a significantly better prognosis than MSS CRC tumors, we were not surprised to observe an increased prevalence of metastatic disease in patients with MSS CRC harboring RTK fusions compared to patients with MMR-D harboring RTK fusions. However, this finding is therapeautically important because the need for novel therapies is arguably more urgent in MSS CRC considering that they are not candidates for immune checkpoint inhibition. Both these factors are exemplified in our RTK fusion cohort, in which only two of eight patients with MMR-D CRC had stage IV disease, as compared with all four patients with MSS CRC. Furthermore, both patients with MMR-D stage IV CRC (one NTRK1 fusion and one ALK fusion) are benefiting from an ongoing response to PD-1 blockade. In contrast, since MSS CRC patients do not benefit from PD1-directed immunotherapy, the two patients with MSS CRC with RTK fusions were treated with targeted therapy directed against their RTK translocation.
One patient with MSS CRC bearing a ROS1-GOPC fusion continues to benefit on entrectinib after 9 months of therapy. To our knowledge, this is the first reported case of a ROS1 fusion being targeted in CRC. The second patient with MSS CRC, whose tumor contained an ALK-CAD fusion, clinically benefited from sequential use of crizotinib and alectinib, which are potent inhibitors of ALK (43,47). Interestingly, we identified a well-characterized gatekeeper mutation in the ALK kinase domain at the time of resistance to alectinib. Gainor et al. have described similar gatekeeper mutations in lung adenocarcinoma biopsy specimens from two patients who progressed on alectinib (48). Yakirevich et al. have described a case of STRN-ALK translocated colon cancer that responded to ceritinib, another oral ALK inhibitor, for 9 months and then developed a KRAS mutation at the time of clinical resistance (14). In addition, Siravegna et al. have described a case of CRC with CAD-ALK fusion that responded to entrectinib for 5 months before relapsing with multiple ALK kinase domain mutations (13). Therefore, multiple paths to resistance to ALK inhibition in CRCs are likely to exist, a phenomenon that has also been noted in patients with ALK+ lung adenocarcinomas (48).
As a group, RTK fusions are found exclusively in KRAS, NRAS and BRAF wild-type CRCs, thus highlighting that these genomic events play critical roles in the oncogenesis of these tumors. EGFR-directed therapy was not effective in the one patient in our cohort treated with this strategy. These findings, together with others in the literature, support the hypothesis that RTK fusions may predict primary resistance to EGFR-directed therapy (49–51). Multi-institutional analysis of a larger RTK fusion cohort is necessary to confirm this hypothesis.
The molecular complexity of translocation events makes their reliable identification difficult through next generation sequencing assays. Given this technological limitation, additional assays are needed to complement the ability of next generation sequencing to identify fusions. One approach is using CLIA approved RNA based assays, which more readily identify the fusion transcripts. However, a limitation of a RNA based sequencing is the extraction of high quality RNA from formalin-fixed archival tissue. Consequently, RNA based approaches can fail to detect fusion events, particularly in older biopsy samples. For example, two NTRK-LMNA fusions that were detected by next generation sequencing failed to be identified with RNA based sequencing but were confirmed by TRK IHC. These findings illustrate the utility of using multiple modalities, including FISH and IHC, to detect the presence of fusions. Using this multi-pronged fusion detection approach in the right genetic subpopulation, such as RAS/RAF wild type CRC, can help identify the small number of patients whose tumor harbors an RTK translocation and might benefit from targeted therapies.
In summary, we provide a comprehensive molecular overview of RTK fusion-associated CRC. The enrichment of RTK fusions in acquired MMR-D cancers is consistent with findings from other reports, and deeper genomic analyses may help elucidate the fundamental mechanisms of oncogenesis in this disease subset. However, our study underscores the need to evaluate all RAS/RAF wild type patients with CRC for targetable RTK fusions. Compared with MMR-D CRCs, RTK fusions are more common in patients with MSS CRC who develop metastatic disease at some point in their disease course. These findings have important therapeutic implications, because these RTK fusions can be therapeutically targeted and also likely predict resistance to EGFR-directed therapies. Our results support the use of a multi-institutional prospective basket trial-like approach to comprehensively establish the role of targeted therapy in this genomically defined patient population.
Supplementary Material
Statement of translational relevance:
Colorectal cancer (CRC) is the second most frequent cancer-related cause of death in the US. We characterize a rare subset of CRCs bearing driver oncogenic fusions in receptor tyrosine kinases (RTKs). RTK fusions were enriched in CRCs with acquired mismatch repair deficiency. However, in our series, 33% of the CRCs with RTK fusions were microsatellite stable (MSS). We present two cases of patients with MSS CRC, whose tumors bore RTK fusions and derived clinical benefit from targeted therapy. One patient with an ALK fusion was treated with two sequential ALK targeted agents, crizotinib and alectinib, whereas the second patient with a ROS1 fusion has received entrectinib for 10 months, with treatment ongoing. RTK fusions are targetable, and clinicians should have a high index of suspicion in CRCs that are BRAF and RAS wild-type, particularly those with MSS disease where molecular therapeutic options for patients with metastatic disease are limited.
Acknowledgements:
This project was supported by the Grateful Foundation, Steve O’Connor Memorial Fund and Jacqueline Fish Memorial Fund. The work was also supported by a grant from the National Institutes of Health (P50CA127003).
J.M.C. is supported by the Dana-Farber Cancer Institute Hale Center for Pancreatic Cancer Research, Stand Up To Cancer and the Lustgarten Foundation.
H.S. is a William Raveis Charitable Fund Physician-Scientist of the Damon Runyon Cancer Research Foundation (PST-15-18).
The research of J.A.M. is supported by the Douglas Gray Woodruff Chair fund, Guo Shu Shi Fund, Anonymous Family Fund for Innovations in Colorectal Cancer, Project P fund and George Stone Family Foundation.
We would like to thank Mr. Dan Shea and Ms. Jaimie Reposa from the Brigham and Womens Hospital Division of Clinical Cytogenetics for their assistance in performing ALK FISH.
Conflicts of Interest
H.S., L.F.S., A.B.S., R.A., E.R., L.B., A.N., M.D., A.T., R.B.K., C.S., R.G., M.K., L.E.M., A.H.L., E.S. report no conflicts of interest.
Y.Y.L. reports equity in g.Root Biomedical Services.
G.I.S. has received research funding from Eli Lilly, Merck KGaA/EMD-Serono, Merck, and Sierra Oncology. He has served on advisory boards for Pfizer, Eli Lilly, G1 Therapeutics, Roche, Merck KGaA/EMD-Serono, Sierra Oncology, Bicycle Therapeutics, Fusion Pharmaceuticals, Cybrexa Therapeutics, Astex, Almac, Ipsen, Bayer, Angiex, Daiichi Sankyo, Seattle Genetics, Boehringer Ingelheim, ImmunoMet, Asana, Artios, Atrin, Concarlo Holdings, Syros and Zentalis. In addition, he holds a patent entitled, “Dosage regimen for sapacitabine and seliciclib,” also issued to Cyclacel Pharmaceuticals, and a pending patent, entitled, “Compositions and Methods for Predicting Response and Resistance to CDK4/6 Inhibition,” together with Liam Cornell.
J.W.C. received research funding to his institution from Pfizer.
A.F.F. is now an employee at Novartis.
J.J.L. has served as a compensated consultant or received honoraria from Chugai Pharma, Boehringer-Ingelheim, Pfizer, C4 Therapeutics, Nuvalent, Turning Point Therapeutics, Blueprint Medicines and Genentech; received institutional research funds from Hengrui Therapeutics, Turning Point Therapeutics, Neon Therapeutics, Relay Therapeutics, Roche/Genentech, Pfizer and Novartis; received CME funding from OncLive, MedStar Health and Northwell Health; and received travel support from Pfizer.
M.M. receives research support from Bayer, Janssen, Novo and Ono; is scientific advisory board chair and a consultant for OrigiMed; and receives patent royalties from LabCorp.
J.A.M. has received institutional research funding from Boston Biomedical, has served as an advisor/consultant to Ignyta and COTA Healthcare and has served on a grant review panel for the National Comprehensive Cancer Network funded by Taiho Pharmaceutical.
A.D.C. received research funding from Bayer.
B.M.W received research funding from Celgene and Eli Lilly, received consulting fees from BioLineRx, Celgene and Grail.
K.N. receives institutional research funding from Janssen, Pharmavite, Revolution Medicine, Evergrande group; serves on the advisory board for Array Biopharma; received consulting fees from X-Biotix Therapeutics and BiomX.
M.G. received research funding from Bristol-Myers Squibb and Merck.
J.L.H. is a consultant to Epizyme, Aadi Biosciences, and TRACON Pharmaceuticals.
J.M.C. received research funding to his institution from AbbVie, Merus, Roche and Bristol Myers Squib. He received research funding from Merck, AstraZeneca, Esperas Pharma and Tesaro, received consulting fees from Bristol Myers Squibb and received travel funding from Bristol Myers Squib.
Data availability:
Genomic data for cases described in our study is available through the AACR GENIE portal. GENIE patient IDs are provided in Suppl. Table 4.
References:
- 1.Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357(20):2040–8 doi 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 2.Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med 2019;381(17):1632–43 doi 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 3.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17(6):738–46 doi 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 4.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372(26):2509–20 doi 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subbiah V, Velcheti V, Tuch BB, Ebata K, Busaidy NL, Cabanillas ME, et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann Oncol 2018;29(8):1869–76 doi 10.1093/annonc/mdy137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378(8):731–9 doi 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 2017;14(12):735–48 doi 10.1038/nrclinonc.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schram AM. Clinical proof-of-concept for MCLA-128, a bispecific HER2/3 antibody therapy, in NRG1 fusion-positive cancers. Triple meeting - AACR-NCI-EORTC. Boston2019. [Google Scholar]
- 9.Chou A, Fraser T, Ahadi M, Fuchs T, Sioson L, Clarkson A, et al. NTRK gene rearrangements are highly enriched in MLH1/PMS2 deficient, BRAF wild-type colorectal carcinomas-a study of 4569 cases. Mod Pathol 2019. doi 10.1038/s41379-019-0417-3. [DOI] [PubMed] [Google Scholar]
- 10.Cocco E, Benhamida J, Middha S, Zehir A, Mullaney K, Shia J, et al. Colorectal Carcinomas Containing Hypermethylated MLH1 Promoter and Wild-Type BRAF/KRAS Are Enriched for Targetable Kinase Fusions. Cancer Res 2019;79(6):1047–53 doi 10.1158/0008-5472.CAN-18-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloosterman WP, Coebergh van den Braak RRJ, Pieterse M, van Roosmalen MJ, Sieuwerts AM, Stangl C, et al. A Systematic Analysis of Oncogenic Gene Fusions in Primary Colon Cancer. Cancer Res 2017;77(14):3814–22 doi 10.1158/0008-5472.CAN-16-3563. [DOI] [PubMed] [Google Scholar]
- 12.Lasota J, Chlopek M, Lamoureux J, Christiansen J, Kowalik A, Wasag B, et al. Colonic Adenocarcinomas Harboring NTRK Fusion Genes: A Clinicopathologic and Molecular Genetic Study of 16 Cases and Review of the Literature. Am J Surg Pathol 2020;44(2):162–73 doi 10.1097/PAS.0000000000001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siravegna G, Sartore-Bianchi A, Mussolin B, Cassingena A, Amatu A, Novara L, et al. Tracking a CAD-ALK gene rearrangement in urine and blood of a colorectal cancer patient treated with an ALK inhibitor. Annals of oncology : official journal of the European Society for Medical Oncology 2017;28(6):1302–8 doi 10.1093/annonc/mdx095. [DOI] [PubMed] [Google Scholar]
- 14.Yakirevich E, Resnick MB, Mangray S, Wheeler M, Jackson CL, Lombardo KA, et al. Oncogenic ALK Fusion in Rare and Aggressive Subtype of Colorectal Adenocarcinoma as a Potential Therapeutic Target. Clin Cancer Res 2016;22(15):3831–40 doi 10.1158/1078-0432.CCR-15-3000. [DOI] [PubMed] [Google Scholar]
- 15.Sato K, Kawazu M, Yamamoto Y, Ueno T, Kojima S, Nagae G, et al. Fusion Kinases Identified by Genomic Analyses of Sporadic Microsatellite Instability-High Colorectal Cancers. Clin Cancer Res 2019;25(1):378–89 doi 10.1158/1078-0432.CCR-18-1574. [DOI] [PubMed] [Google Scholar]
- 16.Crockett SD, Nagtegaal ID. Terminology, Molecular Features, Epidemiology, and Management of Serrated Colorectal Neoplasia. Gastroenterology 2019;157(4):949–66 e4 doi 10.1053/j.gastro.2019.06.041. [DOI] [PubMed] [Google Scholar]
- 17.Sholl LM, Do K, Shivdasani P, Cerami E, Dubuc AM, Kuo FC, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016;1(19):e87062 doi 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med 2017;141(6):751–8 doi 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 19.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303 doi 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31(3):213–9 doi 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015;36(4):E2423–9 doi 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abo RP, Ducar M, Garcia EP, Thorner AR, Rojas-Rudilla V, Lin L, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res 2015;43(3):e19 doi 10.1093/nar/gku1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47(D1):D941–D7 doi 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46(D1):D1062–D7 doi 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017 doi 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol 2015;16(7):30 doi 10.1007/s11864-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papke DJ Jr., Nowak JA, Yurgelun MB, Frieden A, Srivastava A, Lindeman NI, et al. Validation of a targeted next-generation sequencing approach to detect mismatch repair deficiency in colorectal adenocarcinoma. Mod Pathol 2018;31(12):1882–90 doi 10.1038/s41379-018-0091-x. [DOI] [PubMed] [Google Scholar]
- 28.Touat M, Li YY, Boynton AN, Spurr LF, Iorgulescu JB, Bohrson CL, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature 2020;580(7804):517–23 doi 10.1038/s41586-020-2209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics 2015;31(22):3673–5 doi 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500(7463):415–21 doi 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31 doi 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20(12):1479–84 doi 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 33.Nagtegaal ID, Arends MJ, Salto-Tellez M. WHO Classification of Tumours Editorial Board. Digestive System Tumours. Lyon: International Agency for Research on Cancer. 2019. [Google Scholar]
- 34.Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am J Surg Pathol 2017;41(11):1547–51 doi 10.1097/PAS.0000000000000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature 2012;488(7413):660–4 doi 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seeber A, Kocher F, Xiu J, Spizzo G, Puccini A, Swensen J, et al. Molecular landscape of colorectal cancers harboring R-spondin fusions. Journal of Clinical Oncology 2019;37(15_suppl):3588– doi 10.1200/JCO.2019.37.15_suppl.3588. [DOI] [Google Scholar]
- 37.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet 2004;41(9):664–8 doi 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Cunningham JM, Winters JL, Guenther JC, French AJ, Boardman LA, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res 2003;63(17):5209–12. [PubMed] [Google Scholar]
- 40.Cortes-Ciriano I, Lee S, Park WY, Kim TM, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun 2017;8:15180 doi 10.1038/ncomms15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenson JK, Huang SC, Herron C, Moreno V, Bonner JD, Tomsho LP, et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am J Surg Pathol 2009;33(1):126–33 doi 10.1097/PAS.0b013e31817ec2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61(5):759–67 doi 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 43.Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377(9):829–38 doi 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 44.Okada K, Araki M, Sakashita T, Ma B, Kanada R, Yanagitani N, et al. Prediction of ALK mutations mediating ALK-TKIs resistance and drug re-purposing to overcome the resistance. EBioMedicine 2019;41:105–19 doi 10.1016/j.ebiom.2019.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rad R, Cadinanos J, Rad L, Varela I, Strong A, Kriegl L, et al. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell 2013;24(1):15–29 doi 10.1016/j.ccr.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JJ, Park S, Park H, Kim S, Lee J, Lee J, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019;177(7):1842–57 e21 doi 10.1016/j.cell.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368(25):2385–94 doi 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 48.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6(10):1118–33 doi 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bray SM, Lee J, Kim ST, Hur JY, Ebert PJ, Calley JN, et al. Genomic characterization of intrinsic and acquired resistance to cetuximab in colorectal cancer patients. Sci Rep 2019;9(1):15365 doi 10.1038/s41598-019-51981-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clifton KR, Thereasa A; Parseghian Christine; Raymond Victoria M; Dasari Arvind; Pereira Allan Andresson Lima; Willis Jason; Loree Jonathan M; Bauer Todd M; Chae Young Kwang; Sherrill Gary; Fanta Paul; Grothey Axel; Hendifar Andrew; Henry David; Mahadevan Daruka; Nezami Mohammad Amin; Tan Benjamin; Wainberg Zev A; Lanman Richard. Identification of Actionable Fusions as an Anti-EGFR Resistance Mechanism Using a Circulating Tumor DNA Assay. JCO Precis Oncol 2019;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stangl C, Post JB, van Roosmalen MJ, Hami N, Verlaan-Klink I, Vos HR, et al. Diverse BRAF Gene Fusions Confer Resistance to EGFR-Targeted Therapy via Differential Modulation of BRAF Activity. Mol Cancer Res 2020;18(4):537–48 doi 10.1158/1541-7786.MCR-19-0529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data for cases described in our study is available through the AACR GENIE portal. GENIE patient IDs are provided in Suppl. Table 4.