Abstract
The COVID-19 pandemic and the detection of SARS-CoV-2 RNA in sewage has expanded global interest in wastewater surveillance. However, many underserved communities throughout the world lack improved sanitation and use informal combined sanitary and storm sewer systems. Sewage is transported via open channels, ditches, and rivers, where it mixes with surface water and/or stormwater. There is a need to develop better methods for the surveillance of pathogens such as SARS-CoV-2 RNA in this context. We developed a simplified surveillance system and monitored flow rates and concentrations of SARS-CoV-2 RNA in the Tijuana River at two locations downstream of the United States–Mexico border in California, United States. SARS-CoV-2 RNA was detected in the upstream location on six out of eight occasions, two of which were at concentrations as high as those reported in untreated wastewater from California sanitary sewer systems. The virus was not detected in any of the eight samples collected at the downstream (estuarine) sampling location, despite the consistent detection of PMMoV RNA. Synchrony was observed between the number of cases reported in Tijuana and the SARS-CoV-2 RNA concentrations measured with the CDC N1 assay when the latter were normalized by the reported flow rates in the river.
Keywords: combined sewer systems, RNA detection, COVID-19, sewage spill, aging infrastructure, SARS-CoV-2 RNA, wastewater surveillance
Short abstract
This research provides a framework for surveillance of SARS-CoV-2 RNA in surface waters impacted by sewage from households without safely managed sanitation.
Introduction
Wastewater-based epidemiology uses the analysis of wastewater in the interest of public health to provide knowledge about the measurable constituents excreted from the correspondent population. The detection of SARS-CoV-2 RNA in untreated wastewater influent1−4 has led to the more widespread use of wastewater surveillance and wastewater-based epidemiology methods for predicting and mitigating COVID-19 outbreaks.5−8 For example, the surveillance of wastewater for SARS-CoV-2 RNA has been used to predict community outbreaks and positive cases in university residence halls like the University of Arizona, the University of Connecticut, the University of Notre Dame, and Oregon State University.9−13 In addition, other surveillance communities include hospitals,14 nursing homes,15 and urban centers with sanitary sewer collection systems, and wastewater surveillance has even been used to detect positive COVID-19 cases in airplanes.16
In many urban areas, wastewater is conveyed to treatment facilities by centralized sanitary sewer infrastructure. Although separate sewer systems were implemented in cities in the United States (U.S.) in recent decades, millions of people in more than 770 U.S. cities and billions of people in other cities around the world still primarily utilize combined sewer systems (CSS),17 which transport sewage during dry weather and a combination of sewage and stormwater to treatment plants during wet weather. Globally, many communities lack closed pipe collection and conveyance systems, so wastewater may be conveyed away from homes, institutions, and businesses via lateral sewer lines. Those lines may empty into open channels, creeks, or other open conveyance systems, which are mainly dominated by wastewater during dry weather but comprise both wastewater and stormwater during wet weather. These open conveyance systems are often found in low-income urban communities, which means that the residents of these communities do not benefit as much from the wastewater surveillance programs that have been implemented globally at wastewater treatment facilities with closed-pipe sewage conveyance systems.
Many neighborhoods within the city of Tijuana, Mexico fall under the latter category of wastewater conveyance systems, with creeks (arroyos) serving as collectors for large amounts of sewage, sediment, and trash. The creeks and channels ultimately drain into the Tijuana River, which acts as an open conveyance system, where sewage from households not connected to the centralized treatment system mixes with stormwater and other surface waters. The Tijuana River flows northwest across the United States–Mexico border into San Diego County and empties into the Pacific Ocean near the city of Imperial Beach, CA (Figure 1). Flow from the Tijuana River is normally intercepted by the Comisión Internacional de Límites y Aguas (International Boundary and Water Commission) Pump Station (PBCILA), and a portion of that flow is treated at the South Bay International Wastewater Treatment Plant. During high-volume storm events and when the pumps are clogged or out of order, the pump station is bypassed and flow reaches Imperial Beach.
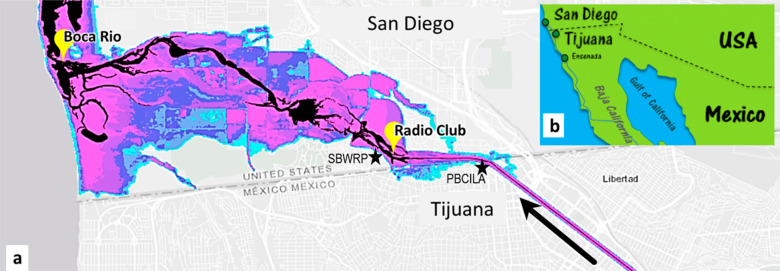
Figure 1.
(a) Map of the San Diego—Tijuana border region generated using the UC Irvine FloodRISE flood hazard visualization interactive viewer with black regions corresponding to the Tijuana River under normal flow conditions and colored regions showing flood depths that correspond to a 1% annual exceedance probability. Yellow markers show sampling points (Radio Club and Boca Rio), and additional markers show the South Bay Water Reclamation Plant (SBWRP), Comisión Internacional de Límites y Aguas (International Boundary and Water Commission) Pump Station (PBCILA).
In cities like Tijuana, the surveillance of contaminated surface waters might provide opportunities for predicting diseases such as COVID-19. However, wastewater surveillance methods appropriate for these settings have not been well developed or studied. The detection of SARS-CoV-2 RNA in surface waters may be more challenging than its detection in sewage. For example, river water was sampled in Japan from April to May 2020 after the peak of the COVID-19 outbreak in this region, but SARS-CoV-2 RNA was not detected in any samples.18 In a study of three rivers in Italy during April 2020, the presence of SARS-CoV-2 RNA was detected but virus concentrations were not quantified.4 SARS-CoV-2 RNA in river samples receiving discharge of untreated wastewater during June 2020 in Ecuador,19 October 2020 through January 2021 in Mexico,20 July 2020 through February 2021 in Nepal,21 and December 2020 in Serbia22 were detected and quantified, but these studies generally relied on discrete or one-time sample collection, have river water samples receiving secondary treated wastewater effluent23 (as opposed to raw sewage), and did not make inference about trends in concentrations. As such, more needs to be discovered about the temporal trends and relationships with community infectivity for wastewater-impaired surface waters and open channel sewage conveyance systems.
Documented cross-border flow events and heavy stormwater flows in combination with its conveyance structure make the Tijuana River an ideal model system to study SARS-CoV-2 RNA surveillance in this setting. The goals of this study were (1) to evaluate concentrations of SARS-CoV-2 RNA in water samples from the Tijuana River, (2) to compare SARS-CoV-2 RNA concentrations with the concentrations of general and host-associated fecal indicators, such as enterococci, E. coli, and pepper mild mottle virus (PMMoV) RNA, and (3) to propose a simplified surveillance model that shows the relationship of SARS-CoV-2 RNA concentrations to transboundary flow data and the number of COVID-19 cases originating in Tijuana, Mexico.
Materials and Methods
Sampling
Surface water samples were collected from two sites during eight different dates between July 2020 and May 2021 (Figure 1). At the first site, which is identified as Radio Club (32°54′85.2″N, −117°06′44.8W″) due to its proximity to the “Chula Vista Model and Radio Control Club”, the Tijuana River flows through a floodplain that regularly receives transboundary flows. The second location, commonly known as Boca Rio (32°55′73.2″N, −117°12′61.4″W) (Spanish for “river mouth”), is an estuary with major tidal influence and saltwater intrusion. The dry season in southern California extends from May to September, while the wet season is from October to April when a few significant storms occur.24 For this study, samples were collected during both dry and wet seasons, and all samples were collected under dry weather conditions. Samples from 10 November 2020, 30 December 2020, 27 January 2021, 14 March 2021, and 5 May 2021 were intentionally collected 72 h after rainfall ended due to site access and safety limitations that occur during wet weather.
Samples were collected using a telescopic dipper sampler equipped with a 1 L sampling container, which was rinsed 3 times with river water prior to sample collection. Triplicate measurements were taken in the field using a portable Oakton 450 meter for pH, conductivity, and total dissolved solids (TDS) and a YSI Pro Solo meter for dissolved oxygen (DO). Samples were transported immediately (within ∼2 h) to San Diego State University (SDSU) laboratories for subsequent analyses. Turbidity was measured in triplicate upon arrival with a HACH TL2300 turbidity meter. Aliquots of 45 mL were acidified to pH 2 and stored in precombusted amber vials at 4 °C for later analysis of chemical oxygen demand (COD), dissolved organic carbon (DOC), and total dissolved nitrogen (TDN). Remaining sample volumes were immediately used for bacterial and viral analyses.
Chemical Analyses
Aliquots (2 mL) of preserved sample were analyzed for COD using the Hach COD TNT+ HR protocol. The remaining volume from the aliquot was filtered through a precombusted 0.7 μm glass-fiber filter for preparation of simultaneous DOC and TDN measurements using the Shimadzu TOC analyzer.
Bacterial Analyses
For total coliforms, E. coli, and enterococcus measurements of unfiltered water samples, the IDEXX Colilert-18 and Enterolert methods were used, following the manufacturer’s recommended protocols. All trays were sealed within 6 h of sample collection; then, the trays were immediately placed in an incubator at 35 ± 0.5 °C for 18 h (for Colilert-18) and at 41 ± 0.5 °C for 24 h (for Enterolert). Using the IDEXX most probable number (MPN) computer generator, the counts of positive wells were converted to a final bacteria concentration for total coliforms, E. coli, and enterococcus. In cases where multiple dilutions were used, the best dilution was defined as one that was neither below the limit of detection nor saturated.
Virus Analysis
Viruses were concentrated from samples using the adsorption-extraction method as described previously.16,25,26 Briefly, 2.5 M MgCl2 was added to samples at a ratio of 1:100 to achieve a final concentration of 25 mM. Depending on the turbidity, samples were filtered until the drip rate reached 2 drips per minute through a 0.45 μm pore size membrane (HAWP type, Millipore), which was also pretreated with 10 mL of 25 mM MgCl2. Effective volumes filtered for each sample (summarized in Table S3) ranged from 198 to 495 mL at Boca Rio and from 40 to 99 mL at Radio Club. As described below, 2 μL of template RNA was used for RT-qPCR; therefore, the equivalent volumes analyzed range from approximately 8 to 20 mL for Boca Rio and from 1.6 to 4 mL for Radio Club. Nucleic acid extraction was performed on each filter using the Qiagen PowerViral kit following the manufacturer’s recommended instructions. The DNA/RNA was eluted with 50 μL of RNase-free water and frozen immediately at −80 °C until RT-qPCR analysis within the same week.
For samples collected on 31 July 2020, acidification to pH 4 was also performed in addition to MgCl2 pretreatment following suggestions from the adsorption-extraction method.16,25,26 Subsequent samples were not acidified due to the potential destruction of SARS-CoV-2 RNA integrity.26 Internal split-sample tests revealed no difference between virus concentrations with acidification or without acidification as long as MgCl2 was added. This method of concentrating SARS-CoV-2 RNA is consistent with a previous study that reported a recovery efficiency of 65.7%.26
In an attempt to filter more volume and prevent clogging due to suspended solids, samples collected on 28 August 2020 were prefiltered using an 8 μm membrane. For these samples, viral RNA was quantified on both the 8 μm membrane and the 0.45 μm membrane; however, it was discovered that up to 50% of PMMoV RNA was retained on the 8 μm membrane. Therefore, prefiltration was not used for subsequent samples.
SARS-CoV-2 RNA and PMMoV RNA were quantified from the RNA that was extracted and purified from all samples. The United States Centers of Disease Control and Prevention (CDC) primers and probes for the N1 and N2 targets were used for the detection of SARS-CoV-2 RNA. PMMoV RNA was quantified using a previously published assay.27 Samples for this study were collected between July 2020 and May 2021, and while there were several variants of SARS-CoV-2 that circulated in Mexico and the United States between those dates, mutations affecting the target regions of the U.S. CDC’s RT-qPCR assays for variants prior to October 2021 were extremely rare.28
2019-CoV plasmids and customized gBlocks (gene fragments) containing the appropriate amplicons (Integrated DNA Technologies) were used as positive controls (standards) for SARS-CoV-2 RNA and PMMoV RNA, respectively. For quality control and avoidance of freeze/thaw cycling, which negatively affects RNA integrity, the 2019-CoV plasmids and gBlocks were aliquoted into 10 μL portions, and any unused thawed standards were disposed of. Starting with the highest working concentration of standards, serial dilutions were analyzed for each run in duplicate. The limit of detection (LOD) for each assay was calculated using a probabilistic approach with the exponential survival model as described previously.29
RT-qPCR was carried out manually using the TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems, 4444432) via QuantStudio 3 (Applied Biosystems, A28137). For the detection of SARS-CoV-2 RNA, each well of the MicroAmp Optical 96-Well Reaction Plate (Applied Biosystems, 4326659) contained 20 μL of reaction mixture with 2 μL of template RNA, 1X master mix, 500 nM primers, and 125 nM probes. For the detection of PMMoV RNA, each reaction well contained 2 μL of template RNA, 1X master mix, 400 nM primers, and 200 nM probes.27 Nuclease-free water was added to bring the total reaction volumes up to 20 μL. Samples were analyzed in duplicate. Negative (no template) controls for each assay were analyzed for each PCR run. A process blank was analyzed alongside a random batch of samples and was negative for N1, N2, and PMMoV assays. Inhibition was checked using 1:10 dilution of template RNA for each sample. Additional information regarding qPCR validation and data analysis can be found in the Supporting Information.
SARS-CoV-2 RNA Simplified Surveillance Model
Equation 1 shows a theoretical model of the concentration of SARS-CoV-2 RNA (Ci) expected at the Radio Club site
| 1 |
where D is the assumed duration of shedding, L is the assumed lag time between the date of the clinical test and the initial date that SARS-CoV-2 RNA is shed in sewage by an infected person, Qi is the flow rate in the Tijuana River at the United States–Mexico border, as reported by the California State Water Board via the International Boundary and Water Commission (IBWC), nk is the daily number of clinically confirmed cases reported by the Baja California government database for the city of Tijuana, ft is the fraction of infected individuals within the Tijuana population for whom the infection was actually recorded by the official Baja California government count), fs is the fraction of COVID-19 positive cases that shed SARS-CoV-2 RNA in the feces, rs is the shedding rate for SARS-CoV-2 RNA for infected shedders (copies/day shed in feces), fsan is the fraction of the Tijuana population in the watersheds/sewersheds who lack improved sanitation (i.e., whose fecal waste is conveyed to the Tijuana River without any treatment), kd is the rate of decay of SARS-CoV-2 RNA in the river prior to reaching the sample site (e.g., Radio Club), and t is the travel time from the point of excretion to the sample collection point.
Because infected individuals shed for an extended period, the number of shedding cases in a region will “accumulate” over time until the virus is cleared from the person’s system. The total number of accumulating shedding cases for a given day was estimated based on an assumed shedding duration of D = 22 days with a lag time of L = 5 days between clinical confirmation and the start of shedding.30 Viral shedding decreases over the course of the infection;30 however, to keep the model simple, it was assumed to be constant for a duration of 22 days. The fraction ft is unknown but is assumed to be constant for the population in the Tijuana River watershed within the time period of this study. Likewise, fsan is assumed to be approximately constant since the fraction of the population without connections to the sewage treatment plant would not have changed within the time frame of this study. Also, it is assumed that the population with improved sanitation has similar rates of COVID-19 infection as the population without access to improved sanitation. There are sanitary sewer overflow events in Tijuana that last for short periods of time during rain events, and a fraction of the fecal waste that normally reaches one of the city’s treatment plants may end up being discharged into the Tijuana River. For this study, it was assumed that the additional SARS-CoV-2 RNA loading to the Tijuana River due to these SSO events would be negligible compared to the much larger loadings to the Tijuana River from households that lack connections to the sewage treatment plant.
The first-order decay rate of SARS-CoV-2 RNA in wastewater has been estimated to be approximately 0.183 day–1 at 25 °C,1 and it has been reported that cultivable SARS-CoV-2 RNA persisted in river water with a T90 value of 1.9 days at 24 °C.31 This decay rate is faster than the median decay rates reported for the RNA gene targets (measured via RT-qPCR) of nonenveloped waterborne viral pathogens (e.g., rotavirus, enterovirus, astrovirus, norovirus) in surface waters under similar conditions.32,33 SARS-CoV-2 RNA also decays more rapidly than PMMoV RNA in river water and seawater at 20 °C.34 This is consistent with previous studies that have shown that RNA from enveloped viruses degrades faster in aquatic environments than the RNA from nonenveloped viruses due to the sensitivity in its thin lipid bilayer enclosing the protein capsid.35 For our proposed model, the fraction of viral RNA that decays (e–kd t) was assumed to be relatively constant as most samples had measurable flow rates (indicating cross-boundary flow) at the Tijuana River International Boundary (Table S2). The exceptions to this (collected under stagnant conditions) occurred on 28 August 2020 and 5 May 2020, when decay could have been greater, potentially leading to lower concentrations at the sample location.
Data reported by the IBWC from transboundary spill event volumes associated with the Tijuana River Main Channel, Stewart’s Drain, and Canyon del Sol (which are all located upstream from the sample collection point (Figure 1) were retrieved from the State of California San Diego Regional Water Quality Control Board (see Supporting Information). The volumes and date ranges were converted to flow rates (liters per day) using dimensional analysis. A flow rate of zero was assumed between the reported cross-border flow events. If more than one cross-border flow event was reported for the same day, the flow rates for all events for that date were summed.
Daily COVID-19 cases originating from Tijuana, Mexico were obtained from the Baja California Secretariat of Health.36 Relative to the SARS-CoV-2 RNA concentrations in the Tijuana River, the number of accumulating COVID-19 cases, the flow rate in the river, and all other variables from eq 1 were assumed to be relatively constant and are represented by B in eqs 2 and 3.
| 2 |
| 3 |
Thus, the measured concentrations should be approximately proportional to the ratio of accumulating COVID-19 shedding cases divided by the flow rate of the Tijuana River as shown in eq 4.
| 4 |
Results and Discussion
Viral Persistence at Upstream and Downstream Sampling Sites
During the study period, a total of 80 dry-weather transboundary spill reports were recorded. These transboundary flows represented 12 billion liters of combined waters including treated and untreated sewage, groundwater, runoff, and stormwater that discharge into the Tijuana River.37 As stated previously, flow from the Tijuana River is normally intercepted by the PBCILA Pump Station, but sewage-laden flows enter the United States due to pump failure, intentional shutoff during wet weather, or overloading of the pump station. Sewage runoff also drains into the Tijuana River from two canyon collectors, Stewart’s Drain and Canyon del Sol, located upstream of the sample sites. The high sewage load in the Tijuana River is evidenced by the high concentrations of fecal indicator bacteria (FIB), total coliforms, E. coli, and enterococci as well as pathogenic bacteria and viruses.38−40
In our study, total coliforms and either E. coli or enterococci were found at high concentrations in all samples at both the upstream Radio Club site and the downstream Boca Rio site, except for on the last sampling date, 5 May 2021, for which E. coli and enterococci concentrations were both below detection limits at Boca Rio (Table 1). On all dates, FIB concentrations were significantly higher at the upstream Radio Club site than those at the Boca Rio site; however, the range of concentrations at Boca Rio were still well above San Diego Water Quality Control Board benchmarks of 400 MPN/100 mL and 104 MPN/100 mL for E. coli and enterococci, respectively, during wet and dry weather.41 The concentrations of E. coli at the Radio Club site ranged from <100 MPN/100 mL to as high as 1.0 × 106 MPN/100 mL and were sometimes similar to concentrations measured in raw wastewater in the region, which on average is 2.2 × 107 MPN/100 mL.42 The lower FIB concentrations at Boca Rio may be due to dilution of riverine water by flood tide flows and the mixing of seawater in the tidal zone at the river mouth. Electrical conductivity and TDS concentrations (Table S1) at the Boca Rio site, which are orders of magnitude higher than those at the Radio Club site and similar to values found in seawater, are evidence of the tidal influence at this site. In addition, Boca Rio is located 6.4 km downstream of Radio Club, and the Tijuana River National Estuarine Research Reserve lies between the two sites; therefore, the lower FIB concentrations at Boca Rio may also reflect the influence of environmental decay due to sunlight exposure, natural decay, and other removal mechanisms (e.g., sorption, sedimentation, or straining) underway in the estuary.
Table 1. Log10-Transformed Concentrations of Total Coliforms (TC), E. coli, Enterococci, PMMoV RNA, and SARS-CoV-2 RNA at the Boca Rio and Radio Club Sampling Sites and in Wastewater from California Wastewater Treatment Plants.
| sample collection date | total coliforms (log-MPN/100 mL) | E. coli (log-MPN/100 mL) | enterococci (log-MPN/100 mL) | SARS-CoV-2 RNA using nCoV-N1 assay (log-copies/L) | SARS-CoV-2 RNA using nCoV-N2 assay (log-copies/L) | PMMoV RNA (log-copies/L) |
|---|---|---|---|---|---|---|
| Radio Club | ||||||
| 31 Jul 2020 | 7.9 | 7.1 | 5.9 | 3.0 | 3.1 | 6.0 |
| 6 Aug 2020 | 7.5 | 7.0 | 4.7 | 3.9 | 3.5 | 7.5 |
| 28 Aug 2020 | 4.4 | <2.0 | 2.6 | <2.7 | <2.7 | 7.0 |
| 10 Nov 2020 | 7.4 | 7.2 | 4.5 | 4.3 | 3.8 | 7.2 |
| 30 Dec 2020 | >7.4 | 7.2 | 5.9 | 4.2 | 3.3 | 7.0 |
| 27 Jan 2021 | 7.1 | 6.3 | 5.7 | 3.6 | <2.7 | 6.2 |
| 14 Mar 2021 | >7.4 | >7.4 | >7.4 | 2.8 | 3.0 | 6.5 |
| 5 May 2021 | 5.6 | 3.4 | 4.2 | <2.7 | <2.7 | 6.8 |
| mean (SD)d | 6.8 (1.2) | 5.7 (2.4) | 5.1 (1.4) | 3.3 (0.8) | 3.0 (0.6) | 6.8 (0.5) |
| Boca Rio | ||||||
| 31 Jul 2020 | 3.0 | 2.2 | 2.0 | <2.7 | <2.7 | 3.2 |
| 6 Aug 2020 | 3.4 | 2.0 | <2.0 | <2.7 | <2.7 | 6.1 |
| 28 Aug 2020 | 3.5 | 2.4 | >3.4 | <2.7 | <2.7 | 4.8 |
| 10 Nov 2020 | 6.7 | 5.5 | 3.3 | <2.7 | <2.7 | 5.5 |
| 30 Dec 2020 | 5.4 | 4.2 | 3.4 | <2.7 | <2.7 | 4.3 |
| 27 Jan 2021 | 5.5 | 4.6 | 4.0 | <2.7 | <2.7 | 3.8 |
| 14 Mar 2021 | 4.8 | 4.0 | 3.0 | <2.7 | <2.7 | 3.7 |
| 5 May 2021 | 2.3 | <2.0 | <2.0 | <2.7 | <2.7 | 5.3 |
| mean (SD)d | 4.3 (1.5) | 3.2 (1.5) | 2.7 (1.0) | <2.7 | <2.7 | 4.6 (1.0) |
| Raw Sewage | ||||||
| 7.3a | 6.5a | 3.4–7.2b | 9.2–10.3c | |||
Geometric mean concentrations listed from untreated wastewater collected from the San Elijo Water Reclamation Facility in Cardiff, CA.43
Range of SARS-CoV-2 RNA concentrations (using mean Cq values from N1 and N2 assays) detected in raw sewage between 18 February and 1 June 2020 in California.44
Range of PMMoV RNA concentrations detected in raw sewage obtained from United States wastewater treatment facilities.45
Maximum likelihood method was used to calculate summary statistics for censored data.46
PMMoV RNA, which is associated with human fecal contamination, was detected in samples from the Radio Club site at concentrations normally found in untreated wastewater.45 PMMoV RNA concentrations at the Boca Rio site were lower than those at the Radio Club site, but the difference was not statistically significant (p = 0.055, n = 8; using a paired one-tailed t test). These results support the high environmental persistence of PMMoV RNA, which has been demonstrated in other studies.47,48 In our study, the high PMMoV RNA concentrations observed at the downstream Boca Rio site, where there is also mixing with seawater, provide evidence of the influence of untreated wastewater in this estuarine environment.
SARS-CoV-2 RNA was detected in six out of eight samples at the Radio Club site with the N1 assay, the N2 assay, or both assays (Table 1). Moreover, the SARS-CoV-2 RNA concentrations were as high as concentrations detected during the pandemic in untreated sewage from several counties in California with sanitary sewer systems.44 These results support the known wastewater dominance of Tijuana River cross-border flows, which has been reported in other studies.38−40 By contrast, SARS-CoV-2 RNA concentrations at the Boca Rio site were below detection limits, despite having evidence of sewage contamination from the high PMMoV RNA concentrations (average of 105 copies/L) and moderate FIB concentrations (average of 105 MPN/100 mL) (Table 1). Therefore, based on detection of RNA, SARS-CoV-2 appears to be less resistant to biological and photochemical degradation than PMMoV.
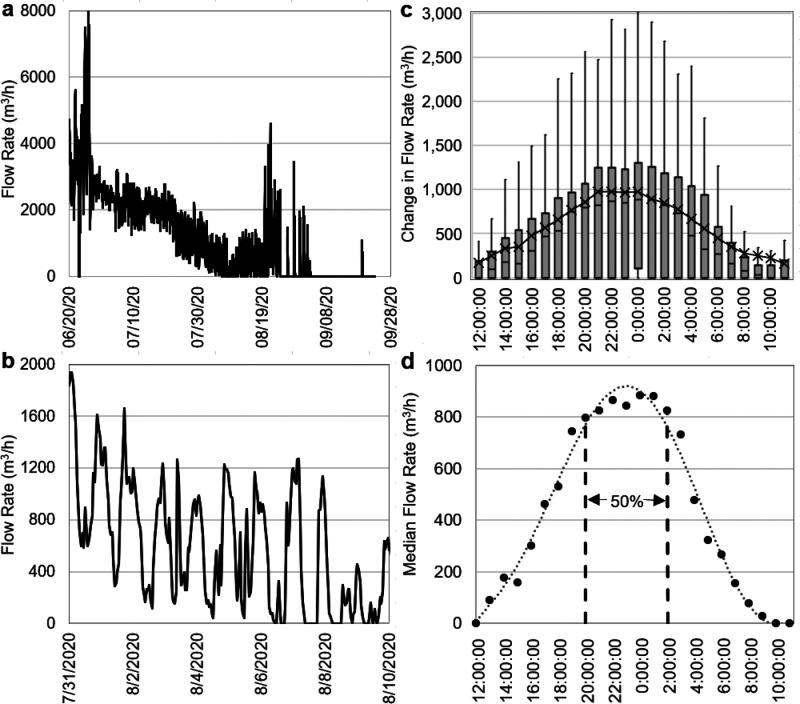
The travel time of wastewater and the resulting decay of the SARS-CoV-2 RNA signal from source areas in Tijuana to the point of surface water surveillance (i.e., the Radio Club site) are potentially important for understanding the persistence of SARS-CoV-2 RNA in sewage-dominated surface water. There are many distributed sources of wastewater to the Tijuana River, including drains and creeks that transport urban runoff, wastewater, and other water types. However, dry weather flows at the United States–Mexico border occurred for 13 weeks during the boreal summer from 20 June 2020 to 22 September 2020 (Figure 2a). During this period, the PBCILA pump station was not operating due to clogging or pump shutdown when flows exceeded 1000 L/s49 (3600 m3/h). The recurring peaks in daily flow provided a way to estimate average wastewater travel times to the U.S. border (which is close to the Radio Club surveillance site). Dry weather flows during this period showed consistent daily peaks in the hydrograph due to the daily “wave” of wastewater derived from the City of Tijuana (Figure 2b). By subtracting the baseflow, superimposing hourly flows for each day during this dry weather flow period (Figure 2c), and calculating median flows for each hour (Figure 2d), we determined that the time period between the hours of 20:00 and 02:00 was when the top 50% of median hourly flow occurred at the IBWC gage at the United States–Mexico border (Figure 2).
Figure 2.
Daily time series of flow rates during (a) summer of 2020 and (b) a 10-day period highlighted to illustrate the regular evening peaks in flow rate. (c) Hourly time series of deviation in flow rate from the lowest daily flow and (d) median hourly flow rate calculated for each day of the summer 2020 period at the Tijuana River International Boundary.
The travel time of this daily “wave” of wastewater-dominated flow that arrived at the Tijuana River International Boundary can be estimated using knowledge about household peak water use and discharge in Tijuana. Peak water demand in households was estimated to occur between the hours of 8:00 and 10:00 on weekdays, based on an analysis of major cities in Mexico, including Tijuana.50 Discharge from the households can be assumed to occur shortly thereafter. Therefore, based on the difference between hours of sewage discharge and median time of arrival of the wastewater “wave” at the border, the travel time of wastewater along the Tijuana River is estimated to range from 6 to 14 h with a mean time of 10 h. This estimate would not apply to wet weather conditions when faster river velocities in the mostly concrete trapezoidal channel would result in faster travel times. Nevertheless, this range of travel times is shorter than the T90 value of 1.9 days at 24 °C reported in ref (31) for cultivable SARS-CoV-2 RNA in river water and much less than the T90 value of 12.6 days reported for SARS-CoV-2 RNA in wastewater.1
These calculations indicate that during the dry weather flows that happened on the 31 July 2020 and 6 August 2020 sampling visits, the genetic material of SARS-CoV-2 RNA was still detected after an average travel time of approximately 10 h under mainly sunlight-exposed, open channel conditions. According to de Oliveira et al.,31 10 h of sunlight exposure would have caused about a 56% reduction in SARS-CoV-2 RNA detection. Therefore, if the decay rate of the surface water surveillance target is known and the travel time between the population center and the point of surveillance can be estimated, then eq 1 can be used to correct for the effect of decay when making inferences about the number of cases based on measured concentrations. It is also important to note that the high turbidity encountered at the Radio Club site (>20 NTU on all sampling visits) may act to shield the virus from sunlight and extend its persistence in water. Nevertheless, SARS-CoV-2 RNA concentrations in the Tijuana River at Radio Club were as high as concentrations detected during the pandemic in untreated sewage from several counties in California with sanitary sewer systems,44 which is, of itself, relevant to management of pollution in this international river.
Relationships between SARS-CoV-2 RNA in River Water and Community Infection Rates
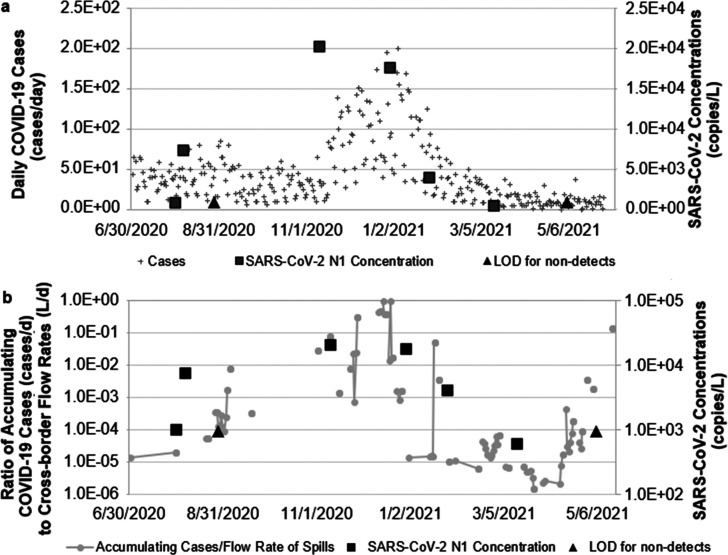
Figure 3a shows SARS-CoV-2 RNA concentrations in the Radio Club samples plotted with the time series of daily COVID-19 cases in Tijuana, Mexico. Consistent with eq 4, SARS-CoV-2 RNA concentrations in the Tijuana River at the Radio Club site were found to be proportional to the ratio of accumulating COVID-19 shedding cases to the IBWC transboundary flow data (Figure 3b).
Figure 3.
Changes in (a) daily COVID-19 cases in Tijuana, Mexico and (b) case:flow ratio (filled gray circles) plotted alongside SARS-CoV-2 RNA concentrations measured using the N1 marker (filled black squares) and LOD (filled black triangles) in the Tijuana River from July 2020 to May 2021. b uses log scale for both the accumulating case:flow rate ratio and SARS-CoV-2 RNA concentrations.
Overall, synchrony was observed between the case:flow ratio and SARS-CoV-2 RNA concentrations measured using the N1 primer. It is plausible that eq 4 could be rearranged mathematically to solve for nk to estimate trends in the case count for the purposes of surveillance. The only data inputs required would be the concentrations and flow rates in the open channel or river. Assumptions about the lag time and duration of fecal shedding for the pathogen of interest would also be needed. The CDC51 indicated that a person who is exposed to SARS-CoV-2 RNA may not display symptoms for several days. Also, it is known that an infected person could shed SARS-CoV-2 RNA through both respiratory and fecal routes before displaying symptoms52−54 and well before the date of their clinical test.30 The simplified model in eq 4 accounts for this lag time between clinical confirmation of a case and the dates of fecal shedding. Due to Mexico’s limitations to COVID-19 testing, there may have been an underestimation of COVID-19 cases in Mexico.55 Therefore, the timing of testing, the availability of testing, and an individuals’ decision whether to get tested could all affect variable ft which, as stated previously, was assumed to be approximately constant relative to other parameters in the model (eq 1).
There are important limitations of the simplified model, especially when flow rates are very low or when the case count is very low. When the prevalence of COVID (or any other infection from a pathogen that is excreted) in a community is high, the concentrations detected in samples (especially grab samples) collected from downstream surveillance points will be more consistent than they are when the prevalence is lower. This effect is evidenced in a study of hourly concentrations of adenovirus (a pathogen) compared to crAssphage (a ubiquitous fecal indicator).56 crAssphage is excreted by most of the population, but a much smaller percentage of the population is likely to have adenovirus infections at any given time. Similarly, when the case counts for a disease like COVID-19 are very low, there will likely be more peaks and valleys in the hourly concentrations, especially for a community with a smaller population, meaning that it would be less likely to see synchrony between the accumulating case count and the river concentrations (as shown in Figure 3), especially when grab samples are used. Nevertheless, communities in low-income regions or developing country settings where wastewater from some neighborhoods is discharged directly to surface waters may lack the resources to purchase autosamplers and personnel to program, deploy, and maintain such systems in the field setting, and manual collection over 24 h periods may be challenging in an urban setting. Therefore, our approach and model, which accounts for flow variability, may further benefit those communities where only grab sampling is possible. Future research comparing concentrations in composite and grab samples for surveillance in surface waters would help provide a better understanding about the limitations associated with the use of grab samples in this context.
Another limitation of the simplified model is that when the flow rate is very low (since Q appears in the denominator of the ratio shown in eq 4), the modeled ratios will be very high. The travel times from households to the sample point will also be longer under these conditions, so increased decay of the target may result in decreased concentrations detected at the sampling point. When flow rates were low in May 2021 (Figure 3b), SARS-CoV-2 RNA concentrations were below the LOD, which was about 2000 copies/L for this sample (see Table S3 for equivalent volumes for each sample), but case load data suggested that disease prevalence was increasing. The more stagnant conditions on both 28 August 2020 and 5 May 2021, when low or undetectable concentrations of FIB and SARS-CoV-2 RNA were measured at both sites, likely increased environmental exposure times and allowed for virus decay en route to the sampling point. This limitation can potentially be corrected for by incorporating the decay term from eq 1 into eq 4.
Conclusions
Many cities throughout the world lack safely managed sanitation, and there is a need for a surveillance approach that can be used to monitor sewage-polluted surface waters in these settings for diseases like COVID-19. Indeed, in Tijuana only about 50% of households have sanitation systems that safely dispose of and treat sewage.57 Furthermore, urban areas in the United States and other countries utilize combined storm and sanitary sewer systems, and wastewater surveillance in these systems should correct for dilution factors when stormwater flows increase. Our study of the temporal trends in concentrations of SARS-CoV-2 RNA in the Tijuana River during the height of the pandemic allowed us to develop simplified surveillance model that can be used in other urban settings without safely managed sanitation or with combined sewer systems.
Our finding that SARS-CoV-2 RNA was present in the Tijuana River at concentrations as high as have been found in untreated wastewater is relevant for management of pollution in this shared international waterway. Furthermore, based on our simplified model, river concentrations were found to be synchronous with the number of COVID-19 cases originating from Tijuana, Mexico, after normalizing by accumulating shedders and divided by measured flow rates in the river. The methodology used here can be applied for billions of people around the world who lack access to piped sanitary sewer infrastructure as well as hundreds of communities in the world with combined sewer systems. Taking into consideration both the concentrations of human pathogens and the flow rates in waterways can help estimate community infection rates in the absence of or with limited clinical data. Therefore, this information could be used to inform community members to seek medical attention or encourage a government or agency administration to increase resources and bring medical support to certain areas.
To apply our surveillance model to other urban regions with a lack of improved sanitation and polluted natural waters, it is necessary to understand the temporal dynamics of sewage inputs to the waterway, the climate and other environmental factors that may impact flow rates and virus decay rates, and the spatial variability in SARS-CoV-2 RNA concentrations in the waterway to determine the appropriate sampling distance for virus surveillance. Ultimately, larger sample sizes and the use of clinically confirmed data is recommended to further refine the model and yield better predictive cases. While the recommended parameters associated with viral shedding of SARS-CoV-2 RNA would benefit from further work, the model suggested in this paper provides a framework for conducting surveillance in surface waters of urban areas with limited sanitation coverage for current and future variants of SARS-CoV-2 as well as other pathogens of interest.
Acknowledgments
This study was funded by grants to N.M., M.E.V., and K.E.S. from the San Diego River Conservancy and the California State University Council on Ocean Affairs, Science, and Technology (COAST) Rapid Response Program. Support for N.M. was also provided by the William E. Leonhard Jr. Endowment. Support for K.E.S. was also provided by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under award number U54MD012397. This material is based upon work supported by the National Science Foundation under Grant No. 1827251 to M.E.V. and N.M. We acknowledge F. Espinosa and C. Mota for their assistance with method development. We acknowledge G. Muñoz Melendez from El Colegio de la Frontera Norte and M. Rogers at the U.S. International Boundary and Water Commission for support with data retrieval and J. Crooks from the Tijuana River National Estuarine Research Reserve for providing site access. A.R. was coadvised by Verbyla and Mladenov. We also acknowledge M. Thornton, S. Arredondo, and staff at the San Elijo Water Reclamation Facility for collection of wastewater for this study.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsestwater.2c00062.
Additional methodological description, figures, and tables of quality control, water quality, and field parameters (PDF)
Author Contributions
Using the CrediT (Contributor Roles Taxonomy) system, the following contributions were made. A.R.: investigation, data curation, formal analysis, visualization, writing (original draft, review, and editing). M.V.: conceptualization, formal analysis, funding acquisition, supervision, methodology (RT-qPCR, modeling), visualization, writing (original draft, review, and editing). K.S.: data curation, methodology (sample collection), writing (review and editing). N.M.: conceptualization, data curation, formal analysis, funding acquisition, project administration, supervision, methodology (sample collection, water quality and hydrologic analysis), visualization, writing (original draft, review, and editing).
The authors declare no competing financial interest.
Supplementary Material
References
- Ahmed W.; Bertsch P. M.; Bibby K.; Haramoto E.; Hewitt J.; Huygens F.; Gyawali P.; Korajkic A.; Riddell S.; Sherchan S. P.; Simpson S. L.; Sirikanchana K.; Symonds E. M.; Verhagen R.; Vasan S. S.; Kitajima M.; Bivins A. Decay of SARS-CoV-2 and Surrogate Murine Hepatitis Virus RNA in Untreated Wastewater to Inform Application in Wastewater-Based Epidemiology. Environ. Res. 2020, 191, 110092. 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K. E.; Loeb S. K.; Wolfe M. K.; Catoe D.; Sinnott-Armstrong N.; Kim S.; Yamahara K. M.; Sassoubre L. M.; Mendoza Grijalva L. M.; Roldan-Hernandez L.; Langenfeld K.; Wigginton K. R.; Boehm A. B. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ. Sci. Technol. 2021, 55 (1), 488–498. 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Peccia J.; Zulli A.; Brackney D. E.; Grubaugh N. D.; Kaplan E. H.; Casanovas-Massana A.; Ko A. I.; Malik A. A.; Wang D.; Wang M.; Warren J. L.; Weinberger D. M.; Arnold W.; Omer S. B. Measurement of SARS-CoV-2 RNA in Wastewater Tracks Community Infection Dynamics. Nat. Biotechnol. 2020, 38 (10), 1164–1167. 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S. G.; Stefani F.; Gigantiello A.; Polesello S.; Comandatore F.; Mileto D.; Maresca M.; Longobardi C.; Mancon A.; Romeri F.; Pagani C.; Cappelli F.; Roscioli C.; Moja L.; Gismondo M. R.; Salerno F. Presence and Infectivity of SARS-CoV-2 Virus in Wastewaters and Rivers. Sci. Total Environ. 2020, 744, 140911. 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. G. Wastewater Surveillance for Population-Wide Covid-19: The Present and Future. Sci. Total Environ. 2020, 736, 139631. 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R.; Curtis K.; Bivins A.; Bibby K.; Weir M. H.; Yetka K.; Thompson H.; Keeling D.; Mitchell J.; Gonzalez D. COVID-19 Surveillance in Southeastern Virginia Using Wastewater-Based Epidemiology. Water Res. 2020, 186, 116296. 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S. P.; Shahin S.; Ward L. M.; Tandukar S.; Aw T. G.; Schmitz B.; Ahmed W.; Kitajima M. First Detection of SARS-CoV-2 RNA in Wastewater in North America: A Study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W.; Angel N.; Edson J.; Bibby K.; Bivins A.; O’Brien J. W.; Choi P. M.; Kitajima M.; Simpson S. L.; Li J.; Tscharke B.; Verhagen R.; Smith W. J. M.; Zaugg J.; Dierens L.; Hugenholtz P.; Thomas K. V.; Mueller J. F. First Confirmed Detection of SARS-CoV-2 in Untreated Wastewater in Australia: A Proof of Concept for the Wastewater Surveillance of COVID-19 in the Community. Sci. Total Environ. 2020, 728, 138764. 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C.; Lambirth K.; Mittal N.; Juel M. A. I.; Barua V. B.; Roppolo Brazell L.; Hinton K.; Lontai J.; Stark N.; Young I.; Quach C.; Russ M.; Kauer J.; Nicolosi B.; Chen D.; Akella S.; Tang W.; Schlueter J.; Munir M. Implementing Building-Level SARS-CoV-2 Wastewater Surveillance on a University Campus. Sci. Total Environ. 2021, 782, 146749. 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. C.; Aubee A.; Babahaji L.; Vigil K.; Tims S.; Aw T. G. Targeted Wastewater Surveillance of SARS-CoV-2 on a University Campus for COVID-19 Outbreak Detection and Mitigation. Environ. Res. 2021, 200 (May), 111374. 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S.; Ronquillo N.; Belda-ferre P.; Alvarado D.; Javidi T.; Longhurst C. A.; Knight R. High-Throughput Wastewater SARS-CoV-2 Detection Enables. mSystems 2021, 6 (2), 1–6. 10.1128/mSystems.00045-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Lovett S.; Nelson K. L.; Beamer P.; Bischel H. N.; Bivins A.; Bruder A.; Butler C.; Camenisch T. D.; De Long S. K.; Karthikeyan S.; Larsen D. A.; Meierdiercks K.; Mouser P. J.; Pagsuyoin S.; Prasek S. M.; Radniecki T. S.; Ram J. L.; Roper D. K.; Safford H.; Sherchan S. P.; Shuster W.; Stalder T.; Wheeler R. T.; Korfmacher K. S. Wastewater Surveillance for Sars-Cov-2 on College Campuses: Initial Efforts, Lessons Learned and Research Needs. Int. J. Environ. Res. Public Health 2021, 18 (9), 4455. 10.3390/ijerph18094455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W. Q.; Schmitz B. W.; Innes G. K.; Prasek S. M.; Pogreba Brown K. M.; Stark E. R.; Foster A. R.; Sprissler R. S.; Harris D. T.; Sherchan S. P.; Gerba C. P.; Pepper I. L. COVID-19 Containment on a College Campus via Wastewater-Based Epidemiology, Targeted Clinical Testing and an Intervention. Sci. Total Environ. 2021, 779, 146408. 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosi L. M.; Barry K.; Kotay S. M.; Porter M. D.; Poulter M. D.; Ratliff C.; Simmons W.; Steinberg L. I.; Wilson D. D.; Morse R.; Zmick P.; Mathers A. J. Development of Wastewater Pooled Surveillance of Severe Congregate Living Settings. Appl. Environ. Microbiol. 2021, 87 (13), e00433. 10.1128/AEM.00433-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davó L.; Seguí R.; Botija P.; Beltrán M. J.; Albert E.; Torres I.; López-Fernández P. Á.; Ortí R.; Maestre J. F.; Sánchez G.; Navarro D. Early Detection of SARS-CoV-2 Infection Cases or Outbreaks at Nursing Homes by Targeted Wastewater Tracking. Clin. Microbiol. Infect. 2021, 27 (7), 1061–1063. 10.1016/j.cmi.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W.; Bertsch P. M.; Angel N.; Bibby K.; Bivins A.; Dierens L.; Edson J.; Ehret J.; Gyawali P.; Hamilton K. A.; Hosegood I.; Hugenholtz P.; Jiang G.; Kitajima M.; Sichani H. T.; Shi J.; Shimko K. M.; Simpson S. L.; Smith W. J. M.; Symonds E. M.; Thomas K. V.; Verhagen R.; Zaugg J.; Mueller J. F. Detection of SARS-CoV-2 RNA in Commercial Passenger Aircraft and Cruise Ship Wastewater: A Surveillance Tool for Assessing the Presence of COVID-19 Infected Travellers. J. Travel Med. 2020, 27 (5), 1–11. 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Environmental Protection Agency . What are Combined Sewer Overflows (CSOs)?; https://www3.epa.gov/region1/eco/uep/cso.html (accessed Jul 31, 2021).
- Haramoto E.; Malla B.; Thakali O.; Kitajima M. First Environmental Surveillance for the Presence of SARS-CoV-2 RNA in Wastewater and River Water in Japan. Sci. Total Environ. 2020, 737, 140405. 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Latorre L.; Ballesteros I.; Villacrés-Granda I.; Granda M. G.; Freire-Paspuel B.; Ríos-Touma B. SARS-CoV-2 in River Water: Implications in Low Sanitation Countries. Sci. Total Environ. 2020, 743, 140832. 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlknecht J.; Padilla Reyes D. A.; Ramos E.; Reyes L. M.; Álvarez M. M. The Presence of SARS-CoV-2 RNA in Different Freshwater Environments in Urban Settings Determined by RT-QPCR: Implications for Water Safety. Sci. Total Environ. 2021, 784, 147183. 10.1016/j.scitotenv.2021.147183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar S.; Sthapit N.; Thakali O.; Malla B.; Sherchan S. P.; Shakya B. M.; Shrestha L. P.; Sherchand J. B.; Joshi D. R.; Lama B.; Haramoto E. Detection of SARS-CoV-2 RNA in Wastewater, River Water, and Hospital Wastewater of Nepal. Sci. Total Environ. 2022, 824, 153816. 10.1016/j.scitotenv.2022.153816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarević S.; Micsinai A.; Szántó-Egész R.; Lukács A.; Kračun-Kolarević M.; Lundy L.; Kirschner A. K. T.; Farnleitner A. H.; Djukic A.; Čolić J.; Nenin T.; Sunjog K.; Paunović M. Detection of SARS-CoV-2 RNA in the Danube River in Serbia Associated with the Discharge of Untreated Wastewaters. Sci. Total Environ. 2021, 783, 146967. 10.1016/j.scitotenv.2021.146967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Reyes D. A.; Álvarez M. M.; Mora A.; Cervantes-Avilés P. A.; Kumar M.; Loge F. J.; Mahlknecht J. Acquired Insights from the Long-Term Surveillance of SARS-CoV-2 RNA for COVID-19 Monitoring: The Case of Monterrey Metropolitan Area (Mexico). Environ. Res. 2022, 210, 112967. 10.1016/j.envres.2022.112967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOAA; NIDIS . DROUGHT EARLY WARNING SYSTEM California-Nevada; https://www.drought.gov/dews/california-nevada (accessed Aug 14, 2021).
- Ahmed W.; Harwood V. J.; Gyawali P.; Sidhu J. P. S.; Toze S. Comparison of Concentration Methods for Quantitative Detection of Sewage-Associated Viral Markers in Environmental Waters. Appl. Environ. Microbiol. 2015, 81 (6), 2042–2049. 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W.; Bertsch P. M.; Bivins A.; Bibby K.; Farkas K.; Gathercole A.; Haramoto E.; Gyawali P.; Korajkic A.; McMinn B. R.; Mueller J. F.; Simpson S. L.; Smith W. J. M.; Symonds E. M.; Thomas K. V.; Verhagen R.; Kitajima M. Comparison of Virus Concentration Methods for the RT-QPCR-Based Recovery of Murine Hepatitis Virus, a Surrogate for SARS-CoV-2 from Untreated Wastewater. Sci. Total Environ. 2020, 739 (June), 139960. 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E.; Kitajima M.; Kishida N.; Konno Y.; Katayama H.; Asami M.; Akiba M. Occurrence of Pepper Mild Mottle Virus in Drinking Water Sources in Japan. Appl. Environ. Microbiol. 2013, 79 (23), 7413–7418. 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Jean S.; Wilson S. A.; Lucyshyn J. M.; McGrath S.; Wilson R. K.; Magrini V.; Leber A. L. A Deletion in the N Gene of SARS-CoV-2 May Reduce Test Sensitivity for Detection of SARS-CoV-2. Diagn. Microbiol. Infect. Dis. 2022, 102 (4), 115631. 10.1016/j.diagmicrobio.2021.115631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla M. E.; Symonds E. M.; Kafle R. C.; Cairns M. R.; Iriarte M.; Mercado Guzmán A.; Coronado O.; Breitbart M.; Ledo C.; Mihelcic J. R. Managing Microbial Risks from Indirect Wastewater Reuse for Irrigation in Urbanizing Watersheds. Environ. Sci. Technol. 2016, 50 (13), 6803–6813. 10.1021/acs.est.5b05398. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Cen M.; Hu M.; Du L.; Hu W.; Kim J. J.; Dai N. Prevalence and Persistent Shedding of Fecal SARS-CoV-2 RNA in Patients With COVID-19 Infection: A Systematic Review and Meta-Analysis. Clin. Transl. Gastroenterol. 2021, 12 (4), e00343. 10.14309/ctg.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira L. C.; Torres-Franco A. F.; Lopes B. C.; Santos B. S. A. d. S.; Costa E. A.; Costa M. S.; Reis M. T. P.; Melo M. C.; Polizzi R. B.; Teixeira M. M.; Mota C. R. Viability of SARS-CoV-2 in River Water and Wastewater at Different Temperatures and Solids Content. Water Res. 2021, 195, 117002. 10.1016/j.watres.2021.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A. B.; Silverman A. I.; Schriewer A.; Goodwin K. Systematic Review and Meta-Analysis of Decay Rates of Waterborne Mammalian Viruses and Coliphages in Surface Waters. Water Res. 2019, 164, 114898. 10.1016/j.watres.2019.114898. [DOI] [PubMed] [Google Scholar]
- Ngazoa E. S.; Fliss I.; Jean J. Quantitative Study of Persistence of Human Norovirus Genome in Water Using TaqMan Real-Time RT-PCR. J. Appl. Microbiol. 2008, 104 (3), 707–715. 10.1111/j.1365-2672.2007.03597.x. [DOI] [PubMed] [Google Scholar]
- Sala-Comorera L.; Reynolds L. J.; Martin N. A.; O’Sullivan J. J.; Meijer W. G.; Fletcher N. F. Decay of Infectious SARS-CoV-2 and Surrogates in Aquatic Environments. Water Res. 2021, 201, 117090. 10.1016/j.watres.2021.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra S.; Menon N. G.; Mohapatra G.; Pisharody L.; Pattnaik A.; Menon N. G.; Bhukya P. L.; Srivastava M.; Singh M.; Barman M. K.; Gin K. Y. H.; Mukherji S. The Novel SARS-CoV-2 Pandemic: Possible Environmental Transmission, Detection, Persistence and Fate during Wastewater and Water Treatment. Sci. Total Environ. 2021, 765, 142746. 10.1016/j.scitotenv.2020.142746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobierno de México Secretaría de Salud (Mexico Government Secretary of Health). Casos Diarios Por Municipio (Daily Cases by Municipality).
- California Regional Water Quality Board San Diego Region. Sewage Pollution within the Tijuana River Watershed; https://www.waterboards.ca.gov/sandiego/water_issues/programs/tijuana_river_valley_strategy/sewage_issue.html (accessed May 24, 2021).
- Gersberg R. M.; Rose M. A.; Robles-Sikisaka R.; Dhar A. K. Quantitative Detection of Hepatitis A Virus and Enteroviruses near the United States-Mexico Border and Correlation with Levels of Fecal Indicator Bacteria. Appl. Environ. Microbiol. 2006, 72 (12), 7438–7444. 10.1128/AEM.01024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoubre L. M.; Love D. C.; Silverman A. I.; Nelson K. L.; Boehm A. B. Comparison of Enterovirus and Adenovirus Concentration and Enumeration Methods in Seawater from Southern California, USA and Baja Malibu, Mexico. J. Water Health 2012, 10 (3), 419–430. 10.2166/wh.2012.011. [DOI] [PubMed] [Google Scholar]
- Zimmer-Faust A. G.; Steele J. A.; Xiong X.; Staley C.; Griffith M.; Sadowsky M. J.; Diaz M.; Griffith J. F. A Combined Digital PCR and Next Generation DNA-Sequencing Based Approach for Tracking Nearshore Pollutant Dynamics Along the Southwest United States/Mexico Border. Front. Microbiol. 2021, 12, 1–15. 10.3389/fmicb.2021.674214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Total Maximum Daily Loads (TMDLs). Water Quality Control Plan For The San Diego Basin; California Regional Water Quality Board San Diego Region, 2021; Chapter 7. [Google Scholar]
- Calderon J.; Verbyla M. E.; Gil M.; Pinongcos F.; Kinoshita A.; Mladenov N. Persistence of Fecal Indicators and Microbial Source Tracking Markers in Water Flushed from Riverbank Soils. Unpubl. Manuscr. Submitt. to Water, Air, Soil Pollut. 2022, 233, 83. 10.1007/s11270-022-05542-8. [DOI] [Google Scholar]
- Mladenov N.; Verbyla M. E.; Kinoshita A. M.; Gersberg R.; Calderon J.; Pinongcos F.; Garcia M.; Gil M.. San Diego River Contamination Study: Increasing Preparedness in the San Diego River Watershed for Potential Contamination Final Report; San Diego River Conservancy, 2020. [Google Scholar]
- Wu F.; Xiao A.; Zhang J.; Moniz K.; Endo N.; Armas F.; Bushman M.; Chai P. R.; Duvallet C.; Erickson T. B.; Foppe K.; Ghaeli N.; Gu X.; Hanage W. P.; Huang K. H.; Lee W. L.; McElroy K. A.; Rhode S. F.; Matus M.; Wuertz S.; Thompson J.; Alm E. J. Wastewater Surveillance of SARS-CoV-2 across 40 U.S. States from February to June 2020. Water Res. 2021, 202 (April), 117400. 10.1016/j.watres.2021.117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K.; Symonds E. M.; Sinigalliano C.; Stewart J.; Breitbart M. Pepper Mild Mottle Virus as an Indicator of Fecal Pollution. Appl. Environ. Microbiol. 2009, 75 (22), 7261–7267. 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Sperling M.; Verbyla M. E.; Oliveira S. M. A. C. Assessment of Treatment Plant Performance and Water Quality Data: A Guide for Students, Researchers and Practitioners. Assess. Treat. Plant Perform. Water Qual. Data A Guid. Students, Res. Pract. 2020, 10.2166/9781780409320. [DOI] [Google Scholar]
- Hamza I. A.; Jurzik L.; Überla K.; Wilhelm M. Evaluation of Pepper Mild Mottle Virus, Human Picobirnavirus and Torque Teno Virus as Indicators of Fecal Contamination in River Water. Water Res. 2011, 45 (3), 1358–1368. 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Breitbart M.; Lee W. H.; Run J. Q.; Wei C. L.; Soh S. W. L.; Hibberd M. L.; Liu E. T.; Rohwer F.; Ruan Y. RNA Viral Community in Human Feces: Prevalence of Plant Pathogenic Viruses. PLoS Biol. 2005, 4 (1), e3. 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijuana River Diversion Study: Flow Analysis, Infrastructure Diagnostic and Alternatives Development; Arcadis U.S. Inc., 2019. [Google Scholar]
- Boletim De Acompanhamento No. 6. Minas Gerais Sanitation Company (Companhia de Saneamento de Minas Gerais), 2020.
- United States Centers for Disease Control and Prevention . Symptoms of COVID-19; https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed Jan 6, 2020).
- Buscarini E.; Manfredi G.; Brambilla G.; Menozzi F.; Londoni C.; Alicante S.; Iiritano E.; Romeo S.; Pedaci M.; Benelli G.; Canetta C.; La Piana G.; Merli G.; Scartabellati A.; Viganò G.; Sfogliarini R.; Melilli G.; Assandri R.; Cazzato D.; Rossi D. S.; Usai S.; Tramacere I.; Pellegata G.; Lauria G. GI Symptoms as Early Signs of COVID-19 in Hospitalised Italian Patients. Gut. 2020, 69, 1547–1548. 10.1136/gutjnl-2020-321434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.; Lau E. H. Y.; Wu P.; Deng X.; Wang J.; Hao X.; Lau Y. C.; Wong J. Y.; Guan Y.; Tan X.; Mo X.; Chen Y.; Liao B.; Chen W.; Hu F.; Zhang Q.; Zhong M.; Wu Y.; Zhao L.; Zhang F.; Cowling B. J.; Li F.; Leung G. M. Temporal Dynamics in Viral Shedding and Transmissibility of COVID-19. Nat. Med. 2020, 26 (5), 672–675. 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Wang D.; Hu B.; Hu C.; Zhu F.; Liu X.; Zhang J.; Wang B.; Xiang H.; Cheng Z.; Xiong Y.; Zhao Y.; Li Y.; Wang X.; Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323 (11), 1061–1069. 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Feldman A.; Heres D.; Marquez-Padilla F. Air Pollution Exposure and COVID-19: A Look at Mortality in Mexico City Using Individual-Level Data. Sci. Total Environ. 2021, 756, 143929. 10.1016/j.scitotenv.2020.143929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W.; Bivins A.; Bertsch P. M.; Bibby K.; Gyawali P.; Sherchan S. P.; Simpson S. L.; Thomas K. V.; Verhagen R.; Kitajima M.; Mueller J. F.; Korajkic A. Intraday Variability of Indicator and Pathogenic Viruses in 1-h and 24-h Composite Wastewater Samples: Implications for Wastewater-Based Epidemiology. Environ. Res. 2021, 193, 110531. 10.1016/j.envres.2020.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO; UNICEF . Joint Monitoring Program WASH Data; https://washdata.org/ (accessed Jul 31, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.