Abstract
Metal nanoparticles have received great attention from researchers across the world because of a plethora of applications in agriculture and the biomedical field as antioxidants and antimicrobial compounds. Over the past few years, green nanotechnology has emerged as a significant approach for the synthesis and fabrication of metal nanoparticles. This green route employs various reducing and stabilizing agents from biological resources for the synthesis of nanoparticles. The present article aims to review the progress made in recent years on nanoparticle biosynthesis by microbes. These microbial resources include bacteria, fungi, yeast, algae and viruses. This review mainly focuses on the biosynthesis of the most commonly studied metal and metal salt nanoparticles such as silver, gold, platinum, palladium, copper, cadmium, titanium oxide, zinc oxide and cadmium sulphide. These nanoparticles can be used in pharmaceutical products as antimicrobial and anti-biofilm agents, targeted delivery of anticancer drugs, water electrolysis, waste water treatment, biosensors, biocatalysis, crop protection against pathogens, degradation of dyes etc. This review will discuss in detail various microbial modes of nanoparticles synthesis and the mechanism of their synthesis by various bioreducing agents such as enzymes, peptides, proteins, electron shuttle quinones and exopolysaccharides. A thorough understanding of the molecular mechanism of biosynthesis is the need of the hour to develop a technology for large scale production of bio-mediated nanoparticles. The present review also discusses the advantages of various microbial approaches in nanoparticles synthesis and lacuna involved in such processes. This review also highlights the recent milestones achieved on large scale production and future perspectives of nanoparticles.
This review paper is a comprehensive outline of biosynthesis of metal nanoparticles by microbes including their mechanistic aspects, large-scale production approaches, mechanism of action, advantages and limitations of bio-mediated approach.
1. Introduction
In recent times, research interest in metal nanoparticles and their production has increased significantly because of their innovative applications in different industrial domains.1,2 Nanoparticles are particulate dispersions of solid particles with at least one dimension between 1 and 100 nm. Nanoparticles have opened various fronts for the design of new materials and evaluation of their properties by modulating particle size, morphology and distribution.3 Metal nanoparticles have been explored widely due to their unique characteristics like antimicrobial, anticancer and catalytic activity, and magnetic and optical properties. The most important property of metal nanoparticles is their large surface area to volume ratio which increases their interaction with other molecules.4 Because of these remarkable and fascinating characteristics, nanoparticles are gaining significant attention in a diverse range of applications such as biochemical sensors, electronic equipment, catalysts, bio-assay, tumor-imaging, drug delivery and pharmaceutical treatment procedures.5,6
Generally nanoparticles are produced and stabilized either by a “top down” or “bottom up” strategy.7 In the “bottom up” strategy, nanoparticles are synthesized via self-assembly of atoms into nuclei which further develop into nanoscale particles. This approach includes chemical and biological methods of production whereas in the “top down” strategy, bulk material is broken down into small particles by size reduction using various physical and chemical techniques.8,9 The physical methods include grinding, milling and thermal ablation etc. On the other hand, the chemical approach of nanoparticles synthesis includes electrochemistry, chemical reduction, and photochemical reduction techniques. The physical approaches require high amount of energy which makes these types of processes more capital intensive. Another drawback of physical methods is the lower production yield of nanoscale materials.8 Over the past years, the chemical processes have been most preferable approach for nanoparticles synthesis because of requirement of less energy during reduction step and formation of homogenous particles with high preciseness in size and shape.3 However, chemical methods are environmentally hazardous because of the use of various perilous chemicals (hydrazine or potassium bitartrate) which are responsible for carcinogenicity, genotoxicity, and cytotoxicity.9,10 The use of chemical methods for synthesis of nanoparticles for biomedical application have been restricted because of toxicity, instability and less biocompatibility.11,12 Therefore, developing an environment friendly approach that effectively modulates the size, morphology, stability and characteristics is presently the main focus of research area on nanoparticle synthesis.13
The bio-mediated synthesis using microbes has evolved as a promising substitute to traditional methods of nanoparticles synthesis.14 Microbial synthesis is an environment-friendly green approach that exploits biological creatures such as bacteria, actinomycetes, fungi, algae, viruses and yeast for nanoparticles production. Microbial route provides a nontoxic, inexpensive and reliable way for nanoparticles synthesis with diversity in size, shape, composition, and physicochemical properties. This “green” route of nanoparticle synthesis is an attractive practice that enables synthesis in aqueous environment with minimum costs and low energy requirement, and can be easily scaled up to higher level.15 Another important property of these microbiological agents is their capability to serve as templates for the synthesis and organisation of nanorange particles into well-defined structures. Although several reviews have been published in past on nanoparticles biosynthesis, the present article aims to review the recent developments and progress made in nanoparticles synthesis in recent five to six years. This review paper mainly emphasizes on the utilization of various microbial agents for the metal nanoparticles synthesis, and understanding the possible mechanisms involved in fabrication of metal nanoscale particles. The present review will discuss various advantages and applications of microbial approaches in nanoparticles synthesis and lacuna involved in such processes. Finally, recent milestones achieved in large scale production and concluding remarks on future perspectives have been summarized.
2. Microbial synthesis of metal nanoparticles
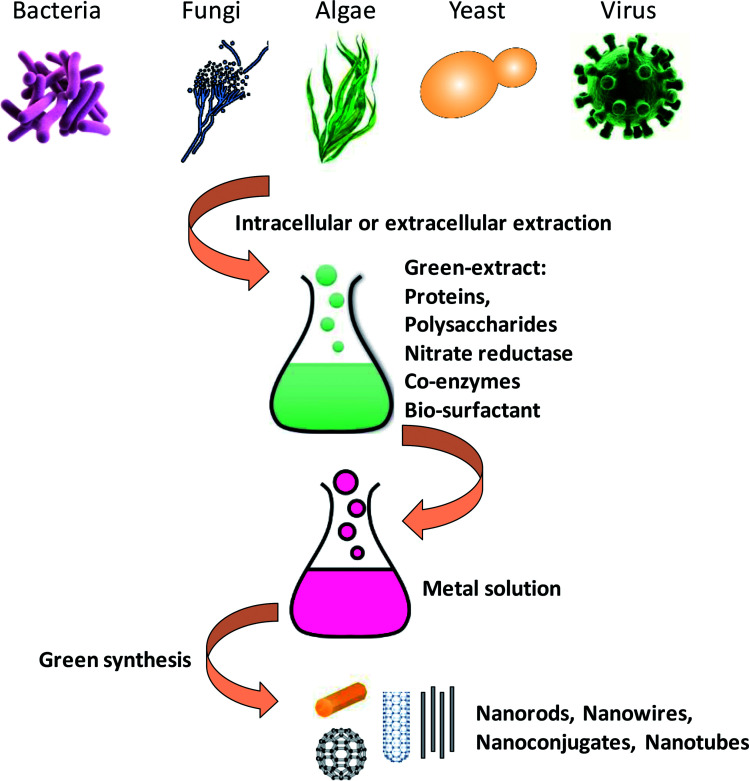
In past few years, biosynthesis of nanoparticles using microbial agents such as bacteria, actinomycetes, fungi, yeast, viruses and marine algae has received tremendous attention in the area of green nanotechnology.14 Microorganisms serve as potential nanofactories for ecofriendly and inexpensive synthesis of different metallic nanoparticles like silver, gold, palladium and copper, and metal oxides such as zinc oxide, titanium oxide etc. These nanoscale structures could exist in different shapes and forms such as nanotubes, nanoconjugates, nanorods and nanowires etc. (Fig. 1).3 These different morphological forms have shown remarkable properties for use in biomedical applications as anticancer and antimicrobial compounds.
Fig. 1. A mechanistic scheme with graphical representation about the synthesis of metal nanoparticles from microbes [this figure has been adapted from ref. 163 with permission from Royal Society of Chemistry].
2.1. Bacteria and actinomycetes
Bacteria possess unique ability to reduce metallic ions into nanoparticles and are one of the most suitable candidates for nanoparticles synthesis because of their ease of handling and high growth rates. As opposed to other microbes, bacteria can be easily moulded and manipulated genetically for the biomineralization of metal ions.16 Bacteria are continuously exposed to harsh and toxic environment conditions resulting from high concentrations of heavy metal ions in their surroundings. However, they have evolved various natural defence mechanisms such as intracellular sequestration, efflux pumps, change in metal ion concentration and extracellular precipitation, to cope up with these stress conditions.17 These defence mechanisms can be efficiently utilized by the bacteria for the synthesis of nanoparticles for different kind of applications. Table 1 shows the list of different bacterial strains adopted for the fabrication of nanoparticles and their respective applications.
A representative list of bacteria and actinomycetes used for the synthesis of nanoparticles and their applications.
| Bacteria | Source of isolation | Metal nanoparticles | Characterization | Applications | Reference | |
|---|---|---|---|---|---|---|
| Size (nm) | Shape | |||||
| Bacillus cereus | Leaf of Garcinia xanthochymus | Silver | 20–40 | Spherical | Antibacterial activity | 34 |
| Stenotrophomonas GSG2 | Coral sample collected from Bay of Bengal | Silver and gold | Gold – 10 to 50; silver – 40 to 60 | Circular, triangular, hexagonal | nd | 30 |
| Alteromonas macleodii | Sediment sample from Kochi back water, India | Silver | 70 | Spherical | nd | 32 |
| Alcaligenes faecalis | Coral from Palk Bay located near Mandapam, Gulf of Mannar | Silver | 30–50 | Spherical | Antimicrobial and anti-biofilm activity | 35 |
| Bacillus sp. CS11 | Soil samples from Cochin, India | Silver | 42–92 | Spherical | nd | 21 |
| Kocuria flava | Kanyakumari coast of India | Copper | 5–30 | Spherical | nd | 31 |
| Deinococcus radiodurans | American Type Culture Collection, Manassas, USA | Silver | 4–50 | Spherical | Antibacterial activity, anti-biofouling agent and anticancer activity | 22 |
| Ochrobactrum rhizosphaerae | Marine water | Silver | 10 | Spherical | Antibacterial activity | 4 |
| Pseudomonas aeruginosa JP-11 | Marine water | Cadmium sulphide | 20–40 | Spherical | Removal of cadmium pollutant from aqueous solution | 33 |
| Klebsiella pneumoniae, Escherichia coli, Pseudomonas jessinii | Isolated from tiger nut, carrot juice and feces | Silver | 50–100 | Cubic and star/flower like shapes | nd | 36 |
| Pseudomonas aeruginosa | Mangrove Water Sample Mandovi Estuary | Silver | 35–60 | Spherical and triangular | Antibacterial activity | 37 |
| Shewanella loihica PV-4 | DSMZ, Germany | Palladium and platinum | 2–7 | Spherical | Degradation of methyl orange dye | 23 |
| Morganella psychrotolerans | — | Silver | 2–5 | Spherical | — | 28 |
| Ochrobactrum sp. MPV1 | Roasted Arsenopyrites, Tuscany, Italy | Tellurium | nd | Roughly spherical and rods | Reduction of toxic compounds | 24 |
| Bacillus subtilis | Hutti gold mine, India | Gold | 20–25 | Spherical | Degradation of methylene blue | 25 |
| Bacillus brevis NCIM 2533 | National Collection of Industrial Microorganism (NCL), Pune, India | Silver | 41–68 | Spherical | Antibacterial activity against multi-drug resistant bacteria | 26 |
| Actinomycetes | ||||||
| Rhodococcus NCIM 2891 | nd | Silver | 10 | Spherical | nd | 39 |
| Rhodococcus sp. NCIM 2891 | National Chemical Laboratory, India | Silver | 10–15 | Spherical | Antimicrobial activity, catalytic reduction of 4-nitrophenol | 45 |
| Streptomyces sp. LK3 | Marine soil sample, Nicobar Island | Silver | 5 | Spherical | Acaricidal activity | 40 |
| Streptacidiphilus durhamensis | Acidic forest soil | Silver | 8–48 | Spherical | Antibacterial and anticancer activity | 41 |
| Streptomyces rochei MHM13 | Sediment samples along Suez Gulf, Red Sea, Egypt | Silver | 22–85 | Spherical | Antimicrobial activity and synergistic effect with antibiotics | 42 |
| Streptomyces griseoruber | Soil Sample, Mercara region | Gold | 5–50 | Spherical, hexagonal and triangular | Degradation of methylene blue | 43 |
| Streptomyces parvulus | DPUA Culture Collection, Brazil | Silver | 1–40 | nd | Antimicrobial activity | 46 |
| Streptomyces capillispiralis Ca-1 | Medicinal plant Convolvulus arvensis | Copper | 3.6–59 | Spherical | Antimicrobial activity | 44 |
| Streptomyces xinghaiensis OF1 | Sediment sample of Lonar Crater, Maharashtra, India | Silver | 5–20 | Spherical | Antimicrobial activity and synergistic effect with antibiotics | 47 |
Bacteria are generally known to synthesize metal nanoparticles either by extracellular or intracellular mechanisms. Beveridge and Murray first time reported the deposition of gold nanoparticles (AuNP) extracellularly on Bacillus subtilis cell wall when gold chloride solution was used to suspend unfixed wall.18 In another report, a silver-resistant strain of Pseudomonas stutzeri AG259 accumulated silver nanoparticles (AgNP) intracellularly within size range of few nm to 200 nm using NADH-dependent reductase enzyme that supplies electrons and itself oxidises to NAD+.19 The transfer of electrons from NADH results in the bioreduction of silver ions into silver nanoparticles. In 2012, Srivastava et al. concluded that Pseudomonas aeruginosa has the capability to synthesize variety of nanoparticles intracellularly such as Pd, Ag, Rh, Ni, Fe, Co, Pt, and Li nanoparticles.20 This work did not include any external stabilizing agent and electron donors and was free from the step of modifying pH during the biomineralization step of different metal ions. In recent reports, different bacterial strains such as Escherichia coli, Bacillus subtilis, Bacillus megaterium, Pseudomonas aeruginosa, Klebsiella pneumoniae, Bacillus cereus, Alteromonas and Ochrobactrum etc. have been extensively used for nanoparticles synthesis (Table 1).
Das et al. revealed the extracellular synthesis of silver nanoparticle at ambient temperature in 24 h using Bacillus cereus isolated from heavy metal contaminated soil.21 Synthesized AgNPs showed surface plasmon resonance properties which could be useful in various application. Kulkarni et al. elucidated the extracellular biosynthesis of AgNPs using radiation resistant Deinococcus radiodurans via reduction of silver chloride solution.22 The AgNPs showed broad-spectrum antibacterial and anti-biofilm activity against both Gram negative and Gram positive bacteria. The AgNPs also showed excellent anticancerous activity against human breast cancer cell lines. The cell viability and cytotoxicity assay demonstrated that AgNPs can inhibit the proliferation of cancer cell lines. D. radiodurans can withstand high radiation and desiccation condition which suggest that it can be used for on-field application for bioremediatizon. Researchers are now shifting towards the synthesis and development of different type of nanoparticles such as palladium, platinum and tellurium etc. specifically targeting them for various end uses. For example, Ahmed et al. reported synthesis of ultra small palladium and platinum nanoparticles by Shewanella loihica PV-4 within the size range of 2–7 nm.23 In this report, electrochemically active biofilms of S. loihica were employed for the successful synthesis of ultra small nanoparticles. The synthesized palladium and platinum nanoparticles have shown excellent catalytic performance in decomposition of methyl orange dye. Zonaro et al. described the synthesis of tellurium nanoparticles using Ochrobactrum sp. and concluded that this strain can serve as a potential nanofactory for the conversion of toxic tellurite oxyanions into useful nanoparticles.24 In recent study, Srinath et al. described the synthesis of AuNPs using Bacillus subtilis isolated from Hatti Gold Mine, India.25 The microorganisms isolated from gold mine have high resistance to gold ions toxicity and can synthesize AuNPs efficiently. The AuNPs served as biocatalyst in degradation of methylene blue and can be used to degrade other toxic dyes in the environment. Saravanan et al. used Bacillus brevis for the synthesis of spherical silver nanoparticles within the size range of 41–62 nm.26 In this work, AgNPs showed remarkable antibacterial activity against multi-drug resistant strains of Salmonella typhi and Staphylococcus aureus.
The reduction of metallic ions into nanoparticles is considered to be dependent on a variety of factors. The first important factor is organic functional molecules present on the cell wall that induce biomineralization, and another essential factor is suitable environmental conditions such as pH, composition of medium, metallic salt concentration and temperature.19 Nanoparticle size, morphology and composition can be significantly affected by these environment parameters.27 Therefore, it is essential to optimize these factors during biosynthesis step to increase the overall efficiency of particles. For example, Ramanathan et al. synthesized silver nanoparticles (AgNPs) using Morganella psychrotolerans and optimized growth kinetics parameters to study their effect on AgNPs morphology.28 At optimum growth temperature of 20 °C, spherical AgNPs were produced with average diameter of 2–5 nm, while a mixture of triangular and hexagonal nanoplates along with spherical nanoparticles were obtained at 25 °C. As temperature was decreased from 20 °C to 15 °C, again a mixture of nanoplates and spherical particles were obtained. Further reduction in bacterial physiological activity and growth by decreasing its optimum growth temperature to 4 °C resulted in a significant increase in the number of nanoplates, with only small number of spherical nanoparticles. Moreover, the spherical nanoparticles formed at 4 °C were larger in size around 70–100 nm. In another study, Yumei et al. studied the AgNP synthesis using Arthrobacter sp. and demonstrated that synthesis of nanoparticle can be modulated by metal ion concentration, temperature and pH.29 Low concentrations of silver nitrate (1 mM) synthesized face-centred-cubic AgNPs with size in between 9 to 72 nm at 70 °C and pH 7 to 8. On increasing the silver nitrate concentration to 3 mM, aggregation of AgNPs was observed at 70 °C. However, no AgNPs synthesis was reported below 5 and above 8 pH, and synthesis was found to be facilitated within the range of 7 to 8 pH. Upon increasing the incubation temperature from 70 °C to 90 °C, there was decrease in synthesis time from 10 min to 2 min. This study concluded that medium pH and metal ion concentration have direct influence on nanoparticle synthesis.
Till few years ago most of the researchers were mainly focused on terrestrial bacteria only. Recently, marine microbial cultures have also been explored extensively as nanofactories for nanoparticles synthesis. Malhotra et al. explored the potential of novel marine bacterium, Stenotrophomonas for the biosynthesis of AuNPs and AgNPs.30 The research work investigated that low molecular weight secretory proteins present in supernatant were responsible for AuNPs and AgNPs biosynthesis. Similarly, Kaur et al. identified a new marine strain, Kocuria flava capable of synthesizing copper nanoparticle with particle size within the range of 5 to 30 nm.31 Various literature reports have explored nanoparticles synthesis using bacterial extracellular polymeric substances (EPS) which act as effective bio-reductant and capping agent.4,32,33 For example, Mehta et al. demonstrated application of EPS, secreted by an osmotolerant marine isolate Alteromonas macleodii, for the production of silver nanoparticle having narrow size distribution.32Table 1 gives an overview of results of various other recent reports on nanoparticles biosynthesis by different bacteria.21–37 The table also highlights the application of these nanoparticles in bioremediation and biomedical field.
Actinomycetes have been generally used for the synthesis of extracellular enzymes and secondary metabolites.38 They have also been adopted for the biosynthesis of nanoparticles as they have unsurpassed capacity for the production of various bioactive compounds and contain high protein content. Actinomycetes synthesize nanoparticles via both intracellular and extracellular pathway, but extracellular reduction is the most common pathway and has more commercial applications in different fields. In 2012, Otari et al. explained the green biosynthesis of silver nanoparticles using actinobacteria Rhodococcus NCIM 2891.39 The TEM graph analysis of AgNPs revealed spherical shape with an average diameter of 10 nm. Intracellular biomineralization of silver ions was thought to be the result of enzymes present on the cell wall, resulting in production of silver nuclei. Karthik et al. adopted the marine bacterium, Streptomyces sp. LK-3 for the reduction of silver ion into AgNPs.40 Their study concluded that nanoparticles were synthesized extracellularly and NADH-dependent nitrate reductase was mainly responsible for the reduction of silver ion into stable AgNPs via an electron transfer reaction. The AgNPs exhibited strong acaricidal or antiparasitic activity against Rhipicephalus microplus and Haemaphysalis bispinosa. Recently, Buszewski et al. employed an acidophilic actinobacteria, Streptacidiphilus durhamensis for the synthesis of silver nanoparticles.41 Their work displayed formation of stable spherical AgNPs within size range of 8 to 48 nm which showed the antibacterial activity against Pseudomonas aeruginosa, Staphylococcus aureus, and Proteus mirabilis. Generally, biosynthesized nanoparticles exhibit higher antimicrobial activity in comparison to traditionally synthesized nanoparticles due to the action of various bioactive molecules involved in capping and stabilization of the nanoparticles. Later on, Abd-Elnaby et al. screened 41 actinomycetes isolates from Suez Gulf, Red Sea and found that only two strains were capable of synthesizing AgNPs.42 Moreover, AgNPs exhibited a strong antibacterial activity against various pathogenic bacteria such as Pseudomonas aeruginosa, Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Salmonella typhimurium, Vibrio damsela, Vibrio fluvialis, Bacillus cereus. It can be concluded that AgNPs remains the most widely studied nanoparticles by actinomycetes. However, there are recent reports which have described synthesis of copper and gold nanoparticles using Streptomyces griseoruber and Streptomyces capillispiralis Ca-1 respectively.43,44 Among actinomycetes, species of Streptomyces are most widely used in pharmaceutical and enzymatic applications because, out of more than 10 000 known antibiotics, 55% are produced by them. Table 1 summarizes list of various recent reports on nanoparticles synthesis by actinomycetes and their applications.39–47
2.2. Fungi and yeast
Fungal biosynthesis of nanoparticles is another simple and straightforward approach which has been explored extensively for fabrication of nanoparticle. In comparison to bacteria, fungi have higher productivity in terms of nanoparticles generation and higher tolerances to metals especially in context of high cell wall binding capacity of metal ions with biomass.14 The downstream processing and biomass treatments are relatively easy in fungi as compared to bacteria and viruses. Moreover, fungi possess higher bioaccumulation ability towards metal ions resulting in an efficient and cost-effective production of nanoparticles. However, the process parameters have a significant effect on the biosynthesis of nanoparticles. An in-depth investigation of different process parameters was carried out by Bhargava et al. to study the effect of pH, salt concentration, and reaction time on the particle size and yield of fungi Cladosporium oxysporum to convert gold ion into nanoparticles.48 The maximum yield of AuNPs was obtained with biomass to water ratio of 1 : 5 at 1 mM salt concentration and 7 pH. Moreover, the synthesized AuNPs exhibited excellent catalytic activity in the degradation of textile dye, rhodamine B within 7 min. Mishra et al. also described the extracellular formation of gold nanoparticles by culture filtrate of Hypocrea lixii and Trichoderma viride, and studied the effect of reaction temperature and incubation time on nanoparticles biosynthesis.49T. viride reported AuNPs biosynthesis within 10 min at 30 °C which further served as biocatalyst and strong antimicrobial agents. Metuku et al. collected a white rot fungus, Schizophyllum radiatum from Eturnagaram forest of Warangal, India and found it capable of producing well-dispersed stable silver nanoparticles.50 Their research work investigated the potential of white rot fungus in the extracellular biomineralization of silver ion to nanoparticles of size 10 to 40 nm. These small size AgNPs demonstrated strong antibacterial activity against various pathogenic Gram negative and Gram positive bacterial strains. Most of the studies reported till date have explained the involvement of extracellular component in the fabrication of nanomaterials. Main advantage of extracellular mediated nanoscale material synthesis is that it is devoid of impurities such as intracellular proteins, and treatment with detergents, ultrasound are not required.
Apart from this, understanding the mechanistic aspects of nanoparticles synthesis has also become indispensable for developing reliable applications. To overcome this knowledge gap, Rajput et al. explored various fungal strains of Fusarium oxysporum for silver nanoparticle synthesis and studied the effect of isolate selection, temperature and pH on nanoparticles morphology.51 Their study summarized that understanding the interactions between organic and interfacial layer will be helpful in developing novel uses, mainly in the area of biosensors. To further explore the bioinspired formation of nanoparticles, Kitching et al. extracted the cell surface proteins of Rhizopus oryzae for in vitro production of gold nanoparticles for biomedical and biocatalytic applications.52 In 2017, Suryavanshi et al. explored the synthesis of aluminium oxide nanoparticles using Colletotrichum sp., and nanoparticles were functionalized by essential oils extracted from the Eucalyptus globulus and Citrus medica.53 The results concluded that nanofunctionalized oil can be used as antimicrobial agents against food-borne pathogens for the prevention of food spoilage. Recently, two filamentous fungi Penicillium citreonigrum and Scopulariopsis brumptii, and an edible mushroom, Pleurotus ostreatus have also been adopted for the synthesis of nanoparticles for anti-cancer and antimicrobial application, respectively.54,55Table 2 depicts the list of various fungi that have been used for the biosynthesis of different metallic nanostructures for various applications.48–66
A representative list of fungi and yeast used to synthesize metal nanoparticles and their applications.
| Fungi | Source of isolation | Metal nanoparticle | Characterization | Applications | Reference | |
|---|---|---|---|---|---|---|
| Size (nm) | Morphology | |||||
| Penicillium diversum | Microbial Type Culture Collection, Chandigarh, India | Silver | 10–50 | Roughly spherical | Antimicrobial activity | 56 |
| Aspergillus foetidus | Kalyani Waste Water Centre, West Bengal India | Silver | 20–40 | Roughly spherical | Antifungal activity | 57 |
| Fusarium oxysporum sp. cubense JT1 | Isolated from wilt infected banana plants | Gold | 22 | nd | Antimicrobial activity | 58 |
| Trichoderma harzianum | Procured from College of Life Sciences, Gwalior, India | Cadmium sulfide | 3–8 | Spherical | Photocatalytic activity | 59 |
| Botrytis cinerea | Isolated form rotten grapes collected from Region IV, Chile | Gold | 1–100 | Triangular, spherical, hexagonal, pyramidal, decahedral | nd | 60 |
| Nigrospora oryzae | nd | Gold | 6–18 | Cubic and spherical | Anthelmintic activity | 61 |
| Schizophyllum radiatum HE863742.1 | Eturnagaram forest, Andhra Pradesh, India | Silver | 10–40 | Irregular shapes | Antimicrobial activity | 50 |
| Trichoderma viride and Hypocrea lixii | Microbial Type Culture Collection, Chandigarh, India | Gold | 61 | Triangles, rods, spheres, hexagons | Biocatalytic and antimicrobial activity | 49 |
| Aspergillus terreus | Microbial Type Culture Collection, Chandigarh, India | Zinc oxide | 28–63 | Spherical | Anticancer activity | 62 |
| Curvularia lunata | Leaves of Catharanthusroeus | Silver | 10–50 | Spherical | Synergistic antimicrobial activity | 63 |
| Metarhizium anisopliae | T-Stanes & Company Limited, Tamil Nadu, India | Silver | 28–38 | Rod-shaped | Mosquitocidal activity against Anopheles culicifacies | 64 |
| Cladosporium oxysporum AJP03 | Soil from Sonshi mining region, Goa, India | Gold | 72 ± 21 | Spherical | Degradation of rhodamine B | 48 |
| Fusarium oxysporum 405 | Obtained from American Research Service, Washington, USA | Silver | 10–50 | Spherical | Colloidal stability | 51 |
| Rhizopus oryzae | National Collection of Industrial Microorganism (NCIM), Pune, India | Gold | 16–43 | Spherical and Flower like | Hemcompatible activity | 52 |
| Trichoderma harzianum | nd | Silver | 20–30 | Spherical | Antifungal activity | 65 |
| Colletotrichum sp. | Amravati University, Amravati, India | Aluminium oxide | 30–50 | Spherical | Antimicrobial activity | 53 |
| Fusarium oxysporum | National Institute of Genetic Engineering and Biotechnology (NIGEB), Tehran, Iran | Silver | 34–44 | Spherical | Antibacterial activity | 66 |
| Pleurotus ostreatus | Biotechnology Center, Cairo University, Egypt | Gold | 10–30 | Spherical and prism shape | Anticancer and synergistic antimicrobial activity | 54 |
| Penicillium citreonigrum | East of Lake Burullus, Egypt | Silver | 6–26 | Spherical | Antibacterial activity | 55 |
| Yeast | ||||||
| Rhodosporidium diobovatum | Isolated from Indian Ocean | Lead sulfide | 2–5 | Spherical | Lead accumulation | 72 |
| Yarrowia lipolytica NCYC789 | National Collection of Yeast Cultures, Norwich, U.K. | Silver | 15 | - | Antibiofilm activity | 67 |
| Candida utilis NCIM 3469 | National Collection of Industrial Microorganism (NCIM), Pune, Maharashtra, India | Silver | 20–80 | Spherical | Antibacterial activity | 68 |
| Cryptococcus laurentii | nd | Silver | 35 ± 10 | Roughly spherical | Antitumor activity | 73 |
| Saccharomyces cerevisiae | Purchased from local market | Silver | 2–20 | Spherical | nd | 74 |
| Magnusiomyces ingens LH-F1 | Sea mud of Harbor Industrial Zone, Dalian, China | Gold | 75 | |||
| Pichia pastoris | Recombinant strain overexpressing Mucor racemosus cytochrome b5 reductase | Silver | 70–180 | Spherical | nd | 69 |
| Candida lusitaniae | Isolated from gut of Cornitermes cumulans termite | Silver and silver chloride | 2–10 | Cubical, cuboctahedral, icosahedral and spherical | Antiproliferative and microbicidal activity | 70 |
| Cryptococcus laurentii; Rhodotorula glutinis | Isolated from apple peel | Silver | 15–35 | Spherical | Antifungal activity | 76 |
| Saccharomyces cerevisiae | AB Mauri (P) Ltd, Bangaluru, Karnataka, India | Gold nanoplates | — | Hexagonal and triangular nanoplates | Surface-plasmon enhanced applications | 77 |
| Candida albicans ATCC 10231 | American Type Culture Collection, Manassas, USA | Silver | 10–20 | Spherical | 78 | |
| Pichia kudriavzevii | Isolated from Sourdoughs and Tanzanian Togwa | Zinc oxide | 10–61 | Hexagonal wurtzite structure | Antioxidant and antibacterial property | 79 |
| Rhodotorula mucilaginosa | Copper Waste Pond at the Sossego Mine, Brazil | Silver | 11 | Spherical | Bioremediation of silver ions | 80 |
| Phaffia rhodozyma | American Type Culture Collection (ATCC), USA | Silver and gold | Silver: 5–9, gold: 4–7 | Spherical and quasi-spherical | Antifungal activity | 81 |
| Magnusiomyces ingens LH-F1 | Sea mud of Harbor Industrial Zone, Dalian, China | Gold | 20.3–28.3 | Spherical and pseudo-spherical | Catalyst for nitrophenols reduction | 82 |
| Rhodotorula glutinis | Soil Sample of Pici Campus, The Federal University of Ceará, Brazil | Silver | 15.45 ± 7.94 | Spherical | Antifungal activity, degradation of nitrophenol and methylene blue | 83 |
| Saccharomyces cerevisiae | Purchased from Pagariya Food Products (P) Ltd, Tamil Nadu, India | Palladium | 32 | Hexagonal | Degradation of textile dyes | 71 |
| Candida glabrata | Oropharyngeal Mucosa of HIV patients | Silver | 2–15 | Spherical | Antimicrobial activity | 84 |
Apart from fungi, some researchers have investigated the use of yeasts for the biogenic synthesis of the nanoparticles. Yeast possess the inherent capability to absorb and accumulate high concentrations of toxic metal ions from their surroundings.11 Yeast cells adapt themselves under metal toxicity conditions using various detoxification mechanisms viz. bio-precipitation, chelation, and intracellular sequestration. This property of yeast cells has been exploited by various researchers. For example, in one study a marine strain of ascomycetous yeast Yarrowia lipolytica was employed for the biomimetic synthesis of silver nanoparticles in a cell associated manner.67 The study concluded that possibly brown pigment (melanin) obtained from the yeast cells was responsible for biomineralization of metal ions. The pigment-derived silver nanoparticles displayed antibiofilm activity against Salmonella paratyphi pathogen. In another research work, Waghmare et al. reported the ecofriendly extracellular biosynthesis of AgNP using Candida utilis NCIM 3469.68 Nanoparticles were circular in shape with size in-between 20 to 80 nm and showed antibacterial activity against pathogenic strains i.e. Staphylococcus aureus, Pseudomonas aeruginosa, and Escherichia coli. In a recent study, Elahian et al. utilized a genetically modified yeast, Pichia pastoris for the biosynthesis of silver nanoparticles.69 Engineered Pichia pastoris strain overexpressed a metal-resistant gene, cytochrome b5 reductase enzyme obtained from Mucor racemosus, for the reduction of metal ion into nanoparticles. The cytochrome b5 reductase enzyme leads to synthesis of stable and well-dispersed metal nanoparticles within size range of 70–180 nm. In 2016, Eugenio et al. isolated a yeast strain, Candida lusitaniae from gut of a termite and demonstrated production of silver nanoparticles with diameter in the range of 2–10 nm.70 The silver nanoparticles showed antiproliferative activity against S. aureus and Klebsiella pneumoniae, and presented a promising alternative to commonly used antibiotics. Sriramulu and Sumathi (2018) employed Saccharomyces cerevisiae aqueous extract for the synthesis of hexagonal palladium nanoparticles (PdNPs) of size 32 nm.71 PdNPs showed photocatalytic degradation of textile azo dye (direct blue 71) to 98% within 60 min under UV light. All these literature reports suggested that the difference in nanoparticle size, shape and properties is because of the different mechanisms adopted by yeast cells to synthesize and stabilize the nanoparticles. Table 2 shows the list of various yeasts used for the synthesis of different metal nanoparticles.67–84
2.3. Algae
Similarly to yeast, there are diverse literature reports on algae being used as a “nanofactory” for biosynthesis of metal nanoparticles (Table 3). Ferreira et al. employed the dried unicellular microalga, Chlorella vulgaris for the biosynthesis of silver nanoparticles within range of 9.8 ± 5.7 nm.85 The spherical shaped nanoparticles were observed to be a promising green alternative for biomedical application as antimicrobial agents. In another study, Arsiya et al. evaluated the synthesis of palladium nanoparticles using Chlorella vulgaris aqueous extract within 10 min.86 TEM results revealed that the nanoparticles were circular and mono-dispersed in nature having size of 5 to 20 nm. This study first time reported synthesis of palladium nanoparticles in a comparatively shorter time duration using C. vulgaris. The biosynthesis of palladium nanoparticles has also been reported using marine alga, Sargassum bovinum which is isolated from Persian Gulf area.87 Dhas et al. explored the synthesis of silver chloride nanoparticles using the aqueous extract of marine alga, Sargassum plagiophyllum.88 Recently, an economical green method has been reported for the synthesis of silver nanoparticles using a marine green alga, Caulerpa racemosa.89 The synthesized nanoparticles exhibited remarkable catalytic activity towards the degradation of methylene blue. Ramakrishna et al. studied the synthesis of gold nanoparticles using aqueous extracts of brown algae, Sargassum tenerrimum and Turbinaria conoides.90 The AuNPs displayed excellent biocatalytic activity in the degradation of aromatic nitro compounds and organic dyes. The metal nanoparticles of zinc oxide (ZnO) have also generated curiosity among researchers due to their unique physicochemical characteristics and wide applications in opto-electronics, sunscreens, biomedicine and food additives etc. Rajeshkumar (2018) adopted two marine brown seaweeds such as Padina tetrastromatica and Turbinaria conoides algal formulation for the biosynthesis of ZnO nanoparticles and evaluated their antimicrobial potential against fish pathogens.91 In another recent work, Sanaeimehr et al. synthesized ZnO nanoparticles using Sargassum muticum extract and established their antiangiogenic and antiapoptotic potential against human liver cancer cell lines.92 All these literature reports indicates that researchers are now exploring marine organisms for the biogenic synthesis of nanoparticles because marine algae contain various biologically active compounds and secondary metabolites that allow them to act as “nanofactories”.93 These marine algae have lots of applications in biomedicine as antioxidants, anticancer, antidiabetic, cardioprotective, hepatoprotective and antiviral agents. Table 3 highlights the results of recent reports on algae-based biosynthesis of metal nanoparticles for different biological applications.85–108
A representative list of algae used for the synthesis of nanoparticles and their applications.
| Algae | Source of isolation | Metal nanoparticle | Characterization | Applications | Reference | |
|---|---|---|---|---|---|---|
| Size (nm) | Morphology | |||||
| Porphyra vietnamensis | nd | Silver | 13 | Spherical | Antibacterial activity | 94 |
| Stoechospermum marginatum | Tuticorin Coast, Tamil Nadu, India | Gold | 19–94 | Spherical, hexagonal and triangular | Antibacterial activity | 95 |
| Tetraselmis kochinensis | nd | Gold | 05–35 | Spherical and triangular | nd | 96 |
| Chaetomorpha linum | Kanyakumari Coast, India | Silver | 03–44 | Clusters | nd | 97 |
| Sargassum plagiophyllum | Rameshwaram Coast, Tamil Nadu, India | Silver | 18–42 | Spherical | Antibacterial potential | 88 |
| Spirogyra varians | Sweet water areas, Kerman, Iran | Silver | 35 | Quasi-sphere | Antibacterial activity | 98 |
| Scenedesmus sp. | CSIR – Institute of Minerals and Materials Technology, Bhubaneswar, India | Silver | 15–20 | Spherical crystalline | Antibacterial assay | 99 |
| Chlorella vulgaris | Algal Culture Collection, Chennai, India | Gold | 02–10 | Spherical self assembled cores | Anti-pathogenic activity | 100 |
| Ecklonia cava | Busan, South Korea | Gold | 30 | Spherical and triangular | Antimicrobial activity | 101 |
| Caulerpa racemosa | Gulf of Mannar, Southeast Coast, India | Silver | 05–25 | Spherical and triangular | Antibacterial assay | 102 |
| Sargassum bovinum | Persian Gulf area, South Western, Iran | Palladium | 05–10 | Octahedral | Hydrogen peroxide sensor | 87 |
| Ulva lactuca | Coastal areas of Rameshwaram, Tamiladu, India | Silver | 20–35 | Cubical | Antiplasmodial activity | 103 |
| Pithophora oedogonia | Fresh Water Pond of Hoogly, West Bengal, India | Silver | 25–44 | Cubical and hexagonal | Antibacterial activity | 104 |
| Caulerpa racemosa | Mandapam Coastal Area, Tamil Nadu, India | Silver | 25 | Distorted spherical | Degradation of methylene blue | 89 |
| Sargassum tenerrimum and Turbinaria conoides | Mandapam Coast, Tamil Nadu, India | Gold | 27–35 | Spherical | Reduction of dyes – rhodamine B and sulforhodamine 101 | 90 |
| Chlorella vulgaris | Culture Collection of Algae, University of Texas, Austin | Silver | 03–15 | Spherical | Antibacterial activity | 85 |
| Cystoseira baccata | Northwest Coast of Spain | Gold | 8.4 | Spherical | Anticancer activity | 105 |
| Chlorella vulgaris | Faculty of Natural Resources and Environment, University of Birjand, Iran | Palladium | 05–20 | Spherical | nd | 86 |
| Galaxaura elongata | Northwest Coast of Red Seashore | Gold | 3.8–77.1 | Spherical, rods, hexagonal and triangular | Antibacterial activity | 106 |
| Laminaria japonica | Local Seaweed Industry in Korea | Silver | 31 | Spherical to oval | Phytotoxicity and seedling growth assay | 107 |
| Gelidium amansii | Coastal Belt of South Korea | Silver | 27–54 | Spherical | Antibacterial and antibiofilm activity | 108 |
| Padina tetrastromatica and Turbinaria conoides | Tuticorin Coast, Tamil Nadu, India | Zinc oxide | 90–120 | Spherical, pentagonal, hexagonal and triangles | Antibacterial activity | 91 |
| Sargassum muticum | Northwest Pacific Region, Iran | Zinc oxide | 30–57 | Hexagonal | Anti-angiogenesis and anti-apoptotic activity | 92 |
2.4. Viruses
An interesting property of viruses is their thick outer surface coating of capsid proteins which provide a highly suitable platform for interaction with metallic ions.109 These protein cages can build monodispersed units that are highly robust and mouldable through genetic engineering. Viruses can be modified to serve as templates for material deposition or engineered to create three-dimensional vessels for targeted drugs delivery.110 Viruses can be employed for the synthesis of nano-conjugates and nanocomposites with metal nanoparticles which are important bioengineering materials in drug delivery and cancer therapy (Table 4). Mao et al. investigated the use of M13 bacteriophage for the nucleation and orientation process of semiconductor nanocrystals.111 This group showed a genetically controllable biogenic synthesis route to a semiconductor nanocrystals of zinc sulfide and cadmium sulfide. The plant viruses are being proved to be safe for nanotechnology applications due to their structural and biochemical stability, ease of cultivation, non-toxicity and non-pathogenicity in animals and humans. In one study, low concentrations of tobacco mosaic virus (TMV) and bovine papilloma virus (BPV) were used as additives along with extracts of various plants e.g. Nicotiana benthamiana, Avena sativa and Musa pradisiaca etc.112 The TMV and BPV not only helped in the reduction of size, but also significantly enhanced the numbers of the nanoparticles in comparison to the non-virus control. Cao et al. employed red clover necrotic mosaic virus (RCNMV) for the synthesis of nanoparticles for the controlled delivery of doxorubicin drug for chemotherapy.113 The unique morphology of RCNMV and structural changes in response to divalent cations removal helps in doxorubicin infusion to the capsid through surface pore formation mechanism. Le et al. investigated the potential of potato virus X nanoparticles for the delivery of doxorubicin drug for cancer treatment.114 Potato virus X has the capability to synthesize elongated filamentous nanoparticles which exhibits enhanced tumor homing and penetration power in comparison to spherical ones. However, synthesis of nanoparticles by viruses still faces various drawbacks such as involvement of host organism for protein expression, under-developed processes for synthesis and limited research on large scale application. Table 4 summarizes the results of recent reports on virus-based synthesis of metal nanoparticles and their applications.109–118
A representative list of viruses used for the synthesis of nanoparticles and their applications.
| Algae | Nanoparticle type | Characterization | Applications | Reference | |
|---|---|---|---|---|---|
| Width (nm) | Morphology | ||||
| Tobacco mosaic virus (TMV) | Palladium | 2.9–3.7 | Multiwalled carbon nanotubes | Catalyst in Suzuki reaction and recyclable | 115 |
| Cucumber mosaic virus | Nanoassemblies | ∼29 | Icosahedral | Anticancer drug delivery | 110 |
| Tobacco mosaic virus (TMV) | Gold | 5 | Spherical | Building block for chiral meta-molecules | 109 |
| Red clover necrotic mosaic virus | Nanocarriers | 36 | Icosahedral | Doxorubicin delivery | 113 |
| M13 virus | Titanium dioxide | 20–40 | Mesoporous nanowires | Photo-electrochemical properties | 116 |
| Potato virus X | Nanoconjugates | 12 | Filamentous rod shaped | Herceptin drug delivery in breast cancer therapy | 117 |
| Potato virus X | Nanocarriers | 13 | Helical | Doxorubicin delivery in cancer therapy | 114 |
| Hepatitis E virus | Nanoconjugates | 27–34 | Icosahedral | Cancer therapy | 118 |
3. Mechanism of nanoparticles synthesis using various bioreducing agents
In literature there are large number of reports which have discussed about the possible mechanistic aspects to explain the complex process of biomineralization of metal ions into nanoparticles. However, our current understanding of the mechanistic aspects is still very limited and need to be explored further. The biosynthesis of metal nanoparticles in microorganisms have been proposed to be dependent on three different mechanisms: reductase enzymes and proteins, exopolysaccharide, and electron shuttle quinones.14
3.1. Role of enzyme and proteins
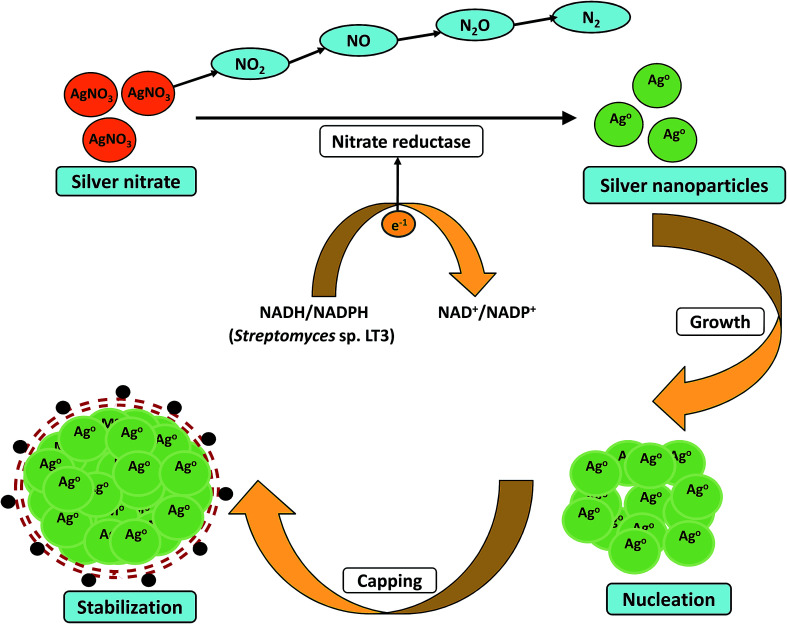
Majority of the studies have suggested possible involvement of enzymes and proteins as the main bioactive moiety which act as reducing and capping agent during the nanoparticles synthesis. Nanoparticles formation have been generally considered as an outcome of the microbes resistance to metal ions. Metal resistance has been seen in most of the bacteria and fungi. Microbes have adopted various defense mechanisms viz. reduction of metal ions, complex formation, precipitation, dissimilatory oxidation to fight against metal toxicity.119 Metal nanoparticles are produced via redox reactions which occur either intracellularly or extracellularly. Earlier reports have indicated the possible involvement of NADH-dependent nitrate reductases in metal bioreduction. In 2014, Karthik et al. proposed a possible mechanism for the bioreduction of silver ions into nanoparticles by using Streptomyces sp. LK3 at room temperature.40 Their study concluded that the nitrate reductase enzyme of actinomycetes was responsible for the bioreduction process (Fig. 2). The culture not only demonstrated nitrate reduction to nitrite, but also showed reduction of nitrite to nitrogenous gases. The AgNPs were found to be stable for months without using any capping agents. Even no particle aggregation was observed in the mixture which further strengthens the stability of AgNPs. Their work provided an easy, cost-effective, and ecofriendly approach for the green synthesis of small size (5 nm) silver nanoparticles. In another study, Divya et al. demonstrated synthesis of monodispersed silver nanoparticles using the bacterial strain Alcaligenes faecalis.35 This report also illustrated that the reducing agents such as NADH and NADH dependent reductases produced by culture in supernatant were mainly responsible for bio-mediated synthesis of silver nanoparticles. The investigators also suggested that the synthesized silver nanoparticles showed antimicrobial activity against urinary tract infection causing clinical isolates such as Bacillus sp., E. coli, K. pneumoniae, S. aureus and C. albicans. Recently, Hamedi et al. studied the synthesis of AgNPs using the cell free culture filtrates of Fusarium oxysporum66 and results showed that the highest synthesis rate of nanoparticles was obtained during stationary phase when activity of extracellular enzyme nitrate reductase was maximum. Moreover, increase in C : N ratio resulted in the induction of nitrate reductase enzyme and synthesized AgNPs with small size and narrow size distribution.
Fig. 2. Proposed mechanistic scheme of the bioreduction and stabilization of nanoscale particles by nitrate reductase enzyme.
Many researchers have investigated the role of conductive pili and cell surface proteins in transfer of electron resulting in extracellular reduction of metal ions.52,120,121 For e.g., the biomineralization of uranium metal ions to uranium nanoparticles by Geobacter sulfurreducens has been reported by Cologgi et al. (2011).120 The importance of pili (bacterial external appendages made up of pilin proteins) in extracellular reduction was validated by performing experiments under pilin-inducing and noninducing conditions. Astonishingly, in the pilin-deficient mutants, uranium nanoparticles were formed inside the periplasmic space, whereas in the pilin-supplemented strains, nanoparticles were produced extracellularly. Pilin expression significantly increased the rate of uranium biomineralization and prevented its periplasmic localization. Furthermore, another pioneer work in the area of uranium reduction explained the phenomenon of electron transport in actively respiring Geobacter spp. using conductive pili.121 It was concluded that pilin proteins in Geobacter were essential for the immobilization of uranium, c-cytochrome organization, and extracellular reduction of uranium. Kitching et al. demonstrated the one-pot synthesis of gold nanoparticles by purified cell surface protein of Rhizopus oryzae.52 This work elucidated the effect of cell surface protein on the size and morphology of AuNPs. It was concluded that different cell surface protein extraction methods could affect stability of protein and the AuNPs biosynthesis. Biosynthesized nanoparticles showed good stability and hemocompatibility for different biomedical applications.
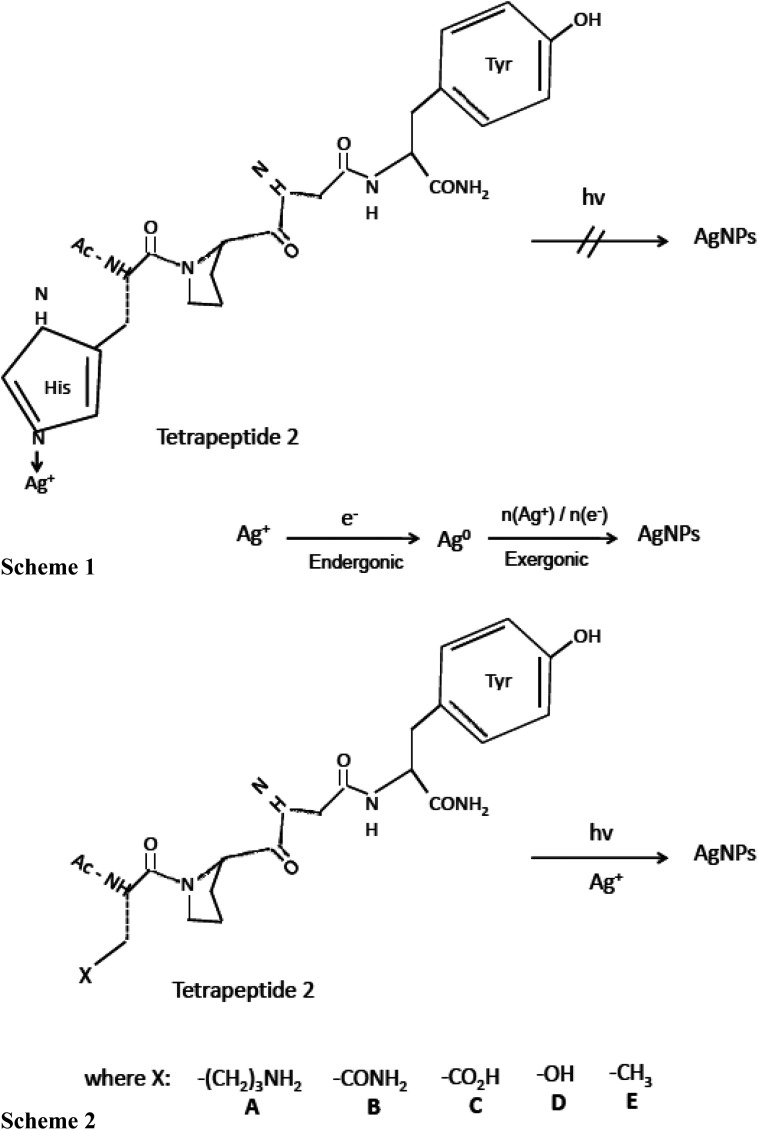
Vasylevskyi et al. carried out an in-depth investigation of biomineralization process of silver ion at molecular level using Geobacter sulfurreducens.122 Vasylevskyi and his group explained that the reduction of Ag+ ion to Ag0 is an endergonic reaction, whereas aggregation to Agn clusters is an exergonic process which further leads to stable AgNPs synthesis. This hypothesis was based on various experiments conducted during AgNPs synthesis by photoinduced electron transfer through tetrapeptide/Ag+ solution (shown in Fig. 3). Electron transfer through tetrapeptide 1 containing histidine at N-terminal did not generate AgNPs by photoinduction. Although, tyrosine amino acid present at C-terminal region of tetrapeptide 1 generated tyrosyl radicals and electrons, Ag+ ions attached to the histidine at N-terminal were not reduced to AgNPs. It was concluded that high binding energy of the imidazole side chain of histidine prevented the aggregation of Ag0 into Agn clusters and hence inhibited the synthesis of AgNP by photoinduction (Fig. 3, Scheme 1). But amino acids containing weaker Ag+ binding side chains such as aliphatic amines, amides, acids, alcohols, or alkyl groups could allow AgNPs synthesis by photoinduction. Therefore, histidine residue of the N-terminal region of tetrapeptide 1 was replaced by lysine, asparagine, aspartate, serine, and alanine, to generate the different tetrapeptides 2 A-E, respectively (Fig. 3). The addition of AgNO3 solution to these tetrapeptides (2A–2E) synthesized AgNPs upon irradiation of the C-terminal tyrosine (Fig. 3, Scheme 2). Thus, it was concluded that successful production of AgNPs essentially requires a rapid exergonic aggregation of Ag0 into Agn clusters.
Fig. 3. Synthesis of silver nanoparticles (AgNPs) by electron transfer in peptides. Scheme (1) Inhibited synthesis of AgNPs by photoinduced electron transfer in tetrapeptide 1/Ag+ solution. Scheme (2) AgNPs synthesis in tetrapeptide 2A–E/Ag+ solution by photoinduced electron transfer [this figure has been adapted from ref. 122 with permission from Wiley].
3.2. Role of electron shuttle quinones (or redox mediators)
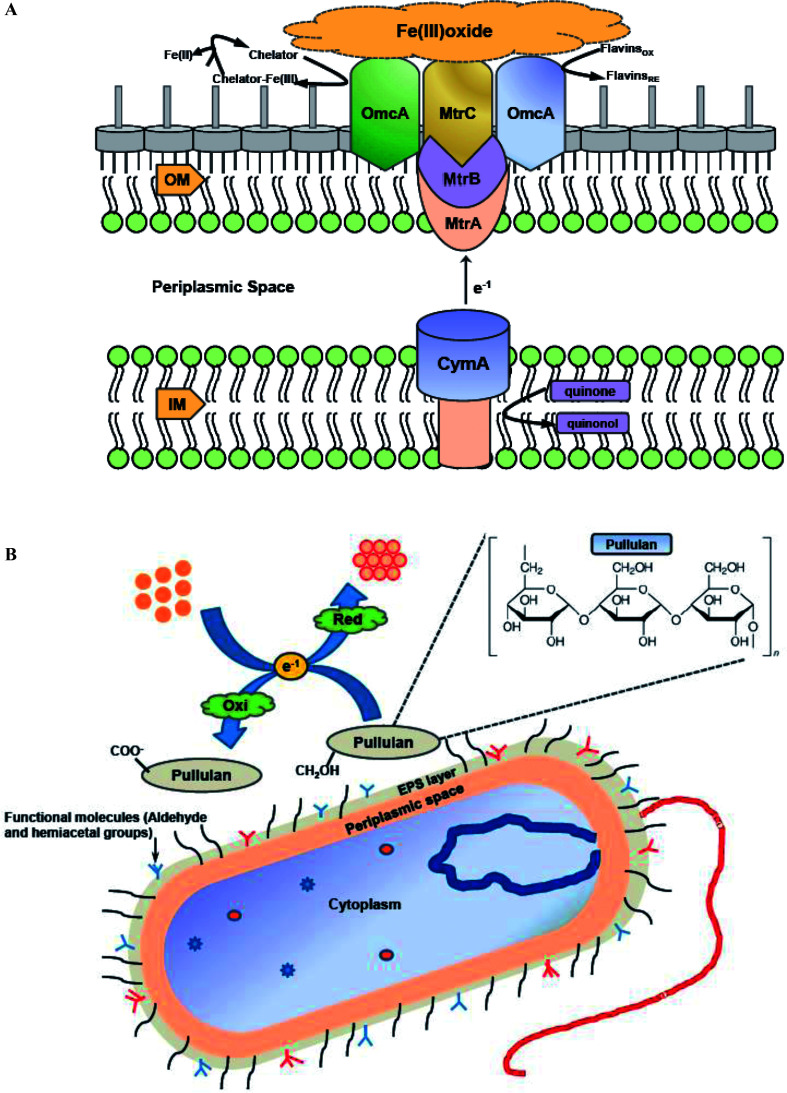
During nanoparticles synthesis, electrons can be transferred via low molecular weight redox mediators like ubiquinol, NADH or oxygen/superoxide or by direct interaction between c-type cytochromes redox proteins and the metal ion.123 Various literature reports have discussed the importance of both cytochromes and redox mediators in promoting extracellular synthesis of metal nanoparticles. Shi et al. explored the possibility of Shewanella oneidensis MR-1 utilizing ferric oxides minerals as terminal electron acceptors during anaerobic respiration,124 and the role of c-type cytochromes in this electron transfer pathway. S. oneidensis MR-1 and related strains of metal-reducing Shewanella have evolved a metal-reducing machinery or Mtr pathway for transferring electrons across cell membranes to the surface. A molecular mechanism was proposed for the involvement of c-type cytochromes in transferring electrons from quinol at the inner membrane (IM), through the periplasmic space (PS), and across the outer membrane (OM) to the metal oxide surface (Fe(iii) oxide) (Fig. 4A). The protein components identified to date for the Mtr pathway include CymA, MtrA, MtrB, MtrC, and OmcA. A similar mechanism can be hypothesized for bacterial metal nanoparticles synthesis outside the bacterial cell surface (Fig. 4A). Shi et al. revealed that CymA is an inner-membrane tetraheme c-type cytochrome (c-Cyt), that belongs to the NapC/NrfH family of quinol dehydrogenases.124 It was hypothesized that CymA oxidizes the quinol in the inner-membrane and transfers the electrons to MtrA either directly or indirectly through other periplasmic proteins. MtrA is a decaheme c-Cyt and observed to be embedded in the trans outer-membrane and porin like protein MtrB. Along with MtrB, MtrA transfers the electrons from the outer-membrane to the MtrC and OmcA located on the outermost surface. OmcA and MtrC are two outer-membrane decaheme c-Cyt which are translocated from the outer-membrane to cell surface by the bacterial type II secretion pathway. MtrC and OmcA function as terminal reducing agents and can bind to the surface of Fe(iii) oxides and transfer electrons directly to these oxides through their exposed hemes part. MtrC and OmcA can also use the flavins secreted by S. oneidensis MR-1 cells as diffusible factors for reduction of Fe(iii) oxides at faster rates. MtrC and OmcA can also serve as the terminal reducing agents for soluble forms of Fe(iii) because of their broad redox potentials and extracellular location.
Fig. 4. (A) The proposed Metal-reducing (Mtr) extracellular electron transfer pathway of S. oneidensis MR-1 which is similar to bacterial metal nanoparticles synthesis outside the bacterial cell surface. CymA, MtrA, MtrC and OmcA are multiheme c-type cytochromes, while MtrB is a porin-like trans-outer membrane protein [this figure has been adapted from ref. 124 with permission from Frontiers]; (B) proposed mechanistic scheme of the biomineralization of gold ions by pullulan exopolysaccharide [this figure has been adapted from ref. 135 with permission from Elsevier].
Earlier, Rodrigues et al. determined the molecular structure of NrfH of Desulfovibrio vulgaris, which is the only available molecular structure for the NapC/NrfH family of quinol dehydrogenases. This model proposes that quinol binds inside the pocket adjacent to heme 1 of NrfH of D. vulgaris, where quinol undergoes oxidation.125,126 The proximal axial ligand of NrfH of D. vulgaris contains a methionine residue (Met49) that is two residues downstream from the histidine residue of the CX2CH motif for binding of heme 1, and an aspartate residue (Asp89) is at the position usually occupied by the distal axial ligand. However, Asp89 is not used for heme coordination but it binds to quinol.125,126 Later on, Hartshorne et al. provided the first molecular model of electron transfer across the bacterial outer-membrane.127 MtrB is a trans outer-membrane spanning β-barrel protein that serves as a pocket to incorporate MtrA inside the membrane. MtrAB function as a trans outer-membrane delivery module for transfer of electrons to MtrC, which serves as an extracellular reductase. MtrA contains a signal peptide that targets the synthesized polypeptide to the periplasmic space via the bacterial secretary system. The MtrA polypeptides can be divided into two pentaheme domains, each of which shares sequence similarity with NrfB of E. coli.128 When it is expressed in E. coli, the truncated MtrA having only one of its pentaheme domains is folded properly and contains five hemes, providing experimental evidence that MtrA contains two repetitive functional domains.128 This type of heme arrangement help in rapid electron transfer from the heme groups of NrfB that form a molecular wire. Clarke et al. (2011) determined the molecular structure of MtrF, an MtrC homolog, and provided the molecular structural evidence in favour of terminal reducing agent for the outer-membrane c-Cyt of S. oneidensis MR-1 in Fe(iii) oxide reduction.129 Results revealed that MtrF is folded into four different domains: domains I (aa 49–186) and III (aa 319–473) each containing seven anti-parallel β-strands and folded together to form a split-β barrel structure, while domains II (aa 187–318) and IV (aa 474–641) each bind five tightly packed hemes. It was thought that domain II binds with solid-phase Fe(iii) oxides by transferring electrons directly to the oxides via the solvent-exposed heme. Domain I and III are proposed to be involved in binding and reduction of flavins and soluble metals such as chelated Fe(iii), while domain IV is involved in physical interaction with the MtrDE (MtrAB homologs) complex and exchange electrons with MtrD. It should be mentioned that the overall shape of MtrF is very similar to that of OmcA;129 which further confirms that MtrF and OmcA can fold in similar way. Like MtrC and OmcA, MtrF also reduces Fe(iii) oxides and flavins.130 Thus, the structural characterization of MtrF supported the fact that cell surface-bound c-Cyts MtrC, MtrF, and OmcA transfer electrons directly to the surface of Fe(iii) oxides through their solvent-exposed hemes.
Later on, Ng et al. elucidated a technique to explain the role of c-type cytochromes in the extracellular synthesis of nanoparticles.131 In this technique, a mutant strain of Shewanella oneidensis lacking cytochrome genes (MtrC and OmcA) was used to perform silver nanoparticles synthesis. Nanoparticles produced by mutant strain were smaller in size and lesser in number as compared to wild-type strain of S. oneidensis. This indicated that c-type cytochromes help in transferring electrons to extracellular metal ions. Liu and co-authors demonstrated the direct involvement of outer membrane c-type cytochrome protein complexes (ombB, omaB and omcB) in extracellular reduction of Fe(iii)-citrate and ferrihydrite using a metal-reducing bacterium Geobacter sulfurreducens PCA.132,133 The similar phenomenon of electron transfer via c-cytochromes was observed in another report on silver nanoparticles synthesis by Geobacter sulfurreducens.122 During this process, electrons generated through respiration were transported by Fe2+/hemes of c-cytochromes (Ppc) via periplasmic space to Fe3+/hemes of the outer membrane cytochromes (Omc). Subsequently, electrons were transferred from protein core of Omc to the surface attached Ag+ ions to synthesize AgNPs while oxidizing the cytochromes again to Fe3+/hemes (Fig. 4A). Overall, these reports concluded that the multiheme complexes help in electrons movement from the inner membrane to the cell wall (or outer membrane) via periplasm, and thus help in the extracellular reduction of metal ions into nanoparticles.
3.3. Role of exopolysaccharides
Bacterial exopolysaccharides (EPSs) are biomolecules which are secreted extracellularly and have different biological roles, such as environmental protection, surface adherence, and cell to cell interactions. EPSs have been explored as agents for greener production of numerous metal nanoparticles, since they have the capability to reduce metal ions to synthesize nanoparticles and stabilize them acting as capping agents.4,30 Kang et al. studied silver nanoparticles formation by exopolysaccharide in Escherichia coli biofilm and concluded that aldehyde and hemiacetal groups present in the exopolysaccharides act as the bioreducing agents.134 FTIR and 13C nuclear magnetic resonance (NMR) spectroscopy results of EPS interactions with metal ion indicated that the hemiacetal groups of rhamnose sugars were involved in synthesis of silver nanoparticles. Moreover, the aldehyde groups in rhamnose and pyranose sugars were oxidized to carboxyl groups by silver ions. Choudhury et al. developed a strategy for the synthesis of gold nanoparticles using pullulan as reducing agent.135 In-depth analysis of thermodynamic parameters further revealed that gold nanoparticles synthesis followed first order kinetics and small size nanoparticles were formed at elevated temperature (100 °C). FTIR analysis clearly showed that the basic structure of pullulan remain unaffected during synthesis of gold nanoparticles by pullulan polymer, and α-1,4 and α-1,6 linkages of polymer were also intact even after the reaction. It was hypothesized that the biomineralization of gold ions was due to the oxidation of side chain aliphatic alcoholic groups of pullulan molecule. Thus, it may be concluded that the free CH2OH groups of pure pullulan molecules were oxidized to COO− carboxyl group, while simultaneous reduction of Au3+ to Au0 resulting in the formation of gold nanoparticles. Fig. 4B demonstrates the pictorial representation of biomineralization of gold ions by pullulan exopolysaccharide.
Similar observations were reported in another work while describing the synthesis of silver nanoparticles using Ochrobactrum rhizosphaerae, and deciphered that the active moiety responsible for synthesis and capping of silver nanoparticles was EPS.4 Later on characterization of the EPS by various biochemical techniques suggested that it is a glycoprotein (GLP) in nature. FTIR spectrum further revealed the involvement of various functional groups responsible for the reduction of metal ions to nano-colloidal solution. It was concluded that the free CH2OH groups of GLP molecules were oxidized to carboxyl groups (COO−) with simultaneous reduction of Ag+ to Ag0 resulting in the formation of silver nanoparticles. In 2016, Raj et al. illustrated that EPS obtained from a marine bacterium, Pseudomonas aeruginosa JP-11, has the potential of forming spherical cadmium sulfide (CdS) nanoparticles which were in 20–40 nm diameter range.33 Surface functionalization of EPS with sulphur groups enhanced the adsorption efficiency of cadmium metal ions. The sulphur groups are mostly preferred for functionalization due to their high stability constant and low solubility products. They easily attach with the cadmium ions in the aqueous solution and synthesize CdS nanoparticles. This work showcased that CdS nanoparticles synthesis using EPS also assists in cadmium metal detoxification from aqueous solution. In 2017, Yumei et al. reported synthesis of silver nanoparticles using EPS derived from Arthrobacter sp. B4.29 Subsequent zeta potential and FTIR analysis revealed that the shift of –C O and –OH absorption peaks in B4-EPS before and after the formation of silver nanoparticles was responsible for reducing Ag+ ion and leading to high stabilization of nanoparticles as capping agents. Furthermore, these nanoparticles showed high stability, excellent antibacterial activity, and low phytotoxicity.
The mechanistic aspects of nanoparticles synthesis by exopolysaccharides further need to be discussed in detail. The structure of EPSs is mainly composed of carbohydrates such as monomers of d-glucose, d-mannose, l-fucose, d-galactose, l-rhamnose, d-galacturonic acid, d-glucuronic acid, l-guluronic acid, d-mannuronic acid, N-acetyl-d-glucosamine, and N-acetyl-d-galactosamine, as well as noncarbohydrate components that confers an anionic nature to the EPSs such as carboxyl, phosphate, sulfate, and pyruvate substituents.136–139 In addition to providing the negative charge, these organic groups increase the lipophilicity of the EPSs and directly influence their interaction with other polysaccharides and cations.136 In general, it has been noted that once metal ions are in contact with EPSs that contain reducing sugars, they are chelated and then reduced and stabilized by various functional groups. Polyanionic groups are the best known chemical moieties to be involved in the reduction and stabilization of metal nanoparticles.140 Moreover, electrostatic interactions between metal cationic ions and anionic groups such as carboxylic and phosphoric functional groups of EPS have been mentioned to be an advantage for the synthesis of metal nanoparticles.141 Among these functional groups, hydroxyl, carboxyl, phosphoric, hemiacetal, and amino end groups have been proposed to reduce metal ions from the precursor salts to obtain the respective nanoparticles.142 In relation with the above, hydroxyl groups have been attributed the ability to coordinate with metal ions.143 In fact, during the reducing process, oxidation of hydroxyl groups to form carbonyl groups as well as oxidation of alcoholic and aldehydic groups to form carboxylic groups has been reported to be an important factor during synthesis of metal nanoparticles.144 Thus, in relation to the EPSs described in this study, the structure of xanthan gum has been described to be helical with many hydroxyl groups that can carry out reduction of ions. In addition, this structure creates a network by hydrogen bonding in which nanoparticles stabilize.145 Further, xanthan gum has negative charge due to acetyl groups and pyruvic acid linked with mannose, and provides this charge to nanoparticle surface creating a steric repulsion among them.146 Dextran, is useful in the synthesis of metal nanoparticles since their hydroxyl, ketone, aldehydes, and carboxyl groups interact and allow the reduction of Ag+ ions to form Ag-NPs, preventing their agglomeration as well.147 At this point, it was mentioned that oxygen from dextran functional groups can donate their pair of electrons to gold ions thereby producing Au-NPs.148 As it is well known, dextran is rich in hydroxyl groups that can interact with magnetic nanoparticles by hydrogen bonds stabilizing them.149 For curdlan, the carboxylic groups of curdlan derivatives such as carboxylic curdlan and carboxymethyl curdlan have the ability to adsorb metal ions by electrostatic attractive forces. These functional groups reduce the metal ions, and the necessary nucleation is created to form stabilized clusters during synthesis of AuNPs and AgNPs.150,151
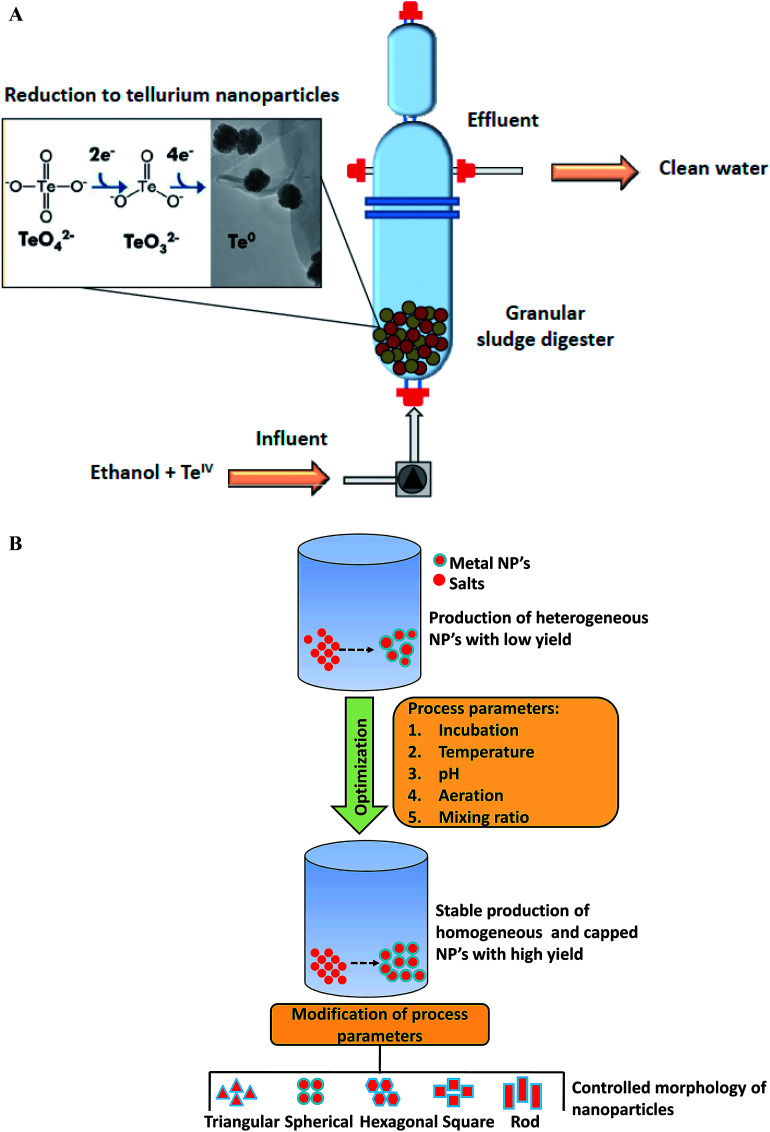
4. Large scale production of biomediated nanoparticles
Microbial fermentation represents state of the art approach for large-scale production of nanoscale structures of different metals. In recent years, researchers have explored large-scale synthesis of nanoparticles using biogenic routes with a narrow size distribution.152,153 In 2010, Moon and his group first time reported large scale production of magnetic and metal-substituted magnetic nanoparticles using Thermoanaerobacter sp. TOR-39.154 This report concluded that magnetic nanoparticles production can be obtained in huge quantities at low cost, similar to traditional chemical synthesis. At the end, about 1 kg (wet weight) of Zn-substituted magnetites were obtained from 30 L fermentations. The magnetic nanoparticles have become focus of recent research as they are promising candidate for magnetic resonance imaging, bioremediation, data storage, catalysis biosensors development, and they can be manipulated very easily under magnetic field influence. In another report, Moon et al. employed the same thermophilic strain Thermoanaerobacter sp. TOR-39 for the extracellular synthesis of cadmium sulfide (CdS) nanoparticles.155 The size of CdS crystallites was less than 10 nm and process was easily scalable up to 24 L. Various factors such as biomass concentration, dosing amount, type of precursors used, and the basal medium composition were found to be crucial for producing tailor-made nanoparticles. Later on, Moon et al. elucidated synthesis of semiconducting zinc sulfide (ZnS) nanoparticles in lab-scale reactor of 24 L capacity using an anaerobic thermophilic metal-reducing bacterium Thermoanaerobacter sp. X513.152 Production of ZnS nanoparticles was scalable, reproducible and controllable (within 2–10 nm range) from 10 mL to 24 L with yields of 5 g per L per month. More recently, Moon et al. demonstrated the scale-up of nanoparticles synthesis from lab scale to pilot-plant level using same bacterium.156 This work investigated the scalability of bacteria mediated ZnS nanoparticles production in 100 L and 900 L scale bioreactors. Repeated 100 L batches using fresh or recycled media produced ZnS nanoparticles with high reproducibility in crystalline size of 2 nm and yields of approximately 0.5 g L−1 which were close to the small-scale batches. The cultivation at 900 L scale yielded around 320 g ZnS nanoparticles powder and this amount was sufficient for the synthesis of ZnS thin film with thickness of 120 nm over 0.5 m width and 13 km length. In another study, Ramos-Ruiz et al. discussed the potential of upflow anaerobic sludge bed reactors for continuous conversion of toxic tellurite oxyanions (TeIV) to non-toxic recoverable tellurium (Te0) nanoparticles using methanogenic microbial consortium.153 This group also evaluated the effect of redox mediating flavonoid compound, riboflavin (RF), with the aim of increasing the reduction of tellurite oxyanions. The presence of riboflavin mediator enhanced the conversion rate of tellurite by approximately 11-fold. This work elucidated that the methanogenic anaerobic granular sludge can be adopted as a bioreactor technology for the continuous production of tellurium nanoparticles in direct recoverable mode (Fig. 5A). Moreover, the sludge was able to sustain the reduction of high loads of toxic tellurite oxyanions. With respect to large-scale production, the fact that only few reports are available on bioreactor cultivation strategies for biosynthesis in last six years is not a good finding, as these strategies are essential for obtaining higher productivity of nanoparticles synthesis. These are indeed important for large-scale level production and thus need to be further investigated.
Fig. 5. (A) Continuous reduction of toxic tellurite (TeIV) oxyanions into recoverable tellurium nanoparticles using anaerobic sludge reactor [this figure has been adapted from ref. 153 with permission from Elsevier]; (B) parameters for producing monodispersed, stable, and high-yield biological nanoparticles [this figure has been adapted from ref. 16 with permission from Elsevier].
5. Advantages and limitations of biological methods in nanoparticles synthesis
There have been tremendous developments in the field of microorganism produced nanoparticles and their applications over the last decade. The biosynthesis of nanoparticles has numerous advantages such as benign and eco-friendly production, cost-effectiveness and the biocompatibility of synthesized nanoparticles.14 As opposed to physicochemical processes, biosynthesized nanoparticles are free from toxic chemical contaminants which is essentially a desirable trait for biomedical applications.7 The another benefit of the biogenic route of synthesis is that it does not require an additional step of capping or attachment of bioactive compounds to the nanoparticle surface to generate stable and pharmacological active particles which is otherwise essential in physicochemical synthesis.4,29,40 Furthermore, the time required for biosynthesis of nanoparticles is much lesser than the physicochemical methods. For example, Arsiya et al. demonstrated one-step biosynthesis of palladium nanoparticles using Chlorella vulgaris.86 The reduction of palladium ions into nanoparticles was achieved within 10 minutes at room temperature. The FTIR analysis of Chlorella vulgaris extract revealed that polyol and amide groups present in extract act as reducing and stabilizing agents. Several other investigators have discovered rapid biosynthetic procedures with high nanoparticles yield using different algal extracts. For example, silver nanoparticles were synthesized by algal extracts within 2 min,106 15 min (ref. 104) and 1 h.103 Gold nanoparticles were also formed within 5 min (ref. 106) and 10 min (ref. 95) highlighting the importance of nanoparticles synthesis using biogenic agents. In spite of various advantages offered by the biological route for the synthesis, the polydispersity and size of the nanoparticles are still big and challenging issues. Further, much work is needed to improve the efficiency of synthesis, the control of particle size and morphology. Thus, several current reports have developed a stable system for nanoparticles biosynthesis with monodispersity in size and shape. Size and shape of metal nanoparticles could be controlled by either optimizing the process parameters or modifying these parameters (Fig. 5B). For example, Hamedi et al. demonstrated synthesis of highly monodispersed silver nanoparticles using F. oxysporum by altering the process conditions such as incubation time, temperature, metal salt concentration and C : N ratio.66 The increase in C : N ratio resulted in synthesis of small size AgNPs with high monodispersity and productivity. In 2018, Domany et al. synthesized stable gold nanoparticles with moderate dispersity using Pleurotus ostreatus extracellular filtrate.54 The AuNPs synthesis rate was found to increase with the increase in HAuCl4 salt concentration, incubation time and agitation, whereas pH and temperature showed negative relation with AuNPs synthesis rate which indicates higher productivity at lower values.
In case of microbes, modification in pH leads to alteration in the overall charge of bioactive molecules, which in turn facilitates their binding affinity and hence biomineralization of metal ions into nanoparticles. For example, Yumei et al. showcased that Arthrobacter sp. promotes synthesis of silver nanoparticles at pH 7.0 and 8.0, whereas no AgNPs synthesis was recorded below pH 5.0 due to strong electrostatic repulsion between silver ion and EPS in acidic conditions.29 Even at higher pH (above 8), no AgNPs formation was observed due to high electronegativity under alkaline conditions, which is not favourable for reduction of Ag+ ion due to the presence of –COO− group. Furthermore, process parameters could also affect the shape and yield of nanoparticles. Ramanathan et al. demonstrated that spherical shaped silver nanoparticles (AgNPs) of small size were synthesized at optimum growth temperature of 20 °C using Morganella psychrotolerans,28 whereas silver nanoplates of large size were observed at growth temperature of 4 °C. Although there are few reports on process parameter optimization, it is clear from the results that optimizing these variables can solve the issue of polydispersity and production yield of nanoparticles. This demands further investigation on bio-mediated synthesis of nanoparticles for their production with high efficiency.
For efficient bio-synthesis of metallic nanoparticles, a number of controlling factors are involved in the nucleation and subsequent formation of stabilized nanoparticles. These factors include pH, reactant concentrations, reaction time, and temperature (as discussed earlier). Apart from optimizing these parameters, the use of biofilms is another prospective approach for efficient biosynthesis of nanoparticles.119 Biofilms have recently been recognized as the most active growth mode of bacteria.157 Biofilms exhibit various interesting properties such as catalyzing activity, highly reducing matrix and ability to control electrochemical reactions which provide a favourable environment for easy and efficient synthesis of nanoparticles than planktonic cells at forming nanoparticles.158 Moreover, the protective nature of biofilms with diffusion limitation for outside materials, keeps the entire synthesis process free from contamination; overall, this makes it a promising approach for biosynthesis of nanoparticles in aqueous systems.159 Nanoparticles synthesis in biofilms offers additional advantages, such as high biomass concentrations and large surface areas, which can lead to more efficient and scalable biosynthesis. Biofilms have up to 600 times higher metal resistance properties than their planktonic counterparts160 and can catalyze electrochemical redox reactions by providing an appropriate environment with natural reducing agents such as proteins, peptides, and heterocyclic compounds for metal reduction to nanoparticles.159,161 However, there has been very limited work on nanoparticles synthesis in biofilms and little is known about the stabilizing mechanism of nanoparticles in biofilms. Therefore, a thorough understanding of the molecular mechanism of nanoparticles synthesis in biofilms as well as their planktonic counterparts would help future researchers to develop more robust microbial systems for rapid and optimized biosynthesis of nanoparticles with desired sizes and shapes. Based on the mechanisms of metal reduction in bacterial biofilms, genetic modification of bacterial strains can be designed to obtain controlled sizes and shapes of nanoparticles and optimize production with high yield.162
Furthermore, the conversion of metal into nanoparticles also brings toxicity issues. Several reports have mentioned adverse effects of these nanomaterials on the biological systems and cellular components. Cytotoxicity of nanoparticles depends on various factors such as their size, shape, capping agent, density of nanoparticles, and the type of pathogens against which their toxicity is evaluated.163 Nanoparticles synthesized from non-biological route are generally more toxic than those synthesized from the biological route. Some pathogens are more prone to nanoparticles, specially AgNPs than others due to the presence of both the Ag+ ions and NPs. They slowly envelope the microbial cell and enter inside it inhibiting their essential metabolic functions. Nanoparticles are comparatively more toxic than bulk materials. They are toxic at cellular, subcellular and molecular levels.164 There are several reasons for the cytotoxicity of nanoparticles such as physicochemical properties, contamination with toxic element, small size, high surface charge and free radical species generation. Oxidative stress and lipid peroxidation have been observed in fish brain tissue on exposure to nanoparticles.165 The cytotoxicity by nanoparticles is thought to be generated through reactive oxygen species (ROS) as a result of which a decrease in glutathione levels and an increase in free radicals occur. Nanoparticles have large surface area which provide a better contact with microbes. Therefore, these nanoparticles are able to penetrate the cell membrane or attach to the cell surface based on their particle size.166 Moreover, they were observed to be highly toxic to the bacterial strains and their antibacterial efficacy is increased with the decrease in particle size. Carlson et al. have demonstrated an increase in ROS generation for 15 nm hydrocarbon coated AgNPs as compared to 55 nm.167 It has been observed by Liu et al. that 5 nm AgNPs were more toxic than 20 and 50 nm nanoparticles to four cell lines, namely, A549, HePG2, MCF-7 and SGC-7901.168 Till date, there have been extensive research on nanoparticles toxicity in order to explain their mechanism of action, and three different mechanisms have been devised so far which include cell wall and membrane damage, intracellular penetration and molecular damage, and oxidative stress. These mode of action have been discussed in detail in next section.
The toxicity concern of nanoparticles can be suitably reduced by coating these nanoparticles with biocompatible agents. Although the main role of coating/capping is to stabilize that nanoparticles and prevent agglomeration, but the biocompatible nature also makes it suitable for various biomedical applications.163 In green synthesis, stabilization of the nanoparticles is achieved by the biocompatible material only, and hence the toxicity issue is reduced in most of the cases. In 2012, one report suggested that the stabilization of AgNPs by different polymer surfactants reduces the toxicity of AgNPs against mouse skin fibroblasts (L929), human hepatocarcinoma cells (HepG2), and mouse monocyte macrophages (J774A1).169 Polymer capped AgNPs at a concentration of 1.5 ppm showed hemocompatible nature. It is a well known fact that the materials with a hemolysis ratio less than 5% are generally regarded as hemocompatible and safe.163,169 Biosynthesis using polymer i.e. glucan resulted in a glucan capped spherical AgNPs of size 2.44 nm.170 These nanoparticles showed only 0.68% hemolysis to human red blood cells (RBCs) at its LD50 dosage. Thus, AgNPs–glucan conjugates were observed to be biocompatible with human RBCs at their LD50 dosage. In a similar study, polysaccharide capped AgNPs of size 2.78 nm were synthesized using a hetero-polysaccharide isolated from Lentinus squarrosulus (Mont.).171 These nanoparticles also showed compatible nature with human RBCs at its LD50 dosage. Thus, the biocompatibility obtained through green synthesis route suggests that it is possible to use nanoparticles in varying field of biomedical application.
6. Mechanism of antimicrobial action
The interaction of nanoparticles with microorganisms starts with adhesion of nanoparticles on the microbial cell wall and membrane, and it is based on electrostatic attraction between the negatively charged microbial cell membrane and positively or less negatively charged nanoparticles.172 The interaction of nanoparticles with bacterial cells is also demonstrated by a significant drop in zeta potential of cell surface in both Gram-positive and Gram-negative bacteria.173,174 After adhesion, morphological changes in the membrane structure are induced by the nanoparticles, and thereby resulting in disruption of membrane permeability and respiratory functions through membrane depolarization, and ultimately disruption of the cell structure and cell death.175 As a result of the disruption of the cell structure, the cellular components including enzymes, proteins, DNA, metabolites start leaking into the environment.175–177 Therefore, this degeneration of the cell wall by the nanoparticle attachment is believed to be first mechanism of the antimicrobial action.178 Moreover, nanoparticles have been reported to cause irregular pit formations on the cell wall, which further help the nanoparticles to enter into the periplasmic space and finally inside the cell.179 After the interaction of nanoparticles with bacterial cells, peripheral damages and dense cavities on the cell surface can be observed by advanced imaging techniques such as transmission electron microscopy (TEM), scanning electron microscopy (SEM) and atomic force microscopy (AFM).180 Gahlawat et al. investigated the morphological changes in bacterial cell caused by AgNPs by scanning electron microscopy (SEM).4 The untreated bacterial cells demonstrated smooth morphology and original structures, whereas the cells treated with AgNP for 5 h showed pores and cavities on the cell membrane and lost their membrane integrity and the original morphology. They also reported the cell lysis and leakage of intracellular contents from the AgNP treated bacterial cells using SEM.4 In addition, it is thought that nanoparticles can also interact with proteins on the outer surface, form complexes with oxygen, phosphorous, nitrogen or sulphur atom containing electron donors, and cause irreversible damages in the cell wall.181 The interaction of protein thiol groups with silver ions and AgNPs is well-understood, and thiol groups in the enzymes of bacterial respiratory chain have been found to be possible sites for Ag+ ion binding.182 In another study, it was elucidated that the interaction of nanoparticles with sulphur containing membrane-bound proteins and enzymes led to inactivation of these molecules.183 McQuillan and Shaw (2014) proposed another possible mechanism in which Ag+ released from AgNPs may enter inside the cell by cation selective membrane transport proteins called porins, and cause damage to cellular machine.184
After disintegrating cell membrane and cell wall, nanoparticles can penetrate inside the cell and affect crucial functions of the cell by interacting with DNA and proteins.185 One of the proposed mechanism for antimicrobial activity of AgNPs is based on silver ion release from the nanoparticles, which has an adverse effect on both DNA and proteins.186 Feng et al. reported that silver ions led to deformation of the bacterial DNA from the natural relaxed state to a condensed state in which the DNA loses its replication ability.187 Moreover, Energy Dispersive X-ray Analysis demonstrated the sulphur existence which indicates that silver ions interacted with thiol groups of proteins and resulted in inactivation of enzymatic activity. Besides this conformational change in DNA, nanoparticles can also cause degradation and/or denaturation of DNA. Toxicity by nanoparticles is generally induced by the formation of free radicals, such as ROS.188 If the ROS is produced it may cause membrane disruption and disturb the permeability. The ROS include superoxide (O2−), hydroxyl (˙OH), peroxy (RCOO˙), hypochlorous acid (HOCl), and hydrogen peroxide (H2O2). These free radicals damage the cell wall and biomolecules such as proteins, enzymes, lipids and DNA. DNA damages include deletions, additions, mutations, single-breaks, double-strand breaks, and cross-linking with proteins. In this regard, Das et al. evaluated the ROS-mediated antibacterial activity of AgNPs against multidrug resistant E. coli and S. aureus, and found that ROS generation significantly contributed to the antibacterial action.189 High ROS levels caused the membrane damage by increasing the permeability, which ultimately resulted in disruption of electron transport chain and leakage of the intracellular content. It is also possible that ROS generation mediated by Ag+ ions released from AgNPs can cause dysfunction of the bacterial electron transport chain and proton motive force as a result of inhibition of enzymes involved in the reactions.190 In addition to disruption of membrane functions, Soo-Hwan et al. found that AgNP-mediated ROS generation caused protein leakage by increasing the membrane permeability.191 Protein leakage from the cells treated with AgNPs finally resulted in cell death.
7. Future perspectives
In recent years, metal nanoparticles have been studied widely for various biomedical, bioremediation and biosensor applications because of their remarkable antibacterial, antioxidant and optical properties, large surface area to volume ratio and higher efficacy. The synthesis of metal nanoparticles by biological mode has evolved as an important branch of nanobiotechnology, and bio-agents serves as potential nanofactories for the production of nanomaterials. However, there are certain gaps and limitations in successful production of these nanoparticles which need to be sorted out by the scientific community. One of the major limitation in biomediated synthesis is complete and thorough understanding of mechanistic aspects of biofabrication of nanoparticles. Although there are reports in literature on identification and isolation of bioactive moiety responsible for biomineralization of metal ions using biological extracts, much detailed analysis of biochemical pathway is further needed for the development of tailor-made nanoparticles. Especially for biomedical purposes, it is indispensable to understand how active moieties from various biological resources bind to the nanoparticle surface to provide stability, and to synthesize nanoparticles with higher biocompatibility. Large-scale production is another major bottleneck in the development and commercialization of biocompatible nanostructures with controlled sizes and shapes. Recently, researchers have focussed on large-scale cultivation methods for nanoparticles synthesis which are scalable and reproducible with narrow size distribution. But these bulk cultivation methods for bio-nanomaterials and downstream processing techniques need to be improved further. Large scale cultivation of nanoparticles is generally hampered by the factors of high cost, high energy requirement, polydispersity and low nanoparticles yield. The production of nanoparticles at room temperatures using natural active biomolecules without any reducing agents would make large-scale fermentation more cost-effective and energy sustainable. Stable production of monodispersed nanoparticles with high yield could be achieved by optimizing various process parameters (pH, temperature, contact time, mixing ratio, salt concentration) and altering the overall charge on functional molecules. Apart from this, issues related to the biomedical applications viz. the distribution profile, release kinetics and clearance of nanostructures in vivo need to be sorted out. In-depth evaluation of the biocompatibility and bioavailability of nanomaterials is still in stage of infancy and considerable research efforts are needed in this area. The collaborative research on fermentation process development along with understanding of mechanistic aspects, scale-up and exploration of other biological agents could expedite the process of cost-effective tailor-made synthesis of nanomaterials.
Conflicts of interest
Authors declare that they have no conflicts of interest.
Supplementary Material
Acknowledgments
All authors duly acknowledge the Council of Scientific and Industrial Research – Institute of Microbial Technology (CSIR-IMTECH) for providing all necessary facilities. One of the authors (Dr. Geeta Gahlawat) acknowledges the University Grants Commission, Government of India for providing Dr. D. S. Kothari fellowship for her research work.
References
- Thakkar K. N. Mhatre S. S. Parikh R. Y. Nanomedicine. 2010;6:257–262. doi: 10.1016/j.nano.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Narayanan K. B. Sakthivel N. Adv. Colloid Interface Sci. 2010;156:1–13. doi: 10.1016/j.cis.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Albanese A. Tang P. S. Chan W. C. Annu. Rev. Biomed. Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- Gahlawat G. Shikha S. Chaddha B. S. Chaudhuri S. R. Mayilraj S. Choudhury A. R. Microb. Cell Fact. 2016;15:25. doi: 10.1186/s12934-016-0422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda C. Chen X. Narayanan R. El-Sayed M. A. Chem. Rev. 2005;105:1025–1102. doi: 10.1021/cr030063a. [DOI] [PubMed] [Google Scholar]
- Daniel M.-C. Astruc D. Chem. Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- Ahmed S. Ahmad M. Swami B. L. Ikram S. J. Adv. Res. 2016;7:17–28. doi: 10.1016/j.jare.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedbalkar U. Singh R. Wadhwani S. Gaidhani S. Chopade B. Adv. Colloid Interface Sci. 2014;209:40–48. doi: 10.1016/j.cis.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Kharisov B. I., Kharissova O. V. and Ortiz-Mendez U., CRC concise encyclopedia of nanotechnology, CRC Press, 2016 [Google Scholar]
- Nath D. Banerjee P. Environ. Toxicol. Pharmacol. 2013;36:997–1014. doi: 10.1016/j.etap.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Shah M. Fawcett D. Sharma S. Tripathy S. Poinern G. Materials. 2015;8:7278–7308. doi: 10.3390/ma8115377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You H. Yang S. Ding B. Yang H. Chem. Soc. Rev. 2013;42:2880–2904. doi: 10.1039/C2CS35319A. [DOI] [PubMed] [Google Scholar]
- Kulkarni N. Muddapur U. J. Nanotechnol. 2014;2014:1–8. doi: 10.1155/2014/510246. [DOI] [Google Scholar]
- Singh P. Kim Y.-J. Zhang D. Yang D.-C. Trends Biotechnol. 2016;34:588–599. doi: 10.1016/j.tibtech.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Dhuper S. Panda D. Nayak P. Nano Trends: J Nanotech. App. 2012;13:16–22. [Google Scholar]
- Faramarzi M. A. Sadighi A. Adv. Colloid Interface Sci. 2013;189:1–20. doi: 10.1016/j.cis.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Iravani S. Int. Sch. Res. Notices. 2014;2014:1–18. doi: 10.1155/2014/359316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge T. Murray R. J. Bacteriol. 1980;141:876–887. doi: 10.1128/jb.141.2.876-887.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus-Joerger T. Joerger R. Olsson E. Granqvist C.-G. Trends Biotechnol. 2001;19:15–20. doi: 10.1016/s0167-7799(00)01514-6. [DOI] [PubMed] [Google Scholar]
- Srivastava S. K. Constanti M. J. Nanopart. Res. 2012;14:831. doi: 10.1007/s11051-012-0831-7. [DOI] [Google Scholar]
- Das V. L. Thomas R. Varghese R. T. Soniya E. Mathew J. Radhakrishnan E. 3 Biotech. 2014;4:121–126. doi: 10.1007/s13205-013-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni R. R. Shaiwale N. S. Deobagkar D. N. Deobagkar D. D. Int. J. Nanomed. 2015;10:963. doi: 10.2147/IJN.S72888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed E. Kalathil S. Shi L. Alharbi O. Wang P. J. Saudi Chem. Soc. 2018;22:919–929. [Google Scholar]
- Zonaro E. Piacenza E. Presentato A. Monti F. Dell'Anna R. Lampis S. Vallini G. Microb. Cell Fact. 2017;16:215. doi: 10.1186/s12934-017-0826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinath B. Namratha K. Byrappa K. Adv. Sci. Lett. 2018;24:5942–5946. doi: 10.1166/asl.2018.12224. [DOI] [Google Scholar]
- Saravanan M. Barik S. K. Mubarak Ali D. Prakash P. Pugazhendhi A. Microb. Pathog. 2018;116:221–226. doi: 10.1016/j.micpath.2018.01.038. [DOI] [PubMed] [Google Scholar]
- Hulkoti N. I. Taranath T. Colloids Surf., B. 2014;121:474–483. doi: 10.1016/j.colsurfb.2014.05.027. [DOI] [PubMed] [Google Scholar]
- Ramanathan R. O'Mullane A. P. Parikh R. Y. Smooker P. M. Bhargava S. K. Bansal V. Langmuir. 2010;27:714–719. doi: 10.1021/la1036162. [DOI] [PubMed] [Google Scholar]
- Yumei L. Yamei L. Qiang L. Jie B. J. Nanomater. 2017;2017:1–8. doi: 10.1155/2017/9703614. [DOI] [Google Scholar]
- Malhotra A. Dolma K. Kaur N. Rathore Y. S. Mayilraj S. Choudhury A. R. Bioresour. Technol. 2013;142:727–731. doi: 10.1016/j.biortech.2013.05.109. [DOI] [PubMed] [Google Scholar]
- Kaur H. Dolma K. Kaur N. Malhotra A. Kumar N. Dixit P. Sharma D. Mayilraj S. Choudhury A. R. Biotechnol. Bioprocess Eng. 2015;20:51–57. [Google Scholar]
- Mehta A. Sidhu C. Pinnaka A. K. Choudhury A. R. PLoS One. 2014;9:e98798. doi: 10.1371/journal.pone.0098798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj R. Dalei K. Chakraborty J. Das S. J. Colloid Interface Sci. 2016;462:166–175. doi: 10.1016/j.jcis.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Sunkar S. Nachiyar C. V. Asian Pac. J. Trop. Biomed. 2012;2:953–959. doi: 10.1016/S2221-1691(13)60006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divya M. George S. K. Saqib H. Joseph S. Biocatal. Agric. Biotechnol. 2019;18:101037. doi: 10.1016/j.bcab.2019.101037. [DOI] [Google Scholar]
- Müller A. Behsnilian D. Walz E. Gräf V. Hogekamp L. Greiner R. Biocatal. Agric. Biotechnol. 2016;6:107–115. doi: 10.1016/j.bcab.2016.02.012. [DOI] [Google Scholar]
- Naik M. M. Prabhu M. S. Samant S. N. Naik P. M. Shirodkar S. Thalassas: an International Journal of Marine Sciences. 2017;33:73–80. doi: 10.1007/s41208-017-0023-4. [DOI] [Google Scholar]
- Manimaran M. Kannabiran K. Lett. Appl. Microbiol. 2017;64:401–408. doi: 10.1111/lam.12730. [DOI] [PubMed] [Google Scholar]
- Otari S. Patil R. Nadaf N. Ghosh S. Pawar S. Mater. Lett. 2012;72:92–94. doi: 10.1016/j.matlet.2011.12.109. [DOI] [Google Scholar]
- Karthik L. Kumar G. Kirthi A. V. Rahuman A. Rao K. B. Bioprocess Biosyst. Eng. 2014;37:261–267. doi: 10.1007/s00449-013-0994-3. [DOI] [PubMed] [Google Scholar]
- Buszewski B. Railean-Plugaru V. Pomastowski P. Rafińska K. Szultka-Mlynska M. Golinska P. Wypij M. Laskowski D. Dahm H. J. Microbiol., Immunol. Infect. 2016;20:1–10. doi: 10.1016/j.jmii.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Abd-Elnaby H. M. Abo-Elala G. M. Abdel-Raouf U. M. Hamed M. M. Egypt. J. Aquat. Res. 2016;42:301–312. doi: 10.1016/j.ejar.2016.05.004. [DOI] [Google Scholar]
- Ranjitha V. Rai V. R. 3 Biotech. 2017;7:299. doi: 10.1007/s13205-017-0930-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad E. Salem S. S. Fouda A. Awad M. A. El-Gamal M. S. Abdo A. M. J. Radiat. Res. Appl. Sci. 2018;30:1–9. [Google Scholar]
- Otari S. Patil R. Nadaf N. Ghosh S. Pawar S. Environ. Sci. Pollut. Res. 2014;21:1503–1513. doi: 10.1007/s11356-013-1764-0. [DOI] [PubMed] [Google Scholar]
- Mara Silva-Vinhote N. Caballero N. E. D. de Amorim Silva T. Quelemes P. V. de Araujo A. R. de Moraes A. C. M. dos Santos Câmara A. L. Longo J. P. F. Azevedo R. B. da Silva D. A. Afr. J. Biotechnol. 2017;16:2072–2082. doi: 10.5897/AJB2017.16148. [DOI] [Google Scholar]
- Wypij M. Czarnecka J. Świecimska M. Dahm H. Rai M. Golinska P. World J. Microbiol. Biotechnol. 2018;34:23. doi: 10.1007/s11274-017-2406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A. Jain N. Khan M. A. Pareek V. Dilip R. V. Panwar J. J. Environ. Manage. 2016;183:22–32. doi: 10.1016/j.jenvman.2016.08.021. [DOI] [PubMed] [Google Scholar]
- Mishra A. Kumari M. Pandey S. Chaudhry V. Gupta K. Nautiyal C. Bioresour. Technol. 2014;166:235–242. doi: 10.1016/j.biortech.2014.04.085. [DOI] [PubMed] [Google Scholar]
- Metuku R. P. Pabba S. Burra S. Gudikandula K. Charya M. S. 3 Biotech. 2014;4:227–234. doi: 10.1007/s13205-013-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput S. Werezuk R. Lange R. M. McDermott M. T. Langmuir. 2016;32:8688–8697. doi: 10.1021/acs.langmuir.6b01813. [DOI] [PubMed] [Google Scholar]
- Kitching M. Choudhary P. Inguva S. Guo Y. Ramani M. Das S. K. Marsili E. Enzyme Microb. Technol. 2016;95:76–84. doi: 10.1016/j.enzmictec.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Suryavanshi P. Pandit R. Gade A. Derita M. Zachino S. Rai M. LWT–Food Sci. Technol. 2017;81:188–194. doi: 10.1016/j.lwt.2017.03.038. [DOI] [Google Scholar]
- El Domany E. B. Essam T. M. Ahmed A. E. Farghali A. A. J. Appl. Pharm. Sci. 2018;8:119–128. doi: 10.7324/JAPS.2018.8918. [DOI] [Google Scholar]
- Hamad M. Int. J. Environ. Sci. Technol. 2018:1–10. [Google Scholar]
- Ganachari S. V. Bhat R. Deshpande R. Venkataraman A. BioNanoScience. 2012;2:316–321. doi: 10.1007/s12668-012-0046-5. [DOI] [Google Scholar]
- Roy S. Mukherjee T. Chakraborty S. Das T. K. Dig. J. Nanomater. Biostruct. 2013;8:197–205. [Google Scholar]
- Thakker J. N. Dalwadi P. Dhandhukia P. C. ISRN Biotechnol. 2012:2013. doi: 10.5402/2013/515091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadwal A. S. Tripathi R. Gupta R. K. Kumar N. Singh R. Shrivastav A. RSC Adv. 2014;4:9484–9490. doi: 10.1039/C3RA46221H. [DOI] [Google Scholar]
- Castro M. Cottet L. Castillo A. Mater. Lett. 2014;115:42–44. doi: 10.1016/j.matlet.2013.10.020. [DOI] [Google Scholar]
- Kar P. K. Murmu S. Saha S. Tandon V. Acharya K. PLoS One. 2014;9:e84693. doi: 10.1371/journal.pone.0084693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskar G. Chandhuru J. Fahad K. S. Praveen A. Chamundeeswari M. Muthukumar T. J. Mater. Sci.: Mater. Med. 2015;26:43. doi: 10.1007/s10856-015-5380-z. [DOI] [PubMed] [Google Scholar]
- Ramalingmam P. Muthukrishnan S. Thangaraj P. J. Nanosci. Nanoeng. 2015;1:241–247. [Google Scholar]
- Amerasan D. Nataraj T. Murugan K. Panneerselvam C. Madhiyazhagan P. Nicoletti M. Benelli G. J. Pest Sci. 2016;89:249–256. doi: 10.1007/s10340-015-0675-x. [DOI] [Google Scholar]
- Guilger M. Pasquoto-Stigliani T. Bilesky-Jose N. Grillo R. Abhilash P. Fraceto L. F. De Lima R. Sci. Rep. 2017;7:44421. doi: 10.1038/srep44421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamedi S. Ghaseminezhad M. Shokrollahzadeh S. Shojaosadati S. A. Artif. Cells, Nanomed., Biotechnol. 2017;45:1588–1596. doi: 10.1080/21691401.2016.1267011. [DOI] [PubMed] [Google Scholar]
- Apte M. Sambre D. Gaikawad S. Joshi S. Bankar A. Kumar A. R. Zinjarde S. AMB Express. 2013;3:32. doi: 10.1186/2191-0855-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare S. R. Mulla M. N. Marathe S. R. Sonawane K. D. 3 Biotech. 2015;5:33–38. doi: 10.1007/s13205-014-0196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahian F. Reiisi S. Shahidi A. Mirzaei S. A. Nanomedicine. 2017;13:853–861. doi: 10.1016/j.nano.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Eugenio M. Müller N. Frasés S. Almeida-Paes R. Lima L. M. T. Lemgruber L. Farina M. de Souza W. Sant'Anna C. RSC Adv. 2016;6:9893–9904. doi: 10.1039/C5RA22727E. [DOI] [Google Scholar]
- Sriramulu M. Sumathi S. Adv. Nat. Sci.: Nanosci. Nanotechnol. 2018;9:025018. [Google Scholar]
- Seshadri S. Saranya K. Kowshik M. Biotechnol. Prog. 2011;27:1464–1469. doi: 10.1002/btpr.651. [DOI] [PubMed] [Google Scholar]
- Ortega F. G. Fernández-Baldo M. A. Fernández J. G. Serrano M. J. Sanz M. I. Díaz-Mochón J. J. Lorente J. A. Raba J. Int. J. Nanomed. 2015;10:2021. doi: 10.2147/IJN.S75835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbekandi H. Mohseni S. Mardani Jouneghani R. Pourhossein M. Iravani S. Artif. Cells, Nanomed., Biotechnol. 2016;44:235–239. doi: 10.3109/21691401.2014.937870. [DOI] [PubMed] [Google Scholar]
- Zhang X. Qu Y. Shen W. Wang J. Li H. Zhang Z. Li S. Zhou J. Colloids Surf., A. 2016;497:280–285. doi: 10.1016/j.colsurfa.2016.02.033. [DOI] [Google Scholar]
- Fernández J. G. Fernández-Baldo M. A. Berni E. Camí G. Durán N. Raba J. Sanz M. I. Process Biochem. 2016;51:1306–1313. doi: 10.1016/j.procbio.2016.05.021. [DOI] [Google Scholar]
- Yang Z. Li Z. Lu X. He F. Zhu X. Ma Y. He R. Gao F. Ni W. Yi Y. Nano-Micro Lett. 2017;9:5. doi: 10.1007/s40820-016-0102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla J. J. A. Guerrero D. J. P. Sáez R. G. T. Ishida K. Fonseca B. B. Rozental S. López C. C. O. J. Nanosci. Nanotechnol. 2017;17:1729–1739. doi: 10.1166/jnn.2017.12822. [DOI] [Google Scholar]
- Moghaddam A. B. Moniri M. Azizi S. Rahim R. A. Ariff A. B. Saad W. Z. Namvar F. Navaderi M. Mohamad R. Molecules. 2017;22:872. doi: 10.3390/molecules22060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvadori M. R. Ando R. A. Nascimento C. A. O. Corrêa B. J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng. 2017;52:1112–1120. doi: 10.1080/10934529.2017.1340754. [DOI] [PubMed] [Google Scholar]
- Rónavári A. Igaz N. Gopisetty M. K. Szerencsés B. Kovács D. Papp C. Vágvölgyi C. Boros I. M. Kónya Z. Kiricsi M. Int. J. Nanomed. 2018;13:695. doi: 10.2147/IJN.S152010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y. You S. Zhang X. Pei X. Shen W. Li Z. Li S. Zhang Z. Bioprocess Biosyst. Eng. 2018;41:359–367. doi: 10.1007/s00449-017-1869-9. [DOI] [PubMed] [Google Scholar]
- Cunha F. A. da CSO Cunha M. da Frota S. M. Mallmann E. J. Freire T. M. Costa L. S. Paula A. J. Menezes E. A. Fechine P. B. World J. Microbiol. Biotechnol. 2018;34:127. doi: 10.1007/s11274-018-2514-8. [DOI] [PubMed] [Google Scholar]
- Jalal M. Ansari M. Alzohairy M. Ali S. Khan H. Almatroudi A. Raees K. Nanomaterials. 2018;8:586. doi: 10.3390/nano8080586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Ferreira V. ConzFerreira M. E. Lima L. M. T. Frasés S. de Souza W. Sant'Anna C. Enzyme Microb. Technol. 2017;97:114–121. doi: 10.1016/j.enzmictec.2016.10.018. [DOI] [PubMed] [Google Scholar]
- Arsiya F. Sayadi M. H. Sobhani S. Mater. Lett. 2017;186:113–115. doi: 10.1016/j.matlet.2016.09.101. [DOI] [Google Scholar]
- Momeni S. Nabipour I. Appl. Biochem. Biotechnol. 2015;176:1937–1949. doi: 10.1007/s12010-015-1690-3. [DOI] [PubMed] [Google Scholar]
- Dhas T. S. Kumar V. G. Karthick V. Angel K. J. Govindaraju K. Spectrochim. Acta, Part A. 2014;120:416–420. doi: 10.1016/j.saa.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Edison T. N. J. I. Atchudan R. Kamal C. Lee Y. R. Bioprocess Biosyst. Eng. 2016;39:1401–1408. doi: 10.1007/s00449-016-1616-7. [DOI] [PubMed] [Google Scholar]
- Ramakrishna M. Babu D. R. Gengan R. M. Chandra S. Rao G. N. J. Nanostruct. Chem. 2016;6:1–13. doi: 10.1007/s40097-015-0173-y. [DOI] [Google Scholar]
- Rajeshkumar S. Res. J. Biotechnol. 2018;13:15–19. [Google Scholar]
- Sanaeimehr Z. Javadi I. Namvar F. Cancer Nanotechnol. 2018;9:3. doi: 10.1186/s12645-018-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D. Verduin J. J. Shah M. Sharma S. B. Poinern G. E. J. J. Nanosci. 2017;2017:1–15. [Google Scholar]
- Venkatpurwar V. Pokharkar V. Mater. Lett. 2011;65:999–1002. doi: 10.1016/j.matlet.2010.12.057. [DOI] [Google Scholar]
- Rajathi F. A. A. Parthiban C. Kumar V. G. Anantharaman P. Spectrochim. Acta, Part A. 2012;99:166–173. doi: 10.1016/j.saa.2012.08.081. [DOI] [PubMed] [Google Scholar]
- Senapati S. Syed A. Moeez S. Kumar A. Ahmad A. Mater. Lett. 2012;79:116–118. doi: 10.1016/j.matlet.2012.04.009. [DOI] [Google Scholar]
- Kannan R. R. R. Arumugam R. Ramya D. Manivannan K. Anantharaman P. Appl. Nanosci. 2013;3:229–233. doi: 10.1007/s13204-012-0125-5. [DOI] [Google Scholar]
- Salari Z. Danafar F. Dabaghi S. Ataei S. A. J. Saudi Chem. Soc. 2016;20:459–464. doi: 10.1016/j.jscs.2014.10.004. [DOI] [Google Scholar]
- Jena J. Pradhan N. Nayak R. R. Dash B. P. Sukla L. B. Panda P. K. Mishra B. K. J. Microbiol. Biotechnol. 2014;24:522–533. doi: 10.4014/jmb.1306.06014. [DOI] [PubMed] [Google Scholar]
- Annamalai J. Nallamuthu T. Appl. Nanosci. 2015;5:603–607. doi: 10.1007/s13204-014-0353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan J. Manivasagan P. Kim S.-K. Kirthi A. V. Marimuthu S. Rahuman A. A. Bioprocess Biosyst. Eng. 2014;37:1591–1597. doi: 10.1007/s00449-014-1131-7. [DOI] [PubMed] [Google Scholar]
- Kathiraven T. Sundaramanickam A. Shanmugam N. Balasubramanian T. Appl. Nanosci. 2015;5:499–504. doi: 10.1007/s13204-014-0341-2. [DOI] [Google Scholar]
- Murugan K. Samidoss C. M. Panneerselvam C. Higuchi A. Roni M. Suresh U. Chandramohan B. Subramaniam J. Madhiyazhagan P. Dinesh D. Parasitol. Res. 2015;114:4087–4097. doi: 10.1007/s00436-015-4638-1. [DOI] [PubMed] [Google Scholar]
- Sinha S. N. Paul D. Halder N. Sengupta D. Patra S. K. Appl. Nanosci. 2015;5:703–709. doi: 10.1007/s13204-014-0366-6. [DOI] [Google Scholar]
- González-Ballesteros N. Prado-López S. Rodriguez-Gonzalez J. Lastra M. Rodríguez-Argüelles M. Colloids Surf., B. 2017;153:190–198. doi: 10.1016/j.colsurfb.2017.02.020. [DOI] [PubMed] [Google Scholar]
- Abdel-Raouf N. Al-Enazi N. M. Ibraheem I. B. Arabian J. Chem. 2017;10:S3029–S3039. doi: 10.1016/j.arabjc.2013.11.044. [DOI] [Google Scholar]
- Kim D.-Y. Saratale R. G. Shinde S. Syed A. Ameen F. Ghodake G. J. Cleaner Prod. 2018;172:2910–2918. doi: 10.1016/j.jclepro.2017.11.123. [DOI] [Google Scholar]
- Pugazhendhi A. Prabakar D. Jacob J. M. Karuppusamy I. Saratale R. G. Microb. Pathog. 2018;114:41–45. doi: 10.1016/j.micpath.2017.11.013. [DOI] [PubMed] [Google Scholar]
- Kobayashi M. Tomita S. Sawada K. Shiba K. Yanagi H. Yamashita I. Uraoka Y. Opt. Express. 2012;20:24856–24863. doi: 10.1364/OE.20.024856. [DOI] [PubMed] [Google Scholar]
- Zeng Q. Wen H. Wen Q. Chen X. Wang Y. Xuan W. Liang J. Wan S. Biomaterials. 2013;34:4632–4642. doi: 10.1016/j.biomaterials.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Mao C. Flynn C. E. Hayhurst A. Sweeney R. Qi J. Georgiou G. Iverson B. Belcher A. M. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6946–6951. doi: 10.1073/pnas.0832310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A. J., Talianski M. E., Chapman S. N. and Shaw J., US Pat., No. 9,688,964, U.S. Patent and Trademark Office, Washington, DC, 2017 [Google Scholar]
- Cao J. Guenther R. H. Sit T. L. Opperman C. H. Lommel S. A. Willoughby J. A. Small. 2014;10:5126–5136. doi: 10.1002/smll.201400558. [DOI] [PubMed] [Google Scholar]
- Le D. H. Lee K. L. Shukla S. Commandeur U. Steinmetz N. F. Nanoscale. 2017;9:2348–2357. doi: 10.1039/C6NR09099K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F. Li Y. Liu T. Xu K. Zhang L. Xu C. Gao J. Chem. Eng. J. 2013;226:52–58. doi: 10.1016/j.cej.2013.04.036. [DOI] [Google Scholar]
- Chen P.-Y. Dang X. Klug M. T. Courchesne N.-M. D. Qi J. Hyder M. N. Belcher A. M. Hammond P. T. Chem. Mater. 2015;27:1531–1540. doi: 10.1021/cm503803u. [DOI] [Google Scholar]
- Esfandiari N. Arzanani M. K. Soleimani M. Kohi-Habibi M. Svendsen W. E. Tumor Biol. 2016;37:1229–1236. doi: 10.1007/s13277-015-3867-3. [DOI] [PubMed] [Google Scholar]
- Chen C. C. Stark M. Baikoghli M. Cheng R. H. J. Visualized Exp. 2018:e57020. doi: 10.3791/57020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzil A. H. Sultana S. T. Saunders S. R. Shi L. Marsili E. Beyenal H. Enzyme Microb. Technol. 2016;95:4–12. doi: 10.1016/j.enzmictec.2016.07.015. [DOI] [PubMed] [Google Scholar]
- Cologgi D. L. Lampa-Pastirk S. Speers A. M. Kelly S. D. Reguera G. Proc. Natl. Acad. Sci. U. S. A. 2011:201108616. doi: 10.1073/pnas.1108616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G. Biochem. Soc. Trans. 2012;40:1227–1232. doi: 10.1042/BST20120162. [DOI] [PubMed] [Google Scholar]
- Vasylevskyi S. I. Kracht S. Corcosa P. Fromm K. M. Giese B. Füeg M. Angew. Chem., Int. Ed. 2017;56:5926–5930. doi: 10.1002/anie.201702621. [DOI] [PubMed] [Google Scholar]
- Bewley K. D. Ellis K. E. Firer-Sherwood M. A. Elliott S. J. Biochim. Biophys. Acta, Bioenerg. 2013;1827:938–948. doi: 10.1016/j.bbabio.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L. Rosso K. M. Clarke T. A. Richardson D. J. Zachara J. M. Fredrickson J. K. Front. Microbiol. 2012;3:50. doi: 10.3389/fmicb.2012.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L. Oliveira T. F. Pereira I. A. Archer M. EMBO J. 2006;25:5951–5960. doi: 10.1038/sj.emboj.7601439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L. Scott K. A. Sansom M. S. Pereira I. A. Archer M. J. Mol. Biol. 2008;381:341–350. doi: 10.1016/j.jmb.2008.05.066. [DOI] [PubMed] [Google Scholar]
- Hartshorne R. S. Reardon C. L. Ross D. Nuester J. Clarke T. A. Gates A. J. Mills P. C. Fredrickson J. K. Zachara J. M. Shi L. Beliaev A. S. Marshall M. J. Tien M. Brantley S. Butt J. N. Richardson D. J. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22169–22174. doi: 10.1073/pnas.0900086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T. A. Holley T. Hartshorne R. S. Fredrickson J. K. Zachara J. M. Shi L. Richardson D. J. Biochem. Soc. Trans. 2008;36:1005–1010. doi: 10.1042/BST0361005. [DOI] [PubMed] [Google Scholar]
- Clarke T. A. Edwards M. J. Gates A. J. Hall A. White G. F. Bradley J. Reardon C. L. Shi L. Beliaev A. S. Marshall M. J. Wang Z. Watmough N. J. Fredrickson J. K. Zachara J. M. Butt J. N. Richardson D. J. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9384–9389. doi: 10.1073/pnas.1017200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursolle D. Gralnick J. A. Mol. Microbiol. 2010;77:995–1008. doi: 10.1111/j.1365-2958.2010.07266.x. [DOI] [PubMed] [Google Scholar]
- Ng C. K. Sivakumar K. Liu X. Madhaiyan M. Ji L. Yang L. Tang C. Song H. Kjelleberg S. Cao B. Biotechnol. Bioeng. 2013;110:1831–1837. doi: 10.1002/bit.24856. [DOI] [PubMed] [Google Scholar]
- Liu Y. Wang Z. Liu J. Levar C. Edwards M. J. Babauta J. T. Kennedy D. W. Shi Z. Beyenal H. Bond D. R. Environ. Microbiol. Rep. 2014;6:776–785. doi: 10.1111/1758-2229.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Fredrickson J. K. Zachara J. M. Shi L. Front. Microbiol. 2015;6:1075. doi: 10.3389/fmicb.2015.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F. Alvarez P. J. Zhu D. Environ. Sci. Technol. 2013;48:316–322. doi: 10.1021/es403796x. [DOI] [PubMed] [Google Scholar]
- Choudhury A. R. Malhotra A. Bhattacharjee P. Prasad G. Carbohydr. Polym. 2014;106:154–159. doi: 10.1016/j.carbpol.2014.01.072. [DOI] [PubMed] [Google Scholar]
- Freitas F. Alves V. D. Reis M. A. M. Trends Biotechnol. 2011;29:388–398. doi: 10.1016/j.tibtech.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Sutherland I. W. Trends Microbiol. 2001;9:222–227. doi: 10.1016/S0966-842X(01)02012-1. [DOI] [PubMed] [Google Scholar]
- Gutierrez T. Morris G. Green D. H. Biotechnol. Bioeng. 2009;103:207–216. doi: 10.1002/bit.22218. [DOI] [PubMed] [Google Scholar]
- Thornton D. C. O. Fejes E. M. DiMarco S. F. Clancy K. M. Limnol. Oceanogr.: Methods. 2007;5:73–87. doi: 10.4319/lom.2007.5.73. [DOI] [Google Scholar]
- Sathiyanarayanan G. Dineshkumar K. Yang Y.-H. Crit. Rev. Microbiol. 2017;43:731–752. doi: 10.1080/1040841X.2017.1306689. [DOI] [PubMed] [Google Scholar]
- Banerjee A. Halder U. Bandopadhyay R. J. Cluster Sci. 2017;28:1803–1813. doi: 10.1007/s10876-017-1219-8. [DOI] [Google Scholar]
- Emam H. E. Ahmed H. B. Carbohydr. Polym. 2016;135:300–307. doi: 10.1016/j.carbpol.2015.08.095. [DOI] [PubMed] [Google Scholar]
- Pandey S. Goswami G. K. Nanda K. K. Int. J. Biol. Macromol. 2012;51:583–589. doi: 10.1016/j.ijbiomac.2012.06.033. [DOI] [PubMed] [Google Scholar]
- Mata Y. N. Torres E. Blázquez M. L. Ballester A. González F. Muñoz J. A. J. Hazard. Mater. 2009;166:612–618. doi: 10.1016/j.jhazmat.2008.11.064. [DOI] [PubMed] [Google Scholar]
- Xu W. Jin W. Lin L. Zhang C. Li Z. Li Y. Song R. Li B. Carbohydr. Polym. 2014;101:961–967. doi: 10.1016/j.carbpol.2013.10.032. [DOI] [PubMed] [Google Scholar]
- Pooja D. Panyaram S. Kulhari H. Rachamalla S. S. Sistla R. Carbohydr. Polym. 2014;110:1–9. doi: 10.1016/j.carbpol.2014.03.041. [DOI] [PubMed] [Google Scholar]
- Davidović S. Lazić V. Vukoje I. Papan J. Anhrenkiel S. P. Dimitrijević S. Nedeljković J. M. Colloids Surf., B. 2017;160:184–191. doi: 10.1016/j.colsurfb.2017.09.031. [DOI] [PubMed] [Google Scholar]
- Medhat D. Hussein J. El-Naggar M. E. Attia M. F. Anwar M. Latif Y. A. Booles H. F. Morsy S. Farrag A. R. Khalil W. K. El-Khayat Z. Biomed. Pharmacother. 2017;91:1006–1016. doi: 10.1016/j.biopha.2017.05.043. [DOI] [PubMed] [Google Scholar]
- Shaterabadi Z. Nabiyouni G. Soleymani M. Mater. Sci. Eng., C. 2017;75:947–956. doi: 10.1016/j.msec.2017.02.143. [DOI] [PubMed] [Google Scholar]
- Yan J. K. Cai P. F. Cao X. Q. Ma H. L. Zhang Q. Hu N. Z. Zhao Y. Z. Carbohydr. Polym. 2013;97:391–397. doi: 10.1016/j.carbpol.2013.05.049. [DOI] [PubMed] [Google Scholar]
- Yan J. K. Liu J. L. Sun Y. J. Tang S. Mo Z. Y. Liu Y. S. Carbohydr. Polym. 2015;117:771–777. doi: 10.1016/j.carbpol.2014.10.048. [DOI] [PubMed] [Google Scholar]
- Moon J.-W. Ivanov I. N. Joshi P. C. Armstrong B. L. Wang W. Jung H. Rondinone A. J. Jellison Jr G. E. Meyer III H. M. Jang G. G. Acta Biomater. 2014;10:4474–4483. doi: 10.1016/j.actbio.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Ramos-Ruiz A. Sesma-Martin J. Sierra-Alvarez R. Field J. A. Water Res. 2017;108:189–196. doi: 10.1016/j.watres.2016.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.-W. Rawn C. J. Rondinone A. J. Love L. J. Roh Y. Everett S. M. Lauf R. J. Phelps T. J. J. Ind. Microbiol. Biotechnol. 2010;37:1023–1031. doi: 10.1007/s10295-010-0749-y. [DOI] [PubMed] [Google Scholar]
- Moon J.-W. Ivanov I. N. Duty C. E. Love L. J. Rondinone A. J. Wang W. Li Y.-L. Madden A. S. Mosher J. J. Hu M. Z. J. Ind. Microbiol. Biotechnol. 2013;40:1263–1271. doi: 10.1007/s10295-013-1321-3. [DOI] [PubMed] [Google Scholar]
- Moon J.-W. Phelps T. J. Fitzgerald Jr C. L. Lind R. F. Elkins J. G. Jang G. G. Joshi P. C. Kidder M. Armstrong B. L. Watkins T. R. Appl. Microbiol. Biotechnol. 2016;100:7921–7931. doi: 10.1007/s00253-016-7556-y. [DOI] [PubMed] [Google Scholar]
- Ikuma K. Decho A. W. Lau B. L. T. Front. Microbiol. 2015;6:1–6. doi: 10.3389/fmicb.2015.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalathil S. Lee J. Cho M. H. Green Chem. 2011;13:1482–1485. doi: 10.1039/C1GC15309A. [DOI] [Google Scholar]
- Khan M. M. Kalathil S. Han T. H. Lee J. Cho M. H. J. J. Nanosci. Nanotechnol. 2013;13:6079–6085. doi: 10.1166/jnn.2013.7666. [DOI] [PubMed] [Google Scholar]
- Teitzel G. M. Parsek M. R. Appl. Environ. Microbiol. 2003;69:2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. M. Ansari S. A. Lee J. H. Ansari M. O. Lee J. Cho M. H. J. Colloid Interface Sci. 2014;431:255–263. doi: 10.1016/j.jcis.2014.06.026. [DOI] [PubMed] [Google Scholar]
- Mandal D. Bolander M. E. Mukhopadhyay D. Sarkar G. Mukherjee P. Appl. Microbiol. Biotechnol. 2006;69:485–492. doi: 10.1007/s00253-005-0179-3. [DOI] [PubMed] [Google Scholar]
- Roy A. Bulut O. Some S. Mandal A. K. Yilmaz M. D. RSC Adv. 2019;9:2673–2702. doi: 10.1039/C8RA08982E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasree L. Janakiram P. Madhavi R. J. World Aquacult. Soc. 2006;37:523–532. doi: 10.1111/j.1749-7345.2006.00066.x. [DOI] [Google Scholar]
- Oberdorster E. Environ. Health Perspect. 2004;112:1058–1062. doi: 10.1289/ehp.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M. Yadav A. Gade A. Biotechnol. Adv. 2009;27:76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Carlson C. Hussain S. M. Schrand A. M. Braydich-Stolle L. K. Hess K. L. Jones R. L. Schlager J. J. J. Phys. Chem. B. 2008;112:13608–13619. doi: 10.1021/jp712087m. [DOI] [PubMed] [Google Scholar]
- Liu W. Wu Y. Wang C. Li H. C. Wang T. Liao C. Y. Cui L. Zhou Q. F. Yan B. Jiang G. B. Nanotoxicology. 2010;4:319–330. doi: 10.3109/17435390.2010.483745. [DOI] [PubMed] [Google Scholar]
- Lin J. J. Lin W. C. Dong R. X. Hsu S. Nanotechnology. 2012;23:065102. doi: 10.1088/0957-4484/23/6/065102. [DOI] [PubMed] [Google Scholar]
- Sen I. K. Mandal A. K. Chakraborti S. Dey B. Chakraborty R. Islam S. S. Int. J. Biol. Macromol. 2013;62:439–449. doi: 10.1016/j.ijbiomac.2013.09.019. [DOI] [PubMed] [Google Scholar]
- Manna D. K. Mandal A. K. Sen I. K. Maji P. K. Chakraborti S. Chakraborty R. Islam S. S. Int. J. Biol. Macromol. 2015;80:455–459. doi: 10.1016/j.ijbiomac.2015.07.028. [DOI] [PubMed] [Google Scholar]
- Abbaszadegan A. Ghahramani Y. Gholami A. Hemmateenejad B. Dorostkar S. Nabavizadeh M. Sharghi H. J. Nanomater. 2015;2015:1–8. doi: 10.1155/2015/720654. [DOI] [PubMed] [Google Scholar]
- Ramalingam B. Parandhaman T. Das S. K. ACS Appl. Mater. Interfaces. 2016;8:4963–4976. doi: 10.1021/acsami.6b00161. [DOI] [PubMed] [Google Scholar]
- Ahmad A. Wei Y. Syed F. Tahir K. Rehman A. U. Khan A. Ullah S. Yuan Q. Microb. Pathog. 2017;102:133–142. doi: 10.1016/j.micpath.2016.11.030. [DOI] [PubMed] [Google Scholar]
- Gomaa E. Z. J. Gen. Appl. Microbiol. 2017;63:36–43. doi: 10.2323/jgam.2016.07.004. [DOI] [PubMed] [Google Scholar]
- Ravichandran A. Subramanian P. Manoharan V. Muthu T. Periyannan R. Thangapandi M. Ponnuchamy K. Pandi B. Marimuthu P. N. J. Photochem. Photobiol., B. 2018;185:117–125. doi: 10.1016/j.jphotobiol.2018.05.031. [DOI] [PubMed] [Google Scholar]
- Yuan Y. G. Peng Q. L. Gurunathan S. Int. J. Mol. Sci. 2017;18:569. doi: 10.3390/ijms18030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M. A. Alzohairy M. A. J. Evidence-Based Complementary Altern. Med. 2018;2018:1–9. doi: 10.1155/2018/1860280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninganagouda S. Rathod V. Singh D. Hiremath J. Singh A. K. Mathew J. ul-Haq M. BioMed Res. Int. 2014;2014:753419. doi: 10.1155/2014/753419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszewski B. Railean-Plugaru V. Pomastowski P. Rafińska K. Szultka-Mlynska M. Golinska P. Wypij M. Laskowski D. Dahm H. J. Microbiol., Immunol. Infect. 2018;51:45–54. doi: 10.1016/j.jmii.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Chopade B. Ghosh S. Patil M. Ahire R. Kitture S. Jabgunde K. Kale S. S. Pardesi J. Cameotra D. D. Bellare A. Dhavale B. A. Int. J. Nanomed. 2012;7:483. doi: 10.2147/IJN.S24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H. S. Batchelor-McAuley C. Tschulik K. Compton R. G. Sci. China: Chem. 2014;57:1199–1210. doi: 10.1007/s11426-014-5141-8. [DOI] [Google Scholar]
- Lok C. N. Ho C. M. Chen R. He Q. Y. Yu W. Y. Sun H. Tam P. K. H. Chiu J. F. Che C. M. J. Proteome Res. 2006;5:916–924. doi: 10.1021/pr0504079. [DOI] [PubMed] [Google Scholar]
- McQuillan J. S. Shaw A. M. Nanotoxicology. 2014;8:177–184. doi: 10.3109/17435390.2013.870243. [DOI] [PubMed] [Google Scholar]
- Gogoi S. K. Gopinath P. Paul A. Ramesh A. Ghosh S. S. Chattopadhyay A. Langmuir. 2006;22:9322–9328. doi: 10.1021/la060661v. [DOI] [PubMed] [Google Scholar]
- Hsueh Y. H. Lin K. S. Ke W. J. Hsieh C. T. Chiang C. L. Tzou D. Y. Liu S. T. PLoS One. 2015;10:e0144306. doi: 10.1371/journal.pone.0144306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q. Wu J. Chen G. J. Biomed. Mater. Res. 2000;52:662–668. doi: 10.1002/1097-4636(20001215)52:4<662::AID-JBM10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Soenen S. J. Rivera-Gil P. Montenegro J. M. Parak W. J. De Smedt S. C. Braeckmans K. Nano Today. 2011;6:446–465. doi: 10.1016/j.nantod.2011.08.001. [DOI] [Google Scholar]
- Das B. Dash S. K. Mandal D. Ghosh T. Chattopadhyay S. Tripathy S. Das S. Dey S. K. Das D. Roy S. Arabian J. Chem. 2017;10:862–876. doi: 10.1016/j.arabjc.2015.08.008. [DOI] [Google Scholar]
- Belluco S. Losasso C. Patuzzi I. Rigo L. Conficoni D. Gallocchio F. Cibin V. Catellani P. Segato S. Ricci A. Front. Microbiol. 2016;7:307. doi: 10.3389/fmicb.2016.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo-Hwan K. Seon Lee H. Seon Ryu D. Jae Choi S. Seok Lee D. Korean J. Microbiol. Biotechnol. 2011;39:77–85. [Google Scholar]