Abstract
The mreA gene from Streptococcus agalactiae COH31 γ/δ, resistant to macrolides and clindamycin by active efflux, has recently been cloned in Escherichia coli, where it was reported to confer macrolide resistance (J. Clancy, F. Dib-Hajj, J. W. Petitpas, and W. Yuan, Antimicrob. Agents Chemother. 41:2719–2723, 1997). Cumulative data suggested that the mreA gene was located on the chromosome of S. agalactiae COH31 γ/δ. Analysis of the deduced amino acid sequence of mreA revealed significant homology with several bifunctional flavokinases/(flavin adenine dinucleotide (FAD) synthetases, which convert riboflavin to flavin mononucleotide (FMN) and FMN to FAD, respectively. High-performance liquid chromatography experiments showed that the mreA gene product had a monofunctional flavokinase activity, similar to that of RibR from Bacillus subtilis. Sequences identical to those of the mreA gene and of a 121-bp upstream region containing a putative promoter were detected in strains of S. agalactiae UCN4, UCN5, and UCN6 susceptible to macrolides. mreA and its allele from S. agalactiae UCN4 were cloned on the shuttle vector pAT28. Both constructs were introduced into E. coli, where they conferred a similar two- to fourfold increase in the MICs of erythromycin, spiramycin, and clindamycin. The MICs of a variety of other molecules, including crystal violet, acriflavin, sodium dodecyl sulfate, and antibiotics, such as certain cephalosporins, chloramphenicol, doxycycline, nalidixic acid, novobiocin, and rifampin, were also increased. In contrast, resistance to these compounds was not detected when the constructs were introduced into E. faecalis JH2–2. In conclusion, the mreA gene was probably resident in S. agalactiae and may encode a metabolic function. We could not provide any evidence that it was responsible for macrolide resistance in S. agalactiae COH31 γ/δ; broad-spectrum resistance conferred by the gene in E. coli could involve multidrug efflux pumps by a mechanism that remains to be elucidated.
Streptococcus agalactiae (group B streptococcus) is responsible for neonatal sepsis and meningitis as well as serious invasive infections in adults, such as postpartum endometritis (6). The first line of therapy for these infections consists of administration of beta-lactam agents. However, macrolides and related drugs are useful alternate therapies in allergic patients.
Until recently, macrolide resistance in streptococci was considered to result only from target modification by 23S rRNA methylases encoded by erm genes, which conferred cross-resistance to macrolides, lincosamides, and streptogramin B components (MLSB phenotype) (21). Another phenotype, called M, related to efflux of only 14- and 15-member ring macrolides, has been reported in various streptococcal species, including Streptococcus pneumoniae, Streptococcus pyogenes, and S. agalactiae. The mechanism of resistance relies on a proton-dependent efflux system encoded by mef(A) class genes: (3, 15, 19). The mef(A) genes belong to the major facilitator superfamily and are believed to encode a hydrophobic membrane protein containing 12-membrane-spanning regions that pumps the antibiotic out of the cell. In addition, a novel efflux system distinct from the Mef pump and encoded by mreA (for macrolide resistance efflux) was recently reported in a unique strain of S. agalactiae COH31 γ/δ by Clancy et al. (4). The strain harboring this gene was resistant to 14-, 15-, and 16-member macrolides and to clindamycin. The results of experiments with radiolabeled erythromycin suggested the presence of a macrolide efflux mechanism. The mreA gene was cloned from total DNA of S. agalactiae COH31 γ/δ into Escherichia coli, where it conferred macrolide resistance. The presence of the gene in E. coli also resulted in a significant decrease in erythromycin accumulation. Sequencing revealed that mreA encodes a 310-amino-acid protein, with a predicted molecular mass of 35.4 kDa. This protein is hydrophilic with interspersed hydrophobic and amphipathic sequences. The protein displayed homology with RibC, a flavokinase/flavin adenine dinucleotide (FAD) synthetase from Bacillus subtilis; however, its function has not been studied (4). The present study demonstrates that the product of the mreA gene displays a flavokinase activity and is responsible for a broad-spectrum resistance to a variety of compounds when cloned in E. coli, but not when expressed in Enterococcus faecalis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Streptococcal strains were grown on Trypticase soy (TS) agar (Bio-Rad, Marnes-la-Coquette, France) supplemented with 5% horse blood. E. coli, Bacillus subtilis, and Enterococcus faecalis strains were cultured in TS broth or agar. All cultures were incubated at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. agalactiae | ||
| COH31 γ/δ | Emr | 4 |
| UCN4 | Ems | Clinical isolate |
| UCN5 | Ems | Clinical isolate |
| UCN6 | Ems | Clinical isolate |
| E. faecalis JH2-2 | Fusr Rifr | 9 |
| B. subtilis Marburg 168 | trpC2 | 23 |
| E. coli DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) Ψ 80dlacZΔ M15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU λ−rpsL nupG | Gibco-BRL |
| Plasmids | ||
| pCR2.1 | Cloning vector, Kmr Amr | Invitrogen |
| pUV6 | pCR2.1 with 1,065-bp mreA insert | This study |
| pUV7 | pCR2.1 with 1,065-bp mreA allele from S. agalactiae UCN4 | This study |
| pUV10 | pCR2.1 with 1,087-bp ribC insert | This study |
| pAT28 | Cloning vector, Spcr | 20 |
| pUV8 | pAT28 with 1,065-bp mreA insert in the SacI-XbaI sites | This study |
| pUV9 | pAT28 with 1,065-bp mreA from S. agalactiae UCN4 insert in the SacI-XbaI sites | This study |
Amr, ampicillin resistant; Emr, erythromycin resistant; Ems, erythromycin susceptible, Fusr, fusidic acid resistant; Kmr, kanamycin resistant; Rifr, rifampin resistant; Spcr, spectinomycin resistant.
Susceptibility testing.
MICs of antibiotics were determined by the agar dilution method with Mueller-Hinton medium (Bio-Rad) supplemented with 5% horse blood inoculated with 104 CFU and incubated at 37°C under aerobic conditions according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (5). The antibiotics and molecules tested were supplied by Sigma Chemical Co. (St. Louis, Mo.) or by their manufacturer.
PCR and cloning experiments.
DNA sequences specific for the mreA gene were amplified by PCR with the primers mre3 (5′-ATA AAG AAA GTC AAT CAT G-3′ [nucleotides 106 to 124]) and mre4 (5′-AT ACA AAA AAT TAA AGA G-3′ [nucleotides 1064 to 1045]). The numbers in brackets refer to the numbers of the mreA sequence in the GenBank database (accession no. U92073). PCRs were performed with a GeneAmp PCR system 2400 cycler (Perkin-Elmer Cetus, Norwalk, Conn.) with Taq DNA polymerase (Eurobio, Les Ullis, France). The mreA gene preceded by a 121-bp sequence containing a putative promoter (4) was amplified from the DNA of three macrolide-susceptible S. agalactiae strains, UCN4, UCN5, and UCN6, by PCR with oligonucleotides mre5 (5′-CTT ATT AGA AAA TGA AGC AG-3′ [nucleotides 1 to 20]) and mre4. The various amplicons were cloned into plasmid pCR2.1 (Invitrogen, Groningen, The Netherlands) in the same orientation. The recombinant plasmids were introduced into competent E. coli DH10B cells by electrotransformation with a Gene Pulser (Bio-Rad) and selected by using TS agar plates containing 50 μg of kanamycin per ml.
The fragments were then subcloned on the multicopy shuttle vector pAT28 (spectinomycin resistance) with the SacI and XbaI restriction sites (20). The plasmid constructs were made with E. coli DH10B prior to transformation into E. faecalis JH2–2, as described previously (12), and were selected by using TS agar plates containing 60 and 150 μg of spectinomycin per ml, respectively.
The ribC and promoter sequences were amplified from B. subtilis Marburg 168 DNA by PCR with oligonucleotides ribc1 (5′-ATT GCC GTC TTT ACT GAA TCC G-3′ [nucleotides 241 to 262]) and ribc2 (5′-AAA CTA TCA TAC TAA AAA TCG TGC C-3′ [nucleotides 1387 to 1363]). The numbers in brackets refer to numbers of the ribC sequence in the GenBank database (accession no. X95312). The amplicon was cloned into plasmid pCR2.1 and introduced into competent E. coli DH10B cells.
Southern blot hybridization.
DNA from S. agalactiae COH31 γ/δ was digested with the restriction endonuclease HindIII. DNA fragments were separated in a 0.7% agarose gel, denatured, and transferred onto a nylon membrane (Hybond-N; Amersham France, Les Ullis, France). The 738-bp mreA-specific PCR product obtained with the primers mre1 (5′-AAT TTG AAA ATT GTC GTC TTA ACG T-3′ [nucleotides 260 to 285]) and mre2 (5′-GTT GTT TTA CAA GAT CGT CAA TAC C-3′ [nucleotides 997 to 974]) was used as a probe. This product was labeled with digoxigenin (Boehringer Mannheim France, Meylan, France), and hybridization was detected by using an anti-digoxigenin-alkaline phosphatase conjugate with a chromogenic enzyme substrate.
The chromosomal location of the mreA gene was determined by restriction of total DNA of S. agalactiae COH31 γ/δ with I-CeuI (New England Biolabs, Beverly, Mass.) an intron-encoded endonuclease specific for rRNA genes, followed by pulsed field gel electrophoresis as previously described (13). DNA fragments were transferred onto a nylon membrane and successively hybridized with 16S rRNA and mreA probes.
Inverse PCR.
In order to sequence the DNA regions located upstream and downstream of mreA, DNA of S. agalactiae COH31 γ/δ was digested with HindIII and ligated with T4 ligase. Inverse PCR was performed with oligonucleotides mre1 and mre in2 (5′-CGC AAT CTT CTT TAG CTT GAA TAT C-3′ [nucleotides 176 to 152]) with Taq polymerase (Eurobio). The reaction consisted of (i) an initial step of 3 min at 94°C; (ii) 35 cycles of PCR, with 1 cycle consisting of 30 s at 94°C, 30 s at 50°C, and 120 s at 72°C; and (iii) a final step of 10 min at 72°C with 2 mM MgCl2. A 3.5-kb amplified fragment was cloned in pCR2.1 and sequenced with an automated ABI PRISM 377 system (Perkin-Elmer Corp.). Nucleotide and amino acid sequences were analyzed by using the software available online over the internet at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/). The microbial databases used were those of The Institute for Genomic Research (TIGR) (http://www.tigr.org), the Doe Joint Genome Institute (JGI) (http://www.jgi.doe.gov), and The University of Oklahoma (http://www.genome.ou.edu).
Preparation of cell extracts, enzyme assay, and HPLC analysis of flavins.
Cell extracts of E. coli DH10B containing various constructs were prepared as follows. Cells of an overnight culture (100 ml) were collected by centrifugation. The cell pellet was washed with a mixture of 100 mM potassium phosphate (pH 7.5), 0.1 mM EDTA, and 1 mM dithiothreitol (buffer A). The cells were resuspended in buffer A and sonicated twice for 10 s. After centrifugation (18,000 × g for 30 min), an aliquot of the supernatant was directly used in the flavokinase assay. All procedures were carried out at 4°C. Protein concentrations were determined by the method of Bradford, with reagents from the Bio-Rad protein assay and with bovine serum albumin as a standard (2).
The β-lactamase activity encoded by the plasmid pCR2.1 was measured to standardize the extract. Assays were performed by UV spectrophotometry with freshly prepared penicillin G solutions in 100 mM phosphate buffer (pH 7.0). The assays were run at 37°C and monitored at 235 nm (22).
Flavokinase activity was measured in a final volume of 1 ml of potassium phosphate (pH 7.5) containing 50 μM riboflavin, 3 mM ATP, 15 mM MgCl2, and 10 mM Na2SO3. A similar FAD synthetase assay containing 50 μM flavin mononucleotide (FMN) instead of riboflavin was performed to measure the formation of FAD from FMN and ATP (14). The mixture was preincubated for 5 min at 37°C; the reaction was started by addition of the cell extract and stopped by boiling after 5 or 30 min of incubation. A centrifugation eliminated the denatured proteins.
The high-performance liquid chromatography (HPLC) analysis required a C8 Satisfaction column (4.6 by 250 mm) (CIL, Cluzeau, France) and fluorescence detector (excitation, 470 nm; emission, 530 nm) (ThermoQuest, Les Ullis, France). The solvent system was composed of 40% methanol in 100 mM potassium phosphate (pH 4) and used at a flow rate of 1 ml/min. Flavokinase activity was expressed as micrograms of FMN formed from riboflavin and ATP per minute and per milligram of total protein. FAD synthetase activity was expressed as micrograms of FAD formed from FMN and ATP per minute and per milligram of total protein.
RESULTS
Detection of mreA in erythromycin-susceptible Streptococcus agalactiae strains.
A 960-bp DNA fragment internal to mreA was amplified from DNA of S. agalactiae COH31 γ/δ and, surprisingly, that of three clinical erythromycin-susceptible S. agalactiae isolates with oligonucleotides mre3 and mre4. To assess if mutations could explain the differences in erythromycin susceptibility of the strains, we have amplified and sequenced a 1,065-bp DNA fragment including the entire mreA gene and a 121-bp upstream region containing a putative promoter (4) and the allelic sequences from the three erythromycin-susceptible strains. Previous cloning of this 1,065-bp fragment in the two opposite orientations by Clancy et al. has shown that it contained the sequences required to confer a two- to fourfold decrease in macrolide susceptibility in E. coli (4). Sequencing revealed complete identity between all of the DNA fragments. In a recent study, the mreA gene was found by PCR in all strains of a collection of 88 clinical isolates of S. agalactiae resistant to macrolides and containing erm or mef genes, whereas it was not found in two strains of group G streptococcus strains (E. Bingen, personal communication). We did not detect by PCR mreA sequences in group A streptococci (10 strains) and pneumococci (10 strains).
mreA confers a broad-spectrum drug resistance when cloned in E. coli
mreA with its putative promoter and the allele mreAS from S. agalactiae UCN4, susceptible to erythromycin, were cloned on plasmid pCR2.1 to generate plasmids pUV6 and pUV7, respectively. The inserts were subsequently subcloned on the shuttle plasmid pAT28 to generate plasmids pUV8 and pUV9, respectively. The recombinant plasmids were introduced into E. coli DH10B. In this host, all constructs conferred the same levels of resistance to erythromycin (MIC = 128 μg/ml), spiramycin (a 16-member macrolide) (MIC = 1,024 μg/ml), and clindamycin (MIC = 128 μg/ml). These MICs corresponded to an increase of a factor 2 or 4, as reported previously by Clancy et al. (4). This increase was repeatedly found in several experiments.
In addition, MreA and its allele conferred in E. coli a similar four- to eightfold increase in MICs of acriflavin, as reported previously (4), as well as an increase in MICs of a variety of other compounds, including cationic dyes, such as crystal violet; detergents, such as sodium dodecyl sulfate; various antibiotics, including cefoxitin, cefepime, and ceftazidime; lipophilic compounds, such as rifampin (zwitterionic) and doxycycline; and hydrophobic agents, such as novobiocin and nalidixic acid, as well as chloramphenicol, an uncharged antibiotic. Except for the cross-resistance to macrolides (MIC of erythromycin = 4 μg/ml) and clindamycin (MIC = 0.5 μg/ml) in S. agalactiae COH31 γ/δ, no other differences in the MICs of the tested compounds for the four tested S. agalactiae strains were found.
The pUV8 and pUV9 constructs were introduced into E. faecalis JH2–2. The stability of the pAT28 derivatives in E. faecalis was verified by confirming the expression of spectinomycin resistance at crucial steps of the experiments. The presence of mreA was also verified by PCR. HPLC experiments showed that flavokinase was expressed in E. faecalis, although at a slightly lower level than in E. coli. Retransformation of E. coli with the recombinant plasmid extracted from E. faecalis led to increased MICs of erythromycin and of the other compounds at the expected level. In contrast, in the E. faecalis background, pUV8 and pUV9 did not confer any increase in the MICs of the compounds tested, including erythromycin.
MreA is a flavokinase.
MreA shared a significant degree of similarity with several members of an enzyme family possessing a bifunctional flavokinase/FAD synthetase activity. Flavokinases (EC 2.7.1.26) catalyze the conversion of riboflavin to FMN, whereas FAD synthetases (EC 2.7.7.2) convert FMN to FAD; these two reactions require ATP as a cofactor (1). The optimal aligment of the amino acid sequence of MreA revealed a 37% identity with RibC, a bifunctional flavokinase/FAD synthetase from B. subtilis (316 amino acids), and 30.4 and 32.7% identity with flavokinases/FAD synthetases RibF from E. coli (313 amino acids) and Haemophilus influenzae (312 amino acids), respectively (Fig. 1) (7, 8; K. Kitatsuji, S. Ishino, S. Teshiba, and M. Arimoto, 1993, European patent application 0 542 240 A2). Two conserved motifs were found in the C-terminus ends of MreA and FAD synthetases (Fig. 1). Furthermore, the hydrophobic cluster analysis showed that MreA and FAD synthetases proteins shared similar presumed secondary structures (data not shown). These results suggested that mreA could encode a bifunctional flavokinase/FAD-synthetase.
FIG. 1.
Alignment of the N-terminus amino acid sequence of RibR from B. subtilis with the C termini of the mreA gene product (MreA), and the bifunctional flavokinase/FAD-synthetase from B. subtilis, E. coli (E.c), and H. influenzae (H.i) with the CLUSTALW program. Dashes indicate gaps introduced to increase the number of matches. Homologous and similar amino acids are represented by double and single dots, respectively. Asterisks represent amino acid residues identical among the five sequences. The boxed blocks of amino acids correspond to motifs of riboflavin kinase/FAD synthetase according to Block Searcher results.
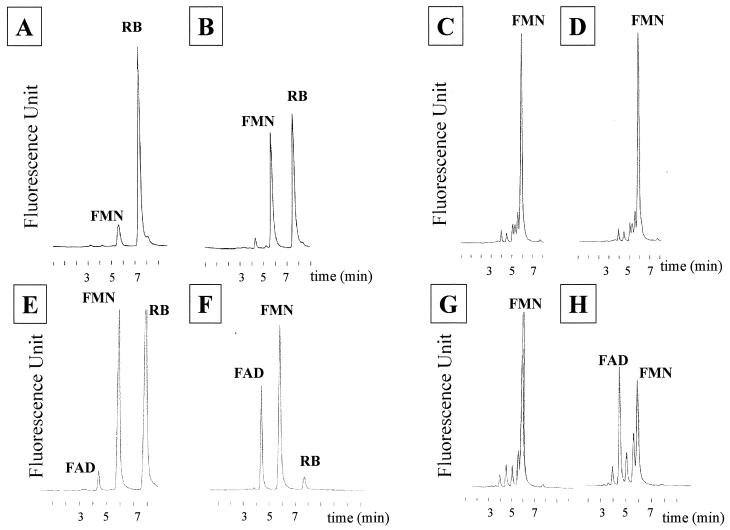
To elucidate the function of MreA, flavokinase and FAD synthetase activities were measured in cell extracts of E. coli containing either pUV6 (mreA cloned in pCR2.1) or pUV7 (mreAS cloned in pCR2.1). The flavokinase and FAD synthetase enzymatic activities were investigated by HPLC with riboflavin and FMN as substrate donors, respectively. The preliminary calibration of HPLC methodology showed that peaks of FAD, FMN, and riboflavin were resolved at 4.3, 5.9, and 7.9 min, respectively, similar to the retention times reported by Mack et al. (14). After 30 min of incubation at 37°C of the cell extracts containing MreA (Fig. 2) or MreAS (data not shown) with riboflavin, there was a decrease in the size of the riboflavin peak concomitant with an increase in FMN production, which demonstrated that both cell extracts displayed a flavokinase activity. Despite the high level of flavokinase activity, no significant FAD peak was detected after 30 min of incubation of cell extracts in the presence of FMN as a donor, indicating the absence of FAD synthetase activity (Fig. 2B). Flavokinase activities measured in cell extracts of E. coli containing mreA or mreAS were similar and corresponded respectively to 0.087 and 0.083 μg of FMN produced per min per mg of protein. Control experiments with extracts from E. coli containing pCR2.1 did not reveal any flavokinase or FAD synthetase activity (data not shown). Probably the level of FMN or FAD produced by the flavokinase of E. coli encoded by a gene present as a single copy in the chromosome was too low to be detected by the HPLC technique. The RibC control showed a flavokinase activity (0.062 μg of FMN produced per min per mg of protein) and a FAD synthetase activity (0.049 μg of FAD produced per min per mg of protein) when riboflavin and FMN, respectively, were used as substrate donors (Fig. 2).
FIG. 2.
HPLC chromatograms of the products of flavokinase/FAD synthetase assays. Flavokinase (A, B, E, and F) and FAD synthetase (C, D, G, and H) activities were evaluated by fluorescence detection in the presence of 50 μM riboflavin (RB) and 50 μM FMN, respectively. Cell extracts of E. coli DH10B/pUV6 containing mreA (A, B, C, and D) or E. coli DH10B/pUV10 containing ribC (E, F, G, and H) were added to the reaction mixture, and the activity assays were incubated for 5 min (A, C, E, and G) or 30 min (B, D, F, and H) and stopped by boiling. Aliquots were removed and separated on an HPLC column. The chromatograms show three clearly resolved peaks of riboflavin (7.9 min), FMN (5.9 min), and FAD (4.3 min). Peak intensity is given in arbitrary fluorescence units.
Location of the mreA gene.
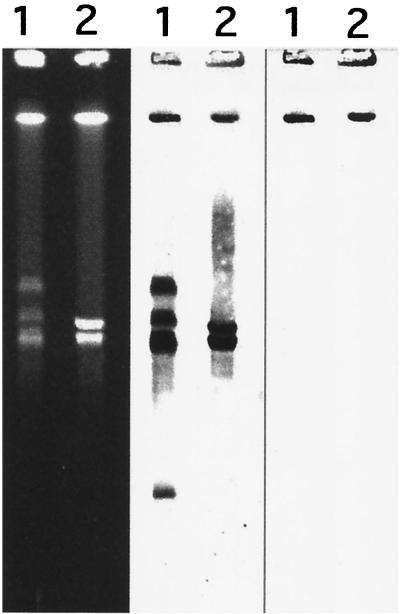
The mreA and the rrs probes hybridized with the same I-CeuI-generated fragment of S. agalactiae COH31 γ/δ DNA (Fig. 3). This observation was strongly in favor of a chromosomal location of the mreA gene. Southern blot experiments with a probe specific for mreA confirmed that only one chromosomal copy of the gene could be detected in S. agalactiae COH31 γ/δ (data not shown). DNA regions located upstream and downstream of the mreA gene were amplified by inverse PCR. A 3.5-kb amplified fragment was cloned in pCR2.1, introduced in E.coli DH10B, and sequenced. Upstream of mreA, sequence analysis identified an open reading frame (ORF) that could encode a 214-amino-acid protein that displayed 48% identity with B. subtilis TruB, a 309-amino-acid protein (17). TruB is a tRNA pseudouridine 55 (psi 55) synthase, an enzyme specific for the conversion of U55 to pseudouridine in the CG loop of most tRNAs (10). Nucleotide sequence analysis showed the presence of an additional 414-bp ORF downstream of mreA, 43 bp after the termination codon of the gene. This ORF could encode a protein of 137 amino acids, which shared 28% identity with arsenate reductase (ArsC) of B. subtilis, a protein of 118 amino acids.
FIG. 3.
Analysis of genomic DNA from S. agalactiae COH31 γ/δ (lane 1) and UCN4 (lane 2), digested with I-CeuI by pulsed-field gel electrophoresis (left) and hybridization (middle and right). The digested fragments were transferred to a nylon sheet and hybridized to an in vitro digoxigenin-labeled 16S probe (middle). After dehybridization, the filter was hybridized to a digoxigenin-labeled mreA probe (right).
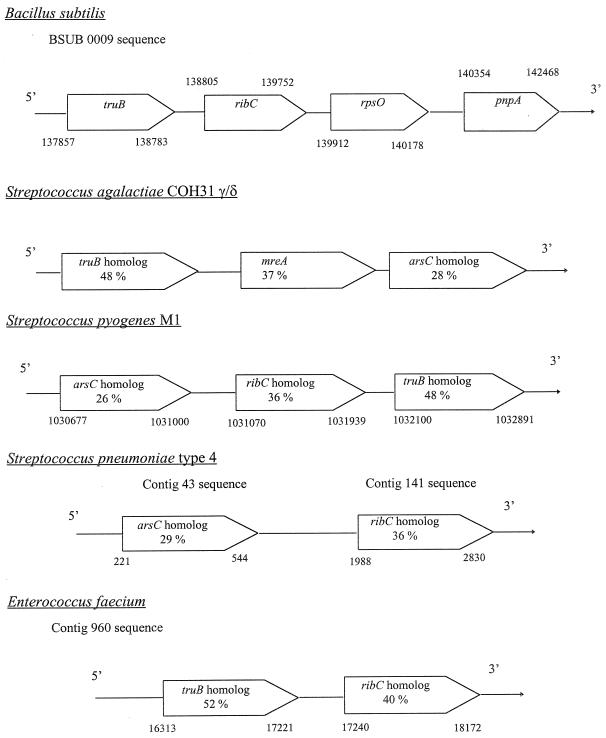
The homologs of truB, ribC, and arsC were identified on the chromosome of gram-positive microorganisms, including S. pyogenes, S. pneumoniae, Enterococcus faecium, and B. subtilis, according to the BLAST program for unfinished and finished bacterial genomes (http://www.tigr.org./tdb/mdb/mdbcomplete.html). The organization of the genes was similar within these organisms, consistent with a likely chromosomal location of the mreA gene in S. agalactiae (Fig. 4).
FIG. 4.
Genetic organization of mreA and ribC homologs in different Streptococcus species, in E. faecium, and in B. subtilis. These results were obtained after analysis with Blast 2 program of sequences of B. subtilis from Kunst et al. (11), S. pneumoniae M1 from Oklahoma University (http://www.genome.ou.edu), S. pneumoniae type 4 from the TIGR database (http://www.tigr.org), and E. faecium from the JGI database (http://www.jgi.doe.gov). The percentages of identity relative to the B. subtilis genes, obtained with the ALIGN program, are indicated within the arrows. rpsO, ribosomal protein S15 gene; pnpA, polynucleotide phosphorylase gene; truB, tRNA pseudouridine 55 synthase gene; arsC, arsenate reductase gene. Numbers refer to genomic position.
DISCUSSION
Analysis of the deduced amino acid sequence of MreA revealed that this protein displayed homology with various bifunctional flavokinases/FAD synthetases. We have shown that cell extracts of E. coli expressing this protein possessed only a monofunctional riboflavin kinase activity and were devoid of FAD synthetase activity. By using site-specific mutagenesis on the flavokinase/FAD synthetase gene from E. coli, it was shown that the flavokinase activity of the enzyme was associated with the C-terminal region, and the FAD synthetase activity was associated with the N-terminal region (Kitatsuji et al., patent application). Recently, a monofunctional flavokinase, RibR, has been reported in B. subtilis (18). This 230-amino-acid protein is about 100 amino acids shorter than the bifunctional FAD synthetases, including RibC from B. subtilis. Similarly to the C terminus of MreA, RibR showed significant homology with the C-terminus regions of RibC and RibF of E. coli and H. influenzae, starting near residue 200. In particular, a conserved motif of 10 amino acid residues, GR(K/T)(L/I)GFPTAN, between residues 15 and 24 (numbering with respect to the ribR gene product) was found.
Cumulative and convergent data strongly suggested that the mreA gene was chromosomal in S. agalactiae COH31 γ/δ. Sequencing of the flanking regions revealed that it was surrounded by truB and arsC homologs. Analysis of the data banks showed that a similar genetic linkage between homologs of truB and arsC and flavokinase genes could be found in the chromosome of other gram-positive organisms, including several Streptococcus species and B. subtilis. The gene organization appeared different in the E. coli chromosome, where ribF was located between infB and rpsO (16).
Taken together, these observations suggested that the mreA gene was resident in S. agalactiae and could putatively encode a metabolic function. Several of our results questioned the role of the mreA gene in conferring resistance to macrolides in S. agalactiae COH31 γ/δ. First, sequences identical to mreA were found in macrolide-susceptible strains of S. agalactiae conferring similar levels of macrolide resistance after cloning in E. coli. No difference in the sequences upstream of the mreA gene, which included a putative promoter, could be detected, and the gene was present in one copy on the chromosome of S. agalactiae COH31 γ/δ. However, we did not compare the transcription of the mreA gene in the erythromycin-susceptible or erythromycin-resistant strains. We could not characterize the macrolide resistance phenotype after disruption of the mreA gene, since we were unable to introduce plasmid DNA by electrotransformation in S. agalactiae COH31 γ/δ. In addition, the phenotype of isolated resistance to macrolides and clindamycin displayed by S. agalactiae COH31 γ/δ could not be reproduced after cloning of the gene either in E. coli or in a gram-positive host. In E. coli, it conferred broad-spectrum resistance, while in E. faecalis, no expression of resistance could be detected. The mechanism by which the flavokinase MreA conferred macrolide resistance in E. coli remains unclear. The enzyme did not inactivate erythromycin (4). Intriguingly, Clancy et al. have shown in studies of [14C]erythromycin accumulation that an energy-dependent macrolide efflux mechanism was associated with the presence of mreA in E. coli (4). This erythromycin efflux was inhibited by uncouplers of oxidative phosphorylation, such as CCCP (carbonyl cyanide-m-chlorophenylhydrazone) and arsenate, proving that efflux was an energy-dependent process. Interestingly, the spectrum of resistance conferred by mreA extends to numerous other compounds, including other flavins, such as acriflavin, and various antibiotics. This effect did not appear to be specifically related to the presence of the mreA gene, but rather to flavokinase activity, since similar MIC increases were observed when ribC was introduced into E. coli (data not shown). Both the pattern of broad-spectrum antibiotic resistance and the energy-dependent efflux of erythromycin suggested that multidrug efflux pumps, possibly belonging to the resistance nodulation division (RND) family, could intervene to confer resistance. This requirement for the presence of a functional gram-negative pump for expression of resistance would be consistent with the lack of expression of mreA in gram-positive hosts. This hypothesis is currently under investigation.
ACKNOWLEDGMENTS
G. Clarebout was the recipient of a FEDER fellowship from the Conseil Régional de Basse-Normandie.
We are grateful to A. Coquerel and D. Debruyne from the Department of Pharmacology for help with the HPLC experiments and J. Clancy for the gift of the S. agalactiae COH31 γ/δ strain.
REFERENCES
- 1.Bacher A. Riboflavin kinase and FAD synthetase. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 1. Boca Raton, Fla: CRC Press; 1991. pp. 349–370. [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 4.Clancy J, Dib-Hajj F, Petitpas J W, Yuan W. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob Agents Chemother. 1997;41:2719–2723. doi: 10.1128/aac.41.12.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comité de l'Antibiogramme de la Société Française de Microbiologie. 1996 report of the Comité de l'Antibiogramme de la Société Française de Microbiologie. Technical recommendations for in vitro susceptibility testing. Clin Microbiol Infect. 1996;2S1:11–25. [Google Scholar]
- 6.Edwards M S, Baker C J. Streptococcus agalactiae (group B streptococcus) In: Mandell G L, Bennett J E, Dolin R, editors. Mandell, Douglas, and Bennett. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1995. pp. 1835–1845. [Google Scholar]
- 7.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J, Dougherty B A, Merrick J M, McKenney K, Sutton G G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L L, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 8.Gusarov I I, Kreneva R A, Rybak K V, Podchernyaev D A, Iomantas Y V, Kolibaba L G, Polanuer B M, Kozlov Y I, Perumov D A. Primary structure and the functional activity of the ribC gene of Bacillus subtilis. Mol Biol. 1997;31:825–830. [PubMed] [Google Scholar]
- 9.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonin E V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq R, Derlot E, Weber M, Duval J, Courvalin P. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1989;33:10–15. doi: 10.1128/aac.33.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S H, Hessel A, Sanderson K E. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mack M, van Loon A P G M, Hohmann H P. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J Bacteriol. 1998;180:950–955. doi: 10.1128/jb.180.4.950-955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sands J F, Regnier P, Cummings H S, Grunberg-Manago M, Hershey J W B. The existence of two genes between infB and rpsO in the Escherichia coli genome: DNA sequencing and S1 nuclease mapping. Nucleic Acids Res. 1988;16:10803–10816. doi: 10.1093/nar/16.22.10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shazand K, Tucker J, Grunberg-Manago M, Rabinowitz J C, Leighton T. Similar organization of the nusA-infB operon in Bacillus subtilis and Escherichia coli. J Bacteriol. 1993;175:2880–2887. doi: 10.1128/jb.175.10.2880-2887.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solovieva I M, Kreneva R A, Leak D J, Perumov D A. The ribR gene encodes a monofunctional riboflavin kinase which is involved in the regulation of the Bacillus subtilis riboflavin operon. Microbiology. 1999;145:67–73. doi: 10.1099/13500872-145-1-67. [DOI] [PubMed] [Google Scholar]
- 19.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Wu P, Livermore D M. Biochemical characterization of a β-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob Agents Chemother. 1990;34:755–758. doi: 10.1128/aac.34.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida K-I, Fujita Y, Ehrlich S D. An operon for a putative ATP-binding cassette transport system involved in acetoin utilization of Bacillus subtilis. J Bacteriol. 2000;182:5454–5461. doi: 10.1128/jb.182.19.5454-5461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]