Abstract
Citrobacter sedlakii 2596, a clinical strain resistant to aminopenicillins, carboxypenicillins, and early cephalosporins such as cephalothin, but remaining susceptible to acylureidopenicillins, carbapenems, and later cephalosporins such as cefotaxime, was isolated from the bile of a patient treated with β-lactam and quinolone antibiotics. The isolate produced an inducible class A β-lactamase of pI 8.6, named Sed-1, which was purified. Characterized by a molecular mass of 30 kDa, Sed-1 preferentially hydrolyzed benzylpenicillin, cephalothin, and cloxacillin. The corresponding gene, blaSed-1, was cloned and sequenced. Its deduced amino acid sequence shared more than 60% identity with the chromosome-encoded β-lactamases from Citrobacter koseri (formerly C. diversus) (84%), Klebsiella oxytoca (74%), Serratia fonticola (67%), and Proteus vulgaris (63%) and 71% identity with the plasmid-mediated enzyme MEN-1. A gene coding for a LysR transcriptional regulator was found upstream from blaSed-1. This regulator, named SedR, displayed 90% identity with the AmpR sequence of the chromosomal β-lactamase from C. koseri and 63 and 50% identity with the AmpR sequences of P. vulgaris and Enterobacter cloacae, respectively. By using DNA-DNA hybridization, a blaSed-1-like gene was identified in two reference strains, C. sedlakii (CIP-105037) and Citrobacter rodentium (CIP-104675), but not in the 18 strains of C. koseri studied. Two DNA fragments were amplified and sequenced from the reference strains of C. sedlakii CIP-105037 and C. rodentium CIP-104675 using two primers specific for blaSed-1. They shared 98 and 80% identity with blaSed-1, respectively, confirming the diversity of the chromosomally encoded class A β-lactamases found in Citrobacter.
The genus Citrobacter, which was defined in 1932, initially encompassed seven species including Citrobacter freundii (type species) and Citrobacter koseri (formerly termed Citrobacter diversus or Levinea malonatica) (60). In 1993, Brenner et al. identified eight new DNA hybridization groups genetically distinct from C. freundii and C. koseri (13). These additional genomospecies included C. sedlakii (genomospecies 8) and C. rodentium (genomospecies 9, a bacterial pathogen of rodents).
In the Citrobacter genus, resistance to β-lactam antibiotics is mainly mediated by production of chromosomally encoded β-lactamases. C. freundii produces an inducible Ambler class C β-lactamase (60). In C. koseri (formerly C. diversus), which is naturally resistant to aminopenicillins and carboxypenicillins, resistance to β-lactams is mediated by a chromosome-encoded class A β-lactamase (4). The cloning and sequencing of the corresponding gene, which has been termed blaCdiA, revealed a high degree of similarity (75%) between CdiA and the class A β-lactamase from Klebsiella oxytoca (33). Previous experiments carried out by DNA amplification with primers specific for blaCdiA showed that this gene is not ubiquitous in C. koseri (32). Accordingly, different chromosome-encoded β-lactamases with distinct isoelectric points (pIs) varying from 4.8 to 9.5 have been identified in C. koseri, but the corresponding genes have not been characterized at the genetic level (27, 31, 41, 42, 47, 53, 55). The LysR-type transcriptional regulator (LTTR) protein CdiR, divergently transcribed from CdiA, has also been characterized. The AmpR regulator protein most closely related to CdiR is the Proteus vulgaris CumR protein, with an amino acid identity of 66%, whereas only 46% identity was found with the AmpR protein from C. freundii, strengthening the idea of a wide genetic diversity of the genes determining resistance to β-lactam antibiotics in the Citrobacter genus.
C. sedlakii has been rarely described since its first identification in 1993 (1, 2, 13, 23, 43). Strains isolated from human stools, blood, and wounds were studied by Brenner et al. (13), and a report of neonatal meningitis and brain abscess involving a strain resistant to ampicillin (MIC, 16 μg/ml), cefazolin (MIC, 16 μg/ml), and cefuroxime (MIC, 16 μg/ml), was made in 1997 by Dyer et al. (23), but the mechanism of resistance to β-lactams in these strains has not been investigated.
In the present study, we report the cloning and sequencing of the gene coding for the class A β-lactamase from C. sedlakii. The biochemical characteristics of this enzyme, named Sed-1, were studied. The transcriptional regulator associated with blaSed-1 was also cloned and sequenced. In order to address the issue of the genetic diversity of the β-lactamases found within the Citrobacter genus, DNA-DNA hybridization, high-stringency PCR, and DNA sequencing were used to study the distribution of blaSed-1 in 21 isolates of Citrobacter spp.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The clinical strain 2596 of C. sedlakii was isolated in 1997 from the bile of a patient treated with β-lactam and quinolone antibiotics. The strain was identified as C. sedlakii by biochemical tests using BIOTYPE 100 (Biomérieux) and the programs RECOGNIZER, ADANSON, and DENDOGRAPH of the Taxotron package (Institut Pasteur Taxolab). The identification was confirmed by amplifying and determining the nucleotide sequence of a PCR-amplified DNA fragment of 568 bp corresponding to the 16S RNA of the strain (the sequences of the primers are given in Table 1). The other bacterial strains and plasmids used in this work are listed in Table 2.
TABLE 1.
Nucleotide sequences of primers
| Primer | Nucleotide sequence (5′ → 3′) | Genes | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| AmpC div sens | AACGAGGTCGTCAGACG | blaCdiA | 49 | 33 |
| AmpC div A sens | CAGAATATCTTTACGCCC | |||
| Class A | AGCGAYAAYACGGCGATG | Class Aa | 55 | 8 |
| Class A rev | TGCKCCGGTYTTATCGCC | |||
| SHV rev | GCGTTGCCAGTGCTCGATCAG | blaSHV | 62 | 6 |
| SHV bis | ATGCGTTATATTCGCCTGTGTATT | |||
| TEM H | TGAGATCGAAGGGCCGTT | blaTEM | 52 | 54 |
| TEM rev | GGTCTGACAGTTACCAAT | |||
| OXA C | AGGTGCCATGAAAACATT | blaOXA group 1b | 45 | |
| OXA C bis | TTAGCCACCAATGATGAA | |||
| OXA A | AAGGAAAAGTTAATGGCA | blaOXA group 2b | 44 | |
| OXA A bis | TTTATCGCGCAGCGTCCG | |||
| OXA E | AGGACTTGGGACATCGAT | blaOXA18 | 47 | 48 |
| OXA E bis | TGACTGGTCAGAAGTTTT | |||
| OXA B | AAGGAGGCTTCCTTGATAA | blaOXA20 | 49 | 38 |
| OXA B bis | TTGGGTGGCAAAGCATTG | |||
| OXA C | As indicated above | blaOXA13 | 49 | 37 |
| OXA O bis | TTATGTGCTTAGTGCATC | |||
| SmeA | CGGTCCTGA GGGGATGAC | blaSme-1 | 53 | 39 |
| SmeB | CGTGATGCTTCCGCAATA | |||
| A1 | GGAATTCCTWTGCTGCGCBCTGCTGCT | blaAmpCc | 60 | 34 |
| A2 | CGGGATCCCTGCCAGTTTTGATAAAA | |||
| B1 | GGAATTCCTCAFCGAGCAGACSCTGTT | blaAmpCc | 60 | 34 |
| B2 | CGGGATCCCCCGCACMTKAYRTAGOTGTGG | |||
| CsA S | GCGCTGATTAATACCGC | blaSed-1 | 49 | Present study |
| CsA AS | GCATCCTGCTGTGGCTGT | |||
| CUM 1 | GGTCGTTTAGGTTAAAAC | blaCUM | 48 | 22 |
| CUM 1 bis | CCAGTGTTTTGGTAAACC | |||
| OXY 1 | GTTCGTGGCGTAAAAC | blaOXY | 49 | 25 |
| OXY 1 rev | TAACACCTCTTTGCGGCT | |||
| RNA-S | AGAGTTTGATCCTGGYTCAG | 16S RNA | 56 | 61 |
| RNA-AS | CTTTACGCCCARTAAWTCCG |
Degenerated oligonucleotide primer designed from the consensus sequence of five class A β-lactamase genes (blaCdiA, blaOXY, blaTOHO-1, blaMEN, and blaSHV).
Group 1 included oxacillinases OXA-5, -7, -11, -14, -16, and -17 and OXA-10 or PSE-2 (16, 18, 19, 21, 28, 29, 52). Group 2 included oxacillinases OXA-2, -3, -15, and -21 (17, 20, 50, 57).
Degenerated oligonucleotide primers were designed from the consensus sequence of the ampC genes of E. coli, E. cloacae, and C. freundii for A1 and A2 and from MOX-1, FOX-1, and ampC of S. marcescens for B1 and B2.
TABLE 2.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| C. sedlakii 2596a | Clinical isolate resistant to β-lactams | Present study |
| E. coli Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC)φ80lacZ ΔM15ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | |
| E. coli K12 | Rifampin | |
| C. koseri CKB | ESBLb | Clinical isolate |
| C. koseri CKC | ESBL | Clinical isolate |
| C. koseri CKD | ESBL | Clinical isolate |
| C. koseri CKE | ESBL | Clinical isolate |
| C. koseri CKG | ESBL | Clinical isolate |
| C. koseri CK1 | Wild type | Clinical isolate |
| C. koseri CK2 | Wild type | Clinical isolate |
| C. koseri CK3 | Wild type | Clinical isolate |
| C. koseri CK4 | Wild type | Clinical isolate |
| C. koseri CK5 | Wild type | Clinical isolate |
| C. koseri CK7 | Wild type | Clinical isolate |
| C. koseri CK10 | Wild type | Clinical isolate |
| C. koseri CIP-82.87 | Wild type (reference strain) | CIP |
| C. koseri CIP-76.3 | Wild type (reference strain) | CIP |
| C. koseri CIP-72.8 | Wild type (reference strain) | CIP |
| C. koseri CIP-72.14 | Penicillinase | CIP |
| C. diversus CIP-82.94 | Wild type (reference strain) | CIP |
| L. malonatica CIP-82.88 | Wild type (reference strain) | CIP |
| C. sedlakii CIP-105037 | Wild type (reference strain) | CIP |
| C. rodentium CIP-104675 | Wild type (reference strain) | CIP |
| Plasmids | ||
| pBC SK+ | Chloramphenicol | Stratagene |
| pBC 2596 | pBC SK+ + 2.78-kb Sau3AI fragment from genomic DNA of C. sedlakii 2596 | Present study |
The strain was deposited in the CIP under the number CIP-106793.
Phenotype corresponding to penicillin and oxyminocephalosporin resistance by production of ESBLs.
Antibiotics, media, and susceptibility testing.
The antibiotics were obtained from the following suppliers: ampicillin, oxacillin, and aztreonam from Bristol-Myers Squibb, Paris, France; chloramphenicol, cefuroxime, and cephalothin from Sigma Chemical Co., St. Louis, Mo.; cefoxitin and imipenem from Merck Sharp & Dohme, Chibret, France; cefotaxime, cefpirome, and rifampin from Hoescht-Marion-Roussel, Paris, France; ceftazidime and nitrocefin from Glaxo, Nanterre, France; ticarcillin, amoxicillin, and clavulanate, from SmithKline Beecham, Paris, France; piperacillin from Lederle, Paris, France; and benzylpenicillin from Sarbach, Suresnes, France.
MICs were determined on Mueller-Hinton (MH) agar by dilution technique (24) with a Steers multiple inoculator and an inoculum of 104 CFU per spot. The plates were incubated at 37°C for 18 h. Brain heart infusion (BHI), Luria-Bertani (LB), and MH media were from Gibco-BRL.
Mating-out assays and plasmid content analysis.
Transfer of of β-lactam resistance to E. coli K-12 was attempted by liquid and solid mating-out assays. The recipient and donor cells were mixed into a ratio of 1:1 or 4:1 and were incubated in BHI with moderate shaking at 37°C for 3 h. After incubation, 200 μl of each mixture was plated out on a Millipore filter disk onto BHI plates and incubated for 18 h at 37°C. Transconjugants were selected on LB agar containing ampicillin (50 or 100 μg/ml) and rifampin (50 or 100 μg/ml). C. sedlakii 2596 was examined for its plasmid DNA content by the procedures of Birnboim (9) and Takahashi and Nagano (56).
DNA amplification.
DNA amplification of β-lactamase genes was carried out with the various specific primers (Eurogentec, Seraing, Belgium) listed in Table 1. The DNA amplifications were performed on 100-μl samples containing DNA (5 μl), deoxynucleoside triphosphate (250 μM), primers (0.4 μM concentrations each), Taq DNA polymerase (1 U), and its buffer. The following cycles were used: 10 min of denaturation at 94°C (1 cycle); 1 min of denaturation at 94°C, 1 min of annealing (see temperatures in Table 1), and 1 min of polymerization at 72°C (35 cycles), followed by 10 min of extension at 72°C. The amplified products were analyzed by electrophoresis of 5-μl aliquots on 1% agarose gels.
Nucleic acid techniques and sequence analysis.
Genomic DNA from C. sedlakii 2596 was extracted as described previously (49). For cloning experiments, the extracted DNA was partially digested with Sau3AI. The fragments were ligated into the dephosphorylated vector pBC SK+ previously digested with BamHI. The ligations were done at 4°C for 16 h with 100 ng of chromosomal DNA, 200 ng of digested plasmid vector pBC SK+, and 1 U of T4 DNA ligase (Amersham). After purification and concentration with the High Pure PCR product purification kit (Boehringer Mannheim), the ligation mixture was transformed by electroporation into Escherichia coli Top10. Transformants resistant to β-lactam antibiotics were selected on LB agar plates supplemented with 50 μg of amoxicillin/ml. Recombinant plasmid DNA was extracted using the rapid procedure of Birnboim (9) from 5-ml aliquots of overnight cultures grown at 37°C in BHI in the presence of amoxicillin (50 μg/ml) and chloramphenicol (50 μg/ml).
The inserted DNA fragment was sequenced on both strands by primer walking using the Taq DyeDeoxy Terminator cycle sequencing kit (Perkin Elmer) and an Applied Biosystems sequencer (PRISM 377). Sequence analysis was performed with the software available on the National Center for Biotechnology Information website. The program ORF Finder was used to determine all the putative open reading frames (ORFs), which were analyzed using the BLASTP program (3). The Sed-1 primary structure was analyzed with the software SignalP and ProtParam, which are available on the ExPASY tools website (Swiss Institute of Bioinformatics [5]).
For the hybridization experiments, genomic DNA was extracted from Citrobacter strains as described previously (49), except that phenol-chloroform was replaced by N-cetyl-N,N,N-trimethylammonium bromide (Merck). DNA was denatured by heating at 100°C for 10 min, and 5 μl of each sample was hybridized onto a nylon membrane (Hybond-N+; Amersham). The DNA was cross-linked on the membrane by 5 min of UV exposure. The hybridization was performed according to the manufacturer's instructions (ECL kit; Amersham) using as a probe an internal fragment of 668 bp obtained by PCR from the blaSed-1 gene.
Preparation of crude extracts and isoelectrofocusing.
Exponentially growing cells were harvested and resuspended in 600 μl of 50 mM phosphate sodium buffer (pH 7.0). The suspensions were disrupted by sonication and the crude extracts were used for β-lactamase detection. Isoelectric focusing was performed with an LKB Multiphor apparatus using polyacrylamide gel plates (Pharmacia Biotech, Saint Quentin en Yvelines, France) at pH 3.5 to 9.5 Gels were focused at 30 W for 90 min at 10°C. β-lactamase activity was revealed by staining the gel with the chromogenic β-lactam nitrocefin (40).
β-Lactamase purification and kinetic assays.
C. sedlakii 2596 was grown overnight at 37°C in 6 liters of BHI broth supplemented with ampicillin (50 μg/ml) and cefoxitin (2 μg/ml) in order to induce β-lactamase production. After centrifugation at 5,000 × g for 10 min at 4°C, the bacterial pellet (30 g) was resuspended in 120 ml of 50 mM Tris (pH 8.0). Bacterial cells were lysed by ultrasonic treatment and the suspension was clarified by centrifugation at 38,000 × g at 4°C. The nucleic acids contained in the supernatant were precipitated by adding spermine (0.2 M) at 4°C, followed by centrifugation for 10 min at 12,000 × g and 60 min at 48,000 × g. The supernatant was then dialyzed overnight against 3 liters of 50 mM Tris (pH 8.0). After an additional centrifugation at 12,000 × g for 30 min, the supernatant was applied onto a 2.5- by 10-cm Q-Sepharose Fast Flow column (Pharmacia Co. Ltd., Uppsala, Sweden) previously equilibrated with the dialysis buffer. β-lactamase activity was detected in the unabsorbed fraction with the chromogenic cephalosporin nitrocefin (40). The active fractions were pooled, dialyzed overnight at 4°C against 2 liters of 40 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [pH 8.0]), and loaded onto a 2.5- by 10-cm S-Sepharose cation exchange column (Pharmacia Co. Ltd.) previously equilibrated with the dialysis buffer. The protein was eluted by a linear gradient of 0 to 1 M NaCl in 40 mM HEPES (pH 8.0). Active fractions were pooled, dialyzed overnight at 4°C against 1 liter of 40 mM Tris (pH 9.0), and then loaded on a Mono Q anion exchange column previously equilibrated with the dialysis buffer. The enzyme was eluted by a linear gradient of 0 to 1 M NaCl in 40 mM Tris (pH 9.0). The active fractions were pooled and dialyzed overnight at 4°C against 1 liter of 40 mM HEPES (pH 8.0) and loaded on a Mono S cation exchange column equilibrated with the dialysis buffer. The β-lactamase activity was eluted in the nonadsorbed fraction. Enzyme purity was assessed by electrophoresis on sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gels. The intensity of the β-lactamase band was measured using a computerized densitometer (Densylab; Bioprobe) from a gel stained with Coomassie blue. The enzyme concentration was determined in reference to a standard bovine serum albumin scale analyzed under the same conditions. β-lactamase activity was renatured by soaking an SDS-polyacrylamide gel in 100 mM Tris-HCl (pH 7.0) for 1 h and was detected by overlaying the gel with nitrocefin (1,000 μg/ml). N-terminal sequences were determined after protein purification using an Applied Biosystems sequencer. The purified protein was stored in 50% glycerol at −20°C.
Kinetic measurements and inhibition of β-lactamase activity.
The kinetic parameters Km and kcat were determined spectrophotometrically at 35°C in 50 mM phosphate buffer (pH 7.0) using an Uvikon 940 spectrophotometer. The absorption coefficients used were those previously described (11). Kinetic parameters were determined by fitting the Henri-Michaelis-Menten equation to the experimental data by using the regression analysis program LEONORA written by Cornish-Bowden (15). The values of kcat and Km were estimated using a nonlinear least-squares regression method with dynamic weights (15).
Enzyme inhibition was studied with benzylpenicillin (100 μM) as the substrate. The different inhibitors, at various concentrations, were preincubated with the enzyme for 5 min at 35°C prior to addition of the substrate. The concentration of inhibitor required to inhibit 50% of the β-lactamase activity (IC50) was determined graphically for clavulanic acid, sulbactam, and cefoxitin.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this report have been deposited in GenBank under accession numbers AF321607 and AF321608.
RESULTS
Antibiotic susceptibility.
Analysis of C. sedlakii 2596 by the conventional disk susceptibility assay indicated that the strain produced an inducible β-lactamase inhibited by clavulanic acid. Indeed, a synergy was detected between clavulanic acid on one hand and aztreonam, cefuroxime, or cefotaxime on the other hand, while an antagonism phenomenon was visible between cefoxitin or imipenem and extended-spectrum cephalosporins like cefotaxime or cefepime (data not shown). Determination of the MICs of β-lactams for strain 2596 showed that it was resistant to penicillins and to early cephalosporins such as cephalothin and cefuroxime, but it remained susceptible to acylureidopenicillins such as piperacillin and to later β-lactams such as cefoxitin, cefotaxime, and aztreonam (Table 3). The MICs of amoxicillin and cephalothin were significantly lowered by clavulanic acid, while that of cefuroxime was increased fourfold when cefoxitin was added at a concentration of 2 μg/ml. Such a phenotype closely resembled that of C. sedlakii CIP-105037, a reference strain from the Collection of the Institut Pasteur (CIP), but it was significantly different from that of the wild-type strain CK1 of C. koseri (Table 3).
TABLE 3.
MICs of β-lactam antibiotics for various strains studied
| β-Lactam | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| C. sedlakii 2596 | E. coli Top10 (pBC2596) | E. coli Top10a | C. sedlakii CIP-105037b | C. koseri CK1c | |
| Amoxicillin | >512 | >512 | 2 | >512 | 64 |
| Amoxicillin + CLAd | 8 | 512 | 2 | 8 | 1 |
| Ticarcillin | >512 | >512 | 1 | 512 | 64 |
| Piperacillin | 8 | 128 | 2 | 2 | 8 |
| Cephalothin | 256 | >512 | 1 | 256 | 2 |
| Cephalothin + CLA | 4 | 1 | 1 | 4 | NDf |
| Cefuroxime | 64 | >512 | 4 | 32 | 4 |
| Cefuroxime + FOXe | 256 | ND | ND | 256 | ND |
| Cefoxitin | 4 | 2 | 2 | 4 | 2 |
| Cefotaxime | 0.25 | 2 | <0.125 | <0.125 | <0.125 |
| Aztreonam | 2 | 16 | <0.125 | 1 | <0.125 |
Reference strain.
Reference strain from CIP.
A wild-type strain.
CLA, clavulanic acid at a fixed concentration of 4 μg/ml.
FOX, cefoxitin at a fixed concentration of 2 μg/ml.
ND, not determined.
Transfer of resistance and cloning of the bla gene from C. sedlakii 2596.
No plasmid was detectable in C. sedlakii 2596, suggesting that the β-lactamase gene was located on the chromosome. Accordingly, repeated mating-out experiments failed to transfer the β-lactam resistance into E. coli. The negative DNA amplification tests obtained with 15 pairs of primers designed to amplify the most frequent class A, C, and D β-lactamases (listed in Table 1) suggested that C. sedlakii 2596 harbored a β-lactamase gene characterized by a nucleotide sequence substantially different from that of the most commonly encountered bla genes.
The genomic DNA from C. sedlakii 2596 was partially digested with the restriction endonuclease Sau3AI and was ligated to the BamHI site of pBC SK+ vector. After electroporation into E. coli Top10, recombinant clones were selected on plates containing amoxicillin (50 μg/ml). Analysis by disk susceptibility assay of E. coli Top10 harboring the recombinant plasmid pBC 2596 revealed an antagonism between cefoxitin and extended-spectrum cephalosporins, as was observed for C. sedlakii 2596 (data not shown). Overall, the β-lactam MICs determined for E. coli(pBC 2596) were higher than those for C. sedlakii 2596, but the resistance profiles of the two strains were similar (Table 3).
Sequence analysis of blaSed-1 and blaSedR.
Restriction analysis of the recombinant plasmid pBC 2596 showed the presence of a 2.78-kb DNA insert. The sequencing of this fragment revealed two ORFs in opposite orientations, SedR and Sed-1 (Fig. 1). The nucleotide sequence of blaSed-1, which is 888 bp long, displayed 82% identity with the chromosomal gene coding for the CdiA β-lactamase from C. koseri and 80% identity with the bla gene from K. oxytoca. The bla Sed-1 gene was found to encode a 31.9-kDa protein comprising 295 amino acid residues. Analysis of the amino acid sequence with the program SignalP suggested that the cleavage site in the precursor protein is located between the first alanine and glutamine residues in the amino acid sequence LHAQATSDVQQ (Fig. 1). This putative site corresponds well to the N-terminal sequence determined experimentally for the mature β-lactamase purified from C. sedlakii 2596, which was QATSDVQQVQKKLAALEKQ. The pI and molecular mass values of 8.86 and 28.6 kDa, respectively, which were predicted from the sequence of the deduced mature protein Sed-1, were in agreement with the values measured by isoelectrofocusing (8.6) and by SDS-PAGE analysis (30 kDa).
FIG. 1.
Nucleotide sequences of blaSed-I and blaSedR from E. coli(pBC2596). The deduced amino acid sequence is indicated in single-letter code above the nucleotide sequence. RBS indicates a potential ribosome-binding site. The Sed-1 signal peptide extends from amino acid 1 to 28, and the postulated cleavage site is indicated by a vertical arrow. The residues conserved in the active site of serine β-lactamases are boxed. Two numbering schemes are indicated: the one in boldface is for the Sed-1 amino acid sequence, and the other one is for the Sed-1 and SedR nucleotide sequences.
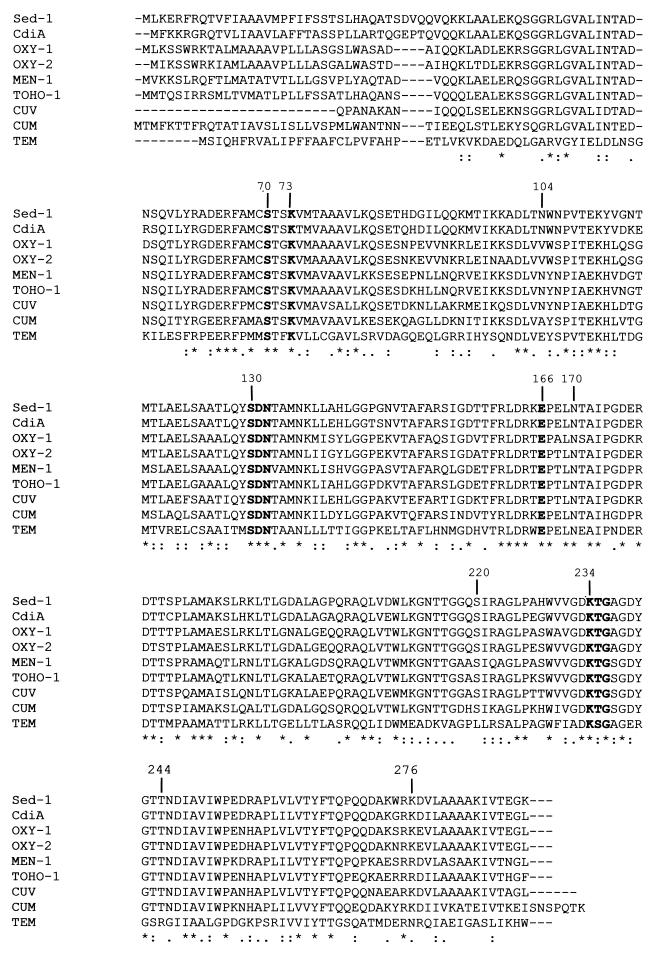
The degree of amino acid sequence identity found between Sed-1 and the TEM and SHV enzymes was rather low (about 40%), while the protein had high identity with the chromosome-encoded β-lactamases of C. koseri (84%), K. oxytoca (74%), Serratia fonticola (67%), P. vulgaris (63%), and the plasmid-mediated enzymes MEN-1 (71%) and TOHO-1 (70%). A multiple amino acid sequence alignment of these class A β-lactamases is shown in Fig. 2. Most of the residues known to be involved in the catalytic mechanism and in substrate binding are conserved in Sed-1, including the consensus sequences 70SXXK73, 130SDN132, 234KTG236, and the highly conserved residue Glu-166 (based on the numbering system of Ambler et al. [4]) (Fig. 2). Other important conserved residues were identified in Sed-1, such as Arg-164, Asn-170, and Asp-179, which are located in the Ω-loop structure. It must be noted here that the sequence of this catalytically important loop is highly conserved in Sed-1, compared to the class A β-lactamases presented in Fig. 2. Regarding the other positions known to contribute significantly to the catalytic activity of class A β-lactamases, we found in Sed-1 a cysteine in position 69, an asparagine in position 104, and an alanine and a glycine in positions 237 and 238, respectively. No arginine was identified in Sed-1 in either position 220 or position 244, but a basic residue (a Lys for Sed-1) is present in position 276, a feature which is shared by the seven class A β-lactamases related to the chromosomal enzyme of K. oxytoca shown in Fig. 2 (OXY-1, OXY-2, CdiA, MEN-1, TOHO-1, CUV, and CUM).
FIG. 2.
Multiple alignment of amino acid sequences for nine class A β-lactamases. Dashes indicate gaps inserted in the alignment; asterisks indicate identical residues. Numbering is according to Ambler (R. P. Ambler et al., Letter, Biochem. J. 276:269–270, 1991). Boldface residues are the highly conserved residues involved in the catalytic site of the protein. The β-lactamases included in the alignment are Sed-1 from C. sedlakii (present study), CdiA from C. diversus (32), OXY-1 and OXY-2 from K. oxytoca (25), MEN-1 from E. coli (8), TOHO-1 from E. coli (30), CUV from S. fonticola (44), CUM from P. vulgaris (22), and TEM from E. coli (54).
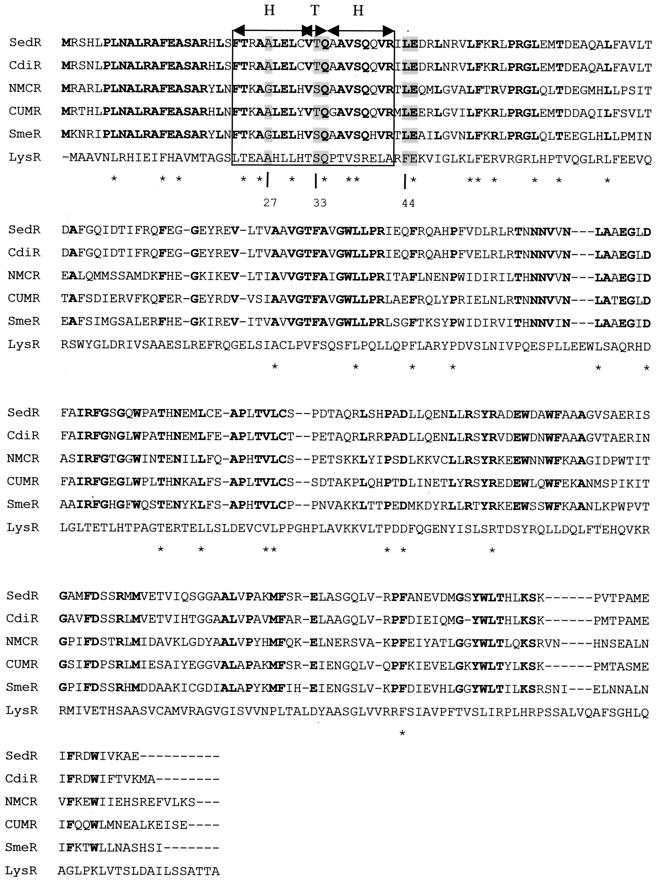
The blaSedR gene found upstream from blaSed-1 is an 861-bp ORF divergently transcribed from blaSed-1 which encodes a 32-kDa protein comprising 286 amino acid residues. This protein displayed an identity of 90% with CdiR, the transcriptional regulator of the class A β-lactamase CdiA from C. koseri, and 68% identity with CumR from P. vulgaris. The identity found with the transcriptional regulator AmpR of class C β-lactamases was lower (47% with the C. freundii AmpR protein). As shown in Fig. 3, the identity is mainly located at the level of the N-terminal part of the sequences, where the helix-turn-helix motif required for binding to the ampR-ampA intercistronic region is found. Regarding the 20 residues involved in this motif, 14 are strictly conserved among the five AmpR sequences of class A β-lactamases shown in Fig. 3.
FIG. 3.
Alignment of LysR-type proteins. The AmpR sequences involved in the class A β-lactamase regulation systems were from C. diversus (CdiR), E. cloacae (NMCR), P. vulgaris (CumR), and Serratia marcescens (SmeR). LysR, the type regulator protein, was from E. coli. The predicted helix-turn-helix domain is boxed. Residues identical in SedR and the AmpR family sequences are indicated in boldface, whereas those identical in SedR and the LysR family sequences are indicated with asterisks. The residues highly conserved in LTTRs are shaded in gray (the amino acid numbering is indicated below the multiple alignment).
Purification and kinetic study of the β-lactamase Sed-1.
After four purification steps, two bands of 30 kDa (major band) and 26 kDa (minor band) were observed on SDS-PAGE analysis of the enzyme preparation. By soaking the polyacrylamide gel in Tris-HCl (pH 7.0), the β-lactamase activity could be renatured and was associated with the 30-kDa band. Isoelectric focusing performed from the partially purified enzyme confirmed the pI of 8.6 initially found for Sed-1 from C. sedlakii 2596.
As shown in Table 4, the highest catalytic activities were measured for benzylpenicillin, ampicillin, cefpirome, cephalothin, and cloxacillin (kcat values between 300 and 165 s−1). Nevertheless, the high Km values found for ampicillin (565 μM) and cefpirome (1,000 μM) reduced the corresponding catalytic efficiencies of these two drugs (kcat/KM = 425 and 215 s−1 · mM−1, respectively) when compared to those of benzylpenicillin (6,650 s−1 · mM−1), cephalothin (6,000 s−1 · mM−1), and cloxacillin (1,270 s−1 · mM−1). Cefuroxime had a relatively low kcat (65 s−1) but a high apparent affinity (KM = 20 μM), resulting in a catalytic efficiency (kcat/KM = 3,250 s−1 · mM−1) higher than that observed for cloxacillin. Ticarcillin, piperacillin, oxyimino-cephalosporins, and aztreonam were hydrolyzed with a lower catalytic efficiency (2 to 625 s−1 · mM−1; Table 4). Imipenem and cefoxitin hydrolysis was not detectable. The IC50s determined with benzylpenicillin as a substrate showed that Sed-1 was well inhibited by clavulanic acid but poorly inhibited by sulbactam (0.065 and 2.5 μM, respectively). These IC50s are comparable to the values of 0.09 μM (for clavulanic acid) and 6.1 μM (for sulbactam) obtained for the TEM-1 β-lactamase (14). Sed-1 is also weakly inhibited by cefoxitin, with an IC50 of 16 μM.
TABLE 4.
Kinetic parameters of various β-lactam antibiotics for the Sed-1 β-lactamase from C. sedlakii 2596
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (s−1 · mM−1) | IC50 (μM) |
|---|---|---|---|---|
| Benzylpenicillin | 300 ± 3 | 45 ± 2 | 6,650 ± 230 | |
| Ampicillin | 240 ± 20 | 565 ± 75 | 425 ± 30 | |
| Ticarcillin | 25 ± 1 | 40 ± 4 | 625 ± 50 | |
| Piperacillin | 20 ± 0.05 | 165 ± 1 | 120 ± 0.04 | |
| Cephalothin | 180 ± 0.3 | 30 ± 2 | 6,000 ± 30 | |
| Cefoxitin | <0.01 | NDb | ND | 16 |
| Cefuroxime | 65 ± 5 | 20 ± 0.3 | 3,250 ± 100 | |
| Cefotaxime | 80 ± 5 | 170 ± 20 | 470 ± 40 | |
| Ceftazidime | 5 ± 0.1 | 2,380 ± 200 | 2 ± 0.1 | |
| Aztreonam | 30 ± 3 | 390 ± 80 | 75 ± 8 | |
| Cefpirome | 215 ± 15 | 1,000 ± 100 | 215 ± 15 | |
| Oxacillin | 85 ± 8 | 340 ± 60 | 255 ± 20 | |
| Cloxacillin | 165 ± 8 | 130 ± 10 | 1,270 ± 40 | |
| Imipenem | <0.01 | ND | ND | |
| Clavulanic acid | 0.065 | |||
| Sulbactam | 2.5 |
Mean ± standard deviation values are indicated.
ND, not determined.
β-Lactamase diversity within the Citrobacter genus.
Twenty strains representing three species (C. koseri, C. sedlakii, and C. rodentium) within the Citrobacter genus were studied. The two reference strains, C. sedlakii CIP-105037 and C. rodentium CIP-104675, were from the CIP. For C. koseri, 12 clinical isolates were studied in addition to three CIP reference strains. All the strains were identified using Api 20E galleries, the identification being confirmed by PCR and sequencing of the hypervariable region in the 16S rRNA. In order to characterize the β-lactamases present in each isolate, the pIs of the β-lactamases produced by each strain were determined by isoelectrofocusing. Genetic characterizations by PCR with primers specific to blaTEM, blaCdiA, and blaSed-1 were also carried out. The results are shown in Table 5.
TABLE 5.
Isoelectric points and results of the hybridization and amplification experiments carried out on the different Citrobacter isolates
| Strain | Phenotype | Isoelectric point(s) | PCR result
|
Hybridization with blaSed-1 probe | ||
|---|---|---|---|---|---|---|
| blaTEM | blaCdiA | blaSed-1 | ||||
| C. sedlakii 2596 | Wild type | 8.6 | − | − | + | + |
| C. sedlakii CIP105037 | Wild type | 8.7 | − | − | + | + |
| C. rodentium CIP104675 | Wild type | 8.7 | − | − | + | + |
| C. koseri CKB | ESBL | 6.8, 6.1, 4.8 | + | − | − | − |
| C. koseri CKC | ESBL | 6.1, 5.7, 4.8 | + | − | − | − |
| C. koseri CKD | ESBL | 6.8, 6.1, 4.8 | + | − | − | − |
| C. koseri CKE | ESBL | 6.8, 6.1, 4.8 | + | − | − | − |
| C. koseri CKG | ESBL | 6.1 | + | − | − | − |
| C. koseri CIP-72.14 | Penicillinase | 5.4 | + | − | − | − |
| C. koseri CK1 | Wild type | 7, 4.7 | − | − | − | − |
| C. koseri CK2 | Wild type | 8.3 | − | − | − | − |
| C. koseri CK3 | Wild type | 5.6, 5.4 | − | − | − | − |
| C. koseri CK4 | Wild type | 5 | − | − | − | − |
| C. koseri CK5 | Wild type | 7 | − | − | − | − |
| C. koseri CK7 | Wild type | 5 | − | − | − | − |
| C. koseri CK10 | Wild type | 8.2, 5.6, 5.3 | − | − | − | − |
| C. koseri CIP-82.87 | Wild type | 5.7 | − | − | − | − |
| C. koseri CIP-76.3 | Wild type | 5 | − | − | − | − |
| C. koseri CIP-82.94 | Wild type | 7.4 | − | − | − | − |
| L. malonatica CIP82.88 | Wild type | 8.1, 5, 4.5 | − | − | − | − |
The clinical strain 2596 and the reference strain CIP-105037 of C. sedlakii displayed very similar phenotypes: they were resistant to aminopenicillins, carboxypenicillins, and early cephalosporins but remained susceptible to piperacillin (Table 3). An antagonism between imipenem and extended-spectrum cephalosporins could be observed for both strains, indicating inducible β-lactamase production in C. sedlakii. The hybridization experiment carried out with a blaSed-1-specific probe gave a positive result for both strains. Accordingly, sequencing of the DNA products amplified with primers specific for blaSed-1 showed that C. sedlakii CIP-105037 harbored a β-lactamase gene, the sequence of which is very similar to that of blaSed-1 (98% identity for 640 bp sequenced). The differences found at the amino acid level between the two β-lactamases accounted for the difference in pI values observed for C. sedlakii 2596 (8.6) and for C. sedlakii CIP-105037 (8.7).
The C. rodentium CIP-104675 strain produced a β-lactamase characterized by a pI value of 8.7 and yielded a positive hybridization signal with the internal probe specific for blaSed-1. The DNA fragment of 640 bp obtained by PCR from C. rodentium CIP-104675 was sequenced and was found to share 80% identity with blaSed-1.
Regarding C. koseri, three different phenotypes of resistance to β-lactams (wild type, penicillinase, and extended-spectrum β-lactamase [ESBL] phenotypes) could be identified among the 17 strains included in the present study. As shown in Table 5, very heterogeneous pI values were found for these different strains of C. koseri which produced different combinations of noninducible β-lactamases, each strain being characterized by one, two, or three distinct pI values. PCR amplification and sequence analysis were made in an attempt to identify the corresponding β-lactamase genes. Amplification with the primers specific for blaTEM confirmed the presence of this gene in the six strains showing a penicillinase or an ESBL phenotype. By contrast, the amplifications with the blaCdiA and blaSed-1 primers remained negative in all the strains tested, and none of the C. koseri strains hybridized with the blaSed-1 internal probe.
DISCUSSION
C. sedlakii 2596, which produces the chromosomally encoded class A β-lactamase Sed-1, was resistant to aminopenicillins, carboxypenicillins, and early cephalosporins but not to acylureidopenicillins such as piperacillin. This resistance profile can be readily explained by the hydrolytic properties of Sed-1 (high catalytic efficiency against benzylpenicillin, cloxacillin, and early cephalosporins such as cephalothin and cefuroxime, but low efficiency for piperacillin), which closely resemble those of the cefuroximases, such as the chromosome-encoded β-lactamase CUM from P. vulgaris or the plasmid-encoded β-lactamases FEC-1, FPM-1, and FUR from E. coli, Proteus mirabilis, and Klebsiella pneumoniae, respectively (36, 45, 58, 59). Cefotaxime, cefpirome, and aztreonam were also hydrolyzed by Sed-1 but with low catalytic efficiencies and moderate apparent affinities (Table 4). Clavulanic acid and sulbactam inhibited the β-lactamase activity of Sed-1 with IC50s similar to those encountered among class A β-lactamases highly susceptible to these inhibitors, such as TEM-1.
With the aim of determining whether the unusual substrate profile of Sed-1, including its significant activities with cefuroxime, cefotaxime, and cefpirome, could be related to the presence of specific amino acid residues in the sequence of the enzyme, we compared its amino acid sequence with those from other class A β-lactamases. A high percentage of identity was found between Sed-1 and the chromosome-encoded β-lactamases of C. koseri (84%), K. oxytoca (74%), S. fonticola (67%), P. vulgaris (63%), and the plasmid-mediated enzymes MEN-1 or CTX-M (71%) and TOHO-1 (70%), suggesting that these class A enzymes may be derived from a common ancestor. All the conserved residues considered to be important for catalysis in class A β-lactamases (70SXXK73, 130SDN132, 234KTG236, and E166) were found in Sed-1, as was the Ω-loop, an important structural element including the amino acid residues 161 to 179 in class A enzymes.
Regarding positions 104, 164, 179, 205, 237, 238, and 240, which are involved in the extended substrate specificity of the mutants of the class A β-lactamases TEM and SHV (35), it is striking that an Asn was found at position 104 in Sed-1. Indeed, according to Petit et al. (46), residue 104 contributes to the precise positioning of the 130SDN132 loop, which is a crucial structural and catalytic element for the binding and hydrolysis of β-lactams. Accordingly, amino acid modifications are found at this position in a large number of ESBLs derived from TEM-1 and TEM-2. Moreover, an asparagine residue is also found at position 104 in the nine CTX-M variants described to date and also in related enzymes such as TOHO-1 from E. coli and CUV from S. fonticola (Fig. 2), which, like Sed-1, are enzymes that efficiently hydrolyze cefuroxime and cefotaxime but not ceftazidime (8, 10, 12, 26). Note here that the residue found at position ABL 237 in Sed-1 is an alanine, whereas a serine has been found at this position in CUV, CUM, MEN-1 (CTX-M), and TOHO-1. In TEM-type ESBLs, as in other class A enzymes with an extended spectrum of activity, the presence of a serine residue in position 237 has been clearly shown to increase the level of β-lactamase activity against expanded-spectrum cephalosporins. Consequently, the presence of Ala 237 in Sed-1 could explain the relatively low level of activity the enzyme displays against cefotaxime, compared to the activities reported for the related enzymes MEN-1 (CTX-M), CUV, CUM, and TOHO-1, which all confer a high level of resistance to this drug (7, 22, 30, 44).
Other amino acid residues of interest, which are not strictly conserved in class A βlactamases, were found in Sed-1. Among them, there is a cysteine at position 69 in Sed-1, which neighbors the active-site serine found at position 70. Cys 69 is present in all the β-lactamases belonging to the K. oxytoca subgroup (Fig. 2), so that this residue could have a significant role in the substrate profile of these enzymes. Also note that there is no arginine residue at positions ABL 220 or 244 in Sed-1. Matagne et al. (35) have suggested that a basic residue is found in position ABL 276 when Arg is absent at positions 220 and 244. This is the case in Sed-1, which presents a Lysine at position 276 that is highly conserved in the cefuroximases group and perfectly aligned with the corresponding arginine residue found in MEN-1 and TOHO-1 (Fig. 2).
The negative results obtained from the conjugation and plasmid extraction experiments strongly suggested a chromosomal location for the blaSed-1 gene. The presence, upstream from blaSed-1, of a gene coding for a transcriptional regulator belonging to the LysR family reinforces this hypothesis. In C. sedlakii, the regulator protein SedR induces the production of Sed-1 when the bacteria are grown in the presence of an inducer β-lactam antibiotic such as cefoxitin and imipenem, as illustrated by the increase of the MIC of cefuroxime observed in the presence of cefoxitin for C. sedlakii 2596 (Table 3). Regarding its amino acid sequence, SedR is more related to the regulators of the class A β-lactamases from C. koseri and P. vulgaris (90% identity with CdiR and 68% identity with CumR) than to the regulators of the class C β-lactamases (47% identity with the C. freundii AmpR protein), suggesting that the relationships among LysR proteins are related to the type of β-lactamase they regulate. The similarities are the highest around the N-terminal region, where the consensus sequence for the helix-turn-helix motif is found. In this region, the residues conserved among the LTTRs which are Ala(Gly)-27, Ser(Thr)-33, Gln-34, Pro-35, Phe(Leu)-44, and Glu-45 (51), are all present in SedR, except for Pro-35.
β-Lactamase distribution within the Citrobacter genus is complex. C. freundii produces an inducible chromosome-encoded class C β-lactamase (62), whereas C. sedlakii, C. rodentium, and C. koseri produce class A β-lactamases (27, 41, 47, also, the present study). On the basis of the sequencing of the DNA fragments amplified in this study, C. sedlakii CIP-105037 and C. rodentium CIP-104675 harbor β-lactamase genes sharing 98 and 80% of identity to blaSed-1, respectively. The expression of blaSed-1, and also that of the closely related gene found in C. sedlakii CIP-105037, is inducible, whereas that of the blaSed-1-like gene found in C. rodentium CIP-104675 seems to be constitutive. Regarding C. koseri, DNA amplification experiments with blaSed-1-specific primers showed that this β-lactamase gene was not present in the 17 C. koseri isolates tested in this study. Moreover, the results obtained by PCR were confirmed by dot-blot hybridization with a blaSed-1-specific probe, strongly supporting the idea that this gene is not present in C. koseri. More surprisingly, no DNA amplification could be obtained using the set of primers designed to amplify specifically the blaCdiA gene initially reported in the strain ULA27 of C. koseri, suggesting that the latter gene is not ubiquitous in this species or, alternatively, that the strain used for the initial characterization of blaCdiA was not a C. koseri strain. Such a hypothesis is confirmed by the fact that the constitutive resistance phenotypes observed for the clinical isolates of C. koseri included in the present study were all clearly different from the inducible resistance phenotype initially reported for the strain ULA27 of C. koseri, which produces CdiA under control of the regulator protein CdiR (33). Moreover, we have characterized 13 distinct isoelectric point values among the 17 C. koseri strains studied, with one to three different pI values detected per strain. The strains with a wild-type phenotype had pI values varying from 4.7 to 8.2. Such values are in agreement with those found in the literature, which vary from 4.8 to 9.5 (27, 47). For the strains displaying an ESBL profile, three pI values of 6.8, 6.1, and 4.8 were found, one corresponding to the pI value of a TEM variant, the identification of which was confirmed by PCR with blaTEM primers. Therefore, it is likely that the β-lactamase genes found in C. koseri encompass a series of genes that have significantly diverged, as previously described by Jones et al. (32), and which are characterized by a nucleotide sequence different from that of blaSed-1 and blaCdiA. Further studies on the genetic characterization of these genes must be done to elucidate the origin of the bla genes in C. koseri.
ACKNOWLEDGMENTS
We thank Jean-Pierre Lecaer for the N-terminal sequencing and Tania Rybkine, Murielle Renard, and Jean-Pierre Lagarde for their excellent technical assistance. We are also grateful to Ekkehard Collatz for constructive comments and helpful discussions. We acknowledge Jacqueline Nguyen for providing bacterial strains.
This work was supported by the Institut National de la Santé et de la Recherche Médicale (grants CRI 950601 and EMI 0004) and by the Ministère de la Recherche (grant UPRES JE-2227). Stéphanie Petrella was a fellow of the Ministère de la Recherche.
REFERENCES
- 1.Aldova E, Schindler J, Nemec A, Sourek J, Urbaskova P. Biochemical indicators of Citrobacter sedlakii. Epidemiol Mikrobiol Imunol. 1995;44:57–64. [PubMed] [Google Scholar]
- 2.Aldova E, Schindler J, Sourek J, Nemec A, Urbaskova P. Detection and identification of Citrobacter sedlakii in the Czech Republic. Zentbl Bakteriol. 1997;285:389–396. doi: 10.1016/s0934-8840(97)80005-9. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Amicosante G, Frere J M, Franceschini N, Oratore A, Strom R. Some molecular properties of Citrobacter diversus beta-lactamases. J Chemother. 1991;3:83–85. doi: 10.1080/1120009x.1991.11739070. [DOI] [PubMed] [Google Scholar]
- 5.Appel R D, Bairoch A, Hochstrasser D F. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 6.Barthelemy M, Peduzzi J, Labia R. Complete amino acid sequence of p453-plasmid-mediated PIT-2 beta-lactamase (SHV-1) Biochem J. 1988;251:73–79. doi: 10.1042/bj2510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 8.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birnboim H C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet R, Sampaio J L, Labia R, De Champs C, Sirot D, Chanal C, Sirot J. A novel CTX-M beta-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob Agents Chemother. 2000;44:1936–1942. doi: 10.1128/aac.44.7.1936-1942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouthors A T, Delettre J, Mugnier P, Jarlier V, Sougakoff W. Site-directed mutagenesis of residues 164, 170, 171, 179, 220, 237 and 242 in PER-1 beta-lactamase hydrolysing expanded-spectrum cephalosporins. Protein Eng. 1999;12:313–318. doi: 10.1093/protein/12.4.313. [DOI] [PubMed] [Google Scholar]
- 12.Bradford P A, Yang Y, Sahm D, Grope I, Gardovska D, Storch G. CTX-M-5, a novel cefotaxime-hydrolyzing beta-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob Agents Chemother. 1998;42:1980–1984. doi: 10.1128/aac.42.8.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner D J, Grimont P A D, Steigerwalt A G, Fanning G R, Ageron E, Riddle C F. Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmeri sp. nov., Citrobacter youngae sp. nov., Citrobacter braakii sp. nov., Citrobacter werkmanii sp. nov., Citrobacter sedlakii sp. nov., and three unnamed Citrobacter genomospecies. Int J Syst Bacteriol. 1993;43:645–658. doi: 10.1099/00207713-43-4-645. [DOI] [PubMed] [Google Scholar]
- 14.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornish-Bowden A. Analysis of enzyme kinetic data. New York, N.Y: Oxford University Press; 1995. [Google Scholar]
- 16.Couture F, Lachapelle J, Levesque R C. Phylogeny of LCR-1 and OXA-5 with class A and class D beta-lactamases. Mol Microbiol. 1992;6:1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 17.Dale J W, Godwin D, Mossakowska D, Stephenson P, Wall S. Sequence of the OXA2 beta-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 1985;191:39–44. doi: 10.1016/0014-5793(85)80989-3. [DOI] [PubMed] [Google Scholar]
- 18.Danel F, Hall L M, Duke B, Gur D, Livermore D M. OXA-17, a further extended-spectrum variant of OXA-10 beta-lactamase, isolated from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1362–1366. doi: 10.1128/aac.43.6.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danel F, Hall L M, Gur D, Livermore D M. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1881–1884. doi: 10.1128/aac.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danel F, Hall L M, Gur D, Livermore D M. OXA-15, an extended-spectrum variant of OXA-2 beta-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother. 1997;41:785–790. doi: 10.1128/aac.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danel F, Hall L M, Gur D, Livermore D M. OXA-16, a further extended-spectrum variant of OXA-10 beta-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother. 1998;42:3117–3122. doi: 10.1128/aac.42.12.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datz M, Joris B, Azab E A, Galleni M, Van Beeumen J, Frere J-M, Martin H H. A common system controls the induction of very different genes. The class-A beta-lactamase of Proteus vulgaris and the enterobacterial class-C beta-lactamase. Eur J Biochem. 1994;226:149–157. doi: 10.1111/j.1432-1033.1994.tb20036.x. [DOI] [PubMed] [Google Scholar]
- 23.Dyer J, Hayani K C, Janda W M, Schreckenberger P C. Citrobacter sedlakii meningitis and brain abscess in a premature infant. J Clin Microbiol. 1997;35:2686–2688. doi: 10.1128/jcm.35.10.2686-2688.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ericsson H M, Sherris J C. Antibiotic sensitivity testing. Report of an international collaborative study. Acta Pathol Microbiol Scand Suppl B. 1971;217:1. [PubMed] [Google Scholar]
- 25.Fournier B, Roy P H, Lagrange P H, Philippon A. Chromosomal β-lactamase genes of Klebsiella oxytoca are divided into two main groups, blaOXY-1 and blaOXY-2. Antimicrob Agents Chemother. 1996;40:454–459. doi: 10.1128/aac.40.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gniadkowski M, Schneider I, Palucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing beta-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42:827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gootz T D, Jackson D B, Sherris J C. Development of resistance to cephalosporins in clinical strains of Citrobacter spp. Antimicrob Agents Chemother. 1984;25:591–595. doi: 10.1128/aac.25.5.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall L M, Livermore D M, Gur D, Akova M, Akalin H E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huovinen P, Huovinen S, Jacoby G A. Sequence of PSE-2 beta-lactamase. Antimicrob Agents Chemother. 1988;32:134–136. doi: 10.1128/aac.32.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A beta-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jepsen O B, Mortensen I, Rosdahl V T. Screening methods for detection of beta-lactamases in gram-negative bacteria. J Antimicrob Chemother. 1979;5:383–389. doi: 10.1093/jac/5.4.383. [DOI] [PubMed] [Google Scholar]
- 32.Jones M E, Avison M B, Damdinsuren E, MacGowan A P, Bennett P M. Heterogeneity at the beta-lactamase structural gene ampC amongst Citrobacter spp. assessed by polymerase chain reaction analysis: potential for typing at a molecular level. J Med Microbiol. 1994;41:209–214. doi: 10.1099/00222615-41-3-209. [DOI] [PubMed] [Google Scholar]
- 33.Jones M E, Bennett P M. Inducible expression of the chromosomal CdiA from Citrobacter diversus NF85, encoding an ambler class A beta-lactamase, is under similar genetic control to the chromosomal AmpC, encoding an ambler class C enzyme, from Citrobacter freundii OS60. Microb Drug Resist. 1995;1:285–291. doi: 10.1089/mdr.1995.1.285. [DOI] [PubMed] [Google Scholar]
- 34.Marchese A, Arlet G, Schito G C, Lagrange P H, Philippon A. Characterization of FOX-3, an AmpC-type plasmid-mediated beta-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob Agents Chemother. 1998;42:464–467. doi: 10.1128/aac.42.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matagne A, Lamotte-Brasseur J, Frère J M. Catalytic properties of class A beta-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto Y, Ikeda F, Kamimura T, Yokota Y, Mine Y. Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob Agents Chemother. 1988;32:1243–1246. doi: 10.1128/aac.32.8.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mugnier P, Podglajen I, Goldstein F W, Collatz E. Carbapenems as inhibitors of OXA-13, a novel, integron-encoded beta-lactamase in Pseudomonas aeruginosa. Microbiology. 1998;144(Pt. 4):1021–1031. doi: 10.1099/00221287-144-4-1021. [DOI] [PubMed] [Google Scholar]
- 38.Naas T, Sougakoff W, Casetta A, Nordmann P. Molecular characterization of OXA-20, a novel class D beta-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:2074–2083. doi: 10.1128/aac.42.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naas T, Vandel L, Sougakoff W, Livermore D M, Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob Agents Chemother. 1994;38:1262–1270. doi: 10.1128/aac.38.6.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Callaghan C H, Morris A, Kirby S M, Shingler A H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliva B, Marinucei M C, Amicosante G, Oratore A, Bennett P M. Citrobacter diversus ULA27 produces two forms of a chromosomal beta-lactamase. J Antimicrob Chemother. 1987;20:23–35. doi: 10.1093/jac/20.1.23. [DOI] [PubMed] [Google Scholar]
- 42.Oliva B, Segatore B, Amicosante G, Franceschini N, Oratore A, Bennett P M. Broad spectrum beta-lactamases of Citrobacter diversus. J Antimicrob Chemother. 1990;25:335–341. doi: 10.1093/jac/25.3.335. [DOI] [PubMed] [Google Scholar]
- 43.Park C H, Martin E A, White E L. Isolation of a nonpathogenic strain of Citrobacter sedlakii which express Escherichia coli O157 antigen. J Clin Microbiol. 1998;36:1408–1409. doi: 10.1128/jcm.36.5.1408-1409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peduzzi J, Farzaneh S, Reynaud A, Barthelemy M, Labia R. Characterization and amino acid sequence analysis of a new oxyimino cephalosporin-hydrolyzing class A beta-lactamase from Serratia fonticola CUV. Biochim Biophys Acta. 1997;1341:58–70. doi: 10.1016/s0167-4838(97)00020-4. [DOI] [PubMed] [Google Scholar]
- 45.Peduzzi J, Reynaud A, Baron P, Barthelemy M, Labia R. Chromosomally encoded cephalosporin-hydrolyzing beta-lactamase of Proteus vulgaris RO104 belongs to Ambler's class A. Biochim Biophys Acta. 1994;1207:31–39. doi: 10.1016/0167-4838(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 46.Petit A, Maveyraud L, Lenfant F, Samama J P, Labia R, Masson J M. Multiple substitutions at position 104 of beta-lactamase TEM-1: assessing the role of this residue in substrate specificity. Biochem J. 1995;305(Pt. 1):33–40. doi: 10.1042/bj3050033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philippon A, Paul G, Barthelemy M, Labia R, Nevot P. Properties of the beta-lactamase (penicillinase) produced by Levinea malonatica. FEMS Microbiol Lett. 1980;8:191–194. [Google Scholar]
- 48.Philippon L N, Naas T, Bouthors A T, Barakett V, Nordmann P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum beta-lactamase. from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sanschagrin F, Couture F, Levesque R C. Primary structure of OXA-3 and phylogeny of oxacillin-hydrolyzing class D beta-lactamases. Antimicrob Agents Chemother. 1995;39:887–893. doi: 10.1128/aac.39.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 52.Scoulica E, Aransay A, Tselentis Y. Molecular characterization of the OXA-7 beta-lactamase gene. Antimicrob Agents Chemother. 1995;39:1379–1382. doi: 10.1128/aac.39.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson I N, Plested S J. The origin and properties of beta-lactamase. satellite bands seen in isoelectric focusing. J Antimicrob Chemother. 1983;12:127–131. doi: 10.1093/jac/12.2.127. [DOI] [PubMed] [Google Scholar]
- 54.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sykes R B, Matthew M. The beta-lactamases of gram-negative bacteria and their role in resistance to beta-lactam antibiotics. J Antimicrob Chemother. 1976;2:115–157. doi: 10.1093/jac/2.2.115. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi S, Nagano Y. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J Clin Microbiol. 1984;20:608–613. doi: 10.1128/jcm.20.4.608-613.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vila J, Navia M, Ruiz J, Casals C. Cloning and nucleotide sequence analysis of a gene encoding an OXA-derived beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 1997;41:2757–2759. doi: 10.1128/aac.41.12.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vuye A, Verschraegen G, Claeys G. Plasmid-mediated beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli resistant to ceftazidime. Antimicrob Agents Chemother. 1989;33:757–761. doi: 10.1128/aac.33.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe Y, Yokota T, Higashi Y, Wakai Y, Mine Y. In vitro and in vivo transferrable beta-lactam resistance due to a new plasmid-mediated oxyiminocephalosporinase from a clinical isolate of Proteus mirabilis. Microbiol Immunol. 1991;35:87–97. doi: 10.1111/j.1348-0421.1991.tb01537.x. [DOI] [PubMed] [Google Scholar]
- 60.Werkman C M, Gillen G F. Bacteria producing trimethyleneglycol. J Bacteriol. 1932;23:167–182. doi: 10.1128/jb.23.2.167-182.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilbrink B, van der Heijden I M, Schouls L M, van Embden J D, Hazes J M, Breedveld F C, Tak P P. Detection of bacterial DNA in joint samples from patients with undifferentiated arthritis and reactive arthritis, using polymerase chain reaction with universal 16S ribosomal RNA primers. Arthritis Rheum. 1998;41:535–543. doi: 10.1002/1529-0131(199803)41:3<535::AID-ART20>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto T, Murayama S Y, Sawai T. Cloning and expression of the gene(s) for cephalosporinase production of Citrobacter freundii. Mol Gen Genet. 1983;190:85–91. doi: 10.1007/BF00330328. [DOI] [PubMed] [Google Scholar]