Abstract
As an anaerobe, Clostridioides difficile relies on the formation of a dormant spore for survival outside of the mammalian host’s gastrointestinal tract. The spore is recalcitrant to desiccation, numerous disinfectants, UV light, and antibiotics, permitting long-term survival against environmental insults and efficient transmission from host to host. Although the morphological stages of spore formation are similar between C. difficile and other well-studied endospore-forming bacteria, the C. difficile genome does not appear to encode many of the known, conserved regulatory factors that are necessary to initiate sporulation in other spore-forming bacteria. The absence of early sporulation-specific orthologs suggests that C. difficile has evolved to control sporulation initiation in response to its unique and specific ecological niche and environmental cues within the host. Here, we review our current understanding and highlight the recent discoveries that have begun to unravel the regulatory pathways and molecular mechanisms by which C. difficile induces spore formation.
Keywords: Clostridium difficile, Clostridioides difficile, sporulation, spore, sigma factor, transcriptional regulator, regulation, anaerobe
Graphical Abstract
INTRODUCTION
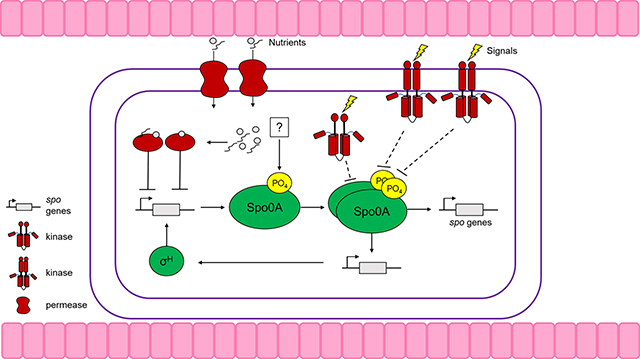
Clostridioides difficile is an anaerobic, Gram-positive, spore-forming bacterium that is a foremost cause of antibiotic-associated diarrhea. C. difficile infection (CDI) symptoms range from mild diarrhea and abdominal distress to life-threatening pseudomembranous colitis. Antibiotic usage causes gut dysbiosis by altering the gut microbiota and sensitizes individuals to C. difficile colonization. A key aspect of C. difficile pathogenesis is its ability to form spores, as they are critical for the infection cycle and also resistant to antibiotics, environmental insults, and disinfectants. While much of the general sporulation pathway in C. difficile is conserved with other Firmicutes, the molecular mechanisms controlling the initiation of sporulation are not conserved and remain poorly understood. This review summarizes our current understanding of regulation of C. difficile sporulation initiation mechanisms and outlines the identified factors that contribute to initiation through the activation of the conserved master regulator of sporulation, Spo0A. Further, we provide a summary of the nutritional and environmental regulations that promote or repress spore formation, as well as an overview of recently characterized factors that contribute to sporulation regulation through undiscovered mechanisms.
Positive Regulators of Sporulation Initiation
Spo0A
Spo0A is the master transcriptional regulator of sporulation in all endospore-forming bacteria [1]. As observed in other spore formers, Spo0A is essential for sporulation in C. difficile. A spo0A null mutant cannot initiate sporulation and was unable to transmit infection in a mouse model due to the absence of spores [2]. In addition to regulating sporulation in C. difficile, Spo0A impacts a myriad of cellular processes, including motility, protein transport, metabolism, cell envelope, and global gene regulation [3].
Once activated by phosphorylation, Spo0A binds to DNA at conserved binding sites known as Spo0A-boxes and induce the expression of genes required for initiating sporulation, including sigH and the sigma factors for early spore development, sigF and sigE [4]. In B. subtilis, Spo0A activity is modulated through a phosphorelay composed of a series of phosphotransfer proteins and accessory phosphatases [5]. Spo0A activation is notably different amongst the Clostridia, with most clostridial species lacking apparent orthologs to the phosphorelay kinases [1]. While C. difficile lacks orthologs to many of the activating proteins of Spo0A in B. subtilis, many of the functional and protein-interaction residues of B.s. Spo0A are conserved in C.d. Spo0A (DiCandia et al., unpublished). This suggests that while the phosphorelay that controls Spo0A is not conserved in C. difficile, there may be factors that perform similar functions to control Spo0A activity. The absence of an obvious phosphorelay in C. difficile led to the hypothesis that Spo0A could be directly activated by orphan sensor histidine kinases in the Clostridia [6]. Further contributing to this view, multiple clostridial species directly phosphorylate and dephosphorylate Spo0A via orphan histidine kinases (i.e., kinases that are not encoded with cognate response regulators) [7–9]. Three predicted orphan histidine kinases, PtpA (CD1492), PtpB (CD2492), and PtpC (CD1579), have been implicated in regulating early sporulation events in C. difficile [10, 11*, 12]. PtpA, PtpB, and PtpC repress spore production, as evidenced by the hypersporuiation pnenotype of their respective null mutants [11*–13]. The sporulation-repressing activities of PtpA, PtpB, and PtpC strongly suggest they do not function as Spo0A kinases, thus, the activating kinase for Spo0A in C. difficile remains unidentified.
SigH
SigH is an alternative sigma factor and a key regulator of transition phase and sporulation initiation in C. difficile and other firmicutes. SigH regulates over 700 genes involved in sporulation, cellular division, motility, virulence, and metabolism [14]. As the primary transition phase sigma factor, SigH allows C. difficile to rewire metabolism to adapt to limited availability of nutrients. SigH regulates hundreds of genes involved in energy production, metabolite transport, amino acid synthesis, and carbon fixation, therein linking metabolism with the initiation of sporulation. SigH-RNA polymerase holoenzyme transcribes several genes known to impact sporulation initiation, including spo0A, the phosphotransfer protein PtpB, and the spo0J, soj, and spoIIP genes involved in chromosomal segregation and prespore engulfment. SigH is required for both the initiation and progression of spore formation, and consequently, a sigH mutant is asporogenic. The transcription of sigH and spo0A are intertwined, with both factors promoting positive feedback expression of each other [4,14]. In addition, SigH positively regulates genes involved in the later stages of sporulation, including the spoIIIA operon, spolllD, spolVA, spoVD, spoVE, spoVG, and spoVS [14].
RstA
Another regulator that modulates Spo0A activity in C. difficile is RstA, a highly conserved and multifunctional protein [15]. RstA shares some sequence similarity to the Bacillus genus’ Rap phosphatases, which directly dephosphorylate Spo0F to inhibit the transfer of phosphate to Spo0A and impede sporulation. RstA belongs to the RRNPP protein family. RRNPP proteins possess an N-terminal DNA-binding domain and/or protein-binding domain, followed by multiple C-terminal tetratricopeptide repeat (TPR) domains that encompass a small quorum-sensing-binding domain. RstA positively influences sporulation and directly represses toxin production and motility through two distinct domains, indicating that RstA employs different molecular mechanisms to regulate these processes [16**]. A null mutation in rstA reduces sporulation frequency by ~20-fold and decreases Spo0A phosphorylation through an unknown mechanism [15,17]. RstA does not appear to directly bind Spo0A, but is hypothesized to interact with PtpA and PtpB to influence Spo0A phosphorylation [13]. Interestingly, there are some strain-dependent effects on RstA regulation between the historical 630Δerm and the epidemic R20291 strains. RstA exhibited stronger regulation of sporulation and toxin production in R02921 compared to 630Δerm, and surprisingly, the loss of rstA did not affect R20291 motility [17].
The DNA-binding and/or protein-binding activities of RRNPP proteins are controlled by the interaction of quorum-sensing peptides with the C-terminus of the protein [18]. It is unclear whether RstA is regulated by quorum-sensing peptides. But, evidence suggests that RstA activity is controlled by a cofactor, as purified RstA is unable to bind DNA and substitution of the C-terminal quorum-sensing-binding domain with other species’ RstA orthologs abolishes RstA DNA-binding activity [17]. Determining how RstA functions to promote C. difficile sporulation may uncover additional sporulation factors and environmental cues that regulate Spo0A phosphorylation and activation.
Negative Regulators of Sporulation Initiation
Spo0E
The Spo0E class of proteins are a family of small aspartyl-phosphate phosphatases that dephosphorylate and inactivate Spo0A in B. subtilis, but their functions are poorly understood in C. difficile or other anaerobes [19,20]. Comparative genomics identified putative Spo0E orthologs in several Clostridia species by probing candidate genomes for the “SQELD” phosphatase motif identified in B. subtilis [21, 22**, 23]. The clostridial Spo0E-like proteins cluster separately from aerobic Spo0E orthologs with no conserved synteny between anaerobic and aerobic Spo0E clusters [22], suggesting that clostridial Spo0E proteins have evolved independently of the Bacillus Spo0E orthologs.
C. difficile encodes at least one putative Spo0E ortholog (CD3271) that contains a loosely conserved functional motif (SKKID). Recent work reveals that a C. difficile spo0E mutant exhibits increased sporulation, similar to observations in B. subtilis (DiCandia et al., unpublished). In addition, co-immunoprecipitation of recombinant Spo0A revealed that Spo0E co-purifies with Spo0A, suggesting that Spo0E in C. difficile directly dephosphorylates Spo0A to negatively regulate sporulation initiation (DiCandia et al., unpublished). However, the C. difficile spo0E mutant has additional phenotypes that were not observed in B. subtilis, including hypermotility, increased toxin expression, and mucoid colony morphology, which are not associated with Spo0A regulation. These results suggest that the Spo0E of clostridial species have evolved additional regulatory functions outside of the sporulation pathway, though the additional mechanisms remain to be determined.
PtpA and PtpB
In other sporulating species, phosphorylation of Spo0A occurs through a phosphorelay initiated by the activation of orphan histidine kinases [1,8,24,25]. C. difficile encodes five predicted orphan histidine kinases; PtpA (CD1492), PtpB (CD2492), PtpC (CD1579), CprK (CD1352), and CD1949. CprK and CD1949 have no involvement in sporulation initiation [13,26,27]. PtpA and PtpB repress sporulation initiation, though it is not known whether PtpA or PtpB control Spo0A phosphorylation directly, as observed in other Clostridia [7,8]. Deletion of ptpA increases sporulation frequency ~2.4-fold (strain 630Δerm), suggesting that PtpA represses sporulation initiation. The hypersporulation phenotype of the ptpA mutant suggests that PtpA does not phosphorylate Spo0A, but instead acts as a phosphatase or an indirect regulator of Spo0A function. The ptpA mutant also produces less toxin and is less virulent than wild-type, likely due to reduced expression of sigD, which induces toxin production, or through reciprocal regulation with RstA [11**].
Results from an early examination of PtpB suggested that this predicted kinase promotes sporulation, since disruption of ptpB appeared to reduce sporulation. However, that study did not complement the mutation, and the assay used to examine sporulation was unconventional and did not include important controls [10]. Further examination of a ptpB mutant revealed a hypersporulation phenotype, similar to that of a ptpA mutant, suggesting that PtpB is also a negative regulator of sporulation initiation [12]. Additionally, a ptpA ptpB double mutant results in the same sporulation phenotype as the single mutants, suggesting both proteins function together in the same regulatory pathway [12]. Surprisingly, the conserved histidine residue necessary for phosphate transfer is required for PtpA function, but not for PtpB function, suggesting that although these proteins function in the same pathway, they may have divergent roles [13]. Finally, the inverse pattern of gene expression and phenotypes for the ptpA, ptpB, and rstA mutants suggests that PtpA, PtpB, and RstA function within the same regulatory pathway to influence sporulation in C. difficile [11**]. Although PtpB and PtpA clearly play a role in sporulation regulation, their direct targets and phosphotransfer functions remain to be determined.
PtpC
PtpC (CD1579) is one of the predicted orphan histidine kinases originally hypothesized to promote sporulation by phosphorylating Spo0A [1,10]. PtpC was shown to phosphorylate Spo0A in vitro; however, it is unclear whether this is the preferential direction of phosphate transfer between these proteins in vivo [10]. Although it was presumed that PtpC would activate sporulation, deletion of ptpC results in increased, but variable, sporulation, indicating that PtpC represses sporulation in C. difficile [13]. The mechanism of PtpC activation is not known, but ptpC transcription is influenced by other sporulation factors, including RstA, PtpA, and PtpB [11**,15]. Further characterization of PtpC is needed to define the role of PtpC in Spo0A phosphoregulation.
SigB
Similar to other Gram-positive species, SigB transcribes factors in C. difficile to aid in survival during stress-inducing conditions, including nitrosative and oxidative conditions, acidic environments, and thiol homeostasis [28–31]. In C. difficile, a sigB mutant displays a 10-fold increase in spore formation relative to wild-type, indicating that SigB has a negative effect on sporulation [29]. However, as a sigma factor, SigB is a direct positive regulator of transcription, thus the negative effects of SigB on sporulation are expected to be indirectly mediated through transcription of genes whose product(s) impedes the initiation process [32]. Candidate SigB-dependent factors that may impede sporulation initiation were identified by transcriptional analysis of a sigB mutant [29]. These factors include the aforementioned phosphotransfer protein PtpA and the Spo0J-soj system, which is involved in chromosome partitioning. Whether through the expression of these or other sporulation suppressing factors, SigB allows C. difficile to repress sporulation under conditions of cellular stress that do not support spore formation.
CcpA
The global regulator catabolite control protein, CcpA, is an acI/GalR transcriptional repressor that is responsible for transcriptional carbon catabolite repression (CCR) in Gram-positive bacteria [33]. Antunes et al. investigated the role of CcpA in the CCR of C. difficile and found that CcpA directly represses expression of genes encoding the key early sporulation regulators spo0A and sigF [34*]. Relief of CcpA repression in a ccpA null mutant resulted in a higher sporulation frequency compared to the parental strain in the absence of glucose, but at later time points, the sporulation frequencies were similar [34*]. The addition of glucose substantially reduced sporulation to a similar level in both the wild-type and ccpA mutant, indicating that glucose-mediated CCR of sporulation is independent of CcpA, as observed in C. perfringens [35]. However, the mechanism for CcpA-independent glucose-mediated repression of sporulation has not been determined.
CodY
CodY is a nutrient-sensing global regulator that was discovered in B. subtilis [36]. CodY acts as a sensor of branched-chain amino acids (BCAAs) and GTP concentrations within the cell and modulates the expression of CodY-dependent genes [37,38**]. CodY acts primarily as a repressor of transcription during exponential phase, when BCAAs and GTP are abundant [37,39]. When GTP and BCAA levels are low in later growth phases, CodY repression is alleviated and the transcription of genes involved in amino acid biosynthesis, virulence, and sporulation increase [39]. CodY represses transcription of the toxin genes, which results in increased toxin production in low nutrient conditions [37]. Deletion of codY in the epidemic strain UK1 (RT027) resulted in more than a 1000-fold increase in spore formation [40*]. The mechanism by which CodY represses sporulation is not clear [38**,40*]. Since CodY represses the expression of several regulatory factors, many genes that are derepressed in a codY mutant are not directly regulated by CodY. However, several genes that impact sporulation initiation have predicted CodY binding sites or are enriched in CodY-DNA binding experiments, including the phosphotransfer protein PtpC (CDR20291_1476), spo0E, and sinR/sinR’ [38**]. Although evidence indicates that CodY represses sporulation initiation in C. difficile, the specific mechanism(s) through which this is accomplished is not clear.
Other factors that influence sporulation frequency
Agr
The accessory gene regulator (Agr) system is a quorum-sensing system found in most Gram-positive bacteria [41–43]. The Agr system controls many cellular processes, including the expression of genes involved in colonization and virulence. The typical Agr system includes an autoinducer peptide (AIP), AgrD, which is processed and exported by AgrB. The AgrD peptide is sensed exogenously by AgrC, a histidine kinase that activates a response regulator, AgrA, to directly regulate gene expression. There are three identified Agr systems in C. difficile: Agr1, Agr2 and Agr3. All sequenced C. difficile strains contain a partial Agr1 system, encoding only agrB1 and agrD1 in a single operon [44,45]. A complete Agr2 system is found in the R20291 (RT027) [46], while the Agr3 system, lacking the response regulator, is encoded in RT078 strains [47]. Deletion of agrB1D1 in C. difficile strain 630 results in significantly reduced sporulation [48**]. Sporulation of the agrB1D1 mutant was recovered by providing the supernatant of stationary phase cultures from the parent strain. These data are the first evidence that quorum sensing regulates C. difficile sporulation; however, because a cognate histidine kinase (AgrC) and response regulator (AgrA) are missing from strain 630, the signaling pathway that responds to the AgrB1D1 AIP remains unknown.
Opp/App Permeases
Opp and App are oligopeptide permease systems that are hypothesized to import small peptides into C. difficile. Loss of Opp and App results in earlier and increased spore formation, increased expression of the SinR orthologs, CD2214 and CD2215, as well as expression of other CodY and CcpA-dependent genes [49]. Loss of Opp and App also results in increased virulence in a hamster model of infection [49]. The data suggest that Opp and App indirectly repress sporulation through importation of small peptides, but the specific cargo that they transport has not been verified. OppA resembles a nickel-uptake receptor, while AppA is structurally similar to the oligopeptide-binding protein CtaP (cysteine transport-associated protein) from L. monocytogenes, which binds a restricted set of peptides, suggesting that C. difficile AppA may also bind a restricted set of peptides [50–52*].
SinR orthologs
The SinR orthologs (CD2214 and CD2215) impact sporulation, motility, and toxin production in C. difficile strain R20291 [53*]. A mutant that does not transcribe either CD2214 or CD2215 is asporogenic [53*]. Specifically, CD2214 promotes sporulation, motility, and toxin production, while CD2215 decreases sporulation, motility, and toxin production indirectly by binding and preventing CD2214 from influencing target genes [53*]. CD2214-CD2215 binding is mediated through the multimerization domain of CD2215, which was shown to complement the sporulation, motility, and toxin production phenotypes seen in the R20291 CD2215 (CDR20291_2122) mutant [54]. However, the CD2215 helix-turn-helix domain was dispensable for complementation, and its function remains unknown [54]. CD2214 promotes sporulation in C. difficile through a currently unknown mechanism, but likely through DNA-binding and regulation of target genes [55].
CsiA (CD2589)
CD2589(0) was identified as a genomic signature for C. difficile sporulation in the gastrointestinal tract [56]. Deletion of CD25890 resulted in increased sporulation in SM broth, but not 70:30 medium, suggesting that CD25890 has a nutritional function or impact [57]. Notably, expression of the sin genes was also increased ~five-fold in the CD25890 mutant (strain 630Δerm). Though the data suggest that CD25890 decreases sporulation in response to nutritional cues, the mechanism is currently unknown.
c-di-GMP
The nucleotide-based second messenger signaling molecule, c-di-GMP, modulates several physiological processes in C. difficile important for pathogenesis and colonization. Two recent studies revealed that overexpression of a diguanylate synthase reduces sporulation and the regulated production of a phosphodiesterase increases sporulation in C. difficile [12,58], Altogether, these studies support that c-di-GMP inhibits C. difficile sporulation, although the molecular mechanisms and regulatory pathways that mediate this response are unknown.
Orthologs of unknown influence
Below is a summary of C. difficile orthologs to sporulation initiation proteins from B. subtilis that have not been investigated: The KipI-KipA system in B. subtilis regulates sporulation initiation by inhibiting phosphorylation of KinA. Orthologs of KipIA may play a similar role in C. difficile, though in the absence of a KinA ortholog, their target kinase is not apparent [59–61].
Soj (ParA) inhibits B. subtilis sporulation by preventing early sporulation gene transcription [62]. Spo0J (ParB) is responsible for chromosome segregation in B. subtilis and allows Soj to dissociate from DNA, to enable transcription of early sporulation genes [63]. C. difficile encodes similar proteins, but they have not been characterized.
Summary
Successful sporulation initiation is critical to C. difficile transmission and survival within an aerobic environment. This review highlights our current understanding of sporulation initiation in C. difficile; however, sporulation initiation in this species is complex, and many details of the process remain to be determined. It is clear that the sporulation initiation mechanisms of Clostridia, including C. difficile, vary considerably from the Bacillus paradigm and from each other [1,7–9,64]. Other Clostridia encode predicted kinases that directly phosphorylate or dephosphorylate Spo0A, rather than indirectly via a phosphorelay. But, each of the clostridial initiation pathways that have been characterized appear to have evolved distinct Spo0A regulatory mechanisms with regulators that bear limited similarity to each other [1]. Thus far, the majority of factors that are characterized in the initiation pathway of C. difficile repress Spo0A activity and spore formation. Although direct activators of Spo0A have not been identified, the presence of multiple Spo0A inactivating factors in C. difficile alludes to the existence of a mechanism for deliberate Spo0A phosphorylation. Most likely, there are unidentified factors involved in Spo0A activation. Identification and characterization of these additional initiation factors will be critical for piecing together the puzzle of the sporulation initiation program.
Table 1.
Sporulation genes of interest within C. difficile.
| Gene name | Locus taga | Known or predicted function | References |
|---|---|---|---|
| spo0A | CD1214 | Master transcriptional regulator of sporulation | [2,3] |
| sigH | CD0057 | Transition/stationary-phase sigma factor | [14] |
| rstA | CD3668 | Multifunctional regulator; promotes sporulation | [14, 15, 16**] |
| spo0E | CD3271 | Putative Spo0A phosphatase; sporulation inhibition | |
| PtpA | CD1492 | Sporulation inhibition | [11*] |
| PtpB | CD2492 | Sporulation inhibition | |
| PtpC | CD1579 | Sporulation inhibition | |
| sigB | CD0011 | Alternative sigma factor; stress responses; sporulation inhibition | [29–31] |
| ccpA | CD1064 | Carbon catabolite control, transcriptional regulator; cofactor: fructose-1,6-bisphosphate; sporulation inhibition | [34*] |
| codY | CD1275 | Transcriptional regulator; cofactors: GTP and BCAA; sporulation inhibition | [38**–40*] |
| agrB1D1 | CD27491–50 | quorum-sensing related sporulation inhibition | [48**] |
| oppA-F | CD0853–57 | Putative peptide permease; sporulation inhibition | [49] |
| appA-F | CD2670–74 | Putative peptide permease; sporulation inhibition | [49] |
| sin | CD2214–15 | sporulation | [53*,54] |
| csiA | CD2589 | Conditional repression of sporulation | [57] |
| kipI-kipA | CD1386–87 | Putative inhibitor of histidine kinase | |
| spo0J-soj-spo0J2 | CD3671–73 | Putative inhibitor of sporulation gene transcription |
Locus tag number based on C. difficile 630 reference genome (GenBank: AM180355)
HIGHLIGHTS.
The master transcriptional regulator of sporulation in C. difficile is Spo0A.
Spo0A activity depends on phosphorylation.
Diverse regulators translate environmental signals to impact Spo0A phosphorylation.
This complex regulatory network ensures sporulation initiates only when required.
ACKNOWLEDGEMENTS
This research was supported by the U.S. National Institutes of Health through research grants AI116933 and AI156052 to SMM, GM008490 to MAD, AI106699 to GVC and CDL, and DK126467 to GVC. The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Statement: Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Paredes CJ, Alsaker KV, Papoutsakis ET: A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol 2005, 3:969–978. [DOI] [PubMed] [Google Scholar]
- 2.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD: The Clostridium difficile spo0A gene is a persistence and transmission factor. Infection and immunity 2012, 80:2704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, Martin MJ, Goulding D, Duncan SH, Flint HJ, et al. : Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC genomics 2014,15:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK: C. difficile 630Deltaerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PIoS one 2012, 7:e48608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang M, Shao W, Perego M, Hoch JA: Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Molecular microbiology 2000, 38:535–42. [DOI] [PubMed] [Google Scholar]
- 6.Quisel JD, Burkholder WF, Grossman AD: In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. Journal of bacteriology 2001, 183:6573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiner E, Dago AE, Young DI, Heap JT, Minton NP, Hoch JA, Young M: Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol Microbiol 2011, 80:641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mearls EB, Lynd LR: The identification of four histidine kinases that influence sporulation in Clostridium thermocellum. Anaerobe 2014, 28:109–19. [DOI] [PubMed] [Google Scholar]
- 9.Freedman JC, Li j, Mi E, McClane BA: Identification of an Important Orphan Histidine Kinase for the Initiation of Sporulation and Enterotoxin Production by Clostridium perfringens Type F Strain SM101. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, Wilcox MH, Stephenson K: Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. Journal of bacteriology 2009, 191:7296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Childress KO, Edwards AN, Nawrocki KL, Woods EC, Anderson SE, McBride SM: The Phosphotransfer Protein CD1492 Represses Sporulation Initiation in Clostridium difficile. Infection and immunity 2016, doi:10.1128/IAI.00735–16. *Showed that CD 1492 represses sporulation initiation in C. difficile.
- 12.Edwards AN, Willams CL, Pareek N, McBride SM, Tamayo R: c-di-GMP inhibits early sporulation in Clostridioides difficile. Microbiology; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards AN, Wetzel D, DiCandia MA, McBride SM: Three orphan histidine kinases inhibit Clostridioides difficile sporulation. Microbiology; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saujet L, Monot M, Dupuy B, Soutourina 0, Martin-Verstraete I: The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. Journal of bacteriology 2011,193:3186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards AN, Tamayo R, McBride SM: A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Molecular microbiology 2016, 100:954–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edwards AN, Anjuwon-Foster BR, McBride SM: RstA Is a Major Regulator of Clostridioides difficile Toxin Production and Motility. mBio 2019,10. **Demonstrated that RstA is a multifunctional protein that controls toxin production and sporulation through separate regulatory domains and molecular mechanisms.
- 17.Edwards AN, Krall EG, McBride SM: Strain-Dependent RstA Regulation of Clostridioides difficile Toxin Production and Sporulation. J Bacteriol 2020, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neiditch MB, Capodagli GC, Prehna G, Federle MJ: Genetic and Structural Analyses of RRNPP Intercellular Peptide Signaling of Gram-Positive Bacteria. Anna Rev Genet 2017, 51:311–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perego M, Hoch JA: Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. Journal of bacteriology 1991, 173:2514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohlsen KL, Grimsley JK, Hoch JA: Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proceedings of the National Academy of Sciences of the United States of America 1994, 91:1756–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perego M: A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Molecular microbiology 2001, 42:133–43. [DOI] [PubMed] [Google Scholar]
- 22. Dubey GP, Narayan A, Mattoo AR, Singh GP, Kurupati RK, Zaman MohdS, Aggarwal A, Baweja RB, Basu-Modak S, Singh Y: Comparative genomic study of spo0E family genes and elucidation of the role of Spo0E in Bacillus anthracis. Arch Microbiol 2009, 191:241–253. *Comparison of aerobic Spo0E-like proteins to anaerobic Spo0E-like proteins shows great variation in the phosphatase motif and independent evolution of Spo0E in Bacillus and Clostridia.
- 23.Diaz AR, Stephenson S, Green JM, Levdikov VM, Wilkinson AJ, Perego M: Functional role for a conserved aspartate in the Spo0E signature motif involved in the dephosphorylation of the Bacillus subtilis sporulation regulator Spo0A. The Journal of biological chemistry 2008, 283:2962–72. [DOI] [PubMed] [Google Scholar]
- 24.Hoch JA: Two-component and phosphorelay signal transduction. Current opinion in microbiology 2000, 3:165–70. [DOI] [PubMed] [Google Scholar]
- 25.Durre P, Hollergschwandner C: Initiation of endospore formation in Clostridium acetobutylicum. Anaerobe 2004, 10:69–74. [DOI] [PubMed] [Google Scholar]
- 26.McBride SM, Sonenshein AL: Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infection and immunity 2011, 79:167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suarez JM, Edwards AN, McBride SM: The Clostridium difficile cpr locus is regulated by a noncontiguous two-component system in response to type A and B lantibiotics. Journal of bacteriology 2013, 195:2621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecker M, Pané-Farré J, Uwe V: SigB-Dependent General Stress Response in Bacillus subtilis and Related Gram-Positive Bacteria. Annu Rev Microbiol 2007, 61:215–236. [DOI] [PubMed] [Google Scholar]
- 29.Kint N, Janoir C, Monot M, Hoys S, Soutourina O, Dupuy B, Martin-Verstraete I: The alternative sigma factor sigma(B) plays a crucial role in adaptive strategies of Clostridium difficile during gut infection. Environmental microbiology 2017, 19:1933–1958. [DOI] [PubMed] [Google Scholar]
- 30.Kint N, Alves Feliciano C, Hamiot A, Denic M, Dupuy B, Martin-Verstraete I: The sigma (B) sign ailing activation pathway in the enteropathogen Clostridioides difficile. Environmental microbiology 2019, 21:2852–2870. [DOI] [PubMed] [Google Scholar]
- 31.Boekhoud IM, Michel A-M, Corver J, Jahn D, Smits WK: Redefining the Clostridioides difficile σ B Regulon: σ B Activates Genes Involved in Detoxifying Radicals That Can Result from the Exposure to Antimicrobials and Hydrogen Peroxide. mSphere 2020, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haldenwang WG: The sigma factors of Bacillus subtilis. Microbiological reviews 1995, 59:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henkin TM, Grundy FJ, Nicholson WL, Chambliss GH: Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a transacting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol 1991, 5:575–584. [DOI] [PubMed] [Google Scholar]
- 34. Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B: Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic acids research 2012, 40:10701–18. *CcpA directly represses sporulation genes. A ccpA null mutant has increased sporulation, while glucose repression of sporulation is independent of CcpA.
- 35.Varga J, Stirewalt VL, Melville SB: The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. Journal of bacteriology 2004, 186:5221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slack FJ, Serror P, Joyce E, Sonenshein AL: A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Molecular microbiology 1995, 15:689–702. [DOI] [PubMed] [Google Scholar]
- 37.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL: Repression of Clostridium difficile toxin gene expression by CodY. Molecular microbiology 2007, 66:206–19. [DOI] [PubMed] [Google Scholar]
- 38. Daou N, Wang Y, Levdikov VM, Nandakumar M, Livny J, Bouillaut L, Blagova E, Zhang K, Belitsky BR, Rhee K, et al. : Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027. PLoS ONE 2019, 14(l):e0206896. **Demonstrated independent regulation of sporulation and toxin expression by CodY
- 39.Dineen SS, McBride SM, Sonenshein AL: Integration of metabolism and virulence by Clostridium difficile CodY. Journal of bacteriology 2010, 192:5350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nawrocki KL, Edwards AN, Daou N, Bouillaut L, McBride SM: CodY-Dependent Regulation of Sporulation in Clostridium difficile. Journal of bacteriology 2016, 198:2113–30. *Showed that CodY represses sporulation in C. difficile.
- 41.Le KY, Otto M: Quorum-sensing regulation in staphylococci—an overview. Front Microbiol 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Autret N, Raynaud C, Dubail I, Berche P, Charbit A: Identification of the agr Locus of Listeria monocytogenes: Role in Bacterial Virulence. Infect Immun 2003, 71:4463–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenberg M, Kuo D, Jankowsky E, Long L, Hager C, Bandi K, Ma D, Manoharan D, Shoham Y, Harte W, et al. : Small-molecule AgrA inhibitors F12 and F19 act as antivirulence agents against Gram-positive pathogens. Sci Rep 2018, 8:14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, et al. : The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 2006, 38:779–786. [DOI] [PubMed] [Google Scholar]
- 45.Darkoh C, Odo C, DuPont HL: Accessory Gene Regulator-1 Locus Is Essential for Virulence and Pathogenesis of Clostridium difficile. mBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP, Pettit L, Dougan G, Lawley TD, Wren BW: The agr locus regulates virulence and colonization genes in Clostridium difficile 027. Journal of bacteriology 2013, 195:3672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hargreaves KR, Kropinski AM, Clokie MRJ: What Does the Talking?: Quorum Sensing Signalling Genes Discovered in a Bacteriophage Genome. PLoS ONE 2014, 9:e85131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ahmed UKB, Shadid TM, Larabee JL, Ballard JD: Combined and Distinct Roles of Agr Proteins in Clostridioides difficile 630 Sporulation, Motility, and Toxin Production. mBio 2020, 11. ** Provided the first evidence that a small quorum-sensing peptide regulates C. difficile sporulation.
- 49.Edwards AN, Nawrocki KL, McBride SM: Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infection and immunity 2014, 82:4276–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xayarath B, Marquis H, Port GC, Freitag NE: Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis: Multifunctional role of CtaP in Listeria virulence. Molecular Microbiology 2009, 74:956–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xayarath B, Alonzo F, Freitag NE: Identification of a Peptide-Pheromone that Enhances Listeria monocytogenes Escape from Host Cell Vacuoles. PLoS Pathog 2015, 11:e1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hughes A, Wilson S, Dodson EJ, Turkenburg JP, Wilkinson AJ: Crystal structure of the putative peptide-binding protein AppA from Clostridium difficile. Acta Crystallogr F Struct Biol Commun 2019, 75:246–253. * Crystal structure of C. difficile AppA and evaluated the specificity of peptide binding for AppA and OppA.
- 53. Girinathan BP, Ou J, Dupuy B, Govind R: Pleiotropic roles of Clostridium difficile sin locus. PLOS Pathogens 2018, 14:e1006940. *Observed that the sin locus promotes sporulation, toxin production, and motility.
- 54.Ciftci Y, Girinathan BP, Dhungel BA, Hasan MK, Govind R: Clostridioides difficile SinR’ regulates toxin, sporulation and motility through protein-protein interaction with SinR. Anaerobe 2019, 59:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poquet I, Saujet L, Canette A, Monot M, Mihajlovic J, Ghigo J-M, Soutourina O, Briandet R, Martin-Verstraete I, Dupuy B: Clostridium difficile Biofilm: Remodeling Metabolism and Cell Surface to Build a Sparse and Heterogeneously Aggregated Architecture. Front Microbiol 2018, 9:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD: Culturing of‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533:543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martins D, DiCandia MA, Mendes AL, Wetzel D, McBride SM, Henriques AO, Serrano M: CD25890, a conserved protein that modulates sporulation initiation in Clostridioides difficile. Sci Rep 2021, 11:7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dhungel BA, Govind R: Phase-variable expression of pdcB, a phosphodiesterase, influences sporulation in Clostridioides difficile. Mol Microbiol 2021, 116:1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacques DA, Langley DP, Hynson RM, Whitten AE, Kwan A, Guss JM, Trewhella J: A novel structure of an antikinase and its inhibitor. Journal of molecular biology 2011, 405:214–26. [DOI] [PubMed] [Google Scholar]
- 60.Wang L, Grau R, Perego M, Hoch JA: A novel histidine kinase inhibitor regulating development inBacillus subtilis. Genes Dev 1997, 11:2569–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jacques DA, Langley DB, Jeffries CM, Cunningham KA, Burkholder WF, Guss JM, Trewhella J: Histidine Kinase Regulation by a Cyclophilin-like Inhibitor. Journal of Molecular Biology 2008, 384:422–435. [DOI] [PubMed] [Google Scholar]
- 62.Quisel JD, Grossman AD: Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB). Journal of bacteriology 2000, 182:3446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Autret S, Nair R, Errington J: Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Molecular microbiology 2001, 41:743–55. [DOI] [PubMed] [Google Scholar]
- 64.Worner K, Szurmant H, Chiang C, Hoch JA: Phosphorylation and functional analysis of the sporulation initiation factor Spo0A from Clostridium botulinum. Molecular microbiology 2006, 59:1000–12. [DOI] [PubMed] [Google Scholar]


