Abstract
Objective:
To determine the effect of hyperthermic intraperitoneal chemotherapy (HIPEC) with carboplatin on the transcriptomic profiles of normal and ovarian cancer (OC) tissues.
Methods:
Normal and tumor samples from four OCs were prospectively collected pre- and immediately post-HIPEC treatment and subjected to RNA-sequencing. Differential gene expression, gene ontology enrichment and pathway analyses were performed. Heat shock protein and immune-response protein expression was assessed using protein arrays and western blotting.
Results:
RNA-sequencing revealed 4,231 and 322 genes significantly differentially expressed between pre- and post-treatment normal and OC tissues, respectively (both adjusted p-value <0.05). Gene enrichment analyses demonstrated that the most significantly upregulated genes in normal tissues played a role in immune as well as heat shock response (both adjusted p<0.001). In contrast, HIPEC induced an increased expression of primarily heat shock response and protein folding-related genes in tumor tissues (both adjusted p <0.001). HIPEC-induced heat shock protein (HSP) expression changes, including in HSP90, HSP40, HSP60, and HSP70, were also observed at the protein level in both normal and tumor tissues.
Conclusions:
HIPEC with carboplatin resulted in an upregulation of heat shock-related genes in both normal and tumor tissue, with an additional immune response gene induction in normal and protein folding in tumor tissue. The findings of our exploratory study provide evidence to suggest that HIPEC administration may suffice to induce gene expression changes in residual tumor cells and raises a biological basis for the consideration of combinatorial treatments with HSP inhibitors.
Keywords: HIPEC, ovarian cancer, gene expression, heat shock, immune response
INTRODUCTION
Ovarian cancer is the leading cause of deaths among gynecologic cancers in developed countries, with an estimated 19,880 new cases and 12,810 deaths in the US in 2022 [1]. This high mortality rate is primarily attributed to the fact that the majority of women are diagnosed with advanced disease. Although spread beyond the ovaries is common, the disease largely remains confined to the abdominal cavity and spreads along the peritoneal lining of the abdominal organs without deeply invading the organs [2]. Furthermore, recurrence is experienced by most patients with advanced-stage cancer and preferentially occurs within the abdominal cavity [3]. This prompted investigators to evaluate postsurgical intraperitoneal (IP) drug delivery as a therapeutic approach which demonstrated promising results, however a substantial number of patients experience adverse effects and are unable to complete IP therapy, hindering its widespread adoption [3–6].
Hyperthermic intraperitoneal chemotherapy (HIPEC) offers the option to deliver drug therapy directly to the site of disease in an intraoperative setting with the addition of hyperthermia. The addition of hyperthermia stems from studies showing its ability to increase the cytotoxic effect of chemotherapeutic agents by increasing tumor penetration and platinum-induced DNA adduct formation [7–12]. Although there is increasing clinical evidence to support the role of HIPEC during the upfront management of advanced ovarian cancer [13–15], the molecular basis of HIPEC is poorly understood [16]. In this exploratory study, we sought to assess the effects of HIPEC on normal and tumor tissues at the transcriptomic level to define mechanisms that could be exploited to potentiate the effects of HIPEC in the treatment of ovarian cancer.
METHODS
Cases
This study was performed within the context of a randomized clinical trial investigating the efficacy and safety of carboplatin administered via HIPEC after surgical resection in patients with recurrent platinum-sensitive ovarian cancer at Memorial Sloan Kettering Cancer Center (MSK; NCT01767675)[16]. This study was approved by the Institutional Review Board and patient consent was obtained. Tumor tissue and normal uninvolved tissue, including from the omentum, peritoneum, or portion of bowel (Supplementary Table 1), were intraoperatively collected pre- and immediately post- HIPEC treatment from four cases that randomized to the treatment arm. All patients had high-grade serous carcinoma of ovarian origin. HIPEC treatment consisted of carboplatin 800mg/m2 diluted in 3L of normal saline administered via IP perfusion for approximately 90 minutes in the hyperthermic phase at approximately 41–43°C, as described [16]. Tissue samples were snap-frozen at the time of resection.
RNA extraction
Frozen sections of tumor and normal tissue were stained with hematoxylin and eosin and reviewed by a pathologist (FP) to define the percentage of neoplastic cells within tumor samples and to verify that normal samples lacked neoplastic cells. Only cases in which the normal tissue was completely devoid of neoplastic cells were included. In all tumors samples, neoplastic cells comprised greater than 75% except for the pre-HIPEC tumor sample of Case 2 which tumor cell content was 50%. RNA was extracted from tumor and matched normal tissues using the RNeasy Mini kit (Qiagen, Germantown, MD), according to the manufacturer’s instructions, and quantified using the Qubit Fluorometer (Invitrogen, Thermo Fisher Scientific, Waltham, MA).
RNA-sequencing and differential gene expression analysis
RNA samples were subjected to polyA PE50 RNA-sequencing using validated protocols at MSK Integrated Genomics Operation (IGO), as previously described [17, 18]. In brief, RNA-sequencing reads were aligned using STAR [19]. Genes with very low/ poor expression levels, defined as less than a 0.2 count per million (CPM), were removed from further analyses. Raw counts were normalized using an upper quartile of M-values approach [20], and dispersion was estimated over all genes using the quantile-adjusted conditional maximum likelihood method [21]. The Benjamini-Hochberg method was used to correct for multiple comparisons [22]. Reads Per Kilobase Million (RPKM) values were estimated from raw counts normalizing for sequencing depth and adjusted for length of genes, as previously described [17].
Differential gene expression (DGE) analyses were performed between pre- and post-treatment tumor and normal tissue samples, using the DESeq2 R package [23], and differentially expressed genes after HIPEC exposure were defined using a 1-versus-all t-test and permutation testing. The statistical design included the subject variable to account for differences within samples from the same case. Batch correction was carried out using ComBat (sva R package) [24]. Pre-filtering was applied to remove genes with less than 10 reads across samples. P-values were adjusted for multiple testing using Benjamini-Hochberg correction [22], and p-values <0.05 were considered statistically significant. The gene expression of HSPE1, which codes for HSP10 protein and was present in the protein array (see below), could not be assessed as it was not identifiable among those genes having undergone alignment during RNA-sequencing data processing. For hierarchical clustering, normalized counts were processed using variance stabilizing transformation, and cluster analysis was performed using Euclidean distance and complete linkage. Gene set enrichment analysis (GSEA) was performed using the GSEA-P R application 1.0 [25] with an input using normalized raw counts. The ontology gene set (C5) was obtained from MSigDB version 7.0 [26].
Protein arrays
Frozen normal and tumor tissue (50–200mg) was resuspended in lysis buffer containing protease inhibitors and homogenized using the Qiagen TissueRuptor II. The homogenized tissue was incubated for 2 hours at 4°C in constant rotation, and then centrifuged at 13,000 rpm for 15 minutes at 4°C. The protein in the supernatant was quantified using the BCA Protein Assay Kit (Abcam, Cambridge, MA), and 250μg of protein of a given sample was incubated on the RayBio Human HSP Array C1 (AAH-HSP-1–2; RayBiotech, Peachtree Corners, GA) membrane following the manufacturer’s instruction. The RayBio Human HSP Array C1 included 9 heat shock proteins (i.e. HSP90, GRP75, HSP27, Ubiquitin+1, HSP32, HSP10, HSP40, HSP60 and HSP70). The signal was detected through HRP-streptavidin chemiluminescence following the manufacturer’s instruction and visualized with high performance chemiluminescence film (GE Healthcare, Chicago, IL) and a Konica SRX-101A medical film processor (Konica Minolta, Wayne, NJ). Images were quantified using the Protein Array Analyzer for ImageJ (NIH).
Western blotting
Forty micrograms of protein lysate from frozen tissue were used for western blotting as described [27]. Primary antibodies against IL-6 (D3K2N #12153, Cell Signaling Technology, Danvers, MA) and tubulin (DM1A Cell Signaling Technology) were used at 1:1000 dilution. A conjugated secondary anti-rabbit antibody (LI-COR, 926–68073; LI-COR Biosciences, Lincoln, NE) was used and signal was detected using the Odyssey Infrared Imaging System (LI-COR). Quantification and analysis were performed using Image Studio Lite (LI-COR) as described [17].
Enzyme-linked immunosorbent assay (ELISA)
The amount of IL-6 protein in the frozen tumor and normal tissue derived protein lysates was determined using the Human IL-6 ELISA kit (SigmaAldrich, St. Louis, MO) according to the manufacturer’s instructions. The values were background-corrected, and results were displayed as pg IL-6 per ml of protein lysate according to the standard curve. Tubulin was used as protein loading control.
Immune deconvolution
Immune cell deconvolution from the RNA-sequencing data was performed using CIBERSORT absolute mode in the Immunedeconv R package [28, 29]. Counts were transformed into Transcripts Per Kilobase Million (TPM). A CIBERSORT score was calculated as an estimate of the enrichment of a specific population of cells within a given sample.
Statistical analysis
Wilcoxon rank test was employed to define statistical differences between tumor and normal pre- and post-HIPEC groups. Comparisons between groups across immune subpopulations in the CIBERSORT analysis were performed using Wilcoxon rank test. A two-tailed p-value was reported and considered statistically significant if p<0.05. Statistical analyses were conducted using R3.5.2.
RESULTS
Gene expression patterns of normal and tumor tissues before and after HIPEC exposure
RNA-sequencing was performed on normal and tumor tissues collected before and after HIPEC with carboplatin from four patients (Supplementary Table 1). Unsupervised cluster analysis of the 47 genes intersecting with a false discovery rate (FDR) <0.1 across all four pre- and post-HIPEC tumor and normal samples revealed that the samples clustered by tissue type (i.e. tumor vs normal) and by the presence or absence of exposure to HIPEC (i.e. pre- vs post-HIPEC; Figure 1). Furthermore, we observed that the normal tissues post-HIPEC treatment formed a more discrete cluster away from the remaining samples. These data provide evidence to suggest that the exposure to HIPEC with carboplatin induces transcriptional changes in both normal and tumor tissues.
Figure 1: Hierarchical cluster analysis of normal and ovarian cancer tissues pre and post HIPEC with platinum.

Heatmap of all 47 genes intersecting with a false-discovery rate (FDR) < 0.1 across all four categories (post HIPEC tumor vs normal, pre HIPEC tumor vs normal, post HIPEC tumor vs pre tumor, post HIPEC normal vs pre normal). Hierarchical clustering was performed using Euclidean distance and complete linkage. Cases are shown in columns and genes in rows. Gene expression levels are color-coded according to the legend. Tissue type and time point are is depicted at the top bar.
As a next step, we identified within the normal tissue samples and within the tumor tissue samples the genes that were significantly up- or down-regulated after HIPEC exposure using a 1-versus-all t-test and permutation testing. This analysis revealed 4,231 genes significantly differentially expressed between pre- and post-treatment in normal tissues and 322 genes differentially expressed between pre- and post-treatment in tumor tissues (p-value<0.05; Figure 2, Supplementary Table 2). Whilst the number of genes differentially expressed between pre- and post-HIPEC samples was higher in normal tissues, the magnitude of the gene expression changes induced by HIPEC with carboplatin were similar between normal and tumor tissues (Figure 2).
Figure 2: Volcano plot of genes differentially expressed in normal and ovarian tumor tissue pre vs post HIPEC with platinum exposure.
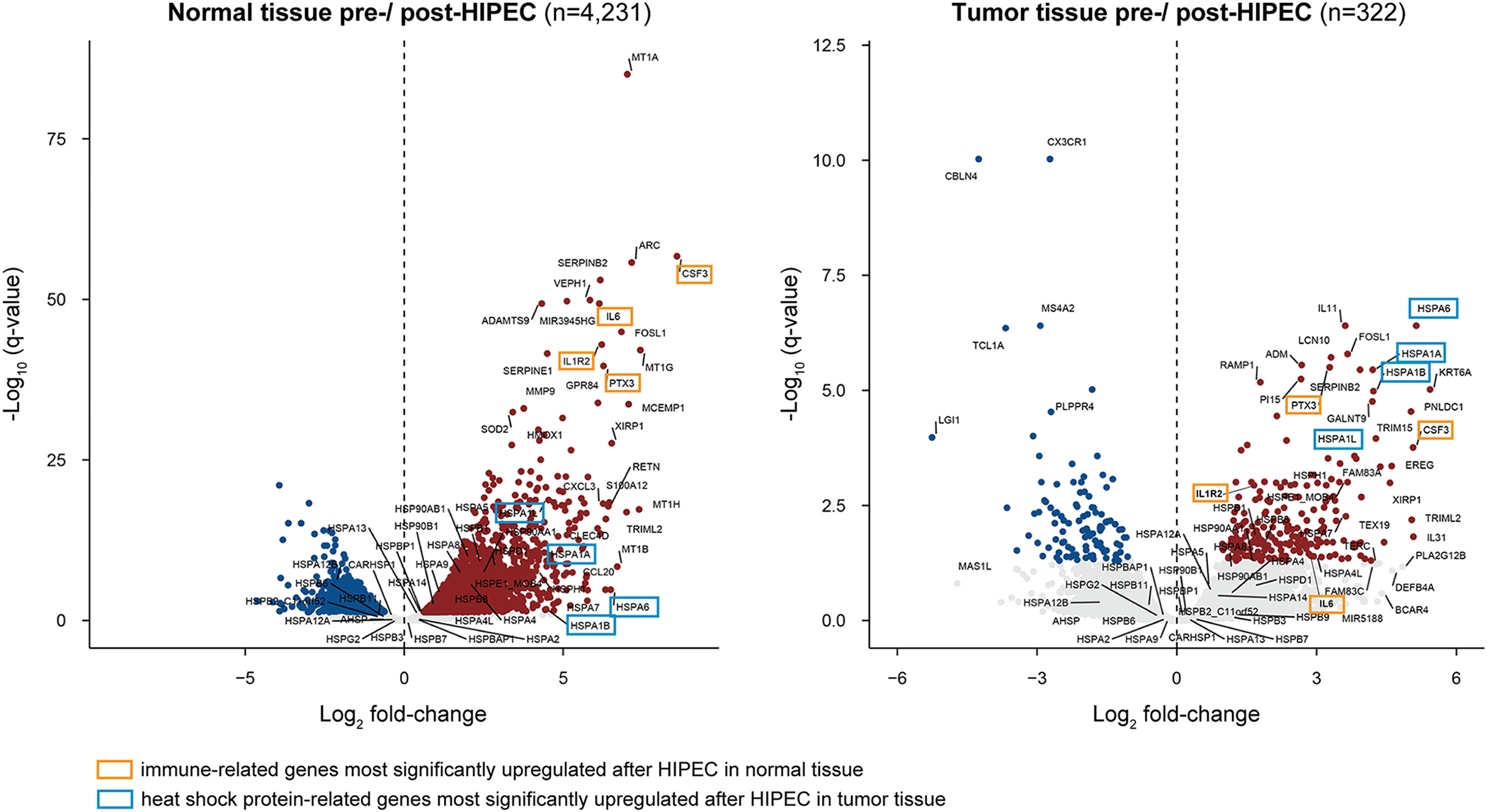
Differentially expressed genes after HIPEC with platinum exposure in normal (left) and tumor (right) tissues, plotted as Log2 fold-change in the x-axis and −Log10 q-value on the y-axis. Expression of genes colored as red (up-regulation) and blue (down-regulation) dots has significantly changed after HIPEC with platinum exposure with a p-value <0.05 and a Log2 fold-change >1.5. Genes highlighted with a yellow box represent the four most significantly upregulated genes related to immune response after HIPEC in normal tissue, whereas those with a light blue box represent the four most significantly upregulated genes after HIPEC in tumor tissue, which are heat shock proteins.
To annotate the differentially expressed genes functionally, a gene set enrichment analysis (GSEA) was performed. Using Gene Ontology classification, we found that the most significantly altered gene sets in normal tissue after exposure to HIPEC were related to heat shock response and cytokine activity (p-value<0.05; Figure 3, Supplementary Table 3). CSF3, IL6, IL1R2 and PTX3 were among the genes with the highest levels of upregulation in normal tissue after HIPEC exposure, all of which play a role in immune response by coding for cytokines and interleukin receptors (adjusted p-value < 0.001; Figure 2, Supplementary Table 2). Heat shock response-related genes were also induced in normal tissue after HIPEC exposure, however to a lesser extent than immune-related genes (Figure 2). Unlike the findings in normal tissue, GSEA analysis revealed that in tumor tissues the most significantly altered gene sets induced by HIPEC exposure were primarily related to heat shock, protein folding and protein binding (p-value<0.05; Figure 3, Supplementary Table 3). This finding corroborates that of the DGE analysis where HSPA6, HSPA1A, HSPA1B and HSPA1L, which are all part of the heat shock response, were among the genes most significantly altered following HIPEC exposure (adjusted p-value <0.001; Figure 2, Supplementary Table 2). These data provide evidence to suggest that transcriptional changes in tumor samples primarily affected heat response- and protein folding-related genes, in contrast to the effect of HIPEC on normal tissue, where immune response-related genes were the most significantly altered.
Figure 3: Gene set enrichment analysis of genes differentially expressed pre- and post-HIPEC with platinum exposure in normal and tumor tissue samples.
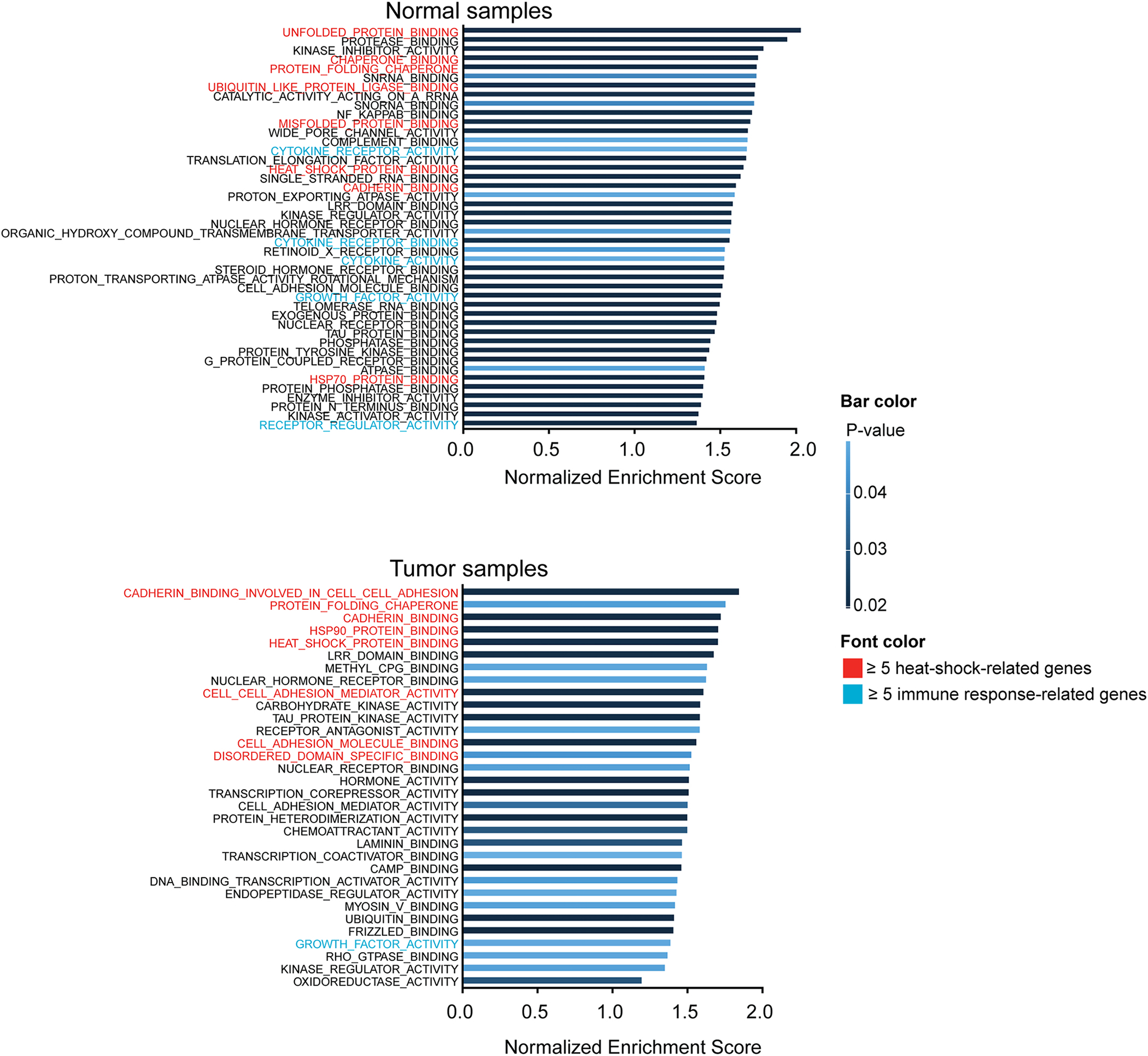
Gene set enrichment analysis of genes differentially expressed pre- and post-HIPEC exposure in normal tissue samples (top) and tumor tissue samples (bottom), illustrated by gene ontology classification and ordered based on the magnitude of the normalized enrichment score (NES). Only gene sets that were significantly altered with a p-value <0.05 are illustrated in this figure. Gene ontology classifications are highlighted in red or green if the classification includes ≥5 genes related to heat-shock or immune response, respectively. If genes from both a heat-shock and immune response were present with the classification, no designation was given.
Heat shock protein expression levels after HIPEC
We next evaluated the impact of HIPEC within normal and tumor tissues at the protein level, through protein antibody array and western blotting analyses. First, we used a heat shock protein antibody array to detect changes in the expression of 9 heat shock proteins (HSPs), including HSP90, GRP75, HSP27, Ubiquitin+1, HSP32, HSP10, HSP40, HSP60, HSP70 (Figure 4A). An overall increase of HSP90, GRP75, and Ubiquitin+1 was observed in normal tissues as compared to a decrease in tumor tissues after HIPEC exposure. On the other hand, an increase in HSP27, HSP32, HSP40, HSP60 protein expression levels was observed in tumor tissues as compared to a decrease in normal tissue after HIPEC exposure. Only HSP70 protein expression increased in both normal and tumor tissues following HIPEC (Figure 4A). These findings at the protein level were consistent with the RNA-sequencing-based gene expression changes detected in tumor tissues, where we observed a decrease in the expression levels of GH1 (GRP75) and RPS27A (Ubiquitin+1) and an increase in the expression levels of HSPB1 (HSP27A), HMOX1 (HSP32), DNAJB5 (HSP40), HSPA14 (HSP60), and HSPA4 (HSP70) (Figure 4B). The only discrepancy between gene and protein expression levels in tumor tissues after HIPEC exposure was related to HSP90, for which was found to be increased at the transcriptomic level (HSP90AA1) but not at the protein level. In normal tissues, while at the transcriptomic level there was an upregulation across all heat shock genes analyzed after HIPEC exposure, increased protein expression levels after HIPEC exposure were only detected for HSP90, GRP75, Ubiquitin+1 and HSP70 (Figures 4A and 4B, Supplementary Table 4).
Figure 4: Effects of HIPEC with platinum on heat-shock and immune-related protein expression in normal and tumor tissue.
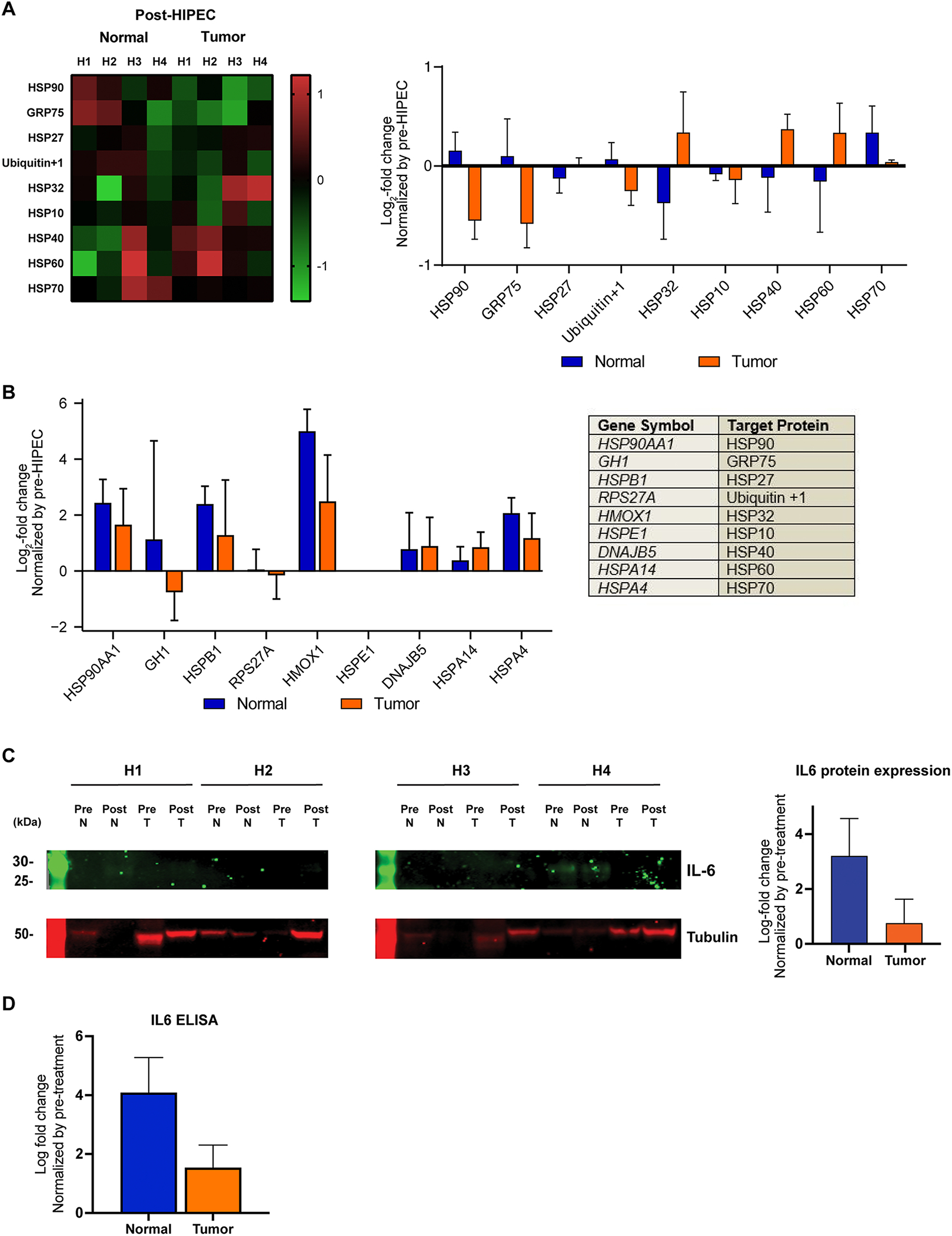
(A) Heatmap of human heat shock protein antibody array demonstrating changes in protein expression levels of normal and tumor tissue samples after HIPEC with platinum exposure, color-coded according to the legend (left). Following background subtraction, the data were normalized to the positive control signals. For each protein, the mean Log2 fold-change post-HIPEC samples compared to pre-HIPEC samples are shown on the right. Error bars, s.d. of mean.
(B) Gene expression Log2 fold-change normalized by gene expression levels pre-HIPEC exposure (Error bars, s.d. of mean). Genes selected for illustration are those that correspond with the 9 heat shock proteins HSP90, GRP75, HSP27, Ubiquitin+1, HSP32, HSP10, HSP40, HSP60 and HSP70 investigated in the protein antibody array (Table, right).
(C) Western blot analysis of IL-6 protein in normal and tumor samples pre- and post-HIPEC with platinum exposure. Tubulin was used as protein loading control (bottom). Mean of Log2 fold-change of post-HIPEC samples as compared to pre-HIPEC samples (right). Error bars, s.d. of mean. Cases are labeled H1 – H4, for normal and tumor tissues.
(D) ELISA of IL-6 protein in normal and tumor samples pre- and post-HIPEC with platinum exposure. Error bars, s.d. of mean.
Finally, we assessed whether the high-level induction of IL6 gene expression in normal tissue after HIPEC was also present at the protein level. We performed two orthogonal assays, western blot analysis and ELISA, which both showed that IL-6 protein expression was increased in normal tissue after HIPEC, and that, akin to the transcriptomic analysis, this increase was significantly higher in normal than in tumor tissues (western blot, p-value=0.03, Figure 4C; IL-6 ELISA, p-value=0.05, Figure 4D).
Comparison of cell type composition across samples
To evaluate the cell composition of the tumor and normal tissue samples based on HIPEC exposure status, we performed a deconvolution analysis using CIBERSORT, assessing 22 types of infiltrating immune cells [28, 29]. The immune cell composition of normal and tumor samples prior to HIPEC exposure was similar (p>0.05). After HIPEC exposure, however, we observed a significant difference in the immune cell composition of normal vs tumor samples, with normal cells showing an increase in CD4 naïve and resting memory cells, macrophage M1 and M2, resting mast cells, and neutrophils (p<0.05; Figure 5). Importantly, there was no significant change observed in the cell composition of the tumor samples before and after HIPEC exposure (p>0.05). In contrast, in the normal tissue samples before and after HIPEC, we found a significant change in the composition of CD8 T cells, activated and resting mast cells, activated NK cells, and neutrophils (p<0.05; Figure 5).
Figure 5: Cell type deconvolution of normal and tumor tissue based on HIPEC exposure.
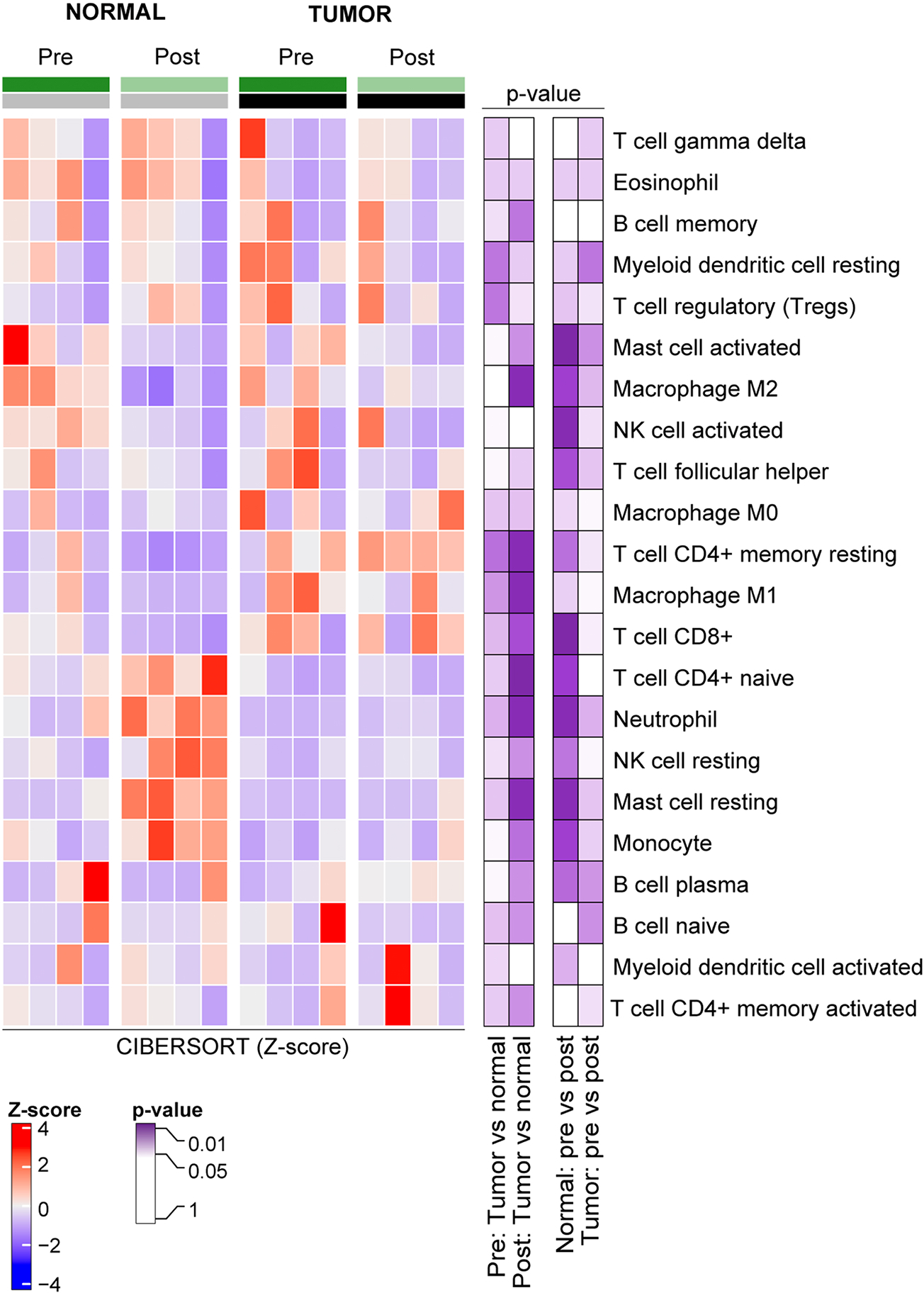
Using RNA-Sequencing data, cell type composition of 22 human leukocyte subtypes using CIBERSORT between normal and tumor tissue samples before and after HIPEC with platinum exposure were defined. Z-score values of the CIBERSORT scores and p-values (Wilcoxon rank test) are color-coded according to the legend.
DISCUSSION
The multimodal approach of cytoreductive surgery and HIPEC is based on both scientific evidence that hyperthermia is selectively cytotoxic to malignant cells and augments the efficacy of the chemotherapeutic agent [30–32] as well as clinical data demonstrating an impact on progression-free and overall survival [14]. Given its combination with surgery, however, delineating the actual effect of a single 90-minute exposure of HIPEC from the effects of surgery on the disease is challenging. The intraoperative setting is an ideal opportunity to sample tissue before and after HIPEC. In this exploratory, hypothesis-generating study, we investigated whether such an exposure would result in measurable changes and demonstrated that a 90-minute single exposure to HIPEC with carboplatin is sufficient to have implications on tumor gene expression as well as protein production.
Gene enrichment analyses demonstrated that the most significantly upregulated genes in normal tissues played a role in immune as well as heat shock response (both adjusted p-value <0.001). In contrast, HIPEC induced an increased expression of primarily heat shock response and protein folding-related genes in tumor tissues (both adjusted p-value <0.001). In addition, changes in gene expression of certain HSP related genes are thought to have occurred relatively quickly during the 90-minute time frame as not only were changes in gene expression observed, but also protein production. Specifically, in tumor samples this time frame proved sufficient to upregulate the gene expression of HSPB1, HMOX1, DNAJB5, HSPA14, and HSPA4, as well as production of the corresponding proteins HSP27, HSP32, HSP40, HSP60, and HSP70. The exception was the increase expression of HSP90AA1 which did not coincide with an observed increase in protein production, indicating that translation was likely still underway.
HSPs act as molecular chaperons and are protective against various cell stressors such as ischemia, heat stress, and oxidative stress [33, 34], properties that have resulted in the development of HSP inhibitors as potential cancer therapeutic agents. Some HSPs, such as HSP90, are also chaperon proteins that inhibit protein misfolding and aggregation and in doing so, promote survival of cancer cells [35]. In ovarian cancer, multiple studies have shown that targeted inhibition of certain HSPs has antitumor effects such as inhibiting cell growth, cell cycle arrest, and induction of apoptosis [36–38]. Specifically, one HSP90 inhibitor that has shown promise across several tumor types is AUY922 [36, 39, 40] and has been tested on ovarian cancer cell lines demonstrating anti-proliferative effects and when administered in combination with carboplatin potentiated platinum-sensitivity [37]. Additionally, the co-inhibition of HSP70/HSP90 has been shown to sensitize cells to hyperthermia [41]. We demonstrated that the most significant changes in gene expressions among ovarian cancer cells exposed to HIPEC was an upregulation of HSP related genes. In addition, on a protein level we also demonstrate increased production of HSP proteins within ovarian cancer cells exposed to HIPEC. This suggests that a specific HSP inhibitor in addition to the chemotherapeutic agent administered during HIPEC could enhance the tumor cells sensitivity to hyperthermia and induce a pro-apoptotic effect. It is important to mention that in addition to the upregulation of HSPs in the tumor tissue, there was a significant increase in the expression of other gene pathways involved in protein folding. These pathways warrant further investigation as they may offer other opportunities for non HSP targeted therapies aimed at inhibiting these rescue pathways instituted by tumor cells in the setting of exposure to HIPEC.
This study has important limitations. First, its small sample size renders it exploratory and hypothesis-generating. Despite the small sample size, a consistent biological signal was observed across patient specimens. Also, with the analysis performed, the relative contribution of platinum versus hyperthermia versus the combination thereof on the observed gene and protein expression changes in the tissues could not be assessed [42]. In a recent study, Javellana et al. reported on the mutational and gene expression changes after neoadjuvant chemotherapy for ovarian cancer [43]. We used these data to assess the effect of chemotherapy in the absence of heat on the transcriptomic profiles of ovarian cancer cells, and found no significant changes in the expression of heat shock-related genes (i.e. with a Log2fold change of >2). This lack of change in the levels of heat shock response gene expression after chemotherapy provides evidence to suggests that the transcriptomic changes we observed in our study post HIPEC are likely due to the heat effect or the combination of heat with chemotherapy. Our findings warrant further studies to investigate the cellular effects of heat versus chemotherapy, the biological impact of HIPEC on cancer cells, as well as the interaction between HIPEC and the genetic make-up of ovarian cancer. Although gene and protein expression changes were observed in post-HIPEC samples, further analyses are required to define the specific HSPs or specific formations of misfolded protein aggregates that are induced by HIPEC across ovarian cancers and whether their targeting could be of potential therapeutic relevance.
Despite its exploratory nature, this study provides an assessment of the biological impact of HIPEC with carboplatin on ovarian cancer cells and normal tissues and raises the hypothesis that the HIPEC-induced increased expression of HSPs and proteins related to protein folding in ovarian cancer cells may provide a biological basis for combinatorial treatments to be deployed in conjunction with HIPEC to target malignant cells while sparing normal tissue.
Supplementary Material
Supplementary Table 1: Patient and sample information.
Supplementary Table 2: Differential gene expression changes between tumor and normal tissues pre- and post-HIPEC treatment.
Supplementary Table 3: Gene set enrichment analysis and gene ontology classification of genes differentially expressed between tumor and normal tissues pre- and post-HIPEC.
Supplementary Table 4: Statistical comparisons of change in gene expression, protein expression and cell type composition.
HIGHLIGHTS.
HIPEC causes transcriptomic changes in normal and tumor tissues
HIPEC induces upregulation of heat shock-related genes in normal and OC tissues
Genes implicated in protein folding are induced in OC tissues post HIPEC
Genes playing a role in immune response are induced in normal tissues post HIPEC
The production of heat shock response proteins changes after HIPEC administration
ACKNOWLEDGEMENTS
J.S. Reis-Filho and B. Weigelt are funded in part by NIH/NCI P50 CA247749 01 and Breast Cancer Research Foundation grants. F. Pareja is funded in part by a NIH/NCI P50 CA247749 01 grant and a K12 CA184746 grant. Research reported in this publication was supported in part by a Cancer Center Support Grant of the NIH/NCI (Grant No. P30CA008748), by the Weickart Ovarian Cancer Postdoctoral Fellowship, Cycle for Survival and the Baird Family Foundation. G. Zoppoli is funded in part by a Fondazione AIRC per la Ricerca sul Cancro Investigator Grant ID 21761, by a 5×1000 IRCCS grant ID C830B, by a University of Genoa CURIOSITY grant, and by anonymous patients’ liberal donations.
Footnotes
CONFLICTS OF INTEREST
G. Zoppoli reports receiving travel grants from Roche, Novartis, and Pfizer, consultation fees from Pfizer, reagents from ThermoFisher Scientific and Cytiva Life Sciences, outside the submitted work. N.R. Abu-Rustum reports Stryker/ Novadaq and GRAIL grants paid to the institution, outside the current study. J.S. Reis-Filho reports receiving personal/consultancy fees from Goldman Sachs, REPARE Therapeutics and Paige.AI, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics and Paige.AI, membership of the Board of Directors of Grupo Oncoclinicas, and ad hoc membership of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Novartis, Genentech and InVicro, outside the scope of this study. J.S. Reis-Filho has stocks or stock options with REPARE Therapeutics and Paige.AI. D.S. Chi reports membership of the medical advisory boards of Bovie Medical Co. (now Apyx Medical), Verthermia Inc and Biom ‘Up, to have/ had stock options of Bovie Medical Co. (now Apyx Medical), Verthermia Inc, Intuitive Surgical Inc and TransEnterix Inc, and to be the Chief Editor and shareholder of C Surgeries. D. Zamarin reports personal/ consulting fees from Agenus, Hookipa Biotech, Western Oncolytics, Synthekine, Mana Therapeutics, Xencor, Memgen and Takeda, grants and personal fees from Merck and from Astra Zeneca, grants and non-financial support from Genentech, grants from Plexxikon and stock options from Calidi Biotherapeutics, outside the submitted work. D. Zamarin has a patent for use of Newcastle Disease Virus for cancer therapy, outside the submitted work. Dr. Aghajanian reports membership of advisory boards/ personal fees from Tesaro, Eisai/Merck, Mersana Therapeutics, Roche/Genentech, Abbvie, AstraZeneca/Merck, Repare Therapeutics, and grants from Clovis, Genentech, AbbVie, Astra Zeneca, all outside the submitted work. R.E. O’Cearbhaill reports receiving honoraria/consulting/advisory role from GlaxoSmithKline, Fresenius, Seagen Inc, Carina Biotech and institutional research funding: Juno Therapeutics, Sellas Life Sciences, Ludwig Institute for Cancer Research, Stem CentRx, TapImmune Inc, TCR2 Therapeutics, Regeneron, Genmab, Atara Biotherapeutics, GlaxoSmithKline, AstraZeneca/Merck, Syndax, Genentech, Kite/Gilead, Gynecologic Oncology Group Foundation. Meal from AstraZeneca. B. Weigelt reports ad hoc membership of the scientific advisory board of Repare Therapeutics. The remaining authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. [DOI] [PubMed] [Google Scholar]
- [2].Zivanovic O, Eisenhauer EL, Zhou Q, Iasonos A, Sabbatini P, Sonoda Y, et al. The impact of bulky upper abdominal disease cephalad to the greater omentum on surgical outcome for stage IIIC epithelial ovarian, fallopian tube, and primary peritoneal cancer. Gynecol Oncol. 2008;108:287–92. [DOI] [PubMed] [Google Scholar]
- [3].Tewari D, Java JJ, Salani R, Armstrong DK, Markman M, Herzog T, et al. Long-term survival advantage and prognostic factors associated with intraperitoneal chemotherapy treatment in advanced ovarian cancer: a gynecologic oncology group study. J Clin Oncol. 2015;33:1460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. [DOI] [PubMed] [Google Scholar]
- [5].Wright AA, Cronin A, Milne DE, Bookman MA, Burger RA, Cohn DE, et al. Use and Effectiveness of Intraperitoneal Chemotherapy for Treatment of Ovarian Cancer. J Clin Oncol. 2015;33:2841–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dedrick RL. Theoretical and experimental bases of intraperitoneal chemotherapy. Semin Oncol. 1985;12:1–6. [PubMed] [Google Scholar]
- [7].Los G, Sminia P, Wondergem J, Mutsaers PH, Havemen J, ten Bokkel Huinink D, et al. Optimisation of intraperitoneal cisplatin therapy with regional hyperthermia in rats. Eur J Cancer. 1991;27:472–7. [DOI] [PubMed] [Google Scholar]
- [8].Los G, van Vugt MJ, den Engelse L, Pinedo HM. Effects of temperature on the interaction of cisplatin and carboplatin with cellular DNA. Biochem Pharmacol. 1993;46:1229–37. [DOI] [PubMed] [Google Scholar]
- [9].Los G, van Vugt MJ, Pinedo HM. Response of peritoneal solid tumours after intraperitoneal chemohyperthermia treatment with cisplatin or carboplatin. Br J Cancer. 1994;69:235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Giovanella BC, Stehlin JS Jr., Morgan AC. Selective lethal effect of supranormal temperatures on human neoplastic cells. Cancer Res. 1976;36:3944–50. [PubMed] [Google Scholar]
- [11].Hettinga JV, Konings AW, Kampinga HH. Reduction of cellular cisplatin resistance by hyperthermia--a review. Int J Hyperthermia. 1997;13:439–57. [DOI] [PubMed] [Google Scholar]
- [12].Reed E, Parker RJ, Gill I, Bicher A, Dabholkar M, Vionnet JA, et al. Platinum-DNA adduct in leukocyte DNA of a cohort of 49 patients with 24 different types of malignancies. Cancer Res. 1993;53:3694–9. [PubMed] [Google Scholar]
- [13].Spiliotis J, Halkia E, Lianos E, Kalantzi N, Grivas A, Efstathiou E, et al. Cytoreductive surgery and HIPEC in recurrent epithelial ovarian cancer: a prospective randomized phase III study. Ann Surg Oncol. 2015;22:1570–5. [DOI] [PubMed] [Google Scholar]
- [14].van Driel WJ, Koole SN, Sikorska K, Schagen van Leeuwen JH, Schreuder HWR, Hermans RHM, et al. Hyperthermic Intraperitoneal Chemotherapy in Ovarian Cancer. N Engl J Med. 2018;378:230–40. [DOI] [PubMed] [Google Scholar]
- [15].Zhang G, Zhu Y, Liu C, Chao G, Cui R, Zhang Z. The prognosis impact of hyperthermic intraperitoneal chemotherapy (HIPEC) plus cytoreductive surgery (CRS) in advanced ovarian cancer: the meta-analysis. J Ovarian Res. 2019;12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zivanovic O, Chi DS, Zhou Q, Iasonos A, Konner JA, Makker V, et al. Secondary Cytoreduction and Carboplatin Hyperthermic Intraperitoneal Chemotherapy for Platinum-Sensitive Recurrent Ovarian Cancer: An MSK Team Ovary Phase II Study. J Clin Oncol. 2021;39:2594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim SH, Da Cruz Paula A, Basili T, Dopeso H, Bi R, Pareja F, et al. Identification of recurrent FHL2-GLI2 oncogenic fusion in sclerosing stromal tumors of the ovary. Nat Commun. 2020;11:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Basili T, Dopeso H, Kim SH, Ferrando L, Pareja F, Da Cruz Paula A, et al. Oncogenic properties and signaling basis of the PAX8-GLIS3 fusion gene. Int J Cancer. 2020;147:2253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–7. [DOI] [PubMed] [Google Scholar]
- [21].McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. [DOI] [PubMed] [Google Scholar]
- [23].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- [25].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pareja F, Brandes AH, Basili T, Selenica P, Geyer FC, Fan D, et al. Loss-of-function mutations in ATP6AP1 and ATP6AP2 in granular cell tumors. Nat Commun. 2018;9:3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].de Bree E, Rosing H, Filis D, Romanos J, Melisssourgaki M, Daskalakis M, et al. Cytoreductive surgery and intraoperative hyperthermic intraperitoneal chemotherapy with paclitaxel: a clinical and pharmacokinetic study. Ann Surg Oncol. 2008;15:1183–92. [DOI] [PubMed] [Google Scholar]
- [31].Sticca RP, Dach BW. Rationale for hyperthermia with intraoperative intraperitoneal chemotherapy agents. Surg Oncol Clin N Am. 2003;12:689–701. [DOI] [PubMed] [Google Scholar]
- [32].Gonzalez-Moreno S, Gonzalez-Bayon LA, Ortega-Perez G. Hyperthermic intraperitoneal chemotherapy: Rationale and technique. World J Gastrointest Oncol. 2010;2:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–72. [DOI] [PubMed] [Google Scholar]
- [34].Lanneau D, Brunet M, Frisan E, Solary E, Fontenay M, Garrido C. Heat shock proteins: essential proteins for apoptosis regulation. J Cell Mol Med. 2008;12:743–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hoter A, Naim HY. Heat Shock Proteins and Ovarian Cancer: Important Roles and Therapeutic Opportunities. Cancers (Basel). 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Abbasi FM, D.; Xiong Y; Bush S; Al Sawah E; Al Rubaish S; Ramirez I; Zgheib NB; Wenham R; Lancaster J HSP90 inhibition decreases ovarian cancer cell proliferation and potentiates platinum sensitivity. Gynecologic Oncology. 2014;133:122–3. [Google Scholar]
- [38].Lianos GD, Alexiou GA, Mangano A, Mangano A, Rausei S, Boni L, et al. The role of heat shock proteins in cancer. Cancer Lett. 2015;360:114–8. [DOI] [PubMed] [Google Scholar]
- [39].Okui T, Shimo T, Hassan NM, Fukazawa T, Kurio N, Takaoka M, et al. Antitumor effect of novel HSP90 inhibitor NVP-AUY922 against oral squamous cell carcinoma. Anticancer Res. 2011;31:1197–204. [PubMed] [Google Scholar]
- [40].Jensen MR, Schoepfer J, Radimerski T, Massey A, Guy CT, Brueggen J, et al. NVP-AUY922: a small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models. Breast Cancer Res. 2008;10:R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cui XB, Yu ZY, Wang W, Zheng YQ, Liu W, Li LX. Co-inhibition of HSP70/HSP90 synergistically sensitizes nasopharyngeal carcinoma cells to thermotherapy. Integr Cancer Ther. 2012;11:61–7. [DOI] [PubMed] [Google Scholar]
- [42].Moser C, Lang SA, Kainz S, Gaumann A, Fichtner-Feigl S, Koehl GE, et al. Blocking heat shock protein-90 inhibits the invasive properties and hepatic growth of human colon cancer cells and improves the efficacy of oxaliplatin in p53-deficient colon cancer tumors in vivo. Mol Cancer Ther. 2007;6:2868–78. [DOI] [PubMed] [Google Scholar]
- [43].Javellana M, Eckert MA, Heide J, Zawieracz K, Weigert M, Ashley S, et al. Neoadjuvant Chemotherapy Induces Genomic and Transcriptomic Changes in Ovarian Cancer. Cancer Res. 2022;82:169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Patient and sample information.
Supplementary Table 2: Differential gene expression changes between tumor and normal tissues pre- and post-HIPEC treatment.
Supplementary Table 3: Gene set enrichment analysis and gene ontology classification of genes differentially expressed between tumor and normal tissues pre- and post-HIPEC.
Supplementary Table 4: Statistical comparisons of change in gene expression, protein expression and cell type composition.


