Abstract
The mechanisms underlying the regulation of a checkpoint receptor, PD-1, in tumor-infiltrating immune cells during the development of colorectal cancer (CRC) are not fully understood. Here we demonstrate that COX-2-derived PGE2, an inflammatory mediator and tumor promoter, induces PD-1 expression by enhancing NF-κB’s binding to the PD-1 promoter via an EP4-PI3K-Akt signaling pathway in both CD8+ T cells and macrophages. Moreover, PGE2 suppresses CD8+ T cell proliferation and cytotoxicity against tumor cells and impairs macrophage phagocytosis of cancer cells via an EP4-PI3K-Akt-NF-κB-PD-1 signaling pathway. In contrast, inhibiting the COX-2-PGE2-EP4 pathway increases intestinal CD8+ T cell activation and proliferation and enhances intestinal macrophage phagocytosis of carcinoma cells accompanied by reduction of PD-1 expression in intestinal CD8+ T cells and macrophages in ApcMin/+ mice. PD-1 expression correlates well with COX-2 levels in human CRC specimens. Both elevated PD-1 and COX-2 are associated with poorer overall survival in colorectal cancer patients. Our results uncover a novel role of PGE2 in tumor immune evasion. They may provide the rationale for developing new therapeutic approaches to subvert this process by targeting immune checkpoint pathways using EP4 antagonists. In addition, our findings reveal a novel mechanism explaining how NSAIDs reduce colorectal cancer risk by suppressing tumor immune evasion.
Keywords: PGE2, PD-1, colorectal cancer, CD8+ T cells, macrophages
Introduction
Chronic inflammation is one of the known risk factors for colorectal cancer (CRC). Clinical and epidemiologic evidence suggests that long-term use of nonsteroidal anti-inflammatory drugs (NSAIDs) reduces the risk for developing colorectal adenomas and CRC and suppresses CRC development (1). Nonselective NSAIDs are known to exert certain anti-inflammatory effects by targeting cyclooxygenase enzymes (COXs) that include COX-1 and COX-2. COX-1 is constitutively expressed in most tissues. In contrast, COX-2 is an immediate-early response gene usually absent in healthy tissues and organs. However, it is highly inducible at sites of inflammation and in the tumor microenvironment of certain cancers, including CRC. For example, COX-2 expression is elevated in approximately 50% of colorectal adenomas and 85% of adenocarcinomas (2,3) and associated with poorer overall survival in patients with CRC (4). Both COX-1 and COX-2 convert arachidonic acid to prostanoids, including prostaglandins (PGs) such as PGE2, PGD2, PGF2α, PGI2, and thromboxane A2 (TxA2) by a two-step process. Among prostanoids, PGE2 is the most abundant found in various types of human malignancies, including CRC, and its presence is often associated with a poor prognosis (5–8). More importantly, only PGE2 and PGI2 levels are elevated in CRC specimens compared to matched normal tissues (9).
Moreover, a urinary PGE2 metabolite (PGE-M) has been evaluated as a biomarker for CRC (10,11). More importantly, a recent epidemiologic study revealed that regular use of NSAIDs, including aspirin, significantly reduced the risk of developing colorectal adenomas in women with high PGE-M levels. This effect was not observed in those with low PGE-M levels (12). PGE2 exerts its cellular effects by binding to specific G-protein coupled receptors, EP1, EP2, EP3, and EP4. Among prostanoids, only PGE2 has been demonstrated to promote intestinal adenoma formation and growth by induction of tumor epithelial cell proliferation and survival, as well as promoting angiogenesis in mouse models of intestinal adenomas (13). However, all the mechanisms that PGE2 accelerates colorectal adenoma development are not fully understood. For example, relatively little is known about the impact of PGE2 on host defenses against tumor cells within the tissue microenvironment.
Tumor immune evasion can occur through several mechanisms, including tumor cell resistance to immune attack, activation of immune checkpoint pathways in effector T cells, suppression of macrophage phagocytosis of tumor cells, and induction of massive infiltration of immunosuppressive cells. For example, tumor cells can evade immunosurveillance by directly impairing CD8+ T cell cytotoxic activity and proliferation through immune checkpoint receptors such as PD-1. Interaction of PD-1 with its ligands, PD-L1 and PD-L2, suppresses CD8+ T cell cytotoxicity and proliferation. It has been documented that a strong and persistent expression of PD-1 is observed in tumor-infiltrating effector T cells (14–16). Tumor-associated macrophages (TAMs) are a significant subpopulation of tumor-infiltrating immune cells (17). Increased infiltration of TAMs is recognized as a poor prognostic sign in CRC patients (18). A recent report revealed that PD-1 expression in macrophages inhibited their phagocytosis of tumor cells (19). However, the mechanisms by which PD-1 is regulated in CD8+ T cells and macrophages in the tumor microenvironment are still largely unknown.
Immunotherapies using checkpoint inhibitors offer great promise for treating some malignancies, but their effectiveness in many solid tumors has been disappointing. For example, the therapeutic effect of a PD-1 monoclonal antibody was mainly observed in patients with MSI CRC but not in patients with MSS CRC (20). However, MSI is only found in about 15% of sporadic CRC, whereas approximately 85% of sporadic CRC is classified as MSS. Interestingly, the elevation of COX-2 expression was observed in 79% of MSS CRC, but only 48% of MSI CRC (21). Other studies also revealed that MSI CRC has a low or absent COX-2 expression (22–24). One potential explanation for the poor efficacy of checkpoint inhibitors in MSS CRC is that the presence of COX-2-derived PGE2 might attenuate their effect by reducing tumor-infiltrating CD8+ T cell abundance and cytotoxicity and/or by impairing TAM phagocytosis of tumor cells. In this study, we investigate whether activation of the COX-2-PGE2 pathway induces tumor immune evasion by reducing tumor-infiltrating CD8+ T cell cytotoxicity and abundance via PD-1 and/or by impairing macrophage phagocytosis of tumor cells via PD-1.
Materials and Methods
Animal experiments
All animal experiments were completed according to our animal protocols approved by the Institutional Animal Care and Use Committee at the Medical University of South Carolina (MUSC). ApcMin/+ mice were purchased from Jackson Laboratory (Cat#002020). For experiments evaluating celecoxib or Ono-AE3–208 treatment, ApcMin/+ male mice at the age of 6 weeks old were randomly divided into two groups fed with a control diet, a diet containing 500ppm of celecoxib, or provided with water containing Ono-AE3–208 (10 mg/kg) for ten weeks. No significant sex variable exists in the intestinal tumor burden of ApcMin/+ mice.
Isolation of immunocytes from organs
All fat and Peyer’s patches were removed from excised intestines under a dissecting microscope for intestinal immune cell preparation. Mouse normal intestinal tissues and adenomas were minced and digested with RPMI 1640 medium containing 5% FBS, 1 mM MgCl2, 1 mM CaCl2, 2.5 mM HEPES, and 200 units/ml collagenase I (Gibco). The immune cells from intestinal tissues were enriched by using a discontinuous (44% and 67%) percoll (GE) separation method. Isolated immune cells were subjected to Flow Cytometry or in vitro culture. Excised spleens were smashed using a 40 μm cell strainer. The red blood cells (RBCs) in the spleen were lysed with RBC lysis buffer (eBioscience). Mouse CD8+ T cells were isolated from the spleen using a mouse CD8+ T Cell Isolation Kit (Miltenyi Biotec Inc., San Diego, CA) according to the manufacturer’s instructions; isolated CD8+ T cells with a purity of 98% were subjected to experiments listed below.
Prostaglandin measurement
Intestinal tissues were homogenized in PBS with 10% 2,6-di-tert-butyl-p-cresol as previously described (25,26). The levels of prostaglandins, including PGE2, were measured using mass spectrometry and normalized with protein concentration as previously described (25,26).
Cell culture and reagents
Isolated mouse splenic CD8+ T cells were cultured in RPMI 1640 medium with 10% FBS (Gibco, Cat#10082147), 25 mM HEPES, 100 IU/ml IL-2 (Peprotech, Cat#200–02), 1:100 100x non-essential amino acids (Sigma-Aldrich) and 1mM sodium pyruvate, and 50 μM β-mercaptoethanol. Mouse CD8+ T cells were activated by adding anti-CD3/CD28 mAb-coated beads (Dynabeads Mouse T-Activator CD3/CD28, Gibco) according to the manufacturer’s instructions. Then cells were starved for 24 hrs. Human monocytic THP-1 (Cat#TIB-202) and HCT-116 (Cat#CCL-24&) cells were obtained from ATCC (Manassas, VA) and maintained in RPMI 1640 with 10% FBS. THP-1 monocytes were treated with 50 mg/ml phorbol 12-myristate 13-acetate (PMA) 24 hrs to stimulate their differentiation into macrophages, followed by incubation for 24 hrs in RPMI medium. Macrophages were treated with 20 ng/ml IL-4 (R&D Systems) and 20 ng/ml IL-13 (R&D Systems) for 24 hrs. THP-1-derived macrophages were starved for 48 hrs followed by treatment with the indicated dose of PGE2 (Cayman Chemical, Ann Arbor, Michigan), 0.1 μM Ly294002 (Calbiochem-Novabiochem Corp., San Diego, CA), 1 μM MK2206 (Selleck Chemicals), and/or 1 μM BAY110785 (ENZO) for the times indicated. For bone marrow-derived macrophages (BMDMs), bone marrow cells were flushed aseptically from the femurs of ApcMin/+ mice (Jackson Laboratory, Cat#000664) and cultured in Falcon™ Petri dishes (BD Biosciences, Cockeysville, MD) with DMEM medium supplemented with 10% FBS and 10 ng/ml of macrophage colony stimulating factor (M-CSF) for 4 days. After reseeding cells, cells were cultured in DMEM with 10% FBS, 10 ng/ml M-CSF (Life Technologies), and 20 ng/ml IL-4 (R&D Systems) for 2 days. BMDMs were cultured in serum-free DMEM medium for 2 days and treated with the indicated dose of PGE2 and/or inhibitors for indicated time for 24 hr. All cell lines were tested using a MycoProbe Mycoplasma Detection Kit (R&D) and authenticated before each experiment using the ATCC STR database.
Quantitative PCR
Total RNA was isolated from indicated cells using an RNeasy Mini Kit (Qiagene), and 1 μg of RNA was reversely transcribed to cDNA using High-Capacity cDNA Reverse Transcription Kits (Bio-Rad). Real-time q-PCR was performed with a SYB® Gene Expression Assay Mix (Bio-Rad) using a QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA). The sequences of the specific PCR primers were as follows (5’ to 3’): mPdcd1 (PD-1), forward: CCTCTGACACTGTGAGCCAG, reverse: GCAGGTACCCTGGTCATTCA; hPDCD1, forward: CTCCGATGTGTTGGAGAAGC, reverse: CGGCCAGGATGGTTCTTAG; β-actin, forward: AGAAAATCTGGCACCACACC; reverse: AGAGGCGTACAGGGATAGCA. The relative expression of each target gene represents an average of triplicates that are normalized against the transcription levels of β-actin.
Western blotting
Whole-cell or nuclear extracts were prepared from the indicated cells in each experiment. Transfer membranes were blocked with 5% dry milk in TBS-T buffer for 1 hr and then incubated with anti-pAkt (1:1000, Cell Signaling, Cat#4058), anti-Akt (1:1000, Cell Signaling, Cat#4691), anti-NF-κB (1:1000, Santa Cruz, Cat#SC372), anti-GAPDH (1:5000, Cell Signaling, Cat#8884S), or anti-LaminA (1:1000, Santa Cruz, Cat#SC56137) antibody for overnight at 4 °C.
Flow cytometry analysis and sorting
For single color flow cytometry immunotypic analyses, each indicated single-cell suspension was incubated with anti-mPD-1-PE (1:100, BioLegend, Cat#135206), anti-hPD-1-PE (1:100), or the corresponding isotype controls in staining buffer (BD Biosciences). For multicolor flow cytometry immunotypic analyses, mouse intestinal immunocyte cells were incubated with the appropriate combination of the following antibodies in staining buffer at the following dilution for 30 min on ice: PD-1-PE (1:100), CD45-PE-Cy7 (1:250, BioLegend, Cat#103114), CD8-FITC (1:50, BioLegend, Cat#100706), CD4-AF700 (1:100, BioLegend, Cat#100536), CD3–Percep5.5 (1:100, Invitrogen, Cat#45–0031-82), and V450 (1:1500, Invitrogen, Cat#65–0863-14) for lymphocytes; PD-1-PE (1:100), CD45-APC (1:100, BioLegend, Cat#103112), CD11b-FITC (1:10, Miltenyi Biotech, Cat#130–081-201), Ly6G-Alexa700 (1:50, BD Biosciences, Cat#561236), CD11c-PE-Cy7 (1:80, BioLegend, Cat#117318), F4/80-Percep5.5 (1:100, Invitrogen, Cat#45–4801-32), V450 (1:1500) for granulocytes/monocytes. Dead cells were excluded using V450 staining. To analyze INFγ expression on CD8+ T cells, immunocyte cells were stained with cell surface markers for CD8+ T cells as described above. Then, the cells were fixed and permeabilized using a Cytofix/Cytoperm kit (BD Biosciences, Cat#554714) followed by intracellular cellular staining with anti-mouse INFγ-FITC antibody (1:50, BD Biosciences, Cat#554411) in permeabilization buffer for 30 min on ice. After incubation of antibodies, the cells were analyzed on a Fortessa X-20 cytometer (BD Biosciences). The flow cytometric profiles were analyzed by counting 30,000 events using FlowJo X software (Tree Star).
Chromatin Immunoprecipitation (ChIP) Assay
The Magna ChIP™ A/G Chromatin IP Kit (EMD Millipore, Cat#MAGNA0017) was used for ChIP assays according to the manufacturer’s instructions. Briefly, cells were treated with 1% formaldehyde for 10 min at room temperature to crosslink proteins to DNA, then quenched by adding glycine to 0.125 M for 5 min at room temperature. Chromatin was sonicated to an average length of 400–600bp. 5 μg of chromatin was immune-precipitated by 0.5 μg IgG or anti-NF-κB p65 antibody (Cell Signaling, Cat#8242). After being reverse crosslinked, the DNA was purified and eluted into 50 μl of elution buffer. Immunoprecipitated DNA was detected by PCR. The primers used for PCR were as follows (5’ to 3’): mPDCD1 (PD-1), forward: 5′-GTGAGACCCACACATCTCATTGC-3′ and reverse: 5′-CCTCACCTCCTGCTTGTCTCTC-3′ and hPDCD1 (PD-1), forward: 5’- AGA GAC ACA GAG GAG GAA GG −3’ and reverse: 5’-AGG GAC TGA GAG TGA AAG GT −3’. All ChIP assays were performed in three independent experiments.
Transient transfection and luciferase assays
The mouse PD-1 luciferase reporter plasmids with WT and mutant NF-κB binding sites (mNF-κB1 or mNF-κB2) were obtained from Dr. Jeremy Boss (27). Mutant NF-κB binding sites were generated by replacing NF-κB binding sites with the scrambled sequences. These scrambled sequences were predicted to have no NF-κB and other transcription factor binding potential (27). The human PD-1 promoter (−884 to +140) was cloned into a PGL3 vector (Promega, Cat#E1751). The NF-κB1 (GGGGATGGGCC) and NF-κB2 (GGGGAGACCCC) elements within the PD-1 promoter were mutated to mutant NF-κB1 (GTTTATGGGAA) and mutant NF-κB2 (GTTTAGACAAA) sites using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). For dual-luciferase reporter assays, mouse CD8+ T cells or THP-1 cells were transfected with 5 μg of a mouse or human PD-1-Luc plasmid and 0.2 μg pRL-TK (Promega, Cat#E2231) DNA as a control using a Cell line Nucleofector Kit V (Lonza, Cat#VCA-1003) according to the manufacturer’s instructions. After transfection, CD8+ T cells were starved for 24 hrs and treated with 0.1μM PGE2 for 24h. Transfected TPH-1 cells were treated with 50 ng/ml PMA for 24 hrs. TPH-1-derived macrophages were treated with 0.1 μM PGE2 for 24 hrs after a 48 hr starvation period. Following treatment, cells were lysed using cell lysis buffer provided in the kit (Promega, Cat#E1960). Luciferase activity was measured using a Dual-luciferase reporter assay kit (Promega) with a Monolight 3010 luminometer (BD Biosciences/Pharmingen, San Diego, CA). The relative luciferase activity was determined and normalized to Renilla luciferase activity.
CFSE cell proliferation
Mouse splenic activated CD8+ T cells were labeled with 5 μM carboxyfluorescein succinimidyl ester (CFSE) (Life Technologies, Cat#C34554) according to the manufacturer’s instructions. CFSE-labeled CD8+ T cells were starved for 24 hrs and treated with 0.1 μM PGE2, 20 μg/ml IgG, 20 μg/ml anti-PD-1 (Bio X Cell, Cat#BE0273), 0.1 μM Ly294002, 1 μM MK2206, and/or 1 μM BAY110785 for 48 hrs. For intestinal CD8+ T cells, immune cells isolated from intestinal normal tissues or tumors were labeled with 0.5 μM CFSE. CFSE-labeled immune cells were cultured in RPMI1640 medium with 10%FBS for 24 hrs. Then the cells were stained with the anti-CD8, anti-CD3 antibodies, and V450. CD8+ T cells proliferation was measured by a Fortessa X-20 cytometer.
Macrophage phagocytosis
THP-1-derived macrophages were starved for 4h and then treated with 0.1 μM PGE2, 20 μg/ml IgG, 20 μg/ml anti-PD-1 (Nivolumab, BioVision, Cat#A137–100), 0.1 μM Ly294002, 1 μM MK2206, and/or 1 μM BAY110785 for 24 hrs. After treatment, 0.5 ×106 macrophages were co-cultured with 1 ×106 GFP-labeled HCT-116 cells for 3 hrs. For mouse intestinal macrophages, 5 × 105 mouse intestinal immune cells were co-cultured with 2 × 105 GFP-labeled MC26 cells for 3 hrs. Human cells were incubated with the antibodies indicated, including hCD45-PE-Cy7 and hCD11b-APC, as well as V450 in flow cytometry buffer. Mouse cells were incubated with antibodies, including mCD45-PE-Cy7, mCD11b-APC, and mF4/80-PerCP/Cy5.5 as well as V450 in flow cytometry buffer. Human macrophages that phagocytose GFP-labeled tumor cells were referred to as CD45+CD11b+GFP+ cells, whereas mouse macrophages that phagocytose GFP-labeled tumor cells were indicated as CD45+CD11b+F4/80+GFP+ cells by flow cytometry analysis. The phagocytic index was calculated as the percentage of CD45+CD11b+GFP+ cells in total CD45+CD11b+ cells for human cells and the percentage of CD45+CD11b+F4/80+GFP+ cells in total CD45+CD11b+F4/80+ cells for mouse cells.
Assays for CD8+ T cell cytotoxicity against tumor cells
Mouse splenic activated CD8+ T cells were treated with 30 U/ml IL-2, 0.1 μM PGE2, 20 μg/ml IgG, 20 μg/ml anti-PD-1 (Bio X Cell), 0.1 μM Ly294002, 1 μM BAY110785, and/or 1 μM MK2206 for 48 hrs. After treatment, CD8+ T cells were co-cultured with 2.5 × 105 MC26 in 6-well plate at ratios (E:T=2:1) for 16 hrs. Then, the cells were incubated with anti-mEpCAM-PE-Cy7 (1:1000, BioLegend, Cat#118216) and anti-mCD3-APC (1:100, BioLegend, Cat#103112) antibodies in staining buffer (Biolegend) for 30 min on ice. The cells were then washed twice with 1 ml of staining buffer and stained with propidium iodide (PI) using a TACS™ Annexin V-FITC Apoptosis Detection Kit according to the manufacturer’s instructions (R&D System). The EpCAM+CD3-PI+ cells were dead epithelial cells and the percentage of dead epithelial cells in total epithelial cells (EpCAM+CD3-) was presented.
Statistical analysis
All in vitro experiments were performed at least 3 times. All animal experiments were repeated at least twice. Results were expressed as mean + SEM. In general, we used the Bonferroni test by factorial analysis of variance to compare outcomes among multiple groups of mice. Student’s t-test or Mann-Whitney U test were used to compare two groups. For all tests, p<0.05 was considered statistically significant.
Data availability statement
The data generated in this study are available within the article and its supplementary data file. Raw data generated in this study are available from the corresponding author upon request. The data analyzed in this study were obtained from the TCGA Colorectal Adenocarcinoma Provisional dataset and in Gene Expression Omnibus (GEO) at GSE17537 and GSE17538.
RESULTS
The COX-2-PGE2-EP4 pathway is involved in the induction of PD-1 expression in intestinal CD8+ T cells and macrophages
To determine whether the COX-2-PGE2 pathway regulates immune checkpoint receptors, we first examined the effect of a COX-2 selective inhibitor, celecoxib, on the expression of immune checkpoint receptors in mouse intestinal immune cells. Treatment of ApcMin/+ mice with celecoxib reduced PD-1 expression in CD8+ T cells, which are resident in both small and large intestinal adenomas and matched normal tissues (Fig. 1A). Interestingly, CD8+ T cell abundance in adenomas is much less than that found in normal tissues; however, celecoxib treatment restored CD8+ T cell abundance in intestinal adenomas as well as matched normal tissues (Fig. 1B). Celecoxib treatment also significantly reduced intestinal prostaglandin levels, including PGE2 (Fig. S1A), providing evidence that PGE2 levels correlated with CD8+ T cell abundance by induction of PD-1 in intestinal tumors and matched normal tissues. Similarly, celecoxib treatment also reduced PD-1 expression in macrophages, resident in both small and large intestinal adenomas, and matched normal tissues (Fig. 1C). In contrast, celecoxib treatment does not appear to affect PD-1 expression in other intestinal immune cells (Fig. S1B). CTLA4 expression in intestinal CD8+ T and CD4+ T cells was not significantly affected following celecoxib treatment as well (Fig. S1C).
Fig. 1. Inhibition of the COX-2-PGE2 pathway reduces PD-1 expression in intestinal CD8+ T cells and macrophages and decreases intestinal CD8+ T cell abundance.
ApcMin/+ mice were treated with vehicle, celecoxib, or Ono-AE3–208 as described in the Methods. (A-B) The percentage of PD-1-positive CD8+ T cells in total CD8+ T cells (A) and the percentage of CD8+ T cells in total T lymphocytes (B) in both small intestinal (SI) and large intestinal (LI) tumors (T) and matched normal tissues (N) were analyzed by Flow Cytometry. (C) The percentage of PD-1-positive macrophages in total macrophages, which are resident in both SI and LI tumors and matched normal tissues, were analyzed by Flow Cytometry. (D-E) The percentage of PD-1-positive CD8+ T cells in total CD8+ T cells (D) and the percentage of CD8+ T cells in total T lymphocytes (E) in both small intestinal and large intestinal tumors and matched normal tissues were analyzed by Flow Cytometry. (F) The percentage of PD-1-positive macrophages in total macrophages, which are resident in both SI and LI tumors and matched normal tissues, were analyzed by Flow Cytometry. The error bar indicates + SEM. * indicates p<0.05.
Expression of EP2 and EP4 receptors is higher in human CRC specimens than in normal colon tissues (28–30). In contrast, the expression of EP3 receptors is reduced in human CRC specimens compared to adjacent normal colon tissues (30). Multiple lines of in vivo evidence further demonstrated that PGE2 promotes intestinal adenoma formation and growth via EP2 and EP4 receptors, but not EP3 (31). The involvement of EP1 in intestinal tumorigenesis remains unclear because different groups have reported conflicting results (31). Importantly, immune cells mainly express EP2 and EP4 receptors. For example, macrophages express EP2 and EP4, but not EP1 or EP3 (32). Moreover, the EP4 receptor has a higher affinity for PGE2 than EP1 or EP2 (33). Indeed, EP2 and EP4 mRNA levels were higher than EP1 and EP3 in splenic CD8+ T cells and bone marrow-derived macrophages (BMDM) isolated from ApcMin/+ mice as well as in THP-1-derived macrophages (Fig. S2A). Notably, only EP4, but not EP2, is expressed on the cell surface of splenic CD8+ T cells and bone marrow-derived macrophages (BMDM) isolated from ApcMin/+ mice (Fig. S2B). In human THP-1-derived macrophages, EP2 is marginally detectable, whereas EP4 is highly expressed (Fig. S2B). Therefore, we examined whether inhibition of PGE2 signaling pathways by targeting its EP4 receptor has a similar effect as that seen following treatment with COX-2 inhibitors. Like celecoxib, treatment of ApcMin/+ mice with an EP4 antagonist (Ono-AE3–208) led to a reduction of PD-1 expression in CD8+ T cells and macrophages, which are resident in both small and large intestinal adenomas and matched normal tissues accompanied by increased intestinal CD8+ T cell abundance (Fig. 1D–F). As expected, Ono-AE3–208 treatment did not affect PD-1 expression in other intestinal immune cells or CTLA-4 expression in intestinal CD8+ T and CD4+ T cells (Fig. S2C–D).
PGE2 directly induces PD-1 expression via an EP4-PI3K-AKT-NF-κB pathway in CD8+ T cells and macrophages
To further evaluate whether PGE2 directly induces PD-1 expression in CD8+ T cells and macrophages, in vitro experiments were performed. PGE2 induces PD-1 mRNA levels in mouse splenic activated CD8+ T cells and THP-1-derived macrophages (Fig. 2A–B) as well as mouse BMDMs (Fig. S3A–B). Significantly, PGE2 induces PD-1 expression levels on the cell surface (Figs. 2C and S3C–D). Moreover, time- and dose-dependent effects of PGE2 on PD-1 expression were observed in these cells (Figs. 2A–C and S3A–D).
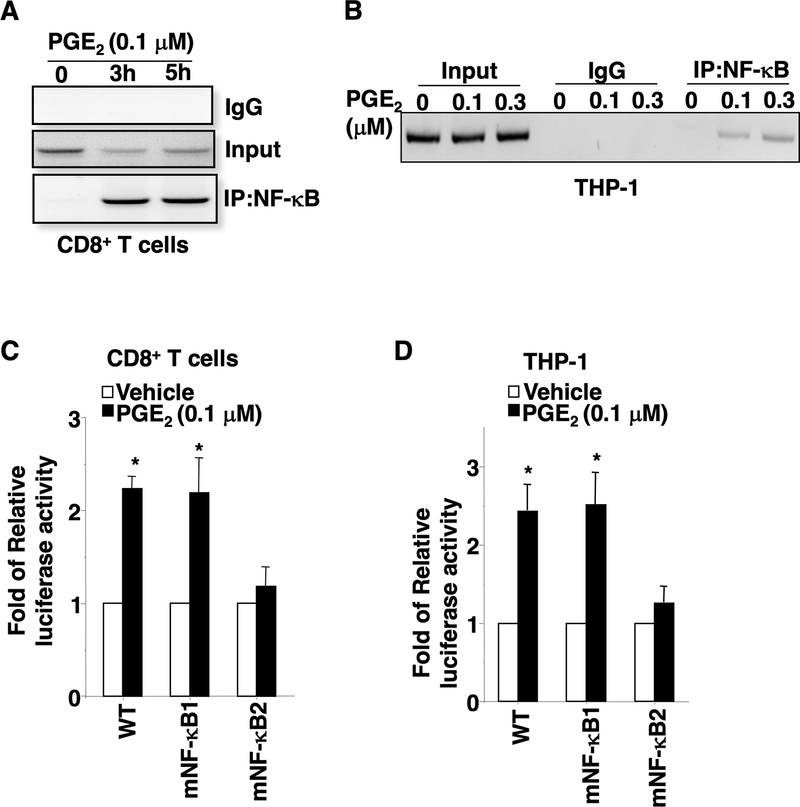
Fig. 2. PGE2 induces PD-1 expression via a PI3K-NF-κB pathway in CD8+ T cells and macrophages.
(A-B) PGE2 induces PD-1 expression at mRNA in a dose- and time-dependent manner in mouse splenic activated CD8+ T cells and THP-1-derived macrophages. (C) After PGE2 treatment, the percentage of PD-1-positive cells in total mouse splenic activated CD8+ T cells or total THP-1-derived macrophages were determined by Flow Cytometry. (D) After PGE2 treatment, the levels of phosphor-AKT and the nuclear NF-κB p65 in mouse splenic activated CD8+ T cells and THP-1-derived macrophages were measured by Western blotting. The images are representative of three independent experiments. The band density of pAkt in CD8+ T cells was normalized by the band density of Gapdh. The results were reported as a mean of fold-induction from three independent experiments (middel panel). (E). The effect of a PI3K inhibitor (Ly294002), an AKT inhibitor (MK2209), and an NF-κB inhibitor (BAY110785) on PGE2 induction of PD-1 expression on the cellular surface. The error bar indicates ± SEM. *p<0.05.
The PD-1 promoter contains two potential NF-κB binding sites located at NF-κB1 (−560 to −551) and NF-κB2 (−240 to −231) for human and NF-κB1 (−1265 to −1256) and NF-κB2 (−974 to −965) for mouse. LPS has been shown to induce PD-1 expression via binding of NF-κB to the NF-κB2 binding site within the PD-1 promoter in mouse macrophages (27). In addition, our previous results demonstrated that PGE2 could activate NF-κB via an EP4-dependent PI3K pathway in colorectal carcinoma cells (34,35). These results prompted us to postulate that PGE2 induces PD-1 expression by activating NF-κB via an EP4-PI3K pathway. To test this hypothesis, we first examined whether PGE2 induces AKT and NF-κB activation. As shown in Fig. 2D, treatment of mouse splenic activated CD8+ T cells and THP-1-derived macrophages with PGE2 led to increased phospho-AKT levels, and nuclear translocation of NF-κB. PGE2 induction of NF-κB nuclear translocation was also observed in mouse BMDMs (Fig. S3E). Moreover, inhibition of PI3K, AKT, or NF-κB by each respective inhibitor attenuated the effect of PGE2 on induction of PD-1 expression in mouse splenic activated CD8+ T cells, THP-1-derived macrophages, and mouse BMDMs (Figs. 2E and S3F). Interestingly, Ly294002 treatment at 0.1 μM inhibited PGE2 induction of PD-1 mRNA expression in these cells (Fig. S3G).
To further examine whether PGE2 induces PD-1 transcription by increasing binding of NF-κB to cis-elements in the PD-1 promoter, ChIP and PD-1 luciferase reporter assays were performed. As shown in Fig. 3A–B, PGE2 induced binding of NF-κB to the PD-1 promoter in both mouse splenic activated CD8+ T cells and THP-1-derived macrophages. Moreover, PGE2 induces PD-1 transcription in both mouse splenic activated CD8+ T cells and THP-1-derived macrophages (Fig. S3H). As expected, site-directed mutations of the NF-κB2 binding element within the PD-1 promoter abolished PGE2 induction of transcription in these cells, whereas mutations of the NF-κB1 binding site within the PD-1 promoter did not affect PGE2 induction of transcription (Fig. 3C–D). These results demonstrate that only the NF-κB2 binding site is required for PGE2 induction of PD-1 transcription. Since EP4, but not EP2, is expressed on the surface of these cells, our results reveal that PD-1 induction is regulated mainly via the EP4-PI3K-AKT-NF-κB pathway.
Fig. 3. PGE2 induces PD-1 transcription by inducing the binding of NF-κB to the PD-1 promoter.
(A-B) Representative image of three independent ChIP assays for NF-κB binding to the PD-1 promoter in mouse splenic activated CD8+ T cells and THP-1-derived macrophages after PGE2 treatment. (C-D) The luciferase activity of WT and mutant PD-1 promoters in mouse splenic activated CD8+ T cells and THP-1-derived macrophages was measured after PGE2 treatment. The error bar indicates ± SEM. *p<0.05.
PGE2 suppresses CD8+ T cell proliferation and cytotoxicity and impairs macrophage phagocytosis of tumor cells via an EP4-PI3K-AKT-NF-κB-PD-1 pathway in vitro
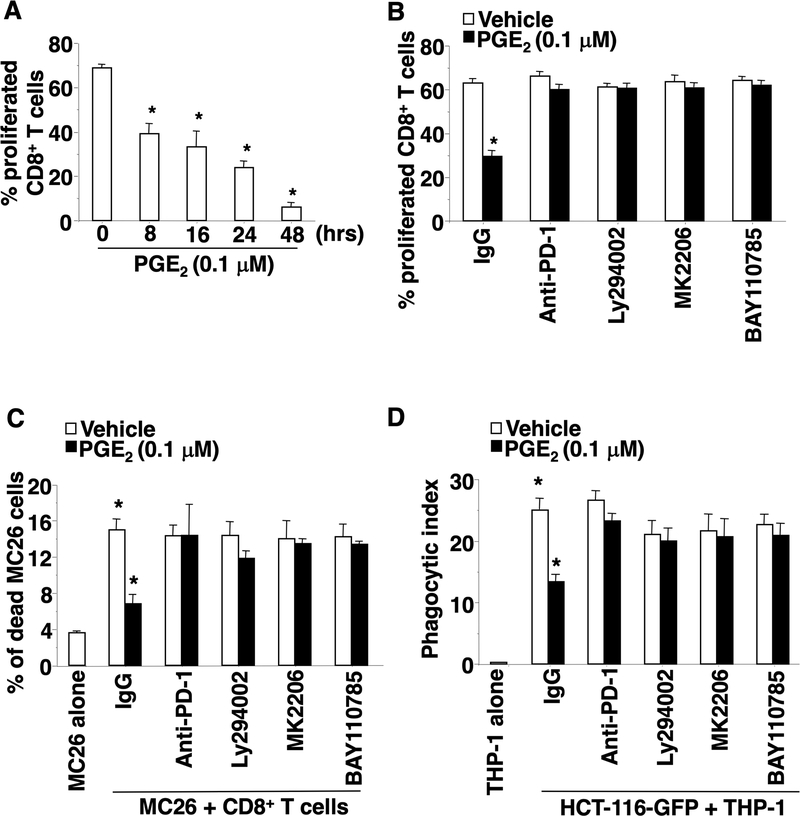
We first evaluated the impact of PGE2 on CD8+ T cell proliferation and cytotoxicity against tumor cells and found that PGE2 inhibited proliferation of mouse splenic activated CD8+ T cells in a time-dependent manner (Figs. 4A). In addition, PGE2 suppressed mouse splenic activated CD8+ T cell cytotoxicity against MC26 cells in a dose-dependent manner (Fig. S4A). Blockage of PD-1 signaling by using a neutralizing antibody attenuated the effect of PGE2 on suppression of CD8+ T cell proliferation and cytotoxicity against MC26 cells (Fig. 4B–C). Moreover, inhibition of PI3K, Akt, or NF-κB by each respective inhibitor also blocked the effect of PGE2 on suppression of CD8+ T cell proliferation and cytotoxicity against MC26 cells (Fig. 4B–C). Like CD8+ T cells, PGE2 reduced THP-1-derived macrophage phagocytosis of HCT-116 cells (Fig. 4D) and mouse BMDM phagocytosis of MC26 cells (Fig. S4B). Treatment with a PD-1 monoclonal antibody, a PI3K inhibitor, an Akt inhibitor, or an NF-κB inhibitor attenuated the effect of PGE2 on inhibition of THP-1-derived macrophage phagocytosis of HCT-116 cells (Fig. 4D) and BMDM phagocytosis of MC26 cells (Fig. S4B). These results demonstrate that PGE2 suppresses CD8+ T cell proliferation and cytotoxicity and impairs macrophage phagocytosis of tumor cells via an EP4-PI3K-AKT-NF-κB-PD-1 pathway in vitro.
Fig. 4. PGE2 suppresses CD8+ T cell proliferation and cytotoxicity and inhibits macrophage phagocytosis via a PI3K-Akt-NF-κB-PD-1 pathway.
(A) After PGE2 treatment, the percentage of proliferating CD8+ T cells was measured by CFSE-based proliferation assays. (B) The effect of a PD-1 neutralizing antibody and an indicated inhibitors on PGE2 reduction of CD8+ T cell proliferation. (C) Mouse splenic activated CD8+ T cells were treated with PGE2, a PD-1 neutralizing antibody, and/or an indicated inhibitor. After treatment, CD8+ T cells were co-cultured with GFP-labeled MC26 cells. The percentage of dead MC26 cells in total MC26 cells was determined by Flow Cytometry. (D) THP-1-derived macrophages were treated with PGE2, a PD-1 neutralizing antibody, and/or an indicated inhibitor. After treatment, THP-1-derived macrophages were co-cultured with GFP-labeled HCT-116 cells. The phagocytic index is the percentage of macrophages that phagocytose GFP-labeled tumor cells in total macrophages. The error bar indicates ± SEM. *p<0.05.
Blocking the COX-2-PGE2-EP4 pathway inhibits tumor immune evasion by increasing intestinal CD8+ T cell activation and inducing macrophage phagocytosis
Since inhibition of the COX-2-PGE2-EP4 pathway reduces PD-1 expression in intestinal CD8+ T cells and macrophages, it was conceivable that inhibition of this pathway could induce intestinal CD8+ T cell activation and macrophage phagocytosis of tumor cells. As shown in Fig. 5A–D, treatment of ApcMin/+ mice with celecoxib or Ono-AE3–208 resulted in increased proliferation and IFNγ production in CD8+ T cells in intestinal adenomas and matched normal tissues, demonstrating that inhibition of the COX-2-PGE2-EP4 pathway increases CD8+ T cell activation in intestinal adenomas and matched normal tissues. Treatment of ApcMin/+ mice with celecoxib or Ono-AE3–208 also enhanced the ability of macrophages isolated from intestinal adenomas and matched normal tissues to phagocytize mouse MC26 cells (Fig. 5E–F). As expected, treatment of ApcMin/+ mice with celecoxib or Ono-AE3–208 reduced both small and large adenoma burden (Fig. 5G–H). These results suggest that PGE2 accelerates colorectal adenoma formation by suppressing CD8+ T cell cytotoxicity and macrophage phagocytosis against transformed epithelial cells via PD-1 in normal tissues and promotes colorectal adenoma growth by suppressing tumor-infiltrating CD8+ T cell cytotoxicity and TAM phagocytosis of tumor cells via PD-1.
Fig. 5. Inhibition of the COX-2-PGE2 pathway promotes intestinal CD8+ T cell activation and macrophage phagocytosis.
ApcMin/+ mice were treated with vehicle, celecoxib, or Ono-AE3–208 as described in the Methods. (A) The percentage of proliferated CD8+ T cells in small intestinal (SI) tumors (T) and matched normal tissues (N) were analyzed by Flow Cytometry. (B) The percentage of INFγ-positive CD8+ T cells in total CD8+ T cells in SI tumors and matched normal tissues were analyzed by Flow Cytometry. (C) The percentage of proliferated CD8+ T cells in small intestinal tumors and matched normal tissues were analyzed by Flow Cytometry. (D) The percentage of INFγ-positive CD8+ T cells in total CD8+ T cells in SI tumors and matched normal tissues were analyzed by Flow Cytometry. (E-F) After macrophages isolated from SI tumors and matched normal tissues were co-cultured with GFP-labeled MC26 cells, the phagocytic index is determined as described in Fig. 4. (G and H). Tumor numbers were counted, and size was measured. The error bar indicates ± SEM. *p<0.05.
PD-1 expression correlates well with COX-2 levels, and high levels of PD-1 and COX-2 are associated with poor survival in CRC patients
To further assess whether our preclinical results may have clinical relevance in humans, we first examined the correlation between mRNA levels of PDCD1 (PD-1) and PTGS2 (COX-2) in a human CRC database. We found a positive correlation between COX-2 and PD-1 mRNA levels in two TCGA and GEO databases (Fig. 6A–B). An evaluation of the GEO database reveals that CRC patients with high levels of both COX-2 and PD-1 in tumor tissues experienced a much poorer overall survival than those with low levels of both COX-2 and PD-1 (Fig. 6C). In the TCGA database, CRC patients with high levels of both COX-2 and PD-1 in tumor tissues tended to have poorer survival compared to patients with low levels of both COX-2 and PD-1. However, the difference does not reach statistical significance (p=0.068) (Fig. 6D).
Fig. 6. Correlations and survival between COX-2 and PD-1 in CRC patients.
(A-B) Pearson correlations between COX-2 and PD-1 mRNA expression in the TCGA Colorectal Adenocarcinoma Provisional dataset (n=381) (A) and the GSE17537 and GSE17538 datasets of the GEO database (n=434) (B). (C-D) Kaplan-Meier survival curves of the GSE17537 and GSE17538 datasets of GEO database (C) and the TCGA Colorectal Adenocarcinoma Provisional dataset (D).
DISCUSSION
Some of the roles of PGE2 in regulating immunity and host defense against viral, fungal, and bacterial pathogens have previously been reported (36). For example, one study showed that PGE2 suppressed CD8+ T cell survival and function during a chronic lymphocytic choriomeningitis virus infection (37). It is not clear whether PGE2 induces tumor immune invasion by inhibiting CD8+ T cell proliferation and cytotoxicity in vivo. This report provides the first in vivo evidence demonstrating that inhibition of the COX-2-PGE2-EP4 pathway increases intestinal CD8+ T cell abundance and activation accompanied by a reduction of PD-1 and tumor burden. Moreover, two in vivo studies suggest that PGE2 might promote intestinal adenoma formation and growth via its effects on macrophages. One study revealed that treatment with celecoxib resulted in a reduction of polyp burden accompanied by conversion of TAMs from the M2 to M1 in the ApcMin/+ mice (38). Another study reported that deletion of EP4 in myeloid cells resulted in reducing adenoma burden in the ApcMin/+ mice (39). However, the mechanisms by which PGE2 promotes colorectal adenoma formation and growth via macrophages were not fully understood or examined. Here our results demonstrate for the first time that inhibition of the COX-2-PGE2-EP4 pathway increases intestinal macrophage phagocytosis of tumor cells accompanied by reduced levels of PD-1 in intestinal macrophages as well as decreased tumor burden. The most striking of our findings is that inhibition of the COX-2-PGE2-Ep4 pathway promotes intestinal CD8+ T cell cytotoxicity and macrophage phagocytosis of epithelial cells via PD-1, which occurs well before tumor formation. This finding provides a potential explanation for the chemopreventive effect of NSAIDs on CRC that occurs very early during a precancer stage.
Following immune activation or infection, PD-1 expression is elevated in effector T cells, macrophages, DCs, B cells, and NKT cells. PD-1 expression is induced in CD8+ T cells following TCR stimulation via NF-AT (40). However, TCR-dependent PD-1 induction is weak and transient. It has been well documented that a strong and persistent expression of PD-1 is observed during chronic viral infection (41). Several cytokines such as INFα, IL-6, and IL-12 have enhanced TCR-induced PD-1 expression in CD8+ T cells (42,43). In addition to CD8+ T cells, PD-1 expression can be regulated by cytokines and TLR ligands such as LPS in macrophages. For example, LPS, TNFα, INFα, and IL-1β induced PD-1 expression in macrophages in vitro (27,44,45). However, how PD-1 is regulated in CD8+ T cells and macrophages in the tumor microenvironment is still largely unknown. Previous studies have shown that PD-1 expression in CD8+ T cells can be upregulated by different transcription factors such as FoxO1, IRF9, Notch, and c-Fos (42,46–48), and we found that activation of NF-κB by PGE2 via an EP4-PI3K-AKT pathway initiates PD-1 transcription via binding to the PD-1 promoter in CD8+ T cells. Our finding extends the scope of our understanding of the underlying mechanisms for PD-1 regulation in CD8+ T cells and macrophages. Moreover, our results also reveal that PGE2 leads to AKT activation in both CD8+ T cells and macrophages. Although AKT has been shown to activate NF-κB by phosphorylation of IKKα (49), further research is needed to determine the molecular mechanisms by which PGE2-induced AKT activation induces NF-κB activation in CD8+ T cells and macrophages. In addition, our results also reveal that PGE2 only induces PD-1, but not CTLA4, in CD8+ T cells. This finding indicates that combined inhibition of PGE2 signaling and CTLA4 may have more anti-tumor efficacy than combined inhibition of PGE2 signaling and PD-1.
Previous in vitro studies showed that PGE2 regulates CD8+ T cell proliferation and function. For example, treatment of CD8+ T cells with PGE2 resulted in suppression of cell proliferation by inducing replicative senescence (50). PGE2 treatment also inhibited CD8+ T cell cytotoxicity by inducing CD94 and the NKG2A C-type lectin receptor complex (51) or by inhibiting IFNγ release induced by TCR (52). Moreover, PGE2 produced by carcinoma cells also blocked initial priming of naïve CD8+ T cells to cytotoxic T cells by tumor cells (53). Here we reveal a novel mechanism by which PGE2 inhibits CD8+ T cell proliferation and cytotoxicity against tumor cells by inducing PD-1 via an EP4-PI3K-AKT-NF-κB pathway. In addition, in vitro studies have shown that PGE2 promotes M2 macrophage polarization via a CREB/CRTC pathway in bone marrow-derived macrophages (54) and the COX-2-PGE2 pathway mediated the effect of bladder tumor cells on induction of PD-L1 in bone marrow-derived macrophages and MDSCs in a co-culture system (55). These PD-L1 positive cells were immunosuppressive (55). Moreover, PGE2 also inhibited alveolar macrophage phagocytosis against bacteria (56). However, the mechanisms underlying PGE2 regulation of macrophage phagocytosis have not been determined. This study found that PGE2 suppresses macrophage phagocytosis of tumor cells by induction of PD-1 via an EP4-PI3K-Akt-NF-κB pathway.
PD-1+ tumor-infiltrating lymphocytes are associated with poor prognosis in human breast cancer (57) and soft tissue sarcomas (58) and correlate with poor survival in patients with high-grade upper tract urothelial carcinomas (59) and gastric cancer (60). PD-1+ tumor-infiltrating immune cells are also associated with advanced tumor-node-metastasis stage and poor survival in patients with renal cell carcinoma (RCC) (61) and operable breast cancer (62). Consistent with these reports, our results showed that COX-2 levels are positively associated with PD-1 in human CRC specimens. A recent study revealed that PGE2 levels are also associated with PD-1 levels in tumor-infiltrating CD8+ T cells in human lung cancer specimens (63), consistent with our results. Moreover, our results reveal that high levels of both COX-2 and PD-1 are associated with poorer overall survival in CRC patients. Recent studies showed that combined inhibition of PGE2 signaling and PD-1 increased CTL proliferation in vitro (64), and EP4 antagonists enhanced antitumor efficacy of PD-1 in a syngeneic mouse model of CRC (65) and in a mouse model of colitis-associated tumorigenesis (66).
In summary, we report that PGE2 promotes tumor immune evasion by suppressing CD8+ T cell cytotoxicity and macrophage phagocytosis against tumor cells by induction of PD-1 via an EP4-PI3K-AKT-NF-κB pathway. In contrast, inhibition of the COX-2-PGE2-EP4 pathway suppresses tumor immune evasion by increasing CD8+ T cell abundance as well as cytotoxicity and enhances macrophage phagocytosis of tumor cells. These findings uncover a novel role of PGE2 in tumor immune evasion and may provide a rationale for the development of new therapeutic approaches to subvert tumor immune evasion by targeting immune checkpoint pathways using EP4 antagonists. Given that EP4 antagonists have similar efficacy as celecoxib in the prevention and treatment of colorectal adenomas and CRC, the use of these agents will likely avoid the serious cardiovascular side effects associated with NSAIDs and COXIB use.
Supplementary Material
Prevention Relevance Statement.
These findings provide a potential explanation underlying the chemopreventive effect of NSAIDs on reducing CRC incidence during premalignancy and provide a rationale for developing EP4 antagonists for CRC prevention and treatment. Simply targeting PGE2 signaling alone may be efficacious in CRC prevention and treatment, avoiding side effects associated with NSAIDs.
Acknowledgments:
This work was supported in part by NIH R01 DK047297 and Flow Cytometry & Cell Sorting Unit, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313).
Financial support:
R.N. DuBois: NIH R01 DK047297 and NIH P30 CA138313
Footnotes
Conflict of interest disclosure statement: All authors have no conflict interests.
References:
- 1.Wang D, Dubois RN. The role of anti-inflammatory drugs in colorectal cancer. Annual review of medicine 2013;64:131–44. [DOI] [PubMed] [Google Scholar]
- 2.Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology 1994;107(4):1183–8. [DOI] [PubMed] [Google Scholar]
- 3.Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer 2001;1(1):11–21. [DOI] [PubMed] [Google Scholar]
- 4.Ogino S, Kirkner GJ, Nosho K, Irahara N, Kure S, Shima K, et al. Cyclooxygenase-2 expression is an independent predictor of poor prognosis in colon cancer. Clin Cancer Res 2008;14(24):8221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigas B, Goldman IS, Levine L. Altered eicosanoid levels in human colon cancer. J Lab Clin Med 1993;122(5):518–23. [PubMed] [Google Scholar]
- 6.Wang D, Dubois RN. Cyclooxygenase-2: a potential target in breast cancer. Semin Oncol 2004;31(1 Suppl 3):64–73. [DOI] [PubMed] [Google Scholar]
- 7.McLemore TL, Hubbard WC, Litterst CL, Liu MC, Miller S, McMahon NA, et al. Profiles of prostaglandin biosynthesis in normal lung and tumor tissue from lung cancer patients. Cancer Res 1988;48(11):3140–7. [PubMed] [Google Scholar]
- 8.Hambek M, Baghi M, Wagenblast J, Schmitt J, Baumann H, Knecht R. Inverse correlation between serum PGE2 and T classification in head and neck cancer. Head Neck 2007;29(3):244–8. [DOI] [PubMed] [Google Scholar]
- 9.Mal M, Koh PK, Cheah PY, Chan EC. Ultra-pressure liquid chromatography/tandem mass spectrometry targeted profiling of arachidonic acid and eicosanoids in human colorectal cancer. Rapid Commun Mass Spectrom 2011;25(6):755–64. [DOI] [PubMed] [Google Scholar]
- 10.Cai Q, Gao YT, Chow WH, Shu XO, Yang G, Ji BT, et al. Prospective study of urinary prostaglandin E2 metabolite and colorectal cancer risk. J Clin Oncol 2006;24(31):5010–6. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JC, Schmidt CR, Shrubsole MJ, Billheimer DD, Joshi PR, Morrow JD, et al. Urine PGE-M: A metabolite of prostaglandin E2 as a potential biomarker of advanced colorectal neoplasia. Clin Gastroenterol Hepatol 2006;4(11):1358–65. [DOI] [PubMed] [Google Scholar]
- 12.Bezawada N, Song M, Wu K, Mehta RS, Milne GL, Ogino S, et al. Urinary PGE-M levels are associated with risk of colorectal adenomas and chemopreventive response to anti-inflammatory drugs. Cancer Prev Res (Phila) 2014;7(7):758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, DuBois RN. Role of prostanoids in gastrointestinal cancer. J Clin Invest 2018;128(7):2732–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mumprecht S, Schurch C, Schwaller J, Solenthaler M, Ochsenbein AF. Programmed death 1 signaling on chronic myeloid leukemia-specific T cells results in T-cell exhaustion and disease progression. Blood 2009;114(8):1528–36. [DOI] [PubMed] [Google Scholar]
- 15.Fourcade J, Kudela P, Sun Z, Shen H, Land SR, Lenzner D, et al. PD-1 is a regulator of NY-ESO-1-specific CD8+ T cell expansion in melanoma patients. J Immunol 2009;182(9):5240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009;114(8):1537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol 2002;23(11):549–55. [DOI] [PubMed] [Google Scholar]
- 18.Isidro RA, Appleyard CB. Colonic macrophage polarization in homeostasis, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol 2016;311(1):G59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017;545(7655):495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine 2015;372(26):2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castells A, Paya A, Alenda C, Rodriguez-Moranta F, Agrelo R, Andreu M, et al. Cyclooxygenase 2 expression in colorectal cancer with DNA mismatch repair deficiency. Clin Cancer Res 2006;12(6):1686–92. [DOI] [PubMed] [Google Scholar]
- 22.Sinicrope FA, Lemoine M, Xi L, Lynch PM, Cleary KR, Shen Y, et al. Reduced expression of cyclooxygenase 2 proteins in hereditary nonpolyposis colorectal cancers relative to sporadic cancers. Gastroenterology 1999;117(2):350–8. [DOI] [PubMed] [Google Scholar]
- 23.Karnes WE Jr., Shattuck-Brandt R, Burgart LJ, DuBois RN, Tester DJ, Cunningham JM, et al. Reduced COX-2 protein in colorectal cancer with defective mismatch repair. Cancer Res 1998;58(23):5473–7. [PubMed] [Google Scholar]
- 24.Ogino S, Brahmandam M, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia 2006;8(6):458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia D, Wang D, Kim SH, Katoh H, DuBois RN. Prostaglandin E2 promotes intestinal tumor growth via DNA methylation. Nat Med 2012;18(2):224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Wang H, Shi Q, Katkuri S, Walhi W, Desvergne B, et al. Prostaglandin E(2) promotes colorectal adenoma growth via transactivation of the nuclear peroxisome proliferator-activated receptor delta. Cancer Cell 2004;6(3):285–95. [DOI] [PubMed] [Google Scholar]
- 27.Bally AP, Lu P, Tang Y, Austin JW, Scharer CD, Ahmed R, et al. NF-kappaB regulates PD-1 expression in macrophages. J Immunol 2015;194(9):4545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chell SD, Witherden IR, Dobson RR, Moorghen M, Herman AA, Qualtrough D, et al. Increased EP4 receptor expression in colorectal cancer progression promotes cell growth and anchorage independence. Cancer Res 2006;66(6):3106–13. [DOI] [PubMed] [Google Scholar]
- 29.Baba Y, Nosho K, Shima K, Goessling W, Chan AT, Ng K, et al. PTGER2 overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Cancer Epidemiol Biomarkers Prev 2010;19(3):822–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoji Y, Takahashi M, Kitamura T, Watanabe K, Kawamori T, Maruyama T, et al. Downregulation of prostaglandin E receptor subtype EP3 during colon cancer development. Gut 2004;53(8):1151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Cabalag CS, Clemons NJ, DuBois RN. Cyclooxygenases and Prostaglandins in Tumor Immunology and Microenvironment of Gastrointestinal Cancer. Gastroenterology 2021;161(6):1813–29. [DOI] [PubMed] [Google Scholar]
- 32.Treffkorn L, Scheibe R, Maruyama T, Dieter P. PGE2 exerts its effect on the LPS-induced release of TNF-alpha, ET-1, IL-1alpha, IL-6 and IL-10 via the EP2 and EP4 receptor in rat liver macrophages. Prostaglandins Other Lipid Mediat 2004;74(1–4):113–23. [DOI] [PubMed] [Google Scholar]
- 33.Abramovitz M, Adam M, Boie Y, Carriere M, Denis D, Godbout C, et al. The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta 2000;1483(2):285–93. [DOI] [PubMed] [Google Scholar]
- 34.Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology 2015;149(7):1884–95 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cen B, Lang JD, Du Y, Wei J, Xiong Y, Bradley N, et al. Prostaglandin E2 Induces miR675–5p to Promote Colorectal Tumor Metastasis via Modulation of p53 Expression. Gastroenterology 2020;158(4):971–84 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Colon GJ, Moore BB. Prostaglandin E2 as a Regulator of Immunity to Pathogens. Pharmacol Ther 2018;185:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen JH, Perry CJ, Tsui YC, Staron MM, Parish IA, Dominguez CX, et al. Prostaglandin E2 and programmed cell death 1 signaling coordinately impair CTL function and survival during chronic viral infection. Nat Med 2015;21(4):327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakanishi Y, Nakatsuji M, Seno H, Ishizu S, Akitake-Kawano R, Kanda K, et al. COX-2 inhibition alters the phenotype of tumor-associated macrophages from M2 to M1 in ApcMin/+ mouse polyps. Carcinogenesis 2011;32(9):1333–9. [DOI] [PubMed] [Google Scholar]
- 39.Chang J, Vacher J, Yao B, Fan X, Zhang B, Harris RC, et al. Prostaglandin E receptor 4 (EP4) promotes colonic tumorigenesis. Oncotarget 2015;6(32):33500–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol 2008;181(7):4832–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006;439(7077):682–7. [DOI] [PubMed] [Google Scholar]
- 42.Terawaki S, Chikuma S, Shibayama S, Hayashi T, Yoshida T, Okazaki T, et al. IFN-alpha directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol 2011;186(5):2772–9. [DOI] [PubMed] [Google Scholar]
- 43.Austin JW, Lu P, Majumder P, Ahmed R, Boss JM. STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. J Immunol 2014;192(10):4876–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Said EA, Dupuy FP, Trautmann L, Zhang Y, Shi Y, El-Far M, et al. Programmed death-1-induced interleukin-10 production by monocytes impairs CD4+ T cell activation during HIV infection. Nat Med 2010;16(4):452–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho HY, Lee SW, Seo SK, Choi IW, Choi I, Lee SW. Interferon-sensitive response element (ISRE) is mainly responsible for IFN-alpha-induced upregulation of programmed death-1 (PD-1) in macrophages. Biochim Biophys Acta 2008;1779(12):811–9. [DOI] [PubMed] [Google Scholar]
- 46.Staron MM, Gray SM, Marshall HD, Parish IA, Chen JH, Perry CJ, et al. The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity 2014;41(5):802–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathieu M, Cotta-Grand N, Daudelin JF, Thebault P, Labrecque N. Notch signaling regulates PD-1 expression during CD8(+) T-cell activation. Immunol Cell Biol 2013;91(1):82–8. [DOI] [PubMed] [Google Scholar]
- 48.Xiao G, Deng A, Liu H, Ge G, Liu X. Activator protein 1 suppresses antitumor T-cell function via the induction of programmed death 1. Proc Natl Acad Sci U S A 2012;109(38):15419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai D, Ueno L, Vogt PK. Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int J Cancer 2009;125(12):2863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou JP, Ramirez CM, Ryba DM, Koduri MP, Effros RB. Prostaglandin E2 promotes features of replicative senescence in chronically activated human CD8+ T cells. PloS one 2014;9(6):e99432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeddou M, Greimers R, de Valensart N, Nayjib B, Tasken K, Boniver J, et al. Prostaglandin E2 induces the expression of functional inhibitory CD94/NKG2A receptors in human CD8+ T lymphocytes by a cAMP-dependent protein kinase A type I pathway. Biochem Pharmacol 2005;70(5):714–24. [DOI] [PubMed] [Google Scholar]
- 52.Ganapathy V, Gurlo T, Jarstadmarken HO, von Grafenstein H. Regulation of TCR-induced IFN-gamma release from islet-reactive non-obese diabetic CD8(+) T cells by prostaglandin E(2) receptor signaling. Int Immunol 2000;12(6):851–60. [DOI] [PubMed] [Google Scholar]
- 53.Basingab FS, Ahmadi M, Morgan DJ. IFNgamma-Dependent Interactions between ICAM-1 and LFA-1 Counteract Prostaglandin E2-Mediated Inhibition of Antitumor CTL Responses. Cancer Immunol Res 2016;4(5):400–11. [DOI] [PubMed] [Google Scholar]
- 54.Luan B, Yoon YS, Le Lay J, Kaestner KH, Hedrick S, Montminy M. CREB pathway links PGE2 signaling with macrophage polarization. Proc Natl Acad Sci U S A 2015;112(51):15642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, Kusmartsev S. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci U S A 2017;114(5):1117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aronoff DM, Canetti C, Peters-Golden M. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol 2004;173(1):559–65. [DOI] [PubMed] [Google Scholar]
- 57.Muenst S, Soysal SD, Gao F, Obermann EC, Oertli D, Gillanders WE. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat 2013;139(3):667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JR, Moon YJ, Kwon KS, Bae JS, Wagle S, Kim KM, et al. Tumor infiltrating PD1-positive lymphocytes and the expression of PD-L1 predict poor prognosis of soft tissue sarcomas. PloS one 2013;8(12):e82870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krabbe LM, Heitplatz B, Preuss S, Hutchinson RC, Woldu SL, Singla N, et al. Prognostic Value of PD-1 and PD-L1 Expression in Patients with High Grade Upper Tract Urothelial Carcinoma. J Urol 2017;198(6):1253–62. [DOI] [PubMed] [Google Scholar]
- 60.Wen T, Wang Z, Li Y, Li Z, Che X, Fan Y, et al. A Four-Factor Immunoscore System That Predicts Clinical Outcome for Stage II/III Gastric Cancer. Cancer Immunol Res 2017;5(7):524–34. [DOI] [PubMed] [Google Scholar]
- 61.Thompson RH, Dong H, Lohse CM, Leibovich BC, Blute ML, Cheville JC, et al. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin Cancer Res 2007;13(6):1757–61. [DOI] [PubMed] [Google Scholar]
- 62.Sun S, Fei X, Mao Y, Wang X, Garfield DH, Huang O, et al. PD-1(+) immune cell infiltration inversely correlates with survival of operable breast cancer patients. Cancer Immunol Immunother 2014;63(4):395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Zhang L, Kang D, Yang D, Tang Y. Activation of PGE2/EP2 and PGE2/EP4 signaling pathways positively regulate the level of PD-1 in infiltrating CD8(+) T cells in patients with lung cancer. Oncol Lett 2018;15(1):552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miao J, Lu X, Hu Y, Piao C, Wu X, Liu X, et al. Prostaglandin E2 and PD-1 mediated inhibition of antitumor CTL responses in the human tumor microenvironment. Oncotarget 2017;8(52):89802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Cui L, Georgiev P, Singh L, Zheng Y, Yu Y, et al. Combination of EP4 antagonist MF-766 and anti-PD-1 promotes anti-tumor efficacy by modulating both lymphocytes and myeloid cells. Oncoimmunology 2021;10(1):1896643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu W, Yu W, He J, Liu W, Yang J, Lin X, et al. Reprogramming immunosuppressive myeloid cells facilitates immunotherapy for colorectal cancer. EMBO Mol Med 2021;13(1):e12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data file. Raw data generated in this study are available from the corresponding author upon request. The data analyzed in this study were obtained from the TCGA Colorectal Adenocarcinoma Provisional dataset and in Gene Expression Omnibus (GEO) at GSE17537 and GSE17538.