Abstract
FGFR inhibitors are approved for the treatment of advanced cholangiocarcinoma harboring FGFR2 fusions. However, the response rate is moderate, and resistance emerges rapidly due to acquired secondary FGFR2 mutations or to other less defined mechanisms. Here, we conducted high-throughput combination drug screens, biochemical analysis, and therapeutic studies using patient-derived models of FGFR2-fusion-positive cholangiocarcinoma to gain insight into these clinical profiles and uncover improved treatment strategies. We found that feedback activation of EGFR signaling limits FGFR inhibitor efficacy, restricting cell death induction in sensitive models, and causing resistance in insensitive models lacking secondary FGFR2 mutations. Inhibition of wild-type EGFR potentiated responses to FGFR inhibitors in both contexts, durably suppressing MEK/ERK and mTOR signaling, increasing apoptosis, and causing marked tumor regressions in vivo. Our findings reveal EGFR-dependent adaptive signaling as an important mechanism limiting FGFR inhibitor efficacy and driving resistance and support the clinical testing of FGFR/EGFR inhibitor therapy for FGFR2-fusion-positive cholangiocarcinoma.
Keywords: Intrahepatic cholangiocarcinoma, FGFR2, EGFR, Targeted therapy, Drug resistance, Apoptosis
Statement of Significance
We demonstrate that feedback activation of EGFR signaling limits the effectiveness of FGFR inhibitor therapy and drives adaptive resistance in patient-derived models of FGFR2-fusion-positive cholangiocarcinoma. These studies support the potential of combination treatment with FGFR and EGFR inhibitors as an improved treatment for patients with FGFR2-driven cholangiocarcinoma.
Introduction
Intrahepatic cholangiocarcinoma (ICC) is a deadly cancer of the liver bile ducts, which has been rising in incidence for several decades and has few therapeutic options (1). Standard of care first-line chemotherapy with gemcitabine/cisplatin for advanced biliary tract cancers provides modest benefit, conferring a median overall survival of 11.7 months and a 5-year survival of <5% (1,2). The recent identification of targetable genomic alterations in subsets of patients with ICC heralds a new era of precision therapy for this disease. In particular, genomically activated isocitrate dehydrogenase 1 (IDH1), FGFR2, BRAF, and HER2 represent promising therapeutic targets in patients with such alterations (1,3-7). The most advanced clinical investigations have centered around the FGFR pathway. Nearly 20% of ICC exhibit chromosomal fusions, point mutations, and in-frame deletions of FGFR2 that result in constitutive kinase activity, a rate that exceeds other malignancies (1,8)(https://genie.cbioportal.org/). The most frequent events (~12% of patients) involve fusions of FGFR2 exons 1-17 (encoding the entire extracellular domain and kinase domain, and only lacking the final exon encoding the final 62 amino acids) fused in-frame with diverse 3’ partners encoding a dimerization domain (3-5,7,9). In addition, point mutations and in-frame deletions in the extracellular domain of FGFR2 are collectively found in ~5% of patients (8). The oncogenic activity of several of these alterations has been demonstrated experimentally (3,4,8).
Multiple selective small molecule FGFR1-3 kinase inhibitors have shown efficacy against FGFR2+ ICC in clinical trials, leading to the regulatory approval of the ATP-competitive FGFR inhibitors, pemigatinib and infigratinib (BGJ398), for this subset of patients who failed standard treatment (10-14). While these FDA approvals represent important landmarks in the advancement of treatment for refractory FGFR2+ ICC, and the great majority of patients derive clinical benefit (>80% disease control rate), overall outcomes remain bleak for these patients. First, the objective response rate (ORR) for each FGFR inhibitor (FGFRi) studied to date in FGFR2+ ICC is <45%, which is modest compared to the >75% ORR seen upon treatment with tyrosine kinase inhibitors targeting epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) rearrangements in non-small cell lung cancers (NSCLCs) harboring these alterations (15-19). Second, disease progression invariably arises within ~6-12 months, often associated with acquired mutations in the FGFR2 kinase domain that impair FGFR inhibitor binding (20-25). The irreversible FGFR1-3 inhibitor, futibatinib (TAS-120) remains active against some, but not all of these secondary FGFR2 mutations (21). A third challenge are the dose-limiting, on-target toxicities of these pan-FGFR inhibitors, including hyperphosphatemia due to inhibition of FGFR1 in the renal tubules (26) as well as ocular toxicity likely relating to FGFR2 inhibition (10-14,26,27). There is considerable promise of emerging next-generation covalent inhibitors with broader coverage of these FGFR2 mutations, including agents that spare FGFR1 and 3 (28). However, it is uncertain whether these new FGFR inhibitors will be more potent leading to a higher initial response rate. Moreover, examination of patient specimens reveals that FGFRi resistance can alternatively arise without evidence of acquired genetic alterations, suggesting potential adaptive resistance processes, or can result from acquired mutations in other MAPK signaling components (8,29). Thus, there is an urgent need to develop strategies that enhance the extent and duration of initial response and that overcome different modes of acquired resistance in patients with FGFR2+ ICC.
In this study, we took advantage of a series of novel patient-derived ICC cell lines and patient-derived xenograft (PDX) models to reveal signaling molecules restricting FGFRi efficacy in FGFR2+ ICC and to identify refined treatment approaches in the setting of both sensitivity and resistance to FGFRi monotherapy. Our work demonstrates a major role of feedback activation of EGFR signaling in maintaining oncogenic signaling and limiting cell death upon FGFR inhibition and highlights the therapeutic potential of dual inhibition of FGFR2 and EGFR.
Results
Signaling feedback upon FGFR inhibition in patient-derived models of FGFR2 fusion+ ICC
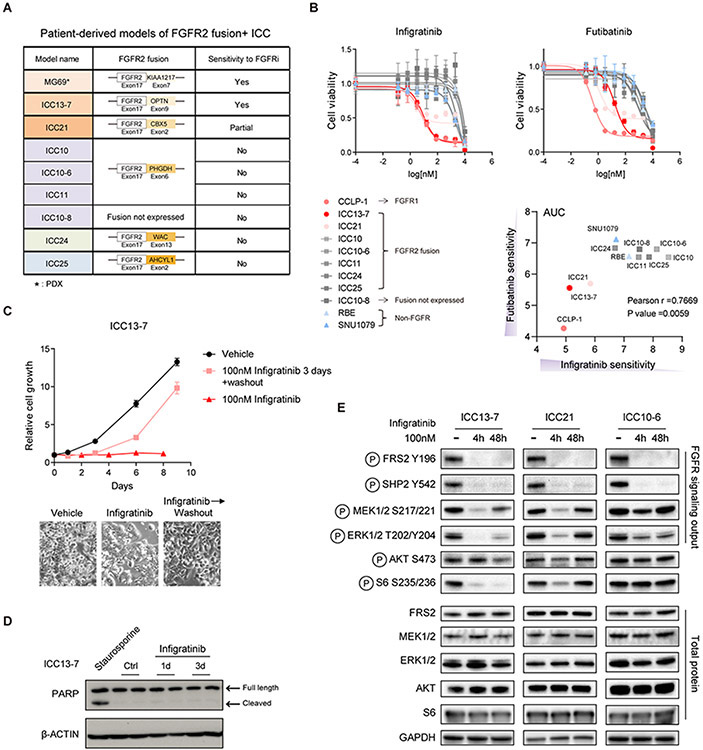
We developed a set of ICC cell lines and PDXs established from patients with FGFR2-fusion+ ICC participating in FGFR inhibitor trials, including models from responsive and resistant tumor nodules (Figure 1A and Supplementary Table 1). Each model expressed the FGFR2 fusion seen at diagnosis, except for ICC10-8, derived post-futibatinib treatment from a metastatic nodule that lacked expression of fusion. Whole exon sequencing showed that none of the models harbored secondary mutations in the FGFR2 kinase domain or mutation of MAPK bypass pathways, although ICC10-6 has an amplification of the FGFR2-PHGDH fusion. Testing the cell lines for sensitivity to the pan-FGFR inhibitor, infigratinib, revealed a range of responses, from highly sensitive—ICC13-7 (IC50 12 nM), partially sensitive—ICC21 (IC50 ~250 nM), or resistant—ICC10, ICC10-6, ICC10-8, ICC11, ICC24, ICC25 (IC50 >1000 nM) (Figure 1B, left panel). As references, we included the FGFR1-overexpressing ICC cell line, CCLP-1 (IC50 8 nM) and two ICC cell lines lacking FGFR alterations (RBE and SNU1079), which were insensitive to FGFR inhibition (IC50 >1000 nM), as we reported previously (21). Similar profiles were observed for the covalent pan-FGFRi, futibatinib and for a second reversible FGFRi, pemigatinib (Figure 1B, right panel and Supplementary Figure 1A).
Figure 1. Characterization of models derived from patients with FGFR2-fusion+ ICC.
A. Summary of ICC models. (*) MG69 is a PDX model. The others are patient-derived cell lines. B. IC50 assay for evaluating the sensitivity of ICC cell lines to infigratinib and futibatinib. Each point on the dose-response curves represents 4 technical replicates (infigratinib) and 2 technical replicates (futibatinib). Data are shown as Mean ± SD. Bottom right: Correlation between infigratinib and futibatinib sensitivity. AUC denotes area under the curve calculated by GraphPad Prism 9. C. Growth curve of ICC13-7 cells treated with vehicle or 100 nM infigratinib for 8 days, or 100 nM infigratinib for 3 days followed by switch to vehicle-containing media (washout) for 5 days. Each point on the curves represents 5 technical replicates. Data are shown as Mean ± SD. Representative photomicrographs are shown below. D. Immunoblot analysis of PARP cleavage in ICC13-7 cells upon treatment with 100 nM infigratinib or vehicle for the indicated times. Staurosporine (1000 nM) served as the positive control. E. Immunoblot analysis of the indicated signaling proteins in ICC13-7 (left), ICC21 (middle), and ICC10-6 cells (right) upon treatment with 100 nM infigratinib for 4 or 48 hours versus vehicle.
While infigratinib treatment of the FGFRi-responsive line, ICC13-7, resulted in impaired proliferation and reduction in S phase of the cell cycle (Figure 1C and Supplementary 1B), proliferation resumed promptly upon washout of infigratinib (Figure 1C, pink line shows 3-day treatment followed by washout). Moreover, there was no evidence of apoptosis as assessed by PARP cleavage in either ICC13-7 or ICC21 cells (Figure 1D and Supplementary 1C), which contrasts with the pronounced induction of apoptotic death seen upon targeting of a number of other RTK-dependent cancer cell lines (30). Thus, FGFR inhibition caused reversible growth arrest rather than induction of senescence or cell death in these models.
The cytostatic effects of FGFR inhibition in sensitive FGFR2-fusion+ ICC lines and the lack of secondary FGFR2 mutations in multiple resistant models suggested a possible role for feedback signaling processes in limiting cell death and/or supporting resistance. Accordingly, we examined the impact of FGFRi treatment on downstream pathways in FGFR2-fusion+ ICC lines after 4- and 48-hours treatment. In the sensitive ICC13-7 cell line, MEK/ERK activity (phospho-MEK1/2 [S217/221] and phospho-ERK1/2 [T202/Y204]) and mTORC1 activity (phospho-S6 [S235/236]) were eliminated by infigratinib treatment at 4 hours and showed a modest rebound at 48 hours, whereas the mTORC2/AKT pathway (phospho-AKT [S473]) was largely unresponsive (Figure 1E, left panel). Moreover, in the partially sensitive ICC21 model and the resistant ICC10-6 model, MEK/ERK and mTORC1 signaling was only transiently inhibited, with full restoration of these pathways within 48 hours (Figure 1E, center and right panels). Importantly, these effects were not due to incomplete inactivation of the FGFR2 kinase, since the phosphorylation of the more proximal targets of FGFR2, FRS2 (P-Y196) and SHP2 (P-Y542) (26) was durably suppressed. Thus, feedback reactivation of downstream signaling upon FGFRi inhibition could be an important factor in limiting treatment efficacy and in driving resistance in FGFR2-fusion+ ICC.
High-throughput drug screen reveals potentiation between EGFR and FGFR inhibitors in FGFR2-fusion+ ICC models
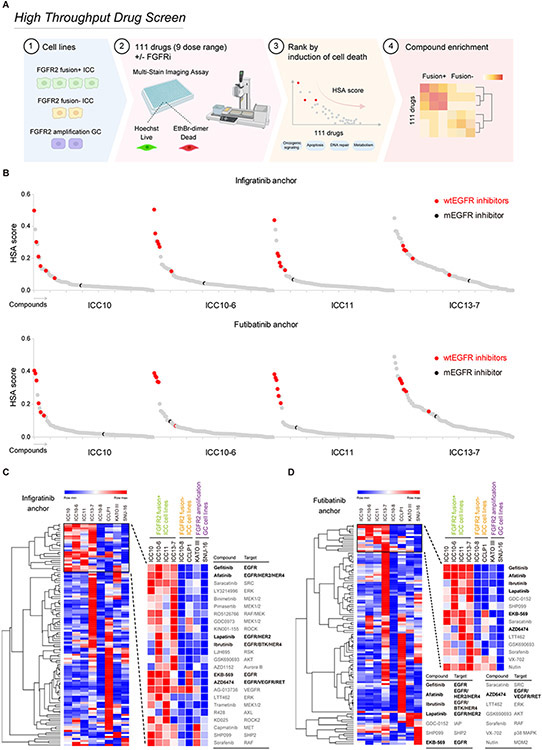
We employed a high-throughput screen to identify targeted small molecule compounds that potentiate response to FGFR inhibitors in FGFR2-fusion+ ICC cells to gain insight into adaptive changes that suppress cell death in this setting and identify improved treatment strategies (Figure 2A). A panel of 111 compounds—including approved agents and advanced tool compounds targeting key oncogenic and stress response pathways (e.g., growth factor signaling, apoptosis, DNA repair, metabolism, Supplementary Table 2) — were screened alone or in combination with different selective FGFR inhibitors (the reversible ATP-competitive inhibitors, infigratinib and rogaratinib, or the covalent inhibitor futibatinib). Six ICC cell lines were tested: FGFR2-fusion+ models that are sensitive (ICC13-7) or resistant to FGFR inhibition (ICC10, ICC10-6, ICC11); the ICC10-8 resistant model, which lost FGFR2 fusion expression; and the FGFR1-dependent CCLP-1 model. We also screened two FGFR2-dependent gastric cancer (GC) cell lines harboring FGFR2 amplifications (KATO III and SNU-16) to uncover possible lineage specificity in response profiles. The cells were treated over 5 days against a 9-point dose range of the compound library combined with a single concentration of an FGFRi (that completely blocks FGFR kinase activity based on signaling analysis) and then tested for cell death and cell number using high-content imaging (Figure 2A, and see Methods). We used a Highest Single Agent (HSA)-based approach (31) to reveal combinations that specifically induced higher cell death than single agents. To identify potential patterns across the combination outcome and visualize the results, we performed hierarchical clustering analysis of the data (see Methods). Most strikingly, the four selective inhibitors of wild type EGFR/ERBB family receptor tyrosine kinases in the screen (afatinib, gefitinib, lapatinib, and EKB-569) were found to strongly potentiate the effects of FGFR inhibitors across all FGFR2 fusion-positive ICC cell lines as did additional receptor tyrosine kinase inhibitors with activity against EGFR/ERBB, ibrutinib and AZD6474 (32,33) (Figure 2B-D, Supplementary Figure 2A, B and Supplementary Table 2). Notably, this potentiation was not seen in the FGFR2 amplified gastric cancer cell lines, KATO-III and SNU-16, the FGFR1-driven CCLP-1 ICC cell line, or in the FGFR2-negative ICC10-8 line. Moreover, the mutant selective EGFR kinase inhibitor CO1686 did not display the same activity as inhibitors capable of inhibiting WT EGFR (highlighted in black in Figure 2B and Supplementary Figure 2A). These results indicate that inhibition of wild type EGFR/ERBB family kinases potentiates the effects of FGFR inhibition in FGFR2-fusion+ ICC, driving cell death both in models that show growth arrest upon FGFRi monotherapy and in those that are resistant. Other hits were found across individual cell lines, including BCL2/MCL1 family inhibitors – where potentiation was restricted to models that are sensitive to single agent FGFRi treatment — as well as inhibitors of PI3K/AKT and MEK signaling (Supplementary Figure 3A and Supplementary Table 2). The multi-kinase inhibitor, saracatinib, which targets SRC family kinases at low nM levels also potentiated the effects of FGFRi, however, only at high concentrations (>1 μM) where a wide target of kinases would be targeted. Given the consistent and potent combinatorial effects seen upon EGFR+FGFR inhibition in FGFR2 fusion+ ICC cells, we focused on this combination for further study.
Figure 2. High-throughput drug screen reveals cooperative induction of cell death by combined EGFR and FGFR inhibition in FGFR2 fusion+ ICC models.
A. Schematic of high-throughput combination drug screen (Created with BioRender.com). The cell lines screened were: FGFR2-fusion+ ICC (ICC10/ICC10-6/ICC11/ICC13-7), FGFR2-fusion- ICC (ICC10-8/CCLP-1), FGFR2 amplified GC (KATO III/SNU-16). B. Screen results for the four FGFR2-fusion+ ICC cell lines tested. The chart shows the cooperative induction of cell death across the 111 drugs screened in combination with infigratinib 100 nM (upper panel), or futibatinib 40 nM (bottom panel), based on second highest single agent (HSA) score. Inhibitors of WT EGFR/ERBB signaling are highlighted red. The mutant selective EGFR inhibitor, CO1686 (Rociletinib) is highlighted black. Other inhibitors are color coded grey. C and D. Heat map and hierarchical cluster analysis generated in Morpheus (https://software.broadinstitute.org/morpheus/) based on second HSA score with infigratinib (C) and futibatinib (D) for cell death across all cell lines screened (color scheme is based on the minimum and maximum HSA values in each row). The top ranked combinations are presented as blown-up images on the right.
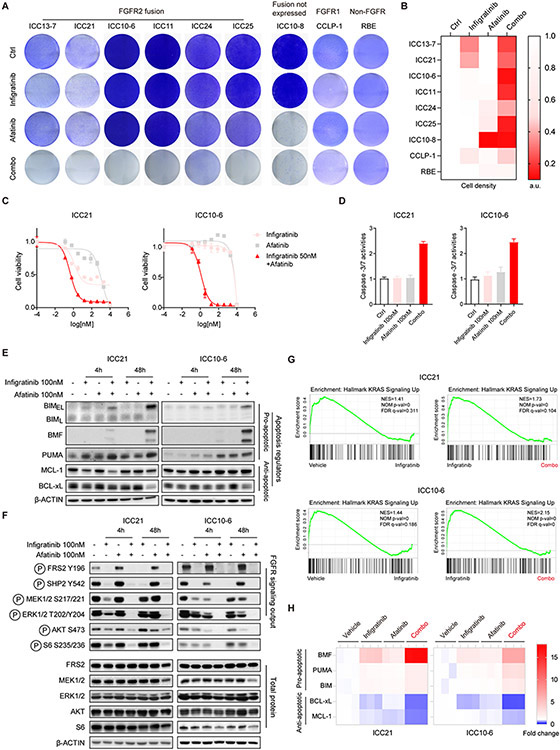
We first validated and extended the screening results across a set of seven ICC cell lines derived from patients with FGFR2-fusion+ ICC, including three lines not included in the initial screen (ICC21, ICC24, and ICC25). By conducting crystal violet staining after 7-10 days treatment using 100 nM infigratinib and 100 nM afatinib (Figure 3A, B) and by examining the cell viability shift across a 9-point dose range of afatinib in the context of 50 nM infigratinib (Figure 3C and Supplementary Figure 4A), we demonstrated the potentiating effect of the combination in all six of the FGFR2+ ICC models tested (ICC13-7, ICC21, ICC10-6, ICC11, ICC24, and ICC25) as well as hypersensitivity to afatinib alone in the ICC10-8 cell line. No combinatorial effects were observed in the FGFR1-driven CCLP-1 cell line or in another ICC cell line lacking FGFR alterations (RBE) (Figure 3A and Supplementary Figure 4A). Moreover, we observed comparable combinatorial effects between infigratinib and either the selective EGFR inhibitor, gefitinib or the EGFR/Her2 inhibitor, EKB569 (Supplementary Figure 4B-E). Equivalent results were observed for combination treatment with the covalent FGFRi, futibatinib (50 nM) and afatinib (100 nM) (Supplementary Figure 4F). Since gefitinib produced similar effects to broader spectrum ERBB family inhibitors, we conclude that this cooperativity with FGFR inhibition was primarily due to suppression of EGFR activity rather than ERBB2 or ERBB4. The identification of hypersensitivity to single agent EGFR inhibitor treatment of ICC10-8 cells suggests that a complete switch to EGFR-dependence may sometimes drive FGFRi resistance in patients with FGFR2 fusion+ ICC.
Figure 3. Combined FGFR and EGFR inhibition induces apoptosis and durably inactivates downstream oncogenic signaling in FGFR2 fusion+ ICC cells.
A and B. ICC cell lines were tested for cell viability via crystal violet staining after 7 days (ICC13-7/ICC21) or 10 days treatment (all other cell lines) with single agent infigratinib 100 nM, afatinib 100 nM or the combination or vehicle control. Quantification of cell density from crystal violet staining is shown in (B). a.u. denotes arbitrary unit. C. IC50 assay evaluating the sensitivity of ICC21 (left) and ICC10-6 cells (right) to single agent infigratinib, afatinib or 50 nM infigratinib combined with a range of doses of afatinib. Each point on the dose-response curves represents 2 technical replicates. Data are shown as Mean ± SD. Experiments were repeated at least twice. D. Induction of apoptosis assessed by Caspase-3/7 activity after 3 days treatment with the indicated single agents or combination. Error bars on the graph represent 4 technical replicates. Data are shown as Mean± SD. E, F. FGFR2-fusion+ ICC cell lines were treated with vehicle, 100 nM infigratinib, 100 nM afatinib or the combination for 4 hours or 48 hours, and lysates were subjected to immunoblot analysis for (E) pro-apoptotic (BIM, BMF, PUMA) and anti-apoptotic proteins (MCL-1, BCL-xL) or (F) the indicated signaling proteins. G and H. ICC21 and ICC10-6 cells were treated with vehicle, 100 nM infigratinib, 100 nM afatinib, or the combination for 4 hours and then profiled by RNA sequencing. (G) Pathway enrichment was determined by Gene Set Enrichment Analysis (GSEA). (H) Heatmap showing relative expression changes of apoptosis regulators under different treatment conditions (normalized to vehicle). Only significantly changed genes are shown. Each column shows a biological replicate (3/condition).
Importantly, afatinib cooperated with infigratinib to substantially increase apoptosis, as indicated by Caspase-3/7 activity, in both sensitive and resistant cell lines (Figure 3D and Supplementary Figure 4G). The intrinsic apoptotic pathway is governed by pro- and anti-apoptotic BCL2 family proteins whose balance determines the integrity of mitochondria, thus commitment to apoptosis (30,34,35). We found that combination treatment markedly induced the pro-apoptotic proteins, BIM and BMF, compared to either inhibitor alone across the FGFR2-fusion+ cell line panel (Figure 3E, Supplementary Figure 4H and Supplementary Figure 5A, see apoptosis regulator panels), consistent with prolonged shut-down of the MEK/ERK pathway (30,35). Other combinatorial effects in a subset of cell lines included suppression of the anti-apoptotic protein BCL-xL (ICC21, ICC13-7, ICC10-6, ICC11: Figure 3E, Supplementary Figure 4H and Supplementary Figure 5A) and induction of the pro-apoptotic protein, PUMA (ICC21: Figure 3E), whereas MCL-1 expression was not significantly affected. Thus, while our set of FGFR2 fusion+ ICC cell lines either show cytostatic responses to FGFR inhibition or are completely resistant, combined EGFR kinase inhibitor treatment results in a highly cytotoxic response. Finally, in ICC10-8 cells, afatinib alone strongly induced the pro-apoptotic response, with marked increases in BIM and BMF (Supplementary Figure 5A), consistent with the sensitivity of these cells to EGFR/ERBB targeted monotherapy.
Dual FGFR/EGFR inhibition blocks signaling feedback in FGFR2-fusion+ ICC
To further explore the potentiating effect of the combination treatment, we evaluated canonical signaling pathways for activating phosphorylation by immunoblot analysis. In ICC21 and ICC13-7 cells (which exhibit partial and strong sensitivity to FGFRi monotherapy, respectively), combined infigratinib plus afatinib treatment (100 nM each) overcame the rebound of signaling seen upon infigratinib monotherapy, durably inhibiting phosphorylation of SHP2, MEK, ERK, AKT (P-S473), and S6 at both 4-hour and 48-hour time points (Figure 3F and Supplementary Figure 5B). Treatment with 100 nM afatinib as a single agent had minimal effects on these signaling outputs in ICC21 and ICC13-7 cells, whereas we observed increased phospho-FRS2 (Y196) in these lines as well as one of resistant lines (ICC10-6), suggesting feedback effects on the activation state of FGFR (Figure 3F and Supplementary Figure 5B). The resistant FGFR2-fusion-positive lines, ICC10-6, ICC24, and ICC11 also showed cooperative effects of infigratinib + afatinib combination treatment, with consistent potent inhibition of MEK and ERK phosphorylation and variable suppression of phospho-AKT (S473) and phospho-S6 (S235/236) (Figure 3F and Supplementary Figure 5B). For each of these lines (ICC13-7, ICC21, ICC10-6, ICC11), infigratinib produced similar results in combination with gefitinib or EKB-569, indicating that suppression of EGFR kinase activity, specifically, was primarily responsible for this cooperative impact on signaling (Supplementary Figure 5C and 5D). In ICC25 cells, responses to EGFR inhibition predominated, with strong reduction of MEK/ERK signaling by afatinib alone, although levels were further reduced in combination with infigratinib (Supplementary Figure 5A). Notably, across the lines analyzed, neither monotherapy with infigratinib or afatinib nor the combination had significant effects on PI3K/AKT signaling (gauged by phospho-AKT [T308]) (Supplementary Figure 6A) or on SRC, PLCγ, or JAK/STAT3 activity (gauged by phospho-SRC [Y416], phospho-PLCγ1 [Y783] and phospho-STAT3 [Y705], respectively) (Supplementary Figure 6B and C). These data are consistent with the MEK/ERK pathway serving as the primary effector of these RTKs in FGFR2 fusion+ ICC.
In the ICC10-8 model (FGFR2 fusion not expressed), each of these EGFR/ERBB family inhibitors alone effectively blocked MEK/ERK and AKT (P-S473) phosphorylation (Supplement Figure 5A, C, and D). Examination of the genomic features of the ICC10-8 model failed to reveal any mutations in EGFR or other ERBB family members or regulators (ERRFI1, CDC25C, and CBL). However, this model was notable for showing highest constitutive levels of activating EGFR (P-Y1173) phosphorylation (Supplement Figure 6D). Collectively, these results reveal important functions of EGFR signaling in FGFR2-fusion+ ICC, serving as a feedback mechanism to restore oncogenic signaling upon FGFR inhibition in multiple sensitive and resistant models, and as a major mediator of growth in other resistant models, including one (ICC10-8) that developed complete FGFR2 independence.
RNA sequencing and pathway analysis of differentially expressed genes further emphasized the cooperative suppression of the MEK/ERK pathway by infigratinib and afatinib in the ICC21 and ICC10-6 models. Gene Ontology (GO) analysis revealed various MEK/ERK-related signatures among the highest ranked alteration in the infigratinib + afatinib condition compared to infigratinib alone (Supplementary Figure 7A, B). Similarly, Gene Set Enrichment Analysis (GSEA) (36,37) demonstrated that infigratinib alone suppressed a KRAS gene signature, whereas this effect was greatly potentiated by combination infigratinib + afatinib treatment (Figure 3G). Consistent with the cooperative effects on viability, apoptosis gene signatures were also highly enriched (Supplement Figure 7A, B), with significant upregulation of pro-apoptotic regulators (BIM/BMF/PUMA) and downregulation of anti-apoptotic regulators (BCL-xL and MCL-1) in the infigratinib + afatinib condition versus infigratinib alone (Figure 3H). Thus, consistent with our signaling and functional studies, these data show that a major outcome of this combination treatment is to potently shut down the RAS/MEK/ERK signaling and induce a pro-apoptotic gene signature in FGFR2+ ICC.
Secondary mutations in the FGFR2 kinase domain are a major mechanism of clinical acquired resistance to FGFR inhibitors in patients with FGFR2-fusion-positive ICC (20-22,24,25). We sought to address whether EGFR inhibitor co-treatment can boost the anti-cancer activity of FGFR inhibitors in this setting. To this end, we ectopically expressed the FGFR2-PHGDH fusion allele with a wild type or mutant kinase domain in the FGFR2-dependent ICC13-7 cell line and tested for cooperativity between futibatinib and afatinib. We examined the recurrent alterations, N550K, V565F, and L618V, which confer a range of resistance to futibatinib monotherapy, with the V565F (gatekeeper) mutation causing complete insensitivity (21). As seen for the parental ICC13-7 line, ICC13-7 cells expressing FGFR2-PHGDH with a wild-type kinase domain (FP-WT) were sensitive to single futibatinib alone (FP-WT, IC50 = 3 nM) and showed further sensitivity to futibatinib in combination with 100 nM afatinib (Supplementary Figure 8A, B). Consistent with previous studies (21), the mutant alleles caused different degrees of resistance to futibatinib, with the IC50 of cells expressing each (N550K, IC50 ~ 33.5 nM; V565F, IC50 ~405 nM; L618V, IC50~20 nM) exceeding the in vitro estimated equivalent of the clinical dose [~12.7~19.1 nM, based on unbound Cmax (12)]. In combination with afatinib, the futibatinib IC50 was reduced to values within this clinical dose range in cells expressing either FP-N550K (11.5 nM) or FP-L618V (5.9 nM), whereas no sensitization was observed for V565F, which is completely refractory to inhibition by futibatinib (Supplementary Figure 8A, B). Immunoblot analyses after 4 hrs of futibatinib monotherapy revealed that the L618V, N550K, and V565F mutations caused a graded resistance to shutdown of ERK, AKT (P-S473), and S6 phosphorylation relative to WT FGFR2 while the combination exhibited improved suppression of downstream signaling by the L618V allele (and to a lesser extent N550K) (Supplementary Figure 8C). Accordingly, analysis at 48 hrs revealed that combination treatment induced apoptotic markers in FP-L618V-expressing cells, but not in cells expressing the other mutations (Supplementary Figure 8D). Thus afatinib + futibatinib co-treatment can cooperatively inhibit oncogenic signaling and drive cell death in context of some FGFR2 kinase domain mutations that confer partial resistance to futibatinib monotherapy.
FGFR2 inhibition causes feedback activation of EGFR signaling associated with transcriptional alteration in intracellular regulators of EGFR/MEK
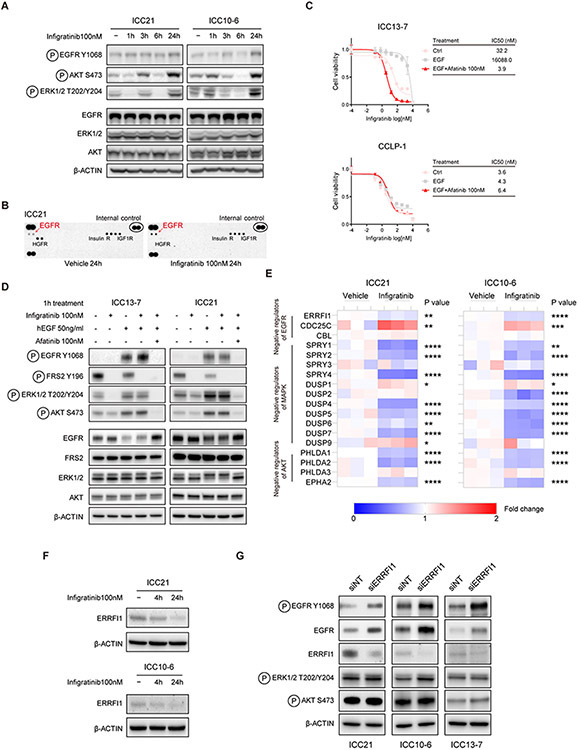
To address the mechanisms underlying the survival pathway reactivation and cooperativity between FGFR and EGFR inhibition in our FGFR2-driven ICC models, we first assessed EGFR signaling status upon FGFRi treatment using an antibody for activating phosphorylation of EGFR (P-Y1068) in the ICC10-6 and ICC21 cell lines. We observed a progressive increase in activating phosphorylation of EGFR (P-Y1068) over 24 hours of treatment with infigratinib in both lines, concomitant with increases in phosphorylation of ERK and AKT (P-S473) (Figure 4A). A more pronounced increase in tyrosine phosphorylation of EGFR in the ICC21 model was detected using phospho-RTK antibody array that probes EGFR phosphorylation at multiple sites (Figure 4B).
Figure 4. Feedback activation of EGFR signaling limits effectiveness of FGFR inhibition.
A. ICC21 and ICC10-6 cells were treated with 100 nM infigratinib or vehicle for the indicated times and lysates were subjected to immunoblot analysis using antibodies against indicated the proteins. B. Phospho-RTK array analysis of ICC21 cells treated with vehicle or 100 nM infigratinib for 24 hours. C. ICC13-7 and CCLP-1 cells were treated with human EGF (50 ng/ml) and/or afatinib (100 nM), or with vehicle control, and tested for sensitivity to infigratinib (IC50 assay). Each point on the dose-response curves represents 4 technical replicates. Data are shown as Mean ± SD. The charts on the right report the IC50 values. D. Immunoblot analysis of ICC13-7 and ICC21 cells treated for 1 hour with human EGF (50 ng/ml), afatinib (100 nM), and/or infigratinib (100 nM) as indicated. E. RNA-seq analysis of FGFR2-fusion+ ICC models. FGFR2-fusion+ ICC cell lines (ICC21 and ICC10-6 cells) were treated with 100 nM infigratinib or vehicle for 4 hours. The heatmaps show the relative expression of negative regulators of EGFR, RTK/MAPK and PI3K/AKT signaling. Each column shows a biological replicate (3/condition). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. F. Immunoblot analysis of ICC21 and ICC10-6 cells treated with vehicle or 100 nM infigratinib for the indicated timepoints. G. FGFR2-fusion+ ICC cell lines (ICC21/ICC10-6/ICC13-7 cells) were transfected with ERRFI1 siRNA or non-targeting control siRNA and then subjected to immunoblot analysis.
Thus, feedback activation of EGFR signaling is elicited by FGFR inhibition with kinetics that matches the rebound activation of AKT and ERK. Moreover, we found that augmentation of EGFR signaling through supplementation of culture media with exogenous EGF ligand was sufficient to overcome sensitivity of ICC13-7 cells to infigratinib, an effect blocked by afatinib co-treatment (Figure 4C). EGF supplementation did not affect infigratinib sensitivity of the FGFR1-dependent CCLP-1 cell line, suggesting a context-dependent role of EGFR signaling in FGFR2 fusion-driven ICC. Consistent with the viability data, EGF supplementation prevented the infigratinib-mediated decrease in phosphorylation of ERK and AKT (P-S473) in ICC13-7 cells, which was reversed by afatinib treatment (Figure 4D); comparable results were seen in ICC21 cells. These data reinforce the potential of EGFR signaling activation to drive FGFRi resistance in FGFR2-fusion-positive ICC.
Transcriptional feedback processes that modulate the expression of regulators of the RTK-MAPK pathway have been observed as one important mechanism for restoring downstream signaling in the context of pharmacologic targeting of pathway components (38-40). To gain insight into how EGFR signaling is activated in response to FGFR inhibition, we examined transcriptional profiles upon 4 hours FGFRi treatment versus vehicle of a set of FGFR2-fusion+ ICC cell lines (ICC21 and ICC10-6 cells, infigratinib). We observed a consistent profile of transcriptional changes in EGFR signaling components across models. Whereas the mRNA expression of different EGF family ligands was either unchanged or decreased upon FGFR inhibition, there were coordinated alterations in expression of intracellular modulators of EGFR/ERBB signaling and increases in ERBB family receptors in each model tested (Supplementary Figure 9A). These changes included decreases in genes encoding key negative regulator of EGFR signaling, ERRFI1 (encoding MIG6) (7,41,42) and as well as increases in ERBB3, and to a lesser extent ERBB2— which can heterodimerize with EGFR and increase downstream signaling (43) (Figure 4E and Supplementary Figure 9A). Moreover, multiple other negative regulators of RTK/MAPK signaling were transcriptionally downregulated, including SPRYs, DUSPs, PHLDAs and EPHA2, suggesting relief of feedback inhibition of MAPK and AKT signaling (Figure 4E).
We explored the potential functional roles of ERRFI1 downregulation and ERBB3 upregulation in EGFR activation in our FGFR2 fusion+ ICC cell lines. Consistent with the transcriptional changes upon FGFR inhibition, we observed a progressive decrease of ERRFI1 protein expression in ICC21 and ICC10-6 cell lines (Figure 4F), concomitant with increases in EGFR signaling (Figure 4A, B). Moreover, siRNA-mediated knockdown of ERRFI1 increased baseline phosphorylation of EGFR (Y1068) and total EGFR protein levels across cell lines tested (Figure 4G). Further linking ERRFI1 levels to EGFR activation in FGFR2+ ICC, we also found that complete genetic knockdown of ERRFI1 provoked FGFR inhibitor resistance in FGFRi-sensitive ICC21 cells (Supplementary Figure 9B, C). On the other hand, genetic knockout of ERBB3 had only modest effects on MEK/ERK and mTORC2/AKT signaling and did not shift the IC50 of FGFR2 fusion+ ICC cells toward FGFR inhibition (Supplementary Figure 9D-F). Thus, the data indicate that the ERRFI1 downregulation observed upon FGFR inhibitor treatment contributes to the adaptive EGFR signaling response, whereas ERBB3 appears dispensable.
In vivo efficacy of combined FGFR and EGFR inhibition in patient-derived FGFR2-fusion+ ICC models
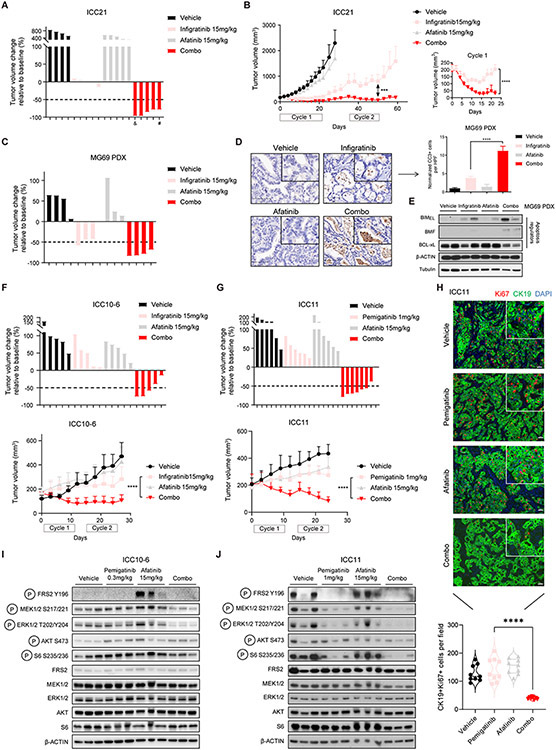
We next tested the cooperative activity of FGFR and EGFR inhibitors in vivo against a set of patient-derived cell line xenograft and PDX models of FGFR2-fusion+ ICC. First, we employed models predicted to be sensitive to FGFRi monotherapy, namely, xenografts formed from ICC21 and ICC13-7 cells and the MG69 PDX model derived from a treatment-naïve patient. For cell line xenograft models, mice were randomized into vehicle, single agent, or combination groups once tumors reached ~200 mm3; PDX studies were commenced with a starting volume of ~600 mm3. As expected, ICC21 xenografts were relatively sensitive to infigratinib (15 mg/kg, daily) alone, exhibiting disease stabilization after an initial 21-day cycle of treatment (Figure 5A). Afatinib alone (15 mg/kg; daily) caused no significant changes in tumor growth, whereas combined infigratinib/afatinib treatment led to dramatic tumor shrinkage, with complete or near complete responses in 5/5 mice. Mice were then left untreated and monitored for recurrence for 2 weeks. Although regrowth was observed, the tumors remained responsive to resumption of combination therapy, again regressing to the limits of detection after a second treatment cycle (Figure 5B shows tumor growth over time; the right panel shows a blow-up of tumor sizes for the first cycle of treatment). Weight loss was observed in 2/5 mice upon prolonged treatment (daily treatment for >2 weeks) in the combination group (Supplementary Figure 10A) and thus cycle 2 was reduced to 14 days. Efficacy of combination infigratinib/afatinib treatment was also observed in the MG69 PDX model with strong regression seen at 10 days treatment (Figure 5C). Correspondingly, there was a marked induction of cell death as demonstrated by cleaved caspase-3 staining and by immunoblot for apoptotic regulators, which showed BMF induction and decreased BCL-xL (Figure 5D and E). Similar responses were seen combining infigratinib with the EGFR-specific RTK inhibitor, gefitinib, in the ICC13-7 xenograft model (Supplementary Figure 10B). Thus, EGFR/ERBB inhibition significantly enhances the efficacy of FGFR inhibition in FGFR2-fusion+ ICC models that exhibit sensitivity to FGFRi monotherapy.
Figure 5. Combination FGFR/EGFR inhibitor treatment is effective against FGFR2+ ICC models in vivo.
A and B. Mice harboring ICC21 subcutaneous xenografts with a starting tumor volume ~200 mm3 were treated with vehicle (n=4), infigratinib 15 mg/kg (n=5), afatinib 15 mg/kg (n=5), or the combination of both drugs (n=5). Mice were treated daily for 21 days, provided an intervening 14-day drug holiday, and then received a second cycle of treatment. (A) Waterfall plot showing tumor volume changes after first treatment cycle (21 days). #: treatment stopped at day-15 and resumed on day 19. &: euthanized after 20-days treatment. (B) Serial monitoring of tumor size. Right panel: Blown up image of tumor growth curves of the infigratinib and combination groups during treatment cycle 1 (21 days). ***P<0.001. ****P<0.0001. Data are shown as Mean ± SD. C-E. Mice bearing subcutaneous tumors from the MG69 PDX model (starting tumor volume ~600 mm3) were treated daily for 10 days with vehicle (n=4), infigratinib 15 mg/kg (n=4), afatinib 15 mg/kg (n=3), or the combination of both drugs (n=4). C: waterfall plot showing tumor volume changes after 10 days treatment. D: representative images of immunohistochemistry staining for cleaved caspase-3 in tumor samples after 10 days treatment. Data are quantified in the chart. Bar: 100 μm. HPF: high power field. One-way ANOVA multiple comparisons with Tukey correction were used to analyze the data. ****P<0.0001. Data are shown as Mean ± SEM. E: immunoblot analysis of apoptosis regulators from tumor lysates after 10 days treatment. F. Mice bearing ICC10-6 subcutaneous xenograft tumors were treated with vehicle, infigratinib 15 mg/kg, afatinib 15 mg/kg, or the combination for cycles of 10 days, with an intervening 4-day drug holiday. Upper: waterfall plot of tumor volume changes at the end of treatment cycle 1 (10 days). Lower: serial monitoring of tumor size. n = 5 mice per group. Two-way ANOVA multiple comparisons with Tukey correction were used to analyze the data. ****P<0.0001. Data are shown as Mean± SD. G. Mice bearing ICC11 subcutaneous xenograft tumors were treated with vehicle (n=6), pemigatinib (1 mg/kg) (n=6), afatinib 15 mg/kg (n=6), or the combination of both drugs (n=8). Treatment was given daily for 10-day cycles with intervening 4-day drug holidays. Upper: Waterfall plot of tumor volume changes at the end of the second cycle of treatment. Lower: Serial monitoring of tumor size. H. Representative immunofluorescence staining for CK19 (green) and Ki67 (red) in tumors from ICC11 dose escalation study after cycle 3 of treatment. Data are quantified in the chart at the bottom. One-way ANOVA multiple comparisons with Tukey correction were used to analyze the data. ****P<0.0001. I. Immunoblot analysis of tumor lysates from the ICC10-6 subcutaneous xenograft model subjected to the indicated treatments for 8 days. Tumors were harvested 6 hours after the last dose of treatment. J. Immunoblot analysis of tumor lysates from ICC11 dose escalation study after cycle 3 of treatment. Tumors were harvested 6 hours after the last dose of treatment.
We next examined the in vivo efficacy of the combination therapy in FGFRi-resistant models. First, whereas neither infigratinib nor afatinib monotherapy prevented progression of ICC10-6 xenografts, the combination led prominent tumor regression after 10 days treatment (Figure 5F, upper panel). These responses continued through two cycles of treatment (Figure 5F, lower panel; treatment cycles were for 10 days, separated by a 4-day break). Weight loss was initially observed in 3/5 mice in the combination group during cycle 1, whereas mice in all groups maintained their weight thereafter (Supplementary Figure 10C). We also sought to obtain further proof of concept data for the FGFRi/EGFRi combination in the context of treatment with pemigatinib, another FGFRi which received FDA approval in April 2020 for patients with FGFR2+ ICC. The FGFRi resistant model, ICC11, was used for these studies. Combination treatment with pemigatinib (1 mg/kg)/afatinib (15 mg/kg) resulted in prominent tumor regression, whereas single agent treatment only slightly slowed down the tumor growth (Figure 5G, upper panel waterfall plot; lower panel tumor growth curve). No weight loss was observed in mice throughout the treatment course (Supplementary Figure 10D). Since current pan-FGFR kinase inhibitors have dose-limiting toxicities and often require dose reductions, we also modeled a dose escalation study, using a pemigatinib dosing scheme that is either below (0.3 mg/kg) or equivalent to the human clinical dose (1 mg/kg) (44). Pemigatinib treatment was given for two 10-day cycles at the 0.3 mg/kg dose and then escalated to 1 mg/kg for a third cycle, with 4-day intervals between cycles. Throughout the first two cycles, whereas both single agent treatments slightly reduced the rate of tumor growth, the inhibitor combination completely abolished any growth. This effect was magnified by the use of 1 mg/kg pemigatinib in cycle 3, which resulted in rapid tumor regression, while the single agent groups continued to progress (Supplementary Figure 10E and F). The combination was well tolerated throughout the treatment course (Supplementary Figure 10G). The ability of afatinib to cooperate with reduced dose pemigatinib (0.3 mg/kg) was also observed in the FGFRi monotherapy resistant ICC10-6 model (Supplementary Figure 10H). Notably, Ki67 staining revealed a virtual elimination of proliferation in the combination treated tumors, while no changes were seen with either single agent (Figure 5H). Moreover, signaling studies demonstrated that the combination led to a more durable shutdown of MEK/ERK/mTOR activity than single agent treatment in both the ICC10-6 and ICC11 xenograft models (Figure 5I, J).
Discussion
The recent approvals of pemigatinib and infigratinib for the treatment of FGFR2+ ICC represent important milestones for targeted therapy development in this disease. However, the response rates are moderate, and resistance emerges rapidly. By employing high-throughput drug screens and signaling studies, we identified signaling feedback via the EGFR pathway as a major mediator of adaptive resistance to FGFR kinase inhibition in a set of patient-derived ICC models. Combination treatment with EGFR/ERBB inhibitors potentiated the effect of FGFR inhibition, overcoming rebound activation of MEK/ERK and mTOR signaling and inducing apoptosis both in models that are sensitive and resistant to single agent treatment. These data support the exploration of dual FGFR/EGFR inhibition as a strategy to improve initial response rates and to extend benefit of patients with acquired FGFRi resistance.
Perturbation of pro- and anti-apoptotic proteins in response to kinase inhibition determines the commitment to cell death in multiple RTK-addicted cancer types (30). Accordingly, our findings highlight the importance of comprehensive shutdown of downstream oncogenic signaling upon FGFR inhibition to induce apoptosis and to promote significant tumor regression. Even in models that are highly sensitive to FGFRi monotherapy [ICC13-7 (Figure 1E, left panel) and MG69 (21)], and show considerable reduction in MEK/ERK phosphorylation upon single agent FGFR inhibition, this treatment alone failed to stimulate pro-apoptotic gene expression or apoptotic cell death in vitro and induced only modest levels in vivo (although we cannot exclude the possibility of a degree of caspase-independent death). Combination FGFR/EGFR inhibitor treatment resulted in complete extinction of MEK and ERK phosphorylation and reduced mTOR activity [phospho-AKT (S473) and phospho-S6 S235/236)] in vitro and induced an apoptotic gene signature (including BMF upregulation and BCL-xL downregulation), marked apoptosis, and significant tumor regressions in vivo. The data suggest that EGFR-dependent feedback processes may contribute to the intermediate response rate and limited tumor shrinkage in responders (typically <50% size change from baseline) seen upon FGFRi treatment in the clinic.
We also studied multiple models that were partially or completely resistant to FGFRi monotherapy despite lacking FGFR2 kinase domain mutations (ICC10-6, ICC11, ICC21, ICC24, ICC25). Here, although FGFRi treatment effectively suppressed FGFR kinase activity (indicated by loss of phospho-FRS2 in ICC10-6 and ICC21), only minimal or transient reduction in phospho-MEK/phospho-ERK was observed. However, these models remained at least partially FGFR-dependent, since FGFR and EGFR inhibitors acted cooperatively to shut down the MEK/ERK pathway and to induce apoptotic gene expression and cell death. The data are consistent with a role for EGFR-mediated adaptive signaling as a mechanism of primary or acquired FGFRi resistance in FGFR2+ ICC. In this regard, a significant subset of patients who progress on FGFRi treatment do not have detectable secondary FGFR2 kinase domain mutations or other clear genetic drivers of resistance (45).
Our studies suggest that combination FGFR/EGFR inhibitor treatment could also potentially provide benefit in the setting of secondary FGFR2 kinase domain mutations, which emerge at the time of progression in ~60-70% patients with FGFR2+ ICC treated with ATP-competitive pan-FGFR kinase inhibitors (45). The irreversible pan-FGFR inhibitor, futibatinib, can overcome some of these mutations, although typically with reduced efficacy as compared to FGFR2 fusions with a wildtype kinase domain (21). We found that co-treatment with afatinib enhanced the ability of futibatinib to suppress downstream oncogenic signaling and reduce viability in ICC cells expressing an FGFR2 fusion harboring the L618V mutation, despite incomplete suppression of FGFR2 kinase activity. Mutations that caused greater resistance to futibatinib were either less sensitive to the combination (N550K) or cells were completely refractory (V565F). However, next-generation FGFR inhibitors with improved efficacy against these mutations (28), may show better cooperativity with EGFR inhibition in the setting of diverse secondary FGFR2 mutations. Notably, FGFRi resistance is often polyclonal and heterogeneous. Patients frequently show several different acquired FGFR2 kinase domain mutations in distinct tumor nodules and these mutations display a range of sensitivity to FGFR inhibitors, including emerging next-generation compounds (21)(28). Moreover, mixed mechanisms of resistance can be seen within the same patient with resistant nodules with and without FGFR2 kinase mutations being observed concurrently (20,24). Thus, the potential of EGFR inhibition to boost FGFRi efficacy in multiple contexts may be beneficial in combating polyclonal and heterogeneous resistance.
Our data indicate that MEK/ERK activation is the primary downstream output of FGFR2 fusions in ICC. Examination of dual FGFR/EGFR inhibition revealed that crosstalk between these pathways also regulates mTORC1 and mTORC2 activity, as reflected by consistent decreases in phosphorylation of S6 and to a lesser extent AKT (P-S473), respectively. Unlike many other RTKs (46,47) and scenarios where FGFR2 is amplified (48), the PI3K pathway does not appear to be strongly stimulated by either FGFR2 or EGFR in FGFR2 fusion+ ICC, since AKT (P-S308) phosphorylation was not significantly decreased by single agent or combination treatment. The identification of MEK/ERK activation as the primary output of FGFR2 fusions in ICC rather than PI3K/AKT signaling is also consistent with the genomic features of these tumors. Prior to treatment, FGFR2+ ICCs show frequent concurrent activating mutations in PIK3Ca or loss of function mutations in PTEN, but very rare concurrent alterations of other RTK-RAS-MEK signaling components (8)(11). Moreover, acquired mutations in RAS genes arise as FGFRi resistance mechanisms in a subset of patients with FGFR2+ ICC (8). Importantly, dual targeting of RTKs may be a more tolerable treatment than combination therapies involving downstream signaling components such as SHP2, MEK/ERK and PI3K, given their selective activity in different tissues and less overlapping toxicity profiles. Nevertheless, further preclinical exploration of efficacy of targeting the SHP2/MEK/ERK axis in FGFR2+ ICC is warranted. In addition, since we used afatinib (which targets EGFR, ERBB2, and ERBB4) for most of our in vivo combination treatment studies, it will be important to determine whether ERBB2 and/or ERBB4 inhibition also contributes to the therapeutic efficacy in some models.
Feedback activation of EGFR/ERBB signaling upon FGFR inhibition has been observed in FGFR3-driven bladder cancer and other contexts, and more broadly, inhibition of oncogenically activated RTK-RAS signaling is frequently offset by EGFR activation (40,49-53). Diverse mechanisms for this interplay have been reported including upregulation of EGF family ligands, altered EGFR trafficking and phosphatases involved in feedback regulation of EGFR (49,52,53). Our data in FGFR2+ ICC suggest a mechanism of adaptive resistance by which FGFR inhibition augments EGFR dependency via negative feedback involving transcriptional downregulation of a network of genes encoding intracellular suppressors of EGFR/ERBB and MEK/ERK signaling. These factors include ERRFI1/MIG6, which binds directly to ERBB family members and limits signaling activity (7,41,42). Our functional data support a role for the observed decrease in ERRFI expression in EGFR activation in FGFR2+ ICC cells upon FGFRi treatment. Notably, a case report demonstrated significant tumor regression in a patient with ICC with an inactivating ERRFI1 mutation in response to treatment with the EGFR inhibitor, erlotinib, consistent with the importance of ERRFI1 loss in driving EGFR dependency in ICC (7). It is likely the broad downregulation of multiple SPRY and DUSP genes— which attenuate downstream signaling by binding at the level of the GRB2-SOS1 complex and by dephosphorylating MAPK, respectively—also contributes to adaptive signaling responses.
The approval of the FGFR inhibitors, pemigatinib and infigratinib, for the treatment of FGFR2 fusion-positive cholangiocarcinoma represents a breakthrough in precision medicine for ICC. However, an urgent need still exists to improve the effectiveness of FGFR targeted therapy, mitigate toxicity, and identify additional treatment options for FGFRi-resistant disease. Given the completion of multiple FGFRi monotherapy trials, now is the ideal time to explore the combination therapy for those patients with primary or acquired resistance to FGFR targeted therapy. Rebound activation of MEK/ERK signaling protects against cell death and maintains proliferation in response to FGFR inhibition across a series of FGFR2+ ICC models. Our data support the use of EGFR inhibitors to potentiate FGFRi efficacy in the treatment-naïve setting and to overcome adaptive resistance.
Materials and Methods
Cell culture
Established cell lines were obtained from the following sources: Riken Bioresource Center (RBE) and Korean Cell Line Bank (SNU-1079). CCLP-1 cell was a kind gift from Dr. P.J. Bosma (Academic Medical Center, Amsterdam, the Netherlands). PC9 and HCC827 cells were kind gifts from Dr. Haichuan Hu (Massachusetts General Hospital). To generate patient-derived biliary tract cancer cell lines (ICC10, ICC10-6, ICC10-8, ICC11, ICC13-7, ICC21, ICC24, and ICC25), we utilized resection or autopsy specimens directly or following growth as patient-derived xenografts, under Institutional Review Board approved protocols (DFCI#13-162, #13-416, and 19-699 ), using previously described methods (21). Samples were minced with sterile razor blades, digested with trypsin for 30 minutes at 37°C, and then resuspended in RPMI supplemented with 20% fetal bovine serum, 1% L-glutamine (Gibco, #25030-081), 1% MEM Non-Essential Amino Acids Solution (Gibco, #11140-050), 1% Sodium Pyruvate (Gibco, #11360-070), 0.5% penicillin/streptomycin, 10 μg/mL gentamicin (Gibco, #15710-064), and 0.2 Units/mL human recombinant insulin (Gibco, #12585-014), seeded on plates coated with rat tail collagen (BD Biosciences), and incubated at 37°C with 5% CO2. Cells were passaged by trypsinization and adapted to uncoated tissue-culture plates in RPMI supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin prior to functional studies. They were routinely checked to be mycoplasma free. Cell lines were authenticated by short tandem repeat (STR) DNA profiling by the cell line bank from which they were obtained (RBE and SNU-1079) or provided by the ATCC between January and September 2020 (ICC21, ICC25) or the Broad Institute between January and December 2018 (ICC10, ICC10-6, ICC10-8, ICC11, ICC13-7). All cell lines were used within 10 passages of establishment from patients or receipt from repositories.
High throughput drug screening
Cells were seeded at a predetermined density based on optimal proliferation for each cell line. The screen was performed in 384 well plates. Cells were seeded using automated dispense (Biotek) and drug added using automated liquid handling (Janus, Perkin Elmer) the next day. A library of 111 drugs was tested as single agents or in combination with FGFR inhibitors. For each library drug, 9 doses were tested covering a range of 0.003 μM-10 μM. FGFR inhibitors were each used at a single dose: infigratinib (100 nM), futibatinib (40 nM), rogaratinib (1000 nM). Cells were incubated for 5 days in the presence of drugs. Cells were stained using Hoechst 33342 (33 μM, all nuclei stain) and ethidium bromide dimer (3 μM, plasma membrane integrity; detection of dead cells). Viability and cell death were measured using high-throughput imaging (ImageXpress Micro_XL, Molecular Devices) using the software provided by the imager manufacturer. Total cell count (Hoechst 33342) and number of dead cells (EthBr) were used to compute the percentile of dead cells in each well. Excess over Highest Single Agent (HSA) was determined by comparing the effect of FGFR inhibitor alone, library drug alone and the combination. For each library drug dose, the percentile of cell death for the most effective single agent (library drug or FGFR inhibitor) was compared to the percentile of cell death obtained with the matched drug dose combination. This yielded nine excess-over-HSA values for each drug. Across these nine values, the second maximum value was then extracted to capture an overall excess over HSA score. The screen was performed in biological duplicates (2 independent cell seeding days), following quality control based on coefficient of variation of the DMSO control cells (CV<25% except for one screen run for the ICC11 cell line with CV= 32%) and effect of the anchor alone (FGFR inhibitor) based on pre-screen data as a control for appropriate drug dispense and cellular response. Average excess over HSA across the replicate runs was used for the ranking of combinations. Heat map and hierarchical cluster analysis were generated in Morpheus (https://software.broadinstitute.org/morpheus/, RRID:SCR_017386) based on second HSA score for cell death across all cell lines screened.
Cell cycle analysis
Cells were seeded on 6 cm dish and incubated overnight. The next day, 100 nM infigratinib was added for a 24h incubation. Before harvesting the cells, 10 μM 5-ethynyl-2′-deoxyuridine (EdU) was added for 1.5h. The staining procedure was performed using the Click-iT® Plus EdU Alexa Fluor® 647 Imaging Kit (ThermoFisher Scientific, C10634) following the manufacturer’s recommended protocol. Finally, cells were incubated in 10 μg/ml RNase A (ThermoFisher Scientific, EN0531) and 20 μg/ml Propidium iodide (Sigma-Aldrich, P4864) solution for 30 min at room temperature. Samples were run on a Cytoflex S flow cytometer (Beckman Coulter Life Sciences). Data were analyzed using FlowJo (Tree Star, Ashland, OR, RRID:SCR_008520) software.
Cell viability assay
Cells were seeded at a density of 3000 cells per well on 96-well plates and incubated overnight. Compounds were added the next day over a 9-point concentration range and then cells were incubated for 10-14 days, depending on the growth rate of individual cell lines. The viability of cells was then measured by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. IC50 values were determined by GraphPad Prism 9 using a 3-parameter dose-response model. Small molecule inhibitors were purchased from Selleckchem: infigratinib (S2183), futibatinib (S8848), pemigatinib (S0088), afatinib (S1011), gefitinib (S1025), EKB-569 (S1392).
Crystal violet staining
Cell numbers and treatment times were adjusted for each line based on growth kinetics. Cells were seeded in an equal number per well (0.5 x 105 – 3 x 105 cells/well) on 6-well plates and allowed to attach overnight. Fresh media with or without drug was replaced the next day and refreshed every 2-3 days until the end of the experiment (10 days treatment unless specifically indicated). Cells were fixed in 4% paraformaldehyde solution for 10 minutes and subsequently stained with 0.5% crystal violet solution for 30 minutes. Plates were washed 3 times with H2O and dried overnight. Relative cell density was then quantified using ImageJ software (RRID:SCR_003070).
Caspase-3/7 Activity
Cells were seeded at a density of 3000 cells per well on 96-well plates and incubated overnight. Compounds were added the next day for an incubation period of 3 days. Caspase-3/7 activity was then assessed using the Caspase-Glo 3/7 Assay (G8090; Promega) using the manufacturer’s recommended protocol.
Phospho-RTK Array
Cells were seeded at 80% confluency in a 6 cm dish and allowed to attach overnight. Compounds were added the next day and incubated for 24 hours. The Proteome Profiler Human Phospho-RTK Array Kit (R&D Systems, ARY001B) was then used to detect activated RTKs according to the manufacturer’s instructions.
SiRNA transfection
Cells were seeded at 60-80% confluency in 6-well plates and allowed to attach overnight. Lipofectamine™ RNAiMAX Transfection Reagent (13778075) was used to perform siRNA transfection according to the manufacturer’s instructions. SiRNAs were purchased from Dharmacon: siGENOME Human ERRFI1 siRNA-SMARTpool (M-017016-01-0010), siGENOME Non-Targeting siRNA (D-001210-01-05).
Generation of ICC13-7 cell derivatives expressing the FGFR2-PHGDH fusion with a wild type or mutated kinase domain
An FGFR2-PHGDH fusion construct, containing exons 1-17 or FGFR2-IIIb isoform fused to PHGDH (NM_006623.3) exons 6-12, was amplified from reverse transcribed cDNAs from an ICC patient sample and inserted into the pMSCV vector using the NEBuilder HiFi DNA Assembly (New England Biolabs). All FGFR2 mutations were introduced into the pMSCV vector using the same kit. Targeted Sanger sequencing was done to confirm mutation generated. Retrovirus was generated by transfecting the pMSCV constructs and packaging plasmids into 293T cells. After collection of retrovirus, transfected 293T cells were collected to confirm protein expression from each construct. Viral infections of ICC13-7 cells were performed in the presence of polybrene. Infected cells were selected in blasticidin (6-15 ug/ml) for one to two weeks.
Generation of ICC21/ICC10-6 cell derivatives using CRISPR/Cas9 gene editing
Cas9 plasmid P473 [LV-EF1a WT Cas9-P2A-Blasticidin] was a kind gift from Dr. Mazhar Adli (University of Virginia, School of Medicine). ERBB3 and non-targeting control gRNAs plasmids were purchased from Addgene: non-targeting control gRNA (#80263), ERBB3 gRNAs (#77479, #77480 and #77481). Lentivirus was generated by transfecting the Cas9/gRNA constructs and packaging plasmids into 293T cells. After collection of lentivirus, ICC21 and ICC10-6 were used for viral infections and selected in blasticidin (25-50 μg/ml) for one week to first establish Cas9 stable cell lines. ERBB3 knockout was then performed in the ICC21/ICC10-6 cell lines with Cas9 expression.
Immunoblot analysis
Snap-frozen tumor tissues were homogenized using a Precellys 24 homogenizer (Bertin Instruments) at 6500 rpm. Tumor and cell protein lysates were prepared in Thermo Scientific™ RIPA Lysis and Extraction Buffer (PI89900) containing Pierce™ Protease Inhibitor (A32965) and Calbiochem phosphatase inhibitor cocktail set I and II. Protein concentration was determined by Pierce BCA Protein Assay. 20μg protein was used to perform analysis by SDS-PAGE, electro-transfer and immunoblotting with specific antibodies. The following primary antibodies were used: from Cell Signaling Technology (all at 1:1000 dilution), PARP (9542, RRID:AB_2160739), phospho-FRS2 Y196 (3864S, RRID:AB_2106222), phospho-MEK1/2 S217/221 (9154S, RRID:AB_2138017), MEK1/2 (4694S, RRID:AB_10695868), phospho-ERK1/2 T202/Y204 (4370S, RRID:AB_2315112), ERK1/2 (4695S, RRID:AB_390779), phospho-AKT S473 (4060S, RRID:AB_2315049), AKT (9272S, RRID:AB_329827), phospho-S6 S235/236 (4858S, RRID:AB_916156), S6 (2217S, RRID:AB_331355), BIM (2933S, RRID:AB_1030947), BMF (50542S, RRID:AB_2892182), PUMA (4976S, RRID:AB_2064551), MCL-1 (94296S, RRID:AB_2722740), BCL-xL (2764S, RRID:AB_2228008), phospho-EGFR Y1068 (3777S, RRID:AB_2096270), phospho-EGFR Y1173 (4407S, RRID:AB_331795), EGFR (4267S, RRID:AB_2246311), ERRFI1 (2440S, RRID:AB_823573), phospho-SRC Y416 (6943T, RRID:AB_10013641), phospho-PLCγ1 Y783 (2821S, RRID:AB_330855), PLCγ1 (2822S, RRID:AB_2163702), phospho-STAT3 Y705 (9131S, RRID:AB_331586), ERBB3 (12708S, RRID:AB_2721919); from Abcam, phospho-SHP2 Y542 (ab62322, RRID:AB_945452) 1:1000, FRS2 (ab183492) 1:1000, Tubulin (ab6160, RRID:AB_305328) 1:5000; from Sigma (1:5000 dilution), β-actin (A5316, RRID:AB_476743); from Invitrogen (1:5000 dilution), GAPDH (AM4300, RRID:AB_437392).
In vivo treatment studies
All mice were housed in a specific pathogen-free environment at the MGH and treated in strict accordance with protocols 2005N000148 and 2019N000116 approved by the Subcommittee on Research Animal Care at MGH. 6- to 10-week-old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, 00557, The Jackson Laboratory) were used in the studies below. All in vivo studies involved subcutaneous implant of ICC tissue or cells into NSG mice, treatments by oral gavage, and serial monitoring by digital calipers. Mice were monitored daily by veterinary services. Treatment was discontinued and dietary supplement provided if mice showed signs of poor body condition (PBC) such as hunched posture, lethargy, 15% weight loss, or reduced grooming. PDXs were used within 5 passages of establishment from patients. For the MG69 PDX model (21), tissue fragments were rinsed in Hank's Balanced Salt Solution and cut into 0.3–0.5 mm3 pieces with sterile razor blades followed by implantation. When tumors reached ~600 mm3, mice were randomized and started on treatment with vehicle, infigratinib 15 mg/kg, afatinib 15 mg/kg or both drugs (infigratinib 15mg/kg+ afatinib 15 mg/kg) daily for 10 days. For all cell line xenograft studies, mice were randomized into treatment groups once tumors reached ~200 mm3. For ICC21 xenografts, 2x10^6 cells were injected and tumor-bearing mice were treated with vehicle, infigratinib 15 mg/kg, afatinib 15 mg/kg or both drugs (infigratinib 15 mg/kg+ afatinib 15 mg/kg) daily for 21 days. After 14 days without treatment, mice received a second, 14-day cycle of treatment. For ICC13-7 xenografts, 2x10^6 cells were injected and tumor-bearing mice were treated with vehicle, infigratinib 30 mg/kg, gefitinib 25 mg/kg or both drugs (infigratinib 30 mg/kg+ gefitinib 25 mg/kg) daily for 10 days. For infigratinib combination studies against ICC10-6 xenografts, 2x10^6 cells were injected and tumor-bearing mice were treated with infigratinib 15mg/kg, afatinib 15 mg/kg or both drugs (infigratinib 15 mg/kg+ afatinib 15 mg/kg) daily for 10 days, followed by 4 days off treatment. For pemigatinib combination studies in ICC10-6 xenografts, 1x10^6 cells were injected. Tumor-bearing mice were treated with vehicle, pemigatinib 0.3 mg/kg, afatinib 15 mg/kg or both drugs (pemigatinib 0.3 mg/kg+ afatinib 15 mg/kg) daily for 10 days (efficacy study) or 8 days (signaling study). Tumor samples were collected post 6 hours at the last dose for biochemical analysis. For ICC11 xenografts, 1x10^6 cells were injected. Tumor-bearing mice were entered into cycles of 10 days on treatment with vehicle, pemigatinib 1 mg/kg, afatinib 15 mg/kg or both drugs (pemigatinib 1 mg/kg+ afatinib 15 mg/kg) followed by 4 days off. In the dose escalation study, pemigatinib treatment was given for two 10-day cycles at the 0.3 mg/kg dose and then escalated to 1 mg/kg for a third cycle, with 4-day intervals between cycles. Tumor samples were collected either 4 hours after the last dose (MG69 PDX model) or 6 hours after the last dose (cell lines xenografts) and apportioned for biochemical analysis (snap frozen in liquid nitrogen) and histology processing.
All drugs were purchased from Selleckchem: infigratinib (S2183), pemigatinib (S0088), afatinib (S1011), gefitinib (S1025). Formulations for each drug were as follows: infigratinib (PEG300/D5W (2:1, v/v)), pemigatinib (5% DMAC in 0.5% methylcellulose aqueous solution), afatinib and gefitinib (a physiological NaCl solution containing 0.5% methylcellulose and 0.4% Tween80).
Immunohistochemistry staining
Immunohistochemistry staining was performed by iHisto Inc. (Salem MA; iHisto.io). Samples were processed, embedded in paraffin, and sectioned at 4 μm. Paraffin sections were then deparaffinized and hydrated using the following steps: 15 min in xylene twice; 5, 5, and 5 min in 100%, 100%, and 75% ethanol, respectively; and 5 min in PBS at room temperature repeated three times. Antigen retrieval was achieved by boiling the sections in 10 mM sodium citrate for 10 min in a microwave oven and 5 min of cooling at room temperature. Sections were then washed with PBS three times, treated with 3% H2O2 for 15 min and 5% bovine serum albumin for 20 min. Primary antibody Cleaved Caspase-3 (Cell Signaling Technology Cat# 9664, RRID:AB_2070042) was used for the staining. Whole slide scanning was performed on an EasyScan infinity (Motic). Five to 12 fields per independent tumor (using the MG69 PDX model) from each treatment condition were used for quantification using ImageJ software (RRID:SCR_003070).
Multiplex immunofluorescence staining
Immunofluorescence staining was performed by iHisto Inc. (ihisto.io). Samples were processed, embedded in paraffin, and sectioned at 4 μm. Paraffin sections were then deparaffinized and hydrated using the following steps: 15 min in xylene twice; 5 min each in 100% ethanol (twice), 75% ethanol, and PBS (thrice) at room temperature. Antigen retrieval was achieved by boiling sections in 10 mM sodium citrate for 10 min in a microwave oven and cooling for 5 min at room temperature. Sections were then washed with PBS three times, treated with 3% H2O2 for 15 min and 5% bovine serum albumin for 20min. The following primary antibodies were used: Keratin 17/19 antibody (Cell signaling technology, 12434, RRID:AB_2797912, 1:200) and KI67 (Servicebio, GB13030-2, 1:200). Stained slides were photographed with a Nikon Eclipse Ti microscope at 10X magnification for representative images and quantitative analysis. Three fields from each independent tumor (in the ICC11 model) per treatment condition were used for quantification using Fiji software (RRID:SCR_002285).
RNA Extraction and RNA sequencing
RNA was extracted from cultured cells under different treatment conditions using the Rneasy Mini Kit (QIAGEN) in accordance with the manufacturer-prescribed protocol. Total RNA was quantified and assessed for quality using a NanodropTM spectrophotometer (Thermofisher). RNA sequencing was performed and analyzed at GENEWIZ (genewiz.com, RRID:SCR_003177). Sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Homo sapiens GRCh38 reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. BAM files were generated as a result of this step. Unique gene hit counts were calculated by using featureCounts from the Subread package v.1.5.2. The hit counts were summarized and reported using the gene_id feature in the annotation file. Only unique reads that fell within exon regions were counted. After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between the defined groups of samples was performed. The Wald test was used to generate p-values and log2 fold changes. Genes with an adjusted p-value < 0.05 and absolute log2 fold change > 1 were called as differentially expressed genes for each comparison. RNA sequencing data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-11324.
Gene set enrichment analysis
To test whether gene sets were enriched in response to different conditions, we utilized GSEA MSigDB gene set enrichment analysis for hallmark gene signatures (Gene Set Enrichment Analysis, RRID:SCR_003199) (36,37). Hallmarks with enrichment scores (ES) greater than 1.4 and possessing an FDR q-value < 0.2 were considered significantly enriched.
Gene ontology analysis
A gene ontology analysis was performed on the statistically significant set of differentially expressed genes by implementing GeneSCF v.1.1-p2 software. The goa_human GO list was used to cluster the set of genes based on their biological processes and determine their statistical significance. A list of genes clustered based on their gene ontologies was generated.
Statistical analysis
Statistics were performed and graphs were generated in GraphPad Prism 9 (RRID:SCR_002798). One-way or two-way ANOVA multiple comparisons with Tukey correction were used to analyze the data between multiple groups (vehicle, single agents and both drugs). Student’s t-test (two tailed) was performed for two groups-only comparison. P < 0.05 was considered to be significant.
Supplementary Material
Acknowledgements:
We thank members of the Bardeesy lab for valuable input.
Funding:
This work was supported by grants from the National Institute of Health (P50CA127003), TargetCancer Foundation, the Wellcome Trust (102696), DOD (CA160216), ProjectLiv, and a V Foundation Translational Research Award. Q. Wu was supported by a Cholangiocarcinoma Foundation Marion U. Schwartz Memorial Research Fellowship. LG was supported by the American Cancer Society Clinical Scientist Development Grant.
Footnotes
Disclosure of Potential Conflicts of Interest:
L. Goyal reports receiving research funding (to institution): Adaptimmune, Bayer, Eisai, Merck, Macrogenics, Genentech, Novartis, Incyte, Eli Lilly, Loxo Oncology, Relay Therapeutics, QED, Servier, Taiho Oncology, Leap Therapeutics, Bristol Meyers Squibb, Nucana; and she serves as an advisor/consultant to Alentis Therapeutics, Astra Zeneca, Exelixis, Genentech, H3Biomedicine, Incyte Corporation, QED Therapeutics, Servier, Sirtex Medical Ltd, and Taiho Oncology Inc. D. Juric reports receiving consulting fees from Novartis, Genentech, Syros, Eisai, Vibliome, Mapkure, and Relay Therapeutics, and commercial research support from Novartis, Genentech, Syros, Pfizer, Eisai, Takeda, Pfizer, Ribon Therapeutics, Infinity, InventisBio, and Arvinas, and has ownership interest in Relay Therapeutics, Transcode Therapeutics and PIC Therapeutics. N. Bardeesy reports receiving a commercial research grant from Taiho Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
Data and Code Availability. RNA sequencing data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-11324.
References
- 1.Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov. 2017;7:943–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus Gemcitabine versus Gemcitabine for Biliary Tract Cancer. N Engl J Med. 2010;362:1273–81. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y-M, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of Targetable FGFR Gene Fusions in Diverse Cancers. Cancer Discov. 2013;3:636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–34. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–10. [DOI] [PubMed] [Google Scholar]
- 6.Farshidfar F, Zheng S, Gingras M-C, Newton Y, Shih J, Robertson AG, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18:2780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, Bryce AH, et al. Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma. PLoS Genet. 2014;10:e1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleary JM, Raghavan S, Wu Q, Li YY, Spurr LF, Gupta HV, et al. FGFR2 Extracellular Domain In-Frame Deletions are Therapeutically Targetable Genomic Alterations that Function as Oncogenic Drivers in Cholangiocarcinoma. Cancer Discov. 2021;11:2488–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sia D, Losic B, Moeini A, Cabellos L, Hao K, Revill K, et al. Massive parallel sequencing uncovers actionable FGFR2 – PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun. 2015;6:6087. [DOI] [PubMed] [Google Scholar]
- 10.Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javle M, Lowery M, Shroff RT, Weiss KH, Springfeld C, Borad MJ, et al. Phase II Study of BGJ398 in Patients With FGFR-Altered Advanced Cholangiocarcinoma. JCO. 2018;36:276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahleda R, Meric-Bernstam F, Goyal L, Tran B, He Y, Yamamiya I, et al. Phase I, first-in-human study of futibatinib, a highly selective, irreversible FGFR1-4 inhibitor in patients with advanced solid tumors. Ann Oncol. 2020;31:1405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzaferro V, El-Rayes BF, Droz dit Busset M, Cotsoglou C, Harris WP, Damjanov N, et al. Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion-positive intrahepatic cholangiocarcinoma. Br J Cancer. 2019;120:165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahleda R, Italiano A, Hierro C, Mita A, Cervantes A, Chan N, et al. Multicenter Phase I Study of Erdafitinib (JNJ-42756493), Oral Pan-Fibroblast Growth Factor Receptor Inhibitor, in Patients with Advanced or Refractory Solid Tumors. Clin Cancer Res. 2019;25:4888–97. [DOI] [PubMed] [Google Scholar]
- 15.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46. [DOI] [PubMed] [Google Scholar]
- 16.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. [DOI] [PubMed] [Google Scholar]
- 17.Shaw AT, Kim D-W, Nakagawa K, Seto T, Crinó L, Ahn M-J, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94. [DOI] [PubMed] [Google Scholar]
- 18.Shaw AT, Bauer TM, de Marinis F, Felip E, Goto Y, Liu G, et al. First-Line Lorlatinib or Crizotinib in Advanced ALK-Positive Lung Cancer. N Engl J Med. 2020;383:2018–29. [DOI] [PubMed] [Google Scholar]
- 19.Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR -Mutated Advanced Non–Small-Cell Lung Cancer. N Engl J Med. 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 20.Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion–Positive Cholangiocarcinoma. Cancer Discov. 2017;7:252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal L, Shi L, Liu LY, Cruz FF de la, Lennerz JK, Raghavan S, et al. TAS-120 Overcomes Resistance to ATP-Competitive FGFR Inhibitors in Patients with FGFR2 Fusion–Positive Intrahepatic Cholangiocarcinoma. Cancer Discov. 2019;9:1064–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varghese AM, Patel J, Janjigian YY, Meng F, Selcuklu SD, Iyer G, et al. Noninvasive Detection of Polyclonal Acquired Resistance to FGFR Inhibition in Patients With Cholangiocarcinoma Harboring FGFR2 Alterations. JCO Precision Oncology. 2021;44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byron SA, Chen H, Wortmann A, Loch D, Gartside MG, Dehkhoda F, et al. The N550K/H Mutations in FGFR2 Confer Differential Resistance to PD173074, Dovitinib, and Ponatinib ATP-Competitive Inhibitors. Neoplasia. 2013;15:975–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krook MA, Bonneville R, Chen H-Z, Reeser JW, Wing MR, Martin DM, et al. Tumor heterogeneity and acquired drug resistance in FGFR2-fusion-positive cholangiocarcinoma through rapid research autopsy. Cold Spring Harb Mol Case Stud. 2019;5:a004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman IM, Hollebecque A, Friboulet L, Owens S, Newton RC, Zhen H, et al. Clinicogenomic analysis of FGFR2-rearranged cholangiocarcinoma identifies correlates of response and mechanisms of resistance to pemigatinib. Cancer Discov. 2021;11:326–39 [DOI] [PubMed] [Google Scholar]
- 26.Touat M, Ileana E, Postel-Vinay S, André F, Soria J-C. Targeting FGFR Signaling in Cancer. Clin Cancer Res. 2015;21:2684–94. [DOI] [PubMed] [Google Scholar]
- 27.Catenacci DVT, Rasco D, Lee J, Rha SY, Lee K-W, Bang YJ, et al. Phase I Escalation and Expansion Study of Bemarituzumab (FPA144) in Patients With Advanced Solid Tumors and FGFR2b-Selected Gastroesophageal Adenocarcinoma. JCO. Wolters Kluwer; 2020;38:2418–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casaletto J, Maglic D, Toure BB, Taylor A, Schoenherr H, Hudson B, et al. Abstract 1455: RLY-4008, a novel precision therapy for FGFR2-driven cancers designed to potently and selectively inhibit FGFR2 and FGFR2 resistance mutations. Cancer Res. 2021;81:1455–1455. [Google Scholar]
- 29.Goyal L, Meric-Bernstam F, Hollebecque A, Morizane C, Valle JW, Karasic TB, et al. Abstract CT010: Primary results of phase 2 FOENIX-CCA2: The irreversible FGFR1-4 inhibitor futibatinib in intrahepatic cholangiocarcinoma (iCCA) with FGFR2 fusions/rearrangements. Cancer Res. 2021;81:CT010–CT010. [Google Scholar]
- 30.Hata AN, Engelman JA, Faber AC. The BCL2 Family: Key Mediators of the Apoptotic Response to Targeted Anticancer Therapeutics. Cancer Discov. 2015;5:475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amzallag A, Ramaswamy S, Benes CH. Statistical assessment and visualization of synergies for large-scale sparse drug combination datasets. BMC Bioinformatics. 2019;20:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Kinoshita T, Sukbuntherng J, Chang BY, Elias L. Ibrutinib Inhibits ERBB Receptor Tyrosine Kinases and HER2-Amplified Breast Cancer Cell Growth. Mol Cancer Ther. 2016;15:2835–44. [DOI] [PubMed] [Google Scholar]
- 33.Horn L, Sandler A. Epidermal Growth Factor Receptor Inhibitors and Antiangiogenic Agents for the Treatment of Non-Small Cell Lung Cancer. Clin Cancer Res. 2009;15:5040–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 Profiling Identifies Three Distinct Classes of Apoptotic Blocks to Predict Response to ABT-737 and Conventional Chemotherapeutic Agents. Cancer Cell. 2007;12:171–85. [DOI] [PubMed] [Google Scholar]
- 35.Faber AC, Ebi H, Costa C, Engelman JA. Apoptosis In Targeted Therapy Responses. Advances in Pharmacology. Elsevier; 2012, Vol.65, p.519–542. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc National Acad Sci. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 38.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of Profound Feedback Inhibition of Mitogenic Signaling by RAF Inhibitors Attenuates Their Activity in BRAFV600E Melanomas. Cancer Cell. 2012;22:668–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin H, Shi Y, Lv Y, Yuan S, Ramirez CFA, Lieftink C, et al. EGFR activation limits the response of liver cancer to lenvatinib. Nature. 2021;1–5. [DOI] [PubMed] [Google Scholar]
- 41.Maity TK, Venugopalan A, Linnoila I, Cultraro CM, Giannakou A, Nemati R, et al. Loss of MIG6 Accelerates Initiation and Progression of Mutant Epidermal Growth Factor Receptor–Driven Lung Adenocarcinoma. Cancer Discov. 2015;5:534–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc National Acad Sci. 2010;107:6912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haikala HM, Janne PA. 30 years of HER3: From basic biology to therapeutic interventions. Clin Cancer Res. 2021;27:3528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu PCC, Koblish H, Wu L, Bowman K, Diamond S, DiMatteo D, et al. INCB054828 (pemigatinib), a potent and selective inhibitor of fibroblast growth factor receptors 1, 2, and 3, displays activity against genetically defined tumor models. PLoS ONE. 2020;15:e0231877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goyal L, Baiev I, Zhang K, Dalgai S, Shroff RT, Kelley RK, et al. , Acquired resistance to selective FGFR inhibitors in FGFR-altered cholangiocarcinoma. European Journal of Cancer, 2020-October, Vol.138, p.S21–S22. [Google Scholar]
- 46.Moritz A, Li Y, Guo A, Villén J, Wang Y, MacNeill J, et al. Akt-RSK-S6-kinase Signaling Networks Activated by Oncogenic Receptor Tyrosine Kinases. Sci Signal. 2010;3:ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engelman JA, Jänne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc National Acad Sci. 2005;102:3788–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer Discov. 2016;6:838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herrera-Abreu MT, Pearson A, Campbell J, Shnyder SD, Knowles MA, Ashworth A, et al. Parallel RNA interference screens identify EGFR activation as an escape mechanism in FGFR3 mutant cancer. Cancer Discov. 2013;3:1058–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR Blockade Reverts Resistance to KRAS G12C Inhibition in Colorectal Cancer. Cancer Discov. 2020;10:1129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Mikse O, Liao RG, Li Y, Tan L, Janne PA, et al. Ligand-associated ERBB2/3 activation confers acquired resistance to FGFR inhibition in FGFR3-dependent cancer cells. Oncogene. 2015;34:2167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.