Abstract
Starting from a clinical isolate of Serratia marcescens that produced a chromosomally encoded AmpC β-lactamase inducibly, we isolated by stepwise selection two laboratory mutants that showed high levels of resistance to some cephalosporins. The 98R mutant apparently overproduced the unaltered β-lactamase constitutively, but the 520R mutant produced an altered enzyme, also constitutively. Ceftazidime and cefpirome MICs for the 520R mutant were much higher (512 and 64 μg/ml, respectively) than those for the 98R mutant (16 and 16 μg/ml, respectively). Yet the MICs of cephaloridine and piperacillin for the 520R mutant were four- to eightfold lower than those for the 98R mutant. Cloning and sequencing of the ampC alleles showed that in the 520R mutant enzyme, the Thr64 residue, about two turns away from the active-site serine, was mutated to isoleucine. This resulted in a >1,000-fold increase in the catalytic efficiency (kcat/Km) of the mutated AmpC enzyme toward ceftazidime, whereas there was a >10-fold decrease in the efficiency of the mutant enzyme toward cefazolin and cephaloridine. The outer membrane permeability of the 520R strain to cephalosporins was also less than in the 98R strain, and the alteration of the kinetic properties of the AmpC enzyme together with this difference in permeability explained quantitatively the resistance levels of both mutant strains to most agents studied.
For many gram-negative bacteria, including Enterobacteriaceae and Pseudomonas spp., the production of the chromosomally encoded, class C β-lactamase, or the AmpC enzyme, represents the intrinsic mechanism of resistance to β-lactam antibiotics. AmpC expression is under the control of a regulatory gene system. Spontaneous mutations affecting the regulatory genes, most frequently ampD (17), cause constitutive overproduction of the enzyme and an increased resistance to agents, such as oxyiminocephalosporins (cefotaxime, cefuroxime, ceftriaxone, and ceftazidime) (38). Most of these compounds are hydrolyzed efficiently because of their high affinity for the AmpC enzyme, which compensates for their low deacylation rates (8, 12, 49); yet against the wild-type strains of Enterobacteriaceae, these compounds are quite effective because they are poor inducers of AmpC (18, 39). Additionally, mutations in β-lactamase structural genes may also confer a modified spectrum of drug resistance to the producing organism. Spontaneous mutations occurring in plasmid-encoded β-lactamases, resulting in the production of expanded-spectrum β-lactamases, are of great concern since they can be spread efficiently through the plasmid transfer process (25).
Until now, mutations in structural genes of chromosomal β-lactamases have been reported in only a few cases. Thus, clinical strains of Enterobacter cloacae (33) and Serratia marcescens (22) showing increased resistance to oxyiminocephalosporins, especially ceftazidime, were found to produce chromosomal AmpC enzymes with alterations in the “omega loop,” located at the entrance of the substrate-binding sites. It is of obvious interest whether mutations in other positions can also produce the chromosomal enzymes of altered specificity. In the work described here we analyzed the biochemical characteristics and the gene sequences of two allelic class C β-lactamases, produced by the S. marcescens clinical isolate S3 and its laboratory-derived 520R mutant, selected for increased resistance to ceftazidime. The mutant enzyme contained a Thr64-to-Ile change, very close to the active-site Ser-Leu-Ser-Lys sequence.
MATERIALS AND METHODS
Bacterial strains.
Strain S3 was a clinical isolate identified as S. marcescens by the use of the API system. Its chromosomal β-lactamase was inducible by typical inducers such as imipenem and cefoxitin (18), and it was susceptible to ceftazidime, cefotaxime, cefpirome, and aztreonam (Table 1). The S. marcescens 520R strain was a mutant of S3, obtained after N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis and after four successive transfers in Luria-Bertani (LB) broth containing increasing concentrations of ceftazidime. At that stage, plating on an LB broth plate containing various concentrations of ceftazidime confirmed the presence of a mutant for which the MIC was higher than 128 μg/ml, and this strain was saved as the 520R mutant. The 98R mutant was a constitutive high-level β-lactamase producer, selected by subculturing strain S3 for several steps in LB broth containing subinhibitory concentrations (0.05 to 0.1 μg/ml) of ceftazidime. When the culture was plated out on LB plates containing 8 to 16 μg of ceftazidime per ml, some colonies were observed to grow on these plates, and one of them was saved as the 98R mutant. Both mutants were resistant to most oxyiminocephalosporins and to aztreonam, but the 520R mutant was much more resistant to ceftazidime and cefpirome than was the 98R mutant (Table 1).
TABLE 1.
MICs of β-lactams for S. marcescens strains and E. coli transformants
| Strain | MIC (μg/ml)a
|
β-Lactamase activityb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFZ | CER | CTX | CAZ | CXM | CRO | CFM | FOX | CPR | PIP | ATM | MEM | Noninduced | Inducedc | |
| S. marcescens | ||||||||||||||
| S3 (wild type) | 1,024 | 32 | 2 | ≤0.25 | 512 | 1 | 2 | 8 | ≤0.5 | 32 | 1 | 0.5 | 3.7 | 151 |
| 98R mutant | >2,048 (2048) | 1,024 (1,024) | 256 (128) | 16 (0.25) | >1,024 | 256 | 256 | 256 | 16 | 512 | 64 | 0.5 | 126 | 260 |
| 520R mutant | >2,048 (128) | 256 (32) | 256 (128) | 512 (512) | >1,024 | 512 | 512 | 256 | 64 | 64 | 128 | 0.5 | 7.0 | 7.0 |
| E. coli | ||||||||||||||
| DH5α | 2 | 2 | ≤0.25 | ≤0.25 | 4 | ≤0.25 | 1 | 1 | ≤0.5 | 1 | ≤0.25 | |||
| DH5α/pGS3d | 1,024 | 128 | 4 | 1 | 256 | 1 | 8 | 4 | 1 | 32 | 2 | |||
| DH5α/pG520Rd | 64 | 8 | 8 | 16 | 256 | 8 | 128 | 16 | 2 | 8 | 4 | |||
Values in parentheses are those theoretically predicted on the basis of the kinetic parameters of the enzymes and of the permeability of the outer membrane, according to reference 30. See the text for details. CFZ, cefazolin; CER, cephaloridine; CTX, cefotaxime; CAZ, ceftazidime; CXM, cefuroxime; CRO, ceftriaxone; CFM, cefixime; FOX, cefoxitin; CPR, cefpirome; PIP, piperacillin; ATM, aztreonam; MEM, meropenem.
Vmax of cefazolin hydrolysis in nanomoles per minute per microgram of cells. It was assumed that 2 mg (dry weight) of cells contains 1 mg of protein.
Maximal activity observed with cefoxitin as the inducer.
E. coli transformant carrying S. marcescens ampC alleles on plasmid pGZ119HE. See Materials and Methods.
Escherichia coli DH5α (37) was used for transformation and propagation of recombinant plasmids. Strains were routinely grown in LB broth and on LB agar plates, unless indicated otherwise.
Susceptibility testing.
MICs were determined by the twofold serial dilution method with Mueller-Hinton broth. Inocula (104 CFU/ml) were from fresh overnight broth cultures. The MICs were read after 18 to 20 h of incubation at 37°C.
β-Lactamase preparation and purification.
The β-lactamases were purified from the S3, 98R, and 520R strains. Overnight cultures grown in LB broth were diluted 10-fold into 2 liters of the prewarmed LB broth and incubated at 37°C with shaking for 3 h. When induction was desired, 90 min before harvest the inducer (either 0.12 to 1 μg of imipenem per ml or 32 to 128 μg of cefoxitin per ml) was added. Although these inducer concentrations often exceeded MICs, the induction of β-lactamase still took place, presumably because at the time of addition of inducers the culture was entering the stationary phase and its density was so high. Cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C, washed once with 0.1 M phosphate buffer (pH 7.2) and once with 20 mM triethanolamine hydrochloride–0.5 M NaCl (pH 7.0; loading buffer), and suspended in the same buffer at 20 times their original density. Cells were disrupted by subjecting the suspension to four 1-min cycles of sonication, and the crude extract was clarified by centrifugation at 10,000 × g for 20 min and then at 135,000 × g for 30 min. The final supernatant was run through a type L aminophenylboronic acid column (2) that had been preequilibrated with loading buffer, and the column was washed with the same buffer until the A280 of washings became virtually 0. The column was then eluted with 0.5 M borate–0.5 M NaCl, pH 7. Active fractions were pooled and dialyzed overnight against 20 mM triethanolamine hydrochloride, pH 7.0, and KCl was added to 100 mM before storage at −20°C.
Isoelectric focusing.
Isoelectric focusing was conducted by the method of Matthew et al. (23) using an LKB 2117 Multiphor II apparatus with native 7% polyacrylamide gel plates containing Ampholine (Pharmacia-LKB) in the pH ranges 3.5 to 10, 8 to 10.5, and 9 to 11. Gels were stained with chromogenic cephalosporin nitrocefin. The following isoelectric-point standards (Bio-Rad) with the indicated isoelectric points were applied together with the sample: cytochrome c, 9.6; human hemoglobin C, 7.5; human hemoglobin A, 7.1; equine myoglobin, 6.8 and 7; and phycocyanin, 4.75, 4.65, and 4.45.
SDS-PAGE analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described earlier (49). Outer membrane protein samples were prepared by extracting inner membrane proteins from crude membrane fractions with N-lauryl sarcosinate (Sarkosyl) (6).
β-Lactamase assays.
Hydrolysis of β-lactam antibiotics was monitored spectrophotometrically at 25°C in 0.1 M phosphate buffer, pH 7.2, using a Uvikon 860 spectrophotometer. The reaction was monitored by tracking the changes in absorbance, usually at 260 nm. For comparison of hydrolysis rates at a fixed concentration (usually 50 μM), cells of the 10-mm light path were used.
For determination of Km values, initial rates of hydrolysis were measured at various substrate concentrations. We tried to include substrate concentrations close to or even exceeding the Km values, in order to avoid errors generated by a long extrapolation. This necessitated the use of cells with a 1-mm light path in most cases, and also the use of off-peak wavelength values (280 nm for cephaloridine, cefotaxime, and ceftazidime and 300 nm for cefazolin). It was possible to use up to 5 and 2 mM concentrations of cefazolin and cephaloridine without problems from the stray light. The Km values were derived by linear-regression analysis of Eadie-Hofstee plots of initial rates. Cefotaxime and ceftazidime, however, produced an initial burst of a decrease in optical density owing to the rapid formation of the acyl enzymes (3). For these substrates, therefore, rates after the establishment of steady states, usually the rates between 2 and 5 min after the initiation of reaction, were used. The kcat values were calculated by using the Km values and hydrolysis rates (at 50 μM) of each substrate and the observed kcat values for cefazolin. The Km for cefotaxime with the S3 enzyme was also estimated as the Ki in competition experiments using cefazolin as a reporter substrate.
Outer membrane permeability.
Coefficients of the permeability of the outer membrane to various cephalosporins were determined from the rates of drug hydrolysis by intact cells, as described previously (31).
Other biochemical methods.
Protein in crude extracts was determined by a bicinchoninic acid assay (42) using the Pierce BCA protein assay reagent. Protein concentrations in purified AmpC preparations were estimated from optical densities at 280 nm on the basis of aromatic-amino-acid content.
Nucleic acid techniques.
Recombinant DNA techniques were essentially standard procedures (37). DNA was sequenced by the dideoxy termination method (40). Plasmid DNA was isolated by using a Qiagen (Florence, Italy) Mini Kit. Restriction fragments and PCR products were recovered from agarose gels with a QIAquick gel extraction kit (Qiagen). T4 DNA ligase reactions and restriction endonuclease digestions were performed under conditions recommended by the manufacturers. Genomic DNA of S. marcescens was purified by the procedure of Marmur (20).
The complete ampC gene was obtained as follows. PCR primers S4up and S3down were chosen to amplify an internal fragment of 1,055 bp with Pfu DNA polymerase (Stratagene, Milan, Italy). Forward primer S4up (5′-CGC ACG CCG CAC AGC AGC AGG ATA-3′) corresponds to nucleotides 59 to 82, while reverse primer S3down (5′-GAT GTG GTA AGC CGC TTC GAC GCG-3′) corresponds to nucleotides 1113 to 1090. (In this as well as subsequent descriptions of primers, the first nucleotide of the coding region of the ampC gene in S. marcescens SR50 [32] was numbered 1). The amplimer was labeled with alkaline phosphatase using the AlkPhos direct labeling system (Amersham, Milan, Italy) and used as a probe in Southern blotting performed on genomic S. marcescens DNA digested with several restriction enzymes. A 3.6-kb fragment from the EcoRV digestion hybridized with the probe, and this fragment was purified and subsequently cleaved with PstI, which recognized a single restriction site inside the fragment. The cleaved, smaller DNA fragments were purified and ligated with T4 DNA ligase. The DNA concentration in the ligation mixture was lowered to 0.5 μg/ml in order to improve the circularization of the DNA fragments. The mixtures of these circularized fragments were used as templates in inverse PCR amplifications with the primer pairs S1-S6R and S3-S5R.
The forward primer S1 (5′-CAG ACG CTG TTT GAA GTG GGC TCG-3′), located between nucleotides 217 and 240, and the reverse primer S6R (5′-CTC GGT GAT CGG TTT GCC GGT GTG-3′), located between nucleotides 216 and 193, amplified a 1-kb fragment that comprised the proximal, or 5′, portion of the bla gene and a short upstream flanking sequence. The forward primer S3R, which was the complement of S3down, and the reverse primer S5R (5′-TTC TTC GCC GGG ATA AAT ACC-3′), located between nucleotides 1043 and 1023, amplified a fragment of 1.2 kb that comprised the 3′ portion of the bla gene and the flanking region downstream.
The sequence of the entire gene was deduced from the sequences of the amplimers. On the basis of this information, two new primers, S13A (5′-CCC TTC TAG ATA AGA GCT TCT ATC ATG ACG-3′) and S14A (5′-CCT CGT CGG AAG CTT TGG CCG TCA GCG CTT-3′), were designed such that, during the amplification, two short sequences containing the XbaI and HindIII restriction sites were added to the ends of a fragment containing the complete gene plus its Shine-Dalgarno sequence. The two ampC alleles amplified from S3 and 520R genomic DNAs were treated with XbaI and HindIII and then ligated into the polylinker region of the pGZ119EH vector plasmid (16), thus producing plasmids pGS3 and the pG520R, respectively, which were used to transform E. coli DH5α. Transformants were selected on plates containing chloramphenicol (30 μg/ml), ampicillin (25 μg/ml), and the inducer IPTG (isopropyl-β-d-thiogalactopyranoside; 0.2 μg/ml).
The nucleotide sequence was obtained by double-strand sequencing of the amplimers of the whole genes generated from the two strains. DNA sequence analysis was performed also on the DNAs of the pGS3 and pG520R plasmids extracted from transformed E. coli.
Nucleotide sequence accession numbers.
The sequences of the ampC alleles from the S3 and 520R strains have been deposited in GenBank (accession no. AF327324 and AF327325, respectively).
RESULTS
β-Lactamase expression in S. marcescens strains.
The S. marcescens strains examined in this study exhibited three different phenotypes of β-lactamase expression and β-lactam susceptibility (Table 1). The enzyme in strain S3 was inducible at a low basal level, which was increased up to 40 times by induction with cefoxitin (128 μg/ml) (see Materials and Methods). MICs of ceftazidime, cefotaxime, cefpirome, ceftriaxone, cefixime, and aztreonam were low in this parent strain.
Both the 98R and 520R strains were mutants derived from S3 that showed increased resistance to some compounds. Strain 98R exhibited a constitutive, high-level β-lactamase activity, although in the presence of inducers there was a further, small increase in activity. As found in the classical AmpC-constitutive mutants (38, 49), the 98R mutant was highly resistant to oxyiminocephalosporins such as cefuroxime, cefixime, cefotaxime, and ceftriaxone, as well as cefoxitin (a cephamycin) and aztreonam (a monobactam). Its resistance to ceftazidime and cefpirome, however, remained at a moderate level (MIC, 16 μg/ml).
The 520R strain also produced the β-lactamase constitutively. It showed high-level resistance to most of the compounds to which the 98R mutant showed resistance and in addition showed much increased levels of resistance to ceftazidime and cefpirome; MICs were 512 and 64 μg/ml, respectively (Table 1). Interestingly, levels of resistance to cephaloridine and piperacillin were greatly decreased in the 520R strain, in comparison with those of the 98R strain (Table 1).
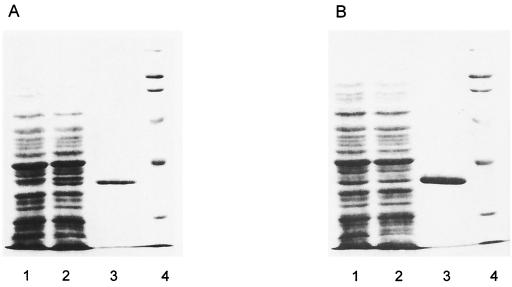
In spite of the increased resistance to several compounds shown by the 520R mutant, its β-lactamase activity, expressed as the Vmax of cefazolin hydrolysis, was only 5% of that in the maximally induced S3 cells (Table 1). When the amounts of β-lactamase proteins produced were estimated from the SDS-PAGE profile of the total cell proteins, the 520R strain was found to have far more than 5% of the level of the AmpC protein in the induced wild-type parent S3 (Fig. 1). This suggested that the specific activity, for cefazolin, of the AmpC enzyme of the 520R strain was decreased in comparison with that of the wild-type enzyme, a conclusion supported by the study of purified enzymes described below.
FIG. 1.
PAGE profile of total cell proteins and purified β-lactamases from S. marcescens strain S3 (A) and the 520R mutant (B). Lane 1, uninduced cells; lane 2, induced cells; lane 3, purified β-lactamase; lane 4, molecular weight markers.
The MICs of agents other than β-lactams, such as tetracycline (4 μg/ml), chloramphenicol (8 μg/ml), and norfloxacin (8 μg/ml) were unaltered for both mutants. This result suggests that the overproduction of a multidrug efflux pump(s) is unlikely in the mutants, as these agents are typical substrates of such pumps (27).
Characterization of β-lactamases.
Enzymes were purified from the S3, 98R, and 520R strains by aminoboronic acid affinity chromatography (2) to near homogeneity (>99%) (Fig. 1). A single protein band was observed in SDS-PAGE for all β-lactamase preparations, and the molecular mass was estimated to be approximately 39 kDa.
An isoelectric-focusing analysis was performed both on crude extracts of the three strains and on the purified β-lactamase preparations from the S3 and 520R strains. All enzymes showed an isoelectric point of 8.75 ± 0.1 (mean ± standard deviation), as expected for chromosomal, AmpC-type enzymes; no additional band with β-lactamase activity was observed (data not shown).
Km values were determined from the initial rates of hydrolysis. The kcat values were calculated from the hydrolysis rates of various substrates at 50 μM and the values of Km. These kinetic parameters, determined for four compounds, are listed in Table 2. With the 520R mutant enzyme, the Km for ceftazidime was too high for precise measurement. In this case, however, v/[S] (where v is hydrolysis rate and [S] is the substrate concentration) could be assumed to be essentially equal to Vmax/Km, because at low substrate concentrations, [S] became negligible in relation to Km in the equation v/[S] = Vmax/(Km + [S]).
TABLE 2.
Kinetic parameters of AmpC β-lactamases from S. marcescens strains
| Source of enzyme | Cefazolin
|
Cephaloridine
|
Cefotaxime
|
Ceftazidime
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) | |
| S3 (wild type) | 890 ± 90 | 3,000 | 3.37 | 700 ± 50 | 1,790 | 2.56 | 8 ± 2 | 2.3 | 0.29 | NDa | ND | ND |
| 98R mutant | 980 ± 140 | 3,060 | 3.12 | 640 ± 30 | 1,680 | 2.63 | 12 ± 2 | 2.2 | 0.18 | 1,350 ± 390 | 0.008 | 6 × 10−6 |
| 520R mutant | 4,600 ± 200 | 1,070 | 0.23 | 3,500 ± 180 | 400 | 0.12 | 1,500 ± 300 | 250 | 0.17 | >5,000 | NDb | 0.012 |
ND, not determined.
Could not be calculated because the Km was too high.
Examination of kinetic parameters of S3 and 98R enzymes showed that there was no significant difference between these enzymes in terms of Km values for several substrates and the relative rates of hydrolysis of these substrates (Table 2). We assume, therefore, that the 98R mutant is a simple AmpC overproduction strain, as seen in the common ampD mutants of various species of Enterobacteriaceae (17, 38, 49).
In contrast, the 520R enzyme indeed appeared to be different from the wild-type enzyme in the preliminary assay with crude extracts, and the kinetic parameters measured using the purified preparations confirmed this conclusion, as detailed below.
As seen with many AmpC β-lactamases from Enterobacteriaceae (see Discussion), the wild-type enzyme (from strains S3 and 98R) was relatively active against older cephalosporins such as cefazolin and cephaloridine in spite of the rather high Km values. In contrast, the Km was much lower for cefotaxime, an oxyiminocephalosporin (Table 2). Ceftazidime was different from other oxyiminocephalosporins in having a higher Km, again as seen with other AmpC enzymes.
The mutant β-lactamase from the 520R mutant exhibited a strongly increased (>1,000-fold) catalytic efficiency (kcat/Km) toward ceftazidime, in comparison to that of the wild-type enzyme. An increase in kcat must have been responsible for this increase in catalytic efficiency, as it occurred in spite of an increase in Km. In contrast, catalytic efficiencies for the older cephalosporins cephaloridine and cefazolin decreased significantly in the mutant enzyme. The kcat value increased about 100-fold for cefotaxime in the mutant enzyme, but this was nearly completely compensated for by a strong increase in Km, and the catalytic efficiency remained about the same in the 520R mutant enzyme.
The catalytic parameters were strongly influenced by the ionic strength of the assay buffer. Thus, the Km value for cefazolin, with the wild-type enzyme, was as low as 200 μM in 20 mM triethanolamine-HCl buffer, pH 7.0, but increased to 980 μM in 0.1 M phosphate buffer, pH 7.4. This result is similar to what has been reported for an S. marcescens AmpC enzyme earlier (12) but appears somewhat more extreme. All kinetic constants reported in Table 2 were those determined with 0.1 M phosphate buffer, pH 7.4.
Some preliminary analysis of kinetic parameters was also carried out with other compounds (cefpirome, cefuroxime, ceftriaxone, and cefixime) (data not shown). Although these assays were carried out under low-ionic-strength conditions (20 mM triethanolamine buffer) and therefore the results are not comparable with those of Table 2, the data showed significant increases in kcat values in the 520R mutant enzyme for other oxyiminocephalosporins (cefuroxime, cefixime, and ceftriaxone) as well as an oxyiminocephalosporin with a quaternary-nitrogen-containing 3-substituent (cefpirome) (not shown).
Cloning of the ampC gene and phenotype of transformants.
On the basis of the published sequence of the S. marcescens SR50 β-lactamase gene (32), oligonucleotide primers were designed and a 1,055-bp fragment was amplified from the genomic DNA of strain S3, as described in Materials and Methods. Its sequence corresponded to an internal fragment of an ampC homolog.
We then identified a 3.6-kb fragment containing this sequence in the EcoRV-digested S3 genomic DNA and finally isolated the complete ampC genes from both S3 (wild type) and the 520R mutant, cloned in the IPTG-inducible expression vector pGZ119EH (16) (see Materials and Methods). The resulting plasmids, named pGS3 and pG520R, containing, respectively, the wild-type and mutant alleles of ampC (see below), were used to transform E. coli DH5α cells.
The introduction of the cloned S. marcescens β-lactamase genes caused decreases in susceptibilities to all β-lactams tested in the transformed E. coli cells (Table 1). Assay of β-lactamase activity showed that similar levels of the enzyme proteins were expressed by the two plasmids (not shown), based on the kinetic parameters described above (Table 2). Nevertheless, the expression of the wild-type enzyme produced strong increases in resistance to cefotaxime, cefixime, cefoxitin, piperacillin, and especially cefazolin, cephaloridine, and cefuroxime but had only a modest effect on resistance to ceftazidime, cefpirome, ceftriaxone, and aztreonam (Table 1). The production of mutant β-lactamase, on the other hand, caused a much stronger increases in resistance to ceftriaxone, cefixime, and, above all, ceftazidime, but it produced decreased levels of resistance, in comparison with that of the strain producing the wild-type enzyme, to cefazolin, cephaloridine, and piperacillin. These results confirm that a major part of the resistance phenotype of the S. marcescens mutants was indeed due to the catalytic properties of the AmpC enzymes produced.
Nucleotide sequence analysis of the β-lactamase genes.
The complete sequences of the ampC alleles were obtained from the PCR amplicons made by using genomic DNAs of S3 and the 520R mutant, as well as by using plasmids pGS3 and pG520R (each extracted from two independent clones of transformants). Analysis of the nucleotide sequence and its translation products showed a translation start codon, ATG, located 7 bases downstream from a likely ribosome-binding site sequence, AAGAG. The determined sequence, which includes only 19 bases upstream of this putative ribosome-binding site sequence, apparently does not include the promoter region. A typical recognition sequence for the signal peptidase (A-X-A) was found between positions 22 and 23 of the translated protein. The open reading frame was 1,137 bp long and coded for a 378-residue polypeptide. The mature protein, apparently 356 residues long, was calculated to have a molecular mass of 38,971 Da (38,983 Da for the 520R variant), which was compatible with the mobility of the protein in SDS-PAGE.
The deduced amino acid sequence of S3 β-lactamase and that of its mutant showed a very high degree of similarity with known AmpC β-lactamases of S. marcescens. It was closest to that of SMA271368 (G. Barnaud, G. J. Arlet, R. Labia, and A. Philippon, GenBank/EMBL accession no. AJ 271368), with only six substitutions, all of which were conservative (Ile for Val, Val for Ile, Glu for Asp, Gln for Glu, Asp for Asn, and His for Arg) (Fig. 2). It was also similar to the other S. marcescens AmpC sequences such as SRT-1 (22), SST-1 (22), and SR50 (32) (98, 96, and 94% identity, respectively) (not shown). The S3 amino acid sequence was also similar to AmpC sequences from other members of Enterobacteriaceae, although more differences (including short deletions and insertions) were seen here (Fig. 2).
FIG. 2.
Multiple alignment of AmpC proteins from Enterobacteriaceae. The active-site Ser residue is double underlined, and the Thr residue that is changed to Ile in the 520R enzyme is underlined. Sources for the sequences are E. coli (11), E. cloacae P99 (9), C. freundii GN346 (41), S. marcescens AJ (SMA271368) (G. Barnaud, G. J. Arlet, R. Labia, and A. Philippon, GenBank/EMBL accession no. AJ 271368), and S. marcescens S3 (this study). The alignment was generated by the CLUSTAL W program (43).
A comparison of the nucleotide sequences revealed that the S3 and 520R mutant ampC alleles were identical at all positions except for a point mutation, a transition from C to T at position 257. This change leads to the replacement of Thr at position 64 of the mature protein with Ile in the 520R β-lactamase (Fig. 2).
Outer membrane permeability.
Having shown that the 520R mutant produced an altered enzyme and exhibited a resistance pattern that was significantly different from that of the 98R mutant, a simple overproducer of the unaltered enzyme, we wanted to see if the properties of the enzymes could explain quantitatively the levels of resistance to various agents, as has been done by the use of E. coli earlier (30). For this analysis, we needed the coefficients of permeability of the outer membrane. Permeability coefficients for cefazolin, cephaloridine, cefotaxime, and ceftazidime were measured from the rates of hydrolysis of these compounds by intact cells (Materials and Methods). Although the permeability to ceftazidime was too low to measure accurately, the 520R mutant was less (between four and six times) permeable to other compounds than the 98R mutant (Table 3).
TABLE 3.
Permeability coefficients of the outer membranes
| Strain | Permeability coefficient (cm s−1)
|
||
|---|---|---|---|
| Cefazolin | Cephaloridine | Cefotaxime | |
| 98R mutant | 3.7 × 10−5 | 5.7 × 10−5 | 4.4 × 10−7 |
| 520R mutant | 0.7 × 10−5 | 1.3 × 10−5 | 7.5 × 10−8 |
In principle, it is possible that the measured permeability coefficients were distorted by an active efflux process. However, multidrug efflux systems such as AcrAB-TolC of enteric bacteria show very little activity toward the compounds tested, with rather hydrophilic side chains or multiple charged groups (28). Furthermore, under the conditions of our assay, efflux is likely to be overwhelmed by the massive influx of drugs present at a 1 mM concentration. We therefore believe that our experiments measured the permeability of the outer membrane rather accurately.
SDS-PAGE analysis of outer membrane proteins.
Because the permeability of the outer membrane of the 520R mutant to the cephalosporins tested was lower than that of the 98R mutant, we suspected that the former strain may produce fewer porins. We thus examined by SDS-PAGE the pattern of outer membrane proteins of samples treated at 100°C for 10 min in the sample buffer. The major protein bands were found at positions corresponding to 37 and 33 kDa, presumably corresponding to porins and OmpA, respectively. The porin band, however, contained more than one component, and the relative levels of abundance of these components differed between the 98R and 520R mutants (data not shown). A more precise analysis, however, was difficult because these putative porin bands migrated very close to each other on PAGE. Each of these porin species may have different channel properties, and a simple interpretation of these results in terms of permeability was not possible.
DISCUSSION
Properties of the wild-type enzyme and their consequences.
The wild-type enzyme had relatively high kcat values for the older cephalosporins such as cefazolin and cephaloridine, similar to the findings of an earlier report (8). Although many class C enzymes (such as the E. cloacae and Citrobacter freundii enzymes) have very high affinities to monoanionic oxyiminocephalosporins such as cefotaxime, as exemplified by Km values as low as 0.005 μM (8), the S. marcescens enzyme is exceptional in showing Km values in the range of 10 to 30 μM for these compounds (8, 35), and our present results were in agreement with these findings (Table 2). As a result, kcat/Km values for cefotaxime are high (in the range of 1 to 4 μM−1 s−1) for the E. cloacae enzyme (8, 35) and much lower (0.14 and 0.19 μM−1 s−1 [8; this study]) for the S. marcescens enzymes.
Ceftazidime, exceptionally among the oxyiminocephalosporins introduced in the 1980s, has a relatively low affinity even to the E. cloacae and E. coli enzymes (with Km values of around 4.0 μM [21, 49] and 16 μM [30] for the two enzymes, respectively). This is probably due at least in part to the presence of the pyridinium side chain at the 3′ position, a feature similar to those uniformly present in the oxyiminocephalosporins developed to exhibit lower affinity toward AmpC enzymes, such as cefpirome and cefepime (29). Ceftazidime also had a very low affinity to the S. marcescens S3 enzyme, which had a Km of 1,350 μM (Table 2). The value of kcat/Km for ceftazidime was therefore orders of magnitude lower in the S. marcescens S3 enzyme (6 × 10−6 s−1 μM−1) (Table 2) than in the E. cloacae P99 enzyme (2.5 × 10−3 s−1 μM−1) (21).
These lower kcat/Km values of the S. marcescens enzyme for oxyiminocephalosporins in general and for ceftazidime in particular lead to the prediction that a simple overproduction of this enzyme may not produce a high level of resistance to these compounds in S. marcescens (in contrast to the situation in other Enterobacteriaceae), and indeed this was borne out by experimental data. For example, Hechler et al. (10) showed that an overproduction alone of the class C enzyme in S. marcescens increased the MICs of cefotaxime and ceftazidime to only 32 and 1 μg/ml, respectively; in contrast, in E. cloacae such an overproduction increased these MICs to 512 and 256 μg/ml, respectively, in one study (49). In another study (5), the cefotaxime and ceftazidime MICs for a laboratory-selected β-lactamase derepression mutant of S. marcescens were only 4 and 0.5 μg/ml, respectively, in contrast to MICs of 32 and 64 μg/ml, respectively, for a similar mutant of E. cloacae. As seen in these examples, this difference between S. marcescens and E. cloacae (and other Enterobacteriaceae) is predictably even more pronounced with ceftazidime; in another recent study, ceftazidime MICs were only 2 μg/ml for S. marcescens strains overproducing their AmpC enzyme, whereas the MICs were ≥16 μg/ml for similar overproducing strains of E. cloacae or C. freundii (44). In the present study, the simple AmpC 98R overproduction strain showed higher levels of resistance to most oxyiminocephalosporins than those of strains described earlier, yet the MIC of ceftazidime was only 16 μg/ml, in contrast to the cefotaxime MIC of 256 μg/ml (Table 1).
Properties of the mutant enzyme and their consequences.
The mutant enzyme had greatly increased kcat values for extended-spectrum cephalosporins, accompanied also by increased Km values (Table 2). This is somewhat reminiscent of the alteration of the chromosomal enzyme observed in a clinical strain of S. marcescens showing modest increases in resistance to cefotaxime, ceftriaxone, and aztreonam (1), although the nature of the mutational alteration of the enzyme sequence is not known in that example.
We can quantitatively analyze the effects of the alterations in the kcat and Km of our enzyme on MICs by using the approach of Nikaido and Normark (30). Although this approach neglects the effect of active efflux, this effect is usually not very significant when enzymatic hydrolysis occurs at high rates (24). The theory indicates that the MIC can be calculated as Cinh{1 + Vmax/[P × A × (Km + Cinh)]} (30), where Cinh, P, and A are the antibiotic concentration that inhibits the target, permeability coefficient of the outer membrane, and area of the outer membrane in unit weight of cells, respectively, and where Vmax and Km are the usual kinetic constants of the β-lactamase. We have determined the coefficients of permeability of the S. marcescens outer membrane to cefazolin, cephaloridine, and cefotaxime and found that the 520R mutant has four- to sixfold less permeability than that of the 98R strain (Table 3). Also, in a given strain, the coefficient of permeability to cefotaxime, containing the potentially diffusion-hindering oxyimino substituent, was nearly 2 orders of magnitude lower than those determined for cefazolin whereas the permeability coefficients for these two compounds differed only by a factor of 3 in E. coli (30). These data suggest that the porin channels of at least these strains of S. marcescens are more restrictive than the OmpF channel of E. coli and that differences in drug structure affect more strongly the diffusion rate through the S. marcescens porins than they do the diffusion rate through the E. coli porin. For ceftazidime, experimental determination of the permeability coefficients was not possible, and therefore we assumed tentatively that the coefficient of permeability to this compound of each strain was 50 times lower than that to cefotaxime, which does not contain the potentially diffusion-hindering second negative charge on the side chain.
Calculation using these values for the permeability coefficients showed (values in parentheses in Table 1) that the MICs predicted from the parameters of the AmpC enzyme were very close to the observed MICs in most cases. Importantly, the theory predicts that the MICs of cefotaxime will be essentially identical for the 98R and 520R mutants (as observed). The theory also predicts correctly that the ceftazidime MIC will increase greatly from a low value for the 98R mutant to about 512 μg/ml for the 520R mutant, largely due to the increase in the catalytic efficiency of the mutant enzyme for this substrate. The prediction also worked quite well for cephaloridine. However, the prediction gave values quite different from the experimentally observed values of the cefazolin MIC for the 520R mutant and of the ceftazidime MIC for the 98R mutant. In the latter case, the discrepancy may have arisen because of the active efflux of the compound, which was totally neglected here. Interestingly, the cefazolin and cephaloridine resistance levels of E. coli DH5α expressing the S3 and 520R enzymes were altered in the predicted manner, with a strong decrease in the MIC for the strain expressing the mutant enzyme (Table 1). One of the reasons why the S. marcescens strains were more resistant than the E. coli strains to cefotaxime and ceftazidime is likely to be the more restrictive porin channels found in S. marcescens, which would severely limit the permeation of these compounds with bulky side chains (see above).
Possible structural effect of the Thr64-to-Ile change.
The crystal structure of S. marcescens β-lactamase is not available, but the possible result of the Thr64-to-Ile change can be examined by referring to the structures of homologous class C enzymes from E. cloacae (4, 19) and E. coli (48), as well as the d-Ala-d-Ala carboxypeptidase from Streptomyces strain R61 (14). The three-dimensional structures of all these enzymes are very similar (13), and we will use the E. cloacae P99 enzyme (19) as an example. The Thr70 residue of this enzyme (which corresponds to the mutated Thr64 residue of the S. marcescens S3 enzyme and the Thr68 enzyme in the E. coli enzyme [Fig. 2]) occurs about two turns further away from the active-site Ser64 residue (Ser58 in S. marcescens) at the beginning of helix 2. Although it is not in the substrate-binding cleft, the side chain hydroxyl oxygen of Thr70 is within a hydrogen-bonding distance of the main chain carbonyl oxygen of Gln219 (Glu213 in the S3 enzyme and Glu216 in E. coli AmpC). This distance is always very short (between 2.55 and 2.74 Å) in all the AmpC enzymes analyzed (4, 19, 48). The replacement of threonine with isoleucine abolishes this hydrogen-bonded structure and also introduces a more bulky side chain. We can thus envisage two possible consequences in the mutated S. marcescens enzyme. (i) The substitution will make helix 2, which contains the active-site serine (and also the threonine now replaced with isoleucine), more movable or pliable, and this will make the attack on compounds like ceftazidime easier. (ii) Alternatively, or additionally, the mutation may shift the positions of Glu213 (corresponding to Gln219 of the E. cloacae enzyme) and its neighbors. Gln219 of the E. cloacae enzyme is located near the end of the long omega loop (from residue 189 to residue 226) at the entrance of the substrate-binding site. This loop is much longer in the class C enzymes than in the class A enzymes, and this added length of the loop is thought to facilitate the accommodation of larger cephalosporins by class C enzymes. It is thus possible that the T64I mutation in the 520R mutant further opens up the entrance of the pocket by moving the omega loop and that it makes the accommodation of ceftazidime easier. Interestingly, when the folding of S3 and 520R mutant AmpC was predicted by the Swiss-Model program (36) using the structure of E. cloacae AmpC (19) as the template, one of the largest differences was predicted to occur in the side chain of Glu213, with some of its atoms being predicted to move as much as by 5 Å in the 520R enzyme in comparison with the positions in the S3 enzyme.
We note also that, in the C. freundii GN346 enzyme, the change of Glu219 (corresponding to Gln219 of the E. cloacae enzyme) to Lys and other bulky residues (46, 47) and the change of a nearby residue, Asp217, to Lys, Thr, or Glu (45) increase the kcat values for oxyiminocephalosporins, somewhat reminiscent of our observation here. In another study, the AmpC enzyme in an oxyiminocephalosporin-resistant clinical isolate of S. marcescens was found to contain a Glu-to-Lys change in residue 213, closer to the center of the omega loop (22). Finally, extension of the omega loop in the middle of both natural and laboratory-generated E. cloacae mutant enzymes was shown to result in an increase in kcat values to oxyiminocephalosporins, including ceftazidime (33, 34). In this case, the crystal structure of the mutant enzyme indeed showed the increased opening of the entrance of the substrate-binding pocket (4).
It should be noted that a BLAST search of the GenBank database revealed that Thr70 of the E. cloacae enzyme is conserved in most species, including S. marcescens, E. cloacae, Enterobacter aerogenes, Acinetobacter baumanii, Aeromonas species, Pseudomonas aeruginosa, E. coli, and Lysobacter lactamgenus, although it is replaced by Asn in C. freundii and Proteus mirabilis AmpC enzymes and by Ala in Hafnia alvei AmpC. The replacement of Thr70 with Asn will produce a similar hydrogen-bonding network, and the replacement with Ala will at least avoid the creation of steric hindrance. Replacement of Thr70 (Thr64 in S. marcescens S3) with an amino acid with a bulky, lipophilic side chain, to our knowledge, has never been observed.
Conclusions.
Among oxyiminocephalosporins introduced in the 1980s, ceftazidime tends to be exceptionally active against Enterobacteriaceae mutants overproducing the chromosomally encoded AmpC β-lactamases, presumably because it has relatively low affinity to these enzymes. This situation was even more pronounced with S. marcescens, whose AmpC enzyme, in comparison with homologous enzymes from other species, has lower affinity to oxyiminocephalosporins in general and to ceftazidime in particular. We have shown in this study that, even with S. marcescens, it is possible to isolate a mutant producing an altered AmpC enzyme, which makes the strain highly resistant to ceftazidime (and also to cefpirome). These observations suggest that increased use of extended-spectrum cephalosporins may eventually select for similar mutant AmpC enzymes. Indeed a similar in vitro selection applied to an E. cloacae strain with cefpirome and cefepime resulted in a mutant producing a mutated AmpC enzyme, which also showed high-level resistance to ceftazidime (although the mutation was located at position 318, far away from the site of mutation observed in this study) (26). This possibility of mutations in the AmpC enzyme is a concern, especially because strains containing plasmid-borne ampC genes are now known (for example, see references 7 and 15), and there is a possibility that the situation may become similar to what has led to the generation of many extended-spectrum β-lactamase mutants of TEM and SHV class A enzymes (for a review, see reference 25).
ACKNOWLEDGMENTS
We thank Rosalia Ticozzi for technical assistance, Emiko Y. Rosenberg for carrying out the analysis of outer membrane proteins, and J. R. Knox for supplying a model of the E. cloacae enzyme bound to ceftazidime as well as for helpful comments on the manuscript.
Work in Berkeley was supported by a grant from the U.S. Public Health Service (AI-09644).
REFERENCES
- 1.Bush K, Flamm R K, Ohringer S, Singer S B, Summerill R, Bonner D P. Effect of clavulanic acid on activity of β-lactam antibiotics in Serratia marcescens isolates producing both a TEM β-lactamase and a chromosomal cephalosporinase. Antimicrob Agents Chemother. 1991;35:2203–2208. doi: 10.1128/aac.35.11.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartwright S J, Waley S G. Purification of β-lactamases by affinity chromatography on phenylboronic acid-agarose. Biochem J. 1984;221:505–512. doi: 10.1042/bj2210505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charnas R L, Then R L. Mechanism of inhibition of chromosomal β-lactamases by third-generation cephalosporins. Rev Infect Dis. 1988;10:752–760. doi: 10.1093/clinids/10.4.752. [DOI] [PubMed] [Google Scholar]
- 4.Crichlow G V, Kusin A P, Nukaga M, Mayama K, Sawai T, Knox J R. Structure of the extended-spectrum class C β-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry. 1999;38:10256–10261. doi: 10.1021/bi9908787. [DOI] [PubMed] [Google Scholar]
- 5.Curtis N A C, Eisenstadt R L, Rudd C, White A J. Inducible type I β-lactamases of Gram-negative bacteria and resistance to β-lactam antibiotics. J Antimicrob Chemother. 1986;17:51–61. doi: 10.1093/jac/17.1.51. [DOI] [PubMed] [Google Scholar]
- 6.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaillot O, Clément C, Simonet M, Philippon A. Novel transferable β-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J Antimicrob Chemother. 1997;39:85–87. doi: 10.1093/jac/39.1.85. [DOI] [PubMed] [Google Scholar]
- 8.Galleni M, Amicosante G, Frère J-M. A survey of the kinetic parameters of class C β-lactamases. Cephalosporins and other β-lactam compounds. Biochem J. 1988;255:123–129. doi: 10.1042/bj2550123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galleni M, Lindberg F, Normark S, Cole S, Honore N, Joris B, Frere J-M. Sequence and comparative analysis of three Enterobacter cloacae ampC beta-lactamase genes and their products. Biochem J. 1988;250:753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hechler U, van den Weghe M, Martin H H, Frère J-M. Overproduced β-lactamase and the outer-membrane barrier as resistance factors in Serratia marcescens highly resistant to β-lactamase-stable β-lactam antibiotics. J Gen Microbiol. 1989;135:1275–1290. doi: 10.1099/00221287-135-5-1275. [DOI] [PubMed] [Google Scholar]
- 11.Jaurin B, Grundstrom T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joris B, De Meester F, Galleni M, Masson S, Dusart J, Frère J-M, van Beeumen J, Bush K, Sykes R. Properties of a class C β-lactamase from Serratia marcescens. Biochem J. 1986;239:581–586. doi: 10.1042/bj2390581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knox J R, Moews P C, Frère J-M. Molecular evolution of bacterial β-lactam resistance. Chem Biol. 1996;3:937–947. doi: 10.1016/s1074-5521(96)90182-9. [DOI] [PubMed] [Google Scholar]
- 14.Kuzin A P, Liu H, Kelly J A, Knox J R. Binding of cephalothin and cefotaxime to d-Ala-d-Ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended spectrum β-lactamases. Biochemistry. 1995;34:9532–9540. doi: 10.1021/bi00029a030. [DOI] [PubMed] [Google Scholar]
- 15.Leiza M G, Perez-Diaz J C, Ayala J, Casellas J M, Martinez-Beltran J, Bush K, Baquero F. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new ampC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob Agents Chemother. 1994;38:2150–2157. doi: 10.1128/aac.38.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lessl M, Balzer D, Lurz R, Vaters V L, Guiney D G, Lanka E. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra 2 by mobilization of the Tra 1 region in trans. J Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindquist S, Galleni M, Lindberg F, Normark S. Signalling proteins in enterobacterial AmpC β-lactamase regulation. Mol Microbiol. 1989;3:1091–1102. doi: 10.1111/j.1365-2958.1989.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 18.Livermore D M, Yang Y J. β-Lactamase lability and inducing power of newer β-lactam antibiotics in relation to their activity against β-lactamase-inducibility mutants of Pseudomonas aeruginosa. J Infect Dis. 1987;155:755–782. doi: 10.1093/infdis/155.4.775. [DOI] [PubMed] [Google Scholar]
- 19.Lobkovsky E, Moews P C, Liu H, Zhao H, Frère J-M, Knox J R. Evolution of an enzyme activity crystallographic structure at 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc Natl Acad Sci USA. 1993;90:11257–11261. doi: 10.1073/pnas.90.23.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 21.Matagne A, Misselyn-Bauduin A-M, Joris B, Erpicum T, Granier B, Frère J-M. The diversity of the catalytic properties of class A β-lactamases. Biochem J. 1990;265:131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumura N, Minami S, Mitsuhashi S. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob Agents Chemother. 1998;42:176–179. doi: 10.1128/aac.42.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthew M, Harris A M, Marshall M. The use of the analytical isoelectric focusing for the detection and the identification of the β-lactamases. J Gen Microbiol. 1975;88:169–175. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 24.Mazzariol A, Cornaglia G, Nikaido H. Contribution of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coli K-12 to β-lactams. Antimicrob Agents Chemother. 2000;44:1387–1390. doi: 10.1128/aac.44.5.1387-1390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medeiros A A. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl. 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 26.Morosini M-I, Negri M-C, Shoichet B, Baquero M-R, Baquero F, Blázquez J. An extended-spectrum AmpC-type β-lactamase obtained by in vitro antibiotic selection. FEMS Microbiol Lett. 1998;165:85–90. doi: 10.1111/j.1574-6968.1998.tb13131.x. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido H, Basina M, Nguyen V, Rosenberg E Y. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J Bacteriol. 1998;180:4686–4692. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikaido H, Liu W, Rosenberg E Y. Outer membrane permeability and β-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob Agents Chemother. 1990;34:337–342. doi: 10.1128/aac.34.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido H, Normark S. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol Microbiol. 1987;1:29–36. doi: 10.1111/j.1365-2958.1987.tb00523.x. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido H, Rosenberg E Y, Foulds J. Porin channels in Escherichia coli: studies with β-lactams in intact cells. J Bacteriol. 1983;153:232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomura K, Yoshida T. Nucleotide sequence of the Serratia marcescens SR50 chromosomal ampC β-lactamase gene. FEMS Microbiol Lett. 1990;70:295–300. doi: 10.1111/j.1574-6968.1990.tb13992.x. [DOI] [PubMed] [Google Scholar]
- 33.Nukaga M, Haruta S, Tanimoto K, Kogure K, Taniguchi K, Tamaki M, Sawai T. Molecular evolution of a class C β-lactamase extending its substrate specificity. J Biol Chem. 1995;270:5729–5735. doi: 10.1074/jbc.270.11.5729. [DOI] [PubMed] [Google Scholar]
- 34.Nukaga M, Taniguchi K, Washio Y, Sawai T. Effect of an amino acid insertion into the omega loop region of a class C β-lactamase on its substrate specificity. Biochemistry. 1998;37:10461–10468. doi: 10.1021/bi980184i. [DOI] [PubMed] [Google Scholar]
- 35.Okonogi K, Sugiura A, Kuno M, Higashide E, Kondo M, Imada A. Effect of β-lactamase induction on susceptibility to cephalosporins in Enterobacter cloacae and Serratia marcescens. J Antimicrob Chemother. 1985;16:31–42. doi: 10.1093/jac/16.1.31. [DOI] [PubMed] [Google Scholar]
- 36.Peitsch M C. ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanders C C. Novel resistance selected by the new expanded-spectrum cephalosporins: a concern. J Infect Dis. 1983;147:585–589. doi: 10.1093/infdis/147.3.585. [DOI] [PubMed] [Google Scholar]
- 39.Sanders C C, Sanders W E. Type I β-lactamase of gram-negative bacteria: interactions with β-lactam antibiotics. J Infect Dis. 1986;154:792–800. doi: 10.1093/infdis/154.5.792. [DOI] [PubMed] [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:2494–2498. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawai T, Yamaguchi A, Tsukamoto K. Amino acid sequence, active-site residue, and effect of suicide inhibitors on cephalosporinase of Citrobacter freundii GN346. Rev Infect Dis. 1988;10:721–725. doi: 10.1093/clinids/10.4.721. [DOI] [PubMed] [Google Scholar]
- 42.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson K S, Sanders C C, Moland E S. Use of microdilution panels with and without β-lactamase inhibitors as a phenotypic test for β-lactamase production among Escherichia coli, Klebsiella spp., Enterobacter spp., Citrobacter freundii, and Serratia marcescens. Antimicrob Agents Chemother. 1999;43:1393–1400. doi: 10.1128/aac.43.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukamoto K, Kikura R, Ohno R, Sawai T. Substitution of aspartic acid-217 of Citrobacter freundii cephalosporinase and properties of the mutant enzymes. FEBS Lett. 1990;264:211–214. doi: 10.1016/0014-5793(90)80250-m. [DOI] [PubMed] [Google Scholar]
- 46.Tsukamoto K, Ohno R, Sawai T. Extension of the substrate spectrum by an amino acid substitution at residue 219 in the Citrobacter freundii cephalosporinase. J Bacteriol. 1990;172:4348–4351. doi: 10.1128/jb.172.8.4348-4351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsukamoto K, Ohno R, Nukaga M, Sawai T. The effect of amino acid substitution at position 219 of Citrobacter freundii cephalosporinase on extension of its substrate spectrum. Eur J Biochem. 1992;207:1123–1127. doi: 10.1111/j.1432-1033.1992.tb17150.x. [DOI] [PubMed] [Google Scholar]
- 48.Usher K C, Blaszczak L C, Weston G S, Shoichet B K, Remington S J. Three-dimensional structure of AmpC β-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design. Biochemistry. 1998;37:16082–16092. doi: 10.1021/bi981210f. [DOI] [PubMed] [Google Scholar]
- 49.Vu H, Nikaido H. Role of β-lactam hydrolysis in the mechanism of resistance of a β-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum β-lactams. Antimicrob Agents Chemother. 1985;27:393–398. doi: 10.1128/aac.27.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]