Abstract
Ecological interactions are ubiquitous on tropical coral reefs, where sessile organisms coexist in limited space. Within these high-diversity systems, reef-building scleractinian corals form an intricate interaction network. The role of biotic interactions among reef corals is well established on ecological timescales. However, its potential effect on macroevolutionary patterns remains unclear. By analysing the rich fossil record of Scleractinia, we show that reef coral biodiversity experienced marked evolutionary rate shifts in the last 3 million years, possibly driven by biotic interactions. Our models suggest that there was an overwhelming effect of staghorn corals (family Acroporidae) on the fossil diversity trajectories of other coral groups. Staghorn corals showed an unparalleled spike in diversification during the Pleistocene. But surprisingly, their expansion was linked with increases in both extinction and speciation rates in other coral families, driving a nine-fold increase in lineage turnover. These results reveal a double-edged effect of diversity dependency on reef evolution. Given their fast growth, staghorn corals may have increased extinction rates via competitive interactions, while promoting speciation through their role as ecosystem engineers. This suggests that recent widespread human-mediated reductions in staghorn coral cover, may be disrupting the key macroevolutionary processes that established modern coral reef ecosystems.
Subject terms: Palaeontology, Evolutionary ecology, Palaeoecology
The effect of biotic interactions among reef corals on macroevolutionary patterns is unclear. Here, the authors study the rich coral fossil record, finding that reef coral diversity experienced potentially biotic interaction-driven evolutionary rate changes, and that Staghorn corals affected fossil diversity trajectories of other coral groups.
Introduction
Biological diversity stems from the interplay between biotic and abiotic controls, operating across temporal and spatial scales1. Among high-diversity systems, coral reefs stand out for harbouring the majority of marine species, despite occupying relatively small areas2. Coral reef biodiversity has been shaped by major changes in climate, ocean chemistry, sea level and nutrient dynamics through geological time3. However, while some of these abiotic factors influenced large-scale patterns of reef biodiversity (through mass-extinctions4, for example), biotic controls have also been suggested as important determinants of the temporal waxing and waning of reefs3. Specifically, competition for space has been hypothesized as a fundamental driver of reef evolution3.
On short ecological timescales, interspecific competition is regarded as one of the key mechanisms shaping the composition of coral reef communities5–8. As the namesake of these ecosystems, reef-building corals (Order Scleractinia) are major contributors to habitat formation and are often one of the most abundant benthic sessile invertebrates on present-day reefs9. Given the limited space available on shallow marine reefs, corals can compete through direct interference and overgrowth, or through indirect pre-emption of space for larval settlers5–8. Alongside tropical rainforests, coral reefs have been used to typify biological systems moulded by interspecific competition, including a foundational work in theoretical ecology5. Yet, it is not known whether competition for space or other biological interactions can scale up to determine large-scale macroevolutionary patterns on coral reefs.
Here we show that the diversification of Acroporidae (commonly known as staghorn corals) is associated with a major disruption in coral evolutionary patterns, suggesting strong diversity-dependent effects (i.e., when diversification in one lineage impacts the evolution of others10). Using recently developed Bayesian modelling techniques11,12, we assessed the diversification dynamics of scleractinian coral fossil lineages at the species level. By focusing on the Cenozoic, we: (a) describe the major temporal changes in evolutionary rates that determined the formation of present-day species richness patterns among scleractinian coral families; (b) explore fossil diversity trajectories of major extant reef-building coral families; and (c) estimate the potential effects of environmental and biological drivers on speciation and extinction rates across reef coral families. Our results reveal a unique mechanism through which biological interactions may shape the evolution of high-diversity systems.
Results and discussion
Recent shifts in diversification
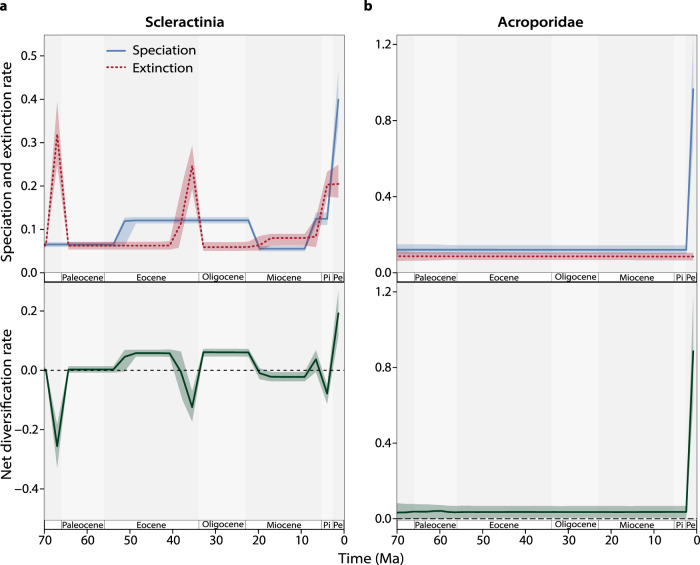
We detected three major episodes of extinction in scleractinian corals during the last 70 million years [Myr]. The first is the mass-extinction event at the Cretaceous–Paleogene boundary (Fig. 1a). This event is known for having affected corals only moderately relative to other marine invertebrates13, however, it was still significant at the species level for Scleractinia. The second, at the Eocene–Oligocene transition (Fig. 1a), coincided with a major cooling event that transformed the world into an icehouse after the hot climate of the Eocene14. Although this transition is not classically recognized as a mass-extinction15, for reef-building corals this cryptic extinction event16 seems to have substantially impacted diversity. Third, we found that coral extinction also increased sharply in the Pliocene–Pleistocene. Remarkably, this extinction also coincided with a striking increase in speciation rates (Fig. 1a).
Fig. 1. Evolutionary rates through time in scleractinian corals.
Rates of speciation (blue) and extinction (red dashed) estimated for fossil lineages of Scleractinia (a; n = 4235) and Acroporidae (b; n = 165). The analyses incorporate all the fossils recorded in the respective groups, although here we only show the last 70 million years. The resulting net diversification rates (speciation minus extinction) are shown at the bottom panels. Solid and dashed lines represent the median rates, while coloured shadings represent 95% posterior credibility intervals. Pi—Pliocene; Pe—Pleistocene. Source data are provided as a Source Data file.
Recent shifts in coral diversification have been noted previously17, but their causes remain elusive. Given that the Pliocene-Pleistocene is not associated with a mass-extinction event, we ask: could these shifts have been driven by biotic interactions? By running the same models across extant reef-building coral families, we found a remarkable spike in speciation rates of Acroporidae in the last 3 Myr (Fig. 1b). But notably the speciation spike was not mirrored by extinction (Fig. 1b). Consequently, we found very high net diversification rates in the Acroporidae close to the present (Fig. 1b), even when compared to the recent diversification of all scleractinians (Fig. 1a). This suggests that acroporids had a markedly different diversity trajectory to most other scleractinian corals, and if diversity dependency was involved, they may have been the winners.
Diversity trajectories and dependency
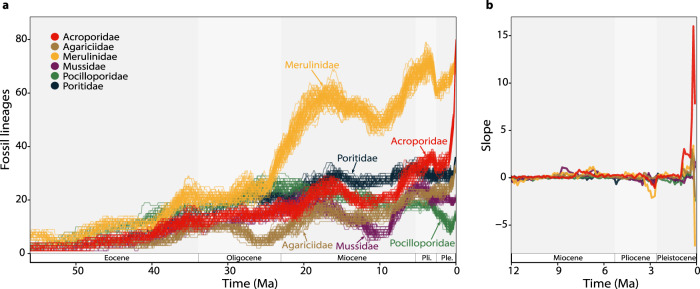
The identified shifts in evolutionary rates point to the Pliocene–Pleistocene as a key period for the formation of modern reef-building coral species richness. Therefore, to further explore this accumulation of diversity, we focused on species that are classified as reef-associated and examined the evolution of coral families that are: (i) abundant on present-day reefs (Acroporidae, Agariciidae, Merulinidae, Mussidae, Pocilloporidae, and Poritidae)18; and (ii) have a high number of occurrences in the fossil record. We estimated fossil diversity trajectories through time12 for each of these six families separately. These models revealed major temporal fluctuations in the number of lineages, particularly in the Miocene (Fig. 2a). But more importantly, they show that for most of its evolutionary history, the Acroporidae was not the most diverse family of corals. It was only very recently, in the Pleistocene, that acroporids became the most species-rich group of reef-building corals (Fig. 2a). Although their diversity almost doubled from the late Miocene to the Pliocene (8–3 Million years ago [Ma]; Fig. 2a), the rate of change was substantially higher in the Pleistocene (Fig. 2b).
Fig. 2. Fossil lineages through time in six extant reef coral families.
a Reconstruction of fossil diversity trajectories estimated through a time-variable Poisson process of preservation. Models were run independently in each family and were replicated fifty times (individual lines) to incorporate uncertainty around the age of the fossil occurrences. b Mean rate of diversity change (slope in species per 0.1 Myr) in the last twelve million years for the same coral families, calculated as the difference in diversity between subsequent time points (see Methods). Acroporidae—red; Agariciidae—brown; Merulinidae—yellow; Mussidae—purple; Pocilloporidae—green; Poritidae—dark blue. Source data are provided as a Source Data file.
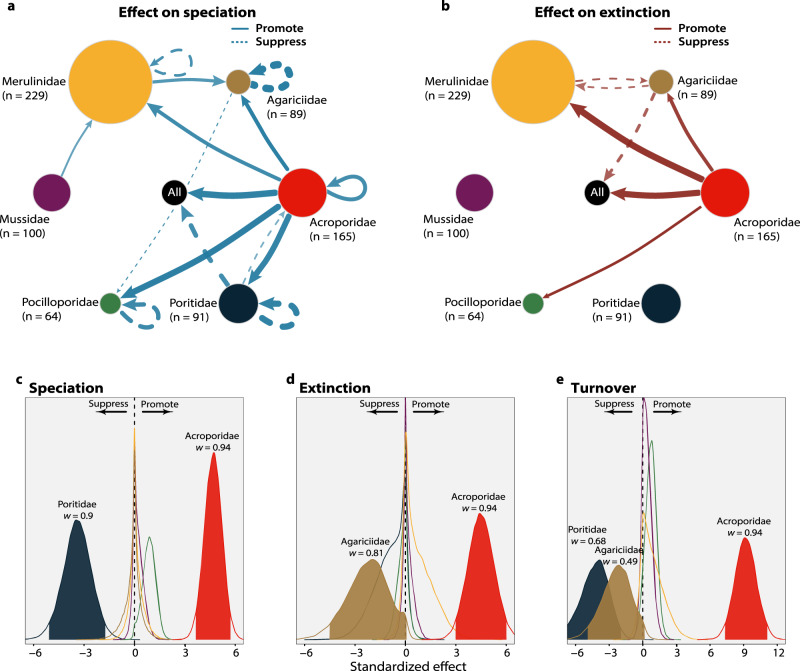
To assess the main factors driving these macroevolutionary trends, we ran models that estimated the correlation between diversity trajectories and both environmental and biological predictors11 (see Methods). Results from these models show that the environmental factors tested—paleotemperature, sea level and rate of sea-level change—were generally less well correlated with fossil trajectories than biological predictors (i.e., the diversity of other coral taxa). The only detectable potential abiotic effects were: (i) a slight increase in speciation rates in the Acroporidae that may have been associated with the rate of sea-level change, and (ii) a negative correlation between paleotemperatures and extinction rates (i.e., higher temperatures associated with less extinction) in the Pocilloporidae (Supplementary Fig. 1). Our models of cross-clade effects, on the other hand, suggest that diversity dependence was widespread throughout reef coral evolution (Fig. 3). But the directionality of effects was not taxonomically homogeneous. The Acroporidae stand out as the only family that may have affected both speciation and extinction in all families combined (Fig. 3a, b; central node). This overwhelming effect had contrasting outcomes; while acroporids promoted extinction in other coral lineages (Fig. 3b; central node), they also seem to have promoted speciation (Fig. 3a; central node). These results were consistent when we performed the same analysis, selecting only sites where Acroporidae species co-occur with the other families (Supplementary Fig. 2), and on Indo-Pacific species only (Supplementary Fig. 3). Family pairwise analyses also showed similar results, with Acroporidae potentially enhancing speciation and extinction in five and three out of six families, respectively (Fig. 3a, b; external nodes). No other family had this many effect links with such a strong overall intensity; and this is despite the fact that the Acroporidae was not the most abundant nor the most diverse coral family before the Pleistocene (i.e., results were not driven by prevalence in the palaeontological record). Moreover, while it is reasonable to expect strong self-diversity dependency (i.e., species within the same family influencing their own diversity trajectories19), given that confamilial species generally share more similar ecologies, this does not seem to be the case for reef corals. We did find self-suppressing effects on speciation in four families (Fig. 3a). However, these effects are negligible when compared to the strong cross-clade influence of Acroporidae on overall speciation (and its own self-promoting effect; Fig. 3a) and extinction.
Fig. 3. The overwhelming cross-clade effect of Acroporidae on the diversity of reef corals.
A network depicts the estimated cross-clade influence on rates of speciation (a) and extinction (b). Arrows represent the directionality of interaction effects, while arrow widths are proportional to the estimated median effect. Continuous or dashed lines indicate promoting or suppressing effects, respectively. Only interactions with a strong signal in the model were included (w > 0.7; see Methods). Loops represent self-diversity dependency; i.e., when confamilial species influence their own diversity trajectories. The size of each external node is proportional to the number of fossil occurrences in each family, and the number of fossil species in each family is shown in parenthesis (n). The central node represents all families combined, for which we also show the posterior distribution of effects on speciation (c), extinction (d) and lineage turnover (e; effect on speciation plus effect on extinction). Filled densities represent effects estimated with a strong signal (w > 0.7). The median shrinkage weights (w) are also shown for these families. Acroporidae—red; Agariciidae—brown; Merulinidae—yellow; Mussidae—purple; Pocilloporidae— green; Poritidae—dark blue. Source data are provided as a Source Data file.
A key feature of the model used herein is that it provides a robust measure to distinguish between noise and signal in the diversity-dependent effects (shrinkage weight [w]—where 0 represents noise and 1 represents signal11; see Methods). This measure reinforces our main results: the median shrinkage effect weight was substantially higher in Acroporidae (wAcro = 0.94; Supplementary Table 1) than in other families. More importantly, the model is blind to species ecological attributes, yet it detected biologically meaningful results. The only family that enhanced overall speciation (Acroporidae; Fig. 3c) is mostly composed of fast-growing branching species9 (Supplementary Fig. 4) that are major ecosystem engineers that contribute to habitat complexity20. On the other hand, the massive corals of the family Poritidae, that generally reduce small-scale habitat heterogeneity8, were found to suppress speciation (Fig. 3c). Today, with their fast growth, acroporids tend to be competitively superior under intermediate disturbance regimes7,21. Competition may, therefore, at least in part, explain why they are the only group to promote extinction in the models (Fig. 3d). The buffering effect of Agariciidae against extinction (Fig. 3d) is the only result that defies a straightforward ecological explanation and should be investigated further. But it is in the sum of these effects that the Acroporidae prevail. The models suggest that acroporids contributed to a nine-fold increase in overall lineage turnover throughout the evolution of reef corals (Fig. 3e).
Our results provide compelling evidence for the presence and potential strength of biological interactions in the evolution of modern coral reefs3. They also suggest that rapid sea-level variation22,23 may only be a partial explanation for recent changes in the relative prevalence of reef coral families. Earlier biological interactions may have shaped both diversity patterns and subsequent prevalence. At first glance, it seems paradoxical that one coral family can simultaneously enhance overall rates of speciation and extinction. However, it is important to note that these effects could vary: 1—between lineages within the same family (i.e., some species might go extinct while others originate during the same time period); and 2—across time (i.e., effects are estimated for the entire duration of the temporal overlap between lineages). Regardless, these seemingly contrasting effects are easily explained by the unique ecology of acroporids. Species within this family are the fastest calcifying corals on reefs24, dominating the benthos and leaving few opportunities for other groups to grow when levels of disturbance are infrequent, but intense21. For example, in one of the few long-term ecological assessments of competition on reefs, branching Acropora species were found to be competitively superior and, subsequently, reduced coral diversity7. Thus, it seems likely that the weedy nature of staghorn corals pushed other coral species to extinction during low sea-level stands in the Pleistocene, a critical time when habitat area was reduced25,26. Despite this inferred effect on extinction, the high accretion rates and reef-building capacity of acroporids24 may have had a double-edged effect on reef coral biodiversity by also promoting speciation in other scleractinian families. As recently demonstrated, acroporids are major contributors to the fractal dimension of reef complexity20, which not only promotes niche differentiation, but may also facilitate coral larvae settlement by creating fine-scale structures27. Since staghorn coral growth forms are more likely to be damaged by intense hydrodynamic disturbances28, they might also have facilitated speciation of opportunistic species that flourish as new space becomes available29. In addition, by tracking sea-level changes, acroporids increased the heterogeneity of reef types in the Pleistocene30, which likely favoured habitat specialization in other coral groups.
The fast and diversified growth forms of acroporids9 were, therefore, key to the development of present-day coral reefs. Ironically, however, their life strategy seems to be most affected by recent human impacts including climate change31–33. Although acroporids thrived with the moderate disturbance levels of the Pleistocene, the current rate of change is severe33 and reef-building corals may struggle to adapt. Framework-building and habitat generation by acroporid corals has already declined globally34, with demonstrated negative consequences for reef-associated faunas35. We show that the ecological attributes of acroporids are not only important for supporting modern reef species, but were also the likely promoters of their diversity. This provides a bitter lesson for the current trajectory of biodiversity on coral reefs: we may be causing irreversible changes in the fundamental evolutionary mechanisms that created these exceptionally rich ecosystems in the first place.
Methods
Fossil data
We downloaded all fossil occurrences recorded for the order Scleractinia at the species level from the Paleobiology Database (PBDB – paleobiodb.org; accessed on 3 August 2021). This is the most comprehensive repository for palaeontological data in reef corals to date. Due to the nature of the data, no ethics approval was required. To minimize identification issues, we excluded taxa with uncertain generic and species assignments (i.e., classified as aff. and cf.) and only selected species that had accepted names. We also selected the variables classification and palaeoenvironment from the output options to facilitate taxonomic and environmental filters applied in downstream analyses. The full dataset consisted of 24,011 occurrences across 4235 species, spanning over 250 Myr of coral evolution from the Triassic to the present. Although our focus here lies on the Cenozoic, we used the complete fossil dataset (i.e., including all of the occurrences) to have estimates of the diversification dynamics in scleractinian corals throughout the whole timespan of their evolution.
Evolutionary rates
With the full palaeontological dataset, we estimated evolutionary rates through time in scleractinian corals using the Bayesian framework of the program PyRate (v3.0)12,36,37. This program uses fossil occurrence data to calculate the temporal variation in rates of preservation, speciation and extinction, while incorporating multiple sources of uncertainty12. At its core implementation, PyRate jointly estimates the times of origination (Ts) and extinction (Te) for each fossil lineage; the fossilization and sampling parameters that determine preservation rates (q); and the overall rates of speciation (λ) and extinction (μ) through time36. Recently, the program has been upgraded to include a reversible jump Markov Chain Monte Carlo (rjMCMC) algorithm to estimate diversification rate heterogeneity, which provides more accurate and precise estimates than other commonly used methods12. Therefore, despite the inherent bias of the fossil record (i.e., estimates are conditioned on sampled lineages), PyRate is a robust method to quantify speciation and extinction rates, and their respective temporal shifts, from fossil occurrence data.
Extant taxa can also be included in the PyRate framework as long as they are also represented in the fossil record. This is done to extend the fossil geologic ranges to the recent times. Hence, the first step in our analysis was to identify which species in our dataset is still alive at the present. To do this, we matched the accepted species names in the PBDB dataset with those from the extant species dataset of Huang et al.38. Subsequently, we split our dataset into eleven independent subsets, with the goal of keeping each subset with an equal number of species. Each data subset included a random selection of species with their respective occurrences, which was enough to calculate Ts and Te (see below). This was done to avoid convergence issues, given the large size of our dataset and the consequent complexity of the model37. For each of our subsets, we generated fifty replicates by resampling the fossil ages from their temporal ranges to account for the uncertainty associated with the age of occurrences. We then used the maximum-likelihood test in PyRate to compare between three models of fossil preservation12: the homogeneous Poisson process (HPP; q is constant through time); the nonhomogeneous Poisson process (NHPP; q varies throughout the lifespan of a species); and the time-variable Poisson process (TPP; q varies across geological epochs). The latter model (TPP) was selected across all of our data subsets (Supplementary Table 2).
After selecting the preservation model, we first focused on assessing the estimates of times of origination and extinction in each data subset, rather than using the full dataset to jointly estimate all parameters at once as in the original implementation of PyRate. This further reduced the complexity of the model and allowed for more precise parameter estimates. For each replicate in all of our data subsets, we approximated the posterior distribution of Ts and Te through a 50 million generation run of the rjMCMC algorithm under the TPP, sampling parameters every 40 thousand iterations. At the end of each run, we discarded 20% of the samples as burn-in and assessed chain convergence through the effective sample sizes of posterior parameter estimates, using the software Tracer39 (v1.7.1).
From the results of this first set of models, we extracted the median estimates of Ts and Te across replicates, and we merged the estimates from the eleven independent data subsets. This merged data frame contained estimated times of origination and extinction for all coral lineages within our fossil dataset. We then used this merged Ts and Te data frame as input for another rjMCMC chain to finally estimate overall λ and μ through time, by applying the option -d in PyRate. In this option, Ts and Te for all fossil lineages are given as fixed values and, therefore, are not estimated by the model. The chain for this model was run for 100 million generations, sampling parameters at every 40 thousand iterations. Once again, we excluded 20% of the initial samples as burn-in and checked model convergence using Tracer. Finally, we calculated net diversification rates through time by subtracting the post burn-in samples of μ from λ.
To explore the taxonomic idiosyncrasies in the evolutionary rates of reef corals, we selected the most abundant families on present-day coral reefs in terms of the number of colonies per area18 (Acroporidae, Agariciidae, Merulinidae, Mussidae, Pocilloporidae, and Poritidae). Altogether, species within these families account for ~40% of the total extant diversity in Scleractinia. These families also account for most of the occurrences in the PBDB fossil dataset (excluding extinct families, which are generally older and had little temporal overlap with extant ones): Acroporidae (1457 occ. in 165 spp.); Agariciidae (722 occ. in 89 spp.); Merulinidae (2464 occ. in 229 spp.); Mussidae (1146 occ. in 100 spp.); Pocilloporidae (615 occ. in 64 spp.); and Poritidae (1149 occ. in 91 spp.). Therefore, from our full dataset, we selected six independent ones encompassing all species in each of the selected families. We also selected only species that are classified as reef-associated within these families, since we were specifically interested in these environments. This selection had a negligible effect on the size of the individual datasets, given that the vast majority of fossil species within these families are reef-associated. In each family, we followed the same modelling steps described above to estimate μ and λ, and diversity trajectories. However, this time it was not necessary to split the datasets into subsets, given that each family has far less occurrences than the full dataset. We started by comparing models of preservation, which showed the TPP as the best supported for all families (Supplementary Table 3). Then we created fifty replicates by resampling fossil ages to accommodate the uncertainty associated with the time of occurrences. For each replicate, we ran the rjMCMC algorithm for 50 million generations under the TPP model, with a sampling frequency of 40 thousand iterations. We discarded initial 20% of the samples as burn-in, and assessed convergence through Tracer. We then combined all replicates, resampling 100 random samples from each replicate to assess the estimates of μ and λ through time for each family. Finally, we extracted diversity trajectories in each family for all of the replicates by applying the -ltt option in PyRate, which generates a table with estimated range-through diversity at every 0.1 Myr. From these trajectories, we calculated the mean difference in diversity (slope in species per 0.1 Myr) between subsequent time samples backwards from the present (i.e., diversity in time t was subtracted from diversity in time t-1) using the diff function in R (v4.0.3).
As an alternative to PyRate, we also calculated the diversity dynamics of reef coral fossils using the R package divDyn40, which combines a range of published methods for quantifying fossil diversification rates. Differently from PyRate, the metrics applied in divDyn require that the fossil occurrences are split into discrete time bins. Therefore, these metrics treat the origination and extinction rates as independent parameters in each bin, while PyRate is designed to detect rate heterogeneity through a continuous time setting12. Our goal here, however, was not to compare models but to assess the robustness of our rate patterns and diversity trajectories using alternative methods. We divided our dataset into one-million-year time bins to have enough temporal resolution for rate calculations. To account for the uncertainty in the assignment of fossil ages, we created 50 binned replicates by sampling the age of each occurrence from a random uniform distribution, with bounds defined by the age ranges provided in the PBDB dataset. We then used the divDyn function to calculate the per capita rates of origination and extinction through time (based on the rate equations by Foote41) for all scleractinians (Supplementary Fig. 5a) and for reef-associated acroporids alone (Supplementary Fig. 5b). We also used the same procedure to generate range-through diversity curves for each of the six families selected previously, to compare with the curves generated by PyRate (Fig. 2a). Although the rate results differed between the PyRate (Fig. 1) and the divDyn (Supplementary Fig. 5) approaches, the general patterns remained unchanged. Rates are more volatile through time in divDyn estimates, with larger confidence intervals, which is expected from the metrics applied in the package12,42. Yet, we found the same peaks in extinction for Scleractinia: at the Cretaceous-Paleogene and Eocene-Oligocene boundaries, and at the Pliocene-Pleistocene (Supplementary Fig. 5a). The recent peak in speciation in Acroporidae was also detected, although less strong (Supplementary Fig. 5b). Despite these slight differences in rate estimates, the diversity curves reconstructed through divDyn (Supplementary Fig. 6) mirrored almost exactly the ones found with PyRate (Fig. 2), demonstrating that the overall macroevolutionary trends described herein (Figs. 1 and 2) are robust to methodological choices.
Diversity-dependent models
To assess the effects of diversity dependency on the evolution of reef coral lineages, we implemented the Multivariate Birth-Death model (MBD)11 within the PyRate framework. This method was first described as the Multiple Clade Diversity Dependence model (MCDD)19, in which rates of speciation and extinction are modelled as having linear correlations with the diversity trajectories of other clades. At its original implementation, the MCDD was developed to assess the effects of negative interactions, where increasing species diversity in one group can suppress speciation rates and/or promote extinction in itself or in other ecologically similar clades19. However, the model also incorporates the possibility of positive interactions, where increasing diversity in one clade can correlate with enhanced rates of speciation or buffered extinction. Through further model developments43, the MCDD was updated to also include a horseshoe prior44 on the diversity-dependence parameters, which helped controlling for overparameterization and enhanced the power of the model to recover true effects43. More recently, this model took its current form as the MBD11, with the additional possibilities of including environmental correlates and setting exponential, rather than just linear, correlations.
We first applied the MBD to estimate the diversity-dependent effects of individual extant coral families (i.e., the ones selected in the previous analysis; see Evolutionary rates) in their combined diversity trajectories. From the rjMCMC model results for individual families, we extracted estimates of Ts and Te in each of the fifty replicates and merged them across families. This merged dataset with fifty replicates of Ts and Te was then used as input for the MBD model, where we set the relative diversity trajectories of each individual family as predictors. We also included three key environmental predictors—paleotemperature, sea level and rate of sea-level change—to assess their influence in overall evolutionary rates. The paleotemperature data was obtained from Westerhold et al.45, and consists of global mean temperature estimates for the last 66 million years, averaged across 0.1 Myr time bins. Eustatic sea-level data was downloaded from Miller et al.46, and contains estimates of sea level for the last 100 million years in comparison to present-day levels, also split in ~0.1 Myr time bins. With this dataset, we calculated the average rate of sea-level change per million years, as measured from the absolute difference between subsequent sea-level values backwards in time (i.e., sea level in time t was subtracted from sea level in time t-1). These environmental factors were rescaled between 0 and 1 to maintain all predictors on the same relative scale.
Under our MBD model, the speciation and extinction rates of all families combined could change through time and through correlations with the relative diversity of individual families or environmental factors. The strength and directionality (positive or negative) of the correlations are also jointly estimated for each predictor within the model11. We ran both linear and exponential correlation models (see formulas in Lehtonen et al.11) in each of our fifty replicates for 25 million generations, sampling parameters at every 25 thousand iterations. We then compared the linear and exponential models through the posterior harmonic means of their log likelihoods, which supported the exponential one as having a better fit. From the posterior estimates, we summarized the speciation (Fig. 3c) and extinction (Fig. 3d) correlation parameters (i.e., the strength of the effect) by calculating their median and 95% Highest Posterior Density (HPD) interval across replicates. Finally, we also summarized the effect of families on lineage turnover (Fig. 3e), which we conceptualize as the sum of the effects on speciation and extinction.
The MBD model also provides posterior samples of the weight of the correlation parameters, which is estimated through the horseshoe prior11. In essence, this prior is able to reliably distinguish correlation parameters that should be considered noise from those that represent a true signal in the data11. The parameterization of the horseshoe prior contains local and global Bayesian shrinkage parameters44 from which shrinkage weights (w) can be calculated (see formulas in Lehtonen et al.11). These shrinkage weights associated with each correlation parameter in the MBD model vary between 0 and 1, with values closer to 0 representing noise and values closer to 1 representing a true signal. Through simulations, it has been shown that values of w > 0.5 indicate that the correlation parameter in question significantly differs from the background noise, being the correlation positive or negative43. However, as a conservative way to infer the weight of correlation parameters, here we use a value of w > 0.7 to detect significance. This value was calculated for each diversity-dependence parameter (speciation, extinction and turnover) from the median values drawn from the model posteriors.
The spatial distribution of reef-associated taxa varied considerably throughout the Cenozoic, with biodiversity hotspots moving halfway across the globe47. Therefore, the best way to capture this dynamic biogeographic history in reef corals is by analysing global diversity patterns like we did in our main MBD model. However, to assess the robustness of our diversity-dependent results against the influence of geographic scale and site co-occurrences, we repeated all the modelling steps described above with two data subsets. First, we selected only fossil species that have occurrences in the Indo-Pacific Ocean (i.e., 30°W–180°W) within the six families. Second, we excluded sites in which the Acroporidae did not co-occur with the other families. In each of these data subsets, we calculated diversity trajectories and used them as predictors in a separate MBD model. These models had a merged dataset of Ts and Te of all species included in each case (Indo-Pacific and co-occurrences) as a response variable.
Finally, we followed the same modelling procedures described above to investigate the diversity-dependent effects in family pairwise analyses. We applied the MBD model to assess the effects of all other families in each individual family at a time, while also estimating correlations with the key environmental predictors. From the rjMCMC model results for individual families, we extracted the fifty replicates of estimated Ts and Te. Each replicate was then used as input for an MBD run using the relative diversity trajectories of each other individual family as predictors, along with the environmental variables. Once again, we ran 25 million generations of the MBD, with a sampling frequency of 25 thousand, using both linear and exponential correlation models in each age replicate. For all families, we found that the exponential model had a better fit. We then summarized the correlation parameters and the shrinkage weights (Supplementary Fig. 7) derived from the exponential models per family by calculating the median and 95% confidence intervals across replicates.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful to the PBDB team for their constant effort in maintaining their database and for making it freely available. We also thank K Quigley and S Tebbett for insightful comments, and D Silvestro for assistance with PyRate. Funding was provided by the Australian Research Council (DRB – LF190100062) and the Deutsche Forschungsgemeinschaft (WK – DFG – Ki 806/17-1). This is Paleobiology Database official publication number #424.
Source data
Author contributions
A.C.S. and D.R.B. conceived the study. W.K. curated the data. A.C.S. performed the analyses and wrote the first draft of the manuscript. D.R.B. and W.K. contributed substantially to revisions.
Peer review
Peer review information
Nature Communications thanks Danwei Huang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The datasets generated and/or analyzed during the current study are available at the Zenodo repository (10.5281/zenodo.6413373)48. There are no restrictions on data availability. All palaeontological data were collected from the publicly available Paleobiology Database (PBDB – paleobiodb.org; accessed on 3 August 2021). Source data are provided with this paper.
Code availability
The PyRate program (v3.0) is written in Python (v3.) and is available at github.com/dsilvestro/PyRate. Tracer (v1.7.1) is available at github.com/beast-dev/tracer/releases/tag/v1.7.1. The R (v4.0.3) packages used were: tidyverse (version 1.3.1); scales (version 1.1.1); igraph (version 1.2.6); HDInterval (version 0.2.2); and shape (version 1.4.6). The codes used during the current study are available at the Zenodo repository (10.5281/zenodo.6413373)48.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-30234-6.
References
- 1.Benton MJ. The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science. 2009;323:728–732. doi: 10.1126/science.1157719. [DOI] [PubMed] [Google Scholar]
- 2.Spalding MD, Grenfell AM. New estimates of global and regional coral reef areas. Coral Reefs. 1997;16:225–230. doi: 10.1007/s003380050078. [DOI] [Google Scholar]
- 3.Kiessling W. Geologic and biologic controls on the evolution of reefs. Annu. Rev. Ecol. Evol. Syst. 2009;40:173–192. doi: 10.1146/annurev.ecolsys.110308.120251. [DOI] [Google Scholar]
- 4.Kiessling W, Simpson C. On the potential for ocean acidification to be a general cause of ancient reef crises. Glob. Change Biol. 2011;17:56–67. doi: 10.1111/j.1365-2486.2010.02204.x. [DOI] [Google Scholar]
- 5.Connell JH. Diversity in tropical rain forests and coral reefs. Science. 1978;199:1302–1310. doi: 10.1126/science.199.4335.1302. [DOI] [PubMed] [Google Scholar]
- 6.Jackson JBC. Competition on marine hard substrata: The adaptive significance of solitary and colonial strategies. Am. Naturalist. 1977;111:743–767. doi: 10.1086/283203. [DOI] [Google Scholar]
- 7.Connell JH, et al. A long-term study of competition and diversity of corals. Ecol. Monogr. 2004;74:179–210. doi: 10.1890/02-4043. [DOI] [Google Scholar]
- 8.Sandin SA, McNamara DE. Spatial dynamics of benthic competition on coral reefs. Oecologia. 2012;168:1079–1090. doi: 10.1007/s00442-011-2156-0. [DOI] [PubMed] [Google Scholar]
- 9.Roff G. Evolutionary history drives biogeographic patterns of coral reef resilience. BioScience. 2021;71:26–39. [Google Scholar]
- 10.Rabosky DL. Diversity-dependence, ecological speciation, and the role of competition in macroevolution. Annu. Rev. Ecol., Evol., Syst. 2013;44:481–502. doi: 10.1146/annurev-ecolsys-110512-135800. [DOI] [Google Scholar]
- 11.Lehtonen S, et al. Environmentally driven extinction and opportunistic origination explain fern diversification patterns. Sci. Rep. 2017;7:4831. doi: 10.1038/s41598-017-05263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silvestro D, Salamin N, Antonelli A, Meyer X. Improved estimation of macroevolutionary rates from fossil data using a Bayesian framework. Paleobiology. 2019;45:576–570. doi: 10.1017/pab.2019.23. [DOI] [Google Scholar]
- 13.Kiessling W, Baron-Szabo RC. Extinction and recovery patterns of scleractinian corals at the Cretaceous-Tertiary boundary. Palaeogeogr., Palaeoclimatol., Palaeoecol. 2004;214:195–223. doi: 10.1016/S0031-0182(04)00421-3. [DOI] [Google Scholar]
- 14.Hutchinson DK, et al. The Eocene-Oligocene transition: a review of marine and terrestrial proxy data, models and model-data comparisons. Climate. 2021;17:269–315. [Google Scholar]
- 15.Raup DM, Sepkoski JJ. Mass extinctions in the marine fossil record. Science. 1982;215:1501–1503. doi: 10.1126/science.215.4539.1501. [DOI] [PubMed] [Google Scholar]
- 16.Cowman PF, Bellwood DR. Coral reefs as drivers of cladogenesis: expanding coral reefs, cryptic extinction events, and the development of biodiversity hotspots. J. Evol. Biol. 2011;24:2543–2562. doi: 10.1111/j.1420-9101.2011.02391.x. [DOI] [PubMed] [Google Scholar]
- 17.Simpson C, Kiessling W, Mewis H, Baron-Szabo RC, Müller J. Evolutionary diversification of reef corals: a comparison of the molecular and fossil records. Evolution. 2011;65:3274–3284. doi: 10.1111/j.1558-5646.2011.01365.x. [DOI] [PubMed] [Google Scholar]
- 18.Dietzel A, Bode M, Connolly SR, Hughes TP. The population sizes and global extinction risk of reef-building coral species at biogeographic scales. Nat. Ecol. Evol. 2021;5:663–669. doi: 10.1038/s41559-021-01393-4. [DOI] [PubMed] [Google Scholar]
- 19.Silvestro D, Antonelli A, Salamin N, Quental TB. The role of clade competition in the diversification of North American canids. Proc. Natl Acad. Sci. USA. 2015;112:8684–8689. doi: 10.1073/pnas.1502803112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres-Pulliza D, et al. A geometric basis for surface habitat complexity and biodiversity. Nat. Ecol. Evol. 2020;4:1495–1501. doi: 10.1038/s41559-020-1281-8. [DOI] [PubMed] [Google Scholar]
- 21.Pratchett MS, McWilliam MJ, Riegl B. Contrasting shifts in coral assemblages with increasing disturbances. Coral Reefs. 2020;39:783–793. doi: 10.1007/s00338-020-01936-4. [DOI] [Google Scholar]
- 22.Renema W, et al. Are coral reefs victims of their own past success? Sci. Adv. 2016;2:e150085. doi: 10.1126/sciadv.1500850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao Y, Economo EP, Satoh N. The roles of introgression and climate change in the rise to dominance of acropora corals. Curr. Biol. 2018;28:3373–3382. doi: 10.1016/j.cub.2018.08.061. [DOI] [PubMed] [Google Scholar]
- 24.Pratchett MS, et al. Spatial, temporal and taxonomic variation in coral growth - implications for the structure and function of coral reef ecosystems. Oceanogr. Mar. Biol.: Annu. Rev. 2015;53:215–295. [Google Scholar]
- 25.Kleypas JA. Modeled estimates of global reef habitat and carbonate production since the last glacial maximum. Paleoceanography. 1997;12:533–545. doi: 10.1029/97PA01134. [DOI] [Google Scholar]
- 26.Ludt WB, Rocha LA. Shifting seas: The impacts of Pleistocene sea-level fluctuations on the evolution of tropical marine taxa. J. Biogeogr. 2015;42:25–38. doi: 10.1111/jbi.12416. [DOI] [Google Scholar]
- 27.Hata T, et al. Coral larvae are poor swimmers and require fine-scale reef structure to settle. Sci. Rep. 2017;7:2249. doi: 10.1038/s41598-017-02402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madin JS, Connolly SR. Ecological consequences of major hydrodynamic disturbances on coral reefs. Nature. 2006;444:477–480. doi: 10.1038/nature05328. [DOI] [PubMed] [Google Scholar]
- 29.Tanner JE, Hughes TP, Connell JH. Species coexistence, keystone species, and succession: a sensitivity analysis. Ecology. 1994;75:2204–2219. doi: 10.2307/1940877. [DOI] [Google Scholar]
- 30.Blanchon P, et al. Postglacial fringing-reef to barrier-reef conversion on Tahiti links Darwin’s reef types. Sci. Rep. 2014;4:4997. doi: 10.1038/srep04997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandolfi JM, et al. Global trajectories of the long-term decline of coral reef ecosystems. Science. 2003;301:955–958. doi: 10.1126/science.1085706. [DOI] [PubMed] [Google Scholar]
- 32.Bellwood DR, Hughes TP, Folke C, Nyström M. Confronting the coral reef crisis. Nature. 2004;429:827–833. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- 33.Hughes TP, et al. Global warming transforms coral reef assemblages. Nature. 2018;556:492–496. doi: 10.1038/s41586-018-0041-2. [DOI] [PubMed] [Google Scholar]
- 34.Wild C, et al. Climate change impedes scleractinian corals as primary reef ecosystem engineers. Mar. Freshw. Res. 2011;62:205–215. doi: 10.1071/MF10254. [DOI] [Google Scholar]
- 35.Stuart-Smith RD, Brown CJ, Ceccarelli DM, Edgar GJ. Ecosystem restructuring along the Great Barrier Reef following mass coral bleaching. Nature. 2018;560:92–96. doi: 10.1038/s41586-018-0359-9. [DOI] [PubMed] [Google Scholar]
- 36.Silvestro D, Salamin N, Schnitzler J. PyRate: a new program to estimate speciation and extinction rates from incomplete fossil data. Methods Ecol. Evol. 2014;5:1126–1131. doi: 10.1111/2041-210X.12263. [DOI] [Google Scholar]
- 37.Silvestro D, Schnitzler J, Liow LH, Antonelli A, Salamin N. Bayesian estimation of speciation and extinction from incomplete fossil occurrence data. Syst. Biol. 2014;63:349–367. doi: 10.1093/sysbio/syu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang D, Goldberg EE, Chou LM, Roy K. The origin and evolution of coral species richness in a marine biodiversity hotspot. Evolution. 2017;72:288–302. doi: 10.1111/evo.13402. [DOI] [PubMed] [Google Scholar]
- 39.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kocsis AT, Reddin CJ, Alroy J, Kiessling W. The R package divDyn for quantifying diversity dynamics using fossil sampling data. Methods Ecol. Evol. 2019;10:735–743. doi: 10.1111/2041-210X.13161. [DOI] [Google Scholar]
- 41.Foote M. Morphological diversity in the evolutionary radiation of paleozoic and post-paleozoic crinoids. Paleobiology. 1999;25:1–115. doi: 10.1017/S0094837300020236. [DOI] [Google Scholar]
- 42.Smiley TM. Detecting diversification rates in relation to preservation and tectonic history from simulated fossil records. Paleobiology. 2018;44:1–24. doi: 10.1017/pab.2017.28. [DOI] [Google Scholar]
- 43.Silvestro D, Pires MM, Quental TB, Salamin N. Bayesian estimation of multiple clade competition from fossil data. Evol. Ecol. Res. 2017;18:41–59. [Google Scholar]
- 44.Carvalho CM, Polson NG, Scott JG. The horseshoe estimator for sparse signals. Biometrika. 2010;97:465–480. doi: 10.1093/biomet/asq017. [DOI] [Google Scholar]
- 45.Westerhold T, et al. An astronomically dated record of Earth’s climate and its predictability over the last 66 million years. Science. 2020;369:1383–1387. doi: 10.1126/science.aba6853. [DOI] [PubMed] [Google Scholar]
- 46.Miller KG, et al. The phanerozoic record of global sea-level change. Science. 2005;310:1293–1298. doi: 10.1126/science.1116412. [DOI] [PubMed] [Google Scholar]
- 47.Renema W, et al. Hopping hotspots: global shifts in marine biodiversity. Science. 2008;321:654–657. doi: 10.1126/science.1155674. [DOI] [PubMed] [Google Scholar]
- 48.Siqueira, A. C., Kiessling, W. & Bellwood, D. R. Data from ‘Fast-growing species shape the evolution of reef corals’. Zenodo. 10.5281/zenodo.6413373 (2022). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available at the Zenodo repository (10.5281/zenodo.6413373)48. There are no restrictions on data availability. All palaeontological data were collected from the publicly available Paleobiology Database (PBDB – paleobiodb.org; accessed on 3 August 2021). Source data are provided with this paper.
The PyRate program (v3.0) is written in Python (v3.) and is available at github.com/dsilvestro/PyRate. Tracer (v1.7.1) is available at github.com/beast-dev/tracer/releases/tag/v1.7.1. The R (v4.0.3) packages used were: tidyverse (version 1.3.1); scales (version 1.1.1); igraph (version 1.2.6); HDInterval (version 0.2.2); and shape (version 1.4.6). The codes used during the current study are available at the Zenodo repository (10.5281/zenodo.6413373)48.