Abstract
With advances in sequencing and instrument technology, bioinformatics analysis is being applied to batches of massive cells at single-cell resolution. High-throughput single-cell sequencing can be utilized for multi-omics characterization of tumor cells, stromal cells or infiltrated immune cells to evaluate tumor progression, responses to environmental perturbations, heterogeneous composition of the tumor microenvironment, and complex intercellular interactions between these factors. Particularly, single-cell sequencing of T cell receptors, alone or in combination with single-cell RNA sequencing, is useful in the fields of tumor immunology and immunotherapy. Clinical insights obtained from single-cell analysis are critically important for exploring the biomarkers of disease progression or antitumor treatment, as well as for guiding precise clinical decision-making for patients with malignant tumors. In this review, we summarize the clinical applications of single-cell sequencing in the fields of tumor cell evolution, tumor immunology, and tumor immunotherapy. Additionally, we analyze the tumor cell response to antitumor treatment, heterogeneity of the tumor microenvironment, and response or resistance to immune checkpoint immunotherapy. The limitations of single-cell analysis in cancer research are also discussed.
Subject terms: Cancer microenvironment, Cancer
Overview
The tumor microenvironment (TME) is a multi-component system composed of tumor cells, stromal cells, and infiltrated immune cells. Accordingly, the high-level complexity of the TME is accompanied by substantial heterogeneity at the intratumoral,1–3 inter-tumoral,4 and inter-individual5 levels. Within such a heterogeneous system, many pro- and antitumor cellular components or signals can regulate tumor progression and affect the efficacy of the antitumor immune response. Bulk-based omic analysis technologies provide insights into the functional mechanisms in the TME. However, owing to this extensive heterogeneity, the bulk sequencing data generated from large numbers of cell lysates provide only a “global” view of TME while obscuring the presence of cells with low abundance and highly specialized functions and ignoring universal intercellular communications.
High-throughput single-cell sequencing approaches refer to characterize a single cell at multiple levels, including their genomic,6 transcriptomic,7 epigenetic,8 and protein products.9 Comparing with traditional bulk sequencing strategies, the significant advantage of sequencing cell sequencing is evaluating heterogeneity among a population of cells, distinguishing cells with a small number and highly specified phenotype, and inferring cell behavior. In the early era of single-cell sequencing, its application is highly limited by unsatisfactory throughput and high detection cost. Nowadays, with the rapid progression in single-cell sequencing technologies, it has been widely applied in the research of various fields, particularly suitable for cancer research. Besides, emerging novel sequencing strategies continues to evolve toward a higher throughput and lower detection cost, such as single-cell combinatorial marker sequencing technique (SCI-seq),10 Topographic Single Cell Sequencing,11 or Split Pool Ligation-based Tranome sequencing.12 In addition, multi-omics sequencing technologies, rather than technologies that focus on single omics, provide multiple features such as DNA, RNA, protein profile for the same individual cell. Single-cell multiple sequencing technique (scCOOL-seq)13 enables the simultaneous examination of copy number variation, ploidy, chromatin, and DNA methylation, providing a broader view of for different cell populations. All these advanced single-cell sequencing technologies allow a broad application in the field of tumor biological and antitumor immunotherapy.
From basic/translational research to clinical practice, single-cell sequencing technology has been widely used to dissect TME composition,14,15 and is particularly promising in the fields of tumor immunology and immunotherapy. Resolution at the single-cell level enables identification of the immune cell population and signaling pathways that are actively involved in modulating tumor immune escape or elimination.16 The success of immune checkpoint blockade (ICB) immunotherapy exhibits potential for the treatment of solid tumors, enabling many patients to achieve long-term survival benefits; however, most patients do not respond well to ICB treatment.17 Understanding the characteristics of the baseline TME and dynamic TME changes during ICB treatment can help clarify the cellular and molecular mechanisms of ICB-driven tumor control and may reveal novel therapeutic targets for overcoming ICB resistance. By performing single-cell RNA sequencing (scRNA-Seq) alone or in combination with multi-omic strategies, the high-dimensional feature matrix at single-cell resolution can be used to infer immune cell identity and functional specification. Annotation of the cell identity merely based on scRNA-Seq may reveal a different result to the gating strategy via flow cytometry analysis. This shortage can be largely compensated by integrating cytometry and proteo-genomic data.18 The high-dimensional properties of scRNA-seq data allow for more refined annotation of cell subpopulations while investigating the activity of signaling pathways and inferring cellular state trajectories. Advanced bioinformatics tools have also been used to reconstruct cellular differentiation potential, determine the events driving distinct cellular states or transitions19,20 and construct a network of intercellular communication by exploring ligand-receptor interaction.21,22 The application of single-cell sequencing technology can be used for precision medicine in the clinic and to improve the outcomes of patients administered with ICB immunotherapies, such as identifying TCF7+CD8+ T cells as a predictor of positive outcomes to anti-PD-1 treatment.23
In this review, we summarize critical progress in single-cell sequencing analysis regarding both tumor cell behavior and tumor immunology, providing insights into tumor cell evolution and heterogeneity of the tumor immune microenvironment. The profiling of single tumor cells and immune cells has great potential to reveal novel mechanisms of resistance, immune tolerance, and relapse in individual patients with cancer, thereby it can be applied to facilitate the development of personalized and effective antitumor treatments.
Principles of single-cell sequencing technologies, analysis pipeline, and data interpretation
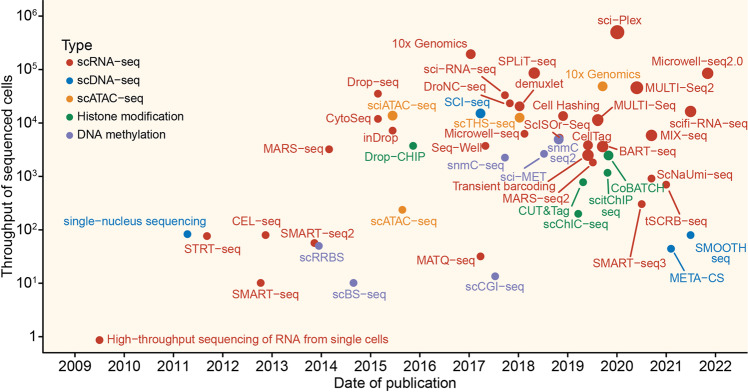
Since Tang et al. successfully developed high-throughput transcriptomic sequencing in 2009,7 many scRNA-seq protocols have been developed in the past decades (Fig. 1). In general, all protocols can be classified into two categories: full-length transcript sequencing approach and 3′/5′-end transcripts sequencing approach. The first approach attempts to produce uniform coverage of each transcript (e.g., Smart-seq224) and therefore has higher sensitivity than the second, which combines unique molecular identifiers (UMIs) with transcripts to reduce technical bias during library construction (e.g., 10× Genomics25). Moreover, all protocols can be classified into plate- or droplet-based approaches, depending on the strategy of cell capture. The droplet-based approach is preferable for capturing large amounts of cells with low sequencing depth.26 However, more technical noise would be present when this strategy is used. Normally, the following steps would be involved: (a) isolate single cells from sample (blood, tissue, etc.); (b) obtain mRNA from single-cell lysis; (c) convert poly(T)-primed mRNA into cDNA with RT; (d) cDNA amplification; (e) library construction; and (f) sequencing.27–29 Recently, in 2017, to reduce the inefficient sample processing and technical batch effect in downstream analysis among multiple samples, a multiplexed cell capturing method for scRNA-seq was successfully developed.30 In brief, there are four strategies: (i) oligo-dA-based barcoding; (ii) combination of mRNA and DNA barcodes; (iii) multiplexing by viral integration; (iv) natural genetic variation.31
Fig. 1.
Timeline and throughput of single-cell sequencing milestones. Timeline of single-cell sequencing milestones. Scatterplot depicts the published date and throughput of sequencing for each technology. Color indicates different sequencing specifications
Sequencing will produce raw data in FASTQ format. The first step is gene count matrix generation, including quality control, read alignment, mapping, and gene count quantification. Cell Ranger pipeline has been developed by 10x Genomics to automatically complete the above steps for Chromium single-cell data. After obtaining the gene count matrix, the next step is processing, which includes quality control, normalization, feature selection, and dimension reduction.32 The goal of quality control in the processing step is to remove low-quality cells (e.g., empty cells and doublet cells). Normalization is used to remove technical bias due to the different cDNA capture efficiency and PCR amplification. In general, UMI counts are transformed to counts per million or transcripts per million.29 Even after removing zero count gene, the feature space for a human sample can include over 15,000 dimensions. To determine the most informative genes from the data, highly variable genes should be defined and selected. After feature selection, dimension reduction can further reduce computational burden and capture comprehensible information from complex data structure. PCA, tSNE, and UMAP are normally used in this step. However, tSNE and UMAP are not only used for dimension reduction but are also the main methods of data visualization. Downstream analysis, including clustering, annotation, trajectory analysis, and cell–cell interaction, can be performed based on well-processing data; therefore, the processing step is the most important step of scRNA-seq analysis. The clustering step involves finding and grouping cells into various populations based on similar expression patterns. Each population can be annotated as a cell type using marker-based or reference-based methods such as singleR.33 After annotation, differential gene expression analysis can be completed at cell-type level to determine detailed differences between cell types. In addition to cell type annotation, cell differentiation trajectory can also be inferred based on variable genes (e.g., monocle series tools34,35 and slingshot36) or RNA velocity (e.g., velocyto20 and scVelo37). In contrast to variable genes strategy, RNA velocity describes the direction and speed automatically without manually setting a root. Cell–cell communication is a common method of analysis to explore the tumor microenvironment. Two widely used platforms, Seurat38 and Scanpy,39 have been developed and integrate various computational methods that can complete most analysis steps. Meanwhile, an increasing number of algorithms are being tested to develop application for scRNA-seq data analysis.
Except transcriptome information, single-cell genome sequencing also provides new perspectives to our understanding of tumors, such as SNS,40 SCI-seq,10 SMOOTH-seq.41 In general, four steps have been implemented to acquire single-cell genomic sequencing data, including cell isolation, whole-genome amplification (WGA), interrogation of WGA products, and error correction.42 Due to only two copies of genomic DNA in human cells, WGA is one of the challenges for single-cell genome sequencing. To uniformly amplify genomic DNA in each cell, three kinds of WGA methods have been developed, including DOP-PCR (PCR-based),43 MDA (isothermal amplification),44 or MALBAC.45 For genomic data, variant calling is one of the most important step in the downstream analysis. Bioinformatics tools, such as SCcaller,46 LiRA,47 and Conbase48 have been developed for SNV detection, in addition, SCNV,49 HMMcopy,50 and Ginkgo51 are tools for CNV calling.
In recent years, single-cell sequencing technologies for epigenomics such as chromatin accessibility, DNA methylation, histone modification have become a possibility as well. Libraries of scATAC-seq are created from single cells that have been exposed to the Tn5 transposase by one of the following protocols: split-and-pool approach,52 Chromium droplet-based approach,53 and C1 approach.54 The typical pipeline includes QC, alignment, peak calling and downstream analysis, such as peak differential analysis, motif enrichment analysis, and footprinting analysis.55 ArchR,56 SnapATAC,57 and Signac,58 which consist of several algorithms, can be used in scATAC-seq data processing and analysis. Another chromatin status investigation technology is single-cell chip-seq. Drop-ChIP combines microfluidics and droplet-based sequencing protocol to obtain chromatin data.59 CUT&Tag uses an enzyme tethering strategy that bounds chromatin protein by appointed antibody and then generates a fusion protein as protein-Tn5 transposase.60 The experimental procedure of scitChIP-seq is similar to traditional ChIP-seq method with tagmentation-based library preparation strategy before canonical ChIP experiment.61 DROMPAplus is a ChIP-seq analysis tools with various algorithms for any species, including QC, normalization, statistical analysis, and visualization.62 Moreover, a few scATAC-seq tools can be used for scChIP-seq as well, such as Signac for CUT&Tag. DNA methylation is also an important aspect of epigenomics that provides information about gene expression regulation, development, and disease. The single-cell DNA methylation sequencing can be simply classified into two categories: bisulfite-based and bisulfite-free.63 One disadvantage of the previous one is that it cannot distinguish 5mC and 5hmC.64 The downstream analysis usually contains methylation calling, visualization, clustering, and methylation segmentation and differentially methylated region detection.65 Several mature statistical algorithms and bioinformatic tools, such as k-means, Epiclonal,66 NMF,67 and BSmooth68 have been applied to data analysis.
Analysis of heterogeneity and response of tumor cells to treatment
The heterogeneity of cancer at the intertumoral and intratumoral levels is consistently among the main obstacles to cancer treatment. A combination of multi-region sampling and bulk sequencing is typically used to study intratumor heterogeneity at the genome level. This method can partly reveal tumor heterogeneity but is insufficient for fully understanding lineage and temporal heterogeneity. Consequently, significant progress has been made in single-cell sequencing. After reconstructing the clonal lineage, the primary clone or subclones in the tumor lineage can be identified using single-cell technology.69–71 Secondly, we used tumor cell phenotypes and their signaling pathways to determine heterogeneity of epithelial–mesenchymal transition (EMT), proliferation, migration, and apoptosis. We also examined spatial heterogeneity of cancer cell clone composition at different spatial sites. During cancer occurrence and development, tumor cells evolve into different clonal lineages in response to selection pressure. Herein, we review the latest advances in the use of single-cell technology to understand the heterogeneity and evolution of tumor cells in different dimensions (Fig. 2 and Table 1).
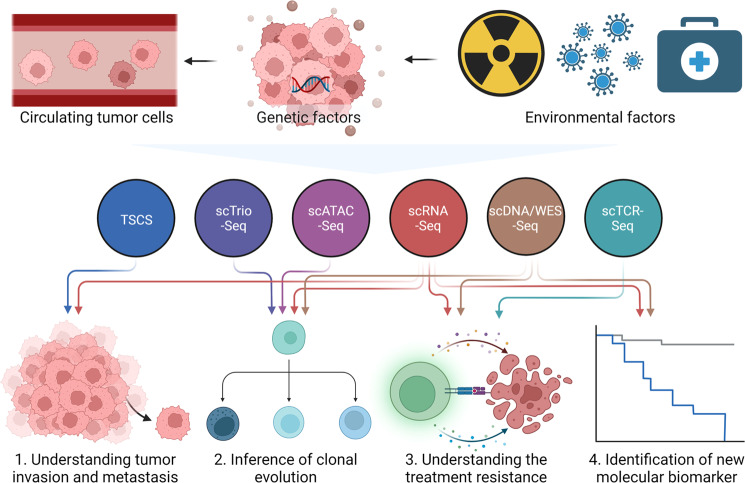
Fig. 2.
Application of single-cell omics in research of tumor cells. Tumor cells are composed of cells with various genomic alterations that influence disease progression and response to environmental perturbations and drug treatment. The characterization of high-dimensional profiling at a single tumor cell resolution facilitates the understanding of complex tumor cell behavior, clonal evolution during tumor progression, and identification of novel biomarkers for clinical application. Colored circle with arrows represents sing cell sequencing technologies and their applications in research of tumor cells
Table 1.
Representative studies of high throughput single-cell sequencing in research of tumor cells
| Tumor | Methods | Cases | Samples | Therapy/treatment | Main findings |
|---|---|---|---|---|---|
| AML | scRNA-seq | 21 | Tumor/normal | - | In primitive AML cells, stemness and myeloid-related genes are co-expressed, which is related to the prognosis of patients. |
| scRNA-seq | 5 | Tumor | - | Apoptosis and chemokine signaling are characteristics of relapsed AML, and co-targeting BCL2 and CXCR4 signaling may benefit patients. | |
| scATAC-seq | 21 | Tumor/normal | - | Chromatin accessibility can reveal the regulatory evolution in AML cells, and HOX is a key regulatory factor in the preleukemia phase. | |
| scDNA-seq | 9 | Tumor | Ivosidenib monotherapy | Ivosidenib resistance in AML patients may be caused by 2-HG restoration. | |
| Liver cancer | scRNA-seq | 2 | Tumor | - | Proposes a method to quantify tumor ITH, revealing the interconnection between different components in the evolution of HCC. |
| scRNA-seq | 1 | Tumor | - | The existence of CD24+ CD44+ subgroups suggests that there may be stemness-related HCC subclones. | |
| scRNA-seq | 6 | Tumor | - | The high expression of MLXIPL in HCC promotes the proliferation of cancer cells and inhibits their apoptosis, which is associated with poor prognosis. | |
| scDNA-seq | 3 | Tumor | - | The copy number of cancer cells only changes significantly in the early stages of cancer and ZNF717 may be the driver gene of HCC. | |
| scRNA-seq | 19 | Tumor | - | The intratumoral heterogeneity of the transcriptome of liver cancer is negatively correlated with the cytolytic activity of T cells and the prognosis of patients. | |
| Breast cancer | scRNA-seq | 6 | Tumor | - | Gene regulatory networks (GRNs) at the single-cell level are of great benefit to the discovery of key regulatory factors and GRNs identified TETV6 as a key gene in TNBC. |
| scRNA-seq | Patient-derived xenograft model | Patient-derived xenograft model | - | Oxidative phosphorylation is a key pathway for breast cancer metastasis, and inhibition of this pathway can significantly reduce the occurrence of metastasis. | |
| TSCS | 10 | Tumor | - | In the process of breast cancer cell invasion, the genome is relatively stable and invasive cancer is established by one or several escaped clones. | |
| scDNA-seq | 18 | Tumor/normal | - | Reveal the role of copy number alterations (CNAs) heterogeneity of breast cancer in therapeutic resistance and cancer recurrence. | |
| scTCR-Seq&scRNA-seq | 40 | Tumor | ICB | Reveal the heterogeneity of breast cancer anti-PD1 treatment response (PD1+ T cells, depleted T cells, and cytotoxic T cells will be cloned and expanded). | |
| Lung cancer | scRNA-seq | 8 | Tumor | platinum | In small cell lung cancer, the emergence of treatment resistance is always accompanied by increased intratumoral heterogeneity. |
| scRNA-seq | 9 | Tumor | - | The heterogeneity of lung cancer genome and transcriptome determine the heterogeneity of tumor-related pathways (including proliferation and inflammation-related), and also determines the heterogeneity of pathological characteristics. | |
| scRNA-seq | Kp1 cell line | Cell line | - | Small cell lung cancer has strong intratumoral heterogeneity, and this heterogeneity increases significantly after metastasis. | |
| scRNA-seq | 7 | Tumor | - | Early-stage lung adenocarcinoma has strong intratumoral heterogeneity, which leads to extremely complex interactions between different types of cells in the tumor microenvironment. | |
| scRNA-seq | 44 | Tumor | - | In the process of lung cancer metastasis, the subclones of metastatic cancer cells dominate, accompanied by the exhaustion of T cells. | |
| scRNA-seq | 30 | Tumor | TKI | TKI treatment can induce the evolution of cancer cells, and the tumors of patients with different clinical responses show different characteristics. | |
| Colorectal cancer | scWES-seq | 2 | Tumor | - | Colorectal cancer is of monoclonal origin and the tumor produces new driver mutations and evolves into different subclones in the process of cancer progression. |
| scWES-seq & scRNA-seq | Mouse model | Mouse model | - | The intratumoral heterogeneity of the genome of advanced colorectal cancer is reduced, and the intratumoral heterogeneity of the transcriptome is increased to adapt to changes in the environment. | |
| scTrio-seq | 10 | Tumor | - | There is strong transcriptome heterogeneity between different subclones of tumors, and DNA demethylation patterns vary greatly. | |
| RCC | scRNA-seq | 3 | Tumor | - | In renal clear cell carcinoma, T cell depletion is one of the main causes of immunosuppression, which is significantly related to poor prognosis. |
| scRNA-seq | 8 | Tumor | ICB | After ICB treatment, the immune effector cells of the responders will simultaneously up-regulate the expression of effector molecules and immunosuppressive markers. | |
| Gastric cancer | scRNA-seq & scDNA-seq | Cell lines | Cell lines | - | The results of single-cell genome and transcriptome sequencing confirmed the strong heterogeneity within tumor cell lines. |
| Uveal melanomas | scRNA-seq | 6 | Tumor | - | HES6 plays a key role in the proliferation and metastasis of uveal melanoma and can be used as a potential therapeutic target. |
- untreated
Acute myeloid leukemia
Acute myeloid leukemias (AML) are complex ecosystems characterized by multilevel heterogeneity.72–74 Single-cell sequencing has revealed that AML cells are diverse, arising from the proliferation and accumulation of immature cells in bone marrow development. Mature blood cells (lymphocytes, erythrocytes, and megakaryocytes) are derived from normal hematopoietic stem cells. Results of single-cell RNA sequencing showed that a series of lineage-committed progenitor states promoted the progression of hematopoietic stem cell commitment.75–77 Galen et al. also found that a group of monocyte-like cells among AML cells express various immunomodulatory genes. In vitro experiments demonstrated that T cell activity was affected by this group of AML cells.78 AML cells can be further subdivided into naïve and differentiated cells. The heterogeneity of initial AML cells (LICs) is considered the origin of AML progression and drug resistance.79–81 Stetson et al. used the RNA expression profiles of 813 LICs to further explain RNA clonal evolution during AML progression. Additionally, LICs exhibited evident apoptosis and chemokine signal evolution in relapsed patients.82 Corces et al. determined the chromatin accessibility map of AML from the perspective of epigenetics, revealing a unique regulatory evolution in cancer cells with a further increased mutation burden. At different stages of development, AML cells also showed distinctive mixed regulome profiles.83
Wang et al. identified single-nucleotide polymorphisms in single tumor cells using single-cell DNA technology. They also found that patients with an IDH1 mutant in leukemia developed complex polyclonal resistance mechanisms after receiving ivosidenib monotherapy.84
Liver cancer
Recent studies have revealed that primary liver cancer is one of the most heterogeneous cancers among all solid tumors.85 There are two main histological subtypes of liver cancer, hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma, which show different degrees of transcriptome heterogeneity.86 The heterogeneity of liver cancer is evident at the intertumoral, intratumoral, interlesional, or even intralesional levels.87–89 Losic et al. performed single-cell RNA sequencing of tumors in different regions, which revealed strong transcriptome heterogeneity among cells in different tumor regions. They also found that most transcription factors are not active or inactive in all tumor areas; however, one or several transcription factors are highly expressed in only one tumor area.90 Ho et al. observed similar intratumoral heterogeneity in liver cancer and identified a set of rare subclones rich in CD24+CD44+, which contain unique carcinogenic characteristics.91 Among the histological subtypes of HCC in liver cancer, Xiao et al. identified five HCC and two hepatocyte subclones with significant differences in gene expression; however, they also observed some common characteristics, including MLXIPL, which is an important marker in HCC cell trajectories.92 Duan et al. determined the copy number of a single HCC via single-cell whole-genome sequencing. These results indicate that HCC shows copy number variations in the early stages of liver cancer, with almost no new copy number variation introduced during tumor progression. In addition, they revealed that the origin of a specific HCC can be monoclonal or polyclonal and the intratumoral heterogeneity of polyclonal tumors is higher than that of monoclonal tumors.93 At the same time, Ma et al. used gene expression profiles to obtain the PCs of each tumor cell by principal component analysis, and then calculated the centroids of tumor cells in the eigenvector space (ie, the arithmetic mean of the PCs of all malignant cells in the tumor). The average distance from each tumor cell to the centroid was calculated, and this value was used as the ITH score of the tumor, which could predict the prognosis of the patient.94
Breast cancer
Breast cancers are clinically stratified based on the expression of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor (HER2). There are four subtypes that correlate with prognosis and define treatment strategies: luminal A, luminal B, HER2-enriched, and triple-negative breast cancer.95 At the transcriptional level, Her2+, luminal A, and luminal B subtypes also exist in malignant epithelial cells of breast cancer. Zhou et al. detected basal-like and normal-like cell subsets and revealed that ETV6 regulated different downstream genes in different subtypes to exert variable cancer-promoting roles.96 In a clinical human patient-derived xenograft model, single-cell RNA sequencing confirmed the transcriptional heterogeneity of primary tumors and micrometastases. Micrometastasis exhibits a specific transcriptome program that is conserved in the patient-derived xenograft model but with significantly upregulated oxidized phosphate metabolism.97 In breast cancer tissue, cancer cells originate from different cell lineages that evolve in parallel and possess different genomic mutations. During this evolution, in situ and aggressive subpopulations are produced.98–100 These subpopulations were comprehensively examined by determining the genome of a single cell. In a study of ductal carcinoma in situ, a direct genomic lineage between in situ and aggressive tumor subgroups was constructed. Before tumor invasion, most of the unique mutations or copy number variant cancer cell subtypes already exist in the duct.101 Copy number variation arises from breast cancer genome evolution and is a type of structural variation.102 Baslan et al. performed single-cell DNA sequencing to obtain genomic information for a single breast cancer cell and mapped the breast cancer genome, specifically copy number variation. Breast cancer cells exhibit both transcriptome and copy number heterogeneity. Additionally, copy number heterogeneity is significantly related to clinical or biological features such as polyploidy or HR-negative status.103
Bassez et al. used scTCR-Seq technology to confirm the presence of clonotype expansion in patients after PD-1 inhibitor treatment, indicating a continuous antitumor immune response; tumor cells from patients with clonotype expansion are enriched in pathways such as cell death, proteolysis, and immune signal transduction.104
Lung cancer
According to histopathological results, lung cancer can be subdivided into non-small cell lung cancer (NSCLC) and small-cell lung cancer (SCLC).105,106 SCLC is a high-grade neuroendocrine lung carcinoma that was once considered a molecularly homogeneous malignancy. Transcriptional diversity exists among SCLC cells and the pathways involved (including EMT and C-MYC) are heterogeneous.107 Similarly, this heterogeneity exists in the genome of lung cancer cells (e.g., copy number variation).108 SCLC cells disseminate early. During cancer cell metastasis, there are dozens of transcriptome heterogeneities, which may be enhanced by metastasis. Schaff et al. revealed that when SCLC cells metastasize to the liver (the common metastatic part), their single-cell regulatory heterogeneity becomes more complicated.109 Lung adenocarcinoma, another subtype of lung cancer, also exhibits high heterogeneity among tumor cells, with the cells expressing high levels of proximal and distal epithelial progenitor markers. This cell heterogeneity is also evident in signaling pathways such as glycolysis, oxidative phosphorylation, and the cell cycle.110 Kim et al. performed scRNA-seq on paired normal or early-stage tumor tissues and metastatic tumor tissues to clarify the transcriptional heterogeneity between metastatic and primary tumor tissues; additionally, a subtype of cancer cell differentiated dissimilarly from normal epithelial cells and may predominate in metastases.111 Different tumor cell subclones exist in different parts of lung cancer. Sharma et al. revealed this heterogeneity and confirmed its presence in the center and edge of the tumor, with proliferation lower in the center of the tumor than at the edge.108 scRNA-seq analysis confirmed that after systemic targeted therapy, patients with lung cancer exhibiting different treatment responses possess cancer cells with varying characteristics. In residual disease, cancer cells exhibit characteristics of alveolar regenerative cells, indicating that the status of cancer cells changes during treatment. In patients with progressive disease, kynurenine, plasminogen, and gap-junction signaling pathways in cancer cells are significantly upregulated.112 In an scRNA-seq study of SCLC, after the development of platinum-based drug resistance, the heterogeneity of the tumor cell transcriptome increased significantly; however, there was almost no change in the genome (copy number).107
Colorectal cancer
The molecular subtypes of colorectal cancer (CRC) are complex and highly heterogeneous. CRC is generally divided into four main consensus molecular subtypes (CMSs). CMS1 is characterized by hypermutation, microsatellite instability, and strong immune activation; CMS2 activates the epithelial, marked WNT, and MYC signal pathways; CMS3 exhibits epithelial and metabolic dysregulation; and CMS4 exhibits significant transforming growth factor-β activation, matrix invasion, and angiogenesis.113 Wu et al. performed single-cell exome sequencing to compare patients with normal or adenoma polyps in CRC. Both adenoma and CRC originate from the gradual accumulation of somatic mutations, particularly abnormalities in key genes such as LAMA1 (P3K-Akt signaling pathway) and ADCY3 (FGFR signaling pathway).114 By combining single-cell transcriptome and single-cell DNA technology, Ono et al. characterized the genome and transcriptome of a single cell; the results showed that at the exome level, tumor cell heterogeneity was not increased. After tumor transplantation, transcriptome heterogeneity increased significantly, resulting in a new subpopulation of cells exhibiting EMT signal activation.115 Bian et al. used single-cell triple omics sequencing to simultaneously detect the genome, transcriptome, and methylome of CRC tissues; epigenetic data showed that DNA methylation was consistent in the same tumor cell subclone but was very different in different genetic lineages.116
Circulating tumor cells
Circulating tumor cells (CTCs) migrate from tumor tissues into blood vessels and play an important role in the formation of metastases. CTCs are one of the main targets of liquid biopsy.117,118 The amount of CTCs in the blood is very low (one part per million) and only a small amount of CTCs can be obtained from a typical blood draw.119 Obtaining large numbers of CTCs is difficult, which limits the utility of bulk sequencing. The application of single-cell sequencing in CTCs has introduced a new research perspective. By characterizing copy number variations or other mutational patterns by single-cell sequencing, tumor metastasis mechanisms may be uncovered. Gao et al. performed single-cell whole-genome sequencing of primary tumor cells, metastatic lymph nodes, and CTCs from colorectal cancer.120 Patterns of copy number variation within primary tumor cells vary greatly, with less variation among CTCs. CTCs had a similar pattern of copy number variation to metastatic lymph nodes and to a subpopulation of primary tumor cells. This suggests that the metastases were derived from a small fraction of primary tumor cells that can enter the circulatory system. Single-cell transcriptome sequencing of CTCs in gastric cancer revealed their transcriptomic heterogeneity and indicated that most gastric CTCs undergo epithelial-mesenchymal transition (EMT).121
Single-cell transcriptome sequencing of CTCs can help researchers understand how patients respond to treatment. For example, in the work of Miyamoto et al., single-cell transcriptome sequencing of 77 CTCs in prostate cancer demonstrated heterogeneity in expression levels of estrogen receptor genes. In a follow-up retrospective study, the non-canonical Wnt signaling pathway was found to be activated in CTCs of patients treated with androgen receptor inhibitors, suggesting potential treatment resistance.122 Furthermore, in small cell lung cancer, Stewart et al. acquired CTCs before platinum–etoposide treatment, at maximum response and following relapse. Subsequent single-cell transcriptome sequencing showed that CTCs from relapsed patients identified more unique clusters, implying increased transcriptomic heterogeneity in CTCs following patient resistance.107
Other cancers
Several single-cell sequencing studies of patients with renal cell carcinoma (RCC) have achieved interesting results. Hu et al. performed single-cell sequencing of 12 RCC samples and nine para-tumor samples from three patients and revealed that the transcriptome of tumor cells was highly heterogeneous.116 In addition, the metabolism of cancer cells is abnormal, including hypoxia, lipid biosynthesis, and enrichment of localization pathways.123 Bi et al. mapped the single-cell RNA atlas of cancer and immune cells in patients with metastatic RCC before and after ICB treatment and revealed that the tumor cells could be divided into two subgroups: angiogenesis signals and the upregulation of immunosuppressive programs.124 In gastric cancer, Andor et al. determined the RNA expression profiles of thousands of single cells from gastric cancer tumor cell lines via scRNA-seq. The atlas revealed at least two subclones in each gastric cancer cell line, indicating strong transcriptome heterogeneity within these cells. The subclones of different cell lines exhibit differences in their enriched signaling pathways; however, all cells contain pathways related to genome mutation or evolution, similar to DNA repair mechanisms and metabolic pathways.124 In primary uveal melanomas, Pandiani et al. revealed intratumoral heterogeneity and identified HES6 as a driver of metastatic disease based on scRNA-seq.125 At the cellular level, single-cell sequencing can comprehensively describe cancer heterogeneity through molecular expression profiling, which cannot be achieved via bulk sequencing.126
Analysis of the complex immune microenvironment
Tumors contain not only malignant tumor cells but also various infiltrating and resident host cells, secreted factors, and extracellular proteins, collectively forming the TME.127 The TME is a complex and dynamic system that directly affects tumor immunity. Therefore, a comprehensive understanding of the TME is necessary for tumor therapy, particularly immunotherapy. The rapid development of single-cell omic technologies, specifically scRNA-seq, provides comprehensive information on the gene expression profile of individual cells, offering insights into the potential role of the TME. Herein, we summarize the key discoveries obtained using scRNA-seq to refine the complex cellular composition of the TME in numerous solid tumor types (Fig. 3 and Table 2).
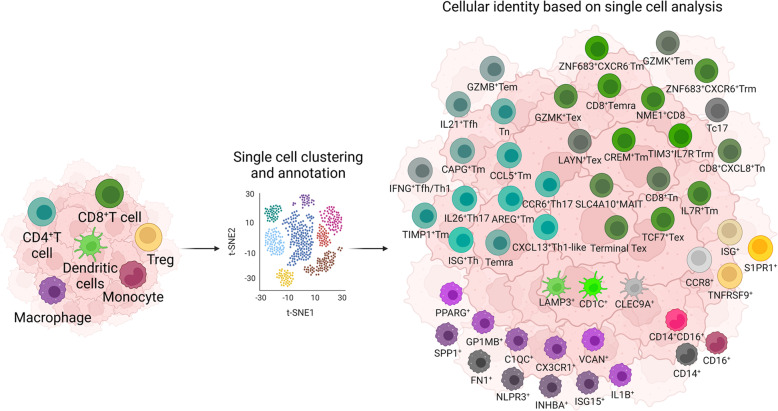
Fig. 3.
High-resolution characterization of tumor microenvironment (TME) by single-cell sequencing. Cellular architecture of the TME-infiltrated immune cells in the TME is broadly grouped using flow cytometry-based markers. Single-cell sequencing has made it possible to characterize the phenotypic heterogeneity of immune cells at the transcriptomic, proteomic, and epigenetic levels
Table 2.
Representative studies of high throughput single-cell sequencing in research of tumor immune microenvironment
| Tumor | Methods | Cases | Samples | Therapy/treatment | Main findings |
|---|---|---|---|---|---|
| NSCLC | scRNA-seq/scTCR-seq | 14 | Tumor/normal/blood | Treatment-naïve | Two pre-exhaustion CD8+ T cells were identified in NSCLC |
| scRNA-seq | 5 | Tumor/normal | Treatment-naïve | By comparing matched normal sites, a tumor environment atlas of NSCLC was constructed | |
| scRNA-seq | 30 | Tumor/normal | Treatment-naïve | PD-1 expressing Trm cells were more proliferative and cytotoxic than PD-1 expression non-Trm cells in NSCLC | |
| scRNA-seq | 44 | Tumor/normal/nLN/mLN/PE/mBrain | Treatment-naïve | In addition to drawing a landscape of LUAD TME, a cancer cell cluster deviated from the normal differential trajectory and was enriched at the metastasis site | |
| scRNA-seq | 42 | Tumor/normal | Treatment-naïve | Several rare cell types in the tumor site, such as follicular dendritic cells and T helper 17 cells, were identified, | |
| scRNA-seq | 30 | Tumor/normal/mLiver/PE/mLN/mBrain | Treatment-naïve/targeted-treatment | scRNA-seq of metastatic lung cancer revealed a rich and dynamic tumor ecosystem | |
| scRNA-seq | 7 | Tumor/blood | Treatment-naïve/chemo-treated | Mapping tumor-infiltrating myeloid cells in patients with NSCLC by scRNA-seq | |
| BC | scRNA-seq/WES/RNA-seq | 11 | Tumor | Treatment-naïve | Highly intratumoral heterogeneity tumor environment was shaped by tumor cells and immune cells in breast cancer |
| scRNA-seq/scTCR-seq | 8 | Tumor/normal/blood/LN | Treatment-naïve | Supported continuous activation model of T cells and disagreed with macrophage polarization model in breast cancer | |
| scRNA-seq/scTCR-seq/RNA-seq | 123 (2 for scRNA-seq) | Tumor | Treatment-naïve | Tissue-resident memory T cells were enriched in breast cancer and expressed high levels of immune molecule and effector proteins | |
| scRNA-seq/scTCR-seq | 14 | Tumor | Treatment-naïve | Drew an atlas of tumor microenvironment of TNBC and defined a novel TCR-expressing macrophage | |
| HCC | scRNA-seq/scTCR-seq | 6 | Tumor/normal/blood | Treatment-naïve | Eleven T cell clusters were defined, and several specific clusters such as exhausted CD8+ T cells were enriched in the HCC tumor site |
| scRNA-seq | 19 | Tumor | Treatment-naïve | Hypoxia-dependent VEGF was associated with tumor diversity and TME polarization. The cytotoxic capacity of T cells was lower in higher heterogeneity HCC | |
| scRNA-seq/scTCR-seq/CyTOF | 13 | Tumor/non-tumor/leading-edge | Treatment-naïve | Defined tumor-associated CD4/CD8 double-positive T cells in HCC and systematically analyzed the function of PD-1+ DPT in HCC | |
| scRNA-seq | 5 | Tumor | Treatment-naïve | Constructed a human liver cancer landscape in single-cell resolution | |
| scRNA-seq/scTCR-seq | 16 | Tumor/normal/blood/ascites/nLN | Treatment-naïve | Drew an immune cell atlas of HCC. A novel DC cluster with high expression of LAMP3 was defined, and it may regulate multiple immune cells | |
| scRNA-seq/RNA-seq | 48 (6 for scRNA-seq) | Tumor/normal/margin tissue | Treatment-naïve | Comprehensively analyzed tumor ILC composition and found that patients with higher IL-33 expression exhibited a higher ILC2/ILC1 ratio, indicating better prognosis | |
| Melanoma | scRNA-seq/WES | 19 | Tumor | Treatment-naïve | Demonstrated the tumor environment ecosystem and how scRNA-seq offers insights into the results |
| scRNA-seq/scTCR-seq | 25 | Tumor | Treatment-naïve | scRNA-seq analysis revealed gradual T cell dysfunction in melanoma and exhausted CD8+ T cells were proliferative and expanded cell cluster | |
| scRNA-seq/scDNA-seq/TCR-seq | 11 | Tumor | Treatment-naïve | Novel CD8+ T cells were observed with predominantly expressing LAG-3, rather than PD-1 or CTLA-4 | |
| CRC | scRNA-seq | 11 | Tumor/normal | Treatment-naïve | Two distinct CAF clusters were identified. Tumor-enriched CAF clusters highly expressed EMT-related genes |
| scRNA-seq/scTCR-seq | 12 | Tumor/normal/blood | Treatment-naïve | Developed STARTRAC indices to analyze the relationship, function, and clonality of 20 identified T cell clusters in CRC | |
| scRNA-seq | 1 | Tumor/normal | Treatment-naïve | Expression clustering identified six gene modules, and functional enrichment was associated with T cells and cancer cells | |
| scRNA-seq/scTCR-seq | 18 | Tumor/normal/blood | Treatment-naïve | Two specific macrophages and cDC clusters, which played a key role of cellular crosstalk in the CRC TME were identified | |
| scRNA-seq | 29 | Tumor/normal | Treatment-naïve | Provided the tumor environment landscape and intercellular communications in CRC | |
| Bladder cancer | scRNA-seq/scTCR-seq | 7 | Tumor/normal | Treatment-naïve | Found multiple tumor-specific CD4 + T cells. Cytotoxic CD4 + T cells in tumor site were highly proliferative and could kill autologous tumors in an MHC class II-dependent manner |
| scRNA-seq | 11 | Tumor/blood | Treatment-naïve | Constructed a cell atlas in bladder cancer and provided deep insights into the tumor microenvironment | |
| ICC | scRNA-seq | 4 | Tumor/adjacent tissue | Treatment-naïve/recurrent | Demonstrated intertumor heterogeneity of human ICC and provided information on intercellular crosstalk between tumor cells and vCAFs |
| Gastric cancer | scRNA-seq | 8 | Tumor/metaplasia/normal/blood | Treatment-naïve | Comparing to normal site, scRNA-seq analysis revealed tumor-enrichment immune cells, transcriptional states, and intercellular interactions in gastric cancer |
| ccRCC | Mass cytometry/RNA-seq | 47 | Tumor/normal | Treatment-naïve/treatment | By profiling 3.5 million single cells, the study developed an in-depth tumor microenvironment atlas of ccRCC and revealed potential biomarkers for therapy strategies |
| Nasopharyngeal | scRNA-seq/scTCR-seq | 3 | Tumor | Treatment-naïve | Provided insights into the tumor microenvironment at single-cell resolution and revealed heterogeneity of immune cells and various functional T cell clusters in NPC |
| Ovarian cancer | scRNA-seq | 9 | Tumor/normal/benign | Treatment-naïve | Identified specific cell clusters enriched in different grades of ovarian cancer |
| Pan-cancer | scRNA-seq/scTCR-seq | 14 | Tumor/normal/blood | Treatment-naïve | Together with published data, demonstrated that non-exhausted T cells from outside of the tumor can replace exhaustion T cells in responsive patients |
| scRNA-seq | 20 | Tumor/normal | Treatment-naïve | Provided an integration immune cell atlas across lung, breast, and ovarian cancers and revealed the complexity of stromal cells in different cancer types | |
| scRNA-seq/RNA-seq/Exome-seq | 48 | Tumor/normal/blood/LN | Treatment-naïve | Drew a pan-cancer myeloid landscape via scRNA-seq. Different sources of LAMP3+ DCs exhibited various transcription expression patterns, and TAMs were also diverse across cancer types |
NSCLC
Non-malignant cells in the NSCLC TME affect both tumor-promoting and tumor-suppressive activities.127–130 The identification of components, particularly the cell populations and their function in the TME, has yielded potential strategies for immunotherapy. Previous studies have revealed that treatment efficacy varies and partly depends on the number and properties of tumor-infiltrating lymphocytes.131–133
Several recent studies have focused on the complexity of T cells in NSCLC. Guo et al. first used a full-length single-cell mRNA-seq technique, Smart-seq2, to evaluate NSCLC biopsies from treatment-naïve patients to determine the baseline landscape of tumor-infiltrating lymphocytes.134 In addition to conventional CD4+ and CD8+ T cell clusters, two novel CD8+ T cell clusters were observed and defined as “pre-exhausted” cells, which exhibited low expression of exhaustion markers. One subset was significantly abundant in NSCLC and exhibited high expression of ZNF683, suggesting that it functions in NSCLC immunity.134 Another pre-exhausted subset was characterized by high expression levels of GZMK, which was associated with the “effector memory” phenotype.135 However, GZMK+CD8+ T cells with intermediate exhaustion markers are likely transferred into exhausted T cells.136 By comparing intratumoral and para-tumoral lung-derived T cells, Lambrechts et al. confirmed elevated glycolysis and suppressed oxidative phosphorylation coherent between T cell clusters.137 Moreover, the authors found that a CD8+ T cell cluster enriched at the tumor site exhibited strong IFN-γ and IFN-α responses, high granzyme expression, and expressed high levels of exhaustion markers (LAG3, TIGIT, PDCD1, and CTLA4). In another study, Clarke et al. reported a TIM3+IL7R-tissue-resident memory T cells (T rm) subset uniquely present at the tumor site and expressing high levels of PD-1 and other molecules linked to inhibitory functions; however, functionality analysis revealed that these cells were not exhausted.138 Compared with non-Trm cells, PD-1-expressing Trm cells were associated with the key effector cytokines IL-2, TNF, and IFN-γ as well as granzyme molecules. In addition to CD8+ T cells, recent studies demonstrated the depletion of natural killer cells139 and increased emergence of Tregs at the primary tumor site compared to at the normal site.128,137,140 Tregs persist in tumors and metastasis sites to suppress antitumor immunity in NSCLC.140 Wu et al. first reported a rare T cell cluster, Th17, in NSCLC with a high expression level of KLRB1 and observed a transitional phenotype from naïve cells to Tregs.141
Myeloid cells also play a pivotal role in tissue homeostasis and inflammation in lungs.140 Unsupervised clustering analysis and t-SNE plotting of myeloid lineage cells revealed a substantial difference between intra-tumoral and non-tumoral lung macrophages, indicating that they have a completely different composition.137 These results correspond with those of Kim et al.140 who revealed that tumor-associated macrophages in tumor sites are primarily derived from monocyte-macrophages rather than from tissue-resident macrophages. Moreover, the upregulation of the transcription factors IRF2, IRF7, IRF9, and STAT2 and downregulation of inflammatory enhancers such as Fos/Jun supported M2 polarization of tumor macrophages in NSCLC. Another study revealed that macrophages in treatment-naïve patients exhibit M2 polarization.142 Zilionis et al. mapped the myeloid landscape in human and murine lung tumors, revealing conserved populations across individuals and species;143 additionally, their work revealed limited overlap of myeloid populations between blood and tumors in patients. One subset of dendritic cells (plasmacytoid dendritic cells [DCs]) was rarely found in normal lung sites compared to tumor sites and metastatic lymph nodes and exhibited an immunosuppressive phenotype with the upregulation of LILR and GZMB but loss of CD86, CD83, CD80, and LAMP3.140 A rare cell type, defined as follicular DCs, was identified by Wu et al.141 Furthermore, Lambrechts et al. detected a fibroblast cluster highly expressing COL10A1 that was strongly enriched at the tumor site and exhibited a strong EMT signal.137
Breast cancer
The function of tumor-infiltrating lymphocytes in breast cancer remains unclear. Some studies have suggested that increasing number of tumor-infiltrating lymphocytes can improve patient survival, whereas other studies revealed contrasting results.144,145 scRNA-seq provided a cell-level landscape that can be used to further investigate the function of tumor-infiltrating lymphocytes in breast cancer.
A previous study of primary breast cancer grouped 175 immune cells into three clusters: T cells, B cells, and M2 macrophages, all exhibiting an immunosuppressive phenotype.146 In contrast, Azizi et al. profiled 47,016 CD45+ cells from treatment-naïve patients with breast cancer and revealed significant heterogeneity for both lymphoid and myeloid cells.147 Moreover, the observed continuum of T cell states indicated that canonical classification of T cell clusters oversimplifies the tumor environment of breast cancer. Treg clusters expressed similar patterns of anti-inflammatory, exhaustion, hypoxia, and metabolism genes, suggesting that the functions of different clusters are inconsistent. Savas et al. identified a Trm-like cluster highly expressing CD103 and occupying a large proportion of CD8+ T cells.148 This cluster expressed granzyme and immune checkpoint molecules, suggesting their cytotoxic ability and proinflammatory potential.
Breast cancer cells secrete various cytokines that influence myeloid cell differentiation and suppress antitumor immunity.149,150 Azizi et al. revealed that gene expression in M1 and M2 macrophages was positively correlated and frequently expressed in the same cells.147 This finding indicates that the polarization model cannot be applied to tumor-associated macrophages. To better understand the characteristics and capacity of myeloid-derived suppressor cells (MDSCs), Alshetaiwi et al. established an MDSC-specific gene set between G- and M-MDSCs. This is unique to normal myeloid counterparts from a murine breast cancer model; however, these results can also be transferred to the human context, with a conserved state of MDSCs between mouse and human suggested.151 IL1B, ARG2, CD84, WFDC17, and chemokine receptors (e.g., CCR2 and CXCR2) were included in this gene set, indicating an immunosuppressive function. Particularly, CD84+ MDSCs capable of T cell suppression and increased reactive oxygen species production were observed. Moreover, the spleen was detected as a major site of MDSC emergence in breast cancer.
Qiu et al. collected 9683 tumor-infiltrating immune cells from treatment-naïve patients with TNBC and identified several novel cell clusters, such as CD8+CXCL8+ naïve T cells. CXCL8 was observed primarily during the production of naïve CD4+ cells in the human peripheral blood or in infants.152 Consistent with its function in CD4+ T cells,153 differential gene expression and pathway enrichment analyses suggest that the cluster mediates neutrophil migration and activates MAPK/extracellular signal-regulated kinase pathways, which contribute to tumor growth. Notably, the number of double-negative T cells (CD3+CD4- CD8-) accounted for ~31.0% of all T cells in breast cancer but only 1–5% in healthy humans.154,155 Double-negative T cells play a key role in inflammation and autoimmunity.152,153 However, three independent clusters with high levels of effector markers (GZMA, GZMB, and IFN-γ), regulatory markers (FOXP3 and IL2RA), and naïve markers (CCR7) indicate that double-negative T cell function is important and complex in the TNBC microenvironment. Notably, a novel cluster of CD3- and T cell receptor (TCR)-positive macrophages was first observed in the TME of breast cancer. The upregulation of TCR signaling and cytotoxic effect of genes compared to TCR- macrophages indicates that these macrophages exert partial T cell functions. At the single-cell level, authors also observed a “pre-exhaustion” T cell cluster and high positive correlation of gene expression between M1 and M2 macrophages, supporting previous results.147
HCC
The functions of specialized immune cells in HCC, such as Kupffer cells (macrophages), innate lymphoid cells, and various T cells, are not well-understood.156,157 Researchers have recently focused on investigating immune cells using scRNA-seq.
Atlas analysis in HCC classified infiltrating T cells into 11 large subsets.158 Most clusters have been reported in other cancers, such as “pre-exhausted” T cells in NSCLC and breast cancer.134 A unique CD8+ cluster with positive expression of FOXP3 was defined as a Treg-like population and exhibited both suppressive and cell-killing characteristics. Zheng et al. also found that LAYN plays a regulatory role not only in Tregs,159 but also in CD8+ T cells in HCC. Furthermore, Ma et al. described the different functions of highly and poorly heterogeneous (Div-high and -low, defined by the authors) HCC based on the analysis of the gene expression pattern, pathway enrichment, and T cell cluster distribution.86 Cytotoxic-related genes (e.g., GZMA and GZMB) and immune checkpoint molecules (IFNG, PDCD1, and NKG7) were highly expressed in Div-low compared to in Div-high HCC. The upregulated pathways in Div-high tumors included EMT and myogenesis, whereas allograft rejection, oxidative phosphorylation, and fatty acid oxidation were upregulated in Div-low tumors. A more detailed analysis revealed that immunosuppressive Tregs were considerably higher in Div-high than in Div-low HCC. By evaluating a combination of tumor, leading edge, and normal samples, Zheng et al. found that the CD4 and CD8 double-positive T (DPT) cell cluster was enriched in the leading-edge region;160 however, this cluster generally exists in the thymus.161 The DPT cluster was divided into 11 subclusters based on canonical markers including cytotoxic DPT, memory DPT, activated DPT, NK-like DPT, MAIT-like DPT, and exhaustion DPT. Trajectory analysis revealed that the DPT was functional and well-differentiated T cells and in conjunction with TCR data, showed a common ancestry of PD-1+DPT and PD-1+CD8+ T cells.160
Several specialized and unique macrophage subsets have been reported in recent studies. MacParland et al. observed two distinct CD68+ macrophages in primary HCC samples, one with high expression levels of inflammatory markers (LYZ, CSTA, and CD74)162 and another with enriched expression of immunoregulator genes (e.g., MARCO, VSIG4, and CD163).163 Aizarani et al. also identified new subsets of Kupffer cells expressing CD1C or LIRB5, which shared gene expression and pathways with endothelial cells, suggesting functional cooperation.164 Zhang et al. found two distinct tumor-associated macrophages165 with high expression levels of SLC40A1 and GPNMB, by combining 10x Genomics and Smart-seq2.166 Consistent with recent studies of iron metabolism in macrophages,167,168 SLC40A1 encodes the iron exporter ferroportin and promotes proinflammatory cytokines, including IL-23 and IL-6, but suppresses IL1β production. In contrast to classical DCs (cDCs), LAMP3+ DCs may represent a common DC cluster with several interesting characteristics.166 Many ligands for T and NK cell receptors were expressed, suggesting that these receptors play immunoregulatory roles in lymphocytes. Moreover, signature genes of LAMP3+ DCs were strongly correlated with the signature of Treg or Tex, indicating that DCs contribute to T cell dysfunction. In contrast, LAMP3+ DCs can migrate from HCC tumors to lymph nodes and prime T cell migration to the tumor site.166
Due to the low abundance of innate immune lymphocytes, the studies on innate lymphoid cells (ILCs) in human liver cancer are limited. Heinrich et al. performed scRNA-seq analysis to draw a landscape and determine the role of ILCs in human HCC.169 Four canonical cell types, including ILC1, ILC2, ILC3, and NK-like, were identified; the authors also defined a CD127-NK-like cluster with an intermediate status between NK cells and ILC1s in HCC. Importantly, the first study that described the details of the conversion of ILC3s to ILC2s and ILC1s to ILC2s169 suggested IL-4 as a cytokine that drives such a transition.170,171 Overall, these findings demonstrate the diversity of immunoregulatory mechanisms in the HCC TME.
Melanoma
Recently, scRNA-Seq began to dissect TME heterogeneity in melanoma. Tirosh et al. identified core exhaustion signatures of T cells, including upregulation of coinhibitory (TIGIT) and costimulatory (TNFRSF9/4-1BB and CD27) receptors.172 In line with the findings for NSCLC and breast cancer,134,147 Li et al. noted that dysfunctional CD8+ T cells transitioned from early effector cells.173 Analysis of the TCR in peripheral blood mononuclear cells revealed dysfunctional processes at the tumor site. Additionally, dysfunctional CD8+ T cells are highly proliferative and dynamic. In contrast to the transitional state of dysfunctional CD8+ T cells, cytotoxic CD8+ T cells formed a discrete state with independent signatures and were unlinked with dysfunctional CD8+ T cells. Durante et al. drew a TME atlas at single-cell resolution in uveal melanoma and identified an unrecognized CD8+ T cell, which primarily expressed LAG3, an exhaustion-associated immune checkpoint molecule in CRC.174
By analyzing 333 individual DCs and monocytes from metastatic melanoma, Nirschl et al. revealed that homeostatic modules were enriched in monocytes and DCs and were positively correlated with IFN-γ signatures.175 Moreover, SOCS2, a member of the SOCS family that uniquely degrade all other members,176,177 was induced by IFN-γ, which is present on monocytes and part of a tissue signature during melanoma formation.
CRC
In contrast to previously summarized cancers, immunotherapy has shown limited advances in the treatment of patients with CRC,178 likely due to incomplete understanding of the TME in CRC.179,180 scRNA-seq is a powerful technique that can improve the capacity to excavate and understand the complexity of the TME in CRC.
Similar to NSCLC134 and HCC,158 Zhang et al. profiled the T cell atlas in CRC using Smart-seq2 and TCR-seq and identified 20 unique T cell clusters, including typical CD8+ and CD4+ T cell clusters. In contrast, additional T cell clusters, including Th17, follicular T helper cells, follicular T regulatory cells, and two CD8+ T cell clusters were identified in CRC.181 Among them, Tex cells, two IFNG+ Th1 cells, and one Treg cluster were enriched in the tumor. Focusing on the tumor subtypes, CXCL13+BHLHE40+ Th1-like cells were enriched in microsatellite instability tumors, whereas Th17 cells primarily existed in microsatellite stable tumors. Lee et al. also found that Th17 cells and Tregs were predominantly present at the tumor site, whereas γδT cells were enriched at the normal tissue site.182 Zhang et al. observed that CD8+ T cells, Th1/Th2 cells, and memory T cells were increased at the tumor site, whereas CD4+ T cells and Tregs were decreased.183 Based on enrichment analysis results, the imbalance in T cell clusters may be affected by T cell proliferation, activation/differentiation, and TCR signaling.
Lee et al. suggested that the immunosuppressive function of myeloid cells is enhanced because of their expansion in CRC.181 SPP1+ macrophages were more abundant in tumor sites than in normal sites. Previous studies have revealed that SPP1+ macrophages play a central role in both the pro- and anti-inflammatory phenotype,184–186 which is consistent with the findings of Lee et al. In addition to T cells, Zhang et al. combined 10x genomics and Smart-seq2 to draw a transcriptome landscape of CRC immune cells, focusing on myeloid cells.187 Two distinct TAM populations were identified: C1QC+ and SPP1+ TAM. Consistent with previous studies of other cancers, these clusters could not be explained by either the M1 or M2 phenotype.147,188 C1QC+ TAM was primarily associated with phagocytosis and antigen presentation, whereas SPP1+ TAM was significantly enriched in angiogenesis regulation. Moreover, compared to the normal site, SPP1+ TAM exhibited greater enrichment at the tumor site. In addition to SPP1, Zhang et al. found that GPNMB, which was reported to mediate MDSCs and inhibit T cells,189 was highly expressed in granulocytes.183 The authors also revealed that IL-17 signaling and ferroptosis pathways were enriched in granulocytes at the tumor site. The IL-17 pathway may play a role in CRC liver metastasis and the ferroptosis pathway has been reported to mediate reactive oxygen species production and p53 downstream effectors during cancer cell death.190,191
Li et al. developed a novel algorithm named as reference component analysis and identified two distinct clusters of cancer-associated fibroblasts (CAFs), CAF-A and CAF-B.192 CAF-A may be an intermediate cluster between normal fibroblasts and CAF-B, as it expresses genes related to extracellular matrix remodeling, whereas CAF-B expresses known markers of activated myofibroblasts. In addition, EMT-related genes are highly expressed in CAFs.
Other cancers
With the widespread use of scRNA-seq technology, an increasing number of TME from different cancer types have been studied. Oh et al. found that the composition of CD8+ T cells did not differ between bladder tumor and non-malignant tissues.193 In contrast, tumor-specific Tregs were identified with high expression levels of IL2RA and immune checkpoint molecules. Multiple cytotoxic CD4+ T cell clusters were clearly defined using canonical markers, suggesting that these cells kill autologous tumor cells. Chen et al. found that monocytes were enriched in the M2 state of the bladder tumor region; moreover, a cluster of DCs with high LAMP3 expression was correlated with Treg recruitment, indicating the regulation of immunosuppressive formation.194 Zhang et al. employed scRNA-seq to create the transcriptomic landscape of intrahepatic cholangiocarcinoma and identified six distinct types of fibroblasts.195 The main fibroblast cluster was vascular CAFs, which expressed high levels of IL-6, thereby enhancing the malignancy of the tumor cells. In gastric cancer, Sathe et al. found that two cytotoxic T lymphocyte clusters highly expressed exhaustion markers, with one cluster exhibiting higher proliferation potential than the other. Macrophages cannot simply be classified as M1 or M2 macrophages because the canonical markers are co-expressed in the same cluster. Additionally, the authors identified a cluster of DCs enriched at the tumor site expressing chemokines such as CCL22, CCL17, CCL19, and IL32, which are associated with naïve T cell recruitment.196 By profiling 3.5 million cells from 73 patients with clear cell RCC (ccRCC), Chevrier et al. constructed an immune atlas with 22T cell clusters and 17 macrophage clusters. Moreover, the authors found that CD38, a mediator of nitric oxide,197 is a potential exhaustion marker of T cells in ccRCC. As the first landscape of TME in nasopharyngeal carcinoma (NPC), Zhao et al. observed that the proportion of B cells was higher than that of T cells, particularly in Epstein–Barr virus-negative patients.198 A study reported that malignant cells secrete mediators (e.g., miR-21) to expand B cells, which suppress CD8+ T cells.199 Thus, B cells may be a useful immunotherapeutic target in NPC.198 Shih et al. recently revealed that myeloid cells of both primary and metastatic ovarian cells and fibroblasts of metastatic ovarian cells produced increased levels of secreted factors, indicating that these cells play important roles in tumor growth.200 Additionally, B cells and T cells did not express high levels of pro-tumor genes such as IL-10, STAT3, and CCL22 but expressed genes that influenced complement pathways.
Pan-cancer analysis
Several studies have recently concentrated on excavating the TME with multiple tumors and integrating scRNA-seq data. Wu et al. identified two T cell clusters that did not match published studies; they collected 141,623 T cells from four different tumor types, including NSCLC, uterine corpus endometrial carcinoma, CRC, and RCC.135 One cluster expressed high levels of the long noncoding RNA MALAT1 and chromatin remodeling enzyme CHD1. Another cluster was enriched in mitosis genes. Neither cluster had high scores on Trm cell signature, which was considered as the origin of exhausted T cells.
Qian et al. collected 233,591 single cells from lung, colon, ovarian, and breast cancer tumors.201 Overall, authors identified 22 tumor-specific clusters, including mast cells, germinal center-independent B cells, and neutrophils. The new cDC2 subset expressed CD1C and Langerhans cell-specific markers, such as CD207 and CD1A. In T cells, clustering was not affected by CCA, suggesting that T cell distribution was not tumor-specific. However, both B cells and T cells were more enriched in LC than in OV and CRC, likely because LC is a hot tumor.
Zhang et al. collected myeloid cells from 15 different types of tumors and identified distinct features of myeloid cells across the tumor types.202 However, NPC possessed a higher proportion of mast cells and was the only tumor with higher TNF+ mast cells than VEGFA+ , indicating a stronger antitumor function. The authors also confirmed a previously reported cDC subset,166 LAMP3+ cDCs, which widely exist across all 15 tumor types. Although both conventional type 1 dendritic cells (cDC1) and cDC2 could differentiate into cDC3, more LAMP3+ cDCs were derived from cDC1, except in pancreatic adenocarcinoma and NPC. cDC1-derived LAMP3+ cDCs highly expressed IL12B and BTLA, which induced the differentiation of T helper 1 cells and Treg cells, respectively.203,204 In contrast, CCL17, a chemokine that recruits Tregs into tumors,205 was specifically expressed by cDC2-derived LAMP3+ DCs. Macrophage clusters from different tumor types were diverse; additionally, SPP1+, C1QC+, ISG15+, and FN1+ TAMs were primarily enriched at the tumor site. The authors also found that both M1 and M2 gene signatures were co-expressed in the TAM clusters of all tumor types, suggesting a limitation of the in vitro polarization model in the TME. Notably, SPP1 can be considered as a marker gene of macrophages with angiogenesis function across eight tumor types, including BRCA, pancreatic adenocarcinoma, lung cancer, CRC, uterine corpus endometrial carcinoma, NPC, OV, and THCA.
Immune microenvironment, dynamic phenotypic changes, and response to immunotherapy by single-cell sequencing
Immune checkpoint blockade (ICB) immunotherapy has introduced a new era of antitumor treatment. It is known that responsiveness to ICB treatment is determined by the preexisting TME and peripheral immuno-compartment of patients. Single-cell analysis is being exploited to investigate whether certain cell populations with multi-parameter defined identities are related to responsiveness or resistance to ICB treatment. Applying single-cell sequencing to predictive biomarker analysis has revealed several translational and clinical insights into ICB-induced tumor control across a range of tumor types. These include melanoma,23,206 NSCLC,207 glioblastoma,208 renal cell carcinoma, squamous cell carcinoma,209 bladder cancer,207 prostate cancer,210 breast cancer,211 and urothelial cancer.212
Several studies in pre-clinical mouse models have indicated the critical role of the CXCR5/TCF1+ subset of CD8+ T cells in sustaining a prolonged response to ICB immunotherapy.213–217 Studies have demonstrated stem cell-like properties in these cells, characterized by lower expression levels of inhibitory checkpoint molecules such as PD-1, LAG3, TIM-3, and 2B4, and potent self-renewal capacity in the tumor niche. From a phenotypic perspective, this CD8+ T cell subset displayed common properties with tumor-residing GZMK+ effector memory CD8+ T cells and MKi67+ expanded CD8+ intratumoral T cells. These support the effector memory origin of these cells.134,158,181 These findings are consistent with the fact that stem cell-like or memory-like intratumoral CD8+ T cells are crucial for effective tumor immunology218 and immunotherapy.219 In response to ICB treatment, while maintaining self-renewal, stem cell-like CD8+ T cells differentiate to yield enough cytotoxic cells with negative TCF1 and high inhibitory checkpoint expression. The CXCR5/TCF1+CD8+ T cell subset is indispensable for successful ICB immunotherapy. In an ICB-treated B16-bearing mouse model, diphtheria toxin-mediated ablation of TCF1+CD8+ T cells could not completely abolish tumor control. This suggests that TCF1+CD8+ T cells are not the only important subset in ICB treatment.216 In addition to TCF1+CD8+ stem cell-like memory cells, tissue-resident memory T cells (Trm) can also be important for supporting T cell reinvigoration during ICB immunotherapy.
CD4 T cells are the hub components of the fine-tuned network of the adaptive immune response. Accumulating evidence indicates that in addition to CD8+ T cells with direct cytolytic activity, CD4+ T cells play an orchestrated role in modulation of tumor immunology and immunotherapy. Facilitated by single T cell analysis by RNA sequencing and TCR tracking (STARTRAC) analysis, Zhang et al. found that Th1 cells are abundant in MSI and sparse in MSS-CRC patients; this is closely associated with responsive or resistant ICB treatment.181 Conversely, Th17 cells, showing a paradoxical function in tumor immunology, were significantly enriched in patients with MSS-, but not in patients with MSI-CRC. The balance between Th1/Th17 cell polarization in bone metastatic TME of castration-resistant prostate cancer is associated with sensitivity to anti-PD-1/anti-CTLA-4 treatment.220 Intraosseous intratumoral CD4+ T cells tend to polarize to the Th17 lineage rather than to the Th1 lineage in the subcutaneous lesion via an osteoclast-associated TGF-β mechanism. In addition to the classical PD-1/PD-L1/CTLA-4-targeting drug, Th1-like CD4+ T cells play a critical role in conferring responsiveness to anti-CD40 immunotherapy in both preclinical models and clinical cohorts.187 Single-cell sequencing analysis suggests crosstalk between these BHLHE40+ Th1-like cells, high in IFN-γ- and CXCL13-producing cells, with cDC1 to synergize with anti-PD-1 administration.
In addition to several helper T cell populations, the presence of MHC-II-restricted cytotoxic CD4+ T cells has been documented in various solid tumor types.134,158,181,193,221–223 In the case of tumor immunotherapy for patients with bladder cancer, recent single-cell analysis demonstrated a critical role of cytotoxic CD4+ T cells in exerting anti-PD-L1-mediated tumor elimination in an MHC-II-restricted manner.193 In tumor-bearing mouse models, CD4+ T cells can harbor the properties of both cytotoxic and helper cells at the same time.224 These multifaceted CD4+ T cells are characterized by high expression of cytolytic mediators (GzmB, IFN-γ, and TNF-α), which are independently controlled by T-bet and Blimp-1.
The classic model of tumor immunology holds that tumor-specific naïve T cells are mainly primed in the tumor-draining lymph node. This leads to expression of activation markers, differentiation to effector phenotypes, and recruitment to the tumor site. In addition to local antitumor immunity in the TME, a systemic immune response was evident following effective ICB therapy.225 In an organism-wide genetically engineered model, Spitzer et al. demonstrated that tumor eradication following ICB therapy requires immune activation in the peripheral compartment.225 Moreover, emerging evidence further supports that T cells from adjacent or peripheral tissues are essential for effective responses to ICB immunotherapies.135,226–228 Single-cell sequencing analysis is being used to investigate the association between the response to ICB therapy, preexisting T-cell reinvigoration, and with recruitment of novel peripheral tumor-specific T cells. Among the contributions from “local reinvigoration” versus “peripheral recruitment,” T cell repertoire tracking strategy indicates a clonal replacement mechanism (“peripheral recruitment”) in response to ICB therapy. This consists of a terminally differentiated phenotype of intratumoral tumor-specific T cells. In terms of the CD4+ T cell subset, peripheral CD4+ T cells are also implicated in mediating antitumor immunity when treated with anti-CTLA-4 antibodies, either from mouse models or patients with metastatic melanoma.225
Intra-tumoral cell–cell communications
Many studies have focused on identifying novel cell types or determining the roles of distinct immune cell types in the TME (Fig. 4 and Table 3). However, tumor microenvironments are composed of heterogeneous tumors and immune cells which interact with each other to modulate the cellular network.229 In addition to analyzing the characteristics and heterogeneity of the TME, scRNA-seq can be used to infer cell–cell communication between different cell types. Several methods, such as CellPhoneDB,22 CellChat,21 and NicheNet,230 were recently developed for evaluating cellular communication. For inferring inter-cellular communication, the algorithms generally compare the expression level of receptor expression from one cell population and corresponding ligand expression on another cell population with a known gene list of ligand and receptor pairs.231–234
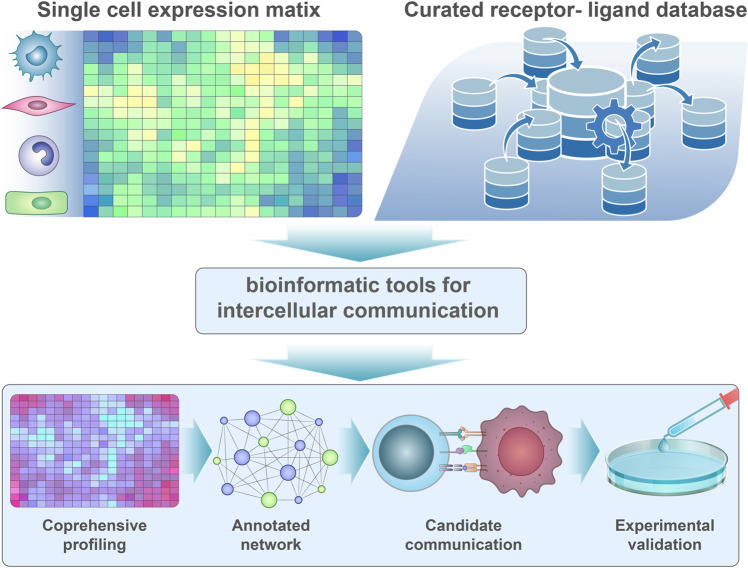
Fig. 4.
Inferring inter-cellular communication by single-cell sequencing. Inter-cellular contact or transfer of informative material is essential for coordinating the antitumor immune response and the malignant phenotype of tumor cells. Dissecting inter-cellular communication with single-cell sequencing analysis is instructive in understanding active signaling pathways between different cell types, which could eventually be applied to construct a communication network in the tumor immune microenvironment
Table 3.
Summary of principle and tools for investigation of intercellular communication by single cell sequencing
| Method | Tools | Platform | Characteristic |
|---|---|---|---|
| Differential combinations | CellTalker | R | 1. Differential ligand–receptor pairs can be calculated. 2. Capture highly abundant ligand–receptor pairs via comparative analysis. |
| iTALK | R | ||
| PyMINEr | Python | ||
| Expression permutation | CellChat | R and Web | 1. Discard random noise results via permutation test. 2. Cluster-to-cluster communication is inferred. |
| CellPhoneDB | Python and Web | ||
| Giotto | R | ||
| ICELLNET | R | ||
| SingleCellSignalR | R | ||
| ProxmID | Software | ||
| CSOmap | Matlab | ||
| Cell2Cell | Python | ||
| MISTY | R | ||
| stLearn | Python | ||
| SVCA | R and Python | ||
| Graph or network | CCCExplorer | Software | 1. With a prior model, the inference is beyond ligand–receptor interactions and incorporates intracellular signaling. 2. Inference of cell-to-cell communication is possible. 3. Signaling pathway information can also be used. |
| NicheNet | R | ||
| SoptSC | Matlab and R | ||
| SpaOTsc | Python | ||
| COMUNET | R | ||
| NATMI | Python | ||
| Tensor based | scTensor | R | Detect many-to-many CCC involving multiple cell clusters rather than one-to-one CCC. |
Based on this database, Ji et al. found that tumor-specific keratinocytes, a subset of keratinocytes in cutaneous squamous cell carcinoma, interact with CAFs, endothelial cells, macrophages, and MDSCs, which influence autocrine and paracrine responses.235 Additional ligands from TAMs and MDSCs to tumor-specific keratinocytes contained ITGA3 and ITGB1, indicating their association with EMT and epithelial tumor invasion.
In bladder cancer, inflammatory CAFs (iCAFs) have the largest number of potential ligand–receptor pairs with other cell types, exhibiting particularly strong interactions with endothelial cells.194 VEGFA, VEGFB, and FGFR1, produced by iCAFs, bind to endothelial and tumor cells, suggesting that iCAFs promote the proliferation of tumor and stromal cells. Additionally, the receptor CXCL12 is highly expressed in iCAFs and affects immune cell infiltration.
Zhang et al. constructed a computational model by combining scRNA-seq and The Cancer Genome Atlas bulk RNA-seq data to examine cell–cell communication in CRC.187 The authors first identified classical cell–cell interactions between fibroblasts and endothelial cells in the tumor and between follicular B cells and Tfh cells in a normal site, indicating that the identification of cell–cell communication using the model was robust and meaningful. In the tumor tissue, TAMs and cDCs became the core of communications, and C1QC+ TAMs and two groups of cDCs interacted with T cell subsets, implicating the regulation of antitumor T cell function. Another study of CRC identified several interactions between tumor cells and myofibroblasts, SPP1+ macrophages, CD8+ T cells, and Treg cells.182 Notably, the connection between IgA+ plasma cells and CD4+ T cells, which supports the mucosal antibody response at the normal site, was weaker than that at the tumor site. Furthermore, the comparison of the CMSs revealed that tumor cells play important roles in the formation of TME.
In gastric tumors, Sathe et al. discovered that the interaction between stromal cells and other cell types exhibited the highest enrichment.196 Ligands on stromal cells were connected to EGFR and MET on epithelial cells to promote tumor proliferation. LGR4-RSPO3 is a paired ligand–receptor on epithelial cells and fibroblasts that may regulate stemness.
Zhang et al. investigated cell–cell communication between lymphocytes in HCC, specifically T cells and DC cells.166 All DC subsets communicated with Tex cells, proliferative T cells, and Treg cells via the CD28/B7 family and IL-15. LAMP3+ interacts with multiple T cells through various ligand–receptor pairs, such as CCL19-CCR7 for CD4+ T cells, PD-L1-PD-1 for Tex cells, and effector memory T cells, suggesting that LAMP3+ DCs influence T cell function. Additionally, LAMP3+ DCs communicate with NK cells through IL-15 and NECTIN2, suggesting that these molecules regulate NK cells.
In head and neck squamous cell carcinoma, Cilo et al. mapped possible cell–cell interactions in human papillomavirus (HPV)- and HPV+ HNSCC, respectively.236 Almost all immune cell types from HPV- tumors showed unique ligand–receptor pairs versus other cell types, whereas unique communications of HPV+ tumors were predicted across pDCs, CD14+ cells, CD16+ cells, and DCs.
Kim et al. demonstrated that a novel cancer cell type, tS2, strongly interacts with myeloid and stromal cells in the progression or metastases of lung adenocarcinoma.140 The most important communication was between tS2 and mo-Macs, whereas the most important communication in the immune cell network was between mo-Macs and CD8+ T cells. In primary lung adenocarcinoma, ligand–receptor pairs of growth factors were the most significant between mo-Macs and tS2 cells. Notably, communication between mo-Macs and exhausted CD8+ T cells was more complex and included both activating pairs and inhibitory pairs.
Overall, an increasing number of studies have focused on cell–cell communication within the tumor microenvironment or between the tumor and immune cells. These studies have improved understanding of the TME and provided opportunities to develop immunotherapy strategies
Limitations of single-cell sequencing in cancer research
With continuous advancements in high-throughput omic technology, various bioinformatic tools have recently emerged. At the single-cell level, high-throughput genomics, transcriptomics and epigenomics have been continuously improved. However, these technical methods have limitations that must be overcome.
First, at the transcriptome level, scRNA-seq data are inherently noisy and sparse. Eukaryotic transcription occurs in a pulsed manner rather than at a consistent basal rate.237,238 Therefore, when a “0” value is obtained in sequencing results, it may not indicate an inactive state in the cell. In fact, the value of “0” may also imply that the gene is activated but not detected due to burst kinetics, improper sampling time, or technical defects.
Secondly, cells captured using single-cell technology may not represent cells in the body. For example, during the processing of brain tissue into a single-cell suspension, neurons are more likely to be lost than glial cells, leading to deviation between cell composition obtained in the analysis and actual cell composition.239,240 In response to this situation, single-nucleus analysis has become a useful alternative. This approach effectively reduces the loss of specific cell types or interference with gene expression but shows lower transcriptome coverage. The specific sample collection and processing procedures also affect the results. scRNA-seq relies on qualified single-cell preparations, which can be obtained from peripheral blood but are typically not easy to isolate from solid tumor mass. In addition, differences in single-cell separation methods and sample storage conditions affect sequencing results.241 For example, in blood samples, inconsistencies in the blood draw time can affect cells transcripts and even lead to the changes in the expression of certain key genes (such as specific markers of immune cells).242
In addition, 10X scRNA-seq is one of the most popular single-cell techniques. It is a high-throughput method and uses 5′ or 3′ primers to capture RNA. Somatic single nucleotide variations (SNVs) far from transcript ends due to end sequencing are often difficult to capture. When bioinformatics tools (Freebayes, copyKAT etc.) are used to infer SNVs or copy number variations (CNVs) in these data, the results may be slightly flawed.243,244 Therefore, methods such as Smart-seq or FLASH-seq that capture the whole transcriptome are better in this regard.245,246 Genotyping of Transcriptomes based on the 10x Genomics platform could be an effective solution to this deficiency.247 The existence of mature single-cell DNA sequencing technologies (such as Tapestri solution from Mission Bio) must also be mentioned; these may be the best way to accurately obtain genotypes (SNVs, CNVs etc.) at the single-cell level.248
The preparation of single-cell suspensions is an essential step in single-cell omics, however cells lose spatial information during this process. Spatial multi-omics (including spatial transcriptomics249,250 and spatial epigenomics251) allows researchers to understand the locational context of these cells in tissues while acquiring omics data. Unfortunately, spatial multi-omics has not yet reached the level of single-cell resolution.252 ST further accelerates the generation of multimodal omics measurements. Multimodal omics implies that multiple modalities up to thousands of single cells can be measured simultaneously in the same experiment.235,253 This technology has significant advantages in study design and bioinformatic analysis. The data coverage of single-cell multi-omics in the measurement is similar to that of scRNA-seq but is sparse and noisy. Therefore, the development of biochemical and molecular-based detection tools is the basis for improving the sensitivity, specificity, and robustness of multimodal omics. Currently, high-throughput multimodal omics analysis remains limited to certain types of omic measurements that are mostly based on RNA. However, high-throughput multimodal omics has great potential, and researchers are developing methods for processing massive numbers of cells in a single experiment.
In both single omics or multimodal omics, powerful computing resources and efficient tools are required to analyze large-scale, high-dimensional data. Different single-cell sequencing technologies have varying sequencing coverage and cell capture efficiencies. Single-cell sequencing platforms and species exhibit substantial variation in heterogeneous resolution, sensitivity, and variability. This greatly limits cross-experiment queries or the establishment of a comprehensive database. At present, more powerful statistical and calculation tools are required so that an increasing number of single-cell data can be better integrated and fully utilized. In addition, rapid and accurate integration of cell metabolomics, proteomics, transcriptomics, epigenetics, and genomics is an extremely complex task. Some existing methods include dimension reduction-based approaches,38 similarity-based approaches,254 and statistical modeling-based approaches,255 a considerable number of which target scRNA-seq. Under the integration requirements of low batch scRNA-seq, the ComBat or Harmony methods perform better,256,257 while on the atlas-level scRNA-seq data integration task, scANVI, Scanorama, scVI, and scGen perform well. Batch effects from scATAC-seq data can be effectively removed with LIGER and Harmony.258 In addition, methods of Single-Cell Multi-omic Integration have been further developed, including LIGER (for gene expression, epigenetic, or spatial data) and WNN (for CITE-seq data).259,260
Finally, the cost of single-cell sequencing is expensive. Before applying single-cell sequencing technology, its technical advantages should be considered. The appropriate number of single-cell sequencing platforms should be selected for the subject, as this is important for supporting hypothesis-driven and well-designed research.
Conclusion
The rapid development of single-cell sequencing technology and analytical tools makes it possible for oncologists to understand the complexity of the tumor immune microenvironment and the resultant antitumor immune response. The application of single-cell sequencing in tumor immunotherapy will substantially enhance the ability of researchers to discover promising targets to overcome immuno-resistance, investigate the signaling pathway and cellular response caused by these drugs and determine the optimal regimen of immuno-combinational therapy in clinical practice. For research on pre-clinical models, single-cell sequencing enables comprehensive characterization of cellular composition and temporal evolution of tumor cells and infiltrated immune and stromal cells. This helps in validating the pathological relevance of disease models and identifying promising targets for drug development. In terms of translation application, the high-dimensional phenotypic information obtained by single-cell sequencing can be used to identify predictive biomarkers in immunotherapy and propose an instructive companion diagnostic strategy for further clinical testing. We anticipate that single-cell sequence analysis will become an indispensable tool in on the fields of tumor immunology and immunotherapy. The massive phenotypic data and biological insights generated from this study will substantially accelerate the progress of antitumor treatment and improve clinical outcomes for patients with cancer.
Acknowledgements
Author contributions
Q.J., H.C., and Z.J. draft the manuscript and draw the figures; B.Z. revised the manuscript. All authors critically reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Qingzhu Jia, Han Chu, Zheng Jin.
Contributor Information
Haixia Long, Email: longhaixialhx@163.com.
Bo Zhu, Email: bo.zhu@tmmu.edu.cn.
References
- 1.Jia Q, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat. Commun. 2018;9:5361. doi: 10.1038/s41467-018-07767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilkington EH, et al. From influenza to COVID-19: lipid nanoparticle mRNA vaccines at the frontiers of infectious diseases. Acta Biomater. 2021;131:16–40. doi: 10.1016/j.actbio.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalli FMG, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–754 e736. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia Q, et al. Tracking neoantigens by personalized circulating tumor dna sequencing during checkpoint blockade immunotherapy in non-small cell lung cancer. Adv. Sci. 2020;7:1903410. doi: 10.1002/advs.201903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navin N, Hicks J. Future medical applications of single-cell sequencing in cancer. Genome Med. 2011;3:31. doi: 10.1186/gm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 8.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 10.Vitak SA, et al. Sequencing thousands of single-cell genomes with combinatorial indexing. Nat. Methods. 2017;14:302–308. doi: 10.1038/nmeth.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casasent AK, et al. Multiclonal invasion in breast tumors identified by topographic single. Cell Seq. Cell. 2018;172:205–217 e212. doi: 10.1016/j.cell.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg AB, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo F, et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 2017;27:967–988. doi: 10.1038/cr.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 15.Zhu C, Preissl S, Ren B. Single-cell multimodal omics: the power of many. Nat. Methods. 2020;17:11–14. doi: 10.1038/s41592-019-0691-5. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Mo S, Li X, He Y, Yang J. Single-cell RNA-seq reveals the immune escape and drug resistance mechanisms of mantle cell lymphoma. Cancer Biol. Med. 2020;17:726–739. doi: 10.20892/j.issn.2095-3941.2020.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Miguel M, Calvo E. Clinical challenges of immune checkpoint inhibitors. Cancer Cell. 2020;38:326–333. doi: 10.1016/j.ccell.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Triana S, et al. Single-cell proteo-genomic reference maps of the hematopoietic system enable the purification and massive profiling of precisely defined cell states. Nat. Immunol. 2021;22:1577–1589. doi: 10.1038/s41590-021-01059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulati GS, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science. 2020;367:405–411. doi: 10.1126/science.aax0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manno GLa, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin S, et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 23.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013 e1020. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 25.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dillies MA, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 27.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haque A, Engel J, Teichmann SA, Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Z. Cui, F. Wang, C. Zhao, L. Zou, Q. Goals and approaches for each processing step for single-cell RNA sequencing data. Brief Bioinform.22, bbaa314 (2021). [DOI] [PubMed]
- 30.Kang HM, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 2018;36:89–94. doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng J, Liao J, Shao X, Lu X, Fan X. Multiplexing methods for simultaneous large-scale transcriptomic profiling of samples at single-cell resolution. Adv. Sci. 2021;8:e2101229. doi: 10.1002/advs.202101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z. et al. Critical downstream analysis steps for single-cell RNA sequencing data. Brief Bioinform.22, bbab105 (2021). [DOI] [PubMed]
- 33.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 38.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan X, et al. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021;22:195. doi: 10.1186/s13059-021-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 43.Telenius H, et al. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-K. [DOI] [PubMed] [Google Scholar]
- 44.Spits C, et al. Whole-genome multiple displacement amplification from single cells. Nat. Protoc. 2006;1:1965–1970. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 45.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong X, et al. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat. Methods. 2017;14:491–493. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bohrson CL, et al. Linked-read analysis identifies mutations in single-cell DNA-sequencing data. Nat. Genet. 2019;51:749–754. doi: 10.1038/s41588-019-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hard J, et al. Conbase: a software for unsupervised discovery of clonal somatic mutations in single cells through read phasing. Genome Biol. 2019;20:68. doi: 10.1186/s13059-019-1673-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Chen H, Zhang NR. DNA copy number profiling using single-cell sequencing. Brief. Bioinform. 2018;19:731–736. doi: 10.1093/bib/bbx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallory XF, Edrisi M, Navin N, Nakhleh L. Assessing the performance of methods for copy number aberration detection from single-cell DNA sequencing data. PLoS Comput. Biol. 2020;16:e1008012. doi: 10.1371/journal.pcbi.1008012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garvin T, et al. Interactive analysis and assessment of single-cell copy-number variations. Nat. Methods. 2015;12:1058–1060. doi: 10.1038/nmeth.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cusanovich DA, et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 2018;174:1309–1324 e1318. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satpathy AT, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buenrostro JD, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell. 2018;173:1535–1548 e1516. doi: 10.1016/j.cell.2018.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan F, Powell DR, Curtis DJ, Wong NC. From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 2020;21:22. doi: 10.1186/s13059-020-1929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Granja JM, et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 2021;53:403–411. doi: 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang R, et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 2021;12:1337. doi: 10.1038/s41467-021-21583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuart T, Srivastava A, Madad S, Lareau CA, Satija R. Single-cell chromatin state analysis with Signac. Nat. Methods. 2021;18:1333–1341. doi: 10.1038/s41592-021-01282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rotem A, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaya-Okur HS, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ai S, et al. Profiling chromatin states using single-cell itChIP-seq. Nat. Cell Biol. 2019;21:1164–1172. doi: 10.1038/s41556-019-0383-5. [DOI] [PubMed] [Google Scholar]
- 62.Nakato R, Shirahige K. Statistical analysis and quality assessment of ChIP-seq data with DROMPA. Methods Mol. Biol. 2018;1672:631–643. doi: 10.1007/978-1-4939-7306-4_41. [DOI] [PubMed] [Google Scholar]
- 63.Karemaker ID, Vermeulen M. Single-cell DNA methylation profiling: technologies and biological applications. Trends Biotechnol. 2018;36:952–965. doi: 10.1016/j.tibtech.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 64.Zhao LY, Song J, Liu Y, Song CX, Yi C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell. 2020;11:792–808. doi: 10.1007/s13238-020-00733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angarica VE, Del Sol A. Bioinformatics tools for genome-wide epigenetic research. Adv. Exp. Med. Biol. 2017;978:489–512. doi: 10.1007/978-3-319-53889-1_25. [DOI] [PubMed] [Google Scholar]
- 66.de Souza CPE, et al. Epiclomal: probabilistic clustering of sparse single-cell DNA methylation data. PLoS Comput. Biol. 2020;16:e1008270. doi: 10.1371/journal.pcbi.1008270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13:R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tarabichi M, et al. A practical guide to cancer subclonal reconstruction from DNA sequencing. Nat. Methods. 2021;18:144–155. doi: 10.1038/s41592-020-01013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salehi S, et al. ddClone: joint statistical inference of clonal populations from single cell and bulk tumour sequencing data. Genome Biol. 2017;18:1–18. doi: 10.1186/s13059-017-1169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eirew P, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woll PS, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25:794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 73.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jan M, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 2012;4:149ra118–149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinreb C, Wolock S, Tusi BK, Socolovsky M, Klein AM. Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl Acad. Sci. 2018;115:E2467–E2476. doi: 10.1073/pnas.1714723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karamitros D, et al. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nat. Immunol. 2018;19:85–97. doi: 10.1038/s41590-017-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Velten L, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 2017;19:271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Galen P, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176:1265–1281. e1224. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akinduro O, et al. Proliferation dynamics of acute myeloid leukaemia and haematopoietic progenitors competing for bone marrow space. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-017-02376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hira VV, Van Noorden CJ, Carraway HE, Maciejewski JP, Molenaar RJ. Novel therapeutic strategies to target leukemic cells that hijack compartmentalized continuous hematopoietic stem cell niches. Biochimica et. Biophysica Acta (BBA)—Rev. Cancer. 2017;1868:183–198. doi: 10.1016/j.bbcan.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 81.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 82.Stetson L, et al. Single cell RNA sequencing of AML initiating cells reveals RNA-based evolution during disease progression. Leukemia. 2021;35:2799–2812. doi: 10.1038/s41375-021-01338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang H, Choe S, DiNardo C D, et al. Complex polyclonal resistance mechanisms to Ivosidenib monotherapy in IDH1-mutant relapsed or refractory acute myeloid leukemia revealed by single cell sequencing analyses. Blood134, 541 (2019).
- 85.Khatib S, Pomyen Y, Dang H, Wang XW. Understanding the cause and consequence of tumor heterogeneity. Trends Cancer. 2020;6:267–271. doi: 10.1016/j.trecan.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 86.Ma L, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019;36:418–430. e416. doi: 10.1016/j.ccell.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Craig AJ, Von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2020;17:139–152. doi: 10.1038/s41575-019-0229-4. [DOI] [PubMed] [Google Scholar]
- 88.Zhang, Q. et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut68, 2019–2031 (2019). [DOI] [PMC free article] [PubMed]
- 89.Nault J-C, Villanueva A. Intratumor molecular and phenotypic diversity in hepatocellular carcinoma. Clin. Cancer Res. 2015;21:1786–1788. doi: 10.1158/1078-0432.CCR-14-2602. [DOI] [PubMed] [Google Scholar]
- 90.Losic B, et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-019-14050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ho DW-H, et al. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and stemness-related subpopulations in liver cancer. Cancer Lett. 2019;459:176–185. doi: 10.1016/j.canlet.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 92.Dong X, et al. Single-cell analysis reveals the intra-tumor heterogeneity and identifies MLXIPL as a biomarker in the cellular trajectory of hepatocellular carcinoma. Cell Death Discov. 2021;7:1–13. doi: 10.1038/s41420-021-00403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duan M, et al. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 2018;28:359–373. doi: 10.1038/cr.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heinrich S, et al. Understanding tumour cell heterogeneity and its implication for immunotherapy in liver cancer using single-cell analysis. J. Hepatol. 2021;74:700–715. doi: 10.1016/j.jhep.2020.11.036. [DOI] [PubMed] [Google Scholar]
- 95.Harbeck N, et al. Breast cancer. Nat. Rev. Dis. Prim. 2019;5:1–31. doi: 10.1038/s41572-018-0051-2. [DOI] [PubMed] [Google Scholar]
- 96.Zhou S, et al. Single-cell RNA-seq dissects the intratumoral heterogeneity of triple-negative breast cancer based on gene regulatory networks. Mol. Ther. Nucleic Acids. 2021;23:682–690. doi: 10.1016/j.omtn.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davis RT, et al. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat. Cell Biol. 2020;22:310–320. doi: 10.1038/s41556-020-0477-0. [DOI] [PubMed] [Google Scholar]
- 98.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 2015;21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miron A, et al. PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res. 2010;70:5674–5678. doi: 10.1158/0008-5472.CAN-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Casasent AK, et al. Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell. 2018;172:205–217. e212. doi: 10.1016/j.cell.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dawson SJ, Rueda OM, Aparicio S, Caldas C. A new genome‐driven integrated classification of breast cancer and its implications. EMBO J. 2013;32:617–628. doi: 10.1038/emboj.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baslan T, et al. Novel insights into breast cancer copy number genetic heterogeneity revealed by single-cell genome sequencing. elife. 2020;9:e51480. doi: 10.7554/eLife.51480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bassez A, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 2021;27:820–832. doi: 10.1038/s41591-021-01323-8. [DOI] [PubMed] [Google Scholar]
- 105.Utada M, Yonehara S, Ozasa K. Historical changes in histological diagnosis of lung cancer. J. Epidemiol. 2019;29:238–240. doi: 10.2188/jea.JE20180037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Travis WD, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 107.Stewart CA, et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat. Cancer. 2020;1:423–436. doi: 10.1038/s43018-019-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sharma A, et al. Non-genetic intra-tumor heterogeneity is a major predictor of phenotypic heterogeneity and ongoing evolutionary dynamics in lung tumors. Cell Rep. 2019;29:2164–2174. e2165. doi: 10.1016/j.celrep.2019.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schaff DL, et al. Fragmentation of small-cell lung cancer regulatory states in heterotypic microenvironments. Cancer Res. 2021;81:1853–1867. doi: 10.1158/0008-5472.CAN-20-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.He D, et al. Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene. 2021;40:355–368. doi: 10.1038/s41388-020-01528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim N, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maynard A, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. 2020;182:1232–1251. e1222. doi: 10.1016/j.cell.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guinney J, et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu H, et al. Evolution and heterogeneity of non-hereditary colorectal cancer revealed by single-cell exome sequencing. Oncogene. 2017;36:2857–2867. doi: 10.1038/onc.2016.438. [DOI] [PubMed] [Google Scholar]
- 115.Ono, H. et al. Single-cell DNA and RNA sequencing reveals the dynamics of intra-tumor heterogeneity in a colorectal cancer model. BMC Biology19.1, 1–17 (2021). [DOI] [PMC free article] [PubMed]
- 116.Bian S, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362:1060–1063. doi: 10.1126/science.aao3791. [DOI] [PubMed] [Google Scholar]
- 117.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 119.Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015;25:1499–1507. doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao Y, et al. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumor cells. Genome Res. 2017;27:1312–1322. doi: 10.1101/gr.216788.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Negishi R, et al. Transcriptomic profiling of single circulating tumor cells provides insight into human metastatic gastric cancer. Commun. Biol. 2022;5:1–12. doi: 10.1038/s42003-021-02937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miyamoto DT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hu J, et al. Single-cell transcriptome analysis reveals intratumoral heterogeneity in ccRCC, which results in different clinical outcomes. Mol. Ther. 2020;28:1658–1672. doi: 10.1016/j.ymthe.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bi K, et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell. 2021;39:649–661. e645. doi: 10.1016/j.ccell.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pandiani C, et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ. 2021;28:1990–2000. doi: 10.1038/s41418-020-00730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Andor N, et al. Joint single cell DNA-seq and RNA-seq of gastric cancer cell lines reveals rules of in vitro evolution. NAR Genom. Bioinform. 2020;2:lqaa016. doi: 10.1093/nargab/lqaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Anderson NM, Simon MC. The tumor microenvironment. Curr. Biol. 2020;30:R921–R925. doi: 10.1016/j.cub.2020.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 130.Altorki NK, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat. Rev. Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang AC, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moore L, et al. The mutational landscape of human somatic and germline cells. Nature. 2021;597:381–386. doi: 10.1038/s41586-021-03822-7. [DOI] [PubMed] [Google Scholar]
- 134.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 135.Wu TD, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature. 2020;579:274–278. doi: 10.1038/s41586-020-2056-8. [DOI] [PubMed] [Google Scholar]
- 136.Chu T, Berner J, Zehn D. Two parallel worlds of memory T cells. Nat. Immunol. 2020;21:1484–1485. doi: 10.1038/s41590-020-00815-y. [DOI] [PubMed] [Google Scholar]
- 137.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 138.Clarke J, et al. Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J. Exp. Med. 2019;216:2128–2149. doi: 10.1084/jem.20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kimani D, et al. Translating the immunogenicity of prime-boost immunization with ChAd63 and MVA ME-TRAP from malaria naive to malaria-endemic populations. Mol. Ther. 2014;22:1992–2003. doi: 10.1038/mt.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim N, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu F, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 2021;12:2540. doi: 10.1038/s41467-021-22801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maynard A, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. 2020;182:1232–1251 e1222. doi: 10.1016/j.cell.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zilionis R, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334 e1310. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Denkert C, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 146.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308 e1236. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Savas P, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 149.Casbon AJ, et al. Invasive breast cancer reprograms early myeloid differentiation in the bone marrow to generate immunosuppressive neutrophils. Proc. Natl Acad. Sci. USA. 2015;112:E566–E575. doi: 10.1073/pnas.1424927112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Alshetaiwi, H. et al. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 5, eaay6017 (2020). [DOI] [PMC free article] [PubMed]
- 152.Gibbons D, et al. Interleukin-8 (CXCL8) production is a signatory T cell effector function of human newborn infants. Nat. Med. 2014;20:1206–1210. doi: 10.1038/nm.3670. [DOI] [PubMed] [Google Scholar]
- 153.Crespo J, et al. Human naive T cells express functional CXCL8 and promote tumorigenesis. J. Immunol. 2018;201:814–820. doi: 10.4049/jimmunol.1700755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ford MS, et al. Peptide-activated double-negative T cells can prevent autoimmune type-1 diabetes development. Eur. J. Immunol. 2007;37:2234–2241. doi: 10.1002/eji.200636991. [DOI] [PubMed] [Google Scholar]
- 155.Young KJ, DuTemple B, Phillips MJ, Zhang L. Inhibition of graft-versus-host disease by double-negative regulatory T cells. J. Immunol. 2003;171:134–141. doi: 10.4049/jimmunol.171.1.134. [DOI] [PubMed] [Google Scholar]
- 156.Johnston MP, Khakoo SI. Immunotherapy for hepatocellular carcinoma: current and future. World J. Gastroenterol. 2019;25:2977–2989. doi: 10.3748/wjg.v25.i24.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chew V, et al. Toll-like receptor 3 expressing tumor parenchyma and infiltrating natural killer cells in hepatocellular carcinoma patients. J. Natl Cancer Inst. 2012;104:1796–1807. doi: 10.1093/jnci/djs436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356 e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 159.Bhairavabhotla R, et al. Transcriptome profiling of human FoxP3+ regulatory T cells. Hum. Immunol. 2016;77:201–213. doi: 10.1016/j.humimm.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zheng B, et al. Trajectory and functional analysis of PD-1(high) CD4(+)CD8(+) T cells in hepatocellular carcinoma by single-cell cytometry and transcriptome sequencing. Adv. Sci. 2020;7:2000224. doi: 10.1002/advs.202000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Overgaard NH, Jung JW, Steptoe RJ, Wells JW. CD4+/CD8+ double-positive T cells: more than just a developmental stage? J. Leukoc. Biol. 2015;97:31–38. doi: 10.1189/jlb.1RU0814-382. [DOI] [PubMed] [Google Scholar]
- 162.Zeiner PS, et al. MIF receptor CD74 is restricted to microglia/macrophages, associated with a M1-polarized immune milieu and prolonged patient survival in gliomas. Brain Pathol. 2015;25:491–504. doi: 10.1111/bpa.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.MacParland SA, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Aizarani N, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Alluhaybi KA, et al. Cellular and humoral immunogenicity of a candidate DNA vaccine expressing SARS-CoV-2 spike subunit 1. Vaccines. 2021;9:852. doi: 10.3390/vaccines9080852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang Q, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845 e820. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 167.Mertens C, et al. Macrophage-derived lipocalin-2 transports iron in the tumor microenvironment. Oncoimmunology. 2018;7:e1408751. doi: 10.1080/2162402X.2017.1408751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mora, J. et al. Strategies to interfere with tumor metabolism through the interplay of innate and adaptive immunity. Cells8, 445 (2019). [DOI] [PMC free article] [PubMed]
- 169.Heinrich, B. et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut, (2021). [DOI] [PMC free article] [PubMed]
- 170.Duerr CU, Fritz JH. Regulation of group 2 innate lymphoid cells. Cytokine. 2016;87:1–8. doi: 10.1016/j.cyto.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 171.Bal SM, Golebski K, Spits H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol. 2020;20:552–565. doi: 10.1038/s41577-020-0282-9. [DOI] [PubMed] [Google Scholar]
- 172.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li H, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2020;181:747. doi: 10.1016/j.cell.2020.04.017. [DOI] [PubMed] [Google Scholar]
- 174.Durante MA, et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020;11:496. doi: 10.1038/s41467-019-14256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nirschl CJ, et al. IFNgamma-dependent tissue-immune homeostasis is co-opted in the tumor microenvironment. Cell. 2017;170:127–141 e115. doi: 10.1016/j.cell.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Greenhalgh CJ, et al. SOCS2 negatively regulates growth hormone action in vitro and in vivo. J. Clin. Investig. 2005;115:397–406. doi: 10.1172/JCI200522710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rico-Bautista E, Flores-Morales A, Fernandez-Perez L. Suppressor of cytokine signaling (SOCS) 2, a protein with multiple functions. Cytokine Growth Factor Rev. 2006;17:431–439. doi: 10.1016/j.cytogfr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 178.Tintelnot J, Stein A. Immunotherapy in colorectal cancer: available clinical evidence, challenges and novel approaches. World J. Gastroenterol. 2019;25:3920–3928. doi: 10.3748/wjg.v25.i29.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 181.Zhang L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 182.Lee HO, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 2020;52:594–603. doi: 10.1038/s41588-020-0636-z. [DOI] [PubMed] [Google Scholar]
- 183.Zhang Y, et al. Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett. 2020;470:84–94. doi: 10.1016/j.canlet.2019.10.016. [DOI] [PubMed] [Google Scholar]
- 184.Castello LM, et al. Osteopontin at the crossroads of inflammation and tumor progression. Mediators Inflamm. 2017;2017:4049098. doi: 10.1155/2017/4049098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Baitsch D, et al. Apolipoprotein E induces antiinflammatory phenotype in macrophages. Arterioscler Thromb. Vasc. Biol. 2011;31:1160–1168. doi: 10.1161/ATVBAHA.111.222745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang KX, Denhardt DT. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev. 2008;19:333–345. doi: 10.1016/j.cytogfr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 187.Zhang L, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459 e429. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 188.Muller S, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18:234. doi: 10.1186/s13059-017-1362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chung JS, Tamura K, Akiyoshi H, Cruz PD, Jr, Ariizumi K. The DC-HIL/syndecan-4 pathway regulates autoimmune responses through myeloid-derived suppressor cells. J. Immunol. 2014;192:2576–2584. doi: 10.4049/jimmunol.1301857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sui X, et al. RSL3 drives ferroptosis through GPX4 inactivation and ROS production in colorectal cancer. Front. Pharm. 2018;9:1371. doi: 10.3389/fphar.2018.01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xie Y, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 192.Li H, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 193.Oh DY, et al. Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181:1612–1625 e1613. doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen Z, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun. 2020;11:5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang M, et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020;73:1118–1130. doi: 10.1016/j.jhep.2020.05.039. [DOI] [PubMed] [Google Scholar]
- 196.Sathe A, et al. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin. Cancer Res. 2020;26:2640–2653. doi: 10.1158/1078-0432.CCR-19-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mayo L, et al. Dual role of CD38 in microglial activation and activation-induced cell death. J. Immunol. 2008;181:92–103. doi: 10.4049/jimmunol.181.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhao J, et al. Single cell RNA-seq reveals the landscape of tumor and infiltrating immune cells in nasopharyngeal carcinoma. Cancer Lett. 2020;477:131–143. doi: 10.1016/j.canlet.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 199.Miao BP, et al. Nasopharyngeal cancer-derived microRNA-21 promotes immune suppressive B cells. Cell Mol. Immunol. 2015;12:750–756. doi: 10.1038/cmi.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Shih AJ, et al. Identification of grade and origin specific cell populations in serous epithelial ovarian cancer by single cell RNA-seq. PLoS One. 2018;13:e0206785. doi: 10.1371/journal.pone.0206785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Qian J, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30:745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cheng S, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809 e723. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 203.Maier B, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 2020;580:257–262. doi: 10.1038/s41586-020-2134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 205.Ishida T, Ueda R. CCR4 as a novel molecular target for immunotherapy of cancer. Cancer Sci. 2006;97:1139–1146. doi: 10.1111/j.1349-7006.2006.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Riaz N, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949 e916. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Banchereau, R. et al. Intratumoral CD103 + CD8 + T cells predict response to PD-L1 blockade. J. Immunother. Cancer9, e002231 (2021). [DOI] [PMC free article] [PubMed]
- 208.Goswami S, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat. Med. 2020;26:39–46. doi: 10.1038/s41591-019-0694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yost KE, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019;25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kfoury Y, et al. Human prostate cancer bone metastases have an actionable immunosuppressive microenvironment. Cancer Cell. 2021;39:1464–1478 e1468. doi: 10.1016/j.ccell.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang Q, et al. Single-cell profiling guided combinatorial immunotherapy for fast-evolving CDK4/6 inhibitor-resistant HER2-positive breast cancer. Nat. Commun. 2019;10:3817. doi: 10.1038/s41467-019-11729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yuen KC, et al. High systemic and tumor-associated IL-8 correlates with reduced clinical benefit of PD-L1 blockade. Nat. Med. 2020;26:693–698. doi: 10.1038/s41591-020-0860-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J. Exp. Med. 2018;215:2520–2535. doi: 10.1084/jem.20180684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cabrita R, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 215.Kurtulus S, et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1(-)CD8(+) tumor-Infiltrating T cells. Immunity. 2019;50:181–194 e186. doi: 10.1016/j.immuni.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Siddiqui I, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50:195–211.e110. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 217.Chen Z, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;51:840–855 e845. doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Amsen D, van Gisbergen K, Hombrink P, van Lier RAW. Tissue-resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 2018;19:538–546. doi: 10.1038/s41590-018-0114-2. [DOI] [PubMed] [Google Scholar]
- 219.Held, W., Siddiqui, I., Schaeuble, K., Speiser, D.E., Intratumoral CD8(+) T cells with stem cell-like properties: Implications for cancer immunotherapy. Sci. Transl. Med.11, eaay6863 (2019). [DOI] [PubMed]
- 220.Jiao S, et al. Differences in tumor microenvironment dictate T helper lineage polarization and response to immune checkpoint therapy. Cell. 2019;179:1177–1190 e1113. doi: 10.1016/j.cell.2019.10.029. [DOI] [PubMed] [Google Scholar]
- 221.Zhou Y, et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020;11:6322. doi: 10.1038/s41467-020-20059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Xie Y, et al. Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J. Exp. Med. 2010;207:651–667. doi: 10.1084/jem.20091921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kitano S, et al. Enhancement of tumor-reactive cytotoxic CD4 + T cell responses after ipilimumab treatment in four advanced melanoma patients. Cancer Immunol. Res. 2013;1:235–244. doi: 10.1158/2326-6066.CIR-13-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sledzinska A, et al. Regulatory T cells restrain interleukin-2- and Blimp-1-dependent acquisition of cytotoxic function by CD4(+) T cells. Immunity. 2020;52:151–166 e156. doi: 10.1016/j.immuni.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Spitzer MH, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502 e415. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang F, et al. Dynamics of peripheral T cell clones during PD-1 blockade in non-small cell lung cancer. Cancer Immunol. Immunother. 2020;69:2599–2611. doi: 10.1007/s00262-020-02642-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Griffiths JI, et al. Circulating immune cell phenotype dynamics reflect the strength of tumor-immune cell interactions in patients during immunotherapy. Proc. Natl Acad. Sci. USA. 2020;117:16072–16082. doi: 10.1073/pnas.1918937117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fairfax BP, et al. Peripheral CD8(+) T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat. Med. 2020;26:193–199. doi: 10.1038/s41591-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 230.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 231.Puram SV, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624 e1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Skelly DA, et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 233.Camp JG, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 234.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ji AL, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182:497–514 e422. doi: 10.1016/j.cell.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Cillo AR, et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity. 2020;52:183–199 e189. doi: 10.1016/j.immuni.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Dar RD, et al. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc. Natl Acad. Sci. 2012;109:17454–17459. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bakken TE, et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PloS One. 2018;13:e0209648. doi: 10.1371/journal.pone.0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Habib N, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.van den Brink SC, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 242.Massoni-Badosa R, et al. Sampling time-dependent artifacts in single-cell genomics studies. Genome Biol. 2020;21:1–16. doi: 10.1186/s13059-020-02032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gao R, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 2021;39:599–608. doi: 10.1038/s41587-020-00795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Liu F, et al. Systematic comparative analysis of single-nucleotide variant detection methods from single-cell RNA sequencing data. Genome Biol. 2019;20:1–15. doi: 10.1186/s13059-018-1612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hahaut, V., Pavlinic, D., Cowan, C., Picelli, S., Lightning fast and highly sensitive full-length single-cell sequencing using FLASH-Seq. bioRxiv, (2021). [DOI] [PMC free article] [PubMed]
- 246.Hagemann-Jensen M, et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 2020;38:708–714. doi: 10.1038/s41587-020-0497-0. [DOI] [PubMed] [Google Scholar]
- 247.Nam AS, et al. Somatic mutations and cell identity linked by genotyping of transcriptomes. Nature. 2019;571:355–360. doi: 10.1038/s41586-019-1367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ruff, D.W., Dhingra, D.M., Thompson, K., Marin, J.A., Ooi, A.T., in Single-Cell Protein Analysis. (Springer, 2022), pp. 171–188. [DOI] [PubMed]
- 249.Rodriques SG, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Eng C-HL, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+ Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Deng Y, et al. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science. 2022;375:681–686. doi: 10.1126/science.abg7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Marx V. Method of the Year: spatially resolved transcriptomics. Nat. Methods. 2021;18:9–14. doi: 10.1038/s41592-020-01033-y. [DOI] [PubMed] [Google Scholar]
- 253.Wang X, et al. BREM-SC: a bayesian random effects mixture model for joint clustering single cell multi-omics data. Nucleic Acids Res. 2020;48:5814–5824. doi: 10.1093/nar/gkaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tran HTN, et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21:1–32. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Büttner M, Miao Z, Wolf FA, Teichmann SA, Theis FJ. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods. 2019;16:43–49. doi: 10.1038/s41592-018-0254-1. [DOI] [PubMed] [Google Scholar]
- 256.Luecken MD, et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods. 2022;19:41–50. doi: 10.1038/s41592-021-01336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e3529. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Welch JD, et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell. 2019;177:1873–1887. e1817. doi: 10.1016/j.cell.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mateo LJ, et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 2019;568:49–54. doi: 10.1038/s41586-019-1035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hao Y. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]