Abstract
Outdoor biting by anopheline mosquitoes is one of the contributors to residual malaria transmission, but the profile of vectors driving this phenomenon is not well understood. Here, we studied the bionomics and genetically characterized populations of An. gambiae and An. funestus complexes trapped outdoors in three selected dryland areas including Kerio Valley, Nguruman and Rabai in Kenya. We observed a higher abundance of Anopheles funestus group members (n = 639, 90.6%) compared to those of the An. gambiae complex (n = 66, 9.4%) with An. longipalpis C as the dominant vector species with a Plasmodium falciparum sporozoite rate (Pfsp) of 5.2% (19/362). The known malaria vectors including An. funestus s.s. (8.7%, 2/23), An. gambiae (14.3%, 2/14), An. rivulorum (14.1%, 9/64), An. arabiensis (1.9%, 1/52) occurred in low densities and displayed high Pfsp rates, which varied with the site. Additionally, six cryptic species found associated with the An. funestus group harbored Pf sporozoites (cumulative Pfsp rate = 7.2%, 13/181). We detected low frequency of resistant 119F-GSTe2 alleles in An. funestus s.s. (15.6%) and An. longipalpis C (3.1%) in Kerio Valley only. Evidence of outdoor activity, emergence of novel and divergent vectors and detection of mutations conferring metabolic resistance to pyrethroid/DDT could contribute to residual malaria transmission posing a threat to effective malaria control.
Subject terms: Ecology, Malaria
Introduction
Malaria case incidence and mortality have fallen in Kenya by 58% and 11%, respectively, since the early 2000s consistent with the global trend1–3. This reduction has been achieved owing to combined interventions of long-lasting insecticidal nets (LLINs), indoor residual spraying (IRS), prompt diagnosis and treatment with artemisinin-based combination therapies and intermittent preventive treatment of pregnant women2. Despite this progress, the disease still exerts a huge health burden accounting for an estimated 19% of all outpatient consultations in Kenya4. This trend is potentially indicative of the disease persistence and facilitated by several factors including house characteristics, use of bednets or other control measures, human activities and behavior, and vector-related factors5. However, entomologic determinants remain critical. For instance, resistance to insecticides has developed and spread among major malaria vectors6. Also noted are changes in Anopheles vector composition following sustained vector control, as well as resting and biting habits especially outdoors7. Collectively, these factors limit the effectiveness of LLINs and IRS mainly deployed indoors resulting in residual malaria transmission (RMT) which is defined as ongoing transmission despite 100% implementation of LLINs and IRS fully susceptible to local vectors7,8
Outdoor biting has received increasing attention regarding control of malaria towards elimination. A recent modeling study predicted that across Africa, outdoor transmission resulted in an estimated 11 million additional malaria cases annually assuming universal LLIN and IRS coverage8. Thus, the contribution of outdoor biting to malaria persistence could be much higher as universal coverage of ITNs and IRS has not been achieved. A multicountry study in Cameroon, Kenya and Ethiopia found high malaria transmission outdoors compared to indoors5. While this phenomenon is not new9–11, the profile of vectors implicated in outdoor transmission could differ in different eco-epidemiologic settings5.
In Kenya, monitoring efforts have assessed the malaria epidemiologic landscape and identified the key vector species. Primary malaria vectors belong to Anopheles gambiae sensu lato (s.l.) and Anopheles funestus group, like most of East Africa9,12,13. The sibling species in these complexes are morphologically similar as adults, although they have distinct ecological preferences, vectorial capacity and behavior, which impact differentially on malaria epidemiology and control efforts14,15. Most comprehensive data on entomologic risk come from the high malaria endemic Lake and Coastal zones. Further, studies in these foci have implicated previously unrecognized species in these complexes in malaria transmission16,17. This trend has been facilitated by application of sensitive molecular techniques including PCR and sequencing in entomologic surveillance and becoming increasingly crucial to identify new threats, availing opportunities for directing interventions12,16–18.
The present study was designed to gain insights into malaria persistence in dryland ecosystems, where malaria is thought to be seasonal and prone to epidemics4. Specifically, we focused on outdoor transmission, and describe the species composition in the An. gambiae s.l. and An. funestus group and natural infection rates with Plasmodium falciparum supported by molecular analyses. To determine if the high prevalence of An. funestus group members was due to insecticide resistance, we screened them for the presence of mutations in the glutathione S-transferase epsilon 2 gene (GSTe2) that confer resistance to pyrethroid-based insecticides and dichloro-diphenyl-trichloroethane (DDT)19. The findings from this study have implications for guiding control of RMT towards elimination.
Results
Anopheles funestus mosquitoes dominate outdoor anopheline catches
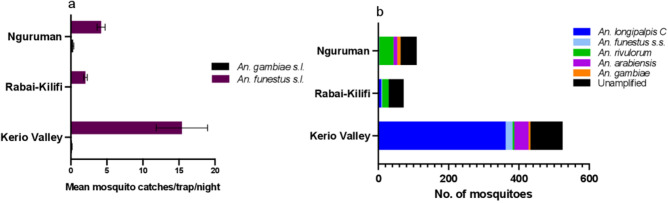
Of the mosquitoes morphologically identified, An. funestus s.l. was predominant across the three study areas (Kerio Valley, Nguruman, Rabai; Fig. 1) comprising 90.6% (639/705) of the total captures. Only 9.4% (66/705) were An. gambiae s.l. The breakdown of the catches by study areas were as follows: Kerio Valley (An. funestus s.l. = 478, An. gambiae s.l. = 46); Rabai (An. funestus s.l. = 72, An. gambiae s.l. = 0) and Nguruman (An. funestus s.l. = 89, An. gambiae s.l. = 20). Mean trap catches were significantly higher for An. funestus s.l. than An. gambiae s.l. in Kerio Valley (χ21,86 = 68.6, p < 0.0001) and Nguruman: (χ21,110 = 102.7, p < 0.0001) (Fig. 2a).
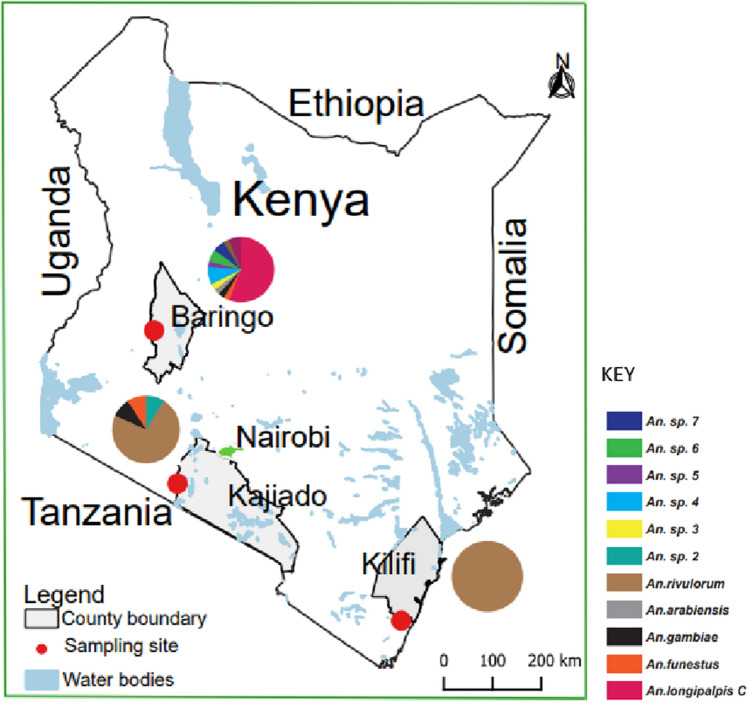
Figure 1.
Map of Kenya showing the study areas and distribution of anopheline species found infected with Plasmodium falciparum sporozoites. Pie-chart indicating the relative abundance of the species infected based on actual numbers is shown for each study area. The map was designed using ArcMap 10.2.2 with the ocean and lakes base layer derived from Natural Earth (http://www.naturalearthdata.com/, a free GIS data source). The sample points were collected using a GPS gadget (garmin etrex 20, https://buy.garmin.com/en-US/US/p/518046), and the county boundaries for Kenya derived from Africa Open data (https://africaopendata.org/dataset/kenya-counties-shapefile, license Creative Commons).
Figure 2.
(a) Outdoor mean catches/trap/day, (b) Species composition, in three study areas in dryland areas of Kenya. The number of trap nights were 44, 50 and 56 in Kerio Valley, Rabai and Nguruman, respectively.
Anopheles longipalpis C dominates sibling species of Anopheles funestus identified
PCR to identify sibling species of An. funestus s.l. revealed variation in the species composition by area (Fig. 2b). In Kerio Valley, An. longipalpis C was predominant (75.7%, 362/478) followed by An. funestus s.s. (4.2%, 20/478) and An. rivulorum 0.8% (4/478). Species in this group were represented by An. rivulorum (26.4%, 19/72), An. longipalpis C (11.1%, 8/72) and An. funestus s.s. (2.8%, 2/72) in Rabai and in Nguruman by An. rivulorum (46.1%, 41/89) followed by 1.1% each of An. longipalpis C (1/89) and An. funestus s.s. (1/89). PCR amplification of DNA extracted from An. funestus s.l specimens was not possible for 28.3% (181/639) of the specimens collected, with Rabai (59.7%) having the highest number of unamplified samples (Fig. 2b). Anopheles arabiensis and An. gambiae were the two species observed in An. gambiae s.l. with the former occurring in higher proportion (41/46) than the latter (5/46) in Kerio Valley. Both species were not detected in samples collected from Rabai (Kilifi), but they were equally represented in the Nguruman samples (9/20 vs 11/20, An. gambiae and An. arabiensis, respectively).
High Plasmodium falciparum sporozoite detected in An. longipalpis C and diverse cryptic species
Forty-six (46) of the 705 female anophelines tested positive for P. falciparum sporozoites (Pfsp) by high resolution melting (HRM-PCR) of the non-coding mitochondrial sequence (ncMS). A representative detection profile is shown in Fig. S1. This translated to an overall Pfsp rate of 6.5% but this finding varied with site and mosquito species (Table 1). In the An. funestus group, high Pfsp rates were recorded in An. longipalpis C (5.2%; 19/362) in Kerio Valley only; An. funestus s.s. in Kerio Valley (5%; 1/20) and Nguruman (100%; 1/1) and An. rivulorum in Nguruman (19.5%; 8/41) and Rabai (5.3%; 1/19). A significant proportion of unamplified cohorts also tested positive for Pfsp in Kerio Valley (12%;11/92) and Nguruman (4.3%; 2/46) (Table 1). Pfsp were detected in An. gambiae in Kerio Valley (20%; 1/5) and Nguruman (11.1%; 1/9), and in An. arabiensis from Kerio Valley (2.4%; 1/41).
Table 1.
Plasmodium falciparum sporozoite rates (%) by species in selected dryland areas, Kenya.
| Species | Kerio Valley | Rabai | Nguruman | Total | |
|---|---|---|---|---|---|
| An. funestus group | An. longipalpis C | 5.2% (19/362) | 0% (0/8) | 0% (0/1) | 5.1% (19/371) |
| An. funestus s.s. | 5% (1/20) | 0% (0/2) | 100% (1/1) | 8.7% (2/23) | |
| An. rivulorum | 0% (0/4) | 5.3% (1/19) | 19.5% (8/41) | 14.1% (9/64) | |
| Unamplified | 12% (11/92) | 0% (0/43) | 4.3% (2/46) | 7.2% (13/181) | |
| An. gambiae s.l. | An. gambiae. | 20% (1/5) | 0% | 11.1% (1/9) | 14.3% (2/14) |
| An. arabiensis | 2.4% (1/41) | 0% | 0% (0/11) | 1.9% (1/52) | |
| Total | 6.3% (33/524) | 1.4% (1/72) | 11% (12/109) | 6.5% (46/705) |
Number in parenthesis indicates proportion of the total that tested positive for each species.
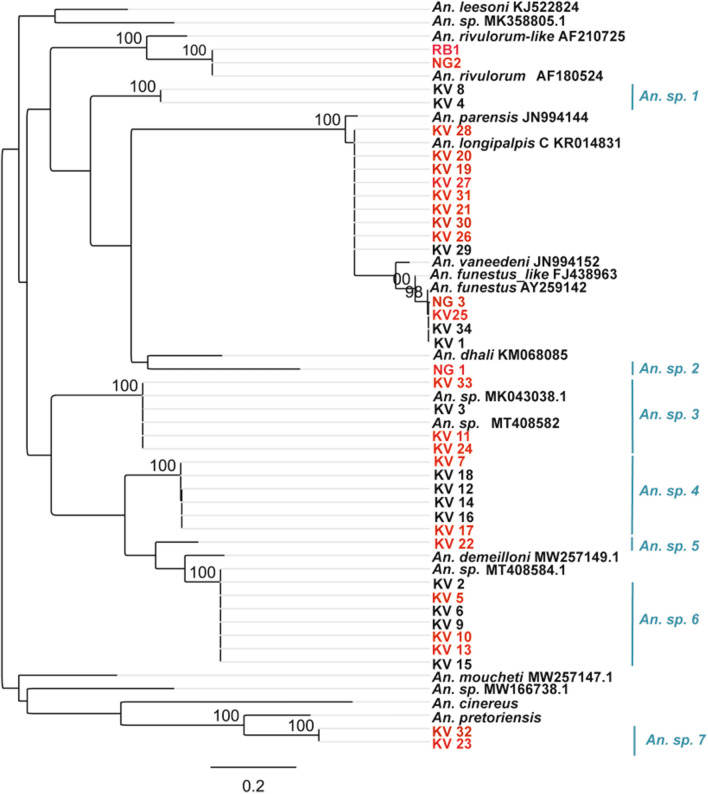
Further confirmation of the species identity of the infected specimens (for An. funestus group only) was conducted by PCR of the ITS2 region and then sequencing. Few samples that tested negative especially for the unamplified cohorts were included. Phylogenetic analysis of the generated sequences showed strong support (bootstrap (BS) = 98% and 89%) for An. funestus s.s. and An. longipalpis C, respectively (Fig. 3); two of the infected specimens clustered with An. rivulorum (BS = 100%; GenBank accession: AF180524). Additionally, sequenced samples of the unamplified cohort (positive and negative for Pfsp) resolved into well distinct clades (BS values = 100%) some clustering with previously reported but unidentified species viz: Anopheles sp.19 DZ 2020 (GenBank accession no: MT408584.1) and Anopheles sp. isolate A (GenBank accession no: MKO43038.1) indicating presence of cryptic species. Some of the positive specimens did not cluster with any referenced species (An. sp. 2, An. sp. 4, An sp. 5, An. sp. 7) (Fig. 3).
Figure 3.
Phylogenetic tree for represetative mosquitoes morphologically scored as An. funestus group infected with P. falciparum sprozoites. Few samples negative for the malaria parasite are also included. Bootstrap support values are indicated above the nodes from 1000 replicates. Highlighted in red are the infected samples.
Low levels of L119F-GSTe2 resistance alleles detected in An. funestus group and polymorphism analysis
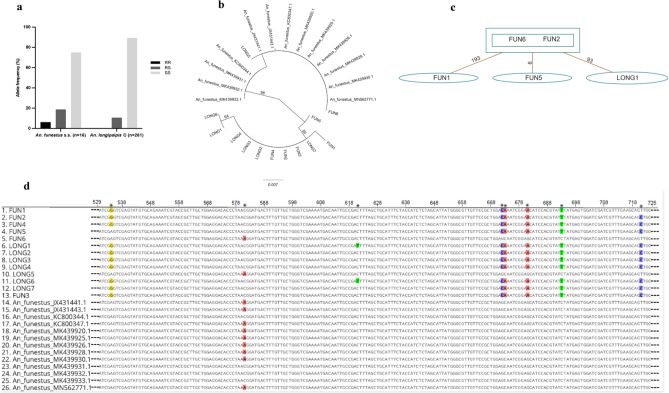
The L119F-GSTe2 mutation conferring metabolic resistance to DDT/pyrethroid19, was detected in An. funestus s.s. and An. longipalpis C. The frequency in the former species was 15.6% (1-RR, 3-RS, 12-SS) and in An. longipalpis C, 3.1% (0-RR, 28-RS, 233-SS) (Fig. 4a). Disaggregation of the data in An. longipalpis C by infection status found no difference in occurrence of the resistance allele (RS) between infected (12.5%) and non-infected (10.6%) cohorts (Fischer’s exact test, p = 0.68). A 240 bp fragment of the GSTe2 gene was successfully sequenced in 17 mosquitoes, An. longipalpis C (7-RS and 3-SS) and An. funestus s.s. (6-RS and 1-RR). Phylogenetic analysis showed most haplotypes resolved as a distinct clade and few clustering with those previously detected in An. funestus s.s. from different parts of Africa (Fig. 4b). The haplotypes from Kenya exhibited genetic diversity with at least three mutational differences from those of An. funestus s.s. in GenBank (Fig. 4b–d) suggesting an independent occurrence, potentially from local selection.
Figure 4.
Analysis of the polymorphism of a portion of the glutathione S-transferase epsilon 2 (GSTe2) gene. (a) Allele frequency distribution, (b) ML phylogenetic tree of GSTe2 gene fragment with previously recorded haplotypes in An. funestus s.s. across Africa, (c) Templeton-Crandall Sing (TCS) haplotype network. The rectangles and ovals represent haplotypes with the lines linking haplotypes showing a single mutation event. (d) Aligned sequence traces showing the mutational differences/polymorphic positions in the haplotypes detected in Kenyan An. longipalpis C/An. funestus s.s.* indicates polymorphic positions.
Discussion
Outdoor malaria transmission is a dynamic process and the vectors involved may vary according to the ecological setting. Here, we report diverse known and cryptic anopheline species collected outdoors and found to harbor P. falciparum sporozoites. These species could play critical roles in residual malaria transmission (RMT), given that parasite infection rate represents the most sensitive indicator in assessing importance of a vector species in this phenomenon20. Furthermore, we detected mutations in the GSTe2 gene conferring resistance to pyrethroid/DDT in An. funestus/An. longipalpis C. This finding in a specific focus (i.e. Kerio Valley) potentially poses a risk to insecticide-based control measures and RMT21. This is the first report of the detection of this resistance marker in any species apart from An. funestus s.s. in the Funestus group. Overall, these findings may be useful in updating risk map of key vectors in Kenya22 and providing evidence-based decision in selection and implementation of appropriate interventions against malaria in the study areas.
We found that An. funestus s.l. was the predominant mosquito collected from the study areas. Molecular methods identified the sibling species An. longipalpis C, An. funestus s.s. and An. rivulorum in the Funestus group and other unidentified cryptic species, which varied with the site. Previous studies have largely implicated An. funestus s.s. as the most important vector in this group and known to highly adapt to dry season survival strategies23. Hence, we posed the question- could the low numbers signal dwindling importance of this species in certain ecological areas? Being a highly anthropophilic and endophilic mosquito12,14,20, the low numbers encountered outdoors is not surprising. It could also be argued that the low numbers reflect LLIN-related impact on their populations. The replacement of An. funestus s.s. by more outdoor resting/biting An. rivulorum after implementation of IRS in Kenya has been reported in the literature24. This could be the likely scenario following its absence in Rabai or enhanced contribution of An. rivulorum to malaria transmission in Nguruman and to a lesser extent in Rabai. Despite the low numbers, this species was still associated with parasite infection, which supports its high susceptibility and efficient role in malaria transmission. To assess its contribution to transmission in the study areas, longer term surveys, both indoors and outdoors, incorporating other trapping methods (e.g. human landing catches) and data on bed net coverage and use patterns in the target communities would be required.
Our findings revealed a high representation of lesser-known species, An. longipalpis C, in the Funestus group. The non-malaria vector status accorded An. longipalpis in southern Africa was attributed to absence of parasite infection and its highly zoophilic yet endophilic habits25. A high human blood meal index observed for this mosquito in Ethiopia26 lend support for its role as a malaria vector supported by previous detection of P. falciparum infection in indoor populations in Kerio Valley12. The present findings of high Pfsp rate and occurrence in high densities, even surpassing those of known malaria vectors, suggest it could assume a major role as a malaria vector in certain foci in Kenya. Understanding its biology and ecological adaption including breeding structure, resting and biting habits, and vectorial capacity are warranted.
Exophilic habits among mosquitoes do not preclude indoor activities and outdoor biting behavior could be a response to indoor insecticidal interventions7. The presence of mutations in the GSTe2 gene conferring metabolic resistance to pyrethroids/DDT could be attributed to selection pressure from pyrethroid insecticide used in LLINs or use of agricultural insecticides. Endophilic behaviour is well documented in An. funestus s.s.14,27 and An. longipalpis C12. Increased vector survival/longevity has been observed as an important phenotype of L119F-GSTe2 resistant An. funestus s.s.28. Thus, its detection could negatively impact vector control in this area through reduction in LLINs effectiveness21. Metabolic resistance produces an order of magnitude higher resistance to pyrethroids (commonly used on bednets) than that associated with kdr gene29. Furthermore, the mutational differences noted in the haplotypes of Kenyan An. longipalpis C/An. funestus s.s. compared to those of An. funestus s.s. elsewhere suggest an independent occurrence of resistance marker of the GSTe2 gene. Further experimental work is needed to correlate the significance of the mutations on phenotypic response through insecticide exposure assays. Comparative transcriptomic and functional analyses are also needed to improve our understanding of the evolution of resistance in Kenyan populations of these species.
Our uncovering of diverse species in the Funestus group highlights the importance of application of molecular tools in routine entomologic surveillance12,18. Not only were diverse species uncovered, but they were also found associated with P. falciparum infection. One of the species in a well-supported clade (BS = 100%) had 100% sequence identity with Anopheles sp. 19 DZ-2020 (GenBank accession number MT408584.1) recently reported in western Kenya17 as a novel cryptic species. Another species shared 99.9% sequence identity with Anopheles sp. Isolate A (GenBank accession number MK043038.1) previously reported in the dryland area of West Pokot Kenya, as a potential secondary malaria vector on the basis of harboring malaria parasite and predominance among outdoor An. funestus mosquitoes16. Its vectoring ability was confirmed in a recent study in western Kenya17. The results suggest an adaption to dry ecological areas but highlight the potential for wider distribution and greater impact of these cryptic species on malaria transmission with growing effect of climate change and global warming. These could provide new, favorable and suitable environmental conditions for the survival of these species, but this needs further investigations. As control targets primary vector species, these species could fill the gap and emerge to become important vectors. Additionally, the observed number of unamplified specimens in the An. funestus group calls for improvement in the existing protocol to further identify and discriminate members of this group other than the commonly known malaria vectors.
In summary, this study has demonstrated the unexpected and role of An. longipalpis C in outdoor malaria transmission in a dryland ecology of Kenya. Furthermore, we detected mutations in the GSTe2 gene conferring resistance to pyrethroid/DDT in An. funestus s.s./An. longipalpis C in a specific focus (Kerio Valley) potentially posing a risk to insecticide-based control measures. The identified outdoor malaria vectors included known species (An. funestus s.s., An. rivulorum, An. gambiae, An. arabiensis), but at least six cryptic potential malaria parasite vectors most of which have not been previously described in Kenya. These species could play crucial roles in RMT, given that parasite presence represents the most sensitive indicator in assessing vector importance in this phenomenon20. Kenya is intensifying its national efforts in malaria control towards achieving elimination and success requires surveillance of all potential vector species. This could be facilitated by integration of molecular tools in routine entomologic surveillance18 to guide malaria control interventions. The outdoor biting and resting behaviour of these vectors underscores the need for complementary malaria control interventions since current conventional vector control tools in Kenya mainly target vectors that rest and bite indoors.
Methodology
Study sites, sample collection and preparation
Adult female mosquitoes used in this study had previously been collected from three areas: Kerio Valley (Baringo county), Rabai (Kilifi county) and Nguruman (Kajiado county) (Fig. 1), as part of vector-borne disease surveillance project and stored at – 80 °C at the International Centre of Insect Physiology and Ecology (icipe). The mosquitoes were surveyed between August 2019 and May 2020. Nguruman is an agropastoral area located in Kajiado county at the southern end of the Kenyan Rift Valley bordering Tanzania. The area has a semi-arid climate characterized by erratic rains, extreme temperatures, and cyclic and prolonged droughts30. The vegetation is dominated by bushland, grassland and open woodlands along seasonal river valleys. Specific indicator data for malaria is not available for Nguruman except for estimates pertaining to the larger Kajiado county which as of 2019 indicates a malaria incidence rate of 5 per 1000 population31. Collections in Kerio Valley (Baringo county within the Rift Valley) were conducted in Kapluk and Barwesa, both agro-pastoral areas with arid and semi-arid ecology. Malaria is a major vector-borne disease in the areas with report of perennially occurrence in neighboring riverine areas32. Rabai is one of the seven administrative sub-counties of Kilifi county in the coastal region of Kenya where malaria is endemic. The main economic activities in the area include subsistence agriculture, casual labor, crafts and petty trading. The weather patterns at the sites during the sampling period were as follows: Kerio Valley (mean daily temperature: 21.2 °C, mean daily rainfall: 4.1 mm, mean relative humidity: 73.4%); Rabai (mean daily temperature: 26.4 °C, mean daily rainfall: 2.1 mm; mean relative humidity: 78.1%) and Nguruman (mean daily temperature: 22.5 °C, mean daily rainfall: 0.9 mm, mean relative humidity: 61.2%).
Mosquito survey and processing
Host seeking mosquitoes were trapped using CDC light traps baited with dry ice (carbon dioxide) attractive to several mosquitoes. Traps were set outdoors about 10–15 m away from randomly selected homesteads from 18:00 h to 06:00 h. After collection, the mosquitoes were anesthetized with trimethylamine and temporarily stored in liquid nitrogen before transportation to the Emerging Infectious Disease (EID) laboratory at icipe and later stored at − 80 °C. Anopheline mosquitoes were morphologically identified to species level using published taxonomic keys15,33.
DNA extraction and Anopheles species discrimination
DNA was extracted from the head/thorax of individual mosquitoes using ISOLATE II Genomic DNA Extraction kit (Bioline, UK) following the manufacturer’s instructions and used for species discrimination and screening for P. falciparum infection and Gste2 mutations (described below).
Cryptic sibling species of the Anopheles funestus and Anopheles gambiae complexes were identified using conventional PCR34,35 and/or sequencing. PCR for An. funestus complex in a 15 µl reaction volume comprised 0.5 µM of each primer targeting: Anopheles funestus s.s, Anopheles vaneedeni, Anopheles rivulorum, Anopheles parensis, Anopheles leesoni, Anopheles longipalpis A and Anopheles longipalpis C, 3 µl of 5X HOT FIREPol Blend Master Mix Ready to Load (Solis BioDyne, Estonia) and 2 µl of DNA template. The cycling conditions were initial denaturation at 95 °C for 15 min, and then 30 cycles of denaturation at 95 °C for 30 s, annealing at 46 °C for 30 s and extension at 72 °C for 40 s and final extension at 72 °C for 10 min. Size fragments of each species were scored after separation in 1.5% agarose gel electrophoresis stained with ethidium bromide against a 1 Kb DNA ladder (HyperLadder, Bioline, London, UK).
For An. gambiae s.l., PCR in a 10 µl volume consisted of 2 µl of 5X Evagreen HRM Master Mix (Solis BioDyne, Estonia), 1 µl of DNA template and 10 µM concentration of each primer targeting An. gambiae s.s and An. arabiensis. The thermal cycling conditions included initial denaturation for 15 min at 95 °C followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 61 °C for 15 s and extension at 72 °C for 20 s followed by final extension at 72 °C for 7 min.
A subset of An. funestus s.l. samples that failed to amplify using the established protocol, was further amplified and sequenced targeting the internal transcribed spacer 2 (ITS2) region of the ribosomal DNA (rDNA)36. This target has shown utility in discriminating closely related mosquito species including anophelines12 and sequences from diverse species for this marker are well represented in reference databases (e.g. GenBank). PCR volumes for rDNA ITS2 were 15 µl containing 0.5 µM of the forward and reverse primers, 3 µl of 5X HOT FIREPol Blend Master Mix Ready to Load (Solis BioDyne, Estonia) and 2 µl of DNA template. The cycling conditions were initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 45 s and final extension at 72 °C for 7 min. ExoSAP IT rapid cleanup kit (Affymetrix Inc., Santa Clara, CA, USA) was used to clean the PCR product as per the manufacturer’s guideline, and then outsourced for bidirectional Sanger sequencing to Macrogen, South Korea.
Detection of malaria parasites
Plasmodium falciparum sporozoites in individual mosquitoes (head/thorax) were detected by analyzing high resolution melting (HRM) profiles generated from real time PCR products of non-coding mitochondrial sequence (ncMS)37. A P. falciparum DNA from National Institute for Biological Standards and Control (NIBSC; London, UK) was used as a reference positive control. PCR was carried out in a 10 µl volume consisting of 2 µl of 5X Evagreen HRM Master Mix (Solis BioDyne, Estonia), 1 µl of DNA template and 10 µM of each primer. PCR cycling conditions were initial denaturation for 15 min at 95 °C followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 61 °C for 15 s and extension at 72 °C for 20 s followed by final extension at 72 °C for 7 min. A fraction of RT-PCR-HRM positive samples were further analyzed using conventional PCR in a 10 µl volume consisting of 2 µl of 5X HOT FIREPol Blend Master Mix Ready to Load (Solis BioDyne, Estonia), 1 µl of DNA template and 10 µM of each primer. The cycling conditions comprised initial denaturation for 15 min at 95 °C followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 61 °C for 15 s and extension at 72 °C for 20 s followed by final extension at 72 °C for 7 min. PCR product of samples positive by RT-PCR were purified using ExoSAP- IT (USB Corporation, Cleveland, OH, USA) and outsourced for sequencing to Macrogen, South Korea. All sporozoite-positive mosquitoes were molecularly identified to species by PCR of the ITS2 region as described above.
Genotyping for L119F-GSTe2 mutation and sequencing
Two outer and two inner primers in a PCR assay were used to genotype the L119F-GSTe2 mutations that confer resistance of An. funestus mosquitoes to pyrethroids/DDT19 as described previously28. Thus, only An. funestus s.l. was screened using this assay. Briefly, PCR in a 15 µl reaction volume consisted of 10 µM of each primer, 3 µl of 5X HOT FIREPol Blend Master Mix Ready to Load (Solis BioDyne, Estonia), and 2 µl of DNA template. The cycling conditions were initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 59 °C for 30 s and extension at 72 °C for 40 s and final extension at 72 °C for 7 min. Amplicons were resolved in a 1.5% agarose gel stained with ethidium bromide (Sigma-Aldrich, GmbH, Germany) against a 1 Kb DNA ladder (HyperLadder, Bioline, London, UK). The amplicons were scored as either homozygous susceptible (SS) at 312 bp, homozygous resistant (RR) at 523 bp or heterozygous (RS) when both bands were visualized.
Representative GSTe2 allele positive samples were sequenced for the GSTe2 gene using the Gste2F and Gste2R primers as described previously38. PCR comprised a reaction volume of 15 µl in MyTaq DNA Polymerase Kit (Bioline, London, UK) containing 10 µM of each primer, 5X My Taq reaction buffer, 2 µl of My taq DNA polymerase and 1 µl of DNA template. PCR conditions were: initial denaturation of 5 min at 95 °C, followed by 30 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min, with a final extension at 72 °C for 10 min. Cleaning and sequencing of amplicons were performed as described above.
Sequence and polymorphism analysis
Sequences (mosquito, P. falciparum, GSTe2) were viewed and cleaned in Geneious Prime39 and queried in GenBank using Basic Local Alignment Search Tool (BLastn). Parasite sequences were assigned as P. falciparum after > 98% percentage identity. MAFFT in Geneious Prime39 was used to perform multiple sequence alignments with default parameters. Maximum likelihood (ML) trees were inferred for mosquito ITS2 sequences using the best fit model of sequence evolution with nodal support for different groupings evaluated through 1000 bootstrap replications. GSTe2 gene polymorphism analysis was performed in Geneious Prime and ML tree reconstructed from MAFFT alignment using PhyML v. 2.2.4. Haplotype distribution network was constructed using Templeton-Crandall Sing (TCS) program v. 1.2140.
Statistical analysis
Relative abundance was used to estimate the outdoor composition of the anopheline mosquitoes. Daily counts of female mosquito/trap/night for An. funestus s.l. and An. gambiae s.l. were compared for each area using generalized linear models (GLM) with negative binomial error structure based on best-fit model residuals. The mean catches/trap/night was computed for each of the species complexes. The P. falciparum sporozoite infection rates (Pfsp) were expressed as the number of positive specimens of the total number of specimens examined. The distribution of L119F-GSTe2 mutations was assessed by determining allelic frequencies in different species. Infection status among the resistant mosquitoes was compared using the Fisher’s Exact Test. Data were analyzed using R v 4.1.0 software at 95% confidence limit.
Ethical considerations
Ethical review and approval of the study was granted by the Scientific and Ethical Review Unit (SERU) of the Kenya Medical Research Institute (KEMRI) (Protocol No. SSC 2787). Prior to data collection, the purpose of the study, procedures and associated benefits/risks were provided to the local leadership at county and community levels. Additionally, informed verbal consent to trap mosquitoes around homesteads was obtained from household heads.
Supplementary Information
Acknowledgements
We thank community members in the study areas who provided access to their homesteads.
Author contributions
D.P.T. conceived the study; G.R., R.S., D.P.T. designed and implemented entomological surveys and morphological identification; F.K. performed experiments; F.K., E.O.O., D.P.T. analysed the data; C.M., R.S., C.S.W., B.T., D.P.T. contributed to resources and funding acquisition; C.M., E.A.O., D.P.T. supervision of the student (F.K.); F.K., B.T., D.P.T. wrote the draft manuscript; All authors reviewed the manuscript and approved the final version.
Funding
FK received an ICIPE MSc-Dissertation Research Internship Programme (DRIP) scholarship through the project Combatting Arthropod Pests for better Health, Food and Climate Resilience (CAP-Africa; project number RAF-3058 KEN-18/0005) funded by Norwegian Agency for Development Cooperation (Norad). Financial support for the study came from the Norad funded CAP-Africa project. This research was funded in part by the Wellcome Trust (International Intermediate Fellowship to DPT (222005/Z/20/Z) and a Senior Research Fellowship to CSW: 217188/Z/19/Z). We also gratefully acknowledge the financial support for this research by the following organizations and agencies: Swedish International Development Cooperation Agency (Sida), Swiss Agency for Development and Cooperation (SDC), Federal Democratic Republic of Ethiopia and the Government of the Republic of Kenya. The views expressed herein do not necessarily reflect the official opinion of the donors. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. For the purpose of open access, the authors have applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Data availability
All data supporting the conclusions of this article are included within the article. Accession codes Sequences of selected haplotypes of anopheline species harboring Plasmodium falciparum sporozoite infection and those without have been deposited in GenBank under accession numbers; An. funestus (OM857956-OMOM857959), An. longipalpis C (OM791454-OM791458), An. rivulorum (OM791451-OM791452) and (OM812265-OM812270 and ON146197-ON146198) unclassified Anopheles spp.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-11333-2.
References
- 1.Division of National Malaria Programme (DNMP) [Kenya] and ICF 2021. 2020 Kenya Malaria Indicator Survey Summary Report. (DNMP and ICF, 2020).
- 2.National Malaria Control Programme. Kenya malaria strategy 2019–2023. http://fountainafrica.org/wp-content/uploads/2020/01/Kenya-Malaria-Strategy-2019-2023 (Ministry of Health, 2018).
- 3.World Health Organisation. World malaria report 2020: 20 years of global progress and challenges. https://apps.who.int/iris/handle/10665/337660. License: CC BY-NC-SA 3.0 IGO (World Health Organization, 2020).
- 4.Ministry of Health (MOH). Malaria epidemic preparedness and response rapid assessment report. (Ministry of Health, 2020). https://www.measuremalaria.org/wp-content/uploads/2020/05/EPR-Rapid-Assessment-Report-Final-Signed.pdf.
- 5.Bamou RM, et al. Entomological and anthropological factors contributing to persistent malaria transmission in Kenya, Ethiopia and Cameroon. J. Infect. Dis. 2021;223:S155–S170. doi: 10.1093/infdis/jiaa774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.PMI. U.S. President’s malaria initiative Kenya: Malaria operational plan FY 2020. Published online (2020).
- 7.Carnevale P, Manguin S. Review of issues on residual malaria transmission. J. Infect. Dis. 2021;223:S61–S80. doi: 10.1093/infdis/jiab084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherrard-Smith E, et al. Mosquito feeding behavior and how it influences residual malaria transmission across Africa. Proc. Natl. Acad. Sci. USA. 2019;116:15086–15095. doi: 10.1073/pnas.1820646116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okello PE, et al. Variation in malaria transmission intensity in seven sites throughout Uganda. Am. J. Trop. Med. Hyg. 2006;75:219–225. doi: 10.4269/ajtmh.2006.75.219. [DOI] [PubMed] [Google Scholar]
- 10.Degefa T, et al. Indoor and outdoor malaria vector surveillance in western Kenya: Implications for better understanding of residual transmission. Malar. J. 2017;16:443. doi: 10.1186/s12936-017-2098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kreppel KS, et al. Emergence of behavioural avoidance strategies of malaria vectors in areas of high LLIN coverage in Tanzania. Sci. Rep. 2020;10:14527. doi: 10.1038/s41598-020-71187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogola EO, et al. Insights into malaria transmission among Anopheles funestus mosquitoes Kenya. Parasit. Vectors. 2018;11:577. doi: 10.1186/s13071-018-3171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakilla C, et al. Malaria vector species composition and entomological indices following indoor residual spraying in regions bordering Lake Victoria, Tanzania. Malar J. 2020;19:383. doi: 10.1186/s12936-020-03452-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dia, I., Guelbeogo, M. W. & Ayala, D. Advances and perspectives in the study of the malaria mosquito Anopheles funestus. Anopheles mosquitoes. In New Insights into Malaria Vectors (ed Manguin, S.) 828 (Intech, 2013).
- 15.Coetzee M. Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae) Malar. J. 2020;19:70. doi: 10.1186/s12936-020-3144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.St. Laurent B, et al. Molecular characterization reveals diverse and unknown malaria vectors in the western Kenyan highlands. Am. J. Trop. Med. Hyg. 2016;94:327–335. doi: 10.4269/ajtmh.15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong D, et al. Extensive new Anopheles cryptic species involved in human malaria transmission in western Kenya. Sci. Rep. 2020;10:16139. doi: 10.1038/s41598-020-73073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogola EO, Chepkorir E, Sang R, Tchouassi DP. A previously unreported potential malaria vector in a dry ecology of Kenya. Parasit. Vectors. 2019;12:80. doi: 10.1186/s13071-019-3332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riveron JM, et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15:R27. doi: 10.1186/gb-2014-15-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okumu F, Finda M. Key characteristics of residual malaria transmission in two districts in South-Eastern Tanzania—Implications for improved control. J. Infect. Dis. 2021;223:S143–S154. doi: 10.1093/infdis/jiaa653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulamba C, et al. Widespread pyrethroid and DDT resistance in the major malaria vector Anopheles funestus in East Africa is driven by metabolic resistance mechanisms. PLoS ONE. 2014;9:e110058. doi: 10.1371/journal.pone.0110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snow, R.W. & Noor, A.M. Malaria risk mapping in Africa. The historical context to the Information for malaria (INFORM) project (2015).
- 23.Charlwood JD, Vij R, Billingsley PF. Dry season refugia of malaria-transmitting mosquitoes in a dry savannah zone of east Africa. Am. J. Trop. Med. Hyg. 2000;62:726–732. doi: 10.4269/ajtmh.2000.62.726. [DOI] [PubMed] [Google Scholar]
- 24.Gillies MT, Smith A. The effect of a residual house-spraying campaign in East Africa on species balance in the Anopheles funestus group. The replacement of An. funestus giles by An. rivulorum leeson. Bull. Entomol. Res. 1960;51:243–252. doi: 10.1017/S0007485300057953. [DOI] [Google Scholar]
- 25.Kent RJ, et al. Feeding and indoor resting behaviour of the mosquito Anopheles longipalpis in an area of hyperendemic malaria transmission in southern Zambia. Med. Vet. Entomol. 2006;20:459–463. doi: 10.1111/j.1365-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adunga N, Peteros B. Determination of the human blood index of some anopheline mosquitoes by using ELISA. Ethiop. Med. J. 1996;34:1–10. doi: 10.1111/mve.12514. [DOI] [PubMed] [Google Scholar]
- 27.Machani MG, et al. Resting behaviour of malaria vectors in highland and lowland sites of western Kenya: Implication on malaria vector control measures. PLoS ONE. 2020;15:e0224718. doi: 10.1371/journal.pone.0224718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tchouakui M, et al. Fitness costs of the glutathione S-transferase epsilon 2 (L119F-GSTe2) mediated metabolic resistance to insecticides in the major African malaria vector Anopheles funestus. Genes. 2018;9:645. doi: 10.3390/genes9120645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemingway J. The role of vector control in stopping the transmission of malaria: Threats and opportunities. Philos. Trans. R. Soc. B. 2014;369:20130431. doi: 10.1098/rstb.2013.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell DJ, Gichohi H, Mwangi A, Chege L. Land use conflict in Kajiado District, Kenya. Land Use Policy. 2000;17:337–348. doi: 10.1016/S0264-8377(00)00038-7. [DOI] [Google Scholar]
- 31.Regional dissemination workshops for the Kenya malaria strategy (2019–2023). https://www.measuremalaria.org/wp-content/uploads/2020/05/KMS-Dissemination-report-April-final-signed.pdf (2020).
- 32.Omondi CJ, et al. Perennial transmission of malaria in the low altitude areas of Baringo County, Kenya. Malar J. 2017;16:257. doi: 10.1186/s12936-017-1904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara (Ethiopian zoogeographical region) S. Afr. Inst. Med. Res. 1987;55:1–146. [Google Scholar]
- 34.Scott JA, Brogdon WG, Collins F. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 1993;49:520–529. doi: 10.1186/s13071-021-05020-w. [DOI] [PubMed] [Google Scholar]
- 35.Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am. J. Trop. Med. Hyg. 2002;66:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- 36.Beebe NW, Saul A. Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction-restriction fragment length polymorphism analysis. Am. J. Trop. Med. Hyg. 1995;53:478–481. doi: 10.4269/ajtmh.1995.53.478. [DOI] [PubMed] [Google Scholar]
- 37.Chaumeau V, et al. Comparison of the performances of five primer sets for the detection and quantification of Plasmodium in anopheline vectors by real-time PCR. PLoS ONE. 2016;11:e0159160. doi: 10.1371/journal.pone.0159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ndo C, Kopya E, Irving H, Wondji C. Exploring the impact of glutathione s-transferase (GST)-based metabolic resistance to insecticide on vector competence of Anopheles funestus for Plasmodium falciparum. Wellcome Open Res. 2019;4:52. doi: 10.12688/wellcomeopenres.15061.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clement DP, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2001;9:1657–1659. doi: 10.4028/www.scientific.net/AMM.571-572.461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the conclusions of this article are included within the article. Accession codes Sequences of selected haplotypes of anopheline species harboring Plasmodium falciparum sporozoite infection and those without have been deposited in GenBank under accession numbers; An. funestus (OM857956-OMOM857959), An. longipalpis C (OM791454-OM791458), An. rivulorum (OM791451-OM791452) and (OM812265-OM812270 and ON146197-ON146198) unclassified Anopheles spp.