Abstract
Clostridium difficile is the etiological agent of antibiotic-associated colitis and the most common cause of hospital-acquired infectious diarrhea. Fluoroquinolones such as ciprofloxacin are associated with lower risks of C. difficile-associated diarrhea. In this study, we have analyzed 72 C. difficile isolates obtained from patients with different clinical courses of disease, such as toxic megacolon and relapses; the hospital environment; public places; and horses. They were investigated for their susceptibilities to moxifloxacin (MXF), metronidazole (MEO), and vancomycin (VAN). Mutants highly resistant to fluoroquinolones were selected in vitro by stepwise exposure to increasing concentrations of MXF. The resulting mutants were analyzed for the presence of mutations in the quinolone resistance-determining regions of DNA gyrase (gyrA), the production of toxins A and B, and the epidemiological relationship of these isolates. These factors were also investigated using PCR-based methods. All strains tested were susceptible to MEO and VAN. Twenty-six percent of the clinical isolates (19 of 72) were highly resistant to MXF (MIC ≥ 16 μg/ml). Fourteen of these 19 strains contained nucleotide changes resulting in amino acid substitutions at position 83 in the gyrA protein. Resistant strains selected in vitro did not contain mutations at that position. These findings indicate that resistance to MXF in a majority of cases may be due to amino acid substitution in the gyrA gene.
Clostridium difficile is the major cause of hospital-acquired infectious diarrhea (29). Strains of C. difficile produce two toxins, an enterotoxin (toxin A) and a cytotoxin (toxin B), which are the largest bacterial toxins known (molecular masses of 308 and 207 kDa, respectively) (2). Most toxigenic strains release both toxins; however, strains deficient in the production of either toxin A or toxin B have been described (6, 33). Antibiotic exposure and changes of environmental conditions affect toxin production (17, 21, 24, 26).
A prolonged course of antibiotic treatment or the use of two or more antibiotics in combination increases the risk of C. difficile-associated diarrhea (CDAD) (14). Vancomycin (VAN) and metronidazole (MEO) are first-line antibiotics for the treatment of CDAD. However, about 15 to 20% of patients relapse after discontinuation of antimicrobial treatment (35). Several studies identified the oral use of VAN in the treatment of CDAD as a risk factor for colonization with VAN-resistant enterococci (13, 27). Therefore, there is still a need for development of new treatments.
Fluoroquinolones are potent, synthetic, antimicrobial agents that are increasingly used in the treatment of human infections. The new-generation quinolones, like gatifloxacin (GAT), trovafloxacin (TVA), and moxifloxacin (MXF), are characterized by improved activity against gram-positive cocci as well as against some gram-positive and -negative anaerobic bacteria (22). MXF, approved by the Food and Drug Administration in 1999, is available only as an oral preparation and shows a high level of activity against many anaerobes (1, 10, 37). There are no data available concerning the genetic basis for quinolone resistance in C. difficile strains. Moreover, a possible relationship between antibiotic treatment and toxin production is obscure.
Since the NCCLS has not yet approved breakpoints for MXF and anaerobes, in this study strains for which MICs were ≤ 2 μg/ml were interpreted as susceptible and those for which MICs were ≥ 8 μg/ml were interpreted as resistant according to breakpoints published for TVA and anaerobic bacteria (23).
Resistance to fluoroquinolones is mediated either by target mutations affecting the genes gyrA and gyrB for the DNA gyrase subunits A and B as well as parC and parE for the respective subunits A and B of DNA topoisomerase IV or by mutations resulting in the reduction of the intracellular accumulation of the drug. Mutations in the highly conserved quinolone-resistance-determining regions (QRDR) occur in a different order in gram-positive and gram-negative organisms: DNA gyrase seems to be the primary target of most quinolones in gram-negative bacteria and mycobacteria (19, 30). However, DNA topoisomerase IV appears to be the primary target in gram-positive bacteria (11). Additionally, depending on the fluoroquinolone, parC and gyrA may be interchangeable targets leading to different levels of resistance (25, 34).
Few data exist about the epidemiology and the mechanisms of resistance to fluoroquinolones in anaerobes, probably due to the low activity of ciprofloxacin (CIP) in contrast to that of newer derivatives, like MXF.
In this study, 72 C. difficile isolates recovered from patients with different clinical courses and from hospital and public environments were investigated for their susceptibility to MXF, MEO, and VAN and for harboring mutations in the QRDR of DNA gyrase (gyrA), toxin production, and genotype. The results of this study provide data on the epidemiology of the antimicrobial susceptibility of C. difficile.
MATERIALS AND METHODS
Antibiotics.
MXF was kindly supplied by Bayer AG, Wuppertal, Germany.
C. difficile strains.
The strains investigated in this study were ATCC 43255 and 17857 and strains recovered from patients (n = 49), the hospital or public environment (n = 12 and n = 5, respectively), and horses (n = 4) (20). The sources of the MXF-resistant isolates are listed in Table 1. All strains were grown anaerobically on the selective medium cycloserine-cefoxitin-fructose agar (12) at 37°C for 48 h. Isolates were identified as C. difficile with the latex agglutination test (Becton Dickinson, Cockeysville, Md.) and the ProDisc test (Remel, Norcross, Ga.). PCR was used to identify toxin A and toxin B genes. Selected strains were subjected to 16S ribosomal DNA (rDNA) PCR. Strains were maintained in cooked meat broth (Hardy Diagnostics, Santa Maria, Calif.).
TABLE 1.
C. difficile strains resistant to MXF investigated in this study and their sources
| Isolate(s) | Source | Reference |
|---|---|---|
| 1542, 2897, 2923, 2518 | Patients with CDAD, toxigenic strains, Leipzig, Germany | This study |
| 1000, 1001, 1002 | Toxic megacolon, UCDMC,a Sacramento, Calif. | 7 |
| 1009 | Relapse patients, toxigenic strains, UCDMC | This study |
| 1111/1, 1111/2 | Relapse patient, toxigenic strains, Mercy Hospital, Sacramento, Calif. | This study |
| E 213, E 214, E 239, E 287, E 496, E 497, E 498, E 510, E 511 | Hospital environment, bathrooms, bedrooms, (all but E 213 and E 214 are toxigenic strains), UCDMC | 5 |
| 6MX4, 6MX8, 9MX16, 9MX32 | In vitro selection | This study |
UCDMC, University of California-Davis Medical Center.
Antimicrobial susceptibility testing.
MICs were determined using the broth microdilution technique performed in brucella broth, according to the recommendations of the NCCLS (23), and with Etest (AB BIODISK, Solna, Sweden). Etest was performed by inoculating the surface of prereduced brucella agar plates containing vitamin K1, hemin, and 5% defibrinated sheep red blood cells with a 1 McFarland standard-matched inoculum. The inoculation was done with cotton-tipped swabs that were dipped into the inoculum, pressed against the inside wall of the test tube to remove excess fluid, and streaked three times, with the plate being rotated approximately 90° each time to ensure an even distribution of inoculum. Etest strips were used according to the manufacturer's instructions.
Selection of quinolone-resistant mutants.
Selection of quinolone-resistant mutants of ATCC strain 43255 was performed in liquid culture. Bacteria collected from five agar plates (after 48 h of growth) were suspended in 10 ml of brain heart infusion (BHI) broth and were centrifuged for 10 min at 2,700 × g. The pellet was resuspended in 2 ml of BHI. Fifty microliters of the suspension was added to 2 ml of BHI containing four different concentrations of MXF (0.5, 1, 2, and 4 μg/ml). After 24 h, cultures which showed visible growth were plated on brucella agar, centrifuged, washed in BHI, and exposed one more time to the same antibiotic concentration. For the next passage, 1-dilution-higher MXF concentrations (1, 2, 4, and 8 μg/ml) were used. That procedure was repeated six and nine times, finally exposing the bacteria to 32 μg of MXF/ml.
DNA extraction.
An isolated colony from each strain was transferred with an inoculating loop into a 0.6-ml tube containing 100 μl of sterile water, boiled at 100°C for 10 min, and centrifuged at low speed (3,000 × g) to remove cell debris. The DNA-containing supernatant was used for amplification and genotyping reactions.
Amplification and sequencing of gyrA.
A 247-bp fragment of gyrA was amplified using degenerated primers developed from consensus regions of Clostridium acetobutylicum (C 94V, C 121V, C 316R, and C 358R) (Table 2). The DNA sequence of C. difficile gyrA was determined and used to design specific primers (CdgaV and CdgaR) (Table 2 and Fig. 1). Thirty cycles of the following PCR profile were run: 30 s at 95°C, 30 s at 48°C, and 60 s at 72°C. The resulting DNA fragments were purified with Amicon Microcon-PCR Centrifugal Filter Devices (Millipore Corporation, Bedford, Mass.). Complementary strands were sequenced on an ABI310 sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.) using either PCR primer.
TABLE 2.
Nucleotide sequence of PCR primers used to amplify the gyrA gene (C 94V, C 121V, C 316R, C 358R, CdgaV, and CdgaR) and 16S rDNA (609V and 699R) of C. difficile
| Primer | Sequence |
|---|---|
| C 94V | 5′-CGTGCACT(G/C/T)CC(A/C/G)GATGT-3′ |
| C 121V | 5′-CTTAAGCC(A/G/T)GTTCAT(A/C)G-3′ |
| C 316R | 5′-GAACCAAAGTT(A/T)CC(G/T/A)TG-3′ |
| C 358R | 5′-GCTTCTGT(A/G)TA(A/T)C(T/G)CAT-3′ |
| CdgaV | 5′-TTTAAAGCCAGTTCATAG-3′ |
| CdgaR | 5′-GAACCAAAGTTACCATG-3′ |
| 609V | 5′-GGATTAGATACCC(C/G/T)(A/G/T)GTA-3′ |
| 699R | 5′-(A/T/G)GGGTTGCGCTCGTT(A/G)C-3′ |
FIG. 1.
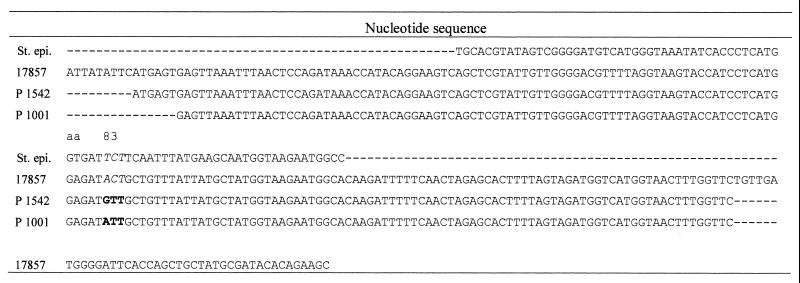
QRDR nucleotide sequence of the amplified fragment of the gyrA gene of C. difficile ATCC 17857, 1542 and 1001 and a part of the published sequence for Staphylococcus epidermidis (St. epi.). Italics indicate the wild-type sequence at amino acid position 83; boldface indicates nucleotide mutation leading to amino acid substitution; E. coli coordinates are given for amino acid sequence.
Primers for 16S rDNA amplification (609V and 699R) (Table 2) were designed using 16S rRNA sequences published by Brosius et al. (3). Sequencing of 16S rDNA identified these strains as C. difficile.
Comparison of sequences and design of primers.
Sequences were compared using ARB software (O. Strunk and W. Ludwig, http://www.mikro.biologie.tu-muenchen.de). This program was primarily designed for phylogenetic analysis of rRNA sequence data but can be used for any other RNA or DNA sequence comparison. Nucleic acid sequences from public databases and those obtained in this study were manually aligned according to the amino acid alignment. The distance-matrix method implemented in the program was used for calculating sequence similarities. In order to amplify parC of C. difficile, the DNA sequence of the following bacteria was used for developing degenerated primers: Bacillus subtilis, Mycoplasma genitalium, Mycoplasma hominis, Staphylococcus aureus, Streptococcus pneumoniae, and Streptococcus pyogenes. These are all so-called “low G+C gram-positive bacteria” which are closely related phylogenetically to C. difficile.
Amplification of toxin A and B genes.
Toxin A and B genes were amplified as described elsewhere (15, 32).
DNA fingerprinting.
Arbitrarily primed PCR (AP-PCR) was performed with the 19-mer oligonucleotide T-7 used as the primer. The DNA banding patterns were compared by running PCR products on the same gel as described previously (9, 29, 31).
RESULTS
Antimicrobial susceptibility.
All 72 strains analyzed were susceptible to MEO. The MIC at which 50% of the isolates tested were inhibited (MIC50) and MIC90 were 0.125 and 0.5 μg/ml, respectively. They were also susceptible to VAN; the VAN MIC50 and MIC90 were 1 μg/ml. The MXF MIC50 and MIC90 were 1 and ≥ 32 μg/ml. For 19 (26%) of the 72 C. difficile isolates studied, MXF MICs were ≥ 16 μg/ml as determined by Etest. Two MXF-resistant isolates were recovered from one patient (1111/1 and 1111/2). They showed differences in growth phenotype and in antimicrobial susceptibility for MEO and VAN by 3 or 4 dilutions.
Nine of twelve isolates from the hospital environment were resistant to MXF, while all strains isolated from the non-hospital environment were susceptible to MXF (five of five). The three isolates from patients with toxic megacolon were highly MXF resistant (1000, 1001, and 1002).
The 19 strains resistant to MXF were tested with the broth microdilution method against 12 other quinolones (one agent each represented groups I to III; all tested group IV fluoroquinolones are shown in Table 3). The MICs of MXF determined by the broth microdilution method were in general 1 or 2 dilutions lower than those determined by Etest (MXF MIC50 and MIC90, 16 and ≥ 32 μg/ml, respectively), which is within acceptable limits of error for the test. Except for clinafloxacin (MIC50 and MIC90, both 2 μg/ml), other quinolones did not show activities against the tested isolates (MIC50 and MIC90 in micrograms per milliliter): norfloxacin (both >64), pefloxacin (both >32), CIP (both ≥32), fleroxacin (both >32), ofloxacin (both ≥32), enoxacin (both 32), grepafloxacin (both 32), sparfloxacin (both >32), GAT (both 32), gemifloxacin (both 32), and TVA (both 32).
TABLE 3.
Antimicrobial susceptibilities, MIC50 and MIC90, gyrA sequence, production of toxins A and B (Tox A/B), and AP-PCR type (AP type) of 19 MXF-resistant C. difficile isolates, 4 in vitro-selected strains, and 3 ATCC strains (ATCC 17857 and ATCC 43255, C. difficile; ATCC 25285, Bacteroides fragilisa,b,e
| Strain | MIC (μg/ml)
|
GyrA aa/ntc | Tox A/B | AP type | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I | Group II | Group III | Group IV | ||||||||||
| NOR | CIP | GRX | GAT | GMX | TVA | CLX | MXF | MEO | VAN | ||||
| ATCC 17857 | 8 | >32 | 16 | 2 | 2 | 2 | 0.5 | 2 | 0.125 | 1.5 | T/ACT | −/− | NTd |
| ATCC 43255 | 8 | >32 | 2 | 1 | 1 | 1 | 0.5 | 0.5 | 0.047 | 1 | T/ACT | +/+ | NT |
| ATCC 25285 | 4 | >32 | 4 | 0.5 | 1 | 1 | 0.5 | 0.5 | 0.5 | NT | NT | NT | NT |
| 1542 | >64 | >32 | 32 | 16 | 32 | 16 | 2 | 16 | 0.38 | 1 | V/GTT | +/+ | 2 |
| 2897 | >64 | >32 | 32 | >32 | 32 | 32 | 4 | >32 | 0.125 | 0.75 | I/ATT | +/+ | 2 |
| 2923 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 16 | 0.25 | 1 | T/ACT | +/+ | 1 |
| 2518 | >64 | 32 | 32 | 32 | 8 | 4 | 2 | >32 | 0.125 | 1 | T/ACT | +/+ | 3 |
| 1000 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 32 | 0.125 | 1 | I/ATT | +/+ | 5A,B |
| 1001 | >64 | >32 | 32 | 16 | 32 | 32 | 2 | 16 | 0.19 | 1 | I/ATT | +/+ | 5A,B |
| 1002 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 16 | 0.125 | 1 | I/ATT | +/+ | 5A,B |
| 1009 | >64 | >32 | 32 | 32 | 16 | 32 | 2 | 16 | 0.125 | 0.75 | I/ATT | +/+ | 5A,B |
| 1111/1 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 16 | 0.125 | 0.5 | IATT | +/+ | 4 |
| 1111/2 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 16 | 1 | 1.5 | IATT | +/+ | 4 |
| E 213 | >64 | >32 | 32 | 32 | 16 | 16 | 2 | 16 | 0.125 | 0.75 | T/ACT | −/− | 5A,B |
| E 214 | >64 | >32 | >32 | 32 | 16 | 32 | 2 | 16 | 0.094 | 0.75 | T/ACT | −/− | 6 |
| E 239 | >64 | 16 | 16 | 16 | 16 | 32 | 2 | >32 | 0.094 | 0.25 | IATT | +/+ | 7 |
| E 287 | >64 | >32 | >32 | 32 | 32 | 32 | 2 | 32 | 0.094 | 1 | IATT | +/+ | 5A,B |
| E 496 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 16 | 0.5 | 0.75 | IATT | +/+ | 5A,B |
| E 497 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 16 | 0.19 | 1 | T/ACT | +/+ | 5A,B |
| E 498 | >64 | >32 | 32 | 32 | 16 | 32 | 2 | 16 | 0.5 | 1 | IATT | +/+ | 6 |
| E 510 | >64 | >32 | 16 | 32 | 16 | 32 | 2 | 16 | 0.125 | 0.75 | IATT | +/+ | 5A,B |
| E 511 | >64 | >32 | 32 | 32 | 32 | 32 | 2 | 16 | 0.094 | 1 | IATT | +/+ | 6 |
| 6MX4 | >64 | >32 | >32 | 8 | 16 | 8 | 2 | 8 | 0.094 | 1.5 | T/ACT | +/+ | NT |
| 6MX8 | >64 | >32 | >32 | 8 | 16 | 4 | 2 | 8 | 0.064 | 1.5 | T/ACT | +/+ | NT |
| 9MX16 | >64 | >32 | 32 | 8 | 8 | 4 | 2 | 4 | 0.023 | 1 | T/ACT | +/+ | NT |
| 9MX32 | >64 | >32 | 32 | 8 | 8 | 4 | 2 | 4 | 0.016 | 0.75 | T/ACT | +/+ | NT |
NOR, norfloxacin; GRX, grepafloxacin; GMX, gemifloxacin; and CLX, clinafloxacin.
Groups I to IV are fluoroquinolone groups (22).
aa, amino acid; nt, nucleotides. E. coli coordinates are given; boldface indicates mutations leading to amino acid substitution.
NT, not tested.
MIC50s for NOR, CIP, GRX, GAT, GMX, TVA, CLX, MXF, MEO, and VAN are >64, >32, 32, 32, 32, 32, 2, 16, 0.125, and 1 μg/ml, respectively. MIC90s for the same drugs are >64, >32, 32, 32, 32, 32, 2, >32, 0.5, and 1 μg/ml, respectively.
ATCC strain 43255 (for which the MXF MIC was 0.5 μg/ml) was exposed six and nine times to increasing concentrations of MXF (starting with 0.5 μg/ml and going to 32 μg/ml). That resulted in MICs increasing from 8 to >32 μg/ml as determined by Etest (isolates 6MX4, 6MX8, 9MX16, and 9MX32) (Table 3). Once again for these four strains, MICs were 1 or 2 dilutions higher when evaluated with Etest than with the microdilution broth procedure.
GyrA sequence.
In Escherichia coli and in the closest related Clostridium species with a known gyrA amino acid sequence, Clostridium acetobutylicum, the amino acid serine was found at position 83 (E. coli coordinates). The wild-type and quinolone-susceptible strains of C. difficile carry the amino acid threonine at position 83.
Fourteen of the 19 MXF-resistant strains contained a mutation at codon 83 (E. coli coordinates) in the gyrA gene resulting in an amino acid exchange. Thirteen isolates showed the same nucleotide transition (ACT→ATT) resulting in a threonine→isoleucine change. In one isolate (1542), two nucleotide changes (ACT→GTT) resulted in a replacement of threonine by valine (Table 3; Fig. 1). All MXF-susceptible strains had the same sequence found in the two MXF-susceptible C. difficile ATCC strains. All four in vitro-selected MXF-resistant strains retained the wild-type sequence at position 83 in the gyrA protein.
Toxin production.
Forty-two (58%) C. difficile isolates produced toxins A and B. Toxin A and B gene sequences were not detected in 30 isolates. Seventeen (89%) of the 19 MXF-resistant strains were toxigenic. The strains recovered from the public environment produced no toxin; however, 9 (75%) of 12 hospital environment strains produced toxins A and B.
Genotyping.
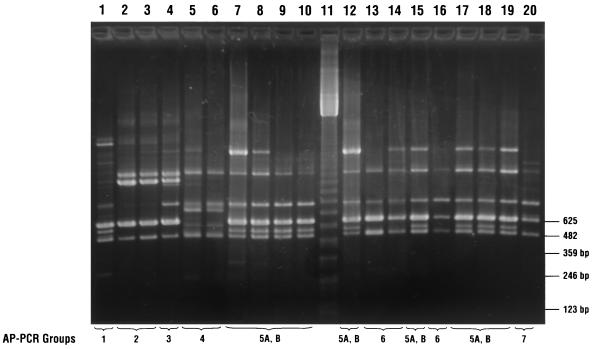
Of the 19 MXF-resistant strains, seven different groups, including two subgroups, were identified using the T-7 primer and the Dice similarity coefficient (Fig. 2) (9, 29, 31). No correlation between genotype and antimicrobial susceptibility was detected, as some resistant and susceptible strains had the same genotype.
FIG. 2.
Genotyping of C. difficile isolates by AP-PCR with a T-7 primer. Lane 11, 123 bp-ladder; lanes 1 to 10 and 12 to 20, AP-PCR banding patterns of MXF-resistant isolates. Lanes and isolates: 1, 2932; 2, 1542; 3, 2897; 4, 2518; 5, 1111/1; 6, 1111/2; 7, 1009; 8, 1002; 9, 1001; 10, 1000; 12, E 214; 13, E 213; 14, E 511; 15, E 510; 16, E 498; 17, E 497; 18, E 496; 19, E 287; and 20, E 239. AP-PCR types are indicated by numbers beneath the lanes.
DISCUSSION
By using degenerated primers developed from conserved QRDR of the gyrA gene of C. acetobutylicum, we were able to amplify and sequence the corresponding region of the gyrA gene from C. difficile. The deduced amino acid sequence shows high homology with other gyrA-like proteins, such as 83% identity with that of B. subtilis (NCBI accession no. Z99104), 81% identity with that of S. pneumoniae (accession no. AB010387), 77% identity with that of S. aureus (accession no. AF044066), and 82% identity with that of C. acetobutylicum (accession no. U35453).
Ser-83 in the QRDR of E. coli seems to play a key role in resistance to fluoroquinolones (4). This study constitutes the first genetic characterization of fluoroquinolone resistance in C. difficile. Since we did not detect base changes in gyrA in 5 of the 19 MXF-resistant strains or in the in vitro-selected strains, resistance in those strains might be due to mutation(s) in topoisomerase IV, the other known target of fluoroquinolones. Additionally, increased drug efflux might be involved in the expression of fluoroquinolone resistance in the mutants. Whether the low level of resistance found in the in vitro-selected mutants is due to the limited drug exposure (considering that clinical isolates may be more consistently exposed to drugs during treatment) is not known and needs further investigation. According to various studies on quinolone resistance, high-level resistance develops in multiple steps involving several targets and pathways of the antibiotic (11, 16, 36). Another possibility is that the in vitro-selected isolates consist of a mixed population (heteroresistance). Due to the finding that the parC gene plays a role as the primary target in gram-positive organisms, further studies will focus on that gene.
Initial attempts to amplify the parC gene using degenerated primers developed from a consensus sequence from published parC sequences of different gram-positive organisms were unsuccessful. The finding that the only PCR fragments obtainable contained the gyrA sequences suggests a very high homology in the nucleotide sequences of the two genes used for the primer design.
The high frequency of MXF resistance among C. difficile isolates (26%) is in contrast to the findings of other studies that reported a prevalence of MXF resistance between 0 and 14% (10, 18, 37). The quinolones of groups I, II, and III showed no activity against the MXF-resistant strains. Only clinafloxacin of group IV was active against the strains tested (22). The sources of the strains investigated in this study may be one explanation for the high rate of resistance and suggest a relationship between antibiotic exposure and antibiotic susceptibility. Almost all clinical isolates were recovered from patients with recurrent CDAD or toxic megacolon (Table 1).
The persistence of environmental strains in hospitals is known to be correlated with endemic and nosocomial CDAD (5, 8, 28). Our discovery of a high number of resistant isolates in the hospital environment and the finding that these hospital isolates contain toxin genes confirm the important role of these strains as a possible source for exogenous infections.
The resistance of C. difficile strains to newer quinolones raises a number of questions regarding the origin and the development of resistance. Since MXF has been approved for use for only about a year in the United States and for a few months in Europe, it is unlikely to be involved in the selection of resistant mutants. Therefore, the role that the wide clinical use of quinolones, such as CIP, OFX, and levofloxacin, might have played in the development of this resistance should be investigated further. The finding of MXF-resistant isolates in the hospital environment but not in public places (parks and public bathrooms) indicates the need for further studies.
ACKNOWLEDGMENT
G.A. was supported by a grant from the Paul-Ehrlich-Society, Frankfurt, Germany.
REFERENCES
- 1.Ackermann G, Schaumann R, Pless B, Claros M C, Goldstein E J C, Rodloff A C. Comparative activity of moxifloxacin in vitro against obligately anaerobic bacteria. Eur J Clin Microbiol Infect Dis. 2000;19:228–232. doi: 10.1007/s100960050465. [DOI] [PubMed] [Google Scholar]
- 2.Banno Y, Kobayashi T, Kono H, Watanabe K, Veno K, Nozawa Y. Biochemical characterisation and biological action of two toxins (D-1 and D-2) from Clostridium difficile. Rev Infect Dis. 1984;6:S11–S20. doi: 10.1093/clinids/6.supplement_1.s11. [DOI] [PubMed] [Google Scholar]
- 3.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Cambau E, Gutmann L. Mechanisms of resistance to quinolones. Drugs. 1993;45:15–23. doi: 10.2165/00003495-199300453-00005. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S H, Tang Y J, Muenzner J, Gumerlock P H, Silva J., Jr Isolation of various genotypes of Clostridium difficile from patients and the environment in an oncology ward. Clin Infect Dis. 1996;24:889–893. doi: 10.1093/clinids/24.5.889. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S H, Tang Y J, Hansen B, Silva J., Jr Isolation of a toxin B-deficient mutant strain of Clostridium difficile in a case of recurrent C. difficile-associated diarrhea. Clin Infect Dis. 1997;26:410–412. doi: 10.1086/516324. [DOI] [PubMed] [Google Scholar]
- 7.Cohen S H, Tang Y J, Silva J., Jr Analysis of the pathogenicity locus in Clostridium difficile strains. J Infect Dis. 2000;181:659–663. doi: 10.1086/315248. [DOI] [PubMed] [Google Scholar]
- 8.Cohen S H, Tang Y J, Rahmani D, Silva J., Jr Persistence of an endemic (toxigenic) isolate of Clostridium difficile in the environment of a general medicine ward. Clin Infect Dis. 2000;30:952–954. doi: 10.1086/313807. [DOI] [PubMed] [Google Scholar]
- 9.Cohen S H, Tang Y J, Silva J., Jr Molecular typing methods for the epidemiological identification of Clostridium difficile strains. Exp Rev Mol Diagn. 2001;1:89–98. doi: 10.1586/14737159.1.1.61. [DOI] [PubMed] [Google Scholar]
- 10.Edlund C, Sabouri S, Nord C E. Comparative in vitro activity of BAY 12-8039 and five other antimicrobial agents against anaerobic bacteria. Eur J Clin Microbiol. 1998;17:193–195. doi: 10.1007/BF01691117. [DOI] [PubMed] [Google Scholar]
- 11.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 12.George W L, Sutter V L, Citron D, Finegold S M. Selective and differential medium for the isolation of Clostridium difficile. J Clin Microbiol. 1979;9:214–219. doi: 10.1128/jcm.9.2.214-219.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerding D N. Is there a relationship between vancomycin-resistant enterococcal infection and Clostridium difficile infection? Clin Infect Dis. 1997;25:206–210. doi: 10.1086/516247. [DOI] [PubMed] [Google Scholar]
- 14.Gorbach S L. Antibiotics and Clostridium difficile. N Engl J Med. 1999;341:1690–1691. doi: 10.1056/NEJM199911253412211. [DOI] [PubMed] [Google Scholar]
- 15.Gumerlock P H, Tang Y J, Weiss J B, Silva J., Jr Specific detection of toxigenic strains of Clostridium difficile in stool specimens. J Clin Microbiol. 1993;31:507–511. doi: 10.1128/jcm.31.3.507-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda T, Hernandez I, Katoh T, Miwatani T. Stimulation of enterotoxin production of Clostridium difficile by antibiotics. Lancet. 1983;i:655. doi: 10.1016/s0140-6736(83)91832-9. [DOI] [PubMed] [Google Scholar]
- 18.Hoogkamp-Korstanje J A A, Roelofs-Willemse J. Comparative in vitro activity of moxifloxacin against Gram-positive clinical isolates. J Antimicrob Chemother. 2000;45:31–39. doi: 10.1093/jac/45.1.31. [DOI] [PubMed] [Google Scholar]
- 19.Hooper D C, Wolfson J S. Mechanisms of bacterial resistance to quinolones. In: Hooper D C, Wolfson J S, editors. Quinolone antibacterial agents. 2nd ed. Washington, D.C.: American Society for Microbiology; 1993. pp. 97–118. [Google Scholar]
- 20.Jang S S, Hansen L M, Breher J E, Riley D A, Magdesian K G, Madigan J E, Tang Y T, Silva J, Jr, Hirsch D C. Antimicrobial susceptibility of equine isolates of Clostridium difficile and molecular characterization of metronidazole-resistant strains. Clin Infect Dis. 1997;25:266–267. doi: 10.1086/516235. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson S, Burman L G, Akerlund T. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology. 1999;145:1683–1693. doi: 10.1099/13500872-145-7-1683. [DOI] [PubMed] [Google Scholar]
- 22.Naber K G, Adam D die Expertengruppe der Paul-Ehrlich-Gesellschaft fuer Chemotherapie e. V. Eint. Fluorchinolone. Chemother J. 1998;2:66–68. [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 24.Onderdonk A B, Lowe B R, Bartlett J G. Effect of environmental stress on Clostridium difficile toxin levels during continuous cultivation. Appl Environ Microbiol. 1979;38:637–641. doi: 10.1128/aem.38.4.637-641.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan X S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. Antimicrob Agents Chemother. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qa'Dan M, Spyres L M, Ballard J D. pH-induced conformational changes in Clostridium difficile toxin B. Infect Immun. 2000;68:2470–2474. doi: 10.1128/iai.68.5.2470-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rafferty M E, McCormick M I, Bopp L H, Baltch A L, George M, Smith R P, Rheal C, Ritz W, Schoonmaker D. Vancomycin-resistant enterococci in stool specimens submitted for Clostridium difficile cytotoxin assay. Infect Control Hosp Epidemiol. 1997;18:343–344. doi: 10.1086/647623. [DOI] [PubMed] [Google Scholar]
- 28.Rojas S, Cohen S H, Tang Y J, Wilson J, Inciardi J, Silva J., Jr Differing epidemiology of Clostridium difficile-associated diarrhea between an oncology ward and a general medicine ward. Infect Control Hosp Epidemiol. 1999;20:14–15. doi: 10.1086/503081. [DOI] [PubMed] [Google Scholar]
- 29.Silva J, Jr, Tang Y T, Gumerlock P H. Genotyping of Clostridium difficile isolates. J Infect Dis. 1994;169:661–664. doi: 10.1093/infdis/169.3.661. [DOI] [PubMed] [Google Scholar]
- 30.Takiff H E, Salazar L, Guerrero C, Philipp W, Huang W M, Kreiswirth B, Cole S T, Jacobs W R, Jr, Telenti A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Y J, Houston S T, Gumerlock P H, Mulligan M E, Gerding D N, Johnson S, Fekety F R, Silva J., Jr Comparison of arbitrarily primed PCR with restriction endonuclease and immunoblot analyses for typing Clostridium difficile isolates. J Clin Microbiol. 1995;33:3169–3173. doi: 10.1128/jcm.33.12.3169-3173.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y J, Gumerlock P H, Weiss J B, Silva J., Jr Specific detection of Clostridium difficile toxin A gene sequences in clinical isolates. Mol Cell Probes. 1994;8:463–467. doi: 10.1006/mcpr.1994.1066. [DOI] [PubMed] [Google Scholar]
- 33.Torres J F. Purification and characterisation of toxin B from a strain of Clostridium difficile that does not produce toxin A. J Med Microbiol. 1991;35:40–44. doi: 10.1099/00222615-35-1-40. [DOI] [PubMed] [Google Scholar]
- 34.Varon E, Janoir C, Kitzis M D, Gutmann L. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:302–306. doi: 10.1128/aac.43.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walters B A J, Roberts R, Stafford R, Seneviratne E. Relapse of antibiotic associated colitis: endogenous persistence of Clostridium difficile during vancomycin therapy. Gut. 1983;24:206–212. doi: 10.1136/gut.24.3.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiedemann B, Heisig P. Mechanisms of quinolone resistance. Infection. 1994;22:S73–S79. doi: 10.1007/BF01793570. [DOI] [PubMed] [Google Scholar]
- 37.Woodcock J M, Andrews J M, Boswell F J, Brenwald N P, Wise R. In vitro activity of BAY 12–8039, a new fluoroquinolone. Antimicrob Agents Chemother. 1997;41:101–106. doi: 10.1128/aac.41.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]