Abstract
BACKGROUND:
A link between the gut microbiome and behavior is hypothesized, but most previous studies are cross-sectional or in animal models. The modifying role of host sex is poorly characterized. We aimed to identify sex-specific prospective associations between the early-life gut microbiome and preschool-age neurobehavior.
METHODS:
In a prospective cohort, gut microbiome diversity and taxa were estimated with 16S rRNA sequencing at six weeks, one year, and two years. Species and gene pathways were inferred from metagenomic sequencing at six weeks and one year. When subjects were three years old, parents completed the Behavioral Assessment System for Children, 2nd edition (BASC-2). 260 children contributed 523 16S rRNA and 234 metagenomics samples to analysis. Models adjusted for sociodemographic characteristics.
RESULTS:
Higher diversity at six weeks was associated with better Internalizing Problems among boys, but not girls [βBoys=−1.86 points/SD Shannon diversity; 95% CI (−3.29,−0.42), pBoys=0.01, βGirls=0.22 (−1.43,1.87), pGirls=0.8, pinteraction=0.06]. Among other taxa-specific associations, Bifidobacterium at six weeks was associated with Adaptive Skills scores in a sex-specific manner. We observed relationships between functional features and BASC-2 scores, including vitamin B6 biosynthesis pathways and better Depression scores.
CONCLUSIONS:
This study advances our understanding of microbe-host interactions with implications for childhood behavioral health.
INTRODUCTION
Neurodevelopmental disorders are detrimental to lifelong health and quality of life,1,2 and carry both social3,4 and financial impacts.5–7 Despite recent advancements in our understanding of the etiology of neurodevelopmental outcomes such as genetics and environmental factors,8–10 questions remain unanswered, especially regarding the mechanisms by which known risk factors act on the vulnerable fetal, infant, and early childhood brain.11,12 The most rapid period of brain development begins in the prenatal period and continues through approximately three years of age,13 mirroring the development of the gut microbiome (the bacteria, viruses, and fungi that reside in the gastrointestinal tract).14,15 We know that enteric bacteria influence brain function through various bidirectional systems of communication collectively known as the microbiome gut-brain axis.16 This connection between the gut and the brain plays a vital role throughout the life course, but particularly in early life when neural circuitry is still forming.17
The gut microbiome has been associated with several neurological outcomes including autism spectrum disorder (ASD),18–24 attention deficit/hyperactivity disorder (ADHD),25,26 anxiety27, and depression28,29 in adult populations. However, most studies to date have described these relationships in a cross-sectional setting where reverse causation is likely given the behavioral nature of these outcomes (i.e., the behaviors may result in changes to the microbiome, rather than the microbiome affecting behaviors). Additionally, only a few studies have captured the critical window of the infant microbiome that parallels the most rapid period of brain development in the lifespan, including our prior work on ASD-related social behaviors.30,31 The infant microbiome is less stable than the that of the adult,14 and early-life exposures influence neurodevelopment and in turn later behavioral outcomes. Furthermore, the prevalence of many of these behavioral outcomes differs by child sex, and the previous epidemiologic studies on the microbiome present aggregate estimates, potentially missing unique associations that exist specifically in either boys or girls.
To address these gaps, we examined the relationships between the infant and early-childhood microbiome and continuous measures of internalizing, externalizing, and social behaviors at three years of age, which inform clinical diagnoses of anxiety, depression, ADHD, and autism in preschool-aged children. Specifically, the Behavioral Assessment System for Children, 2nd edition (BASC-2) is a dimensional instrument that measures clinical and adaptive behaviors in children and young adults, including scales standardized for use in preschool-aged (two to five years) populations.32 When children’s scores fall outside the normal range, the BASC-2 can be informative in pursuing further diagnostic instruments to implement treatment plans including in-school services and medications. In addition to calculating aggregate associations across the population, we explored differences in associations among boys and girls using interaction models.
METHODS
Population Description
The New Hampshire Birth Cohort Study (NHBCS) is a pregnancy cohort with ongoing recruitment and follow up. Women were recruited at early pregnancy visits and were eligible if they were 18–45 years of age, used a private water system (e.g., well) at their residence during pregnancy, and did not plan on moving before delivery. In a subset, parents collected a stool sample from their child at six weeks, one year, and two years postpartum as previously described.30 Parents regularly completed interviews and questionnaires pertaining to sociodemographic data, their child’s health, and other relevant covariate information. When their child was approximately three years old, parents completed neurobehavioral assessments including the BASC-2 preschool form.32 A participant selection flow-chart and a table of exact sample sizes for each analysis are included in Table 1 and the supplement (Supplementary Figure S1, Supplementary Table S1). Parents of participants provided written informed consent. All study protocols were approved by the Center for the Protection of Human Subjects at Dartmouth, and all methods were carried out in accordance with relevant guidelines and regulations.
Table 1.
Sample sizes for analysis
| Microbiome Age | Sequencing Type | Complete Cases | Imputation Seta | ||||
|---|---|---|---|---|---|---|---|
| All | Boys | Girls | All | Boys | Girls | ||
| Six Weeks | 16S Metagenomics |
158 | 84 | 74 | 190 | 102 | 88 |
| 97 | 55 | 42 | 116 | 66 | 50 | ||
| One Year | 16S Metagenomics |
153 | 88 | 65 | 187 | 108 | 79 |
| 98 | 53 | 45 | 118 | 64 | 54 | ||
| Two Years | 16S | 121 | 77 | 44 | 146 | 92 | 54 |
Participants in the imputation set had complete microbiome and outcome data.
Microbiome Sequencing
Details of the protocols for microbial DNA extraction from stool samples using the Zymo Fecal DNA extraction kit (Irvine, CA) have been described elsewhere.33,34 Sequencing of the V4-V5 hypervariable region of the 16S rRNA gene (forward primer 518F: CCAGCAGCYGCGGTAAN, pooled reverse primers: 926R1: CCGTCAATTCNTTTRAGT, 926R3: CCGTCAATTTCTTTGAGT, 926R4: CCGTCTATTCCTTTGANT) was performed at Woods Hole Marine Biological Laboratory, MA (MBL) on the Illumina MiSeq platform (San Diego, CA) using established Illumina protocols.35 DADA2 (v1.6) was used to clean and process reads to determine amplicon sequence variants (ASVs).34,36 Bacteria were classified using the SILVA database (v138).37 Additionally, a smaller set of DNA samples from six-week and one-year stools underwent shotgun metagenomic sequencing at MBL using the NextSeq Illumina platform. As described elsewhere,30 sequences were annotated to the species level using MetaPhlAn238 and metabolic functional capacity was inferred through the HUMAnN239 algorithm using the MetaCyc database (v19.1).40 Alpha diversity was calculated from 16S rRNA data with the Shannon Index to capture both richness and evenness41 using the vegan package.42 Secondary analyses considered the Simpson Index43 and the count of observed ASVs. Beta diversity was computed using generalized UniFrac (GUniFrac) distances with 16S rRNA relative abundance data.44. Taxa inferred from both 16S rRNA and shotgun metagenomic sequencing data were considered in relation to the outcomes.
Behavior Assessment System for Children, 2nd edition
Parents assessed their three-year-old child’s behavioral development using the BASC-2,32 an instrument with a validated preschool version (ages 2–5 years) that captures adaptive and maladaptive behaviors. Internal validity metrics were used to exclude one subject for whom the consistency metric indicated the data should be used with extreme caution (> 21).32 This subject would have only contributed to the analysis of the two-year-old microbiome. A priori, we selected five primary clinical or adaptive scales and one content scale that measure phenotypes that have previously been associated with the microbiome in either animal models or in an epidemiological setting. These included internalizing behaviors (Anxiety and Depression)45, behaviors related to ADHD (Hyperactivity and Attention Problems)46 and behaviors that are related to autism (Social Skills and Developmental Social Disorders).47 To capture broader phenotypes, we included the four BASC-2 composite scales (Internalizing Problems, Externalizing Problems, Behavioral Symptoms Index, and Adaptive Skills). Primary clinical and adaptive scales contribute to the composites and have overlapping items with content scales, meaning that the ten outcomes are not independent (Supplementary Figure S2). Scores are standardized to a mean of 50 with a standard deviation of 10. For adaptive scales (Social Skills and Adaptive Skills Composite) a higher score indicates better behavior, whereas for the maladaptive scales an increased score indicates worse behavior.
Covariates
Potential covariates were selected a priori based on their potential to confound the association between the microbiome and BASC-2 scales (Supplementary Figure S3). After examining the univariate relationships of each variable with alpha diversity and BASC-2 scales, we selected gestational age, maternal education (any graduate education or none), child sex, parity (nulliparous or parous), maternal and paternal age, child age at follow-up, delivery mode, early life exclusive breastfeeding (EBF) versus mixed or exclusive formula feeding, maternal smoking during pregnancy, and breastfeeding duration as variables in our final model. As in our previous work,30 we examined models excluding variables for which the microbiome may act as a mediator (delivery mode,48 EBF and breastfeeding duration,49–51 and maternal smoking during pregnancy)52 as sensitivity analyses.
Multiple Imputation
To maintain sample size and reduce selection we imputed missing covariate data using multivariate imputation by chained equations (mice package in R).53,54 Variables with missing data are indicated in Table 2. We imputed 40 data sets with 20 iterations with all other covariates, outcomes, and bacterial alpha diversity as predictors of the missing covariates, as recommended.55 Predictive mean matching was used for continuous variables (paternal age and breastfeeding duration) and logistic regression with bootstrapping was used for binary variables (EBF delivery mode, parity, maternal education status, and peripartum antibiotic exposure, which was imputed for effect modification models). Where possible in downstream models (i.e., in linear regression models) the mice package was used to run 40 separate models and pool results. For more complex models, a single data set was created by using the median/mode of the imputed data for each subject. We conducted a sensitivity analysis among subjects with no missing covariate data.
Table 2.
Characteristics of participants included in any analysis [mean ± SD (% missing) or n (%)]
| Overall (n =260) |
Boys (n = 144) |
Girls (n = 116) |
p-valuea | |
|---|---|---|---|---|
| Parental Characteristics | ||||
| Maternal Age (years) | 32.2 ± 4.4 (0) |
32 ± 4.6 (0) |
32.3 ± 4.2 (0) |
0.4 |
| Paternal Age (years) | 33.5 ± 5.5 (2.3) |
33.1 ± 5.9 (2.8) |
34.1 ± 5 (1.7) |
0.063 |
| Maternal Smoking Status | >0.9 | |||
| No Smoking During Pregnancy | 246 (94.6) | 135 (93.8) | 111 (95.7) | |
| Any Smoking During Pregnancy | 11 (4.2) | 7 (4.9) | 4 (3.4) | |
| Missing | 3 (1.2) | 2 (1.4) | 1 (0.9) | |
| Maternal Education | 0.3 | |||
| No Postgraduate Education | 155 (59.6) | 91 (63.2) | 64 (55.2) | |
| Any Postgraduate Education | 99 (38.1) | 49 (34) | 50 (43.1) | |
| Missing | 6 (2.3) | 4 (2.8) | 2 (1.7) | |
| Parity | 0.8 | |||
| Nulliparous | 122 (46.9) | 70 (48.6) | 52 (44.8) | |
| Parous | 135 (51.9) | 72 (50) | 63 (54.3) | |
| Missing | 3 (1.2) | 2 (1.4) | 1 (0.9) | |
| Child Characteristics | ||||
| Gestational Age (weeks) | 39.5 ± 1.7 (0) |
39.4 ± 1.8 (0) |
39.6 ± 1.5 (0) |
0.4 |
| Breast Feeding Duration (months) | 11 ± 8.5 (5) |
10.6 ± 7.9 (5.6) |
11.5 ± 9.3 (4.3) |
0.7 |
| Child Age at BASC-2 (years) | 3.2 ± 0.3 (0) |
3.2 ± 0.3 (0) |
3.1 ± 0.2 (0) |
0.8 |
| Delivery Mode | 0.2 | |||
| Vaginal Delivery | 178 (68.5) | 92 (63.9) | 86 (74.1) | |
| Caesarean Delivery | 78 (30) | 49 (34) | 29 (25) | |
| Missing | 4 (1.5) | 3 (2.1) | 1 (0.9) | |
| Early-life Exclusive Breast Feeding | >0.9 | |||
| Not Exclusively Breastfed at Six Weeks | 105 (40.4) | 58 (40.3) | 47 (40.5) | |
| Exclusively Breastfed at Six Weeks | 121 (46.5) | 67 (46.5) | 54 (46.6) | |
| Missing | 34 (13.1) | 19 (13.2) | 15 (12.9) | |
| BASC-2 Scales | ||||
| Hyperactivity | 47.2 ± 6.9 | 48.3 ± 6.6 | 45.8 ± 7.1 | 0.003* |
| Externalizing Problems | 46.6 ± 6.8 | 47.6 ± 6.4 | 45.3 ± 7 | 0.002* |
| Anxiety | 50.4 ± 8.4 | 50.1 ± 8.4 | 50.8 ± 8.5 | 0.4 |
| Depression | 48 ± 7.5 | 48.2 ± 7.3 | 47.6 ± 7.7 | 0.5 |
| Internalizing Problems | 47.6 ± 7.6 | 47.8 ± 7.5 | 47.3 ± 7.7 | 0.7 |
| Attention Problems | 47.4 ± 6.8 | 48.6 ± 6.2 | 45.9 ± 7.2 | 0.005* |
| Behavioral Symptoms Index | 46 ± 6.7 | 46.8 ± 6.4 | 44.9 ± 7 | 0.015* |
| Social Skills | 56.6 ± 7.4 | 55 ± 7.5 | 58.6 ± 6.8 | <0.001* |
| Developmental Social Disorders | 44.9 ± 6.4 | 46.3 ± 6.6 | 43.1 ± 5.6 | <0.001* |
| Adaptive Skills Composite | 54.9 ± 7.3 | 53.1 ± 7.2 | 57.2 ± 6.7 | <0.001* |
P-value for difference between boys and girls. Calculated with Wilcoxon rank sum and Fisher’s exact tests.
Indicates statistically significant difference between boys and girls.
Models
Linear regression was used to determine the association between alpha diversity indices and BASC-2 scores. Using the pool() function of the mice R package,54 parallel regressions were run on each of the imputed datasets before the estimates and their errors were combined. For all other models, a single data set of the median/mode of imputed variables was used. To examine bacterial community structure related to each of the BASC-2 scales (beta diversity), we modeled GUniFrac distances as the outcome in adonis2 models with 10,000 permutations (vegan package in R).42 Marginal p-values are reported. For both alpha and beta diversity analyses a nominal p < 0.05 was considered statistically significant. MaAsLin2 (v1.6) was used to determine the relationships between bacterial ASVs (16S), species (metagenomic sequences), metabolic pathways, and BASC-2 scores.56,57 The minimum abundance and prevalence parameters were set so that a feature had to have at least 0.0001% abundance in at least 10% of samples to be assessed. Otherwise, default parameters were used (i.e., minimum variance: 0, normalization method: total sum-scaling, transformation: log, analysis method: linear, standardized metadata). The Benjamini-Hochberg procedure to adjust for multiple comparisons was applied to all ASVs/species/pathways in a given set of models, and a false discovery rate (FDR) of 10% (q < 0.1) was considered statistically significant for ASVs and pathways,58 Because the functional pathway relative abundance is more exploratory, a nominal p-value of 0.01 was applied as a cut off. Features that were associated with Social Responsiveness Scale, 2nd edition (SRS-2) scores in our previous analysis were examined for similar associations with autism-related BASC-2 scales.30 MaAsLin2 models feature relative abundance as the outcome of models, and because it is the opposite of the hypothesized microbiome-BASC-2 relationship requires careful interpretation.
Our main analysis examined the associations of the microbiome on BASC-2 scores in the overall population, but we hypothesized that the microbiome may have differential effects on BASC-2 scores in certain subgroups, specifically depending on child sex, which is relevant in the context of neurodevelopment and can alter susceptibility to exposures.59–61 To explore this possibility, we fit all models described above with an interaction between sex and the exposure of interest (microbiome measurement or BASC-2 in the cases where the microbiome was modeled as the outcome) to estimate sex-specific effects and interaction p-values (pinteraction). Similarly, in secondary analyses we explored the possibility that certain sensitive or healthy populations respond differently to perturbances in the microbiome. These analyses included models with interactions for peripartum antibiotic exposure and EBF status, as well as models conducted among only infants who were breastfed for at least six months and those who were vaginally delivered. Because these strata are the majority of our population, it was not feasible to conduct parallel analyses in the other strata (i.e., Caesarean delivered infants or those breastfed less than six months). All analyses were conducted in R 4.0.262 and R Studio 1.3.959.63
Availability of Materials and Data
The sequencing data used in this study are available through the National Center for Biotechnology Information (NCBI) Sequence Read Archive: https://ncbi.nlm.nih.gov/sra under accession number PRJNA296814. Epidemiologic data are not publicly available due to their sensitive and identifiable nature. Code is available upon request from Hannah E. Laue (Hannah.E.Laue@dartmouth.edu). Requests to work with the New Hampshire Birth Cohort Study should be directed to Margaret R. Karagas (Margaret.R.Karagas@Dartmouth.edu).
RESULTS
Population characteristics
Of the 260 infants in our analysis, the majority were born by a vaginal delivery (68.5 %), roughly half were exclusively breastfed at six weeks postpartum, and the average duration of any breastfeeding was close to one year (mean ± SD, 11 ± 8.5 months; Table 2). Approximately 95% of mothers were non-smokers during their pregnancy and were an average of 32.2 years old at delivery. When their children were approximately three years of age (mean ± SD, 3.2 ± 0.3 years), parents of children in the New Hampshire Birth Cohort Study (NHBCS) completed the BASC-2. From the full range of scales, we selected ten primary clinical, adaptive, and composite scales for investigation that we hypothesized may be related to the microbiome (Supplementary Figure S4). These captured both maladaptive (higher score is worse, Anxiety, Depression, Internalizing Problems, Attention Problems, Hyperactivity, Behavioral Symptoms Index, Externalizing Problems, Developmental Social Disorders) and adaptive (higher score is better, Social Skills and Adaptive Skills) behaviors. NHBCS participants were rated slightly better than the normative population (50 ± 10), with slightly lower scores on maladaptive scales and higher scores on adaptive behavioral scales. Notably, this was not the case with the Anxiety scale, where children’s scores were similar to those of the normative population. On average, boys were rated worse than girls on most scales with the exception of the three scales related to internalizing behaviors (Anxiety, Depression, and the composite Internalizing Problems; Table 2). As expected, BASC-2 scales were correlated, particularly composite scales and those that contribute to that composite (Supplementary Figures S2, S4). BASC-2 scores were also correlated with scores on the Social Responsiveness Scale, 2nd edition (SRS-2), a separate instrument which measures autism-related social behaviors and was previously examined in relation to the microbiome in this cohort.30 Boys and girls were similar with respect to covariate information, but fathers of girls were marginally older than fathers of boys. Although boys and girls were equally likely to be exposed to antibiotics in the peripartum period, boys were more likely to receive antibiotics within the first year of life (Supplementary Table S2). As expected, boys were heavier than girls at birth, but were similar according to World Health Organization sex- and gestational age-specific expectations (z-scores) for birthweight.
Higher Alpha Diversity Related to Internalizing Behaviors Among Boys
Within-subject bacterial diversity increased with age in both boys and girls, with no notable differences between the sexes (Supplementary Table S3). Increased Shannon diversity at six weeks was associated with lower Depression scores, particularly among boys [βBoys= −1.81 points/std increase Shannon diversity, 95% CI: (−3.22, −0.41), pBoys = 0.01, βGirls = −0.72, 95% CI: (−2.34, 0.89), pGirls = 0.38, pinteraction = 0.346; Figure 1, Supplementary Table S4]. The relationship between increased early-life diversity and fewer internalizing behaviors among boys was also captured with the Anxiety scale and Internalizing Problems composite scale (Figure 1, Supplementary Tables S5-S6). Six-week alpha diversity was not significantly associated with any other BASC-2 scales, nor were there significant differences between boys and girls (Supplementary Tables S7-S13). Secondary interaction models found the relationship between early-life alpha diversity and depression was consistent across most populations (Supplementary Table S4). Alpha diversity at one year was not associated with three-year-old BASC-2 scores (Figure 1, Supplementary Tables S4-S13).
Figure 1.
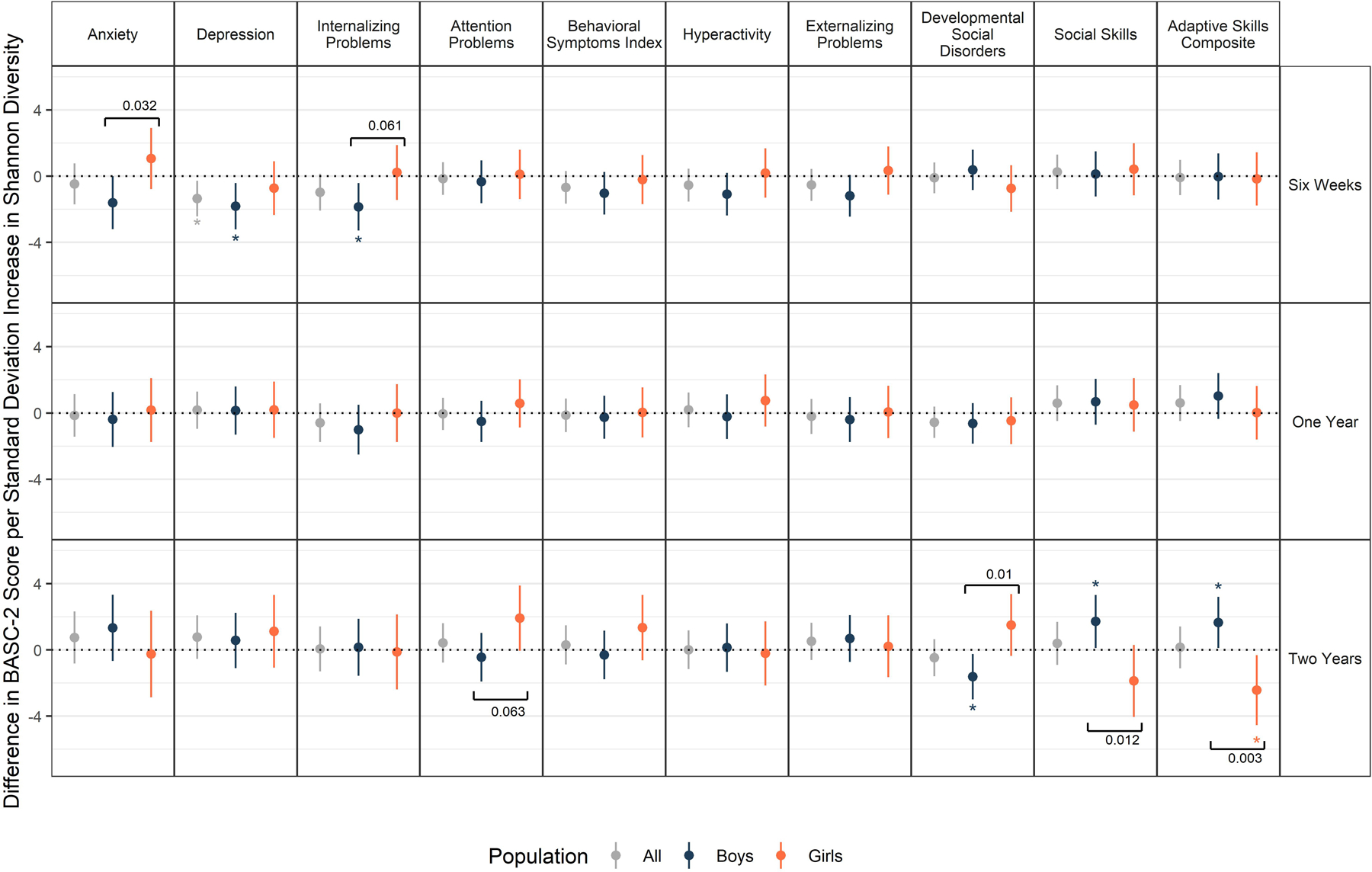
Effect estimates and 95% confidence intervals for a standard deviation increase in Shannon Index on BASC-2 scores (SDSixWeeks = 0.54, SDOneYear = 0.55, SDTwoYears = 0.46). Sex-specific estimates derive from models with sex-specific variables for Shannon z-scores. Each outcome was considered in a separate model adjusting for the main effect child sex, gestational age, maternal education, parity, delivery mode, maternal and paternal age at delivery, maternal smoking during pregnancy, early life exclusive breastfeeding, duration of any breastfeeding, and child age at follow up. *indicates an estimate is statistically significant (p < 0.05), brackets highlight where the sex-specific estimates are different from one another (p < 0.1), with the exact p-value of the interaction term in text near the bracket.
Divergent Associations at Two Years by Subgroups According to Sex and Perinatal Antibiotic Exposure
At two years increased alpha diversity was associated with better Developmental Social Disorders, Social Skills, and Adaptive Skills Composite scales among boys, but worse scores on these same scales among girls [e.g., βBoys,SocialSkills = 1.66 points/std increase Shannon diversity, 95% CI: (0.11, 3.2), βGirls,SocialSkills = −2.43, 95% CI: (−4.54, −0.32), pinteraction = 0.003 ; Figure 1, Supplementary Table S13]. In subgroup analyses, children exposed to peripartum antibiotics performed better on these scales when their Shannon Index was higher at two years (Supplementary Tables S11-S13). In contrast, children who were not exposed to antibiotics in the peripartum period had worse Behavioral Symptoms Index Scores with increased alpha diversity ([β = 1.62 points/std increase Shannon diversity, 95% CI: (0.04, 3.19), p = 0.05; Supplementary Table S8]; a similar pattern was observed on the Depression scale [β = 1.91 points/std increase Shannon diversity, 95% CI: (0.11, 3.72), p = 0.04, Supplementary Table S4].
No Overall Association Between Bacterial Community Structure and BASC-2 Scores
Overall, beta diversity was unrelated to any BASC-2 scale in fully adjusted models. In models excluding factors for which the microbiome may mediate an association, six-week beta diversity was associated with Attention Problem scores (R2 = 0.013, p = 0.01, Supplementary Table S14). Among boys, significant differences in beta diversity occurred with Anxiety scores (R2 = 0.012, p = 0.019). At one year community structure was associated with Attention Problems and Adaptive Skills Composite, but only among those exposed to peripartum antibiotics. In models excluding factors for which the microbiome may mediate an association, we also found an association with Developmental Social Disorders (R2 = 0.011, p = 0.023), Hyperactivity (R2 = 0.009, p = 0.048), and Adaptive Skills Composite (R2 = 0.01, p = 0.028) scores among boys. We found few associations between beta diversity at two years and BASC-2 scores in our main models, although we observed differences by Attention Problems among those who were exclusively breast fed in early life (R2 = 0.012, p = 0.047). Results were similar among complete cases (Supplementary Table S15). Infant sex was not associated with beta diversity at any age (Supplementary Figure S5). However, early life exclusive breastfeeding was associated with beta diversity at all ages and paternal and maternal age were associated with two-year-old beta diversity.
Key Taxa (16S) Relate to Outcomes in a Sex-specific Manner
The relative abundances of several taxa at six weeks were associated with Adaptive Skills Composite scores in a sex-specific manner (FDR q < 0.1, Figure 2, Supplementary Table S16). Notably, this included Bifidobacterium, one of the most abundant taxa in the infant gut (Supplementary Table S17), for which a single point increase in Adaptive Skill Composite score was associated with a 1.91% increase in relative abundance among boys [95% CI: (0.7, 3.13), q = 0.08], but had no association among girls [−1.54 %/point, 95% CI: (−3.14, 0.06), q = 0.89]. Due to the methods used, where bacterial species abundance was modeled as the outcome, the units for the estimates imply behaviors influence the microbiome. However, temporality of data collection suggests the reverse (i.e., microbiome influences behavior). Similarly, Bacteroides vulgatus, and a Streptococcus taxon at six weeks related to better Adaptive Skills Composite scores among boys, but not among girls. In contrast, three ASVs in the Klebsiella, Clostridium, and Haemophilus genera at six weeks related to worse Adaptive Skills Composite scores among boys. Tyzzerella nexilis was associated with better Depression scores among boys [−0.89 %/point, 95% CI: (−1.4, −0.38), q = 0.07]. At one year, the relative abundance of Faecalitalea was associated with worse Hyperactivity scores, with no discernable sex-specific effects [0.16 %/point, 95% CI: (0.08, 0.23), q = 0.05]. At two years, the relative abundance of four Blautia ASVs were associated with worse measures of hyperactivity. These associations were stronger among girls, but q-values did not meet the FDR threshold after imputing missing covariates. None of these ASVs was prevalent enough at earlier time points to be examined. ASVs that were previously associated with SRS-2 scores (one-year Blautia producta and an unknown Lachnospiraceae; two-year Coprococcus, Ruminococcus gnavus, Bifidobacterium, and Sutterella)30 mostly had the same direction of association with autism-related BASC-2 scores, and the association between the Coprococcus ASV relative abundance and worse Social Skills scores did reach nominal significance (−0.18%/point, p = 0.007, Supplementary Table S18).
Figure 2.
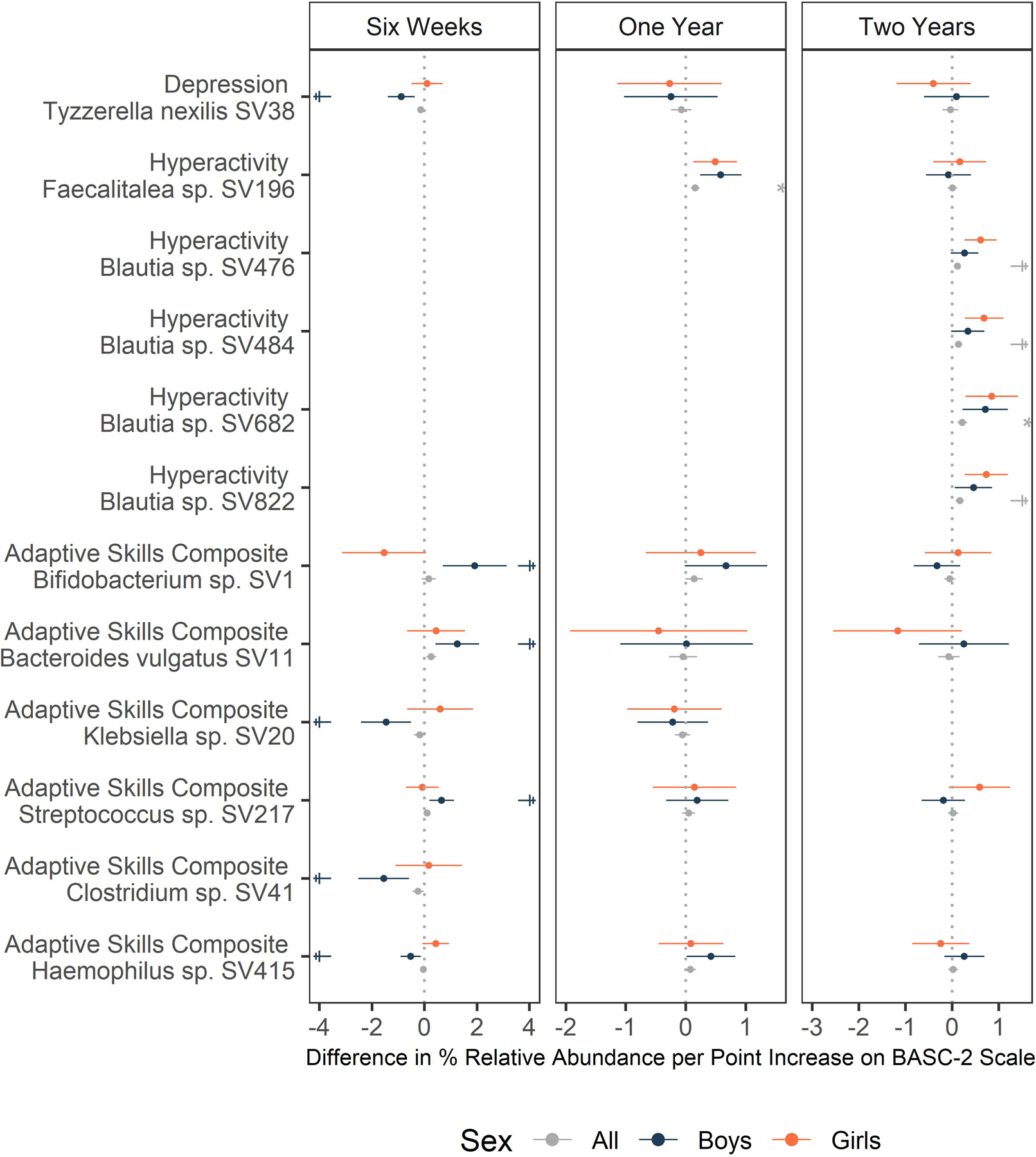
Effect estimates and 95% confidence intervals for the difference in percent relative abundance per point increase on the given BASC-2 scale. Sex-specific estimates derive from models with sex-specific variables for BASC-2 scores adjusting for the main effect child sex, gestational age, maternal education, parity, delivery mode, maternal and paternal age at delivery, maternal smoking during pregnancy, early life exclusive breastfeeding, duration of any breastfeeding, and child age at follow up. * indicates FDR q < 0.05 ; † indicates FDR < 0.1.
Metagenomic Species Related to BASC-2 Scores
Metagenomic sequencing allowed for more precise annotation of bacterial species associated with BASC-2 scores. While no bacterial taxa were strongly associated with any BASC-2 scales, we identified a weak association between Eggerthella lenta at six weeks and Depression (Figure 3, Supplementary Table S19). Additionally, we found evidence of sex-specific associations between several other bacteria at six weeks and BASC-2 scores. Specifically, Klebsiella oxytoca was adversely associated with Adaptive Skills Composite and Developmental Social Disorders scores among boys (−2.98 %/point Adaptive Skills and 2.57 %/point Developmental Social Disorders), but no evident relationship among girls (pinteraction = 0.052 and 0.023 for Adaptive Skills and Developmental Social Disorders, respectively). In contrast, Granulicatella was associated with worse Anxiety scores among girls (0.58 %/point), but not boys (pinteraction = 0.257). Similar to the findings at six-weeks, no strong associations were observed at one year. Weak adverse associations were observed between Anaerofustis stericohominis and Adaptive Skills Composite scores (−0.31 %/point) and Lachnospiraceae bacterium 1_1_57FAA and Externalizing Problems scores (0.21 %/point). Streptococcus peroris relative abundance at one year was associated with lower Depression and Internalizing Problems scores, but only among girls [(−1.38 %/point Depression, pinteraction = 0.098; −1.16 %/point Internalizing Problems, pinteraction = 0.169), Figure 3, Supplementary Table S19].
Figure 3.
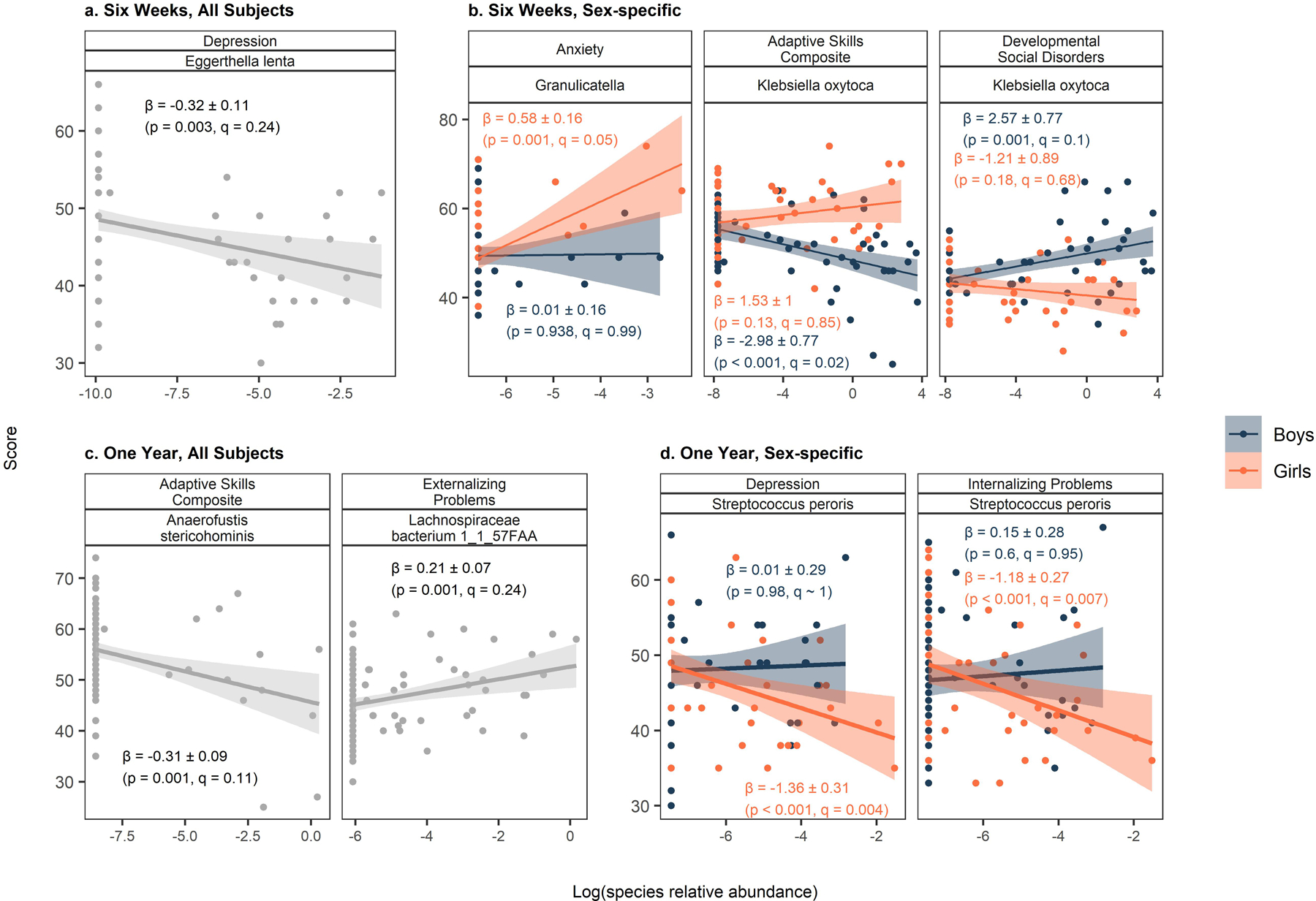
Associations between select bacterial taxa (shotgun metagenomics) and BASC-2 scores. Estimates, standard errors, p-values, and FDR q-values derive from MaAsLin2 models where the species relative abundance is modeled as the outcome and the given BASC-2 is treated as the exposure. Sex-specific estimates derive from models with sex-specific variables for BASC-2 scores. All models adjust for the main effect child sex, gestational age, maternal education, parity, delivery mode, maternal and paternal age at delivery, maternal smoking during pregnancy, early life exclusive breastfeeding, duration of any breastfeeding, and child age at follow up.
Bacteria that were previously identified as being related to worse SRS-2 scores (six-week Flavonifactor plautii and one-year Adlercreutzia equolifaciens, Ruminococcus torques, Eubacterium dolichum, and bacterium 6 1 63 FAA in the Lachnospiraceae)30 were found to have similar relationships (i.e., in the same direction) with the relevant BASC-2 scales (Social Skills and Developmental Social Disorders). Although the effect estimates were further from the null, they were not statistically significant in the context of multiple testing (Supplementary Figure S6). The relationship between Adlercreutzia equolifaciens and Social Skills scores was nominally significant [−0.17 %/point, 95% CI: (−0.33, −0.01), p = 0.04, q = 0.588)], and this relationship was found to be primarily among boys although the interaction term was not statistically significant [−0.98 %/point, 95% CI: (−1.72, −0.24), p = 0.01, pinteraction = 0.187]. Similarly, the association between Lachnospriraceae bacterium 6_1_63FAA and Developmental Social Disorder scores was only significant among boys [0.9 %/point, 95% CI: (0.14, 1.67), p = 0.02] and not girls [−0.03 %/point, 95% CI: (−0.91, 0.85), p = 0.94, pinteraction = 0.64].
Bacterial Functional Pathways Related to BASC-2 Scores
The relative abundance of several metabolic pathways at six weeks and one year were associated with BASC-2 scales (Figure 4). At six weeks, increased relative abundance PWY-4981 [L-proline biosynthesis II (from arginine)] and PWY-7399 (methylphosphonate degradation II) genes were associated with better scores on the Hyperactivity scale and the two composite scales to which it contributes – Behavioral Symptoms Index and Externalizing Problems (Supplementary Table S20). PWY-5910 (superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate)) and PWY-7560 (methylerythritol phosphate pathway II) were also associated with better Externalizing Problems scores. The NAD salvage pathway (PNC VI cycle) was associated with worse Internalizing Problems scores, whereas catechol degradation I (meta-cleavage pathway) was associated with lower Attention Problems scores.
Figure 4.
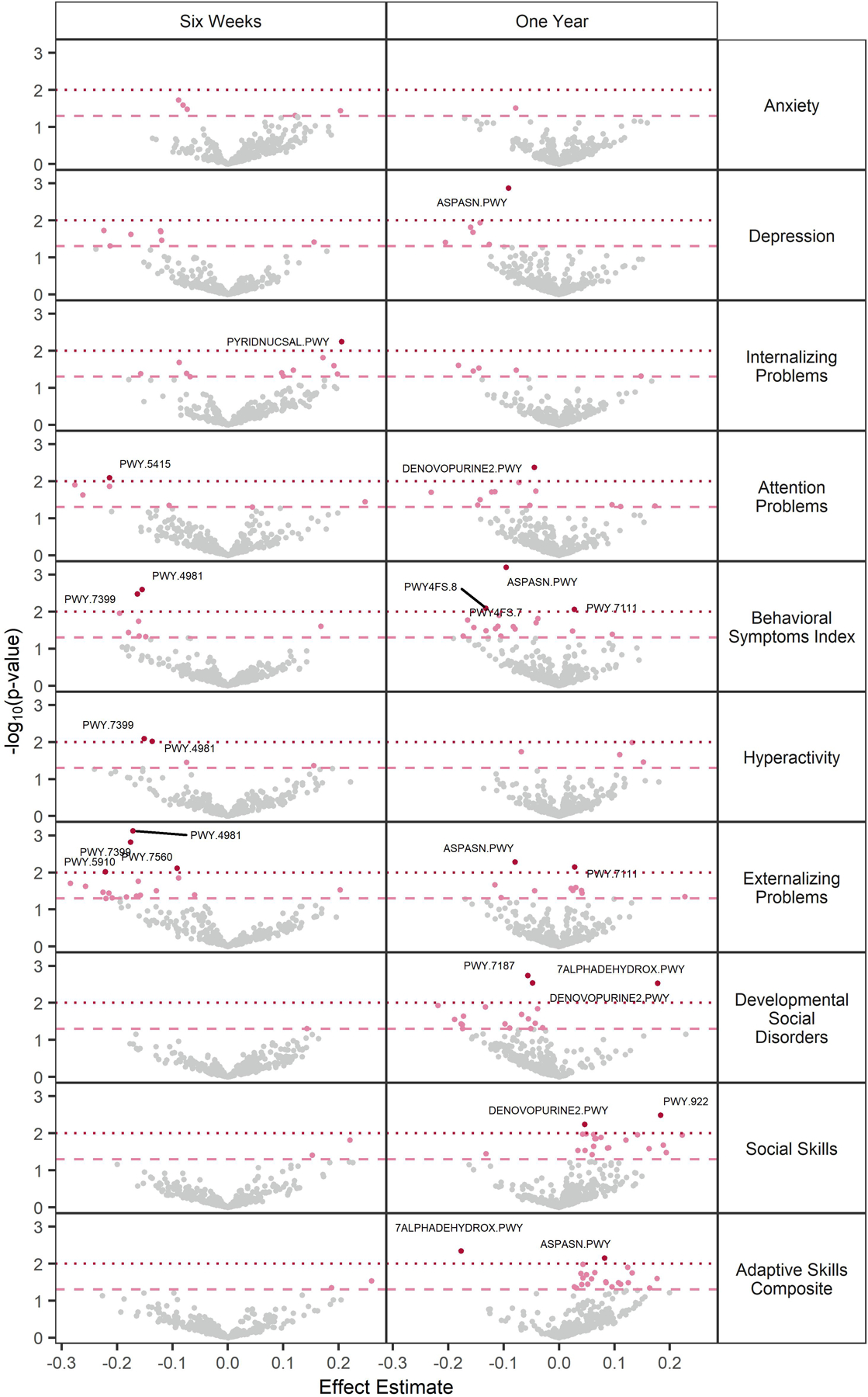
Volcano plots of associations between metabolic functional pathways and BASC-2 scores. Estimates, standard errors, p-values, and FDR q-values derive from MaAsLin2 models where the pathway relative abundance is modeled as the outcome and the given BASC-2 is treated as the exposure. All models adjust for the main effect child sex, gestational age, maternal education, parity, delivery mode, maternal and paternal age at delivery, maternal smoking during pregnancy, early life exclusive breastfeeding, duration of any breastfeeding, and child age at follow up. ASPASN.PWY: superpathway of L-aspartate and L-asparagine biosynthesis, PYRIDNUCSAL.PWY: NAD salvage pathway I (PNC VI cycle), PWY.5415: catechol degradation I (meta-cleavage pathway), DENOVOPURINE2.PWY: superpathway of purine nucleotides de novo biosynthesis II, PWY.4981: L-proline biosynthesis II (from arginine), PWY.7399: methylphosphonate degradation II, PWY4FS.7: phosphatidylglycerol biosynthesis I (plastidic), PWY4FS.8: phosphatidylglycerol biosynthesis I (non-plastidic), PWY.7111: pyruvate fermentation to isobutanol (engineered), PWY.5910: superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate), PWY.7580: phycoerythrobilin biosynthesis II, PWY.7187: pyrimidine deoxyribonucleotides de novo biosynthesis II, 7ALPHADEHYDROX.PWY: bile acid 7α-dehydroxylation, PWY.922: mevalonate pathway I (eukaryotes and bacteria).
At one year, the superpathway of L-aspartate and L-asparagine biosynthesis was associated with better Depression scores and better scores on the Behavioral Symptoms Index (to which the Depression scale contributes), as well as better scores on the Externalizing Problems and Adaptive Skills Composite scales. The superpathway of purine de novo biosynthesis II was associated with better Attention Problems, Developmental Social Disorders, and Social Skills scores, and de novo pyrimidine biosynthesis was also associated with better Developmental Social Disorders scores. PWY-7111, an engineered pathway of pyruvate fermentation, was associated with worse Externalizing Problems and Behavioral Symptoms Index scores. Both plastidic and non-plastidic phosphatidylglycerol biosynthesis were associated with better Behavioral Symptoms Index scores. Bile acid dihydroxylation was associated with worse Developmental Social Disorders and Adaptive Skills Composite scores. Finally, a mevalonate pathway was associated with better Social Skills scores.
Different pathways were associated with BASC-2 scores among boys and girls (Supplementary Figures S7-S8). Generally, pathways that were significantly associated in one sex had null associations in the other, as opposed to being significantly associated in the opposite direction. More associations were observed among girls with one-year functional capacity compared to six weeks, and with the Social Skills scale compared to other BASC-2 scales. In contrast, there were more associations among boys at six weeks compared to one year, and with the Internalizing and Externalizing Problems scales. However, the Depression scores were also associated with the relative abundance of several microbial functional pathways at one year among boys, including PWY0–845 (superpathway of pyridoxal 5’-phosphate biosynthesis and salvage), the superpathway of L-aspartate and L-asparagine biosynthesis, and L-ornithine biosynthesis II. Notably, several pathways whose relative abundances were associated with better Depression scores among boys related to vitamin B6 biosynthesis or salvage (PWY0–845 at one year, PYRIDOXSYN-PWY at one year, and PWY-7204 at six weeks). A high correlation between PWY0–845 and PYRIDOXSYN-PWY may be responsible for their concordant results, but PWY-7204 is not correlated with either of the other pathways (Supplementary Figure S9).
DISCUSSION
This prospective study of the concurrent development of the microbiome and brain function measured by later behaviors is one of the first to be conducted at this young age. We found that at the youngest ages, higher diversity was related to better internalizing behaviors at three years of age, especially among boys. Sex-specific differences in behavioral sensitivities to Shannon diversity were larger with microbiome samples at older ages but were also apparent with six-week microbiome samples. Our analysis of beta diversity found that microbiome differences related to BASC-2 scores did not lead to differences in community structure, with the notable exception of Attention Problems scores in certain models. However, there was some indication that boys may be more sensitive at the beta-diversity level. In taxon-level analyses, using both 16S rRNA data and shotgun metagenomics data, we found time- and sex-specific effects. Similarly, in exploring the functional capacity of the gut microbiome we found time and sex-specific effects with biologically relevant gene pathways.
Our findings regarding the sensitivity of the development of specific internalizing behavioral outcomes, such as anxiety and depression, to early differences in gut bacterial diversity are supported by some prior studies of these outcomes in other populations64–67 and animal models.68 However, it was not expected that this would be detectable as early as six weeks, and our findings suggest there may be an early opportunity to intervene upon asymptomatic at-risk infants to lower the burden of these disorders in early childhood, adolescence, and young adulthood. In addition to the relationship between diversity and internalizing behaviors, specific bacteria were associated with BASC-2 scores. To date, sex-specific differences have not been examined or noted in early life, whereas we found the relative abundance of a Granulicatella species at six weeks was associated with worse Anxiety scores among girls. Conversely, at one year, Streptococcus peroris was associated with better Internalizing Problems scores and specifically scores on the Depression scale, but only among girls. These findings may indicate a potential window of vulnerability for female neurodevelopment related to internalizing behaviors or may be due to chance. Additional research is required to confirm this finding.
While some epidemiological studies have considered the microbiomes of children with ADHD compared to controls, few large-scale longitudinal epidemiological studies have considered ADHD symptoms such as inattention and hyperactivity.69 Although we did not observe large shifts in beta diversity related to most BASC-2 outcomes, there was an association between Hyperactivity scores and GUniFrac distances at six weeks in minimally adjusted models. This is likely due to a strong association between maternal smoking during pregnancy, which was not included in the minimal model, and Hyperactivity scores but more research is needed into whether the microbiome, which is also affected by smoke exposure,52 could mediate this association. Our analysis of bacteria sequenced by 16S also uncovered novel associations between several Blautia ASVs and worse Hyperactivity scores. To date, no other studies have described a similar relationship between Blautia and ADHD symptoms. Some Blautia species have been linked to conditions like depression,70 whereas in other studies some anti-inflammatory Blautia are reduced in subjects with ASD.71–73 Lack of prior evidence related to hyperactivity may be due to few prospective studies of ADHD symptoms in young pediatric populations.
Shotgun metagenomic sequencing allows for annotation of genes across the genome, which permits inference of the metabolic functional capacity of the bacteria in the gut. In this study, we found several associations between functional pathways and BASC-2 measures of externalizing behaviors that are biologically relevant. For example, proline, biosynthesis of which was associated with better Hyperactivity, Externalizing Problems, and Behavioral Symptoms Index scores, has been found to be lower among unmedicated subjects with ADHD, albeit not statistically significantly.74 Another example is that catechol degradation was associated with better Attention Problems scores. Catechol is a toxic compound that naturally occurs in some fruits and vegetables, but is also synthesized for use as a pesticide.75 Interestingly, studies have found that SNPs in the human catechol-O-methyltransferase gene are linked to diagnosis and severity of ADHD symptoms, anxiety, and depression.76–79 It is of note that the scales that had the most associations with bacterial functional pathway relative abundance were related to the Externalizing Problems scale. More research is needed to determine whether the neural mechanisms that underlie these behaviors are especially vulnerable to microbial influences at this early-life stage.
We found that boys were more sensitive to early-life (i.e., six weeks of age) differences in their microbiomes as evaluated with 16S sequencing, with several bacteria, including Bifidobacterium a common beneficial genus, associated with better Adaptive Skills Composite scores.80 Notably, this included a positive association between a Klebsiella ASV and Adaptive Skills Composite scores that was supported by a similar association of Klebsiella oxytoca and Adaptive Skills Composite scores among boys in our metagenomics analysis. This commensal species has been reported to be depleted in patients with ASD,81 although not consistently across studies.18–24 The Adaptive Skills Composite score includes the Social Skills scale, which is relevant to autistic behaviors. In our prior work with an autism-specific outcome (SRS-2) we did not find strong associations between bacterial diversity and autism-related behaviors,30 whereas here we found relationships between the two-year-old microbiome and autism-related BASC-2 scales (i.e., Developmental Social Disorders, Social Skills, and Adaptive Skills Composite) when examining sex-dependent effects not explored previously. Notably, these scales were where we observed the greatest difference between boys and girls, with boys performing significantly worse. It is possible that early manifestations of socially impaired behaviors related to autism spectrum disorder (ASD) affect the two-year old microbiome (i.e., there is reverse causation). Nevertheless, more clinical studies are required to identify when therapies targeting the microbiome may be most effective for ASD, and who may respond best to such interventions.
Our novel finding of sex-specific associations in the relationship between the microbiome and BASC-2 scores highlights the importance of considering populations that may be more sensitive to alterations in the microbiome and host characteristics that may modulate the relationships between gut bacteria and human health. There is strong evidence that microglial abundance, morphology, and gene expression differ between boys and girls in the neonatal period,61 suggesting that the microbiome, which interacts with microglia,82 could have sex-specific effects. We also observed several differences in the relationships between bacterial diversity, relative abundance, and functional capacity and BASC-2 scores between boys and girls. These could result from underlying differences in BASC-2 score distributions between boys and girls, from sex differences in the timing and course of neurodevelopmental maturation, or from chance due to small sample sizes contributing to sex-specific estimates. However, at least some of the sex-specific findings are supported by biological evidence. For example, three pathways related to vitamin B6 biosynthesis and salvage at either six weeks or one year were associated with better Depression scores among boys. Vitamin B6 is essential in the synthesis of several neurotransmitters,83 and low dietary intake of B vitamins has been linked to adolescent depression.84 Additional research is required to understand the interplay between host sex, early-life diet, microbiome, and later depressive symptoms.
Several studies have examined the relationship between the gut microbiome and psychobehavioral outcomes in a case control setting in children,18–26,85,86 in adults,27,87,88 or in specific contexts (e.g.. comorbid inflammatory bowel disease and internalizing behaviors).89 However, many such outcomes develop at an early age, and are best managed through early interventions.90–94 The neonatal and early pediatric period may be particularly important for hypothalamic-pituitary-adrenal system stress response development due to rapid, synchronous development of the microbiome and brain circuitry.95 Notably, the landmark study describing the importance of the early-life microbiome in anxiety-related behaviors compared germ-free and specific pathogen free mice, which better mimics a change in alpha diversity rather than differences in specific taxa, in agreement with our findings for internalizing behaviors.96 The variation in our results depending on microbiome sampling age may reflect differences in the bacteria present in the infant gut as it matures over the first two years of life or could reflect different windows of susceptibility in neurodevelopment related to the relevant BASC-2 scales.
Differences in 16S rRNA compared to metagenomic sequencing results may be explained by several methodological factors. First, 16S rRNA sequencing relies on PCR amplification of a hypervariable region of a highly conserved gene. The primers used likely create some bias in the relative abundances of observed taxa depending on their alignment to the primers.97 Additionally, the reference databases used to infer taxa identities are different for the two methods. For whole genome sequencing we used MetaPhlAn2, which infers taxonomy based on unique clade-specific marker genes and has been considered conservative to many other available tools,98,99 likely contributing to differences. Finally, participants in the 16S rRNA and metagenomic analyses overlap but are not identical. Given the unique composition of individual’s microbiomes results may be subject to population differences. Thus, replication studies are essential to confirm our findings.
Our study is limited by its observational nature and limited sample size, particularly for our population-specific estimates. However, ours is one of the largest studies to prospectively examine the connection between the gut microbiome and neurodevelopment and suggests several novel associations that can be explored in mechanistic models. Available microbiome methods preclude efficient modeling of bacterial species and functions as the exposure, and thus our results should be interpreted carefully and not causally. While we adjusted for many covariates, we did not include dietary factors except for breastfeeding. Although diet has been shown to be important to the adult microbiome, little is known about what nutritional components can alter the infant/toddler microbiome.100 Thus, more research is necessary in this arena to investigate the role of diet as a confounder in the association between the early-life microbiome and neurodevelopment. Further, because antibiotic usage was not associated with our outcomes it was not included in our analysis. Our secondary analysis calculating population-specific estimates for those exposed and unexposed to peripartum antibiotics accounted for the possibility that exposure to peripartum antibiotics may prime certain individuals to be more sensitive to differences in their microbiomes. NHBCS participants have robust neurodevelopment and are rated better than the normative population on most BASC-2 scales, potentially decreasing the sensitivity of our analysis for identifying correlates of adverse behavioral development. Lastly, our assessment of neurobehavioral outcomes at an early age may not fully capture later adverse behaviors.
This is one of the first prospective studies to examine the relationship between the microbiome and a broad range of behavioral outcomes. Given the likelihood for behaviors to affect the microbiome (i.e., reverse causation), our ability to establish temporality of the relationship is particularly important. Additionally, our novel use of multiple imputation to increase sample size and reduce selection bias improves the precision and accuracy of our estimates. We conducted our analysis in healthy individuals that reflect the general population in the geographic region from which they derive, rather than a clinical population, increasing the generalizability of our findings. Notably, while this population performed better on many of the BASC-2 scales than the normative population, they were similar with respect to internalizing behaviors (e.g., anxiety and depression), which are of growing concern in pediatric populations.101,102 Finally, our extensive analysis of modifiers of the relationship between the microbiome and behavior uncovered important differences depending on host sex.
Our findings highlight the importance of prospective data and consideration of host characteristics in studies relating the microbiome to health outcomes. Specifically, host sex may modify the relationship between bacteria and neurodevelopment, even during windows when differences in gonadal steroid production do not occur (one and two years). Future studies should consider sex differences in results. The results of this research suggest the early-life microbiome is relevant to behavioral outcomes. Interventional studies could clarify the causality of this relationship.
Supplementary Material
Consent Statement:
Parents of participants provided written informed consent. All study protocols were approved by the Center for the Protection of Human Subjects at Dartmouth, and all methods were carried out in accordance with relevant guidelines and regulations.
Impact:
This is one of the first studies to examine the early-life microbiome and neurobehavior, and the first to examine prospective sex-specific associations.
Infant and early-childhood microbiomes relate to neurobehavior including anxiety, depression, hyperactivity, and social behaviors in a time- and sex-specific manner.
Our findings suggest future studies should evaluate whether host sex impacts the relationship between the gut microbiome and behavioral health outcomes.
Financial Support:
This study was supported by grants from the National Institute of Environmental Health (P01ES022832, P20ES018175, P42ES007373), the National Institute of General Medical Sciences (P20GM104416), and the U.S. Environmental Protection Agency (RD-83544201).
Footnotes
Disclosure Statement: The authors declare no conflict of interest.
Category of Study: Population study.
REFERENCES
- 1.Romero M. et al. Psychiatric comorbidities in autism spectrum disorder: A comparative study between DSM-IV-TR and DSM-5 diagnosis. International Journal of Clinical and Health Psychology 16, 266–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobanski E. Psychiatric comorbidity in adults with attention-deficit/hyperactivity disorder (ADHD). Eur Arch Psychiatry Clin Neurosci 256, i26–i31 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Brod M, Schmitt E, Goodwin M, Hodgkins P & Niebler G. ADHD burden of illness in older adults: a life course perspective. Qual Life Res 21, 795–799 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Philipp-Wiegmann F, Retz-Junginger P, Retz W & Rösler M. The intraindividual impact of ADHD on the transition of adulthood to old age. Eur Arch Psychiatry Clin Neurosci 266, 367–371 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Matza LS, Paramore C & Prasad M. A review of the economic burden of ADHD. Cost Effectiveness and Resource Allocation 3, 5 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robb JA et al. The Estimated Annual Cost of ADHD to the U.S. Education System. School Ment Health 3, 169–177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kogan MD et al. A National Profile of the Health Care Experiences and Family Impact of Autism Spectrum Disorder Among Children in the United States, 2005–2006. Pediatrics 122, e1149–e1158 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Rylaarsdam L & Guemez-Gamboa A. Genetic Causes and Modifiers of Autism Spectrum Disorder. Front Cell Neurosci 13, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai D. et al. Association of Genetic and Environmental Factors With Autism in a 5-Country Cohort. JAMA Psychiatry 76, 1035–1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sciberras E, Mulraney M, Silva D & Coghill D. Prenatal Risk Factors and the Etiology of ADHD-Review of Existing Evidence. Curr Psychiatry Rep 19, 1 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Modabbernia A, Velthorst E & Reichenberg A. Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Molecular Autism 8, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rock KD & Patisaul HB Environmental Mechanisms of Neurodevelopmental Toxicity. Curr Envir Health Rpt 5, 145–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmore JH et al. Regional Gray Matter Growth, Sexual Dimorphism, and Cerebral Asymmetry in the Neonatal Brain. J Neurosci 27, 1255–1260 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yassour M. et al. Natural history of the infant gut microbiome and impact of antibiotic treatments on strain-level diversity and stability. Sci Transl Med 8, 343ra81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ et al. The Human Microbiome Project. Nature 449, 804–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cryan JF et al. The Microbiota-Gut-Brain Axis. Physiological Reviews 99, 1877–2013 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Osadchiy V, Martin CR & Mayer EA The Gut–Brain Axis and the Microbiome: Mechanisms and Clinical Implications. Clinical Gastroenterology and Hepatology 17, 322–332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Angelis M. et al. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS One 8, e76993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang D-W et al. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS One 8, e68322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S. et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Scientific Reports 9, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma B. et al. Altered Gut Microbiota in Chinese Children With Autism Spectrum Disorders. Front. Cell. Infect. Microbiol 9, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose DR et al. Differential immune responses and microbiota profiles in children with autism spectrum disorders and co-morbid gastrointestinal symptoms. Brain Behav Immun 70, 354–368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strati F. et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 5, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai Q. et al. Disturbance of trace element and gut microbiota profiles as indicators of autism spectrum disorder: A pilot study of Chinese children. Environ. Res. 171, 501–509 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Aarts E. et al. Gut microbiome in ADHD and its relation to neural reward anticipation. PLOS ONE 12, e0183509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prehn-Kristensen A. et al. Reduced microbiome alpha diversity in young patients with ADHD. PLOS ONE 13, e0200728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H. et al. Altered gut microbiota profile in patients with generalized anxiety disorder. Journal of Psychiatric Research 104, 130–136 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Jiang H. et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 48, 186–194 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Naseribafrouei A. et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil 26, 1155–1162 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Laue HE, Korrick SA, Baker ER, Karagas MR & Madan JC Prospective associations of the infant gut microbiome and microbial function with social behaviors related to autism at age 3 years. Scientific Reports 10, 15515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sordillo JE et al. Association of the Infant Gut Microbiome With Early Childhood Neurodevelopmental Outcomes: An Ancillary Study to the VDAART Randomized Clinical Trial. JAMA Netw Open 2, e190905–e190905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds CR & Kamphaus R Behavior Assessment System for Children. (Pearson, 2004). [Google Scholar]
- 33.Antosca K. et al. Reliability of stool microbiome methods for DNA yields and sequencing among infants and young children. Microbiologyopen 9, e1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madan JC et al. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. JAMA Pediatr 170, 212–219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison HG, Grim SL, Vineis JH & Sogin ML 16S amplicon fusion primers and protocol for Illumina platform sequencing. (2013)
- 36.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12, 902–903 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Franzosa EA et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15, 962–968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caspi R. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res 42, D459–D471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon C & Weaver W The Mathematical Theory of Communication. (The University of Illinois Press, 1964). [Google Scholar]
- 42.Oksanen J et al. vegan: community ecology package. https://CRAN.R-project.org/package=vegan (2020).
- 43.Simpson EH Measurement of Diversity. Nature 163, 688 (1949). [Google Scholar]
- 44.Chen J GUniFrac: Generalized UniFrac Distances. https://CRAN.R-project.org/package=GUniFrac (2018).
- 45.Foster JA & McVey Neufeld K-A Gut–brain axis: how the microbiome influences anxiety and depression. Trends in Neurosciences 36, 305–312 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Hiergeist A, Gessner J & Gessner A. Current Limitations for the Assessment of the Role of the Gut Microbiome for Attention Deficit Hyperactivity Disorder (ADHD). Front. Psychiatry 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vuong HE & Hsiao EY Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biological Psychiatry 81, 411–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yip BHK et al. Caesarean section and risk of autism across gestational age: a multi-national cohort study of 5 million births. Int J Epidemiol 46, 429–439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boucher O. et al. Association between breastfeeding duration and cognitive development, autistic traits and ADHD symptoms: a multicenter study in Spain. Pediatr. Res 81, 434–442 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Julvez J. et al. A cohort study on full breastfeeding and child neuropsychological development: the role of maternal social, psychological, and nutritional factors. Developmental Medicine & Child Neurology 56, 148–156 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Tseng P-T et al. Maternal breastfeeding and autism spectrum disorder in children: A systematic review and meta-analysis. Nutritional Neuroscience 22, 354–362 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Levin AM et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep 6, 31775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azur MJ, Stuart EA, Frangakis C & Leaf PJ Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res 20, 40–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Buuren S & Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. Journal of Statistical Software 45, 1–67 (2011). [Google Scholar]
- 55.Harel O. et al. Multiple Imputation for Incomplete Data in Epidemiologic Studies. Am J Epidemiol 187, 576–584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mallick H, Rahnavard A & McIver L Maaslin2: Maaslin2. (2020).
- 57.Mallick H. et al. Multivariable Association in Population-scale Meta’omic Surveys. In Submission.
- 58.Benjamini Y & Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 59.Breedlove S & Arnold A. Hormonal control of a developing neuromuscular system. II. Sensitive periods for the androgen-induced masculinization of the rat spinal nucleus of the bulbocavernosus. J Neurosci 3, 424–432 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forger NG et al. Deletion of Bax eliminates sex differences in the mouse forebrain. Proc Natl Acad Sci U S A 101, 13666–13671 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanamsagar R & Bilbo SD Sex differences in neurodevelopmental and neurodegenerative disorders: Focus on microglial function and neuroinflammation during development. The Journal of Steroid Biochemistry and Molecular Biology 160, 127–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Core Team. R: A language and environment for statistical computing. https://www.R-project.org (2020).
- 63.RStudio Team. RStudio: Integrated Development for R. http://www.rstudio.com (2020).
- 64.Barandouzi ZA, Starkweather AR, Henderson WA, Gyamfi A & Cong XS Altered Composition of Gut Microbiota in Depression: A Systematic Review. Front Psychiatry 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y. et al. Similar Fecal Microbiota Signatures in Patients With Diarrhea-Predominant Irritable Bowel Syndrome and Patients With Depression. Clin Gastroenterol Hepatol 14, 1602–1611.e5 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Madan A. et al. The gut microbiota is associated with psychiatric symptom severity and treatment outcome among individuals with serious mental illness. Journal of Affective Disorders 264, 98–106 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Morales Marroquin E. et al. Association between the Distal Gut Microbiome and Anxiety in Highly Active Individuals. Research Square (2020)
- 68.Zheng P. et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Molecular Psychiatry 21, 786–796 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Checa-Ros A, Jeréz-Calero A, Molina-Carballo A, Campoy C & Muñoz-Hoyos A. Current Evidence on the Role of the Gut Microbiome in ADHD Pathophysiology and Therapeutic Implications. Nutrients 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheung SG et al. Systematic Review of Gut Microbiota and Major Depression. Front Psychiatry 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golubeva AV et al. Microbiota-related Changes in Bile Acid & Tryptophan Metabolism are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 24, 166–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inoue R. et al. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem 80, 2450–2458 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Liu F. et al. Altered composition and function of intestinal microbiota in autism spectrum disorders: a systematic review. Transl Psychiatry 9, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oades RD, Dauvermann MR, Schimmelmann BG, Schwarz MJ & Myint A-M Attention-deficit hyperactivity disorder (ADHD) and glial integrity: S100B, cytokines and kynurenine metabolism - effects of medication. Behav Brain Funct 6, 29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Re-evaluation of Some Organic Chemicals, Hydrazine and Hydrogen Peroxide. (International Agency for Research on Cancer, 1999). [PMC free article] [PubMed] [Google Scholar]
- 76.Antypa N, Drago A & Serretti A. The role of COMT gene variants in depression: Bridging neuropsychological, behavioral and clinical phenotypes. Neurosci Biobehav Rev 37, 1597–1610 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Lee LO & Prescott CA Association of the Catechol-O-Methyltransferase (COMT) Val158Met Polymorphism and Anxiety-Related Traits: A Meta-Analysis. Psychiatr Genet 24, 52–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martínez-Levy GA et al. Association of the catechol-O-methyltransferase gene and attention deficit hyperactivity disorder: results from an epidemiological study of adolescents of Mexico City. Psychiatr Genet 23, 90–91 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Stein MB, Fallin MD, Schork NJ & Gelernter J. COMT Polymorphisms and Anxiety-Related Personality Traits. Neuropsychopharmacology 30, 2092–2102 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Turroni F. et al. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front. Microbiol 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams JB, Johansen LJ, Powell LD, Quig D & Rubin RA Gastrointestinal flora and gastrointestinal status in children with autism -- comparisons to typical children and correlation with autism severity. BMC Gastroenterol 11, 22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abdel-Haq R, Schlachetzki JCM, Glass CK & Mazmanian SK Microbiome–microglia connections via the gut–brain axis. J Exp Med 216, 41–59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mikkelsen K, Stojanovska L & Apostolopoulos V. The Effects of Vitamin B in Depression. Curr Med Chem 23, 4317–4337 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Herbison CE et al. Low intake of B-vitamins is associated with poor adolescent mental health and behaviour. Preventive Medicine 55, 634–638 (2012). [DOI] [PubMed] [Google Scholar]
- 85.Hicks SD et al. Oral microbiome activity in children with autism spectrum disorder. Autism Research 11, 1286–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang C, Geng H, Liu W & Zhang G. Prenatal, perinatal, and postnatal factors associated with autism. Medicine (Baltimore) 96, e6696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y. et al. Association between fecal microbiota and generalized anxiety disorder: Severity and early treatment response. Journal of Affective Disorders 259, 56–66 (2019). [DOI] [PubMed] [Google Scholar]
- 88.Evans SJ et al. The gut microbiome composition associates with bipolar disorder and illness severity. Journal of Psychiatric Research 87, 23–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Humbel F. et al. Association of Alterations in Intestinal Microbiota With Impaired Psychological Function in Patients With Inflammatory Bowel Diseases in Remission. Clinical Gastroenterology and Hepatology 18, 2019–2029.e11 (2020). [DOI] [PubMed] [Google Scholar]
- 90.Dawson G. et al. Randomized, controlled trial of an intervention for toddlers with autism: the Early Start Denver Model. Pediatrics 125, e17–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.DuPaul GJ, Kern L, Gormley MJ & Volpe RJ Early Intervention for Young Children with ADHD: Academic Outcomes for Responders to Behavioral Treatment. School Mental Health 3, 117–126 (2011). [Google Scholar]
- 92.Lai M-C, Lombardo MV & Baron-Cohen S. Autism. The Lancet 383, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Luby JL, Belden A, Harms MP, Tillman R & Barch DM Preschool is a sensitive period for the influence of maternal support on the trajectory of hippocampal development. PNAS 113, 5742–5747 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mazurek MO, Curran A, Burnette C & Sohl K. ECHO Autism STAT: Accelerating Early Access to Autism Diagnosis. J Autism Dev Disord 49, 127–137 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Warner BB The contribution of the gut microbiome to neurodevelopment and neuropsychiatric disorders. Pediatric Research 85, 216–224 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Sudo N. et al. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol 558, 263–275 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tremblay J. et al. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol 0, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ye SH, Siddle KJ, Park DJ & Sabeti PC Benchmarking Metagenomics Tools for Taxonomic Classification. Cell 178, 779–794 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McIntyre ABR et al. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biology 18, 182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mohammadkhah AI, Simpson EB, Patterson SG & Ferguson JF Development of the Gut Microbiome in Children, and Lifetime Implications for Obesity and Cardiometabolic Disease. Children (Basel) 5, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bitsko RH et al. Epidemiology and Impact of Health Care Provider–Diagnosed Anxiety and Depression Among US Children. J Dev Behav Pediatr 39, 395–403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ghandour RM et al. Prevalence and Treatment of Depression, Anxiety, and Conduct Problems in US Children. The Journal of Pediatrics 206, 256–267.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


