Abstract
RNA, with a structure similar to DNA, should exhibit similar behaviors when it interacts with graphene. In this work, we designed a sensing platform of theophylline based on the interaction of an RNA aptamer with graphene oxide (GO) using the fluorescence as a sensing signal. Firstly, quantum dots (QDs) were modified with the selected ssRNA that can be used as an aptamer to recognize the theophylline. The fluorescence of QDs will be quenched in the presence of GO due to the noncovalent assembly between ssRNA aptamer and GO, leading to fluorescence resonance energy transfer (FRET) from QDs to GO, fluorescence “turn-off”. Then, in the presence of theophylline, the ssRNA aptamer recognizes theophylline to form a dsRNA–theophylline complex. The weak affinity between the complex and GO makes QDs move away from the GO surface, leading to the fluorescence recovery of QDs, fluorescence “turn-on”. Because of the high fluorescence quenching efficiency, unique structure of GO and specificity of the RNA aptamer, the proposed sensing platform exhibits high sensitivity and excellent selectivity for the determination of theophylline. The excellent performance of the sensor based on GO provides new opportunities for sensitive and selective detection of biorecognition events.
A fluorescent sensing platform of theophylline based on the interaction of an RNA aptamer with GO and CdTe as the signal.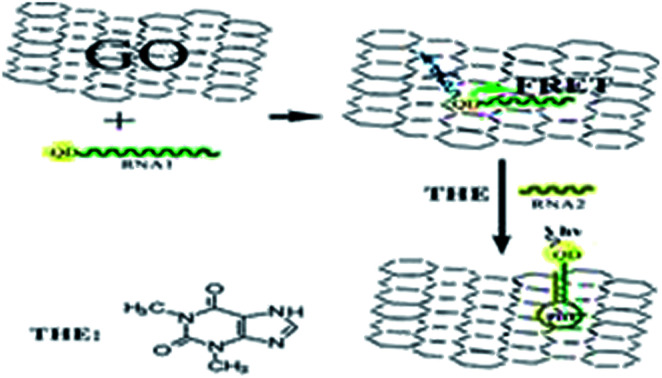
1. Introduction
Theophylline (1,3-dimethylxanthine) is one of the most commonly used bronchodilators and respiratory stimulators for the treatment of the symptoms of acute and chronic asthmatic conditions.1 The plasma levels of theophylline useful for effective bronchodilation action are within a narrow 20–100 μM concentration range. However, it is toxic at higher concentrations which can be lethal or lead to permanent neurological damage.2 At present, measurements of serum or plasma theophylline are carried out routinely in many clinical laboratories using gas/liquid chromatography methods and commercial immunoassays.3,4 Both approaches suffer from interference primarily from structurally closely related caffeine and theobromine resulting in overestimated serum/plasma theophylline concentrations. Despite that increased specificity of theophylline analysis has been achieved with theophylline oxidizing enzyme assays,5 restricted availability and stability of the enzyme impedes their routine clinical application.
Aptasensors based on the recognition of aptamer and target has been explored and widely applied in bioanalysis because of its fast response time, good specificity and selectivity. Up to now, combining with different signal readouts, various aptasensors were developed including electrochemical,6 fluorescent,7 colorimetric,8 electrochemiluminescence9 and surface enhanced Raman scattering (SERS) etc., which have displayed a good future in the detection of small molecules, proteins and drugs, etc. Among these aptasensors, fluorescent aptasensors have attracted special attention because their excellent features such as high sensitivity, high efficiency and simple operation etc. In fluorescent aptasensors, conformational changes of the aptamer upon binding of the target usually occur with the change of fluorescence of fluorophore.10 Despite of some fluorescent aptasensors for theophylline have been developed, the fluorescent probes are still limited in organic dyes such as Cy3,11 FAM (fluorescent amidite),12etc. as probes. For example, Ling et al. used FAM labeled RNA aptamer to prepare selectively turn-on fluorescent aptasensor for theophylline detection in serum.12 However, due to the organic dye showed poor stability and less resistance to photobleaching, short fluorescence lifetime, and long assay time, their further application was limited.
Currently, quantum dots (QDs) have attracted extensive attention in a wide variety of potential applications, because of their unique properties including high efficiency, high purity, high solubility and low cost.13 As fluorescent probes, QDs also have the advantages of long fluorescent life, various colors and high stability, which make them have low overlapping signals and high accuracy when marking the target object.14,15 Therefore, it is still desired to explore the probe of QDs for the fabrication of theophylline aptasensor.
Furthermore, graphene oxide (GO) has attracted great attention in fluorescent detection because it may be a good energy acceptor originated from its peculiar electronic properties,16–18 which has been applied in various fluorescence resonance energy transfer system as an outstanding quencher of electronic excited states of fluorophores.19 In addition, GO interacts strongly with single-stranded oligonucleotides through π-stacking interactions between the ring structures in the nucleobases and the hexagonal cells of Gr. Based on these features, recently, several groups have fabricated a variety of Gr-DNA biosensing platform sensors for the detection of CD63 and EpCAM in colorectal cancer exosomes (various types of cancer exosomes)20 through fluorophore-labeled DNA released from the GO upon the recognition binding with the target analytes.21–23 Despite of these booming progress in GO-DNA based fluorescent aptasensors, there are less aptasensors using the RNA aptamer as the biorecognition unit to develop a selective and sensitive fluorescent sensor, probably as a result of the higher susceptibility of RNA in degradation than DNA. Just like DNA, RNA is also particularly useful as a high-affinity RNA aptamer to recognize target molecules.24 RNA-based aptamers have also offered important applications in clinical diagnostics and treatment of viral and immune diseases.25
In this work, due to the features of quantum dots, CdTe QDs were chosen as fluorescent label with RNA which contained 19 nts RNA aptamer sequence of theophylline, and a fluorescent turn-on aptasensor for theophylline was fabricated based on the quenching capability towards CdTe QDs and the selective adsorption of RNA on GO platform. This novel detection platform shows high sensitivity and specificity, which may be explored to clinical treatment and object detection.
2. Experimental
2.1. Materials and reagents
Graphite power and sodium borohydride of analytical grade were from Sinopharm Chemical Reagent Co. Ltd. (China). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and theophylline et al. were purchased from Sigma-Aldrich, Inc. (U.S.A.). Thioglycolic acid (TGA) and cadmium chloride hemipentahydrate (CdCl2·2.5H2O) were from Alfa Aesar China Ltd. Ultrapure water obtained from a Millipore water purification system (≥18 MΩ, Milli-Q, Millipore, Billerica, MA) was used in all runs. All other reagents were of analytical grade.
The theophylline aptamers (Shanghai Sangon, China) used in this study have the following sequences: 5′-HS-(CH2)6-CCU UGG AAG CC-3′ (RNA1), 5′-GG AUA CCA-(CH2)6-SH-3′ (RNA2). RNA hybridization buffer was Tris–HCl (100 mM NaCl, 2.5 mM Mg2+, pH 7.4). RNA was stored in Tris–HCl buffer (TB) (10 mM, pH 8.2). TB (50 mM, pH 7.4) was used for modification of QDs with RNA.
2.2. Synthesis of graphene oxide
According to Hummer's method, GO was synthesized from graphitic power.26 Generally, graphite powders (1 g) were put into 0 °C solution consisting of concentrated H2SO4 (25 mL), NaNO3 (0.5 g), KMnO4 (3 g) under stirring and reacted for 0.5 h. The mixture was kept at 35 °C for several hours. The mixture was then diluted with 50 mL water and kept at 95 °C for 2 h. After the resulting mixture was stirred for 2 h, 150 mL of water and 10 mL of 30% H2O2 were added to end the reaction. The mixture was filtered and washed with HCl aqueous solution and water. Finally, the product was further purified by dialysis for 1 week to remove the remaining metal species. By sonicating dispersion under ambient conditions for 30 min, the homogeneous GO suspension (0.01 mg mL−1) was obtained, which was stable for several months.
2.3. Preparation of QDs-RNA1
The TGA-capped CdTe QDs were obtained according to literature procedure.27 QDs-RNA1 were prepared by mixing the QDs with 6 equiv. of RNA1 in TB containing EDC (0.1 mg mL−1). The mixture was allowed to react for 12 h at room temperature. Successively, the unreacted RNA1 and excess EDC were removed by precipitation of the QDs, and the purified particles were re-dissolved in TB solution and stored at 4 °C. It is diluted to appropriated concentration before detection.
2.4. Apparatus
The morphology of GO and QDs-RNA1 adsorbed GO were examined with an Agilent 5500 atomic force microscopy (AFM, U.S.A.) and a JEM 2100 high-resolution transmission electron microscope (HRTEM). The ultraviolet-visible (UV-Vis) absorption spectrum of QDs was recorded with a U-3010 UV/vis spectrometer (Hitachi). The photoluminescence (PL) spectra were recorded at room temperature with F-4500 PC (Hitachi). The sample used for AFM observation was prepared by depositing a droplet of GO dispersion (2 μL, 0.01 mg mL−1) on a freshly cleaved mica surface and dried under vacuum at room temperature.
3. Results and discussion
3.1. Design of the sensing procedure
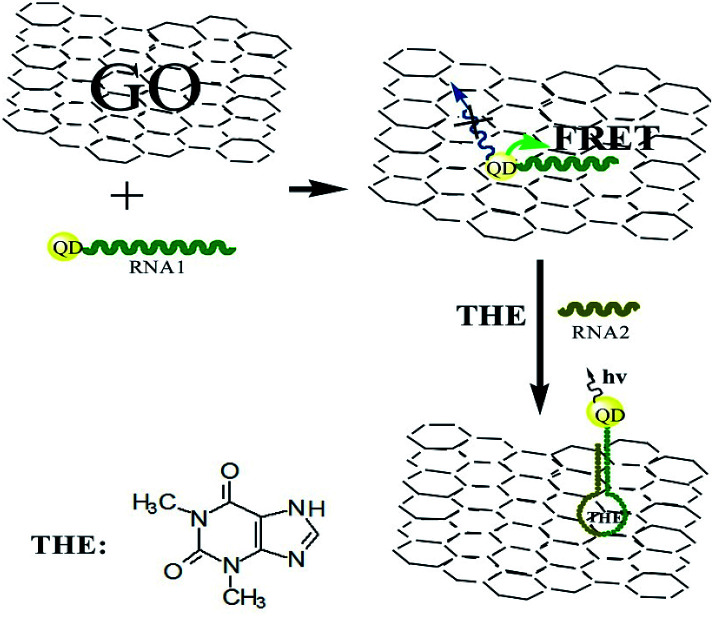
Firstly, a theophylline ssRNA aptamer with –SH group (RNA1) was designed and labeled with CdTe QDs to form QDs labeled RNA aptamer. Then, in the presence of GO, due to the noncovalent assembly of RNA1 chain on GO by π–π stacking, QDs labeled ssRNA were adsorbed on the GO surface. Consequentially, the FRET from QDs to GO can take place, which leads to the decrease of QDs fluorescence, as shown in Scheme 1. In the presence of RNA2 and target, the RNA1 recognizes the target and occurs a conformational change to form dsRNA with RNA2 to detach from the surface of GO. The FRET from QDs to GO is hindered because of the weak affinity of dsRNA with GO which leads to the leaving of QDs from GO and the increasing QDs-GO distance. The fluorescence of QDs can be recovered and the recovery is dependent on the concentration of target. Based on this, the detection of target analyte can be performed. The high quenching efficiency of GO to the fluorescence of QDs will ensure the designed sensing platform with high sensitivity.
Scheme 1. Schematic representation of GO-induced fluorescence quenching of ssRNA-QDs and biosensing mechanism.
3.2. Characterization of GO
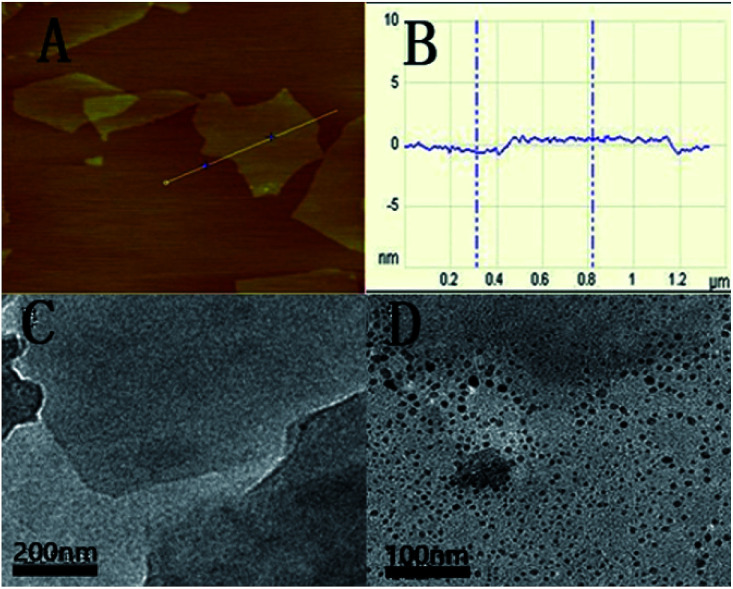
The as-prepared GO was characterized with AFM and HRTEM. Fig. 1A shows the AFM image of GO. From the image, the average thickness of GO sheet was about 1.1 nm. The result is similar to a previous report.28 The typical HRTEM image of as-prepared GO showed that the GO sheet was about 1 × 1 μm in size with occasional folds, crinkles and rolled edges (Fig. 1C). After the QDs-RNA1 adsorbed on the GO (QDs-RNA1-GO), it was found that well-dispersed QDs on GO were observed in Fig. 1D, suggesting that the aggregation of QDs does not take place.
Fig. 1. Tapping mode AFM image of GO sheets deposited on mica substrate (A), the average thickness of GO sheet (B), HRTEM images of GO sheets (C), and QDs-RNA1 adsorbed on GO (D).
3.3. PL and absorption spectra of CdTe QDs
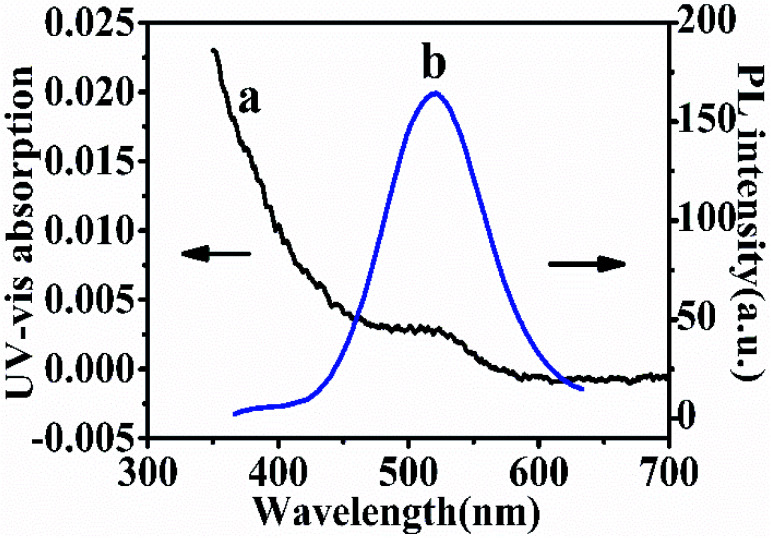
Photoluminescence and UV-Vis absorption spectra are used to characterize the prepared CdTe QDs. Fig. 2 shows the PL and UV-Vis spectra of the thioglycolic acid-capped QDs in 0.05 M TB (pH 7.4). The PL emission peak at 564 nm (λex = 380 nm) and the maximum absorption wavelength at 515 nm indicated the consequence of quantum confinement. According to the literature,29 the prepared particle size may also be calculated in virtue of the following expression:D = (−6.6521 × 10−8) λ3 + (1.9557 × 10−4) λ2 − (9.2352 × 10−2) λ + 13.29where, λ is the UV-Vis peak wavelength (515 nm); the result shows that the particle diameter of the as-prepared QDs is 8.5 nm, which is consistent with the result of the HRTEM.
Fig. 2. UV-Vis absorption spectrum of as-prepared CdTe QDs (a) and PL spectra of QDs (b).
3.4. Fluorescence quenching efficiency
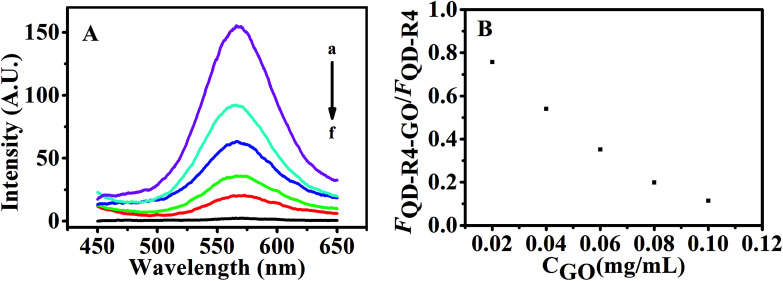
The fluorescence of QDs labeled DNA aptamer can be efficiently quenched by GO due to FRET between QDs and GO.30 Here, CdTe QDs is labeled with the RNA aptamer and its fluorescence quenching by GO is the key step to realize the sensing design. Fig. 3 shows the fluorescence spectra of QDs-RNA1 at various concentrations of GO. The fluorescence intensity of QDs-RNA1 decreased rapidly with increasing GO concentration. The fluorescence of QDs-RNA1 can be quenched fully while GO concentration increases up to 0.1 μg mL−1, implying that GO is an excellent fluorescence quencher. Because the aggregation of QDs does not take place, the fluorescence quenching of QDs-RNA1 is not caused by the aggregation of QDs. It is reasonable that the high efficiency quenching is considered as the direct consequence of noncovalent binding of RNA aptamer on the GO surface and the energy transfer from QDs to GO. The noncovalent binding of ssDNA to GO surface has been reported,31 in which the π–π stacking of DNA bases with aromatic rings of GO guarantees the assembly of ssDNA on the GO surface. Such strong binding between aptamer and GO will cause the labeled fluorophores approach to GO, and high efficiency energy transfer between the fluorophores and GO will occur. For a RNA aptamer, the similar interaction between RNA and GO should be taken place. In addition, high fluorescence efficiency CdTe QDs was used to replace organic fluorophore, obtaining stable, sensitive fluorescence signal. In addition, we carried out the UV-Vis adsorption (Fig. S1†) and zeta potential (Fig. S2†) to further characterize the conjugation of QDs-RNA1 with GO.
Fig. 3. (A) Fluorescence emission spectra of QDs-RNA1 (120 nM) at GO concentrations of 0, 0.02, 0.04, 0.06, 0.08, 0.1 μg mL−1 (a–f). (B) Fluorescent intensity ratio of QDs-RNA1 upon different concentrations of GO.
3.5. Fluorescence recovery of QDs-RNA1-GO aptasensor induced by theophylline
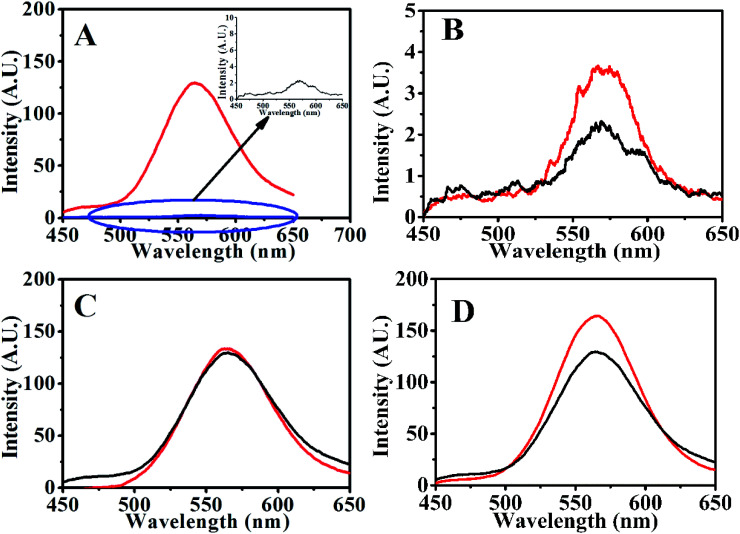
With a fixed amount of the aptasensor (120 nM QDs-RNA1 plus 0.1 μg mL−1 GO, QDs-RNA1-GO), the fluorescence recovered rapidly with the addition of theophylline and RNA2 to the system containing QDs-RNA1-GO (Fig. 4A) due to the RNA aptamer was induced to form hairpin–theophylline.
Fig. 4. Fluorescence emission spectra of (A) QDs-RNA1-GO (black) and QDs-RNA1-GO incubated with target and RNA2 (red), (B) QDs-RNA1-GO (black) and QDs-RNA1-GO incubated with RNA2 (red), (C) QDs-hairpin–theophylline before (red) and after (black) incubated with GO for 10 min, (D) QDs-RNA1 (120 nM, red) and QDs-RNA1-GO incubated with targets (black).
3.6. Theophylline detection with QDs-RNA1-GO
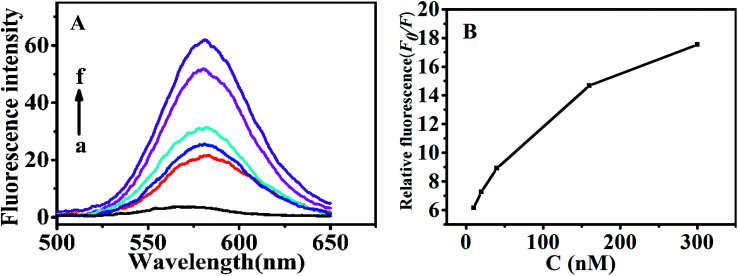
Under the optimal conditions (Fig. S3†), determination of theophylline with the above QDs-RNA1-GO was conducted in 50 mM TB (pH 7.4). As shown in Fig. 5, with a fixed amount of the QDs-RNA1-GO (120 nM QDs-RNA1 plus 0.1 μg mL−1 GO), the fluorescence emission intensity was enhanced depending on theophylline concentration. The fluorescence intensity ratio F0/F was linearly related to the concentration of theophylline within the range from 10 to 300 nM (inset, Fig. 5). The limit of detection (LOD) for theophylline was 4 nM according to three times the signal-to-noise level.
Fig. 5. (A) Fluorescence spectra of the aptasensor via different concentrations of theophylline (0, 10, 20, 40, 160 and 300 nM) in 50 mM TB (pH 7.4). (B) Relative fluorescence changes of the aptasensor via theophylline concentrations. Relative fluorescence changes are calculated by F0/F, where F0 and F are the fluorescence intensity without and with theophylline, respectively.
3.7. Specificity of the QDs-RNA1-GO for theophylline
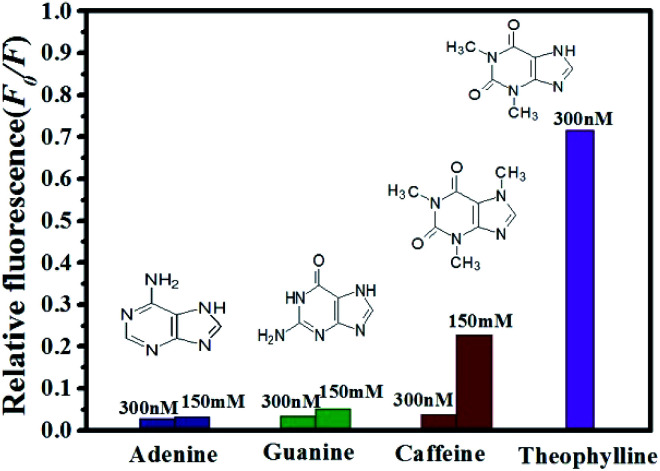
To assess the specificity of the GO based RNA aptasensor for theophylline, the influences of three kinds of biomolecules including caffeine, adenine and guanine were studied in our experiments. In a typical experiment, the above constructed aptasensor was incubated with 300 nM inspected species, 150 mM inspected species, and 300 nM theophylline in TB, individually. As shown in Fig. 6, fluorescence of the FRET aptasensor changed less for 300 nM caffeine, 300 nM adenine and 300 nM guanine, while a significant fluorescence increase was observed for 300 nM theophylline. It is also seen from Fig. 6 that none of the three biomolecules caused obvious fluorescence alteration even with a concentration as high as 150 mM. Therefore, the results have clearly illustrated the constructed aptasensor can be applied in detection of theophylline with high sensitivity and specificity.
Fig. 6. Relative fluorescence of QDs-RNA1 at different concentrations of analytes: theophylline (purple); caffeine (wine); guanine (olive) and adenine (royal).
4. Conclusions
In conclusion, a novel sensing platform for theophylline based on the fluorescence quenching of QDs-RNA1 by GO was proposed. The fluorescence recovery may be explained by the relative weaker binding of hairpin–theophylline complexes to GO compared with RNA. The proposed system showed low detection limit and good specificity. On the basis of their excellent performance, it may also provide a platform for detection of other specific biomolecules, thus extending application of the detection platform and opening new opportunities for sensitive detection of biorecognition events.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported financially by the National Nature Science Foundation of China (20975001) and the Educational Administration of Anhui Province (KJ2016A262).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra02475a
References
- Kawai M. Kato M. Methods Find. Exp. Clin. Pharmacol. 2000;22:309–320. doi: 10.1358/mf.2000.22.5.796650. [DOI] [PubMed] [Google Scholar]
- Owe D. J. Watson I. D. Williams J. Berry D. J. Ann. Clin. Biochem. 1988;25:4–26. doi: 10.1177/000456328802500102. [DOI] [PubMed] [Google Scholar]
- Howard C. E. Capers C. C. Bess D. T. Anderson R. J. Am. J. Hosp. Pharm. 1994;51:1672. [PubMed] [Google Scholar]
- Herrmann G. Aynesworth M. B. Martin J. J. Lab. Clin. Med. 1937;23:135–148. [PubMed] [Google Scholar]
- Ferapontova E. E. Shipovskov S. V. Gorton L. Biosens. Bioelectron. 2007;22:2508–2515. doi: 10.1016/j.bios.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Bostan H. B. Taghdisi S. M. Bowen J. L. Demertzis N. Rezaee R. Panahi Y. Tsatsakis A. M. Karimi G. Biosens. Bioelectron. 2018;119:110–118. doi: 10.1016/j.bios.2018.08.003. [DOI] [PubMed] [Google Scholar]
- Wong R. C. H. Lo P.-C. Ng D. K. P. Coord. Chem. Rev. 2019;379:30–46. doi: 10.1016/j.ccr.2017.10.006. [DOI] [Google Scholar]
- Abnous K. Danesh N. M. Ramezani M. Taghdisi S. M. Microchim. Acta. 2017;184:4451–4457. doi: 10.1007/s00604-017-2491-x. [DOI] [Google Scholar]
- Cao Z. Y. Su B. Electrochem. Commun. 2019;98:47–52. doi: 10.1016/j.elecom.2018.11.016. [DOI] [Google Scholar]
- Park K. S. Oh S. S. Soh H. T. Park H. G. Nanoscale. 2014;6(17):9977–9982. doi: 10.1039/C4NR00625A. [DOI] [PubMed] [Google Scholar]
- Jiang H. Y. Ling K. Tao X. J. Zhang Q. Q. Biosens. Bioelectron. 2015;70:299–303. doi: 10.1016/j.bios.2015.03.054. [DOI] [PubMed] [Google Scholar]
- Ling K. Jiang H. Y. Li Y. Tao X. J. Qiu C. Li F. R. Biosens. Bioelectron. 2016;86:8–13. doi: 10.1016/j.bios.2016.06.024. [DOI] [PubMed] [Google Scholar]
- Yang Z. W. Gao M. Y. Wu W. J. Yang X. Y. Sun X. W. Zhang J. H. Wang H. C. Liu R. S. Han C. Y. Yang H. S. Li W. W. Mater. Today. 2018;24:69–93. doi: 10.1016/j.mattod.2018.09.002. [DOI] [Google Scholar]
- Cunha C. R. A. Oliveira A. D. P. R. Firmino T. V. C. Tenorio D. P. L. A. Pereira G. Carvalho Jr L. B. Santos B. S. Correia M. T. S. Fontes A. Biochim. Biophys. Acta, Gen. Subj. 2018;1862(3):427–439. doi: 10.1016/j.bbagen.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Stanisavljevoc M. Krizkova S. Vaculovicova M. Kizek R. Adam V. Biosens. Bioelectron. 2015;74:562–574. doi: 10.1016/j.bios.2015.06.076. [DOI] [PubMed] [Google Scholar]
- Li W. Yang C. X. Yan X. P. Chem. Commun. 2017;53:11469–11471. doi: 10.1039/C7CC06244C. [DOI] [PubMed] [Google Scholar]
- Zhao Q. Zhang Z. Q. Tang Y. L. Chem. Commun. 2017;53:9414–9417. doi: 10.1039/C7CC04293K. [DOI] [PubMed] [Google Scholar]
- Ying Z. M. Wu Z. Tu B. Tan W. H. Jiang J. H. J. Am. Chem. Soc. 2017;139:9779–9782. doi: 10.1021/jacs.7b04527. [DOI] [PubMed] [Google Scholar]
- Su F. X. Yang C. X. Yan X. P. Anal. Chem. 2017;89:7277–7281. doi: 10.1021/acs.analchem.7b01348. [DOI] [PubMed] [Google Scholar]
- Wang H. Chen H. Huang Z. P. Li T. D. Deng A. M. Kong J. L. Talanta. 2018;184:219–226. doi: 10.1016/j.talanta.2018.02.083. [DOI] [PubMed] [Google Scholar]
- Liu T. T. Gao Y. Q. Zhang X. M. Wan Y. C. Du L. P. Fang H. Li M. Y. Anal. Chem. 2017;89:11173–11177. doi: 10.1021/acs.analchem.7b01148. [DOI] [PubMed] [Google Scholar]
- Qian H. F. Dong C. Q. Weng J. F. Ren J. C. Small. 2006;2:747. doi: 10.1002/smll.200500530. [DOI] [PubMed] [Google Scholar]
- Jiang L. Chen X. Yang W. S. Jin J. Yang B. Q. Xu L. Li T. J. Chem. J. Chin. Univ. 2001;22:1397. [Google Scholar]
- Yu W. W. Qu L. Guo W. Peng X. Chem. Mater. 2003;15:2854. doi: 10.1021/cm034081k. [DOI] [Google Scholar]
- Dong H. F. Gao W. C. Yan F. Ji H. X. Ju H. X. Anal. Chem. 2010;82:5511. doi: 10.1021/ac100852z. [DOI] [PubMed] [Google Scholar]
- Hummers W. S. Offeman R. E. J. Am. Chem. Soc. 1958;80:1339. doi: 10.1021/ja01539a017. [DOI] [Google Scholar]
- Cui R. J. Pan H. C. Zhu J. J. Chen H. Y. Anal. Chem. 2007;79:8494. doi: 10.1021/ac070923d. [DOI] [PubMed] [Google Scholar]
- Shen S. L. Zhang X. F. Ge Y. Q. Zhu Y. Cao X. Q. RSC Adv. 2017;7:55296–55300. doi: 10.1039/C7RA11086C. [DOI] [Google Scholar]
- Wang Z. Q. Zhang J. X. Chen F. Cai K. Y. Analyst. 2017;142:2796–2804. doi: 10.1039/C7AN00528H. [DOI] [PubMed] [Google Scholar]
- Wei M. J. Zhang Y. Y. Li H. T. Yao S. Z. Anal. Methods. 2017;9:3956–3961. doi: 10.1039/C7AY01097D. [DOI] [Google Scholar]
- Tuerk C. Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.