Abstract
Overproduction of reactive oxygen species (ROS) and superlative lipid peroxidation promote tumorigenesis, and mitochondrial aldehyde dehydrogenase 2 (ALDH2) is associated with the detoxification of ROS-mediated lipid peroxidation-generated reactive aldehydes such as 4-hydroxy-2-nonenal (4-HNE), malondialdehyde, and acrolein due to tobacco smoking. ALDH2 has been demonstrated to be highly associated with the prognosis and chemoradiotherapy sensitivity of many types of cancer, including leukemia, lung cancer, head and neck cancer, esophageal cancer, hepatocellular cancer, pancreatic cancer, and ovarian cancer. In this study, we explored the possible relationship between ALDH2 and urological cancers from the aspects of ferroptosis, epigenetic alterations, proteostasis, mitochondrial dysfunction, and cellular senescence.
Keywords: mitochondrial acetaldehyde dehydrogenase 2, urological cancers, ferroptosis, epigenetic alterations, proteostasis, mitochondrial dysfunction, cellular senescence
ALDH2: Structure, Function and Molecular Genetics
Acetaldehyde dehydrogenase-2 (ALDH2), encoded by a nuclear gene located on chromosome 12q24 (Raghunathan et al., 1988), is an enzyme rich in mitochondria with the activity of dehydrogenase, reductase and esterase (Mukerjee and Pietruszko, 1992; Chen and Stamler, 2006). ALDH2 is composed of 4 ∼56 kDa identical subunits, each of which mainly consists of the oligomerization domain, the catalytic domain, and the NAD+ -binding domain. ALDH2 provides protective effects not only by mediating ethanol metabolism and catalyzing the decomposition of acetaldehyde into nontoxic acetic acid but also by metabolizing other toxic short-chain fatty aldehydes and aromatic aldehydes (Klyosov, 1996). Furthermore, biochemical processes including decomposing endogenous aldehydic products, such as 4-HNE, malondialdehyde (MDA) and acrolein, derived from lipid peroxidation and tobacco smoking are associated with ALDH2 (Chen et al., 2010). The gene encoding ALDH2 is highly polymorphic. The most frequent point mutation, called ALDH2*2, occurs when an adenine is replaced by a guanine (rs671) at nucleotide 1459. Mutation of the ALDH2*2 enzyme disturbs the stabilization of hydrogen bonds and leads to damage to the structure of the NAD+ -binding site and several other catalytically important residues, ultimately resulting in enzyme inactivation (Li et al., 2016). Moreover, the combination of the mutant type (ALDH2*2) and the wild type, called AlDH2 *1/*2, can also result in a decrease in the catalytic activity of ALDH2 (Wang et al., 2002). It was reported that the activity of AlDH2 *1/*2 varied with the amount of AlDH2 *1/*2 in the ALDH2 tetramer, possibly reducing ALDH2 activity by 60%–80% compared to the wild-type (Weiner et al., 2001).
ALDH2 and Cancer
Increasing body of evidence has revealed a link between the ADLH2*2 genotype and many age-related, life-shortening diseases, such as cancers and osteoporosis (Chen et al., 2014). In upper aerodigestive track (UADT) cancers, Yokoyama et al. first reported a significant high risk of esophageal cancer in ALDH2*2 carriers (Yokoyama et al., 1996). Multiple original studies, reviews and meta-analyses and from Asia also found that the mutation of ALDH2*2 and the deficiency of ALDH2 enzyme activity were commonly implicated in UADT cancers due to the increased DNA damage induced by acetaldehyde cancers (Matsuda et al., 2006; Asakage et al., 2007; Yang et al., 2007; Hiyama et al., 2008; Ding et al., 2009; Lee et al., 2009; Cadoni et al., 2012). The ALDH2 polymorphism was also reported to be linked with an increased risk of gastrointestinal cancer including gastric cancer, pancreatic cancer, hepatocellular cancer, and colorectal cancer (Miyasaka et al., 2005; Li et al., 2016). Unexceptionally, among urological cancers, it was found that heterozygous ALDH2 carriers had a higher risk of bladder cancer (BCa) (Masaoka et al., 2016). And low expression of ALDH2 was related to lower overall survival in upper tract urothelial carcinoma (Wu et al., 2014). Furthermore, as an age-related disease, prostate cancer (PCa) is one of the most common malignant tumors in European and American senior citizens and is also the main cause of death (Siegel et al., 2021). In particular, it was found that the rs671 allele of the ALDH2 gene was associated with human longevity (Park et al., 2009), indicating a potential link between PCa and ALDH2. We summarized the reaches investigating the effects of ALDH2 in urological cancers in this study in Table 1.
TABLE 1.
The effects of ALDH2 on urological cancers in this study.
BCa: bladder cancer; RCa: renal cancer; PCa: prostate cancer.
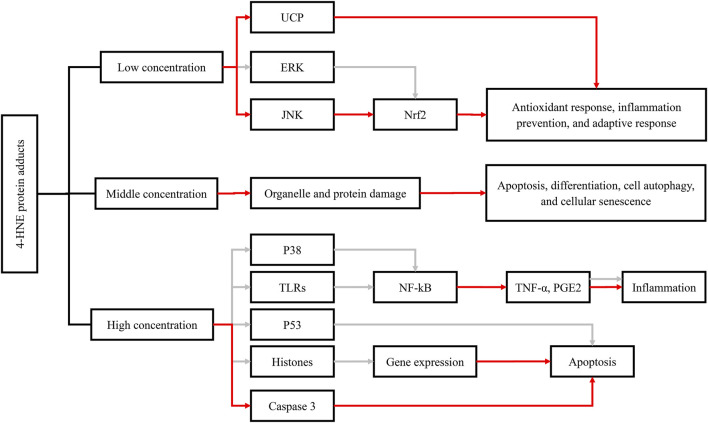
In addition to the direct effect of ALDH2 polymorphism, ALDH2-related aldehyde metabolites are also closely associated with cancers. 4-HNE was found to exhibit both endogenous carcinogenesis and antitumor effects (Zhang and Fu, 2021). Elevated oxidative stress and increased ROS generation have been confirmed to be related to a majority of cancer types by a large amount of strong evidence (DeNicola et al., 2011; Bellot et al., 2013), and 4-HNE generated from oxidative stress-induced lipid peroxidation plays a major role in the carcinogenic effects of lipid peroxidation (Zhong and Yin, 2015). In addition, p53 mutation induced by 4-HNE-DNA adducts is also one of the carcinogenetic mechanisms (Hu et al., 2002). Remarkably, the association between 4-HNE and early-stage carcinogenesis was first described in the case of renal cancer (RCa) (Segura-Aguilar et al., 1990). Interestingly, the antitumor effects of 4-HNE are linked to its concentration and cell type (Ayala et al., 2014). At a physiological or lower concentration, especially when it is similar to those in human tissues (Esterbauer et al., 1991), 4-HNE stimulates gene expression (especially Nrf2) to strengthen the antioxidant capacity of cells, prevent inflammation, and promote the adaptive response of immune cells for maintenance of homeostasis (Marantos et al., 2008). However, with concentration increasing, 4-HNE will tend to promote organelle and protein damage and thus inhibit cell proliferation and angiogenesis (Stagos et al., 2009). Simultaneously, it will induce apoptosis, differentiation, autophagy, and cellular senescence (Barrera et al., 2008; Pizzimenti et al., 2010). At a much higher level, 4-HNE will promote apoptosis or necrosis programmed cell death to avoid cancerization and eventually lead to cell death (Ayala et al., 2014). In this process, 4-HNE elicited its antitumor effect by regulating oncogenic signaling pathways and the expression of key genes, such as oncogenes c-myc (Fazio et al., 1992; Rinaldi et al., 2000; Pizzimenti et al., 2006), c-myb (Barrera et al., 1996), cyclin D, cyclin A (Skorokhod et al., 2010), and Notch1 (Pizzimenti et al., 2008). Moreover, the Doorn team confirmed that 4-HNE was not only a substrate but also an inhibitor of ALDH2, the inhibitory effect of which was reversible at a low concentration until 10 µM (Doorn et al., 2006). Therefore, ALDH2 inactivation induced by 4-HNE may play an essential role in the progression of some cancer species. As mentioned above, the effects of 4-HNE also depend on the cell type. For example, in hepatic cells, 4-HNE was found to promote p53 mutation and result in tumorigenesis, while it induced p53 expression in neuroblastoma cells, thus regulating cell cycle arrest or apoptosis induction and ultimately playing an antitumor role (Hu et al., 2002; Laurora et al., 2005). A similar phenomenon occurred in the regulation of NF-κB signaling pathway by 4-HNE (Ayala et al., 2014). Meanwhile, Lee et al. reported that long-term therapy with 0.1 µM 4-HNE led to an increase in cell growth in young smooth muscle cells (SMCs) but showed cytotoxicity to aged SMCs (Lee et al., 2006). In particular, 4-HNE is now believed to help normal cells defend cancer invasion and is more toxic to a variety of both hematological and solid tumor cells than normal cells. The differential effects of 4-HNE may be the consequence of changes in aldehyde-metabolizing enzymes, antioxidant defenses, and mitochondrial function (Pizzimenti et al., 2010; Barrera, 2012; Gasparovic et al., 2017).
Moreover, other reactive aldehydes, such as MDA and acrolein, also have similar bioactivities to 4-HNE. MDA was found to contribute to DNA damage and mutation (Niedernhofer et al., 2003; VanderVeen et al., 2003), and MDA-DNA adducts may ultimately lead to cell cycle arrest (Ji et al., 1998) and apoptosis (Willis et al., 2004) when DNA repair mechanism is lacking. Furthermore, acrolein seemed to play similar roles in carcinogenesis as HNE. Strikingly, acrolein was found to be associated with PCa progression and biochemical recurrence after prostatectomy and could be regarded as an excellently predictive biomarker of PCa relapse with an accuracy of approximately 90% (Custovic et al., 2010) (Figure 1).
FIGURE 1.
Functions of 4-HNE-protein adduct formation and their effects on cellular metabolic pathways, including the antioxidant response, inflammation, and apoptosis. The red arrow represents activation; the gray arrow represents inhibition. Abbreviations: 4-HNE, 4-hydroxynonenal; UCP, uncoupling protein; ERK, extracellular signal–regulated kinase; JNK, c-Jun N-terminal kinase; NFκB, nuclear factor kappa B; Nrf2, nuclear factor (erythroid-derived 2)-like 2; p38, protein 38; p53, protein 53; TLRs, toll-like receptors; TNF-α, tumor necrosis factor-α; PGE2, prostaglandin E.
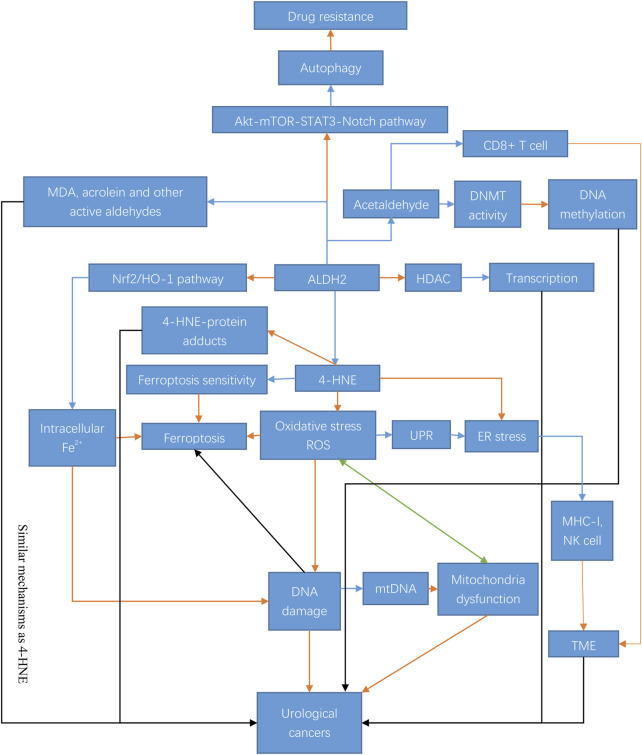
In summary, both ALDH2 and its associated metabolites are more or less directly or potentially related to cancer (Figure 2).
FIGURE 2.
Mechanisms and regulation of ALDH2 in urological cancer tumorigenesis. ALDH2 exerts its antitumor effects in urological tumors through a variety of mechanisms. ALDH2 can play a regulatory role through epigenetic modifications and some pathways, such as the Akt-Notch pathway, and it can also regulate the generation of its metabolites, including 4-HNE, MDA, and acrolein, and play an antitumor role through ferroptosis, oxidative stress, autophagy and many other pathways. The blue arrow indicates inhibition; the orange arrow indicates activation; the black arrow indicates both activation and inhibition; and the green arrow indicates mutual interaction. Abbreviations: STAT3, signal transducer and activator of transcription-3; mTOR, mammalian target of rapamycin; DNMT, DNA methyltransferase; HO-1, heme oxygenase; HDAC, histone deacetylase; ROS, reactive oxygen species; UPR, unfolded protein response; ER stress, endoplasmic reticulum stress; MHC-I, major histocompatibility complex-I; mtDNA, mitochondrial DNA; TME, tumor microenvironment.
Ferroptosis
As aging progresses, a disintegrated genome will lead to intracellular iron retention, resulting in DNA and epigenome damage, and contribute to genomic destabilization, which is one of the hallmarks of both aging and cancer. At the same time, DNA repair process induced by p53 is also blocked, and incomplete DNA further accelerates aging. This vicious cycle is called ferrosenescence (Sfera et al., 2018). In the process of ferrosenescence, retained iron leads to lipid peroxidation and endoplasmic reticulum (ER) stress, accompanied by cellular antioxidation failure, and ultimately results in ferroptosis (Sfera et al., 2018). However, a recent study showed that senescent cells with excessive accumulation of iron did not initiate ferroptosis but protected themselves from ferroptosis by avoiding the body’s self-renewal mechanism and accelerating body aging (Masaldan et al., 2018). Iron has been proven to be associated with RCa, PCa and BCa (Torti and Torti, 2013), in which the intracellular free iron concentration increased abnormally. An RCa model also described the carcinogenesis of iron overload by promoting DNA damage (Ebina et al., 1986). In aging and cancer, ALDH2 may be a double-edged sword. On the one hand, ALDH2 favor the antineoplastic effect of ferroptosis by lowering the 4-HNE concentration, and was also found to activate the Nrf2/HO-1 cascade to decrease intracellular iron (Ma et al., 2018). These effects may reveal the protective effect of ALDH2 at the initial stage of aging and carcinogenesis. On the other hand, it was reported that urological cancer was sensitive to ferroptosis inducers (FIN) (Yu et al., 2017), which indicates that ALDH2 may be a curative target for treatment-resistant PCa (Ghoochani et al., 2021). Therefore, reasonable activation of ferroptosis may be beneficial for both antiaging and antitumor effects. (Tao et al., 2019) reported that ALDH2*2 (rs671) male carriers had lower serum ferritin levels and that rs671 was significantly associated with ferritin concentrations. Accordingly, ALDH2 may regulate iron metabolism and thus contribute to ferroptosis. Different genotypes of ALDH2 among different populations may correspond to tumorigenic effect of alcohol in different degree, which could partly explain the contradictions on whether alcohol is a risk factor for urinary cancers. Furthermore, as a metabolite of lipid peroxidation, 4-HNE also has a strong connection with ferroptosis. Wang et al. reported that compared to low-stage cancers and normal tissues, the level of 4-HNE was relatively lower in high-stage cancer, and lower 4-HNE level was also associated with higher ferroptosis sensitivity in tumors (Wang et al., 2021). Previous studies also confirmed that tumors with a higher degree of malignancy were more sensitive to ferroptosis, manifesting higher propensity to metastasis and drug resistance, which indicated a potential antitumor role of 4-HNE (Viswanathan et al., 2017; Wu et al., 2019; Elgendy et al., 2020).
Given that ferroptosis acts specifically on malignant cells and protects normal cells, ferroptosis-targeted treatment has great therapeutic prospects for clinical applications. Precise therapy targeting ALDH2 and 4-HNE could be potential treatment approach for promoting ferroptosis sensitivity and inducing cell death and eventually achieve better prognosis among urological cancer patients.
Epigenetic Alterations
DNA Methylation
DNA methylation, as a common epigenetic mechanism of gene regulation, has been widely studied for the past few years. It is mainly mediated by DNA methyltransferases (DNMTs). Hypermethylation and hypomethylation in the promoter region is associated with gene silencing and transcription, respectively. Aging is the leading reason for the increased frequency of hypermethylation in CpG islands and promoters in many genes. Increasing global hypomethylation is associated with an increase in age, while many tumor suppressor genes are still hypermethylated (Maegawa et al., 2010; López-Otín et al., 2013). In addition, it is well established that local inflammation can predispose normal tissues to cancer, in which DNA methylation may have been involved (Easwaran et al., 2014). Modeled according to DNA methylation level, DNA methylation (DNAm) age, i.e., epigenetic age, is a more accurate predictor of human aging and biological age than telomere length (Jylhävä et al., 2017). DNAm age is strongly related to the occurrence and development of cancer. A meta-analysis found that accelerated DNAm age might increase the risk of death and adversely affect survival outcomes of RCa, PCa, and urothelial cancer (Dugué et al., 2018). Another study also found that DNAm age acceleration was highly correlated with genetic mutations in RCa and PCa (Horvath, 2013). For RCa, hypermethylation in genes such as TGF-β/RUNX3, NELL1, and ECRG4 can promote cell proliferation, EMT, tumor progression, migration, invasion, and metastasis of RCa (Ito and Miyazono, 2003; Mabuchi et al., 2010; Luo et al., 2016; Kim et al., 2017; Peters et al., 2018; Zheng et al., 2018). In PCa, hypermethylation of RARβ, cyclin D2 (CCND2), GSTP1, MGMT (Tang et al., 2013; Dumache et al., 2014; Litovkin et al., 2015; Gurioli et al., 2016) and hypomethylation of MYC, uPA, PLAU, S100P (Wang et al., 2007; Gurel et al., 2008; Hagelgans et al., 2013), may promote cancer cell proliferation, progression and metastasis and lead to poor clinical outcomes. (Jerónimo et al., 2004) showed that retinoic acid receptor beta2 (RARβ2) was hypermethylated in more than 90% of PCa and prostate intraepithelial neoplasia (PIN) cases while in only 20% of benign prostatic hyperplasia patients. Therefore, hypermethylation of RA receptor genes may affect PC progression by mediating gene expression. Additionally, it was also reported that DNA methylation is highly expressed in patients with BCa and might be relevant for bladder carcinogenesis (Jordahl et al., 2020).
DNA methylation plays a significant role in both aging and urological cancer, and ALDH2 may be involved in aging and urological cancer by regulating DNA methylation directly or indirectly. Firstly, ALDH2 may directly regulate the expression of methylated genes at the transcriptional level to exert its antitumor effect. For example, the ALDH2 promoter was found to contain a retinoid response element, which might contribute to gene regulation (Pinaire et al., 2003), which may be one of the antitumor mechanism for ALDH2 in PCa. Furthermore, ALDH2 also regulates gene methylation levels through its metabolites. Acetaldehyde was found to inhibit DNMT activity (Garro et al., 1991), and compared to healthy controls, long-term drinkers had significantly reduced mRNA levels of DNMT3a and DNMT3b (Bönsch et al., 2006). Metabolites of ALDH2, such as acetaldehyde and 4-HNE, have been proven to influence the clinical characteristics of liver cancer, colorectal cancer, breast cancer and upper aerodigestive tract cancer by regulating DNA methylation levels (Varela-Rey et al., 2013). Therefore, we speculated that a similar mechanism existed in urological cancer. Metabolites of ALDH2 may also regulate downstream signaling pathways of methylated genes since a low concentration of 4-HNE was found to stimulate cell proliferation and cell migration by promoting the nuclear factor kappa B (NF-κB) signaling pathway, and the expression of cyclin D1 and c-Myc (Xu et al., 2017) and 4-HNE also induced the production of TGF-β (Yang et al., 2019), which meant ALDH2 activation might decrease cell proliferation, migration, invasion, and EMT by reducing the concentration of 4-HNE.
Histone Modification
Changes in histone modifications are important in the aging process (Muñoz-Najar and Sedivy, 2011). The acetylation levels of histones are affected by the catalytic equilibrium of histone acetyltransferases (HATs) and histone deacetylases (HDACs). In HDACs, the activity of the sirtuin family is directly linked to biological life cycle control. In aging cells, decreased SIRT1 activity leads to increased genomic aneuploidy and instability (Fatoba and Okorokov, 2011). In addition, SIRT6 depletion is associated with telomere dysfunction, resulting in chromosomal fusions and premature cellular senescence (Michishita et al., 2008). Further study showed that SIRT6 could participate in the antiaging process by reversing the effect of the NF-κB pathway (Kawahara et al., 2009). Additionally, as a component of DNA-dependent protein kinase, SIRT6 can directly participate in DNA damage repair by exerting homologous recombination and nonhomologous terminal junctions, maintaining genome stability and inhibiting the aging process (Mao et al., 2011). In RCa, it was reported that the overexpression of HDAC1 and HDAC6 both increased cell invasion and motility through increasing the expression of matrix metalloproteinase (Ramakrishnan et al., 2016). It was also reported that combining an HDAC inhibitor with an HIV protease inhibitor was effective for RCa cells (Sato et al., 2012). For BCa, HDAC1 mRNA was significantly overexpressed in BCa compared with normal tissues according to a small-scale study (Pinkerneil et al., 2017). While another study confirmed that HDAC1 and HDAC2 double knockout impaired cell proliferation function and led to apoptosis of urothelial carcinoma cells, which indicated the vital contribution of HDAC to BCa tumorigenesis and development (Pinkerneil et al., 2016). HDAC inhibition could induce a strong response of drug-resistant BCa cells (Juengel et al., 2017) and could exert adhesion-blocking properties on BCa cells (Juengel et al., 2013), which might also reveal a tumor-promoting effect of HDAC in BCa. Many studies have also described the oncogenic role of SIRT1-3 and the tumor suppressor role of SIRT4 and 6 in BCa (Giannopoulou et al., 2019). In PCa, it was observed that increased activity of HDACs was associated with elevated levels of serum PSA and increased invasiveness of tumor cells (Damaschke et al., 2013).
ALDH2 also participates in the process of aging and urological cancers by regulating histone modification. Several studies confirmed that ALDH2 could modulate SIRT1-mediated senescence by reducing the amount of 4-HNE (Gu et al., 2013; Xue et al., 2018). Furthermore, Choi et al. reported that ALDH2 could play an epigenetic regulatory role in gene translocation. It could bind to HDACs and result in higher HDAC activity, suggesting that ALDH2 would induce transcriptional repression (Choi et al., 2011). To sum up the above, we come to the conclusion that ALDH2 can inhibit the proliferation and development of urological cancer cells.
As a hallmark of aging, epigenetic alteration occurs not only in tumor cells but also in immune and stromal cells located in the tumor microenvironment (TME) by immune editing and reprogramming (Liu et al., 2017). Tumor cells escape elimination from the immune response through numerous mechanisms, including inhibiting the expression of genes related to tumor-associated antigens and antigen processing by DNA methylation and histone deacetylation (Dunn et al., 2017). In our previous study, we found that the number of CD8+ T cells, B cells, neutrophils and macrophages was positively correlated with the expression of ALDH2 in PCa and that ALDH2 could regulate the immune TME and decrease the inhibition of CD8+ T-cell activation and proliferation by reducing acetaldehyde accumulation. Here, we speculated that ALDH2 could also participate in immune TME regulation by not only its metabolites but also epigenetic alterations including DNA methylation and histone acetylation, while the latter is associated with human aging.
Proteostasis
It is well accepted that aging and some aging-related diseases including varies urological cancers are associated with the loss of proteostasis (Powers et al., 2009). The heat shock response (HSR), unfolded protein response (UPR) and other mechanisms all help maintain protein homeostasis by assisting protein folding correctly and eliminating proteins that fail to fold. Lab animals overexpressing molecular chaperones was reported to possess an extensive life span (Walker and Lithgow, 2003; Morrow et al., 2004). Meanwhile, autophagy and other processes play an equally important role in protein homeostasis by cleaning and recycling garbage proteins through protein degradation. Significantly upregulated heat shock protein levels were observed in RCa (Santarosa et al., 1997), PCa (Rappa et al., 2012), and BCa (Ischia and So, 2013). However, the exact effect of ALDH2 on HSR in both aging and urological cancers is still unknown. It was reported that lipid peroxidation products could bind to HSPs to inhibit their degeneration; therefore, ALDH2 might act as an aging-promoting factor. However, ALDH2 may play a role in the antitumor process by inhibiting the function-enhancing extracellular release of HSP70 of 4-HNE (Yang et al., 2019). 4-HNE can bind to a variety of peptides and proteins, including glutathione, carnosine, enzymes, proteins on membranes and cytoskeleton, chaperones, uncoupling proteins (UCPs) in mitochondria, and antioxidant proteins to form 4-HNE-protein adducts (Poli et al., 2008; Ischia and So, 2013; Zhao et al., 2014). Approximately one-third of the binding target proteins are located in mitochondria (Poli et al., 2008; Zhao et al., 2014). 4-HNE-protein adducts affect the functions and bioactivities of these proteins. After that, adducts of 4-HNE and cyclin-dependent kinases alter enzyme activity, contributing to cell cycle delay (Camarillo et al., 2016). Adducts of 4-HNE with extracellular signal–regulated kinases also changed the function of 4-HNE, resulting in a decrease in Nrf2 activity and a loss of signal transduction, resulting in disorders in cellular homeostasis and cell proliferation (Lin et al., 2014). 4-HNE also adheres to histones and elongation factor-2, modifying gene expression at the transcriptional and translational levels, respectively (Gęgotek and Skrzydlewska, 2019). Furthermore, when bound to other proteins, such as Toll-like receptors, 4-HNE could mediate immune regulation (Gęgotek and Skrzydlewska, 2019). Previous studies have confirmed that 4-HNE protein adducts formed in RCa tissues are related to cancer proliferation and progression (Shoeb et al., 2014).
The TME, partly characterized by high metabolism, hypoxia, nutrition limitation, and acidosis, changes the protein processing capacity of the ER of both cancer cells and infiltrating immune cells, thus leading to accumulation of errant proteins and eventually ER stress. A persistent, yet moderate ER stress response promotes cancer cell proliferation, invasion, metastasis, drug resistance, angiogenesis and immune escape through several mechanisms (Chen and Cubillos-Ruiz, 2021), and cells respond to ER stress by activating the UPR. As a marker of ER stress, GRP78 is overexpressed in PCa and BCa, which is related to tumor development, recurrence and a poor survival outcome (LeRoy et al., 2007; Delie et al., 2013; Li et al., 2013). ALDH2 may inhibit urological cancers by regulating ER stress. For instance, it was found to strengthen the UPR by reducing oxidative stress (Long et al., 2020), and increased expression of ALDH2 was associated with reduced ER stress (Yan et al., 2020). The ER stress of cancer cells can also influence tumor progression by changing the function of the immune TME. It was reported that induction of ER stress impaired major histocompatibility complex class I-peptide (MHC I-peptide) presentation (Granados et al., 2009), and it altered NK-cell-mediated recognition of tumors (Obiedat et al., 2019). Therefore, ALDH2 may also possess an antitumor effect by regulating the TME.
Currently, as a nexus of aging and cancers, autophagy can determine key physiological decisions from cell fate to body lifespan through metabolism and proteostasis pathways. Autophagy works by degrading defective mitochondria and thus provides extra energy, including fatty acids and glutamine, for tumor cells (White, 2015). The blockade of autophagy significantly decreased drug-induced resistance in BCa (Li et al., 2019) and PCa (Farrow et al., 2014). The mTOR and AMP kinase (AMPK) signaling pathways are also significantly involved in aging and cancer. Upregulated mTOR signaling was confirmed to be associated with the development, progression and metastasis of PCa (López-Otín et al., 2013), and downregulated mTOR extended the lifespan of many kinds of laboratory animals (Zaytseva et al., 2012). It was reported that overexpression of ALDH2 suppresses autophagy (Wang and Wu, 2019), possibly by restoring the Akt-mTOR-STAT3-Notch signaling cascade (Ge and Ren, 2012), and ALDH2 might play a repressive role in transcriptional control by AMPK activation (Choi et al., 2011). Thus, ALDH2 could be a possible therapeutic target for both aging and urological cancer.
Mitochondrial Dysfunction
A decline in mitochondrial function occurs with aging. Many studies have shown that mtDNA increases with age in the human body (Corral-Debrinski et al., 1992; Fayet et al., 2002). Experimental evidence revealed the exact relationship between mtDNA and aging, which showed that the accumulation of mtDNA mutations could result in a premature aging phenotype (Trifunovic et al., 2004). In addition to primary mitochondrial dysfunction, abnormal mitochondrial biogenesis, secondary to abnormalities in nuclear genes and mitochondrial metabolism-dependent factors, such as ROS, NO, NAD+/NADH, ATP, and Ca2+, also promotes aging (Ryan and Hoogenraad, 2007). SIRT3 can help maintain mitochondrial and cellular homeostasis by regulating the ROS impacts of numerous physiologies linked with aging (van de Ven et al., 2017). The PI3K/Akt/mTOR signaling pathway is also involved in the regulation of mitochondrial biosynthesis, autophagy and apoptosis. The increase in mTOR activity in aging cells not only results in the accumulation of damaged mitochondria by inhibiting mitochondrial autophagy but also leads to mismatched production of mitochondria and metabolites by improving cell metabolic activity, thus aggravating the damage to mitochondrial function (Mollo et al., 2020).
Compared to normal tissues, there was a tendency of depletion in mtDNA in RCa and BCa (Reznik et al., 2016), especially in PCa (Parr et al., 2013). For example, the mtDNA G10398A polymorphism was related to a higher risk of PCa (Singh and Kulawiec, 2009). Many studies have also shown the significance of oxidative stress in the malignant transformation of kidney epithelial cells (Mahalingaiah et al., 2017), PCa tumorigenesis (Oh et al., 2016; Roumeguère et al., 2017), and the etiology and progression of BCa (Islam et al., 2019). It is widely known that since 1993,4-HNE has been regarded as a “toxic product of lipid peroxidation” and a “second toxic messenger of free radicals,” taking part in several signaling pathways related to proliferation, cell cycle arrest, apoptosis, and the regulation of the expression of a large number of genes (Cheng et al., 2001; Yang et al., 2001; Sharma et al., 2004; Vatsyayan et al., 2011), and has become a reliable biomarker of oxidative stress (Zarkovic, 2003). 4-HNE is also involved in the progression of RCa through oxidative stress-associated mechanisms (Shoeb et al., 2014). Therefore, modulation of 4-HNE-associated oxidative stress and 4-HNE-associated mitochondrial dysfunction can be utilized as a treatment method in cancer prevention and treatment. Existing studies have suggested that 4-HNE may decrease ROS and its related production and reduce oxidative damage in cancer by activating UCPs (Echtay et al., 2003; Esteves and Brand, 2005; Valle et al., 2010). Moreover, strong 4-HNE formation can affect mitochondrial function and ultimately lead to cell death (Zhao et al., 2014).
Overactivation of PI3K-Akt-mTOR signaling also promoted RCa cell proliferation, migration and metastasis, as well as angiogenesis and treatment resistance (Pal and Quinn, 2013) through the same mechanism as PCa (Shorning et al., 2020) and BCa (Xu et al., 2020). A decrease in mitochondrial detoxification induced by lipid peroxidative aldehydes has been observed and was hypothesized as a potential mechanism of aging in animal models (Yu, 2005; Ohsawa et al., 2008). Since ALDH2 is a mitochondrial chaperone (Chen et al., 2014; Wang et al., 2020) confirmed that ALDH2 deficiency might be linked to mtDNA accumulation in the cytoplasm, which indicated that mtDNA damage and ALDH2 recruitment preserve mitochondrial integrity. Elevated oxidative stress products (such as MDA), secondary to ALDH2 loss, not only mediate mitochondrial dysfunction, leading to mtDNA damage and contributing to cellular senescence and aging-associated phenotypes (Gosselin et al., 2009) but also promote cancer development through damaged mtDNA in hepatocellular carcinoma (Seo et al., 2019). It was shown in a preclinical model that the ability of ALDH2 to degrade 4-HNE declines with aging (Chen and Yu, 1996). Therefore, ALDH2 activation may slow down the aging process. Although the role of this mechanism of ALDH2 in the occurrence and development of urological cancers is still unknown, we assumed a similarity here. The interaction between SIRT3 and ALDH2 affects ALDH2-NAD+ binding (Harris et al., 2017), which indicates that ALDH2 might inhibit urological cancer by regulating ROS and oxidative stress.
Cellular Senescence
Cellular senescence was first described by (Hayflick, 1965) as cell proliferative activity decreased with an increase in division number and ultimately cell cycle arrest and cell death, which was called replicative senescence. Recently, it has been pointed out that cell senescence is a kind of cell state generated by pressure signal stimulation and exists in specific physiological processes with four typical characteristics: cell cycle arrest, senescence-associated secretion phenotype (SASP), macromolecular damage and metabolic disorder (Gorgoulis et al., 2019). Cellular senescence is the basis of biological aging, and most single-cell senescence also has the same characteristics as aging. In addition to epigenetic alterations, proteostasis, and mitochondrial dysfunction, cellular senescence is involved in altered intracellular communication through SASP. Senescent cells play a dual role in promoting and attenuating cancer. On the one hand, senescent tumor cells were found in prostate intraepithelial neoplasia but not in the corresponding malignant stage (Chen et al., 2005). This evidence suggests that senescence is an obstacle to the malignant development of tumors and can effectively inhibit the malignant transformation of tumors. Activation of specific oncogenes or inactivation of tumor suppressor genes can induce the senescence process to help cell cycle arrest. For instance, senescent hallmarks were also detected in early-stage prostate tumors (Courtois-Cox et al., 2006). SASP also has antitumor effects. Some SASPs, such as IL-6 and IL-8, promote senescence through a positive feedback loop of autocrine signaling and reduce cell transformation and metastasis by inhibiting the activation of some oncogenes (Wajapeyee et al., 2008). However, on the other hand, SASP is more likely to play a role in carcinogenesis in PCa. It is mainly reflected in the following two aspects: 1) SASPs provide an immunosuppressive microenvironment for tumors and promote tumor immune escape. 2) SASPs promote tumor invasion and metastasis. Senescent cells suppress the antitumor immune response by inducing granulocytic myeloid-derived suppressor cell infiltration and thus inhibiting T cell activity (Toso et al., 2014). Levels of both IL-6 and sIL-6R were found to be strongly associated with bone metastases, tumor volume, and risk of progression in prostatectomy patients (Nguyen et al., 2014). IL-8 was found to induce FGF2 expression, promoting abnormal proliferation in the transition zone (Giri and Ittmann, 2001). Other SASPs, such as TGF-β1, CXCL12, MMP2, and MAPK, are also involved in this process (Fiard et al., 2021). Furthermore, SASPs have also been shown to decrease EMT in cancer cells (Coppé et al., 2008), and a phenotype has been shown to be resistant to chemotherapy and radiation in BCa (McConkey et al., 2009).
ALDH2 may inhibit stress-induced senescence by reducing some aldehyde metabolites of oxidative reactions. It was found that ALDH2 impairment accelerated the acquisition of a premature senescent phenotype mainly due to the impairment of mitochondrial bioenergetic functions and cellular communication (Nannelli et al., 2018). This evidence indicates a potential association between ALDH2 and senescence. NF-κB was activated and enriched in the chromatin portion of senescent cells (Acosta et al., 2008; Kuilman et al., 2008; Chien et al., 2011), regulating senescence by directly regulating SASPs, including IL-8 and IL-6, which in turn controlled SASP transcription and expression. mTOR also regulates SASPs by mediating the translation of MAP kinase–activated protein kinase 2 (MAPKAPK2) (Herranz et al., 2015). Therefore, for urological cancer, as mentioned before, ALDH2 may play an antitumor role in regulating SASPs such as TGF-β, IL-6, and IL-8 by regulating the NF-κB and mTOR pathways.
Conclusion
ALDH2 might be involved in the development and progression of urological cancers through multiple cellular processes.
Author Contributions
DF, WZ, and XS, proposed the project, and wrote the manuscript; QW and LY, supervised the project; All authors reviewed and edited the manuscript.
Funding
This program was supported by the National Natural Science Foundation of China (Grant Nos. 81974099, 82170785, 81974098, 82170784), programs from Science and Technology Department of Sichuan Province (Grant Nos. 21GJHZ0246), Young Investigator Award of Sichuan University 2017 (Grant No. 2017SCU04A17), Technology Innovation Research and Development Project of Chengdu Science and Technology Bureau (2019-YF05-00296-SN), Sichuan University--Panzhihua science and technology cooperation special fund (2020CDPZH-4).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Acosta J. C., O'Loghlen A., Banito A., Guijarro M. V., Augert A., Raguz S., et al. (2008). Chemokine Signaling via the CXCR2 Receptor Reinforces Senescence. Cell 133 (6), 1006–1018. 10.1016/j.cell.2008.03.038 [DOI] [PubMed] [Google Scholar]
- Asakage T., Yokoyama A., Haneda T., Yamazaki M., Muto M., Yokoyama T., et al. (2007). Genetic Polymorphisms of Alcohol and Aldehyde Dehydrogenases, and Drinking, Smoking and Diet in Japanese Men with Oral and Pharyngeal Squamous Cell Carcinoma. Carcinogenesis 28 (4), 865–874. 10.1093/carcin/bgl206 [DOI] [PubMed] [Google Scholar]
- Ayala A., Muñoz M. F., Argüelles S. (2014). Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid Med. Cell Longev 2014, 360438. 10.1155/2014/360438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera G. (2012). Oxidative Stress and Lipid Peroxidation Products in Cancer Progression and Therapy. ISRN Oncol. 2012, 137289. 10.5402/2012/137289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera G., Pizzimenti S., Dianzani M. U. (2008). Lipid Peroxidation: Control of Cell Proliferation, Cell Differentiation and Cell Death. Mol. Aspects Med. 29 (1-2), 1–8. 10.1016/j.mam.2007.09.012 [DOI] [PubMed] [Google Scholar]
- Barrera G., Pizzimenti S., Serra A., Ferretti C., Fazio V. M., Saglio G., et al. (1996). 4-hydroxynonenal Specifically Inhibits C-Myb but Does Not Affect C-Fos Expressions in HL-60 Cells. Biochem. Biophysical Res. Commun. 227 (2), 589–593. 10.1006/bbrc.1996.1550 [DOI] [PubMed] [Google Scholar]
- Bellot G. L., Liu D., Pervaiz S. (2013). ROS, Autophagy, Mitochondria and Cancer: Ras, the Hidden Master? Mitochondrion 13 (3), 155–162. 10.1016/j.mito.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Bönsch D., Lenz B., Fiszer R., Frieling H., Kornhuber J., Bleich S. (2006). Lowered DNA Methyltransferase (DNMT-3b) mRNA Expression Is Associated with Genomic DNA Hypermethylation in Patients with Chronic Alcoholism. J. Neural Transm. (Vienna) 113 (9), 1299–1304. 10.1007/s00702-005-0413-2 [DOI] [PubMed] [Google Scholar]
- Cadoni G., Boccia S., Petrelli L., Di Giannantonio P., Arzani D., Giorgio A., et al. (2012). A Review of Genetic Epidemiology of Head and Neck Cancer Related to Polymorphisms in Metabolic Genes, Cell Cycle Control and Alcohol Metabolism. Acta Otorhinolaryngol. Ital. 32 (1), 1–11. [PMC free article] [PubMed] [Google Scholar]
- Camarillo J. M., Rose K. L., Galligan J. J., Xu S., Marnett L. J. (2016). Covalent Modification of CDK2 by 4-Hydroxynonenal as a Mechanism of Inhibition of Cell Cycle Progression. Chem. Res. Toxicol. 29 (3), 323–332. 10.1021/acs.chemrestox.5b00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Ferreira J. C. B., Gross E. R., Mochly-Rosen D. (2014). Targeting Aldehyde Dehydrogenase 2: New Therapeutic Opportunities. Physiol. Rev. 94 (1), 1–34. 10.1152/physrev.00017.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Sun L., Mochly-Rosen D. (2010). Mitochondrial Aldehyde Dehydrogenase and Cardiac Diseases. Cardiovasc. Res. 88 (1), 51–57. 10.1093/cvr/cvq192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. J., Yu B. P. (1996). Detoxification of Reactive Aldehydes in Mitochondria: Effects of Age and Dietary Restriction. Aging Clin. Exp. Res. 8 (5), 334–340. 10.1007/bf03339590 [DOI] [PubMed] [Google Scholar]
- Chen X., Cubillos-Ruiz J. R. (2021). Endoplasmic Reticulum Stress Signals in the Tumour and its Microenvironment. Nat. Rev. Cancer 21 (2), 71–88. 10.1038/s41568-020-00312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Stamler J. S. (2006). Bioactivation of Nitroglycerin by the Mitochondrial Aldehyde Dehydrogenase. Trends Cardiovasc. Med. 16 (8), 259–265. 10.1016/j.tcm.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Chen Z., Trotman L. C., Shaffer D., Lin H.-K., Dotan Z. A., Niki M., et al. (2005). Crucial Role of P53-dependent Cellular Senescence in Suppression of Pten-Deficient Tumorigenesis. Nature 436 (7051), 725–730. 10.1038/nature03918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.-Z., Sharma R., Yang Y., Singhal S. S., Sharma A., Saini M. K., et al. (2001). Accelerated Metabolism and Exclusion of 4-hydroxynonenal through Induction of RLIP76 and hGST5.8 Is an Early Adaptive Response of Cells to Heat and Oxidative Stress. J. Biol. Chem. 276 (44), 41213–41223. 10.1074/jbc.m106838200 [DOI] [PubMed] [Google Scholar]
- Chien Y., Scuoppo C., Wang X., Fang X., Balgley B., Bolden J. E., et al. (2011). Control of the Senescence-Associated Secretory Phenotype by NF-Κb Promotes Senescence and Enhances Chemosensitivity. Genes Dev. 25 (20), 2125–2136. 10.1101/gad.17276711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.-W., Kim J.-H., Cho S.-C., Ha M.-K., Song K.-Y., Youn H.-D., et al. (2011). Malondialdehyde Inhibits an AMPK-Mediated Nuclear Translocation and Repression Activity of ALDH2 in Transcription. Biochem. Biophysical Res. Commun. 404 (1), 400–406. 10.1016/j.bbrc.2010.11.131 [DOI] [PubMed] [Google Scholar]
- Coppé J. P., Patil C. K., Rodier F., Sun Y., Muñoz D. P., Goldstein J., et al. (2008). Senescence-associated Secretory Phenotypes Reveal Cell-Nonautonomous Functions of Oncogenic RAS and the P53 Tumor Suppressor. Plos Biol. 6 (12), 2853–2868. 10.1371/journal.pbio.0060301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski M., Horton T., Lott M. T., Shoffner J. M., Flint Beal M., Wallace D. C. (1992). Mitochondrial DNA Deletions in Human Brain: Regional Variability and Increase with Advanced Age. Nat. Genet. 2 (4), 324–329. 10.1038/ng1292-324 [DOI] [PubMed] [Google Scholar]
- Courtois-Cox S., Genther Williams S. M., Reczek E. E., Johnson B. W., McGillicuddy L. T., Johannessen C. M., et al. (2006). A Negative Feedback Signaling Network Underlies Oncogene-Induced Senescence. Cancer Cell 10 (6), 459–472. 10.1016/j.ccr.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custovic Z., Zarkovic K., Cindric M., Cipak A., Jurkovic I., Sonicki Z., et al. (2010). Lipid Peroxidation Product Acrolein as a Predictive Biomarker of Prostate Carcinoma Relapse after Radical Surgery. Free Radic. Res. 44 (5), 497–504. 10.3109/10715761003636831 [DOI] [PubMed] [Google Scholar]
- Damaschke N. A., Yang B., Bhusari S., Svaren J. P., Jarrard D. F. (2013). Epigenetic Susceptibility Factors for Prostate Cancer with Aging. Prostate 73 (16), 1721–1730. 10.1002/pros.22716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delie F., Petignat P., Cohen M. (2013). GRP78-targeted Nanotherapy against Castrate-Resistant Prostate Cancer Cells Expressing Membrane GRP78. Targ Oncol. 8 (4), 225–230. 10.1007/s11523-012-0234-9 [DOI] [PubMed] [Google Scholar]
- DeNicola G. M., Karreth F. A., Humpton T. J., Gopinathan A., Wei C., Frese K., et al. (2011). Oncogene-induced Nrf2 Transcription Promotes ROS Detoxification and Tumorigenesis. Nature 475 (7354), 106–109. 10.1038/nature10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J.-H., Li S.-P., Cao H.-X., Wu J.-Z., Gao C.-M., Su P., et al. (2009). Polymorphisms of Alcohol Dehydrogenase-2 and Aldehyde Dehydrogenase-2 and Esophageal Cancer Risk in Southeast Chinese Males. Wjg 15 (19), 2395–2400. 10.3748/wjg.15.2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorn J. A., Hurley T. D., Petersen D. R. (2006). Inhibition of Human Mitochondrial Aldehyde Dehydrogenase by 4-Hydroxynon-2-Enal and 4-Oxonon-2-Enal. Chem. Res. Toxicol. 19 (1), 102–110. 10.1021/tx0501839 [DOI] [PubMed] [Google Scholar]
- Dugué P. A., Bassett J. K., Joo J. E., Jung C.-H., Wong E. M., Moreno-Betancur M., et al. (2018). DNA Methylation-Based Biological Aging and Cancer Risk and Survival: Pooled Analysis of Seven Prospective Studies. Int. J. Cancer 142 (8), 1611–1619. 10.1002/ijc.31189 [DOI] [PubMed] [Google Scholar]
- Dumache R., Puiu M., Motoc M., Vernic C., Dumitrascu V. (2014). Prostate Cancer Molecular Detection in Plasma Samples by Glutathione S-Transferase P1 (GSTP1) Methylation Analysis. Clin. Lab. 60 (5), 847–852. 10.7754/clin.lab.2013.130701 [DOI] [PubMed] [Google Scholar]
- Dunn J., RaoEpigenetics S. (2017). Epigenetics and Immunotherapy: The Current State of Play. Mol. Immunol. 87, 227–239. 10.1016/j.molimm.2017.04.012 [DOI] [PubMed] [Google Scholar]
- Easwaran H., Tsai H.-C., Baylin S. B. (2014). Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell 54 (5), 716–727. 10.1016/j.molcel.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina Y., Okada S., Hamazaki S., Ogino F., Li J. L., Midorikawa O. (1986). Nephrotoxicity and Renal Cell Carcinoma after Use of Iron- and Aluminum-Nitrilotriacetate Complexes in Rats. J. Natl. Cancer Inst. 76 (1), 107–113. [PubMed] [Google Scholar]
- Echtay K. S., Esteves T. C., Pakay J. L., Jekabsons M. B., Lambert A. J., Portero-Otín M., et al. (2003). A Signalling Role for 4-Hydroxy-2-Nonenal in Regulation of Mitochondrial Uncoupling. EMBO J. 22 (16), 4103–4110. 10.1093/emboj/cdg412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy S. M., Alyammahi S. K., Alhamad D. W., Abdin S. M., Omar H. A. (2020). Ferroptosis: An Emerging Approach for Targeting Cancer Stem Cells and Drug Resistance. Crit. Rev. Oncology/Hematology 155, 103095. 10.1016/j.critrevonc.2020.103095 [DOI] [PubMed] [Google Scholar]
- Esterbauer H., Schaur R. J., Zollner H. (1991). Chemistry and Biochemistry of 4-hydroxynonenal, Malonaldehyde and Related Aldehydes. Free Radic. Biol. Med. 11 (1), 81–128. 10.1016/0891-5849(91)90192-6 [DOI] [PubMed] [Google Scholar]
- Esteves T. C., Brand M. D. (2005). The Reactions Catalysed by the Mitochondrial Uncoupling Proteins UCP2 and UCP3. Biochim. Biophys. Acta (Bba) - Bioenerg. 1709 (1), 35–44. 10.1016/j.bbabio.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Farrow J. M., Yang J. C., Evans C. P. (2014). Autophagy as a Modulator and Target in Prostate Cancer. Nat. Rev. Urol. 11 (9), 508–516. 10.1038/nrurol.2014.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatoba S. T., Okorokov A. L. (2011). Human SIRT1 Associates with Mitotic Chromatin and Contributes to Chromosomal Condensation. Cell Cycle 10 (14), 2317–2322. 10.4161/cc.10.14.15913 [DOI] [PubMed] [Google Scholar]
- Fayet G., Jansson M., Sternberg D., Moslemi A.-R., Blondy P., Lombès A., et al. (2002). Ageing Muscle: Clonal Expansions of Mitochondrial DNA point Mutations and Deletions Cause Focal Impairment of Mitochondrial Function. Neuromuscul. Disord. 12 (5), 484–493. 10.1016/s0960-8966(01)00332-7 [DOI] [PubMed] [Google Scholar]
- Fazio V. M., Barrera G., Martinotti S., Farace M. G., Giglioni B., Frati L., et al. (1992). 4-Hydroxynonenal, a Product of Cellular Lipid Peroxidation, Which Modulates C-Myc and Globin Gene Expression in K562 Erythroleukemic Cells. Cancer Res. 52 (18), 4866–4871. [PubMed] [Google Scholar]
- Fiard G., Stavrinides V., Chambers E. S., Heavey S., Freeman A., Ball R., et al. (2021). Cellular Senescence as a Possible Link between Prostate Diseases of the Ageing Male. Nat. Rev. Urol. 18 (10), 597–610. 10.1038/s41585-021-00496-8 [DOI] [PubMed] [Google Scholar]
- Garro A. J., McBeth D. L., Lima V., Lieber C. S. (1991). Ethanol Consumption Inhibits Fetal DNA Methylation in Mice: Implications for the Fetal Alcohol Syndrome. Alcohol. Clin. Exp. Res. 15 (3), 395–398. 10.1111/j.1530-0277.1991.tb00536.x [DOI] [PubMed] [Google Scholar]
- Gasparovic A. C., Milkovic L., Sunjic S. B., Zarkovic N. (2017). Cancer Growth Regulation by 4-hydroxynonenal. Free Radic. Biol. Med. 111, 226–234. 10.1016/j.freeradbiomed.2017.01.030 [DOI] [PubMed] [Google Scholar]
- Ge W., Ren J. (2012). mTOR-STAT3-notch Signalling Contributes to ALDH2-Induced protection against Cardiac Contractile Dysfunction and Autophagy under Alcoholism. J. Cell Mol Med 16 (3), 616–626. 10.1111/j.1582-4934.2011.01347.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gęgotek A., Skrzydlewska E. (2019). Biological Effect of Protein Modifications by Lipid Peroxidation Products. Chem. Phys. Lipids 221, 46–52. 10.1016/j.chemphyslip.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Ghoochani A., Hsu E.-C., Aslan M., Rice M. A., Nguyen H. M., Brooks J. D., et al. (2021). Ferroptosis Inducers Are a Novel Therapeutic Approach for Advanced Prostate Cancer. Cancer Res. 81 (6), 1583–1594. 10.1158/0008-5472.can-20-3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulou A. F., Velentzas A. D., Konstantakou E. G., Avgeris M., Katarachia S. A., Papandreou N. C., et al. (2019). Revisiting Histone Deacetylases in Human Tumorigenesis: The Paradigm of Urothelial Bladder Cancer. Int. J. Mol. Sci. 20 (6), 1291. 10.3390/ijms20061291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri D., Ittmann M. (2001). Interleukin-8 Is a Paracrine Inducer of Fibroblast Growth Factor 2, a Stromal and Epithelial Growth Factor in Benign Prostatic Hyperplasia. Am. J. Pathol. 159 (1), 139–147. 10.1016/s0002-9440(10)61681-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V., Adams P. D., Alimonti A., Bennett D. C., Bischof O., Bishop C., et al. (2019). Cellular Senescence: Defining a Path Forward. Cell 179 (4), 813–827. 10.1016/j.cell.2019.10.005 [DOI] [PubMed] [Google Scholar]
- Gosselin K., Martien S., Pourtier A., Vercamer C., Ostoich P., Morat L., et al. (2009). Senescence-associated Oxidative DNA Damage Promotes the Generation of Neoplastic Cells. Cancer Res. 69 (20), 7917–7925. 10.1158/0008-5472.can-08-2510 [DOI] [PubMed] [Google Scholar]
- Granados D. P., Tanguay P.-L., Hardy M.-P., Caron É., de Verteuil D., Meloche S., et al. (2009). ER Stress Affects Processing of MHC Class I-Associated Peptides. BMC Immunol. 10, 10. 10.1186/1471-2172-10-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Xing Y., Jiang L., Chen M., Xu M., Yin Y., et al. (2013). Impaired Cardiac SIRT1 Activity by Carbonyl Stress Contributes to Aging-Related Ischemic Intolerance. PLoS One 8 (9), e74050. 10.1371/journal.pone.0074050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel B., Iwata T., M Koh C., Jenkins R. B., Lan F., Van Dang C., et al. (2008). Nuclear MYC Protein Overexpression Is an Early Alteration in Human Prostate Carcinogenesis. Mod. Pathol. 21 (9), 1156–1167. 10.1038/modpathol.2008.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurioli G., Salvi S., Martignano F., Foca F., Gunelli R., Costantini M., et al. (2016). Methylation Pattern Analysis in Prostate Cancer Tissue: Identification of Biomarkers Using an MS-MLPA Approach. J. Transl Med. 14 (1), 249. 10.1186/s12967-016-1014-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelgans A., Menschikowski M., Fuessel S., Nacke B., Arneth B. M., Wirth M. P., et al. (2013). Deregulated Expression of Urokinase and its Inhibitor Type 1 in Prostate Cancer Cells: Role of Epigenetic Mechanisms. Exp. Mol. Pathol. 94 (3), 458–465. 10.1016/j.yexmp.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Harris P. S., Gomez J. D., Backos D. S., Fritz K. S. (2017). Characterizing Sirtuin 3 Deacetylase Affinity for Aldehyde Dehydrogenase 2. Chem. Res. Toxicol. 30 (3), 785–793. 10.1021/acs.chemrestox.6b00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. (1965). The Limited In Vitro Lifetime of Human Diploid Cell Strains. Exp. Cell Res. 37, 614–636. 10.1016/0014-4827(65)90211-9 [DOI] [PubMed] [Google Scholar]
- Herranz N., Gallage S., Mellone M., Wuestefeld T., Klotz S., Hanley C. J., et al. (2015). mTOR Regulates MAPKAPK2 Translation to Control the Senescence-Associated Secretory Phenotype. Nat. Cell Biol 17 (9), 1205–1217. 10.1038/ncb3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama T., Yoshihara M., Tanaka S., Chayama K. (2008). Genetic Polymorphisms and Head and Neck Cancer Risk (Review). Int. J. Oncol. 32 (5), 945–973. 10.3892/ijo.32.5.945 [DOI] [PubMed] [Google Scholar]
- Horvath S. (2013). DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 14 (10), R115. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Feng Z., Eveleigh J., Iyer G., Pan J., Amin S., et al. (2002). The Major Lipid Peroxidation Product, Trans-4-hydroxy-2-nonenal, Preferentially Forms DNA Adducts at Codon 249 of Human P53 Gene, a Unique Mutational Hotspot in Hepatocellular Carcinoma. Carcinogenesis 23 (11), 1781–1789. 10.1093/carcin/23.11.1781 [DOI] [PubMed] [Google Scholar]
- Ischia J., So A. I. (2013). The Role of Heat Shock Proteins in Bladder Cancer. Nat. Rev. Urol. 10 (7), 386–395. 10.1038/nrurol.2013.108 [DOI] [PubMed] [Google Scholar]
- Islam M. O., Bacchetti T., Ferretti G. (2019). Alterations of Antioxidant Enzymes and Biomarkers of Nitro-oxidative Stress in Tissues of Bladder Cancer. Oxid Med. Cell Longev 2019, 2730896. 10.1155/2019/2730896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Miyazono K. (2003). RUNX Transcription Factors as Key Targets of TGF-β Superfamily Signaling. Curr. Opin. Genet. Development 13 (1), 43–47. 10.1016/s0959-437x(03)00007-8 [DOI] [PubMed] [Google Scholar]
- Jerónimo C., Henrique R., Hoque M. O., Ribeiro F. R., Oliveira J., Fonseca D., et al. (2004). Quantitative RARbeta2 Hypermethylation: a Promising Prostate Cancer Marker. Clin. Cancer Res. 10 (12 Pt 1), 4010–4014. 10.1158/1078-0432.CCR-03-0643 [DOI] [PubMed] [Google Scholar]
- Ji C., Rouzer C. A., Marnett L. J., Pietenpol J. A. (1998). Induction of Cell Cycle Arrest by the Endogenous Product of Lipid Peroxidation, Malondialdehyde. Carcinogenesis 19 (7), 1275–1283. 10.1093/carcin/19.7.1275 [DOI] [PubMed] [Google Scholar]
- Jordahl K. M., Phipps A. I., Randolph T. W., Tinker L. F., Nassir R., Hou L., et al. (2020). Mediation by Differential DNA Methylation of Known Associations between Single Nucleotide Polymorphisms and Bladder Cancer Risk. BMC Med. Genet. 21 (1), 228. 10.1186/s12881-020-01172-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juengel E., Najafi R., Rutz J., Maxeiner S., Makarevic J., Roos F., et al. (2017). HDAC Inhibition as a Treatment Concept to Combat Temsirolimus-Resistant Bladder Cancer Cells. Oncotarget 8 (66), 110016–110028. 10.18632/oncotarget.22454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juengel E., Santos S. M. d., Schneider T., Makarević J., Hudak L., Bartsch G., et al. (2013). HDAC Inhibition Suppresses Bladder Cancer Cell Adhesion to Collagen under Flow Conditions. Exp. Biol. Med. (Maywood) 238 (11), 1297–1304. 10.1177/1535370213498975 [DOI] [PubMed] [Google Scholar]
- Jylhävä J., Pedersen N. L., Hägg S. (2017). Biological Age Predictors. EBioMedicine 21, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T. L. A., Michishita E., Adler A. S., Damian M., Berber E., Lin M., et al. (2009). SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-κb-dependent Gene Expression and Organismal Life Span. Cell 136 (1), 62–74. 10.1016/j.cell.2008.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-J., Jeon Y., Kim T., Lim W.-C., Ham J., Park Y. N., et al. (2017). Combined Treatment with Zingerone and its Novel Derivative Synergistically Inhibits TGF-Β1 Induced Epithelial-Mesenchymal Transition, Migration and Invasion of Human Hepatocellular Carcinoma Cells. Bioorg. Med. Chem. Lett. 27 (4), 1081–1088. 10.1016/j.bmcl.2016.12.042 [DOI] [PubMed] [Google Scholar]
- Klyosov A. A. (1996). Kinetics and Specificity of Human Liver Aldehyde Dehydrogenases toward Aliphatic, Aromatic, and Fused Polycyclic Aldehydes. Biochemistry 35 (14), 4457–4467. 10.1021/bi9521102 [DOI] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Vredeveld L. C. W., Douma S., van Doorn R., Desmet C. J., et al. (2008). Oncogene-induced Senescence Relayed by an Interleukin-dependent Inflammatory Network. Cell 133 (6), 1019–1031. 10.1016/j.cell.2008.03.039 [DOI] [PubMed] [Google Scholar]
- Laurora S., Tamagno E., Briatore F., Bardini P., Pizzimenti S., Toaldo C., et al. (2005). 4-Hydroxynonenal Modulation of P53 Family Gene Expression in the SK-N-BE Neuroblastoma Cell Line. Free Radic. Biol. Med. 38 (2), 215–225. 10.1016/j.freeradbiomed.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Lee C.-H., Wu D.-C., Wu I.-C., Goan Y.-G., Lee J.-M., Chou S.-H., et al. (2009). Genetic Modulation of ADH1B and ALDH2 Polymorphisms with Regard to Alcohol and Tobacco Consumption for Younger Aged Esophageal Squamous Cell Carcinoma Diagnosis. Int. J. Cancer 125 (5), 1134–1142. 10.1002/ijc.24357 [DOI] [PubMed] [Google Scholar]
- Lee T.-J., Lee J.-T., Moon S.-K., Kim C.-H., Park J.-W., Kwon T. (2006). Age-related Differential Growth Rate and Response to 4-hydroxynonenal in Mouse Aortic Smooth Muscle Cells. Int. J. Mol. Med. 17 (1), 29–35. 10.3892/ijmm.17.1.29 [DOI] [PubMed] [Google Scholar]
- LeRoy B., Painter A., Sheppard H., Popiolek L., Samuel-Foo M., Andacht T. M. (2007). Protein Expression Profiling of normal and Neoplastic Canine Prostate and Bladder Tissue. Vet. Comp. Oncol. 5 (2), 119–130. 10.1111/j.1476-5829.2006.00121.x [DOI] [PubMed] [Google Scholar]
- Li F., Guo H., Yang Y., Feng M., Liu B., Ren X., et al. (2019). Autophagy Modulation in Bladder Cancer Development and Treatment (Review). Oncol. Rep. 42 (5), 1647–1655. 10.3892/or.2019.7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zoubeidi A., Beraldi E., Gleave M. E. (2013). GRP78 Regulates Clusterin Stability, Retrotranslocation and Mitochondrial Localization under ER Stress in Prostate Cancer. Oncogene 32 (15), 1933–1942. 10.1038/onc.2012.212 [DOI] [PubMed] [Google Scholar]
- Li R., Zhao Z., Sun M., Luo J., Xiao Y. (2016). ALDH2 Gene Polymorphism in Different Types of Cancers and its Clinical Significance. Life Sci. 147, 59–66. 10.1016/j.lfs.2016.01.028 [DOI] [PubMed] [Google Scholar]
- Lin M.-H., Yen J.-H., Weng C.-Y., Wang L., Ha C.-L., Wu M.-J. (2014). Lipid Peroxidation End Product 4-Hydroxy-Trans-2-Nonenal Triggers Unfolded Protein Response and Heme Oxygenase-1 Expression in PC12 Cells: Roles of ROS and MAPK Pathways. Toxicology 315, 24–37. 10.1016/j.tox.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Litovkin K., Van Eynde A., Joniau S., Lerut E., Laenen A., Gevaert T., et al. (2015). DNA Methylation-Guided Prediction of Clinical Failure in High-Risk Prostate Cancer. PLoS One 10 (6), e0130651. 10.1371/journal.pone.0130651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhou J., Chen Z., Cheng A. S.-L. (2017). Understanding the Epigenetic Regulation of Tumours and Their Microenvironments: Opportunities and Problems for Epigenetic Therapy. J. Pathol. 241 (1), 10–24. 10.1002/path.4832 [DOI] [PubMed] [Google Scholar]
- Long P., He M., Yan W., Chen W., Wei D., Wang S., et al. (2020). ALDH2 Protects Naturally Aged Mouse Retina via Inhibiting Oxidative Stress-Related Apoptosis and Enhancing Unfolded Protein Response in Endoplasmic Reticulum. Aging 13 (2), 2750–2767. 10.18632/aging.202325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G. (2013). The Hallmarks of Aging. Cell 153 (6), 1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Wu J., Xie J., Xia L., Qian X., Cai Z., et al. (2016). Downregulated ECRG4 Is Associated with Poor Prognosis in Renal Cell Cancer and Is Regulated by Promoter DNA Methylation. Tumor Biol. 37 (1), 1121–1129. 10.1007/s13277-015-3913-1 [DOI] [PubMed] [Google Scholar]
- Ma X., Luo Q., Zhu H., Liu X., Dong Z., Zhang K., et al. (2018). Aldehyde Dehydrogenase 2 Activation Ameliorates CC L 4 ‐induced Chronic Liver Fibrosis in Mice by Up‐regulating Nrf2/ HO ‐1 Antioxidant Pathway. J. Cell. Mol. Medi 22 (8), 3965–3978. 10.1111/jcmm.13677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi M., Kataoka H., Miura Y., Kim T.-S., Kawaguchi M., Ebi M., et al. (2010). Tumor Suppressor, at Motif Binding Factor 1 (ATBF1), Translocates to the Nucleus with Runt Domain Transcription Factor 3 (RUNX3) in Response to TGF-β Signal Transduction. Biochem. Biophysical Res. Commun. 398 (2), 321–325. 10.1016/j.bbrc.2010.06.090 [DOI] [PubMed] [Google Scholar]
- Maegawa S., Hinkal G., Kim H. S., Shen L., Zhang L., Zhang J., et al. (2010). Widespread and Tissue Specific Age-Related DNA Methylation Changes in Mice. Genome Res. 20 (3), 332–340. 10.1101/gr.096826.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingaiah P. K. S., Ponnusamy L., Singh K. P. (2017). Oxidative Stress-Induced Epigenetic Changes Associated with Malignant Transformation of Human Kidney Epithelial Cells. Oncotarget 8 (7), 11127–11143. 10.18632/oncotarget.12091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., et al. (2011). SIRT6 Promotes DNA Repair under Stress by Activating PARP1. Science 332 (6036), 1443–1446. 10.1126/science.1202723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marantos C., Mukaro V., Ferrante J., Hii C., Ferrante A. (2008). Inhibition of the Lipopolysaccharide-Induced Stimulation of the Members of the MAPK Family in Human Monocytes/macrophages by 4-hydroxynonenal, a Product of Oxidized omega-6 Fatty Acids. Am. J. Pathol. 173 (4), 1057–1066. 10.2353/ajpath.2008.071150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaldan S., Clatworthy S. A. S., Gamell C., Meggyesy P. M., Rigopoulos A.-T., Haupt S., et al. (2018). Iron Accumulation in Senescent Cells Is Coupled with Impaired Ferritinophagy and Inhibition of Ferroptosis. Redox Biol. 14, 100–115. 10.1016/j.redox.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaoka H., Ito H., Soga N., Hosono S., Oze I., Watanabe M., et al. (2016). Aldehyde Dehydrogenase 2 (ALDH2) and Alcohol Dehydrogenase 1B (ADH1B) Polymorphisms Exacerbate Bladder Cancer Risk Associated with Alcohol Drinking: Gene-Environment Interaction. Carcin 37 (6), 583–588. 10.1093/carcin/bgw033 [DOI] [PubMed] [Google Scholar]
- Matsuda T., Yabushita H., Kanaly R. A., Shibutani S., Yokoyama A. (2006). Increased DNA Damage in ALDH2-Deficient Alcoholics. Chem. Res. Toxicol. 19 (10), 1374–1378. 10.1021/tx060113h [DOI] [PubMed] [Google Scholar]
- McConkey D. J., Choi W., Marquis L., Martin F., Williams M. B., Shah J., et al. (2009). Role of Epithelial-To-Mesenchymal Transition (EMT) in Drug Sensitivity and Metastasis in Bladder Cancer. Cancer Metastasis Rev. 28 (3-4), 335–344. 10.1007/s10555-009-9194-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E., McCord R. A., Berber E., Kioi M., Padilla-Nash H., Damian M., et al. (2008). SIRT6 Is a Histone H3 Lysine 9 Deacetylase that Modulates Telomeric Chromatin. Nature 452 (7186), 492–496. 10.1038/nature06736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka K., Kawanami T., Shimokata H., Ohta S., Funakoshi A. (2005). Inactive Aldehyde Dehydrogenase-2 Increased the Risk of Pancreatic Cancer Among Smokers in a Japanese Male Population. Pancreas 30 (2), 95–98. 10.1097/01.mpa.0000147084.70125.41 [DOI] [PubMed] [Google Scholar]
- Mollo N., Cicatiello R., Aurilia M., Scognamiglio R., Genesio R., Charalambous M., et al. (2020). Targeting Mitochondrial Network Architecture in Down Syndrome and Aging. Int. J. Mol. Sci. 21 (9), 3134. 10.3390/ijms21093134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G., Samson M., Michaud S., M. Tanguay R. (2004). Overexpression of the Small Mitochondrial Hsp22 extendsDrosophilalife Span and Increases Resistance to Oxidative Stress. FASEB j. 18 (3), 598–599. 10.1096/fj.03-0860fje [DOI] [PubMed] [Google Scholar]
- Mukerjee N., Pietruszko R. (1992). Human Mitochondrial Aldehyde Dehydrogenase Substrate Specificity: Comparison of Esterase with Dehydrogenase Reaction. Arch. Biochem. Biophys. 299 (1), 23–29. 10.1016/0003-9861(92)90239-s [DOI] [PubMed] [Google Scholar]
- Muñoz-Najar U., Sedivy J. M. (2011). Epigenetic Control of Aging. Antioxid. Redox Signal. 14 (2), 241–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannelli G., Terzuoli E., Giorgio V., Donnini S., Lupetti P., Giachetti A., et al. (2018). ALDH2 Activity Reduces Mitochondrial Oxygen Reserve Capacity in Endothelial Cells and Induces Senescence Properties. Oxid Med. Cell Longev 2018, 9765027. 10.1155/2018/9765027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. P., Li J., Tewari A. K. (2014). Inflammation and Prostate Cancer: the Role of Interleukin 6 (IL-6). BJU Int. 113 (6), 986–992. 10.1111/bju.12452 [DOI] [PubMed] [Google Scholar]
- Niedernhofer L. J., Daniels J. S., Rouzer C. A., Greene R. E., Marnett L. J. (2003). Malondialdehyde, a Product of Lipid Peroxidation, Is Mutagenic in Human Cells. J. Biol. Chem. 278 (33), 31426–31433. 10.1074/jbc.m212549200 [DOI] [PubMed] [Google Scholar]
- Obiedat A., Seidel E., Mahameed M., Bernani O., Tsukerman P., Voutetakis K., et al. (2019). Transcription of the NKG2D Ligand MICA Is Suppressed by the IRE1/XBP1 Pathway of the Unfolded Protein Response through the Regulation of E2F1. FASEB j. 33 (3), 3481–3495. 10.1096/fj.201801350rr [DOI] [PubMed] [Google Scholar]
- Oh B., Figtree G., Costa D., Eade T., Hruby G., Lim S., et al. (2016). Oxidative Stress in Prostate Cancer Patients: A Systematic Review of Case Control Studies. Prostate Int. 4 (3), 71–87. 10.1016/j.prnil.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa I., Nishimaki K., Murakami Y., Suzuki Y., Ishikawa M., Ohta S. (2008). Age-dependent Neurodegeneration Accompanying Memory Loss in Transgenic Mice Defective in Mitochondrial Aldehyde Dehydrogenase 2 Activity. J. Neurosci. 28 (24), 6239–6249. 10.1523/jneurosci.4956-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. K., Quinn D. I. (2013). Differentiating mTOR Inhibitors in Renal Cell Carcinoma. Cancer Treat. Rev. 39 (7), 709–719. 10.1016/j.ctrv.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. W., Ji Y. I., Choi Y.-H., Kang M.-Y., Jung E., Cho S. Y., et al. (2009). Candidate Gene Polymorphisms for Diabetes Mellitus, Cardiovascular Disease and Cancer Are Associated with Longevity in Koreans. Exp. Mol. Med. 41 (11), 772–781. 10.3858/emm.2009.41.11.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr R. L., Mills J., Harbottle A., Creed J. M., Crewdson G., Reguly B., et al. (2013). Mitochondria, Prostate Cancer, and Biopsy Sampling Error. Discov. Med. 15 (83), 213–220. [PubMed] [Google Scholar]
- Peters I., Dubrowinskaja N., Hennenlotter J., Antonopoulos W. I., Von Klot C. A. J., Tezval H., et al. (2018). DNA Methylation of Neural EGFL like 1 (NELL1) Is Associated with Advanced Disease and the Metastatic State of Renal Cell Cancer Patients. Oncol. Rep. 40 (6), 3861–3868. 10.3892/or.2018.6732 [DOI] [PubMed] [Google Scholar]
- Pinaire J., Chou W.-Y., Morton M., You M., Zeng Y., Cho W. K., et al. (2003). Identification of a Retinoid Receptor Response Element in the Human Aldehyde Dehydrogenase-2 Promoter. Alcohol. Clin. Exp. Res. 27 (12), 1860–1866. 10.1097/01.alc.0000100941.86227.4f [DOI] [PubMed] [Google Scholar]
- Pinkerneil M., Hoffmann M. J., Schulz W. A., Niegisch G. (2017). HDACs and HDAC Inhibitors in Urothelial Carcinoma - Perspectives for an Antineoplastic Treatment. Curr. Med. Chem. 24 (37), 4151–4165. 10.2174/0929867324666170207142740 [DOI] [PubMed] [Google Scholar]
- Pinkerneil M., Hoffmann M. J., Deenen R., Köhrer K., Arent T., Schulz W. A., et al. (2016). Inhibition of Class I Histone Deacetylases 1 and 2 Promotes Urothelial Carcinoma Cell Death by Various Mechanisms. Mol. Cancer Ther. 15 (2), 299–312. 10.1158/1535-7163.mct-15-0618 [DOI] [PubMed] [Google Scholar]
- Pizzimenti S., Barrera G., Calzavara E., Mirandola L., Toaldo C., Dianzani M., et al. (2008). Down-regulation of Notch1 Expression Is Involved in HL-60 Cell Growth Inhibition Induced by 4-hydroxynonenal, a Product of Lipid Peroxidation. Mc 4 (6), 551–557. 10.2174/157340608786242098 [DOI] [PubMed] [Google Scholar]
- Pizzimenti S., Briatore F., Laurora S., Toaldo C., Maggio M., De Grandi M., et al. (2006). 4-Hydroxynonenal Inhibits Telomerase Activity and hTERT Expression in Human Leukemic Cell Lines. Free Radic. Biol. Med. 40 (9), 1578–1591. 10.1016/j.freeradbiomed.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Pizzimenti S., Toaldo C., Pettazzoni P., Dianzani M. U., Barrera G. (2010). The "Two-Faced" Effects of Reactive Oxygen Species and the Lipid Peroxidation Product 4-hydroxynonenal in the Hallmarks of Cancer. Cancers 2 (2), 338–363. 10.3390/cancers2020338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli G., Schaur R. J., Siems W. G., Leonarduzzi G. (2008). 4-hydroxynonenal: a Membrane Lipid Oxidation Product of Medicinal Interest. Med. Res. Rev. 28 (4), 569–631. 10.1002/med.20117 [DOI] [PubMed] [Google Scholar]
- Powers E. T., Morimoto R. I., Dillin A., Kelly J. W., Balch W. E. (2009). Biological and Chemical Approaches to Diseases of Proteostasis Deficiency. Annu. Rev. Biochem. 78, 959–991. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Raghunathan L., Hsu L. C., Klisak I., Sparkes R. S., Yoshida A., Mohandas T. (1988). Regional Localization of the Human Genes for Aldehyde Dehydrogenase-1 and Aldehyde Dehydrogenase-2. Genomics 2 (3), 267–269. 10.1016/0888-7543(88)90012-2 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S., Ku S., Ciamporcero E., Miles K. M., Attwood K., Chintala S., et al. (2016). HDAC 1 and 6 Modulate Cell Invasion and Migration in clear Cell Renal Cell Carcinoma. BMC Cancer 16, 617. 10.1186/s12885-016-2604-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappa F., Farina F., Zummo G., David S., Campanella C., Carini F., et al. (2012). HSP-molecular Chaperones in Cancer Biogenesis and Tumor Therapy: an Overview. Anticancer Res. 32 (12), 5139–5150. [PubMed] [Google Scholar]
- Reznik E., Miller M. L., Şenbabaoğlu Y., Riaz N., Sarungbam J., Tickoo S. K., et al. (2016). Mitochondrial DNA Copy Number Variation across Human Cancers. Elife 5, 769. 10.7554/eLife.10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi M., Barrera G., Aquino A., Spinsanti P., Pizzimenti S., Farace M. G., et al. (2000). 4-Hydroxynonenal-induced MEL Cell Differentiation Involves PKC Activity Translocation. Biochem. Biophysical Res. Commun. 272 (1), 75–80. 10.1006/bbrc.2000.2691 [DOI] [PubMed] [Google Scholar]
- Roumeguère T., Sfeir J., El Rassy E., Albisinni S., Van Antwerpen P., Boudjeltia K. Z., et al. (2017). Oxidative Stress and Prostatic Diseases. Mol. Clin. Oncol. 7 (5), 723–728. 10.3892/mco.2017.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. T., Hoogenraad N. J. (2007). Mitochondrial-nuclear Communications. Annu. Rev. Biochem. 76, 701–722. 10.1146/annurev.biochem.76.052305.091720 [DOI] [PubMed] [Google Scholar]
- Santarosa M., Favaro D., Quaia M., Galligioni E. (1997). Expression of Heat Shock Protein 72 in Renal Cell Carcinoma: Possible Role and Prognostic Implications in Cancer Patients. Eur. J. Cancer 33 (6), 873–877. 10.1016/s0959-8049(97)00002-6 [DOI] [PubMed] [Google Scholar]
- Sato A., Asano T., Ito K., Asano T. (2012). 17-Allylamino-17-demethoxygeldanamycin and Ritonavir Inhibit Renal Cancer Growth by Inhibiting the Expression of Heat Shock Factor-1. Int. J. Oncol. 41 (1), 46–52. 10.3892/ijo.2012.1419 [DOI] [PubMed] [Google Scholar]
- Segura-Aguilar J., Cortés-Vizcaino V., Llombart-Bosch A., Ernster L., Monsalve E., Romero F. J. (1990). The Levels of Quinone Reductases, Superoxide Dismutase and Glutathione-Related Enzymatic Activities in Diethylstilbestrol-Induced Carcinogenesis in the Kidney of Male Syrian golden Hamsters. Carcinogenesis 11 (10), 1727–1732. 10.1093/carcin/11.10.1727 [DOI] [PubMed] [Google Scholar]
- Seo W., Gao Y., He Y., Sun J., Xu H., Feng D., et al. (2019). ALDH2 Deficiency Promotes Alcohol-Associated Liver Cancer by Activating Oncogenic Pathways via Oxidized DNA-Enriched Extracellular Vesicles. J. Hepatol. 71 (5), 1000–1011. 10.1016/j.jhep.2019.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfera A., Bullock K., Price A., Inderias L., Osorio C. (2018). Ferrosenescence: The Iron Age of Neurodegeneration? Mech. Ageing Development 174, 63–75. 10.1016/j.mad.2017.11.012 [DOI] [PubMed] [Google Scholar]
- Sharma R., Brown D., Awasthi S., Yang Y., Sharma A., Patrick B., et al. (2004). Transfection with 4-Hydroxynonenal-Metabolizing Glutathione S-Transferase Isozymes Leads to Phenotypic Transformation and Immortalization of Adherent Cells. Eur. J. Biochem. 271 (9), 1690–1701. 10.1111/j.1432-1033.2004.04067.x [DOI] [PubMed] [Google Scholar]
- Shoeb M., Ansari N. H., Srivastava S. K., Ramana K. V. (2014). 4-Hydroxynonenal in the Pathogenesis and Progression of Human Diseases. Curr. Med. Chem. 21 (2), 230–237. 10.2174/09298673113209990181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorning B. Y., Dass M. S., Smalley M. J., Pearson H. B. (2020). The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 21 (12), 4507. 10.3390/ijms21124507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. (2021). Cancer Statistics, 2021. CA A. Cancer J. Clin. 71 (1), 7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- Singh K. K., Kulawiec M. (2009). Mitochondrial DNA Polymorphism and Risk of Cancer. Methods Mol. Biol. 471, 291–303. 10.1007/978-1-59745-416-2_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorokhod O. A., Caione L., Marrocco T., Migliardi G., Barrera V., Arese P., et al. (2010). Inhibition of Erythropoiesis in Malaria Anemia: Role of Hemozoin and Hemozoin-Generated 4-hydroxynonenal. Blood 116 (20), 4328–4337. 10.1182/blood-2010-03-272781 [DOI] [PubMed] [Google Scholar]
- Stagos D., Zhou H., Ross D., Vasiliou V. (2009). 4-HNE Inhibits Tube Formation and Up-Regulates Chondromodulin-I in Human Endothelial Cells. Biochem. Biophysical Res. Commun. 379 (3), 654–658. 10.1016/j.bbrc.2008.11.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Kryvenko O. N., Mitrache N., Do K. C., Jankowski M., Chitale D. A., et al. (2013). Methylation of the RARB Gene Increases Prostate Cancer Risk in Black Americans. J. Urol. 190 (1), 317–324. 10.1016/j.juro.2013.01.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Huang X., Xie Y., Zhou X., He X., Tang S., et al. (2019). Genome-wide Association and Gene-Environment Interaction Study Identifies Variants in ALDH2 Associated with Serum Ferritin in a Chinese Population. Gene 685, 196–201. 10.1016/j.gene.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Torti S. V., Torti F. M. (2013). Iron and Cancer: More Ore to Be Mined. Nat. Rev. Cancer 13 (5), 342–355. 10.1038/nrc3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A., Revandkar A., Di Mitri D., Guccini I., Proietti M., Sarti M., et al. (2014). Enhancing Chemotherapy Efficacy in Pten -Deficient Prostate Tumors by Activating the Senescence-Associated Antitumor Immunity. Cell Rep. 9 (1), 75–89. 10.1016/j.celrep.2014.08.044 [DOI] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., et al. (2004). Premature Ageing in Mice Expressing Defective Mitochondrial DNA Polymerase. Nature 429 (6990), 417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Valle A., Oliver J., Roca P. (2010). Role of Uncoupling Proteins in Cancer. Cancers 2 (2), 567–591. 10.3390/cancers2020567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven R. A. H., Santos D., Haigis M. C. (2017). Mitochondrial Sirtuins and Molecular Mechanisms of Aging. Trends Mol. Med. 23 (4), 320–331. 10.1016/j.molmed.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderVeen L. A., Hashim M. F., Shyr Y., Marnett L. J. (2003). Induction of Frameshift and Base Pair Substitution Mutations by the Major DNA Adduct of the Endogenous Carcinogen Malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 100 (24), 14247–14252. 10.1073/pnas.2332176100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Rey M., Woodhoo A., Martinez-Chantar M. L., Mato J. M., Lu S. C. (2013). Alcohol, DNA Methylation, and Cancer. Alcohol. Res. 35 (1), 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsyayan R., Chaudhary P., Sharma A., Sharma R., Rao Lelsani P. C., Awasthi S., et al. (2011). Role of 4-hydroxynonenal in Epidermal Growth Factor Receptor-Mediated Signaling in Retinal Pigment Epithelial Cells. Exp. Eye Res. 92 (2), 147–154. 10.1016/j.exer.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan V. S., Ryan M. J., Dhruv H. D., Gill S., Eichhoff O. M., Seashore-Ludlow B., et al. (2017). Dependency of a Therapy-Resistant State of Cancer Cells on a Lipid Peroxidase Pathway. Nature 547 (7664), 453–457. 10.1038/nature23007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajapeyee N., Serra R. W., Zhu X., Mahalingam M., Green M. R. (2008). Oncogenic BRAF Induces Senescence and Apoptosis through Pathways Mediated by the Secreted Protein IGFBP7. Cell 132 (3), 363–374. 10.1016/j.cell.2007.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. A., Lithgow G. J. (2003). Lifespan Extension in C. elegans by a Molecular Chaperone Dependent upon Insulin-like Signals. Aging Cell 2 (2), 131–139. 10.1046/j.1474-9728.2003.00045.x [DOI] [PubMed] [Google Scholar]
- Wang L.-S., Wu Z.-X. (2019). ALDH2 and Cancer Therapy. Adv. Exp. Med. Biol. 1193, 221–228. 10.1007/978-981-13-6260-6_13 [DOI] [PubMed] [Google Scholar]
- Wang Q., Williamson M., Bott S., Brookman-Amissah N., Freeman A., Nariculam J., et al. (2007). Hypomethylation of WNT5A, CRIP1 and S100P in Prostate Cancer. Oncogene 26 (45), 6560–6565. 10.1038/sj.onc.1210472 [DOI] [PubMed] [Google Scholar]
- Wang R.-S., Nakajima T., Kawamoto T., Honma T. (2002). Effects of Aldehyde Dehydrogenase-2 Genetic Polymorphisms on Metabolism of Structurally Different Aldehydes in Human Liver. Drug Metab. Dispos 30 (1), 69–73. 10.1124/dmd.30.1.69 [DOI] [PubMed] [Google Scholar]
- Wang S., Wang L., Qin X., Turdi S., Sun D., Culver B., et al. (2020). ALDH2 Contributes to Melatonin-Induced protection against APP/PS1 Mutation-Prompted Cardiac Anomalies through cGAS-STING-TBK1-Mediated Regulation of Mitophagy. Sig Transduct Target. Ther. 5 (1), 119. 10.1038/s41392-020-0171-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Qiu S., Wang H., Cui J., Tian X., Miao Y., et al. (2021). Transcriptional Repression of Ferritin Light Chain Increases Ferroptosis Sensitivity in Lung Adenocarcinoma. Front. Cell Dev. Biol. 9, 719187. 10.3389/fcell.2021.719187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H., Wei B., Zhou J. (2001). Subunit Communication in Tetrameric Class 2 Human Liver Aldehyde Dehydrogenase as the Basis for Half-Of-The-Site Reactivity and the Dominance of the oriental Subunit in a Heterotetramer. Chemico-Biological Interactions 130-132 (1-3), 47–56. 10.1016/s0009-2797(00)00221-0 [DOI] [PubMed] [Google Scholar]
- White E. (2015). The Role for Autophagy in Cancer. J. Clin. Invest. 125 (1), 42–46. 10.1172/jci73941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis M. S., Klassen L. W., Carlson D. L., Brouse C. F., Thiele G. M. (2004). Malondialdehyde-acetaldehyde Haptenated Protein Binds Macrophage Scavenger Receptor(s) and Induces Lysosomal Damage. Int. Immunopharmacology 4 (7), 885–899. 10.1016/j.intimp.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Wu J., Minikes A. M., Gao M., Bian H., Li Y., Stockwell B. R., et al. (2019). Intercellular Interaction Dictates Cancer Cell Ferroptosis via NF2-YAP Signalling. Nature 572 (7769), 402–406. 10.1038/s41586-019-1426-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Chen J., Dong P., Zhang S., He Y., Sun L., et al. (2014). Global Gene Expression Profiling Identifies ALDH2, CCNE1 and SMAD3 as Potential Prognostic Markers in Upper Tract Urothelial Carcinoma. BMC Cancer 14, 836. 10.1186/1471-2407-14-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Gu M., Zhou J., Da J., Wang Z. (2020). Interaction of YAP1 and mTOR Promotes Bladder Cancer Progression. Int. J. Oncol. 56 (1), 232–242. 10.3892/ijo.2019.4922 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xu T., Liu S., Ma T., Jia Z., Zhang Z., Wang A. (2017). Aldehyde Dehydrogenase 2 Protects against Oxidative Stress Associated with Pulmonary Arterial Hypertension. Redox Biol. 11, 286–296. 10.1016/j.redox.2016.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Yang F., Han Z., Cui S., Dai S., Xu F., et al. (2018). ALDH2 Mediates the Dose-Response protection of Chronic Ethanol against Endothelial Senescence through SIRT1/p53 Pathway. Biochem. Biophysical Res. Commun. 504 (4), 777–783. 10.1016/j.bbrc.2018.08.081 [DOI] [PubMed] [Google Scholar]
- Yan W., Long P., Wei D., Yan W., Zheng X., Chen G., et al. (2020). Protection of Retinal Function and Morphology in MNU-Induced Retinitis Pigmentosa Rats by ALDH2: an Iin-Vvivo Study. BMC Ophthalmol. 20 (1), 55. 10.1186/s12886-020-1330-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.-j., Hu R., Sun H., Chen B., Li X., Chen J.-B. (2019). 4-HNE Induces Proinflammatory Cytokines of Human Retinal Pigment Epithelial Cells by Promoting Extracellular Efflux of HSP70. Exp. Eye Res. 188, 107792. 10.1016/j.exer.2019.107792 [DOI] [PubMed] [Google Scholar]
- Yang S.-J., Wang H. Y., Li X. Q., Du H.-Z., Zheng C.-J., Chen H.-G., et al. (2007). Genetic Polymorphisms ofADH2andALDH2association with Esophageal Cancer Risk in Southwest China. Wjg 13 (43), 5760–5764. 10.3748/wjg.v13.i43.5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Cheng J.-Z., Singhal S. S., Saini M., Pandya U., Awasthi S., et al. (2001). Role of Glutathione S-Transferases in Protection against Lipid Peroxidation. J. Biol. Chem. 276 (22), 19220–19230. 10.1074/jbc.m100551200 [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Muramatsu T., Ohmori T., Higuchi S., Hayashida M., Ishii H. (1996). Esophageal Cancer and Aldehyde Dehydrogenase-2 Genotypes in Japanese Males. Cancer Epidemiol. Biomarkers Prev. 5 (2), 99–102. [PubMed] [Google Scholar]
- Yu B. P. (2005). Membrane Alteration as a Basis of Aging and the Protective Effects of Calorie Restriction. Mech. Ageing Development 126 (9), 1003–1010. 10.1016/j.mad.2005.03.020 [DOI] [PubMed] [Google Scholar]
- Yu H., Guo P., Xie X., Wang Y., Chen G. (2017). Ferroptosis, a New Form of Cell Death, and its Relationships with Tumourous Diseases. J. Cell. Mol. Med. 21 (4), 648–657. 10.1111/jcmm.13008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkovic N. (2003). 4-hydroxynonenal as a Bioactive Marker of Pathophysiological Processes. Mol. Aspects Med. 24 (4-5), 281–291. 10.1016/s0098-2997(03)00023-2 [DOI] [PubMed] [Google Scholar]
- Zaytseva Y. Y., Valentino J. D., Gulhati P., Mark Evers B. (2012). mTOR Inhibitors in Cancer Therapy. Cancer Lett. 319 (1), 1–7. 10.1016/j.canlet.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Zhang H., Fu L. (2021). The Role of ALDH2 in Tumorigenesis and Tumor Progression: Targeting ALDH2 as a Potential Cancer Treatment. Acta Pharmaceutica Sinica B 11 (6), 1400–1411. 10.1016/j.apsb.2021.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Miriyala S., Miao L., Mitov M., Schnell D., Dhar S. K., et al. (2014). Redox Proteomic Identification of HNE-Bound Mitochondrial Proteins in Cardiac Tissues Reveals a Systemic Effect on Energy Metabolism after Doxorubicin Treatment. Free Radic. Biol. Med. 72, 55–65. 10.1016/j.freeradbiomed.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Mei Y., Xiang P., Zhai G., Zhao N., Xu C., et al. (2018). DNA Methylation Affects Metastasis of Renal Cancer and Is Associated with TGF-β/RUNX3 Inhibition. Cancer Cell Int 18, 56. 10.1186/s12935-018-0554-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., Yin H. (2015). Role of Lipid Peroxidation Derived 4-hydroxynonenal (4-HNE) in Cancer: Focusing on Mitochondria. Redox Biol. 4, 193–199. 10.1016/j.redox.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]