Abstract
A multidrug-resistant plasmid encoding TEM-1, SHV-12, and a variant of IMP-2 metallo-β-lactamase, designated IMP-8, was identified from a clinical isolate of Klebsiella pneumoniae. There are four nucleotide differences between blaIMP-2 and blaIMP-8, resulting in two amino acid differences. blaIMP-8 was also found to be carried by an integron-borne gene cassette similar to the blaIMP-2 cassette.
The emergence of carbapenem-hydrolyzing metallo-β-lactamases in gram-negative bacteria has raised serious concern, since the enzymes usually possess a broad hydrolysis profile that includes carbapenems and extended-spectrum β-lactams (6, 8, 12, 13, 15). The genetic determinants of the metallo-β-lactamases are usually carried on mobile gene cassettes inserted in plasmid- or chromosome-borne integrons (1, 8, 13, 15). Since 1991, the IMP-1 metallo-β-lactamase has spread among isolates of various members of the family Enterobacteriaceae, Pseudomonas aeruginosa, and other nonfastidious, gram-negative nonfermenters in Japan (5, 6, 17, 18). Several novel metallo-β-lactamases have been described more recently. VIM-1 and VIM-2 were found to be produced by P. aeruginosa isolates from Italy (8) and France (13), respectively. IMP-2 was detected in an Italian isolate of Acinetobacter baumannii and was chromosomally encoded (15). IMP-3 was detected in a Shigella flexneri isolate in Japan and was plasmid mediated (7). Among these metallo-β-lactamases, only IMP-1 has been described in Klebsiella pneumoniae in Japan (5) and Singapore (T. H. Koh, G. S. Babini, N. Woodford, L.-H. Sng, L. M. C. Hall, and D. M. Livermore, Letter, Lancet 353: 2162, 1999). In this report we describe a plasmid encoding TEM-1, the SHV-12 extended-spectrum β-lactamase, and a variant of the IMP-2 enzyme (designated IMP-8) from a clinical isolate of K. pneumoniae.
Bacterial strains and vectors.
K. pneumoniae KPO787 was recovered from a central venous catheter tip that was removed from a patient with diabetes mellitus and acute pancreatitis at the intensive care unit of the National Cheng Kung University Medical Center in 1998. The isolate could be a colonizer, and the patient died of multiple organ system failure unrelated to the infection after 40 days of hospitalization. The recipient strain was Escherichia coli C600 (2), which is resistant to streptomycin. E. coli HB101 was the host for the cloning experiments (16). An E. coli C600 strain carrying a blaSHV-12-containing plasmid (pEKPB657) that was transferred from a clinical isolate of K. pneumoniae was used for comparison in susceptibility tests (20). The cloning vectors used were pUC19 and pHP13 (4, 21). pHP13 is a bifunctional multicopy vector with erythromycin and chloramphenicol-resistance markers (4).
Susceptibility tests.
MICs of antimicrobial agents were determined by the agar dilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (11). The antibiotics used in the study were obtained from the following sources: amoxicillin, SmithKline Beecham Pharmaceuticals, Surrey, United Kingdom; aztreonam, Bristol-Myers Squibb, New Brunswick, N.J.; cefotaxime and cefuroxime, Hoechst-Roussel Pharmaceuticals, Inc., Somerville, N.J.; ceftazidime, Glaxo Group Research, Ltd., Greenford, United Kingdom; ceftriaxone, Hoffmann-La Roche, Inc., Nutley, N.J.; cefoxitin and imipenem, Merck Sharp & Dohme, West Point, Pa.; meropenem, Sumitomo Pharmaceuticals, Ltd., Osaka, Japan; cephalothin and cefaclor, Eli Lilly & Co., Indianapolis, Ind.; piperacillin, Lederle Laboratories, Pearl River, N.Y.; and streptomycin and chloramphenicol, Sigma Chemical Co., St. Louis, Mo.
Transfer of resistance determinants.
Conjugation experiments were performed by broth mating as described previously (14, 20). Transconjugants were selected on tryptic soy agar plates supplemented with streptomycin (500 μg/ml) and ceftazidime (10 μg/ml).
Plasmid DNA preparation.
Plasmid preparation was performed by the rapid alkaline lysis method (19). Plasmid DNA obtained from the transconjugant was restricted with EcoRI or PstI (Roche Molecular Biochemicals, Mannheim, Germany), and its size was estimated by adding up the restriction fragment lengths.
Detection of the blaSHV and blaTEM genes.
PCR was used to amplify the entire sequences of the blaSHV and blaTEM genes in plasmid preparations as described previously (20). The amplicons were purified with PCR clean up kits (Roche Molecular Biochemicals) and were sequenced on an ABI PRISM 377 sequencer analyzer (Applied Biosystems, Foster City, Calif.).
Cloning and sequencing of the blaIMP-8 gene.
The resistance plasmid (pEKPO787) obtained from the transconjugant was digested with PstI. A 5.6-kb fragment was cloned into vector pUC19 (pEKPO787-1). The insert was further digested with BamHI and KpnI, and a 1.9-kb fragment was subcloned into vector pUC19 (pEKPO787D). Since there is no KpnI site on pHP13, a 1.9-kb BamHI-EcoRI fragment, which included the 1.9-kb BamHI-KpnI insert and the EcoRI site of pUC19, from pEKPO787D, was further ligated to pHP13, giving rise to pEKPO787D1. The insert was sequenced. Nucleotide and derived amino acid sequences were analyzed with the GCG program (Genetics Computer Group, Inc., Madison, Wis.). Related β-lactamases were identified by comparison with the GenBank and SWISS-PROT databases.
Isoelectric focusing of β-lactamases.
Crude homogenates of β-lactamases were prepared as described previously (3). Isoelectric focusing analysis was performed by the method of Matthew et al. (10) as described previously (20).
An approximately 150-kb plasmid was successfully transferred from K. pneumoniae KPO787 to E. coli C600. PCR and sequence analyses showed that both the clinical isolate and its transconjugant carried SHV-12 and TEM-1.
The nucleotide sequence of a 1.9-kb fragment cloned into vector pUC19 and pHP13 contained an open reading frame of 741 nucleotides which corresponded to a putative protein of 246 amino acids (Fig. 1). The sequence of the gene is identical to that of blaIMP-2, except for four nucleotide substitutions which resulted in two amino acid changes. Substitutions of a G for a C and a C for a G at nucleotide positions 61 and 62, respectively, and substitution of a G for a T at nucleotide position 617 resulted in the replacements of an Arg by an Ala and a Val by a Gly at amino acid positions 21 and 206, respectively, in the mature metallo-β-lactamase. The blaIMP-2-related gene, now designated blaIMP-8, was found to be flanked by nucleotide sequences identical to partial sequences of the intII and aac(6′)Ib genes (Fig. 1), indicating that blaIMP-8 is also carried on an integron-borne gene cassette.
FIG. 1.
Nucleotide sequence of the blaIMP-8 gene and flanking regions. The deduced amino acid sequence of blaIMP-8 is indicated in the single-letter code below the nucleotide sequence of blaIMP-8. The start codons of intII, blaIMP-8, and aac(6′)Ib are indicated by horizontal arrows, and the stop codon of blaIMP-8 is indicated by three asterisks. The nucleotides and amino acids that differ between blaIMP-8 and blaIMP-2 are marked by bold letters, and the nucleotides and amino acids for blaIMP-2 and IMP-2 that differ from the sequences for blaIMP-8 and IMP-8 are shown above and below the sequences. The blaIMP-8 cassette boundaries are indicated by vertical arrows. The conserved core and inverse core sites located at the blaIMP-8 cassette boundaries are boxed.
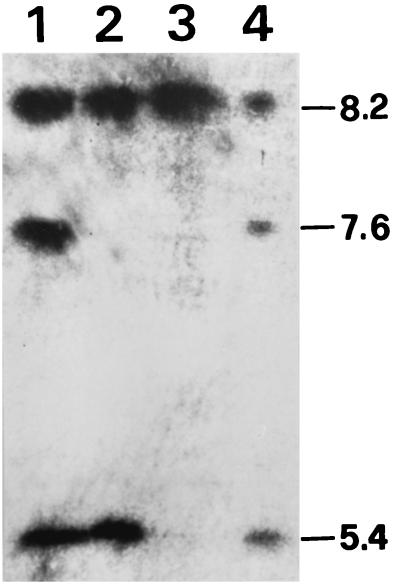
Isoelectric focusing revealed production of β-lactamases with pIs of 5.4, 7.6, and 8.2 by K. pneumoniae KPO787 (Fig. 2). The β-lactamases with pIs of 5.4 and 8.2 were transferred to E. coli C600. Thus, the pI 7.6 band probably represented the chromosomal SHV-1 β-lactamase of the K. pneumoniae strain (9). The E. coli HB101 strain transformed by pEKPO787D1 had a pI 8.2 band only, indicating that the pI of IMP-8 is 8.2, as is that of SHV-12.
FIG. 2.
Results of isoelectric focusing analysis. Lane 1, K. pneumoniae KPO787; lane 2, E. coli C600(pEKPO787); lane 3, E. coli HB101(pEKPO787D1); lane 4, K. pneumoniae KPT986, a clinical isolate known to produce TEM-1 (pI 5.4), SHV-1 (pI 7.6), and SHV-12 (pI 8.2) (18). The numbers on the right are pIs.
The susceptibilities of K. pneumoniae KPO787, its transconjugant, and the transformant E. coli HB101 to various β-lactams are shown in Table 1. Compared with the quality control strain E. coli ATCC 25922 (11), the K. pneumoniae strain showed increased resistance to imipenem (MIC, 0.25 versus 8 μg/ml) and meropenem (MIC, 0.06 versus 16 μg/ml) and high-level resistance to all the other β-lactam agents tested. The resistance phenotype of the transconjugant is quite similar to that of the clinical isolate, except that the MICs of carbapenems were relatively low for the transconjugant, as was expected from previous findings (6). Unlike the clinical strain and its transconjugant, the transformant showed susceptibility to aztreonam.
TABLE 1.
MICs of β-lactams for strains testeda
| Antibiotic(s)b | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| KPO787 | C600(pEKPO787) | HB101(pEKPO787D1) | C600(pEKPB657) | HB101(pUC19) | HB101 | |
| Amoxicillin | >256 | >256 | >256 | >256 | >256 | 4 |
| Amoxicillin + CLA | >256 | 64 | 64 | 8 | 8 | 2 |
| Piperacillin | >256 | >256 | 16 | 64 | 256 | 1 |
| Piperacillin + TZB | 128 | 64 | 8 | 4 | 32 | 1 |
| Cephalothin | >256 | >256 | >256 | >256 | 256 | 2 |
| Cefuroxime | >256 | >256 | >256 | >256 | 8 | 0.5 |
| Cefaclor | >256 | >256 | >256 | >256 | 4 | 0.5 |
| Cefotaxime | 64 | 32 | 32 | 8 | 0.13 | 0.06 |
| Cefotaxime + CLA | 32 | 32 | 32 | 0.03 | 0.03 | 0.03 |
| Ceftazidime | >256 | >256 | >256 | 128 | 0.25 | 0.13 |
| Ceftazidime + CLA | >256 | 256 | 256 | 0.13 | 0.03 | 0.03 |
| Ceftriaxone | 64 | 32 | 32 | 16 | 0.13 | 0.13 |
| Ceftriaxone + CLA | 32 | 32 | 16 | 0.03 | 0.03 | 0.03 |
| Cefoxitin | >256 | >256 | >256 | 4 | 4 | 2 |
| Aztreonam | 256 | 64 | 0.13 | 64 | 0.13 | 0.13 |
| Aztreonam + CLA | 0.13 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 |
| Imipenem | 8 | 2 | 2 | 0.13 | 0.13 | 0.13 |
| Imipenem + CLA | 2 | 2 | 2 | 0.03 | 0.03 | 0.03 |
| Meropenem | 16 | 2 | 1 | 0.13 | 0.13 | 0.06 |
| Meropenem + CLA | 2 | 1 | 0.5 | 0.03 | 0.03 | 0.03 |
K. pneumoniae KPO787 and E. coli C600(pEKPO787) expressed IMP-7, SHV-12, and TEM-1. E. coli HB101(pEKPO787D1), C600(pEKPB657), and HB101(pUC19) expressed IMP-7, SHV-12, and TEM-1, respectively.
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
IMP-2 was first reported on the chromosome of an A. baumannii strain in Italy (15). The blaIMP-2 gene shares an 88% nucleotide identity with blaIMP-1 and is carried by a gene cassette unrelated to the blaIMP-1 cassette. The blaIMP-8 gene found in this study is very closely related to blaIMP-2, with only four nucleotide differences from blaIMP-2. blaIMP-8 was also found to be carried by a gene cassette related to the blaIMP-2 cassette; however, in contrast to the blaIMP-2 cassette, the blaIMP-8 cassette was located on the plasmid, which would facilitate the spread of the resistance gene.
Production of IMP-8 in the E. coli transformant caused reduced susceptibilities to almost all β-lactams, including penicillins, cephalosporins, and carbapenems. Only aztreonam was unaffected, in agreement with the properties of other metallo-β-lactamases (6, 8, 12, 13, 15). Resistance to aztreonam in the K. pneumoniae isolate and its transconjugant should be due to the effects of SHV-12, the most prevalent type of extended-spectrum β-lactamase among clinical isolates of K. pneumoniae at the National Cheng Kung Medical Center (20). The blaIMP-8 containing integron was also found to carry an aminoglycoside resistance gene, aac(6′)Ib. Thus, antibiotics that can be used to treat infections with the microorganisms containing this multidrug-resistant plasmid are very limited. Since K. pneumoniae is a notorious host of resistance plasmids, the prevention of the spread of the multidrug-resistant plasmid and strain is critical and has become a big challenge to the hospital staff.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been submitted to GenBank and assigned accession no. AF322577.
Acknowledgments
This work was partially supported by grants NCKUH89-054 from National Cheng Kung University Hospital and NSC89-2320-B-006-149 from the National Science Council, Republic of China.
REFERENCES
- 1.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B J, Low K B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980;44:1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 4.Haima P, Bron S, Venema G. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol Gen Genet. 1987;209:335–342. doi: 10.1007/BF00329663. [DOI] [PubMed] [Google Scholar]
- 5.Hirakata Y, Izumikawa K, Yamaguchi T, Takemura H, Tanaka H, Yoshida R, Matsuda J, Nakano M, Tomono K, Maesaki S, Kaku M, Yamada Y, Kamihira S, Kohno S. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1998;42:2006–2011. doi: 10.1128/aac.42.8.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. 1995;39:824–829. doi: 10.1128/aac.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyobe S, Kusadokoro H, Ozaki J, Matsumura N, Minami S, Haruta S, Sawai T, O'Hara K. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob Agents Chemother. 2000;44:2023–2027. doi: 10.1128/aac.44.8.2023-2027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauretti L, Riccio M L, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini G M. Cloning and characterization of blaVIM, a new integron borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore D M. β-lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthew M, Harris M, Marshall M J, Rose G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; 9th informational supplement. M100–S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 12.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–897. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Provence D L, Curtiss R., III . Gene transfer in gram-negative bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 317–347. [Google Scholar]
- 15.Riccio M L, Franceschini N, Boschi L, Caravelli B, Cornaglia G, Fontana R, Amicosante G, Rossolini G M. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob Agents Chemother. 2000;44:1229–1235. doi: 10.1128/aac.44.5.1229-1235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Kato N, Ohta M. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob Agents Chemother. 1996;40:349–353. doi: 10.1128/aac.40.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol. 1996;34:2909–2913. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi S, Nagano Y. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J Clin Microbiol. 1984;20:608–613. doi: 10.1128/jcm.20.4.608-613.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J-J, Wu S-M, Tsai S-H, Wu J-J, Su I-J. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in southern Taiwan. Antimicrob Agents Chemother. 2000;44:1438–1442. doi: 10.1128/aac.44.6.1438-1442.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]