Abstract
Recent genome-wide association and transcriptome-wide association studies have identified an association between the PALMD locus, encoding palmdelphin, a protein involved in myoblast differentiation, and calcific aortic valve disease (CAVD). Nevertheless, the function and underlying mechanisms of PALMD in CAVD remain unclear. We herein investigated whether and how PALMD affects the pathogenesis of CAVD using clinical samples from CAVD patients and a human valve interstitial cell (hVIC) in vitro calcification model. We showed that PALMD was upregulated in calcified regions of human aortic valves and calcified hVICs. Furthermore, silencing of PALMD reduced hVIC in vitro calcification, osteogenic differentiation, and apoptosis, whereas overexpression of PALMD had the opposite effect. RNA-Seq of PALMD-depleted hVICs revealed that silencing of PALMD reduced glycolysis and nuclear factor-κB (NF-κB)–mediated inflammation in hVICs and attenuated tumor necrosis factor α–induced monocyte adhesion to hVICs. Having established the role of PALMD in hVIC glycolysis, we examined whether glycolysis itself could regulate hVIC osteogenic differentiation and inflammation. Intriguingly, the inhibition of PFKFB3-mediated glycolysis significantly attenuated osteogenic differentiation and inflammation of hVICs. However, silencing of PFKFB3 inhibited PALMD-induced hVIC inflammation, but not osteogenic differentiation. Finally, we showed that the overexpression of PALMD enhanced hVIC osteogenic differentiation and inflammation, as opposed to glycolysis, through the activation of NF-κB. The present study demonstrates that the genome-wide association– and transcriptome-wide association–identified CAVD risk gene PALMD may promote CAVD development through regulation of glycolysis and NF-κB–mediated inflammation. We propose that targeting PALMD-mediated glycolysis may represent a novel therapeutic strategy for treating CAVD.
Keywords: calcific aortic valve disease, human valve interstitial cells, PALMD, glycolysis, inflammation, calcification, NF-kappa B, cardiovascular disease
Abbreviations: 2-DG, 2-deoxyglucose; ALPL, alkaline phosphatase; BMP2, bone morphogenetic protein 2; CAVD, calcific aortic valve disease; DCA, dichloroacetate; GWAS, genome-wide association study; hVIC, human valve interstitial cell; IL6, interleukin-6; MOI, multiplicity of infection; MSX2, Msh homebox 2; NF-κB, nuclear factor-κB; RUNX2, runt-related transcription factor 2; TNFα, tumor necrosis factor-α; TWAS, transcriptome-wide association study; VECs, valve endothelial cells; VICs, valve interstitial cells
Calcific aortic valve disease (CAVD) is the most common valvular heart disease, which is characterized by progressive fibrocalcific valve thickening and ventricular function impairment, followed by aortic stenosis and subsequent heart failure (1). There are currently no effective medications to slow or stop the progression of CAVD. Surgical or transcatheter aortic valve replacement is the most common and widely used approach for CAVD. However, aortic valve replacement is risky for elderly adults, which may be associated with significant mortality and morbidity due to serious surgical complications including blood clots, infections, and heart attack (2). A comprehensive understanding of the pathogenic mechanisms underlying CAVD is crucial for developing novel therapeutic strategies for this disease.
The aortic valve is composed of three valve leaflets, which can fully open during systole and close during diastole to maintain unidirectional blood flow. In CAVD, the aortic valve cannot open to their full extent due to thickened and calcified valve leaflets. The valve leaflet consists of three distinct extracellular matrix layers including the fibrosa, spongiosa, and ventricularis (3). Valve interstitial cells (VICs) are distributed throughout all of these layers, while the surfaces of valve leaflets are lined by a monolayer of valve endothelial cells (VECs). The communication between VICs and VECs regulates the function of each cell type and is critical to maintain valve homeostasis (4).
CAVD has long been considered as a passive degenerative process associated with aging. However, accumulating evidence supports that it is an actively regulated process that shares many similarities with physiological bone mineralization (5). VICs are predominant cellular components in the aortic valve leaflet that have a crucial role in the regulation of healthy valve tissue homeostasis. Under physiological conditions, VICs exhibit a fibroblastic phenotype and can be transiently activated to myofibroblast phenotype to remodel the microenvironment, thereby maintaining the valve's structure (6). During CAVD progression, most VICs are persistently activated to myofibroblasts, leading to aortic valve fibrosis and valvular thickening (6). VICs can also differentiate into osteoblast-like cells, contributing to the mineralization process associated with CAVD (7). Osteoblastic differentiation of VICs mirrors physiological osteogenesis and is mainly orchestrated by runt-related transcription factor 2 (RUNX2) and bone morphogenetic protein 2 (BMP2) (7, 8). Many other factors have also been shown to promote osteogenic differentiation of VICs including Wnt signaling, msh homebox 2 (MSX2), and the proinflammatory cytokine tumor necrosis factor-α (TNFα) and interleukin-6 (IL6) (7, 9). VECs have also been reported to play an important role in CAVD. VEC-derived nitric oxide regulates Notch signaling in VICs to inhibit calcification (10). Further studies have demonstrated that nitric oxide activates Notch signaling in VICs through regulation of S-nitrosylation of ubiquitin specific peptidase 9, X-linked (11). Valvular osteoblast-like cells have been suggested as promising targets for the development of a novel pharmacological intervention for CAVD. Nevertheless, the molecular mechanisms and cues that drive osteoblastic differentiation of VICs in CAVD are not fully understood.
Familial aggregation and genetic epidemiology studies have demonstrated a strong genetic component being involved in CAVD (12, 13). Over the past years, genome-wide association (GWAS) studies have achieved significant progress in identifying common genetic variations that contribute to increased risk of CAVD. Many of these GWAS genetic variations involve genes with known or expected roles in the pathogenesis of CAVD, such as RUNX2 (14), lipoprotein(a) (15), and alkaline phosphatase (ALPL) (16). In addition, novel genetic variations near genes without a known functional role in CAVD have also been reported. For instance, recent GWAS and transcriptome-wide association (TWAS) studies have simultaneously reported the association of genetic variations at the PALMD locus with CAVD (17, 18), indicating a potential role of PALMD in CAVD.
PALMD is a predominant cytosolic isoform of the Paralemmin families, lipid-anchored proteins implicated in cell shape control and cell membrane dynamics (19). PALMD is ubiquitously expressed in several organs, including testis, heart, and skeletal muscle (20). Previous studies have reported that PALMD promotes myoblast differentiation and muscle regeneration (21) and regulates cell death in response to DNA damage (22). Although these studies increase our current knowledge of the molecular functions of PALMD, our understanding of how PALMD regulates the initiation and progression of CAVD remains largely unknown. Functional and mechanistic analysis of the role of PALMD in CAVD may shed new light on the molecular mechanisms underlying CAVD and provide a novel therapeutic target for pharmacological treatment of CAVD.
In this study, we performed detailed analyses of clinical samples from patients with CAVD, as well as hVIC calcification studies, to investigate the functional role of PALMD in CAVD and explore the underlying mechanisms through which PALMD may regulate CAVD.
Results
PALMD expression is upregulated in calcified hVICs and calcified regions of human aortic valves
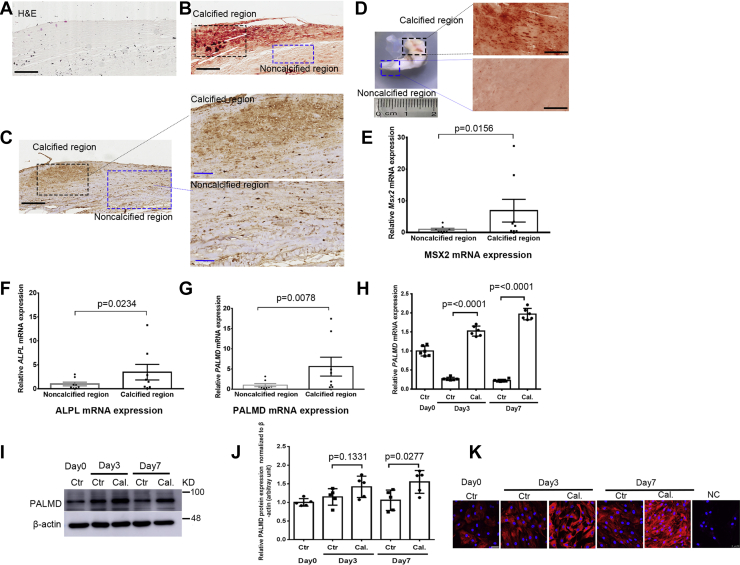
To characterize the role of PALMD in CAVD, we initially evaluated PALMD expression in calcified aortic valves from CAVD patients. Calcification of the aortic valve was confirmed by H&E staining and alizarin red staining (Fig. 1, A and B). Our immunohistochemistry results showed increased PALMD expression in calcified regions compared to adjacent noncalcified regions of the tissue (Fig. 1C). No positive staining was seen in control sections incubated with IgG (Fig. S1). A double immunofluorescence staining showed that PALMD was colocalized with the VIC maker Vimentin in the aortic valve tissue (Fig. S2). In addition, we compared PALMD expression in noncalcified and calcified tissue derived from the same aortic valve. The noncalcified and calcified tissue was confirmed by alizarin red staining (Fig. 1D). RT-qPCR results showed that mRNA expression of the osteogenic gene MSX2 and ALPL was significantly increased in calcified tissue (Fig. 1, E and F). Intriguingly, PALMD mRNA expression was significantly higher in calcified regions than noncalcified regions of the aortic valves (Fig. 1G).
Figure 1.
PALMD expression is upregulated in calcified regions of aortic valves from CAVD patients and calcified hVICs.A, H&E staining and B, alizarin red staining for calcified human aortic valves from CAVD patients. C, immunohistochemistry for PALMD expression in calcified human aortic valves from CAVD patients, and the scale bar (left panel) represents 200 μm; enlarged immunohistochemistry images of PALMD expression in calcified regions (right upper panel) and noncalcified regions (right lower panel), and the scale bar (right panel) represents 80 μm. D, aortic valves from CAVD patients were dissected into noncalcified and calcified tissue (left panel), and alizarin red staining for noncalcified and calcified regions (right panel). E–G, RT-qPCR showing MSX2, ALPL, and PALMD mRNA expression between noncalcified and calcified aortic valve tissue, n = 8. Wilcoxon matched-pairs signed rank test. H, RT-qPCR showing PALMD mRNA expression during hVIC in vitro calcification, n = 6. I, representative Western blotting images for PALMD and β-actin protein expression during hVIC in vitro calcification, n = 5. J, semi-quantification of PALMD expression using image J software, n = 5. K, PALMD immunofluorescence staining during hVIC in vitro calcification. IgG only served as negative controls (NC), and the scale bar represents 50 μm. Data are presented as mean ± SEM, and statistical significance was analyzed by a two-tailed unpaired Student’s t test. CAVD, calcific aortic valve disease; hVICs, human valve interstitial cells.
Isolated human valve interstitial cells (hVICs) were positively stained with SM22-α and Vimentin (Fig. S3A). hVICs cultured under calcifying conditions showed significantly increased calcium deposition, as revealed by alizarin red staining (Fig. S3B). This was accompanied by a significant increase in expression of the osteogenic gene MSX2, RUNX2, BMP2, and ALPL (Fig. S3, C–F), confirming previous reports from our laboratory and others (23, 24). Notably, a significant increase in PALMD mRNA and protein expression was observed during hVIC calcification (Fig. 1, H–J). Immunofluorescence staining also confirmed upregulated PALMD protein expression in calcified hVICs during the calcification process (Fig. 1K). These data demonstrate an increased expression of PALMD in calcified hVICs and calcified regions of human aortic valves, thus indicating a potential role of PALMD in CAVD.
Silencing of PALMD attenuates hVIC calcification and osteogenic differentiation
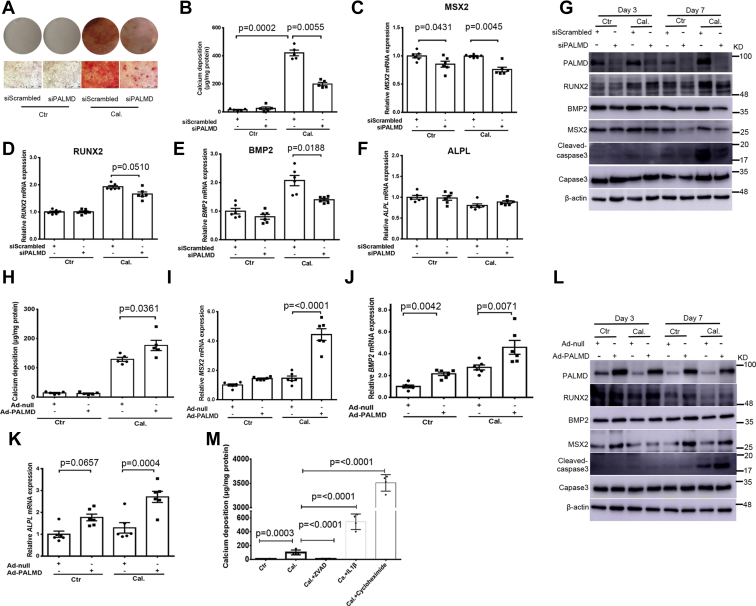
To determine the functional role of PALMD in CAVD, gain- and loss-of-function studies were undertaken using a hVIC calcification model. siRNA-mediated knockdown and adenovirus-mediated overexpression of PALMD were used. The knockdown and overexpression efficiency of PALMD in hVICs were confirmed by RT-qPCR and Western blotting (Fig. S4, A–D). Alizarin red staining and calcium quantitative assay showed that silencing of PALMD expression significantly reduced calcium deposition in hVICs cultured under calcifying conditions at day 7 (Fig. 2, A and B). In line with these results, knockdown of PALMD expression significantly attenuated mRNA expression of the osteogenic gene MSX2, RUNX2, and BMP2 in hVICs cultured with calcifying conditions at day 2 (Fig. 2, C–E), while no significant difference in ALPL mRNA expression was seen in hVICs with a depleted expression of PALMD (Fig. 2F). Western blotting results confirmed that silencing of PALMD in hVICs reduced MSX2 and BMP2 protein expression at day 3 and day 7 (Figs. 2G and S5, A–C). siRNA-mediated knockdown of PALMD expression in hVICs did not alter RUNX2 protein expression at day 3, but a significant reduction in RUNX2 protein expression was seen in hVICs with depleted PALMD expression at day 7 (Figs. 2G and S5D). Previous studies have reported that apoptosis is involved in the pathogenesis of CAVD (25). We evaluated the effects of PALMD on cleaved-caspase3 expression, a well-established indicator of apoptosis in hVICs. We observed that PALMD depletion decreased cleaved-caspase3 expression in hVICs at day 7 (Figs. 2G and S5E).
Figure 2.
PALMD accelerates hVIC in vitro calcification, osteogenic differentiation, and apoptosis. hVICs were transfected with 20 nM siScrambled or siPALMD and treated with control medium (Ctr) or calcifying medium (Cal.) for 48 h or for up to 7 days. A, alizarin red staining for calcium deposition in hVICs at day 7. Plate view (upper) and microscopic view (lower), and the scale bar represents 500 μm. B, quantitative calcium assay showing calcium deposition in hVICs at day 7, n = 5. C–F, RT-qPCR showing MSX2, RUNX2, BMP2, and ALPL mRNA expression in hVICs at 48 h, n = 6. G, representative Western blotting images for PALMD, RUNX2, BMP2, MSX2, Cleaved-caspase3, Caspase3, and β-actin protein expression in hVICs at day 3 and day 7, n = 3 or 5. H, hVICs were infected with Ad-null (MOI = 100) or Ad-PALMD (MOI = 100) and treated with control medium (Ctr) or calcifying medium (Cal.) for up to 7 days. Quantitative calcium assay showing calcium deposition in hVICs at day 3, n = 5. I–K, RT-qPCR showing MSX2, BMP2, and ALPL mRNA expression in hVICs for 48 h, n = 6. L, representative Western blotting images for PALMD, RUNX2, BMP2, MSX2, Cleaved-caspase3, Caspase3, and β-actin protein expression in hVICs at day 3 and day 7, n = 3, 4 or 5. M, hVICs were treated with control medium (Ctr) or calcifying medium (Cal.) in the presence or absence of 20 μM caspase inhibitor ZVAD, 25 ng/ml IL1β, or 10 μg/ml cycloheximide for up to 7 days. Quantitative calcium assay showing calcium deposition at day 7, n = 4. Data are presented as mean ± SEM, and statistical significance was analyzed by a two-tailed unpaired Student’s t test or one-way ANOVA followed by Tukey's multiple comparisons test. hVIC, human valve interstitial cell.
Conversely, adenovirus-mediated overexpression of PALMD induced a small but significant increase in calcium deposition in hVICs cultured with calcifying conditions at day 3 (Fig. 2H). However, the inductive effects of PALMD overexpression on calcium deposition in hVICs were not observed at day 7 (Fig. S6). These data suggest that PALMD may act at the early stage of CAVD. Overexpression of PALMD also significantly increased mRNA expression of the osteogenic gene MSX2, BMP2, and ALPL in hVICs cultured under control and calcifying conditions at day 2 (Fig. 2, I–K). Western blotting results showed that overexpression of PALMD in hVICs enhanced MSX2, RUNX2, and BMP2 protein expression at day 7 (Figs. 2L and S7, A–D). PALMD overexpression also increased cleaved-caspase3 expression in hVICs at day 7 (Figs. 2L and S7E). Taken together, these data support that PALMD is a novel regulator of hVIC calcification, osteogenic differentiation, and apoptosis. In addition, we found that inhibition of apoptosis by the caspase inhibitor ZVAD attenuated calcium deposition in hVICs at day 7, whereas induction of apoptosis by IL1β or cycloheximide promoted hVIC in vitro calcification (Fig. 2M).
Silencing of PALMD modulates the glycolytic and inflammatory gene expression profile in hVICs
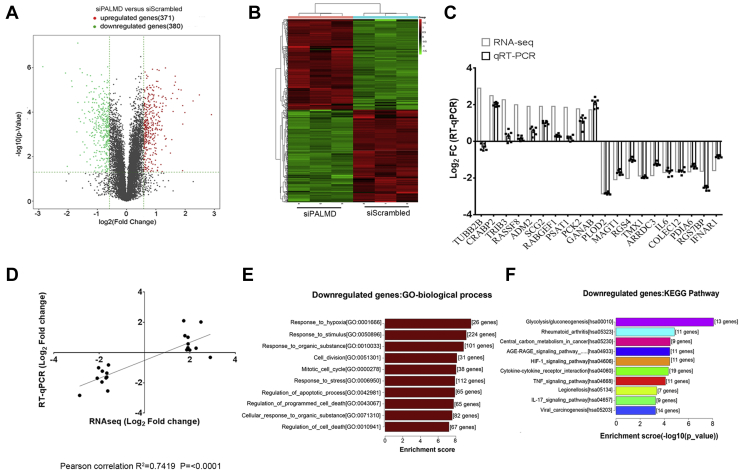
To gain mechanistic insights into the effects of PALMD on hVIC calcification, we performed RNA-Seq–based transcriptomic profiling in hVICs with depleted PALMD expression by siRNA. Our RNA-Seq analysis revealed 751 altered genes (1.5 fold change, p < 0.05), of which 380 genes were significantly downregulated, and 371 genes were significantly upregulated in hVICs with depleted PALMD (Fig. 3, A and B). A complete gene list is provided in Table S1. The RNA-Seq results were validated by examining the expression of the top ten upregulated genes including TUBB2B, CRABP2, and TRIB3 and the top ten downregulated genes including PLOD2, MAGT1, and RGS4 using RT-qPCR in hVICs with depleted PALMD. The expression level changes of all these genes except TUBB2B were consistent with the RNA-Seq results (Fig. 3C). A strong correlation between the log2 fold change of the RNA-Seq results and RT-qPCR was observed (Fig. 3D). Gene ontology and pathway analysis of the downregulated genes indicated that PALMD regulates multiple biological processes such as responses to hypoxia and stress, cell apoptosis, and several important pathways including glycolysis and TNFα signaling pathway (Fig. 3, E and F). These data are consistent with our previous results showing that silencing of PALMD reduces cleaved-caspase3, a well-known indicator of apoptosis. In addition, gene ontology and pathway analysis showed that the upregulated genes are associated with the biological processes such as extracellular matrix and structure organization and multiple pathways, including biosynthesis of amino acids and carbon metabolism (Fig. S8). These data suggest that PALMD may be a novel regulator of glycolysis and TNFα signaling in hVICs.
Figure 3.
PALMD regulates the glycolytic and inflammatory gene expression profile in hVICs. hVICs were transfected with 20 nM siScrambled or siPALMD for 48 h. RNA-Seq–based transcriptomic profiling was performed. The RNA-seq data is deposited in NCBI Gene Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number of GEO: GSE165524. A, volcano plot with green dots representing significantly downregulated protein-coding genes (n = 380) and red dots representing significantly upregulated protein-coding genes (n = 371) in PALMD-silencing and control hVICs. B, hierarchical clustering heat map of 751 protein-coding genes differentially expressed between PALMD-silencing and control hVICs (1.5 fold change, p < 0.05), n = 3. C, top ten upregulated genes and downregulated gene identified by RNA-Seq was validated using RT-qPCR, n = 6. D, correlation between RNA-Seq and RT-qPCR. Comparison of log2 fold change of 20 genes obtained by RNA-Seq and RT-qPCR. E, gene ontology and F, KEGG pathway analysis of the downregulated genes in PALMD-depleted hVICs identified by RNA-Seq. hVIC, human valve interstitial cell.
PALMD regulates glycolysis in hVICs
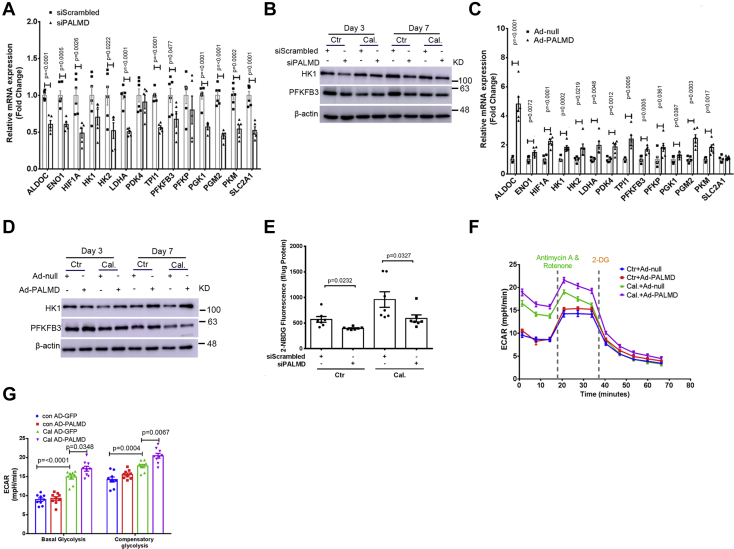
Based on our RNA-Seq data, we next performed a series of experiments to address the functional role of PALMD in the regulation of hVIC glycolysis. Notably, siRNA-mediated PALMD depletion in hVICs significantly reduced mRNA expression of several key glycolytic genes, including ALDOC, ENO1, HIF1A, HK2, LDHA, TPI1, PFKFB3, PGK1, PGM2, PKM, and SLC2A1 at day 2 (Fig. 4A). A comparable decrease in HK1 and PFKFB3 protein expression was observed in hVICs with PALMD depletion at day 7 (Figs. 4B and S9). Conversely, adenovirus-mediated overexpression of PALMD in hVICs significantly increased mRNA expression of key regulators of glycolysis including ALDOC, ENO1, HIF1A, HK1, HK2, LDHA, PDK4, TPI1, PFKFB3, PFKP, PGK1, PGM2, and PKM at day 2 (Fig. 4C), while SLC2A1 mRNA expression remained unchanged (Fig. 4C). Additionally, overexpression of PALMD in hVICs increased HK1 and PFKFB3 protein expression at day 7 (Figs. 4D and S10). In agreement with these results, silencing of PALMD expression in hVICs significantly reduced the uptake of the fluorescent glucose analog 2-NBDG (Fig. 4E). Seahorse flux analysis corroborated these findings, revealing that overexpression of PALMD in hVICs significantly increased both basal and compensatory glycolysis under calcifying conditions (Fig. 4, F and G). However, PALMD overexpression had no significant effect on basal respiration, ATP production-linked respiration, or maximum respiration in hVICs (Fig. S11). Taken together, these data demonstrate that PALMD regulates glycolysis in hVICs and indicates a potential novel role of glycolysis in the regulation of hVIC calcification.
Figure 4.
PALMD enhances glycolysis in hVICs. hVICs were transfected with 20 nM siScrambled or siPALMD and treated with control medium (Ctr) or calcifying medium (Cal.) for 48 h. A, RT-qPCR showing expression of the key glycolytic genes in hVICs with PALMD depletion after 48 h, n = 5. B, representative Western blotting images for HK1, PFKFB3, and β-actin protein expression in hVICs with PALMD depletion at day 3 and day 7, n = 4 or 5. C, RT-qPCR showing expression of the key glycolytic genes in hVICs infected with Ad-null (MOI = 100) or Ad-PALMD (MOI = 100) for 48 h, n = 6. D, representative Western blotting images for HK1, PFKFB3, and β-actin protein expression in hVICs with PALMD overexpression at day 3 and day 7, n = 3. E, normalized fluorescence of the glucose analog 2-NBDG (fl/μg protein) determined after 48 h in hVICs with silencing of PALMD, n = 7. F and G, the glycolysis rate assay using the Seahorse in hVICs with PALMD overexpression after 48 h, n = 9. Data are presented as mean ± SEM, and statistical significance was analyzed by a two-tailed unpaired Student’s t test or one-way ANOVA followed by Tukey's multiple comparisons test. hVIC, human valve interstitial cell.
Inhibition of PFKFB3-mediated glycolysis attenuates osteogenic differentiation and inflammation in hVICs
Having established the key role of PALMD in the regulation of hVIC glycolysis, we examined whether glycolysis itself could regulate hVIC osteogenic differentiation and inflammation, which are key regulators of CAVD. We observed that calcified hVICs showed increased basal and compensatory glycolysis (Fig. 4, F and G). This observation is consistent with previous studies reporting increased aerobic glycolysis during osteogenic differentiation and calcification in vascular smooth muscle cells (26). In line with previous reports (26), calcifying conditions also significantly increased basal respiration, ATP production-linked respiration, and maximum respiration in hVICs cultured in vitro (Fig. S11). These data suggest that hVICs acquire increased energy demand during the calcification process. Further studies were performed to examine the role of glycolysis in the regulation of hVIC calcification in vitro using the glycolysis inhibitor 2-deoxyglucose (2-DG) and dichloroacetate (DCA). We showed that 2-DG increased calcium deposition in hVICs under calcifying conditions at day 7, whereas DCA inhibited hVIC calcification in vitro (Fig. S12).
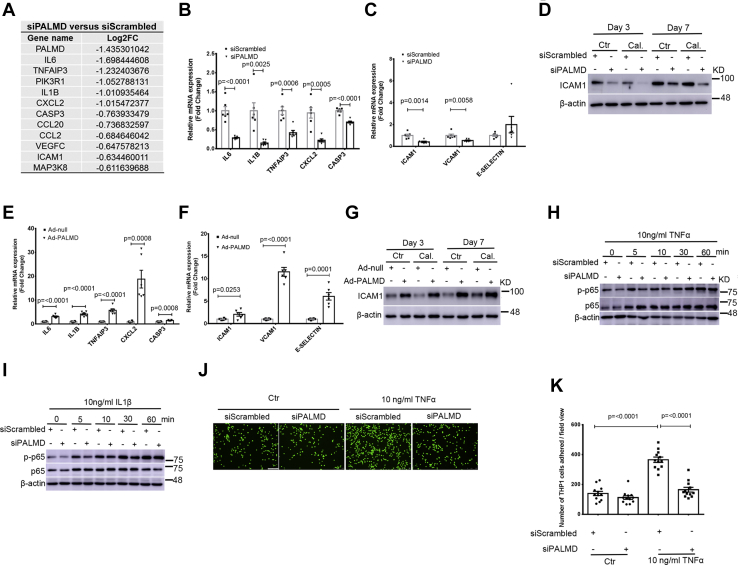
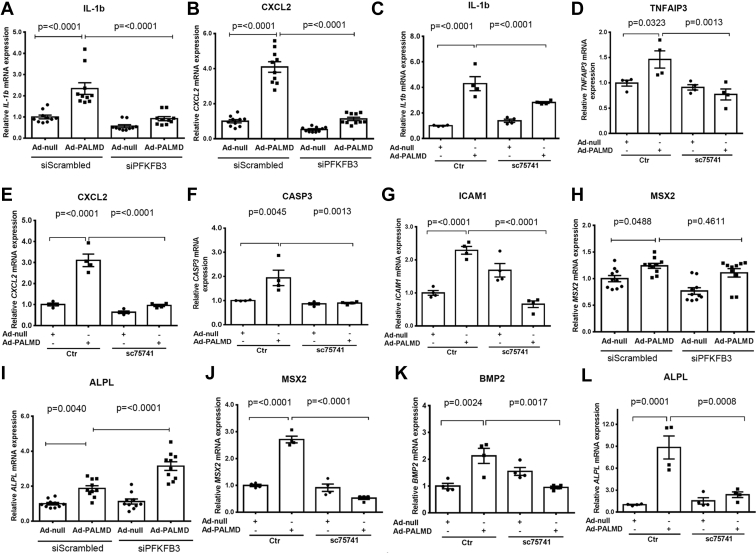
The glycolysis inhibitor 2-DG and DCA produced opposing effects on hVIC calcification, therefore, we used a more specific approach to inhibit glycolysis. Since PFKFB3 acts as a key glycolytic regulator, we depleted PFKFB3 expression in hVICs using siRNA. The siRNA-mediated knockdown efficiency of PFKFB3 was validated by RT-qPCR and Western blotting (Fig. S13). Consistent with these results, knockdown of PFKFB3 in hVICs significantly reduced both basal and compensatory glycolysis under control and calcifying conditions at day 2 (Fig. S14, A and B). Silencing of PFKFB3 expression in hVICs significantly reduced mRNA expression of the osteogenic gene MSX2, BMP2, and ALPL at day 2 (Fig. S14, C, E and F), but not the expression of RUNX2 mRNA (Fig. S14D). Decreased MSX2 and BMP2 protein expression were also observed in hVICs with silencing of PFKFB3 in the presence of control and calcifying conditions at day 3 and day 7 (Fig. S14G). Silencing of PFKFB3 did not alter RUNX2 protein expression at day 3 but decreased RUNX2 protein expression at day 7 (Fig. S14G). We showed that PFKFB3 depletion in hVICs significantly reduced mRNA expression of inflammation-associated genes such as IL6, IL1B, TNFAIP3, CXCL2, and CASP3 (Fig. S14H). The hallmark of an early stage of CAVD is valvular inflammation, which enhances the expression of cell adhesion molecules such as ICAM1, VCAM1, and E-SELECTIN to recruit immune cells including monocytes (27). As expected, we showed that depletion of PFKFB3 in hVICs significantly reduced mRNA expression of ICAM1, VCAM1, and E-SELECTIN at day 2 (Fig. S14I). Reduced ICAM1 protein expression was also seen in hVICs with the silencing of PFKFB3 cultured with control and calcifying conditions at day 3 and day 7 (Fig. S14J). These data demonstrate a crucial role of PFKFB3-mediated glycolysis in the regulation of hVIC osteogenic differentiation and inflammation.
PALMD regulates inflammation in hVICs through PFKFB3 and NF-κB signaling
Our RNA-Seq–based transcriptomic profiling and subsequent pathway analysis of hVICs with PALMD depletion revealed that PALMD regulates a cluster of genes involved in the TNFα signaling pathway (Figs. 3F and 5A). Our RT-qPCR results confirmed that PALMD depletion in hVICs significantly decreased mRNA expression of IL6, IL1B, TNFAIP3, CXCL2, and CASP3 at day 2 (Fig. 5B). A significant decrease in mRNA expression of the adhesion molecules, including ICAM1 and VCAM1, was also observed in hVICs with PALMD depletion at day 2 (Fig. 5C). The adhesion molecule E-SELECTIN remained unchanged (Fig. 5C). A comparable decrease in ICAM1 protein expression was also seen in hVICs with PALMD depletion at day 3 and day 7 (Figs. 5D and S15A). Conversely, overexpression of PALMD in hVICs dramatically increased mRNA expression of IL6, IL1B, TNFAIP3, CXCL2, and CASP3 at day 2 (Fig. 5E). Overexpression of PALMD in hVICs also significantly increased the expression of adhesion molecules, including ICAM1, VCAM1, and E-SELECTIN at day 2 (Fig. 5F). Moreover, a comparable increase in ICAM1 protein expression was seen in hVICs with PALMD overexpression at day 3 and day 7 (Figs. 5G and S15B). As nuclear factor-κB (NF-κB) is the major downstream effector of the TNFα signaling pathway, we next investigated whether silencing of PALMD could alter the activation of NF-κB. We observed that PALMD depletion attenuated the phosphorylation of p65 in hVICs treated with 10 ng/ml TNFα and IL1β (Figs. 5, H and I and S15, C and D). Having demonstrated that PALMD regulates ICAM1 protein expression in hVICs, we next examined whether knockdown of PALMD could reduce monocyte adhesion to hVICs. As expected, we observed that siRNA-mediated knockdown of PALMD significantly reduced TNFα-induced monocyte adhesion to hVICs (Fig. 5, J and K). Taken together, these data suggest that PALMD accelerates NF-κB signaling–mediated inflammation within hVICs.
Figure 5.
PALMD regulates PFKFB3 and NF-κB–mediated inflammation in hVICs. hVICs were transfected with 20 nM siScrambled or siPALMD and treated with control medium (Ctr) or calcifying medium (Cal.) for 48 h. A, selected NF-κB target genes regulated by PALMD in hVICs revealed by RNA-Seq. B, validation of NF-κB target genes revealed by RNA-Seq in hVICs with PALMD depletion at 48 h using RT-qPCR including IL6, IL1B, TNFAIP3, CXCL2, and CASP3, n = 6. C, RT-qPCR showing mRNA expression of the adhesion molecule ICAM1, VCAM1, and E-SELECTIN in hVICs with depleted PALMD at 48 h, n = 6. D, representative Western blotting images for ICAM1 and β-actin protein expression in hVICs with depleted PALMD at day3 and day 7, n = 5. E, RT-qPCR showing the TNFα–NF-κB signaling-relevant genes in hVICs with PALMD overexpression at 48 h, n = 6. F, RT-qPCR showing mRNA expression of ICAM1, VCAM1, and E-SELECTIN in hVICs with PALMD overexpression at 48 h, n = 6. G, representative Western blotting images for ICAM1 and β-actin protein expression in hVICs with PALMD overexpression at day3 and day 7, n = 3. H and I, hVICs with depleted PALMD expression were treated with 10 ng/ml TNFα or IL1β at the indicated time. Representative Western blotting images for p-p65, p65, and β-actin protein expression, n = 4. J, representative fluorescence microscopic images of the calcein-AM labeled THP1 cells that adhered to hVICs with depleted PALMD expression. The scale bar represents 100 μm. K, quantitative assay of the THP1 cells that adhered to hVICs with depleted PALMD expression, n = 12. Data are presented as mean ± SEM, and statistical significance was analyzed by a two-tailed unpaired Student’s t test. hVIC, human valve interstitial cell; NF-κB, nuclear factor-κB.
We next evaluated whether PALMD promotes hVIC inflammation through regulation of PFKFB3 and NF-κB. Our RT-qPCR results revealed that silencing of PFKFB3 expression in hVICs significantly decreased PALMD-induced mRNA expression of the inflammatory gene IL1B and CXCL2 at day 2 (Fig. 6, A and B). In addition, PALMD-induced mRNA expression of the inflammation-associated genes IL1B, TNAIP3, CXCL2, and CASP3 was significantly abolished in hVICs treated with the NF-κB inhibitor SC75741 at day 2 (Fig. 6, C–F). Consistent with these results, the NF-κB inhibitor SC75741 significantly attenuated PALMD-induced mRNA expression of the adhesion molecule ICAM1 in hVICs at day 2 (Fig. 6G). These data suggest that PFKFB3 and NF-κB play important roles in PALMD-induced hVIC inflammation. Interestingly, PALMD-induced mRNA expression of the glycolytic gene HK1, PFKFB3, and PGM2 was not significantly altered in hVICs treated with the NF-κB inhibitor SC75741 at day 2 (Fig. S16).
Figure 6.
Role of PFKFB3 and NF-κB in PALMD-induced hVIC inflammation and osteogenic differentiation. hVICs were infected with Ad-null (MOI = 100) or Ad-PALMD (MOI = 100) and transfected with 20 nM siScrambled or siPFKFB3 or treated with 1 μM of the NF-κB inhibitor SC75741 for 48 h. A and B, RT-qPCR showing mRNA expression of IL1B and CXCL2, n = 10. C–F, RT-qPCR showing mRNA expression of IL1B, TNFAIP3, CXCL2, and CASP3, n = 4. G, RT-qPCR showing mRNA expression of ICAM1, n = 4. H and I, RT-qPCR showing mRNA expression of MSX2 and ALPL, n = 10. J–L, RT-qPCR showing mRNA expression of MSX2, BMP2, and ALPL, n = 4. Data are presented as mean ± SEM, and statistical significance was analyzed by one-way ANOVA followed by Tukey's multiple comparisons test. hVIC, human valve interstitial cell; NF-κB, nuclear factor-κB.
PALMD promotes osteogenic differentiation of hVICs through the activation of NF-κB rather than PFKFB3
We showed that both TNFα and IL1β treatment significantly increased mRNA expression of the osteogenic gene MSX2 and BMP2 expression in hVICs at day 2 (Fig. S17). These data suggest an important role of inflammation in the regulation of hVIC osteogenic differentiation. Having established that PALMD promotes hVIC inflammation through PFKFB3 and NF-κB, we next examined whether inhibition of PFKFB3 or NF-κB could attenuate PALMD-induced hVIC osteogenic differentiation. Our RT-qPCR results revealed that siRNA-mediated knockdown of PFKFB3 did not alter the PALMD-induced osteogenic gene MSX2 or ALPL mRNA expression in hVICs at day 2 (Fig. 6, H and I). Interestingly, the NF-κB inhibitor SC75741 significantly attenuated PALMD-induced osteogenic gene MSX2, BMP2, and ALPL mRNA expression in hVICs at day 2 (Fig. 6, J–L). Taken together, our data demonstrate that PALMD regulates osteogenic differentiation at least in part through the activation of NF-κB in hVICs.
Discussion
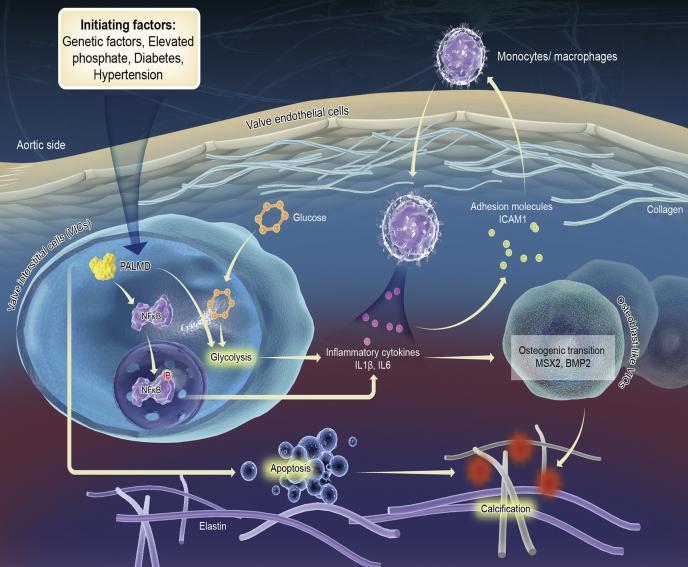
Understanding the function and mechanisms of GWAS-identified CAVD associated genes advances our knowledge of the pathogenesis of CAVD and may yield novel therapeutic strategies. Previous GWAS and TWAS studies have identified PALMD as a novel CAVD risk gene (17, 18). Herein, by using hVIC in vitro calcification model and by analyzing clinical samples from CAVD patients, we provided direct evidence that PALMD regulates hVIC calcification and CAVD. Our genome-wide RNA-Seq profiling data and subsequent experiments showed that PALMD regulates hVIC calcification and osteogenic differentiation by altered glycolysis and NF-κB–mediated inflammation (Fig. 7). This study explained the functional role and the underlying mechanisms of PALMD in the regulation of hVIC calcification and CAVD.
Figure 7.
Scheme illustrating the role of PALMD in CAVD. The working models of the potential mechanisms through which PALMD regulates CAVD are shown. PALMD enhances glycolysis and NF-κB–mediated inflammation in hVICs, thereby leading to increased osteogenic differentiation and calcification of hVICs. On the other hand, PALMD may promote hVIC calcification at least in part through regulation of apoptosis. CAVD, calcific aortic valve disease; hVIC, human valve interstitial cell; NF-κB, nuclear factor-κB.
Previous GWAS and TWAS studies have reported that the lead CAVD risk variant rs6702619 at the PALMD locus is strongly associated with decreased mRNA expression levels of PALMD in human aortic valve tissues (17, 18). Intriguingly, our immunohistochemistry and RT-qPCR results showed that PALMD expression was significantly higher in calcified regions than adjacent noncalcified regions of the aortic valve tissues. Using a well-established hVIC in vitro calcification model of CAVD, we confirmed that PALMD mRNA and protein expression were significantly upregulated during the calcification process. These contrasting findings may be due to ethnic differences. In fact, the minor allele frequency of rs6702619 at the PALMD locus varies significantly between different ethnic groups. In the 1000 Genomes Project, the risk allele “G” of rs6702619 has a frequency of 48% in Europeans, 8% in Africans, 7% in East Asians, and 25% in South Asians, thus suggesting that the genetic architecture at this locus is notably different between Europeans (where the association has been discovered) and East Asians. Our data may also explain some of the discrepancies in the risk of CAVD among ethnic groups (28). Future GWAS studies in East Asians are required to confirm whether rs6702619 is associated with CAVD and may identify novel DNA variants at the PALMD locus associated with CAVD. However, it should be noted that noncalcified and calcified tissues used in this study were isolated from diseased aortic valves with established calcification, we therefore cannot exclude the possibility of some nonspecific effects exist due to the secondary pathologic changes. Moreover, noncalcified and calcified tissues used in this study may contain different leaflet layers including fibrosa, spongiosa, and ventricularis. It has been reported that distinct aortic valve microlayers exhibit unique proteome profiles in CAVD (29). Further studies would be interesting to establish expression profile of PALMD in each of the three leaflet layers during CAVD. This study also included bicuspid aortic valves, which are subjected to higher mechanical stress and may develop calcification at an earlier age (30). This could be the possible reason that the mean age of CAVD patients in this study is lower than a European population with typical calcific valve disease without bicuspid valve.
Our gain- and loss-of-function studies clearly showed that PALMD regulates hVIC calcification and osteogenic differentiation. Previous studies have reported that osteogenic differentiation of hVICs plays a critical role in the pathogenesis of CAVD (7). We observed that knockdown or overexpression of PALMD significantly decreased or increased the osteogenic gene MSX2, RUNX2, and BMP2 expression in hVICs, respectively. To the best of our knowledge, this is the first report showing that PALMD directly regulates the osteogenic differentiation of hVICs. In this study, we showed that adenovirus-mediated overexpression of PALMD induced a small significant increase in calcium deposition in hVICs at day 3, and this inductive effect was not seen at day 7. A previous TWAS study has reported that the CAVD risk alleles at the PALMD locus conferred susceptibility by lowering the mRNA expression levels of PALMD in valve tissues (17). However, a large GWAS study including 6942 individuals of European ancestry did not observe the association between the variant rs6702619 at the PALMD locus and aortic valve calcification, which appear only in later stages of CAVD (15). Moreover, a recent study has reported that PALMD regulates the fibrogenic activity of VICs, an early step in the development of CAVD (31). These studies together with our results support that PALMD may regulate CAVD at early stages. However, it should be noted that the hVICs used in this study were isolated from calcified human aortic valves, which may be more sensitive to calcification and osteogenic differentiation in vitro. This may attenuate the inductive effects of adenovirus-mediated overexpression of PALMD on calcium deposition and osteogenic gene expression in these cells. Apoptosis of hVICs also contributes to the initiation and progression of CAVD. This is consistent with our findings showing that inhibition of apoptosis by ZVAD attenuated hVIC calcification, whereas induction of apoptosis by IL1β or Cycloheximide promoted hVIC calcification. Apoptotic bodies expose phosphatidylserine on the outer membranes and generate a potential calcium-binding site suitable for hydroxyapatite deposition (32, 33). We found that knockdown of PALMD reduced the expression of cleaved-caspase3 in hVICs, a well-known indicator of apoptosis, whereas overexpression of PALMD promoted the expression of cleaved-caspase3. These data confirm and extend previous studies showing that PALMD is a proapoptotic gene induced by p53 in response to DNA damage in osteosarcoma cell lines (22). Taken together, our data suggest that PALMD may promote hVIC calcification through the direct regulation of osteogenic differentiation and apoptosis.
In the present study, we observed that glycolysis is increased during the hVIC calcification process. Osteocalcin, an important energy metabolism-regulating hormone, has been reported to be upregulated in calcified human aortic valves (34). Moreover, previous studies have shown that osteocalcin locally shifts vascular smooth muscle cells and chondrocytes toward the glycolytic breakdown of glucose and promotes calcification (35). It is therefore plausible that the metabolic shift toward glycolytic breakdown of glucose during hVIC in vitro calcification may be regulated by osteocalcin. Intriguingly, we found that the glycolysis inhibitor 2-DG and DCA generated contrasting effects on hVIC in vitro calcification. 2-DG is a glucose analog that competitively inhibits hexokinase, the critical enzyme for the first step of glucose metabolism. Although it is commonly used as a glycolysis inhibitor, 2-DG actually disrupts both glycolysis and mitochondrial oxidative phosphorylation (36). In addition, 2-DG is well characterized as an inducer of endoplasmic reticulum stress (37), which has previously been reported to promote vascular calcification (38). DCA is an inhibitor of pyruvate dehydrogenase kinase, which promotes a shift from glycolysis to oxidative phosphorylation, leading to indirect inhibition of glycolysis (39). The possible reasons for this discrepancy may be explained by their nonspecific inhibitory effects on glycolysis.
To specifically investigate the direct causal link between glycolysis and hVIC calcification, we suppressed PFKFB3 expression using siRNA in hVICs. PFKFB3 generates fructose-2,6-bisphosphate, which in turn activates 6-phosphofructo-1-kinase to enhance glycolysis (40). PFKFB3-mediated glycolysis has been well documented in pathogenesis of cardiovascular disease, including atherosclerosis (41) and pulmonary hypertension (42). Interestingly, we showed that inhibition of PFKFB3-mediated glycolysis reduced the expression of the osteogenic gene MSX2, BMP2, and ALPL in hVICs. Our data extend on these previous reports and report for the first time that PFKFB3-mediated glycolysis may play a critical role in cardiovascular calcification. Furthermore, we found that inhibition of PFKFB3-mediated glycolysis attenuated the expression of the inflammatory gene, such as IL6 and IL1β, and the adhesion molecule, ICAM1, VCAM1, and E-SELECTIN. These results are consistent with previous reports showing that PFKFB3-mediated glycolysis drives vascular endothelial inflammation (43). Inflammation is the hallmark at the early stage of CAVD, characterized by the leaflet endothelium’s activation via enhanced expression of cell adhesion molecules such as ICAM1 and VCAM1 (44). These cell adhesion molecules function to recruit inflammatory cells, such as monocytes and macrophages, into the valve tissue, which in turn produce proinflammatory mediators to regulate hVIC osteogenic differentiation and calcification (45). The presence of leukocytes in pathologic samples from CAVD patients has been previously reported, thus suggesting that an active inflammatory process is involved in the initiation and progression of CAVD (46). Interestingly, our genome-wide RNA-Seq and subsequent pathway analysis revealed that PALMD regulated a cluster of genes involved in glycolysis. We showed that adenovirus-mediated overexpression of PALMD in hVICs significantly increased the expression of several key glycolytic genes including HK1 and PFKFB3. Moreover, our Seahorse analysis confirmed that overexpression of PALMD significantly enhanced basal and compensatory glycolysis in hVICs. Taken together, our data indicate that PALMD is a novel regulator of glycolysis in hVICs and highlight the crucial role of PFKFB3-mediated glycolysis in the regulation of hVIC osteogenic differentiation and inflammation. Intriguingly, silencing of PFKFB3 attenuated the inductive effects of PALMD on mRNA expression of the inflammatory gene IL1B and CXCL2 but did not alter PALMD-induced mRNA expression of the osteogenic gene MSX2 or ALPL in hVICs. These data suggest that PFKFB3 may not be the key regulator of PALMD-induced hVIC osteogenic differentiation.
NF-κB is a key transcription factor that has a critical role in many cardiovascular pathologies. Activated NF-κB subunits, including p65, translocate into the nucleus and directly regulate gene transcription, thus controlling cell proliferation, differentiation, and inflammation (47). Recent studies have demonstrated that activation of NF-κB promotes hVIC inflammation, osteogenic differentiation, and calcification in the pathogenesis of CAVD (48). Our RNA-Seq data and subsequent experiments showed that silencing of PALMD inhibited the NF-κB signaling pathway and reduced NF-κB target gene expression including IL6, IL1β, ICAM1, and VCAM1 in hVICs. To the best of our knowledge, this is the first report demonstrating that PALMD directly regulates NF-κB–mediated inflammation in hVICs. It has been previously reported that inflammation enhances the expression of cell adhesion molecules such as ICAM1 to recruit inflammatory cells into the valve tissue (44). Consistent with these results, we found that knockdown of PALMD expression in hVICs significantly reduced TNFα-induced monocyte adhesion to hVICs. We further showed that inhibition of NF-κB using the chemical inhibitor SC75741 abolished PALMD-induced osteogenic differentiation and inflammation of hVICs. These data suggest that PALMD promotes hVIC inflammation and osteogenic differentiation through the activation of the NF-κB. Intriguingly, inhibition of NF-κB did not alter PALMD-induced glycolysis in hVICs. Further studies are needed to identify the underlying mechanisms through which PALMD regulates glycolysis in hVICs.
It should be noted that the inductive effects of PALMD on the osteogenic gene expression, glycolysis, and inflammation was seen based on the robust overexpression of PALMD in hVICs transduced with Ad-PALMD at an multiplicity of infection (MOI) of 100. To further confirm the physiological relevance of PALMD in these processes, we transduced hVICs at a lower MOI. Our results showed that adenovirus-mediated overexpression of PALMD at an MOI of 50 also significantly increased mRNA expression of the osteogenic gene MSX2, the glycolytic gene ALDOC, HK1, PFKFB3, and PGM2, and the inflammatory gene CXCL2 and VCAM1 at day 2 (Fig. S18). These data support our findings showing that PALMD has an important role in CAVD.
Limitations of the study
This work was performed with hVIC in vitro calcification model to investigate the functional role and the underlying mechanisms of PALMD in CAVD. Whether PALMD promotes the development of CAVD in preclinical animal models remains to be established. Nonetheless, the present findings in human aortic valve tissues and hVICs generate novel hypotheses about the role and mechanisms of PALMD in CAVD. The inclusion of bicuspid aortic valves may be another limitation of the present study. CAVD patients with bicuspid aortic valves may be driven by distinct mechanisms compared to those patients with tricuspid aortic valves. Bicuspid CAVD has a strong genetic basis, while tricuspid CAVD is largely determined by nongenetic factors (49). Indeed, mutations in the transcriptional regulator NOTCH1 have been identified to be associated with human bicuspid aortic valves (50).
Conclusions
This study demonstrates a functional role of GWAS- and TWAS-identified CAVD risk gene PALMD in the pathogenesis of CAVD. We show that PALMD promotes hVIC inflammation, osteogenic differentiation, and calcification. Mechanistically, PALMD enhances hVIC glycolysis and induces NF-κB–mediated inflammation, thereby promoting the osteogenic differentiation and calcification of hVICs. Interestingly, inhibition of PFKFB3-mediated glycolysis attenuates hVIC inflammation and osteogenic differentiation. These findings suggest that PALMD- or PFKFB3-mediated glycolysis may be used as a novel potential therapeutic target for the inhibition of CAVD.
Experimental procedures
Human samples
This study was approved by the Research Ethics Committee of Guangdong Provincial People’s Hospital and Guangzhou Medical University (Ref No: KY-2020-503-01) and was performed in accordance with the Declaration of Helsinki. Thirty-eight aortic valve leaflets were anonymously collected from patients with CAVD undergoing valve replacement surgery at Guangdong Provincial People’s Hospital. Patients with a history of rheumatic disease, congenital valve disease, and infective endocarditis were excluded from the study. Bicuspid valves were also included in this study due to the limited numbers of human samples available. Clinical characteristics of the patients used in the present study are summarized in Table S2. Informed consent was obtained from all patients.
hVIC in vitro calcification
hVIC in vitro calcification was induced and detected as previously described (51).
Adenovirus-mediated overexpression of PALMD
Recombinant adenoviruses expressing flag-tagged PALMD (Ad-PALMD) and adenoviruses containing empty plasmids (Ad-null) that served as negative controls were constructed and purchased from Hanheng Bioscience Incorporation. The schematic of plasmid construction for Ad-PALMD are shown in Fig. S19. Western blotting was performed to confirm the overexpression efficiency of PALMD. hVICs transduced with Ad-PALMD at an MOI of 100 after 3 days resulted in 3.71 fold increase in PALMD protein expression, as shown in Fig. S7A.
Gene expression analysis
RT-qPCR and Western blotting were used to analyze gene expression as previously described (23). The primer sequences for the gene of interest examined are listed in Table S3. The primary antibodies used in the present study are summarized in Table S4.
Seahorse analysis
hVICs were seeded into Seahorse XFe96 FluxPak cell culture microplates (102601-100, Agilent Technologies) at a density of 1.0 × 104 cells/well. Cells were then infected with Ad-null and Ad-PALMD at an MOI of 100 for 48 h. Glycolytic function or oxidative phosphorylation was examined as previously reported (26). N stands for the number of replicates per treatment used for the Seahorse assay from one hVIC isolation.
RNA sequencing
hVICs were transfected with 20 nM siScrambled or siPALMD for 48 h. The sequences of siRNAs used in this study are outlined in Table S5. The cell samples were then sent to KangChen Biotech for RNA sequencing and subsequent bioinformatical analysis.
Statistical analysis
All values are expressed as mean ± SEM. Statistical analysis was performed using GraphPad Prism 6 software. After confirming a normal distribution using the Shapiro-Wilk test, data were analyzed using unpaired Student’s t test for comparison of two groups or one-way ANOVA followed by Tukey's multiple comparisons test for comparison of multiple groups or a suitable nonparametric test. p value < 0.05 was considered to be statistically significant.
Data availability
The data underlying this article will be provided on reasonable request to the corresponding author. The RNA-seq data reported in this article is deposited in NCBI Gene Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number of GEO: GSE165524 on January 26, 2021. Raw data are public available after March 29, 2022.
Supporting information
This article contains supporting information (23, 24, 51, 52, 53, 54).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
S.W., X. F., L. J., and D. Z. conceptualization; S. W., H. Y., J. G., J. C., P. H., H. Z., X. T., K. A. S., V. E. M., and D. Z. methodology; S. W. data curation; S. W. and D. Z. writing–original draft; S. W., H. Y., J. G., J. C., P. H., H. Z., and X. T. investigation; S. W., H. Y., J. G., and D. Z. formal analysis; H. Y. and J. G., validation; H. Y., J. G., J. C., P. H., H. Z., X. T., K. A. S., V. E. M., X. F., and L. J. writing–review and editing; P. H., X. F., and L. J. resources; P. H., L. J., and D. Z. funding acquisition; D. Z. supervision; D. Z. project administration.
Funding and additional information
This study was supported by funding from the National Natural Science Foundation of China (No. 82170428, 81800428 to D. Z.), The ‘Yangcheng Scholar’ Grant of Guangzhou (No. 202032768 to D. Z.), Guangdong Natural Science Foundation (No. 2018A030310178 to D. Z.), and Science and Technology Projects of Guangzhou (No. 201904010289 to D. Z.). L. J. is supported by the National Natural Science Foundation of China (No. 81800262) and Science and Technology Planning Project of Guangzhou (No. 201903010005). P. H. is supported by Natural Science Foundation of Guangdong Province (No. 2021A1515011121).
Edited by Qi Qun Tang
Contributor Information
Xiaodong Fu, Email: fuxiaod@mail.gzhmu.edu.cn.
Lei Jiang, Email: jianglei@smu.edu.cn.
Dongxing Zhu, Email: dongxing.zhu@gzhmu.edu.cn.
Supporting information
References
- 1.Rajamannan N.M., Evans F.J., Aikawa E., Grande-Allen K.J., Demer L.L., Heistad D.D., Simmons C.A., Masters K.S., Mathieu P., O'Brien K.D., Schoen F.J., Towler D.A., Yoganathan A.P., Otto C.M. Calcific aortic valve disease: Not simply a degenerative process: A review and agenda for research from the National Heart and Lung and Blood Institute Aortic Stenosis Working Group. Executive summary: Calcific aortic valve disease-2011 update. Circulation. 2011;124:1783–1791. doi: 10.1161/CIRCULATIONAHA.110.006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindman B.R., Clavel M.A., Mathieu P., Iung B., Lancellotti P., Otto C.M., Pibarot P. Calcific aortic stenosis. Nat. Rev. Dis. Primers. 2016;2:16006. doi: 10.1038/nrdp.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoen F.J. Evolving concepts of cardiac valve dynamics: The continuum of development, functional structure, pathobiology, and tissue engineering. Circulation. 2008;118:1864–1880. doi: 10.1161/CIRCULATIONAHA.108.805911. [DOI] [PubMed] [Google Scholar]
- 4.Hjortnaes J., Shapero K., Goettsch C., Hutcheson J.D., Keegan J., Kluin J., Mayer J.E., Bischoff J., Aikawa E. Valvular interstitial cells suppress calcification of valvular endothelial cells. Atherosclerosis. 2015;242:251–260. doi: 10.1016/j.atherosclerosis.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yutzey K.E., Demer L.L., Body S.C., Huggins G.S., Towler D.A., Giachelli C.M., Hofmann-Bowman M.A., Mortlock D.P., Rogers M.B., Sadeghi M.M., Aikawa E. Calcific aortic valve disease: A consensus summary from the Alliance of Investigators on Calcific Aortic Valve Disease. Arterioscler. Thromb. Vasc. Biol. 2014;34:2387–2393. doi: 10.1161/ATVBAHA.114.302523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu A.C., Joag V.R., Gotlieb A.I. The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology. Am. J. Pathol. 2007;171:1407–1418. doi: 10.2353/ajpath.2007.070251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bostrom K.I., Rajamannan N.M., Towler D.A. The regulation of valvular and vascular sclerosis by osteogenic morphogens. Circ. Res. 2011;109:564–577. doi: 10.1161/CIRCRESAHA.110.234278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X., Meng X., Su X., Mauchley D.C., Ao L., Cleveland J.C., Jr., Fullerton D.A. Bone morphogenic protein 2 induces Runx2 and osteopontin expression in human aortic valve interstitial cells: Role of Smad1 and extracellular signal-regulated kinase 1/2. J. Thorac. Cardiovasc. Surg. 2009;138:1008–1015. doi: 10.1016/j.jtcvs.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 9.Galeone A., Paparella D., Colucci S., Grano M., Brunetti G. The role of TNF-alpha and TNF superfamily members in the pathogenesis of calcific aortic valvular disease. ScientificWorldJournal. 2013;2013:875363. doi: 10.1155/2013/875363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosse K., Hans C.P., Zhao N., Koenig S.N., Huang N., Guggilam A., LaHaye S., Tao G., Lucchesi P.A., Lincoln J., Lilly B., Garg V. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J. Mol. Cell. Cardiol. 2013;60:27–35. doi: 10.1016/j.yjmcc.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majumdar U., Manivannan S., Basu M., Ueyama Y., Blaser M.C., Cameron E., McDermott M.R., Lincoln J., Cole S.E., Wood S., Aikawa E., Lilly B., Garg V. Nitric oxide prevents aortic valve calcification by S-nitrosylation of USP9X to activate NOTCH signaling. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Probst V., Le Scouarnec S., Legendre A., Jousseaume V., Jaafar P., Nguyen J.M., Chaventre A., Le Marec H., Schott J.J. Familial aggregation of calcific aortic valve stenosis in the western part of France. Circulation. 2006;113:856–860. doi: 10.1161/CIRCULATIONAHA.105.569467. [DOI] [PubMed] [Google Scholar]
- 13.Bella J.N., Tang W., Kraja A., Rao D.C., Hunt S.C., Miller M.B., Palmieri V., Roman M.J., Kitzman D.W., Oberman A., Devereux R.B., Arnett D.K. Genome-wide linkage mapping for valve calcification susceptibility loci in hypertensive sibships: The Hypertension Genetic Epidemiology Network Study. Hypertension. 2007;49:453–460. doi: 10.1161/01.HYP.0000256957.10242.75. [DOI] [PubMed] [Google Scholar]
- 14.Guauque-Olarte S., Messika-Zeitoun D., Droit A., Lamontagne M., Tremblay-Marchand J., Lavoie-Charland E., Gaudreault N., Arsenault B.J., Dube M.P., Tardif J.C., Body S.C., Seidman J.G., Boileau C., Mathieu P., Pibarot P., et al. Calcium signaling pathway genes RUNX2 and CACNA1C are associated with calcific aortic valve disease. Circ. Cardiovasc. Genet. 2015;8:812–822. doi: 10.1161/CIRCGENETICS.115.001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thanassoulis G., Campbell C.Y., Owens D.S., Smith J.G., Smith A.V., Peloso G.M., Kerr K.F., Pechlivanis S., Budoff M.J., Harris T.B., Malhotra R., O'Brien K.D., Kamstrup P.R., Nordestgaard B.G., Tybjaerg-Hansen A., et al. Genetic associations with valvular calcification and aortic stenosis. N. Engl. J. Med. 2013;368:503–512. doi: 10.1056/NEJMoa1109034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theriault S., Dina C., Messika-Zeitoun D., Le Scouarnec S., Capoulade R., Gaudreault N., Rigade S., Li Z., Simonet F., Lamontagne M., Clavel M.A., Arsenault B.J., Boureau A.S., Lecointe S., Baron E., et al. Genetic association analyses highlight IL6, ALPL, and NAV1 as 3 new susceptibility genes underlying calcific aortic valve stenosis. Circ. Genom. Precis. Med. 2019;12 doi: 10.1161/CIRCGEN.119.002617. [DOI] [PubMed] [Google Scholar]
- 17.Theriault S., Gaudreault N., Lamontagne M., Rosa M., Boulanger M.C., Messika-Zeitoun D., Clavel M.A., Capoulade R., Dagenais F., Pibarot P., Mathieu P., Bosse Y. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat. Commun. 2018;9:988. doi: 10.1038/s41467-018-03260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helgadottir A., Thorleifsson G., Gretarsdottir S., Stefansson O.A., Tragante V., Thorolfsdottir R.B., Jonsdottir I., Bjornsson T., Steinthorsdottir V., Verweij N., Nielsen J.B., Zhou W., Folkersen L., Martinsson A., Heydarpour M., et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat. Commun. 2018;9:987. doi: 10.1038/s41467-018-03252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B., Petrasch-Parwez E., Laue M.M., Kilimann M.W. Molecular characterization and immunohistochemical localization of palmdelphin, a cytosolic isoform of the paralemmin protein family implicated in membrane dynamics. Eur. J. Cell Biol. 2005;84:853–866. doi: 10.1016/j.ejcb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Andreu N., Escarceller M., Feather S., Devriendt K., Wolf A.S., Estivill X., Sumoy L. PALML, a novel paralemmin-related gene mapping on human chromosome 1p21. Gene. 2001;278:33–40. doi: 10.1016/s0378-1119(01)00719-3. [DOI] [PubMed] [Google Scholar]
- 21.Nie Y., Chen H., Guo C., Yuan Z., Zhou X., Zhang Y., Zhang X., Mo D., Chen Y. Palmdelphin promotes myoblast differentiation and muscle regeneration. Sci. Rep. 2017;7:41608. doi: 10.1038/srep41608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dashzeveg N., Taira N., Lu Z.G., Kimura J., Yoshida K. Palmdelphin, a novel target of p53 with Ser46 phosphorylation, controls cell death in response to DNA damage. Cell. Death Dis. 2014;5 doi: 10.1038/cddis.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui L., Rashdan N.A., Zhu D., Milne E.M., Ajuh P., Milne G., Helfrich M.H., Lim K., Prasad S., Lerman D.A., Vesey A.T., Dweck M.R., Jenkins W.S., Newby D.E., Farquharson C., et al. End stage renal disease-induced hypercalcemia may promote aortic valve calcification via Annexin VI enrichment of valve interstitial cell derived-matrix vesicles. J. Cell. Physiol. 2017;232:2985–2995. doi: 10.1002/jcp.25935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadji F., Boulanger M.C., Guay S.P., Gaudreault N., Amellah S., Mkannez G., Bouchareb R., Marchand J.T., Nsaibia M.J., Guauque-Olarte S., Pibarot P., Bouchard L., Bosse Y., Mathieu P. Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation. 2016;134:1848–1862. doi: 10.1161/CIRCULATIONAHA.116.023116. [DOI] [PubMed] [Google Scholar]
- 25.Galeone A., Brunetti G., Oranger A., Greco G., Di Benedetto A., Mori G., Colucci S., Zallone A., Paparella D., Grano M. Aortic valvular interstitial cells apoptosis and calcification are mediated by TNF-related apoptosis-inducing ligand. Int. J. Cardiol. 2013;169:296–304. doi: 10.1016/j.ijcard.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Rashdan N.A., Sim A.M., Cui L., Phadwal K., Roberts F.L., Carter R., Ozdemir D.D., Hohenstein P., Hung J., Kaczynski J., Newby D.E., Baker A.H., Karsenty G., Morton N.M., MacRae V.E. Osteocalcin regulates arterial calcification via altered Wnt signaling and glucose metabolism. J. Bone Miner. Res. 2020;35:357–367. doi: 10.1002/jbmr.3888. [DOI] [PubMed] [Google Scholar]
- 27.Alushi B., Curini L., Christopher M.R., Grubitzch H., Landmesser U., Amedei A., Lauten A. Calcific aortic valve disease-natural history and future therapeutic strategies. Front. Pharmacol. 2020;11:685. doi: 10.3389/fphar.2020.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel D.K., Green K.D., Fudim M., Harrell F.E., Wang T.J., Robbins M.A. Racial differences in the prevalence of severe aortic stenosis. J. Am. Heart Assoc. 2014;3 doi: 10.1161/JAHA.114.000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlotter F., Halu A., Goto S., Blaser M.C., Body S.C., Lee L.H., Higashi H., DeLaughter D.M., Hutcheson J.D., Vyas P., Pham T., Rogers M.A., Sharma A., Seidman C.E., Loscalzo J., et al. Spatiotemporal multi-omics mapping generates a molecular atlas of the aortic valve and reveals networks driving disease. Circulation. 2018;138:377–393. doi: 10.1161/CIRCULATIONAHA.117.032291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robicsek F., Thubrikar M.J., Cook J.W., Fowler B. The congenitally bicuspid aortic valve: How does it function? Why does it fail? Ann. Thorac. Surg. 2004;77:177–185. doi: 10.1016/s0003-4975(03)01249-9. [DOI] [PubMed] [Google Scholar]
- 31.Chignon A., Rosa M., Boulanger M.C., Argaud D., Devillers R., Bon-Baret V., Mkannez G., Li Z., Rufiange A., Gaudreault N., Gosselin D., Theriault S., Bosse Y., Mathieu P. Enhancer-associated aortic valve stenosis risk locus 1p21.2 alters NFATC2 binding site and promotes fibrogenesis. iScience. 2021;24:102241. doi: 10.1016/j.isci.2021.102241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proudfoot D., Skepper J.N., Hegyi L., Bennett M.R., Shanahan C.M., Weissberg P.L. Apoptosis regulates human vascular calcification in vitro: Evidence for initiation of vascular calcification by apoptotic bodies. Circ. Res. 2000;87:1055–1062. doi: 10.1161/01.res.87.11.1055. [DOI] [PubMed] [Google Scholar]
- 33.Skrtic D., Eanes E.D. Membrane-mediated precipitation of calcium phosphate in model liposomes with matrix vesicle-like lipid composition. Bone Miner. 1992;16:109–119. doi: 10.1016/0169-6009(92)90881-d. [DOI] [PubMed] [Google Scholar]
- 34.Rajamannan N.M., Subramaniam M., Rickard D., Stock S.R., Donovan J., Springett M., Orszulak T., Fullerton D.A., Tajik A.J., Bonow R.O., Spelsberg T. Human aortic valve calcification is associated with an osteoblast phenotype. Circulation. 2003;107:2181–2184. doi: 10.1161/01.CIR.0000070591.21548.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idelevich A., Rais Y., Monsonego-Ornan E. Bone Gla protein increases HIF-1alpha-dependent glucose metabolism and induces cartilage and vascular calcification. Arterioscler. Thromb. Vasc. Biol. 2011;31:e55–e71. doi: 10.1161/ATVBAHA.111.230904. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D., Li J., Wang F., Hu J., Wang S., Sun Y. 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 2014;355:176–183. doi: 10.1016/j.canlet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 37.Yu S.M., Kim S.J. Endoplasmic reticulum stress (ER-stress) by 2-deoxy-D-glucose (2DG) reduces cyclooxygenase-2 (COX-2) expression and N-glycosylation and induces a loss of COX-2 activity via a Src kinase-dependent pathway in rabbit articular chondrocytes. Exp. Mol. Med. 2010;42:777–786. doi: 10.3858/emm.2010.42.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furmanik M., van Gorp R., Whitehead M., Ahmad S., Bordoloi J., Kapustin A., Schurgers L.J., Shanahan C.M. Endoplasmic reticulum stress mediates vascular smooth muscle cell calcification via increased release of Grp78 (glucose-regulated protein, 78 kDa)-loaded extracellular vesicles. Arterioscler. Thromb. Vasc. Biol. 2021;41:898–914. doi: 10.1161/ATVBAHA.120.315506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie J., Wang B.S., Yu D.H., Lu Q., Ma J., Qi H., Fang C., Chen H.Z. Dichloroacetate shifts the metabolism from glycolysis to glucose oxidation and exhibits synergistic growth inhibition with cisplatin in HeLa cells. Int. J. Oncol. 2011;38:409–417. doi: 10.3892/ijo.2010.851. [DOI] [PubMed] [Google Scholar]
- 40.Qi T., Chen Y., Li H., Pei Y., Woo S.L., Guo X., Zhao J., Qian X., Awika J., Huo Y., Wu C. A role for PFKFB3/iPFK2 in metformin suppression of adipocyte inflammatory responses. J. Mol. Endocrinol. 2017;59:49–59. doi: 10.1530/JME-17-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poels K., Schnitzler J.G., Waissi F., Levels J.H.M., Stroes E.S.G., Daemen M., Lutgens E., Pennekamp A.M., De Kleijn D.P.V., Seijkens T.T.P., Kroon J. Inhibition of PFKFB3 hampers the progression of atherosclerosis and promotes plaque stability. Front. Cell Dev. Biol. 2020;8:581641. doi: 10.3389/fcell.2020.581641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y., Zhang X., Wang L., Yang Q., Ma Q., Xu J., Wang J., Kovacs L., Ayon R.J., Liu Z., Zhang M., Zhou Y., Zeng X., Xu Y., Wang Y., et al. PFKFB3-mediated endothelial glycolysis promotes pulmonary hypertension. Proc. Natl. Acad. Sci. U. S. A. 2019;116:13394–13403. doi: 10.1073/pnas.1821401116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnitzler J.G., Hoogeveen R.M., Ali L., Prange K.H.M., Waissi F., van Weeghel M., Bachmann J.C., Versloot M., Borrelli M.J., Yeang C., De Kleijn D.P.V., Houtkooper R.H., Koschinsky M.L., de Winther M.P.J., Groen A.K., et al. Atherogenic lipoprotein(a) increases vascular glycolysis, thereby facilitating inflammation and leukocyte extravasation. Circ. Res. 2020;126:1346–1359. doi: 10.1161/CIRCRESAHA.119.316206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun L., Rajamannan N.M., Sucosky P. Defining the role of fluid shear stress in the expression of early signaling markers for calcific aortic valve disease. PLoS One. 2013;8 doi: 10.1371/journal.pone.0084433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss R.M., Miller J.D., Heistad D.D. Fibrocalcific aortic valve disease: Opportunity to understand disease mechanisms using mouse models. Circ. Res. 2013;113:209–222. doi: 10.1161/CIRCRESAHA.113.300153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cote N., Mahmut A., Bosse Y., Couture C., Page S., Trahan S., Boulanger M.C., Fournier D., Pibarot P., Mathieu P. Inflammation is associated with the remodeling of calcific aortic valve disease. Inflammation. 2013;36:573–581. doi: 10.1007/s10753-012-9579-6. [DOI] [PubMed] [Google Scholar]
- 47.Hoffmann A., Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 48.Gee T., Farrar E., Wang Y., Wu B., Hsu K., Zhou B., Butcher J. NFkappaB (nuclear factor kappa-light-chain enhancer of activated B cells) activity regulates cell-type-specific and context-specific susceptibility to calcification in the aortic valve. Arterioscler. Thromb. Vasc. Biol. 2020;40:638–655. doi: 10.1161/ATVBAHA.119.313248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bravo-Jaimes K., Prakash S.K. Genetics in bicuspid aortic valve disease: Where are we? Prog. Cardiovasc. Dis. 2020;63:398–406. doi: 10.1016/j.pcad.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garg V., Muth A.N., Ransom J.F., Schluterman M.K., Barnes R., King I.N., Grossfeld P.D., Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z., Gordillo-Martinez F., Jiang L., He P., Hong W., Wei X., Staines K.A., Macrae V.E., Zhang C., Yu D., Fu X., Zhu D. Zinc ameliorates human aortic valve calcification through GPR39 mediated ERK1/2 signalling pathway. Cardiovasc. Res. 2021;117:820–835. doi: 10.1093/cvr/cvaa090. [DOI] [PubMed] [Google Scholar]
- 52.Plunde O., Larsson S.C., Artiach G., Thanassoulis G., Carracedo M., Franco-Cereceda A., Eriksson P., Back M. FADS1 (fatty acid desaturase 1) genotype associates with aortic valve FADS mRNA expression, fatty acid content and calcification. Circ. Genom. Precis. Med. 2020;13 doi: 10.1161/CIRCGEN.119.002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin C., Zhu D., Markby G., Corcoran B.M., Farquharson C., Macrae V.E. Isolation and characterization of primary rat valve interstitial cells: A new model to study aortic valve calcification. J. Vis. Exp. 2017 doi: 10.3791/56126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehrhardt C., Ruckle A., Hrincius E.R., Haasbach E., Anhlan D., Ahmann K., Banning C., Reiling S.J., Kuhn J., Strobl S., Vitt D., Leban J., Planz O., Ludwig S. The NF-kappaB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell. Microbiol. 2013;15:1198–1211. doi: 10.1111/cmi.12108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be provided on reasonable request to the corresponding author. The RNA-seq data reported in this article is deposited in NCBI Gene Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number of GEO: GSE165524 on January 26, 2021. Raw data are public available after March 29, 2022.