Abstract
All cells possess an internal stress response to cope with environmental and pathophysiological challenges. Upon stress, cells reprogram their molecular functions to activate a survival mechanism known as the heat shock response, which mediates the rapid induction of molecular chaperones such as the heat shock proteins (HSPs). This potent production overcomes the general suppression of gene expression and results in high levels of HSPs to subsequently refold or degrade misfolded proteins. Once the damage or stress is repaired or removed, cells terminate the production of HSPs and resume regular functions. Thus, fulfillment of the stress response requires swift and robust coordination between stress response activation and completion that is determined by the status of the cell. In recent years, single-cell fluorescence microscopy techniques have begun to be used in unravelling HSP-gene expression pathways, from DNA transcription to mRNA degradation. In this review, we will address the molecular mechanisms in different organisms and cell types that coordinate the expression of HSPs with signaling networks that act to reprogram gene transcription, mRNA translation, and decay and ensure protein quality control.
Keywords: heat shock response, heat shock proteins, heat shock factor 1, stress-regulated translation, mRNA decay, proteostasis, gene expression, acclimation
Abbreviations: AFD, amphid neurons with finger-like ciliated ending; ER, endoplasmic reticulum; HSE, heat shock element; HSF1, heat shock factor 1; HSP, heat shock protein; HSR, heat shock response; IRESs, internal ribosome entry sites; ISR, integrated stress response; mTOR, mammalian target of the rapamycin; ND, neurodegenerative disease; PBs, processing bodies; PKR, protein kinase R; RNAPII, RNA polymerase II; RNP, ribonucleoprotein; SGs, stress granules; TC, ternary complex; UTR, untranslated region
For organisms to grow and function properly, they must maintain specific internal cellular conditions that allow proteins to acquire their functional conformations and cells to achieve protein homeostasis (proteostasis) (1). Maintaining proteostasis becomes critical when facing abrupt changes in the external conditions, such as an increase in temperature, which can lead to protein misfolding and aggregation, and consequently, cellular dysfunction (2). Thus, organisms must sense, rapidly respond, and adapt to new environmental conditions for survival. Organisms from bacteria to mammals have evolved similar and varying stress responses to cope with protein misfolding and maintain proteostasis successfully. Some of these strategies include modulations of signaling cascades, changes in transcriptional programs, and regulation of translation, posttranslational modifications, and the dynamic assembly of RNA and protein condensates (ribonucleoprotein [RNP] granules) through liquid–liquid phase separation (1, 3, 4, 5, 6, 7). Several of these molecular mechanisms converge to sustain proteostasis in response to sudden and acute changes in environmental conditions.
Increases in the environmental temperature is a universal proteostasis challenge encountered by most organisms. For historical reasons, thermal stress has been used as a paradigm to study the stress response. Nowadays, these studies have an additional relevance due to the increased exposure of organisms to heatwaves derived from climate change (8, 9, 10). Increased thermal energy in the cells can result in heat-induced denaturation of proteins and thermally altered metabolic activity leading to an increase in reactive oxygen species, which can damage all biological macromolecules, including proteins (11). Cells cope with an increased load of unfolded and misfolded proteins by modulating the expression of specific molecular chaperones, also known as heat shock proteins (HSPs) (12, 13, 14, 15). The heat shock response (HSR) refers to the activation of the expression of HSPs, and it is the most common and widely studied cell response to thermal stress. HSPs play a central role in the lifecycle of proteins because they promote the folding of nascent polypeptides into their native/functional configurations and prevent protein misfolding and aggregation (12, 15, 16). HSPs also collaborate with the quality control mechanisms, the ubiquitin-proteasome system, and autophagy, to target misfolded proteins and aggregates whose native functional state cannot be recovered for degradation (5, 17).
Given that HSPs are central to the cellular proteostasis network, cells undertake several gene expression adaptations to favor the synthesis of HSPs at the expense of decreasing most cellular functions (Fig. 1). Biochemical and molecular biology approaches highlight the unique regulation of HSP gene expression. The spatiotemporal resolution of such precise regulation is now being uncovered using high-resolution quantitative fluorescence microscopy. Gene expression adaptions during stress act together to protect macromolecules and promptly resume the cytoplasmic and nuclear activities once permissive conditions are restored (3). The regulation of HSP expression coordinates with other cell protective mechanisms, like the formation of RNP condensates and the activation of the integrated stress response (ISR) to repress translation initiation. The ISR and HSR also coordinate their actions with the unfolded protein response in the endoplasmic reticulum (ER) and the mitochondria to preserve proteostasis across cellular compartments. All organisms ranging from bacteria to plants and mammals have genes encoding for HSPs. HSPs are grouped into families based on an apparent molecular weight (18, 19). The HSP70 and HSP90 families are the most functionally relevant HSPs in the cell (15, 20). They are ATP-dependent chaperones that cooperate with small HSPs and HSP110. Cochaperones of the J-domain family of proteins modulate HSP70 activity by accelerating ATP hydrolysis, participating in substrate recognition and substrate folding or refolding (Fig. 2) (21, 22, 23, 24, 25, 26, 27). HSPs are further categorized as constitutive or inducible based on their steady-state expression levels. The expression of all inducible and some constitutive HSPs is upregulated to some extent upon heat stress. Among them, the inducible HSP70 genes are the fastest and most upregulated (23, 24, 27). Interestingly, they are highly conserved among species having an amino acid similarity of 50% between Homo sapiens and Escherichia coli, while some domains are 96% similar, which highlights its vital role in cell adaption to changing environmental conditions (28).
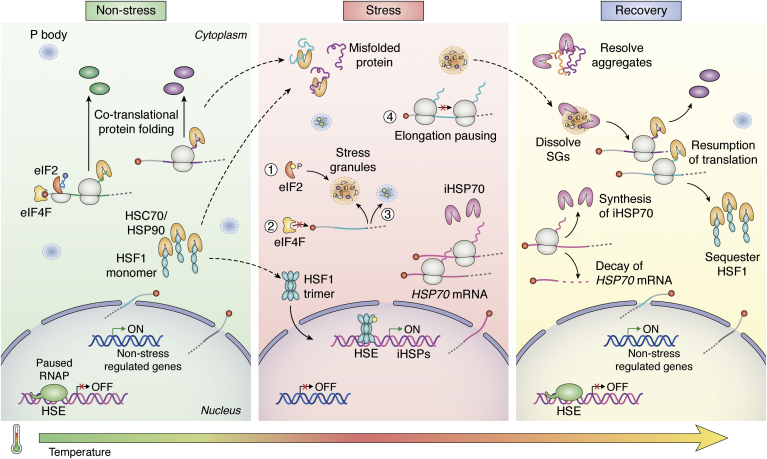
Figure 1.
Overview of the cellular response to heat stress. Cells under nonstress conditions keep the transcription of the inducible HSPs inactive. A paused polymerase occupies their promoter, and the transcription factor HSF1 is sequestered in monomeric form by constitutive chaperones HSC70/HSP90 in the cytoplasm. Constitutive chaperones also assist in protein folding and preserving protein homeostasis. Under physiological conditions, nonstress–regulated genes are transcribed, and their mRNAs undergo canonical cap-dependent translation. Exposure to heat stress induces protein misfolding, which titrates out the HSC70/HSP90 and allows HSF1 to trimerize and translocate to the nucleus, where it binds to the HSE in the promoter of HSPs and activates transcription. Concomitant to the HSR activation, there is a global transcriptional and translational repression. The translation is repressed by (1) phosphorylation of eIF2α (2); inhibition of eIF4F complex formation (3); recruitment of untranslated mRNAs and regulatory proteins in stress granules (SGs) and processing bodies (PBs) (4); and translation arrest at the stage of elongation. The inducible HSP mRNAs, especially HSP70, skip translation repression and are translated through a cap-independent mechanism to increase the number of available chaperones needed to cope with the abundant misfolded proteins and prevent their toxic aggregation. Once the temperature returns to being permissive, the newly synthesized HSPs favor recovering proteostasis and functionality by folding misfolded proteins and disabling SGs. The resumption of regular translation and transcription coincides with the decay of HSP mRNAs and silencing of their transcription. HSEs, heat shock elements; HSF1, heat shock factor 1; HSP, heat shock protein.
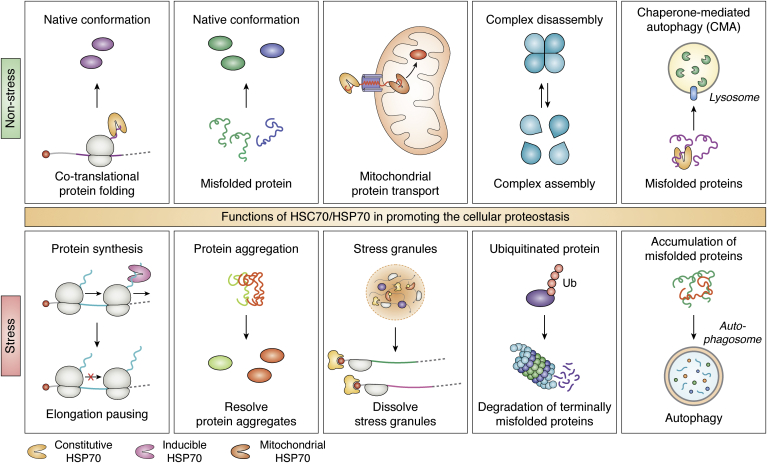
Figure 2.
The function of HSC70/HSP70 in retaining the cellular proteostasis. The illustration depicts the significant tasks of the HSP70 chaperone network inside the cell to maintain proteostasis. (Starting from the top left tile) Under nonstress conditions, HSC70 provides cotranslational folding of the nascent polypeptide to obtain native conformation; helps to refold misfolded proteins; transports nascent polypeptide from the cytoplasm to the mitochondria where it is assisted by mitochondrial HSP70 (mtHSP70) and HSP60 to attain functional conformation; involved in protein complex assembly and/or disassembly; and leads specific proteins for their degradation by the lysosome through chaperone-mediated autophagy (236, 237). (Continuing bottom left tile) During stress, the lack of HSP70 at the exit of the ribosome tunnel represses the translation at the elongation stage. HSP70 and HSP90 prevent protein aggregation, and HSP70 also resolves stress granules so that the sequestered mRNAs can resume their translation during recovery from stress; targets terminally misfolded protein for proteasomal degradation; and mediates autophagy by autophagosome. HSP, heat shock protein.
In this review, we frame the molecular regulation of the HSR to the context of the gene expression changes undertaken by eukaryotic cells in response to an increase in temperature. We compare the response mounted by different organisms and cell types and suggest new technological approaches to overcome the gap in our knowledge on the HSPs expression.
Transcriptional regulation of inducible HSPs versus constitutive genes
Transcriptional upregulation of stress-inducible HSPs
The robust transcriptional induction of genes of the HSP70 family is one of the main and fastest response to heat stress. Their transcriptional induction occurs at the expense of a general transcriptional downregulation of constitutively expressed genes. Most inducible HSP70 genes are short (around 2500 nucleotides) and intronless, and their promoter contains one or more binding sites, known as heat shock elements (HSEs), for the association of the master transcription factor heat shock factor 1 (HSF1) (29). Under physiological conditions, the inducible HSP70 genes are not expressed. However, their loci are neither present in a compact heterochromatin domain nor marked by repressive epigenetic histone modification. The promoter and 3′ end of HSP70 gene is nucleosome-free while its gene body is covered by nucleosomes. The promoter is bound by a paused RNA polymerase II (RNAPII) (30). These characteristics prevent the stable transcriptional repression of HSP70 genes and facilitate their prompt activation in response to the binding of HSF1.
Under physiological conditions, HSF1 shuttles between the nucleus and cytoplasm, and it is kept as an inactive monomer by constitutive members of HSP90 and HSP70 families. Upon stress, HSF1 is released from HSPs, trimerizes, and localizes in the nucleus where it binds to the HSE, which is comprised of at least three nGAAn repeats organized head to tail in the promoters of genes encoding HSPs and other gene products (31, 32) (Fig. 3). HSF1 has three domains, an oligomerization domain next to the DNA binding domain at the N terminus, a trans-activation domain at the C terminus that induces transcription initiation and elongation, and a regulatory domain in the middle that negatively regulates the function of the trans-activation domain in nonstress conditions. By forming a trimer, the affinity of HSF1 for the HSE increases as each HSF1 of the trimer binds to a nGAAn repeat through its DNA binding domain. The binding of HSF1 to HSE is not sufficient to activate transcription and has to be accompanied by extensive posttranslational modifications. HSF1 undergoes hyperphosphorylation of serine and threonine residues that cover up to 90% of the regulatory domain (33, 34, 35, 36). However, only a few of these phosphorylation sites, like serines 230 or 326, are necessary for the activity of HSF1 (35, 37). Concomitantly, sumo groups that have an inhibitory effect on transcription are removed from HSF1 (38). HSF1 acetylation at lysines 116 and 118 favors its transcriptional activity, whereas acetylation at several other lysine residues regulates its nuclear localization and oligomerization (31). Acetylation of HSF1 occurs a few hours after heat shock to decrease its DNA affinity and the transcriptional response (39). In summary, HSF1 undergoes extensive posttranslational modifications, which are regulated under various stresses. Although the function of some of these modifications has been identified, the role of many others, as well as the proteins responsible for their regulation, remains to be elucidated.
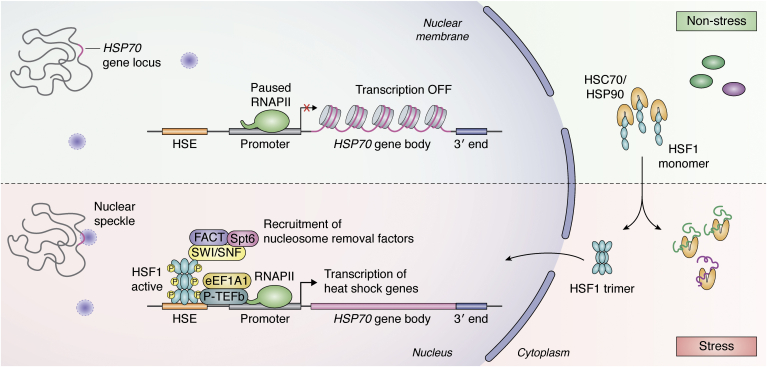
Figure 3.
Chromatin remodeling and transcriptional activation of HSP genes. The figure represents the changes in the chromatin region and the promoter of heat shock genes under nonstress and stress conditions in mammalian cells. Under physiological conditions, HSF1 is sequestered in the cytoplasm by constitutive chaperones HSP90 and HSC70. RNAPII is bound to the open promoter region of HSP genes and remains paused/transcriptionally inactive, and the HSP70 gene locus is located close to the membrane. Under stress, the HSP70 locus moves to the nuclear speckle. The chaperones bound to HSF1 now bind misfolded protein, thereby releasing HSF1, which trimerizes and localizes to the nucleus where it binds to the heat shock elements (HSEs) in the HSP gene promoter. Multiple posttranslational modifications activate the HSF1 trimer, resulting in the recruitment of transcription factors (P-TEFb) and nucleosome removal factors (FACT, SWI/SNF, Spt6) to the site causing chromatin remodeling and favoring transcription elongation. HSF1, heat shock factor 1; HSP, heat shock protein; RNAPII, RNA polymerase II.
The combination of posttranslational modifications and titration of HSPs by misfolded proteins have been demonstrated to activate HSF1. Recent work in the yeast, Saccharomyces cerevisiae, has allowed building a simple mathematical model that points to the dissociation of HSP70/HSP90 from HSF1 as the first “switch on” step to activate HSP70 transcription, which feedback to HSF1 to switch it off or repress it (40, 41). Zheng et al. (41) identified 70 phosphorylation sites on HSF1 upon heat shock and were able to model that these phosphorylations have no effect on HSF1 activation but instead increase its transcriptional activity by favoring its association with the mediator complex. Additionally, the translation factor eEF1A and the noncoding RNA HSR1 are among the factors activating HSF1. They act together to form a nucleoprotein complex with HSF1 and stimulate HSF1 trimerization (42). Following heat shock, HSF1 recruits multiple cofactors to HSE (43, 44, 45, 46), including SGO2, which recruits the subunit mediator complex MED12, essential for the strong transcriptional induction of HSPs genes (47). SGO2 binding to hypophosphorylated RNAPII targets it to the promoter of HSP genes by forming a complex with HSF1. Transcription is then induced by other transcription factors like P-TEFb, recruitment of which are mediated by HSF1 (48). P-TEFb is sufficient to induce the phosphorylation of the serine 2 in the C-terminal domain of RNAPII, which leads to transcription elongation (48, 49). However, a strong transcriptional induction requires the nucleosomes positioned along the HSP70 gene body to be removed. The chromatin remodelers SWI/SNF in mammals and FACT together with the histone chaperone Spt6 in Drosophila melanogaster (D. melanogaster) are recruited by HSF1 to the HSP70 genes within minutes after heat shock to remove the nucleosomes (31).
Besides the activation of HSF1, heat shock induction of HSP70 in mammalian cells depends on the relocation of the HSP70 loci from the nuclear membrane to speckles (50, 51). The rapid, active, and unidirectional movement of HSP70 loci is mediated by nuclear actin polymerization. The association of the HSP70 locus with speckles depends on the promoter sequence and determines the robust transcriptional activation of HSP70 upon heat shock stress. Although speckles contain serine 2–phosphorylated RNAPII and other components of the transcriptional machinery, the specific speckle factors critical for the transcriptional activation of HSP70 have not been yet identified. In yeast, genes encoding for different HSPs coalesce in discrete spots in the nucleus upon transcription stimulation. This interallelic clustering leads to the interaction between HSP104 and HSP12 loci and depends on the activation of their transcription. This result suggested the presence of specific transcriptional factories formed in response to heat stress, which could be coregulated by HSF1 (52).
Transcriptional downregulation of nonstress genes
The transcriptional induction of HSP genes during heat stress is accompanied by the upregulation of other non-HSP genes encoding for cytoskeleton and oxidative stress proteins and a massive downregulation of thousands of genes (For review: (53)). Detailed analysis of the position of the RNA polymerases, chromatin modifications, and domains in D. melanogaster and mammalian cells suggest that changes in the chromatin landscape cannot explain the rapid changes in transcriptional preferences upon heat shock (54, 55).
Heat shock does not induce a global chromatin remodeling nor modifications of topology associated domains in human or D. melanogaster S2 cells (56) or rearrangements of topology associated domain borders in D. melanogaster Kc167 cells (57). The addition of DTT to induce protein unfolding in D. melanogaster S2 cells did not trigger a global decrease in nucleosome occupancy over the induced genes, their promoters, or enhancers, as detected by micrococcal nuclease sequencing (54). However, the accessibility of the chromatin is increased at upregulated genes as measured with ATAC-seq. These results could be explained by nucleosomes undergoing an increased turnover due to their increased acetylation and ongoing transcription. Mueller et al. also reported a decrease in nucleosome occupancy and an increase in accessibility in a few genes, like the constitutively expressed chaperone HSC70. Similar observations were done in human K562 cells following 30 minutes of heat shock. The level of histone 3 lysine 27 acetylation (H3K27Ac) increased at the promoter of all transcribed genes, which also experienced an increase in RNAPII occupancy in the promoter and to a lesser extent in the gene body (55). Conversely, a decrease of polymerase occupancy is observed along the gene body of constitutively expressed genes whose transcription is downregulated upon stress. Hence, it is suggested that the extra available RNAPII quickly replaced the RNAPII undertaking transcription elongation in stress-regulated genes.
Additionally, noncoding RNAs have been shown to be a transcriptional repressor of non-HSP genes during heat shock. For instance, noncoding transcripts such as Alu RNA derived from short interspersed nuclear elements bind to RNAPII during heat shock to inhibit transcription of other mRNAs, such as actin (58, 59, 60). Interestingly, it has recently been shown that the long noncoding RNA, heat-enhanced antisense transcript (Heat) binds to HSF1 in vitro and in vivo via a trans-acting manner to attenuate the expression of stress genes. Experiments on mouse embryonic fibroblasts suggest that Heat uses HSF1 as a carrier by forming an RNP complex to target stress genes. While the exact mechanism by which Heat suppresses transcription is not known, Ji et al. (61) suggest the attenuation of the HSR by Heat involves an m6A modification and the nuclear m6A reader protein YTHDC1. Furthermore, the global downregulation of transcription induced by heat shock is suggested to be caused by activation of cryptic intronic polyadenylation sites in introns. Intronic polyadenylation sites led to premature transcriptional termination and new short mRNAs accumulating in the nucleus (62). Thirty percent of the stress-induced genes have HSF1 bound to the promoter. Several genes upregulated by heat stress but not bound by HSF1 are frequently contacted by distal regions bound by HSF1 as shown by chromosome conformation capture techniques, Hi-C performed in human and Drosophila cells (56). However, the regulatory mechanisms that coordinate the transcriptional induction of HSF1-dependent and -independent genes with the transcriptional repression of more than 6000 genes in humans remains to be uncovered. Overall, heat shock and other stresses dramatically affect global transcriptional regulation, leading to potent induction of genes encoding for prosurvival proteins.
Translational regulation during the HSR
To preferentially synthesize HSPs during stressful events, cells have adapted a mechanism whereby non-HSP transcripts are retained in the nucleus, and HSP transcripts are selectively exported to and translated in the cytoplasm. The exact mechanism of this selective process is not known. However, work conducted on S. cerevisiae has shown that nuclear export of non-HSP mRNA involves RNA adaptor proteins, Npl3, Gbp2, Hrb1, and Nab2, cotranscriptionally loaded onto the pre-mRNA, which then recruits Mex67–Mtr2 (TAP–p15 in humans), an essential heterodimeric receptor mRNA export factor (63). These adaptor proteins have an mRNA quality control function that prevents the nuclear export of incorrect, possibly improperly processed or assembled mRNAs (64). During stress, Mex67 and the adaptor proteins are dissociated from non-HSP transcripts to prevent nuclear export. However, HSP transcripts do not require adaptive proteins and are loaded directly with Mex67 via HSF1 (64). Thus, HSP transcripts bypass the adaptor–protein–mediated quality control mechanism to be rapidly exported and translated.
The newly synthesized HSP mRNAs encounter a cytoplasmic environment in which translation is repressed. Cells sense the load of misfolded proteins and repress translation by different pathways to decrease the load of unfolded and misfolded proteins. However, the translation of mRNA encoding for specific HSPs, such as the inducible HSP70, HSP82, and HSP27, is specifically favored (65). Regulation of translation enables the cells to rapidly adapt their proteome to stress conditions (66). From the three stages of translation: initiation, elongation, and termination, stress conditions repress cap-dependent translation initiation and elongation (67). The association of translation initiation factors (eIF4A, eIF4B, Ded1) and ribosomal proteins with mRNAs is decreased immediately upon stress (68). Additionally, stress conditions promote the recruitment of translation factors and regulatory RNA binding proteins to stress-induced cytoplasmic structures known as stress granules (SGs) and processing bodies (PBs), limiting their availability (69, 70, 71) (Fig. 1).
Regulation of translation initiation and elongation
Eukaryotic translation initiation is a highly regulated multistep process and the rate-limiting step in translation. It involves the assembly of the ternary complex (TC) and binding of the eIF4F complex to the 5′ m7G cap structure in the mRNA. Both steps are downregulated during stress by different signaling pathways (72).
The TC, GTP-eIF2α-initiator methionine tRNA, preloaded with eIF1, eIF1A, eIF3, and eIF5, assembles on the 40S ribosomal subunit to form the 43S preinitiation complex (73, 74, 75). Stress precludes the formation of the TC by the reversible phosphorylation of the Ser51 in eIF2α. This phosphorylation event limits the pool of available eIF2α for the TC formation. (Fig. 1). The phosphorylation of eIF2α in response to stress is part of the ISR (76). Depending on the stress stimulus, one of four serine-threonine kinase enzymes catalyzes the phosphorylation of Ser51. These kinases are the heme-regulated inhibitor and the general control nonderepressible 2, which are conserved in eukaryotes, the protein kinase R (PKR), which is specific to vertebrates, and the PKR-like ER kinase, which is absent in fungi. Heme-regulated inhibitor is activated to cope with heme deficiency in red blood corpuscles, heat and osmotic shock, oxidative and mitochondrial stress, cytosolic protein aggregation, and arsenite treatment in cells other than red blood corpuscles (66, 77, 78, 79, 80, 81, 82). General control nonderepressible 2 phosphorylates eIF2α in conditions of nutrient depletion (83) and UV irradiation (84, 85). PKR mediates the phosphorylation of eIF2α in response to the detection of double-stranded viral RNA and hyperosmotic stress (dsRNA) (82, 86). PKR-like ER kinase activates upon ER stress, aggregation of misfolded proteins in the ER lumen, hypoxia, hypoglycemia, ischemia, oxidative stress, and perturbation in Ca2+ levels (87, 88). The conversion of the eIF2-GDP binary complex to the translation competent eIF2-GTP is mediated by the eIF2-specific guanine nucleotide exchange factor, eIF2B (80, 89). Two copies of eIF2B forms an active decameric complex that interacts with eIF2 and loads a molecule of GTP on eIF2. Phosphorylated eIF2α acts as a noncompetitive inhibitor by sterically hindering the access of eIF2 to the catalytic domain of eIF2B and sequestering it. Consequently, the recycling of eIF2α is decreased, which in turn decreases the abundance of TC (80, 90, 91) and impairs cap-dependent translation promoting metabolic dormancy to survive through the stress (92).
In addition to eIF2α phosphorylation, the binding of the eIF4F complex to the 5′ m7G cap structure is impaired by different means. The eIF4F complex is made of the cap recognizing factor eIF4E, the scaffold protein eIF4G, and the ATP-driven RNA helicase eIF4A that unwinds secondary structures in the 5′ untranslated region (UTR) of mRNAs (75, 93). The binding of eIF4E to the cap is partially regulated by the mammalian target of the rapamycin (mTOR) pathway (93, 94, 95, 96). mTOR is a kinase that phosphorylates the downstream targets eIF4E binding protein 1, preventing its binding to eIF4E, which allows its binding to the cap under favorable conditions (93, 97). During stress, mTOR is inactivated by TSC1/2, leading to the dephosphorylation of eIF4E binding protein 1, which readily sequesters eIF4E and suppresses the eIF4F complex formation (98, 99, 100, 101). Additionally, the newly synthesized HSP27 binds eIF4G with high affinity preventing the formation of the eIF4F complex (102). Together, these mechanisms prevent the assembly and binding of eIF4F to the m7G cap structure and cap-dependent translation.
The elongation step of translation is also regulated to enforce the translation repression of nonstress mRNAs during conditions that challenge protein homeostasis. During translation elongation, the GTP-bound elongation factor eEF1A brings the aminoacyl-tRNA corresponding to the codon in the ribosomal A site, and eEF1A-GDP is released upon codon–anticodon base pairing. The ribosomal RNA in the peptidyl transfer center catalyzes the peptide bond formation, and the translocase eEF2–GTP triggers the mRNA–tRNA movement with the expense of GTP, and eEF2-GDP is released (103, 104, 105). Stress regulates translation elongation by a major downstream effector of mTORC1, the S6 kinase. The S6 kinase phosphorylates Ser366 of the eEF2 kinase and inactivates it (106). Inhibition of mTORC1 upon stress leads to the activation of eEF2 kinase that phosphorylates eEF2 at Thr56 of the GTP binding domain, pausing translation elongation (107, 108, 109, 110, 111, 112, 113). It should be noted that the regulation of eEF2 and its kinase is more complex than what we have described here, as they can be phosphorylated at different residues through different pathways (111, 114).
Besides eEF2 regulation, studies from two independent laboratories have identified chaperone-mediated regulation of translation elongation during stress. The cytoplasmic chaperone HSP70 and HSP90 interact with nascent polypeptides, favoring their cotranslational folding as they emerge out the ribosome exit tunnel (115, 116) (Fig. 1). During severe stress, the prevalence of unfolded proteins titrates out the chaperones leaving the nascent polypeptides unassisted for cotranslational folding. Hence, an arrest in elongation was observed after the synthesis of the first 65 amino acids, which corresponds to the length of the nascent polypeptide that fits in the ribosome exit tunnel. Consequently, stalled ribosomes were observed in several different mRNAs at nucleotide position 195. The lack of chaperone–ribosome interaction impairs the cotranslational folding of nascent polypeptides accounting for the elongation pausing and global translation repression during severe heat stress (117, 118). Overall, HSP encoding mRNAs should be equipped to overcome the several steps at which translation is shut by the cell.
SGs and PB formation
Translation repression is accompanied by changes in the physical properties of the cytoplasmic milieu. Stress triggers RNAs and proteins to phase separate and form SGs and PBs that contain untranslated, long, and highly unstructured mRNAs and proteins that participate in translation, transcription, splicing, and decay (71, 119, 120, 121). SGs and PBs are dynamic membraneless structures assembled by liquid–liquid phase transition that favor stress tolerance and promote cellular fitness (122, 123, 124, 125, 126). While the number and size of PBs increase upon stress, SGs are formed under stress conditions that invoke arrest in translation initiation (127). SGs and PBs have different mRNA and protein compositions. PBs are made of RNA processing factors (e.g., eIF4E, DDX6, and Ded1p), decapping and deadenylation enzymes, exoribonucleases, and factors mediating mRNA stability (For review: (124, 125)). Based on their enrichment in decay-related factors, PBs were believed to be mRNA degradation sites. However, mRNAs retained in PBs can return to the cytoplasm and engage in translation (124, 128). Thus, it is widely accepted that PBs could serve as a reservoir of nontranslating mRNAs and inactive decay enzymes (129, 130).
SGs form under stress conditions that induce eIF2α phosphorylation (131, 132), but they can also form independently of eIF2α phosphorylation. For example, puromycin treatment and inhibition of eIF4A with hippuristanol stimulate SG assembly (133, 134, 135, 136, 137). SGs are enriched in translation initiation factors, mRNA binding proteins, 40S ribosomal subunit, and mRNAs encoding for house-keeping genes (131, 132, 138, 139). mRNAs recruited to SGs are translationally repressed and undergo compaction as the elongating ribosomes are released from the transcripts (140, 141). The SGs were speculated to function in blocking protein synthesis by trapping several initiation factors and stabilizing translationally inactive constitutive mRNAs so that they can reengage in translation upon recovery (136, 142, 143, 144). Certain mRNAs are shown to have paused ribosomes at the start codon to quickly re-initiate the protein synthesis as the cells recover from stress (145). Even so, the lack of SGs does not change mRNA translational repression or stability (142). Additionally, mRNAs-encoding stress-regulated proteins like activating transcription factor 4 have been shown to translate inside the SGs (146). HSP mRNAs skip the localization in SGs and PBs as their ongoing translation prevents them from condensating (119, 147).
Translation of HSP70 mRNAs under stress conditions
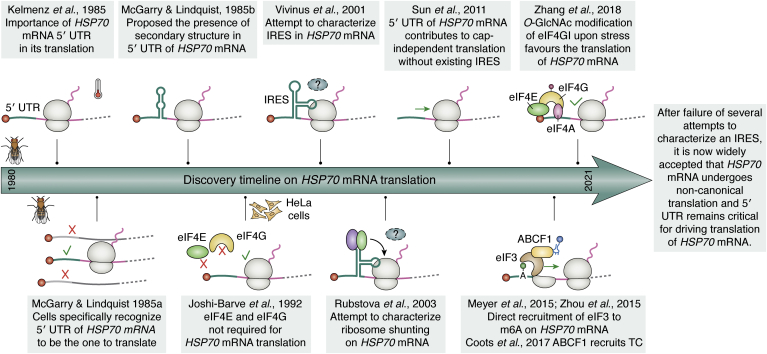
Several inducible HSP mRNAs are highly translated in spite of the general repression of translation during heat stress (148). Among them, the inducible HSP70 is the most synthesized HSP, as shown in 3H-leucine pulse labeling experiments (65). Hence, the HSP70 mRNA should have specific features favoring its translation. In 1985, Klemenz et al. (149) were the first to indicate that the 5′UTR of the HSP70 mRNA is required for its preferential translation during heat stress. McGarry and Lindquist also reported that the 5′ leader sequence of the HSP70 mRNAs is the region detected by D. melanogaster cells to preferentially translate it during heat stress (150). Given that both deletions and insertions within the 5′ UTR rendered HSP70 mRNA untranslated during heat stress and actively translated during recovery from stress, the authors suggested the presence of a secondary structure in the 5′ UTR (150). However, it was not until 1988 that the internal ribosome entry sites (IRESs) were discovered to mediate the translation of picornaviral mRNAs (151, 152), and until 1991 that the first cellular IRES was identified in the mRNA encoding for the immunoglobulin heavy chain binding-protein BiP or GRP78 (153). GRP78 is indeed an HSP70 that localizes and functions in the ER. Unlike the picornavirus mRNAs, the GRP78 mRNAs are capped, and their translation is favored when cap-dependent translation is halted under conditions of stress (149, 154, 155). Studies in HeLa cells also indicated that the factors eIF4E and eIF4G are dispensable for translation of HSP70 mRNA, suggesting a cap-independent mechanism of translation initiation (156). Following these discoveries, several labs have attempted to characterize the IRES structure and trans-regulatory factors required to translate HSP70 mRNA in different organisms (157, 158, 159). However, those attempts were unsuccessful in finding an IRES in the 5′ UTR of HSP70 mRNA and provided early evidence of an HSP70 mRNA cap-independent translation mechanism (160). Overall, these studies indicated that the mammalian HSP70 mRNA translation has reduced dependence on the eIF4F cap-binding complex during stress, HSP70 mRNA is translated in a cap-independent manner upon mTOR inhibitions, and the 5′ UTR is required for HSP70 mRNA translation during heat stress (157, 161) (Fig. 4).
Figure 4.
Milestones on the discovery of HSP70 mRNA translation. Timeline of the discoveries made toward elucidating the translation mechanism of HSP70 mRNA. Since the discovery of internal ribosome entry site (IRES)–mediated cap-independent translation, several studies have attempted to characterize an IRES in 5′ UTR of HSP70 mRNA. While no studies have reported an IRES so far, they have emphasized the significance of the 5′ UTR of HSP70 mRNA. It is now widely accepted that HSP70 mRNA undergoes IRES-independent noncanonical translation. UTR, untranslated region.
Two different translational initiation control mechanisms have been suggested for the mammalian HSP70 mRNA: ribosome shunting (157) and recruitment of eIF3 by N6-methyladenosine (m6A) modification (161, 162). During ribosome shunting, the 40S ribosomal subunit skips a large portion of the HSP70 5′ UTR and shunts to a region proximal to the canonical start codon when the cap-dependent translation is inhibited during heat shock (157, 159). More recently, it was found that HSP70 mRNAs are cotranscriptionally imprinted at adenosine 103 of their 5′UTR by methylation, m6A (161, 162). This methylation supports translation initiation by binding to the initiation complex eIF3, which recruits the ribosome to the mRNA. A follow-up paper concluded that cells activate m6A-mediated translation through the factor ABCF1. ABCF1 serves as an alternative recruiter for the TC to HSP70 mRNA during noncanonical translation upon heat shock (163). Further, heat-stress–mediated O-GlcNACylation of eIF4GI has been reported essential for the translation of HSP70 mRNAs (164). Additionally, the escape of HSP70 mRNA from the shutoff of global protein synthesis was explained by the existence of specialized stress ribosomes. These ribosomes bear the cytoplasmic version of a mitochondrial protein, MRPL18, synthesized upon stress and might facilitate the recruitment of factors involved in translation elongation (165). The main caveat of these publications is the timing at which translation of HSP70 mRNA was studied. They used 4 h of recovery following 1 h of heat stress. At this time point, cap-dependent translation resumes, and SGs are resolved. Hence, they provide solid evidence on the regulation of HSP70 mRNA translated during recovery. Whether the same factors participate in its translation during stress remains to be elucidated (Fig. 4).
A relevant outcome of this research is the suggestion of cotranscriptionally imprinting of the mRNA that provides an advantage for its translation in the cytoplasm (163, 165). Paradigm shifting studies in yeast support the role of the HSE sequence in the promoter of HSP genes in determining the translation of inducible HSP mRNAs in the cytoplasm of glucose-starved yeast (166). We have previously reported that eEF1A1 links HSP70 transcription to translation in mammals, implicating the evolutionary conservation of this “remote control” mechanism of translational regulation (167). Hence, newly synthesized HSP mRNAs might arrive at the cytoplasm equipped for translation. These factors, together with the intrinsic characteristics of the 5′UTR, allow HSP mRNAs to engage in translation at the expense of housekeeping mRNAs. For example, the Ded1 helicase is recruited to condensates upon heat stress, precluding the translation of housekeeping mRNAs with secondary structures and favoring the translation of HSP mRNAs that have little structure in yeast (168). However, the 5′UTR sequence varies among HSP70 inducible genes and species. While D. melanogaster contains mostly AU-rich (70%) sequence and no secondary structure formation, like in yeast (158), the mammalian sequence has higher GC content (63%) which is likely to favor a formation of stable secondary structures. Even though we have not yet put together all the regulatory elements and the cascade of events that lead to the preferential synthesis of the inducible HSP70 during stress, they might differ among species.
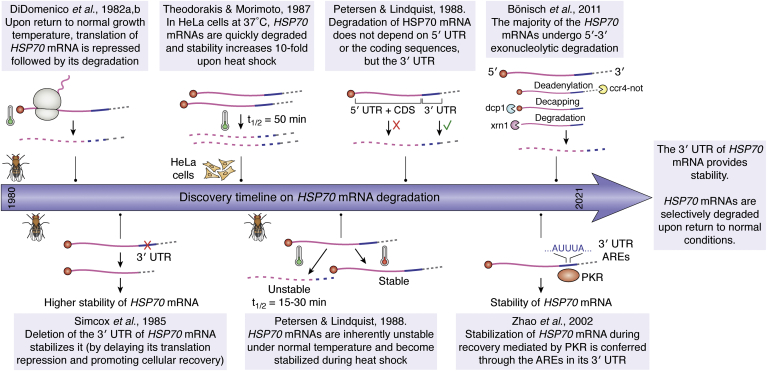
Recovery from stress and the degradation of HSP70 mRNA
The newly synthesized HSP70 allows cells to resume their normal functions gradually by recovering proteostasis through the folding of misfolded proteins, which also prevents their aggregation, and by participating in the disassembly of SGs (169, 170). Accordingly, the synthesis of a nonfunctional HSP70 or inhibition of HSP70 mRNA translation during stress delays the recovery of global translation in D. melanogaster cells (148, 171, 172, 173). However, the synthesis of HSP70 should be repressed when cells return to optimal conditions. The persistent expression of the inducible HSP70 under nonstress conditions causes growth defects in D. melanogaster (174) and promotes malignancy in mammalian cells (175). Hence, the HSP transcription repression is accompanied by the rapid degradation of their mRNAs during recovery (171, 176). HSP70 mRNAs are the first to undergo translational repression and degradation. The timing and rate of its degradation during recovery depends on the severity and duration of heat treatment (176, 177). When cells are returned to optimal conditions following heat shock, they restore global protein synthesis, and constitutive mRNAs engage in translation (176, 177).
In mammalian cells, HSP70 mRNA transitions from being a stable mRNA during heat stress to a short-lived mRNA with a half-life of around 50 min during recovery from heat shock (172, 178, 179, 180). While the instability of HSP70 mRNA is also characteristic of nonstress cells, other stresses like incubation of cells with sodium arsenite or inhibition of protein synthesis initiation with pactamycin also stabilize HSP70 mRNA. Therefore, it has been suggested that a trans-regulator synthesized by the cells under nonstress or recovery conditions could destabilize HSP70 mRNA by binding to its 3′ UTR. Deletion of the HSP70 mRNA 3′UTR or substitution with that of the alcohol dehydrogenase (adh) 3′UTR sequence stabilizes the transcript during recovery from stress (173, 177, 181). In mammalian cells, PKR directly or indirectly, for example through AUF1 protein, associates to the 3′UTR of HSP70 mRNA through its AU-rich elements (AUUUA) that destabilize the transcript during recovery (182). However, AU-rich elements become dispensable for the fast deadenylation of HSP70 transcripts during recovery in D. melanogaster cells, which suggests that RNPs mediating HSP70 mRNA stability might differ among species (183). Like most cellular mRNAs, most HSP70 transcripts undergo a fast deadenylation mediated by the CCR4-NOT complex followed by decapping and degradation by the exonuclease Xrn1 in the 5′-to-3′ direction, as described in D. melanogaster. HSP70 mRNA fragments shorter than the full-length mRNAs were identified by northern blot suggesting that some molecules might undergo degradation after an endonucleolytic cleavage by the exosome in the 3′-to-5′ direction (183) (Fig. 5).
Figure 5.
Milestones on the discovery of HSP70 mRNA degradation. Stress stabilizes the HSP70 mRNA. However, soon after the removal of stress stimulus, the cells rapidly and selectively degrade the HSP70 mRNA. The figure indicates the crucial discoveries made toward elucidating the mechanism of degradation of HSP70 mRNA. Various studies have reported that the 3′ UTR of HSP70 mRNA coordinates its stability or turnover. AREs, AU-rich elements; HSP, heat shock protein; UTR, untranslated region.
How does the cell recognize the HSP70 transcripts as the ones to be degraded? Within minutes of heat shock, hundreds of these mRNAs are synthesized, and when the conditions become optimal, they are selectively degraded with high efficiency. Is there a link between the translational status of the cell and HSP mRNA turnover? Polysome fractionation to study the translational profile of HSP70 mRNA in D. melanogaster and mammalian cells show a fraction of transcripts retained in higher polysomes while a subset of them is translationally inactivated (177). Are both populations degraded by the exact decay mechanism? It might also be possible that RNPs associated with HSP70 mRNA undergo posttranslational modifications, like arginylation, mediated by ATE-1, to regulate the stability of HSP transcripts, providing protection to cells upon heat shock (184).
Proteostasis on specific organisms, cell types, and conditions
While the HSR is a universal survival response to changes in the environment, there are variations in the regulation of the HSR among species and even within cell types of the same organisms. Additionally, the adaptation to abrupt or long-term changes influence the cellular response differently (8, 16, 181, 182, 183).
Variation of the HSR among organisms
The number of genes encoding for HSPs and cochaperones has increased over evolution, probably reflecting the increased number of proteins and complexity of functions undertaken by more evolved cells (185, 186). Multicellular organisms have not only expanded on the HSPs encoded in the genome of unicellular organisms but also adapted them to their proteostasis needs. For example, the mammalian genome lacks the disaggregase HSP104 present in S. cerevisiae (in eubacteria ClpB), which reverses stress-induced protein aggregation (187, 188, 189) and has been suggested to serve as a potential therapeutic agent to disaggregate toxic misfolded aggregates characteristic of neurodegenerative diseases (NDs) (187). While it is unknown why mammalians lack HSP104, in vitro data have shown that protein disaggregation in humans (and other metazoans such as nematodes) relies on the molecular machinery comprised of HSP70, HSP110, and J-proteins (188, 190, 191). These J-proteins are critical in driving HSP70–HSP110–based disaggregase by concomitantly interacting with both substrates and HSP70 partner proteins via single or mixed cooperating J-protein cochaperones of class A and B which relocalize to protein aggregates following heat shock promote specific or broad-range aggregate targeting (188, 190, 191, 192). Overall, the core chaperones of the HSP70 and HSP90 families are similar among species and their organization as constitutive or inducible HSPs depends on their basal expression level (Table 1).
Table 1.
Comparison of HSP genes and ranges of temperatures across different organisms
| Comparison of HSP genes and ranges of heat shock temperatures across different organisms | Yeast 
|
C. elegans
|
Drosophila
|
Mammals 
|
E. coli
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Temperature range of HSR | 30–37 °C (238) DT: 7 °C |
29–35 °C (239) DT: 6 °C |
29–38 °C (35–37 °C) (239) DT: 9 °C (2 °C) |
41–45 °C (240, 241) DT: 4 °C |
37–50 °C DT: 9 °C (13 °C) |
|||||
| Number of HSP genes | 63 (238) | 21 (242) | 87 | >142 (18) | ∼3 (243) | |||||
| Transcription of HSPs | Initiation (244) | Peak (241) | Initiation (239) | Peak (239) | Initiation (245) | Peak (246) | Initiation (247) | Peak (167) | Initiation (248) | Peak (248) |
| 1–5 min at 37 °C | 15 min at 37 °C | Hsp70 and Hsp80: 60 min at 35 °C | Hsp70 and Hsp80: 2 h at 35 °C | Hsp70 4 min at 37 °C | 1–2 min at 37 °C | ∼2.5–12 min at 42 °C | 1 h at 43 °C | Hsp70 (DnaK) 1–4 min at 42 °C | Hsp70 (DnaK) 5–6 min at 42 °C | |
| Translation of HSPs | Temp Initiation (249) | Temp Peak (250) | Temp Initiation (239) | Temp Peak (239) | Temp Initiation (65) | Temp Peak (65) | Temp Initiation (251) | Temp Peak (251) | Temp Initiation (252) | Temp Peak |
| Hsp38, 100, and 90 ∼ 4 min at 36 °C | 10–15 min at 37 °C |
Hsp16: 29 °C Hsp18: 29 °C Hsp19: 35 °C Hsp29: 35 °C Hsp81: 35 °C Hsp70: 29 °C |
Hsp16: 33 °C Hsp18: 33 °C Hsp70: 35 °C |
Hsp23: 33 °C Hsp26: 33 °C Hsp82: 26 oC Hsp70: 26 °C |
Hsp23: 35 °C Hsp26: 35°C Hsp82: 33 °C Hsp70 37 °C |
Hsp70 ∼6 min at 42 °C |
Hsp70 30 min at 42 °C |
Hsp60 (GroEL) and Hsp70 (DnaK): ∼3–5 min at 42 °C |
Hsp60 (188) (GroEL) and Hsp70 (253) (DnaK): 5–10 min at 42 oC |
|
HSP, heat shock protein.
There is a variation in the temperature threshold needed to activate the HSF1 and the HSR across multicellular and unicellular eukaryotic species (193, 194) (Table 1). For example, while D. melanogaster induces the HSR at 30 °C, humans do so at 40 °C. This variability in temperature threshold has been suggested to be based on the environmental temperature of the organism and its capacity to maintain a constant body temperature (195, 196). Indeed, organisms occupying moderate variable thermal environments can modify the constitutive levels of HSPs and adjust their HSR to a higher onset temperature. In contrast, those organisms from a stable or highly variable thermal environment are not as capable of readjusting the levels of their HSPs or onset temperature as they are already meeting their maximum thermal limit.
An interesting example is multicellular organisms lacking the induction of an HSR, like the Antarctic marine invertebrate ciliates and the Antarctic fish of the suborder Notothenioidei (8, 197). The absence of the HSR is suggested to be due to these animals living in a highly stabilized cold environment; thus, they evolved to adapt to this nontransient environment. However, this is the opposite of the Notothenioidei cold-temperate relatives in New Zealand, which have been shown to induce an HSR following heat stress. Another example of an animal with no HSR is the freshwater cnidarian species Hydra oligactis, which is also highly sensitive to minor thermal variations. Its congener, Hydra vulgaris, can tolerate greater thermal ranges because it induces thermal tolerance following induction of HSPs synthesis (8, 197). Despite the lack of expression of inducible HSPs, these organisms express constitutive HSPs. They might have adapted the expression of their constitutive HSPs to help them overcome challenges faced in their environmental niche (8, 198). Thus, constitutive HSPs might play an essential role in overcoming proteostasis challenges without an inducible HSR. Examining the potential coordinated network of constitutive and inducible HSPs in promoting a competent HSR would provide a better understanding of the network of HSPs participating in the adaptation to changes in temperature.
Multicellular endothermal organisms tolerate higher ranges in temperature than unicellular or stenothermal multicellular organisms. However, endothermal organisms required a lower increase over their body temperature to induce the HSR (Table 1). There are unicellular organisms from the Archaeal species that can tolerate extreme temperatures such as those between 50 °C to 70 °C (thermophiles) and 80 °C or higher (hyperthermophiles) before the HSR is induced (199). In addition to the HSR, bacteria rely on a structural liability in response to temperature changes which create biological temperature sensors, such as DNA- and RNA-based environmental temperature sensors. Temperature changes will alter gene expression at transcriptional and posttranscriptional steps via DNA and RNA thermosensors to maintain proteostasis. Furthermore, changes in the secondary or tertiary structure can also be used by “RNA thermometers” in bacteria to regulate the translation efficiency of heat shock mRNAs. In this RNA ‘zipper-like’ thermosensory mechanism, the mRNA will adopt a thermolabile stem-loop structure in the 5′UTR which will either close to block translation at low temperatures or open at high temperatures to favor ribosome binding and translation (3, 10). These RNA thermosensing mechanisms are also important in the translation control of some HSPs in eukaryotic cells when cap-dependent translation initiation is inhibited (3, 10). For example, HSP90 mRNA in D. melanogaster becomes actively translated in response to heat shock but is inefficient in normal growth temperatures. It has been suggested that the HSP90 mRNA translation is substantially activated by heat shock due to the presence of a long stem in the AUG initiation proximal half of the 5′UTR, which serves as a heat-sensitive inhibitory element in the UTR that impedes access to the initiation codon (200). However, in heat stress, the stem undergoes a thermal destabilization, which allows the ribosomal subunits to recognize the region (200). While the expression of HSP90 has been shown to be essential for restoring folding yield when HSP70 levels are high, excess HSP90 (as well as HSP70) produced during heat shock may be detrimental to folding (201). Thus, the presence of a thermosensor is critical in mediating the translation of HSPs that are important for protein folding during heat shock. This preferential heat shock translation occurs in a similar translation mechanism as in bacteria wherein the start codon (AUG) will respond to differences in temperature similarly to bacterial RNA thermosensors. A similar mechanism of translation control has been proposed for HSP70 mRNA in human cells but has yet to be examined (3). These examples suggest that multicellular organisms conserved some of the modes of adaptation to heat stress used by bacteria, but they have changed the regulatory factors sustaining them.
Additionally, endothermal organisms readjust their physiology to increasing temperatures (197). The repeated exposure to elevations in core temperature due to natural environmental heat stress causes heat acclimatization (202). An increase in temperature is associated with a change in the pattern of HSPs expressed is due to proteins being able to adapt their structures to varying temperatures, e.g., altering hydrophobicity, charge, noncovalent interactions, volume, and cooperativity. Indeed, patterns of adaptive variation in the structural and functional properties of proteins from organisms that have adapted to different temperatures have been reported (203, 204). A similar phenomenon has been recently described in the yeast S. Cerevisiae in response to a long-term temperature shift (205). Interestingly, a key feature of long-term temperature adaptation is the disappearance of protein aggregates, whereas acute heat shock induces protein aggregates. Thus, yeast may have adapted to persistent high temperatures by reducing the load of thermolabile proteins and relocating some proteins to minimize protein misfolding/unfolding at high temperatures (205). These findings have important implications in the context of global climate change as the current temperature changes we are experiencing can be considered as long-term temperature adaptation to the constant heat.
Variation of the HSR among cell types
The requirements of cellular proteostasis also vary across cell types in multicellular organisms, which maintain a relatively stable internal environment to sustain proteostasis among specialized tissues and organs (1, 2, 206). These multicellular organisms have highly specialized functions performed by distinct proteomes, in which proteostasis, assisted by a network of core and cell-specific chaperones, becomes challenging (186, 206). There are two hypotheses as to how multicellular organisms maintain proteostasis following an environmental perturbation like temperature. The first is that molecular chaperones are expressed in all cell types to guide folding and prevent misfolding, and thus, they can buffer unexpected folding challenges. This hypothesis requires all cells to invest energy to have a reserve of chaperones for emergencies (1, 197, 205, 206, 207, 208). As described in this review, the constitutive HSPs provide immediate assistance in coping with misfolded proteins upon acute stress (197, 206). Whereas de novo synthesized HSPs favor recovery from stress and fit the cell to overcome subsequent and more detrimental stress stimuli. This phenomenon, known as stress-preconditioning, is used for medical purposes and suggests that cells have a certain buffer of HSPs to handle mild changes in environmental conditions (205, 207, 209, 210, 211, 212).
The second hypothesis states that cells do not store excess chaperones. Instead, the cellular concentration of the chaperones is regulated precisely according to the immediate cellular requirements. Hence, the folding environment in the cell is delicate, with little capacity for a flux of non-native species. This hypothesis requires the HSR to be rapidly tailored to the proteostatic demands of the cell (206). Additionally, cell types and tissues would need to exchange information on the status of internal cellular proteostasis to coordinate proteostasis at the organismal level (2). The second hypothesis is demonstrated in Caenorhabditis elegans, wherein the thermosensory amphid neurons with finger-like ciliated endings (AFDs) detect changes in the ambient temperatures and coordinate the response. This coordinated response involves communication between the different tissues of C. elegans and is regulated by neurons. More specifically, AFDs sense heat shock stress in the environment; these stress signals are then sent via neuroendocrine fashion to tissues such as muscle and intestinal cells to regulate their HSR by activating HSF1 and promoting the induction of HSP70 (C12C8) (2, 213). At the same time, muscle and intestine cells have a transcellular chaperone signaling between nonneuronal tissues that sense local proteotoxic stress and enhance chaperone signaling at a distance by signaling back to the neurons. These AFD neurons along with their postsynaptic cells, AIY interneurons, further regulate the temperature-dependent behavior of these organisms, such as growth and reproduction (206, 213). Similar activation of HSF1 through neuroendocrine signaling from the hypothalamic–pituitary–adrenal axis operates in rats (214).
As neurons play an important role in this organismal-level coordination in eukaryotes, it is important to mention the current literature on neuronal proteostasis. Neurons are highly polarized cells that have the capacity to tune their proteome locally, at axons, dendrites, and synapses, through the regulation of local protein synthesis, degradation, and posttranslational modifications (215). However, it is not fully understood how proteostasis is sustained in different neuronal subcompartments under heat stress conditions. Rodent hippocampal and motor neurons exhibit a lower HSR activation than nonneuronal cells (206, 216). This impaired HSR could make neurons vulnerable to the toxic accumulation of misfolded proteins that underlie age-related NDs (216, 217, 218, 219). Hence, a potential therapeutic strategy for NDs is to promote the activation of the HSR in neurons (220, 221, 222, 223). The overexpression of HSP70 in a mouse model of Alzheimer’s Disease exerted cytoprotective roles and ameliorated physiological and behavioral deficits (220, 221, 222, 223). Promoting the activation of HSF1 has long been considered a promising treatment for NDs. However, the threshold to activate HSF1 in motor neurons is higher than in somatic nonneuronal cells (216). The chromatin environment of HSP genes in neurons does not favor the binding of HSF1, and the treatment with histone deacetylases can enhance the transcriptional activity of HSF1 in motor neurons undergoing specific stresses (224). Hence, neurons might have been wired differently to handle proteostasis challenges and rely on other quality control mechanisms, like ubiquitin-proteasome system and autophagy, to sustain a healthy and functional proteome (218, 225, 226, 227, 228, 229).
Conclusion and perspectives
The induction of HSPs represents the first line of defense toward an increase in protein unfolding (230). To date, the regulation of HSPs expression has been mostly studied by ensemble measurements in cultured cell lines and yeast. This extensive research has provided detailed information on the kinetics of HSPs transcription and mRNA translation and degradation and has identified regulatory factors involved in each step of the life cycle of HSP mRNAs. In the last decade, single-cell microscopy approaches have granted the spatial resolution needed to investigate changes in the localization of HSP loci upon stress (50) as well as the subcellular localization of newly synthesized mRNAs (166, 167). These latest studies indicate that the fate of HSP mRNAs is decided cotranscriptionally. Therefore, a finely tuned communication between the nucleus and the cytoplasm under stress conditions could enable cells to identify HSP mRNAs as the ones to be translated. These results also suggest that each step in the life cycle of HSP mRNAs highly influences the next one. Given that hundreds of HSP mRNAs molecules are rapidly synthesized, translated, and degraded, the use of single-molecule fluorescence microscopy techniques to detect single mRNAs and de novo protein synthesis will mind the gap of our knowledge in the impact of HSP mRNAs translation on decay and the localization of mRNA degradation (231, 232, 233).
Activation of the HSR requires a mechanism to sense the damage and gene expression reprogramming to prioritize the expression of HSPs. The transcription factor HSF1 directs the upregulation of HSPs transcription. Activation of HSF1 occurs in all eukaryotes under a wide range of stresses (31, 230, 234). Together with HSF1 activation, cells attempt to minimize protein unfolding by blocking general translation elongation (230, 235). It is possible that besides the increased load of unfolding proteins, this general ribosome stalling signals to HSF1 by a still unknown mechanism (166). Identifying the mechanisms that act to orchestrate a competent HSR will provide the means to interrogate neurons in their stunt HSR and relate it with the neuronal vulnerability to accumulate misfolded proteins. In this case, using pathophysiological conditions relevant to the neuronal activity will provide a better understanding of the neuronal response to proteostasis challenges. Expanding the pioneering research done in C. elegans to other multicellular organisms will provide the means to integrate the response of the nervous system into the organism effort to sustain proteostasis (213). A comprehensive analysis of the cellular response to stress in the context of the organism will open new windows to study the etiology of diseases derived from the loss of protein homeostasis, like cancer and neurodegeneration.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
Author contribution
L. R. A. B. and S. J. T. conceptualization; C. A., L. R. A. B, and S. J. T. writing-original draft; M. V. supervision; M. V. writing-reviewing and editing.
Funding and additional information
This work is supported by NSERC grant RGPIN-2019 to 04767 to M. V., FRQS through CRBS fellowship to L. R. A. B., Vanier Canada Graduate Scholarship CGV 1757 to S. J. T., and FRQS postdoctoral fellowship 300232 to C. A.
Edited by Ursula Jakob
References
- 1.Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Oosten-Hawle P., Morimoto R.I. Organismal proteostasis: Role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev. 2014;28:1533–1543. doi: 10.1101/gad.241125.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Fuente M., Valera S., Martínez-Guitarte J.L. ncRNAs and thermoregulation: A view in prokaryotes and eukaryotes. FEBS Lett. 2012;586:4061–4069. doi: 10.1016/j.febslet.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Protter D.S.W., Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pohl C., Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818–822. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 6.Pomatto L.C.D., Davies K.J.A. The role of declining adaptive homeostasis in ageing. J. Physiol. 2017;595:7275–7309. doi: 10.1113/JP275072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasuda S., Tsuchiya H., Kaiho A., Guo Q., Ikeuchi K., Endo A., Arai N., Ohtake F., Murata S., Inada T., Baumeister W., Fernández-Busnadiego R., Tanaka K., Saeki Y. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature. 2020;578:296–300. doi: 10.1038/s41586-020-1982-9. [DOI] [PubMed] [Google Scholar]
- 8.Tomanek L. The importance of physiological limits in determining biogeographical range shifts due to global climate change: The heat-shock response. Physiol. Biochem. Zool. 2008;81:709–717. doi: 10.1086/590163. [DOI] [PubMed] [Google Scholar]
- 9.Kassahn K.S., Crozier R.H., Pörtner H.O., Caley M.J. Animal performance and stress: Responses and tolerance limits at different levels of biological organisation. Biol. Rev. Camb. Philos. Soc. 2009;84:277–292. doi: 10.1111/j.1469-185X.2008.00073.x. [DOI] [PubMed] [Google Scholar]
- 10.Sengupta P., Garrity P. Sensing temperature. Curr. Biol. 2013;23:R304–R307. doi: 10.1016/j.cub.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somero G.N. The cellular stress response and temperature: Function, regulation, and evolution. J. Exp. Zool A Ecol. Integr. Physiol. 2020;333:379–397. doi: 10.1002/jez.2344. [DOI] [PubMed] [Google Scholar]
- 12.Parsell D.A., Lindquist S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 13.Kültz D. Molecular and evolutionary basis OF the cellular stress response. Annu. Rev. Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- 14.Daugaard M., Rohde M., Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 15.Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019;20:665–680. doi: 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- 16.Morimoto R.I. The heat shock response: Systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Le W.D. Autophagy and ubiquitin-proteasome system. Adv. Exp. Med. Biol. 2019;1206:527–550. doi: 10.1007/978-981-15-0602-4_25. [DOI] [PubMed] [Google Scholar]
- 18.Jayaraj G.G., Hipp M.S., Hartl F.U. Functional modules of the proteostasis network. Cold Spring Harb. Perspect. Biol. 2020;12 doi: 10.1101/cshperspect.a033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampinga H.H., Hageman J., Vos M.J., Kubota H., Tanguay R.M., Bruford E.A., Cheetham M.E., Chen B., Hightower L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abisambra J.F., Blair L.J., Hill S.E., Jones J.R., Kraft C., Rogers J., Koren J., Jinwal U.K., Lawson L., Johnson A.G., Wilcock D., O'Leary J.C., Jansen-West K., Muschol M., Golde T.E., et al. Phosphorylation dynamics regulate Hsp27-mediated rescue of neuronal plasticity deficits in tau transgenic mice. J. Neurosci. 2010;30:15374–15382. doi: 10.1523/JNEUROSCI.3155-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamerdinger M., Hajieva P., Kaya A.M., Wolfrum U., Hartl F.U., Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P., Ambasta R.K., Veereshwarayya V., Rosen K.M., Kosik K.S., Band H., Mestril R., Patterson C., Querfurth H.W. CHIP and HSPs interact with beta-APP in a proteasome-dependent manner and influence Abeta metabolism. Hum. Mol. Genet. 2007;16:848–864. doi: 10.1093/hmg/ddm030. [DOI] [PubMed] [Google Scholar]
- 23.Lindquist S. The heat-shock response. Annu. Rev. Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 24.Lindquist S., Craig E.A. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q., Liang C., Zhou L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020;29:378–390. doi: 10.1002/pro.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A., De Lucia M., McGowan E., Lewis J., Prihar G., Kim J., Dillmann W.H., Browne S.E., Hall A., Voellmy R., et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 27.Ritossa F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia. 1962;18:571–573. [Google Scholar]
- 28.Sørensen J.G., Kristensen T.N., Loeschcke V. The evolutionary and ecological role of heat shock proteins: Heat shock proteins. Ecol. Lett. 2003;6:1025–1037. [Google Scholar]
- 29.Brocchieri L., Conway de Macario E., Macario A.J. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008;8:19. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petesch S.J., Lis J.T. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anckar J., Sistonen L. Regulation of HSF1 function in the heat stress response: Implications in aging and disease. Annu. Rev. Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 32.Vihervaara A., Sistonen L. HSF1 at a glance. J. Cell Sci. 2014;127:261–266. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 33.Björk J.K., Sistonen L. Regulation of the members of the mammalian heat shock factor family. FEBS J. 2010;277:4126–4139. doi: 10.1111/j.1742-4658.2010.07828.x. [DOI] [PubMed] [Google Scholar]
- 34.Gomez-Pastor R., Burchfiel E.T., Thiele D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guettouche T., Boellmann F., Lane W.S., Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakai A. Molecular basis of HSF regulation. Nat. Struct. Mol. Biol. 2016;23:93–95. doi: 10.1038/nsmb.3165. [DOI] [PubMed] [Google Scholar]
- 37.Boellmann F., Guettouche T., Guo Y., Fenna M., Mnayer L., Voellmy R. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4100–4105. doi: 10.1073/pnas.0304768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hietakangas V., Anckar J., Blomster H.A., Fujimoto M., Palvimo J.J., Nakai A., Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U. S. A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westerheide S.D., Anckar J., Stevens S.M., Sistonen L., Morimoto R.I. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krakowiak J., Zheng X., Patel N., Feder Z.A., Anandhakumar J., Valerius K., Gross D.S., Khalil A.S., Pincus D. Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. Elife. 2018;7 doi: 10.7554/eLife.31668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng X., Krakowiak J., Patel N., Beyzavi A., Ezike J., Khalil A.S., Pincus D. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. Elife. 2016;5 doi: 10.7554/eLife.18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shamovsky I., Ivannikov M., Kandel E.S., Gershon D., Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y., Chen J., Yu J., Yang G., Temple E., Harbinski F., Gao H., Wilson C., Pagliarini R., Zhou W. Identification of mixed lineage leukemia 1(MLL1) protein as a coactivator of heat shock factor 1(HSF1) protein in response to heat shock protein 90 (HSP90) inhibition. J. Biol. Chem. 2014;289:18914–18927. doi: 10.1074/jbc.M114.574053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mason P.B., Lis J.T. Cooperative and competitive protein interactions at the hsp70 promoter. J. Biol. Chem. 1997;272:33227–33233. doi: 10.1074/jbc.272.52.33227. [DOI] [PubMed] [Google Scholar]
- 46.Park J.M., Werner J., Kim J.M., Lis J.T., Kim Y.J. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- 47.Takii R., Fujimoto M., Matsumoto M., Srivastava P., Katiyar A., Nakayama K.I., Nakai A. The pericentromeric protein shugoshin 2 cooperates with HSF1 in heat shock response and RNA Pol II recruitment. EMBO J. 2019;38 doi: 10.15252/embj.2019102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lis J.T., Mason P., Peng J., Price D.H., Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall N.F., Peng J., Xie Z., Price D.H. Control of RNA polymerase II elongation potential by a novel Carboxyl-terminal domain kinase∗. J. Biol. Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 50.Khanna N., Hu Y., Belmont A.S. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. 2014;24:1138–1144. doi: 10.1016/j.cub.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vera M., Singer R.H. Gene regulation: The HSP70 gene jumps when shocked. Curr. Biol. 2014;24:R396–R398. doi: 10.1016/j.cub.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chowdhary S., Kainth A.S., Gross D.S. Heat shock protein genes undergo dynamic alteration in their three-dimensional structure and genome organization in response to thermal stress. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00292-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vihervaara A., Duarte F.M., Lis J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018;19:385–397. doi: 10.1038/s41576-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mueller B., Mieczkowski J., Kundu S., Wang P., Sadreyev R., Tolstorukov M.Y., Kingston R.E. Widespread changes in nucleosome accessibility without changes in nucleosome occupancy during a rapid transcriptional induction. Genes Dev. 2017;31:451–462. doi: 10.1101/gad.293118.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vihervaara A., Mahat D.B., Guertin M.J., Chu T., Danko C.G., Lis J.T., Sistonen L. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat. Commun. 2017;8:255. doi: 10.1038/s41467-017-00151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray J., Munn P.R., Vihervaara A., Lewis J.J., Ozer A., Danko C.G., Lis J.T. Chromatin conformation remains stable upon extensive transcriptional changes driven by heat shock. Proc. Natl. Acad. Sci. U. S. A. 2019;116:19431–19439. doi: 10.1073/pnas.1901244116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L., Lyu X., Hou C., Takenaka N., Nguyen H.Q., Ong C.T., Cubeñas-Potts C., Hu M., Lei E.P., Bosco G., Qin Z.S., Corces V.G. Widespread rearrangement of 3D chromatin organization underlies Polycomb-mediated stress-induced silencing. Mol. Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allen T.A., Von Kaenel S., Goodrich J.A., Kugel J.F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 59.Espinoza C.A., Allen T.A., Hieb A.R., Kugel J.F., Goodrich J.A. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 60.Mariner P.D., Walters R.D., Espinoza C.A., Drullinger L.F., Wagner S.D., Kugel J.F., Goodrich J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Ji Q., Zong X., Mao Y., Qian S.-B. A heat shock–responsive lncRNA Heat acts as a HSF1-directed transcriptional brake via m 6 A modification. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2102175118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cugusi S., Mitter R., Kelly G.P., Walker J., Han Z., Pisano P., Wierer M., Stewart A., Svejstrup J.Q. Heat shock induces premature transcript termination and reconfigures the human transcriptome. Mol. Cell. 2022 doi: 10.1016/j.molcel.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tutucci E., Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat. Rev. Mol. Cell Biol. 2011;12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- 64.Zander G., Hackmann A., Bender L., Becker D., Lingner T., Salinas G., Krebber H. mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature. 2016;540:593–596. doi: 10.1038/nature20572. [DOI] [PubMed] [Google Scholar]
- 65.Lindquist S. Varying patterns of protein synthesis in Drosophila during heat shock: Implications for regulation. Dev. Biol. 1980;77:463–479. doi: 10.1016/0012-1606(80)90488-1. [DOI] [PubMed] [Google Scholar]
- 66.Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 67.Weissbach H., Brot N. The role of protein factors in the biosynthesis of proteins. Cell. 1974;2:137–143. doi: 10.1016/0092-8674(74)90088-9. [DOI] [PubMed] [Google Scholar]
- 68.Bresson S., Shchepachev V., Spanos C., Turowski T.W., Rappsilber J., Tollervey D. Stress-induced translation inhibition through rapid displacement of scanning initiation factors. Mol. Cell. 2020;80:470–484.e8. doi: 10.1016/j.molcel.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alluri R.K., Li Z., McCrae K.R. Stress granule-mediated oxidized RNA decay in P-body: Hypothetical role of ADAR1, Tudor-SN, and STAU1. Front. Mol. Biosci. 2021;8:672988. doi: 10.3389/fmolb.2021.672988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hubstenberger A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M., Daunesse M., Bertrand E., Pierron G., Mozziconacci J., Kress M., et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017;68:144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 72.Aitken C.E., Lorsch J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 73.Hinnebusch A.G., Lorsch J.R. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Preiss T., W Hentze M. Starting the protein synthesis machine: Eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 75.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taniuchi S., Miyake M., Tsugawa K., Oyadomari M., Oyadomari S. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci. Rep. 2016;6:32886. doi: 10.1038/srep32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonanou-Tzedaki S.A., Smith K.E., Sheeran B.A., Arnstein H.R. The high-temperature inactivation of rabbit reticulocyte lysates by a haemin-independent mechanism. Eur. J. Biochem. 1978;84:591–600. doi: 10.1111/j.1432-1033.1978.tb12202.x. [DOI] [PubMed] [Google Scholar]
- 78.Chen J.J. Translational control by heme-regulated eIF2α kinase during erythropoiesis. Curr. Opin. Hematol. 2014;21:172–178. doi: 10.1097/MOH.0000000000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chung J., Chen C., Paw B.H. Heme metabolism and erythropoiesis. Curr. Opin. Hematol. 2012;19:156–162. doi: 10.1097/MOH.0b013e328351c48b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costa-Mattioli M., Walter P. The integrated stress response: From mechanism to disease. Science. 2020;368 doi: 10.1126/science.aat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suragani R.N., Zachariah R.S., Velazquez J.G., Liu S., Sun C.W., Townes T.M., Chen J.J. Heme-regulated eIF2α kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood. 2012;119:5276–5284. doi: 10.1182/blood-2011-10-388132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farabaugh K.T., Majumder M., Guan B.J., Jobava R., Wu J., Krokowski D., Gao X.H., Schuster A., Longworth M., Chan E.D., Bianchi M., Dey M., Koromilas A.E., Ramakrishnan P., Hatzoglou M. Protein kinase R mediates the inflammatory response induced by hyperosmotic stress. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00521-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimball S.R. Regulation of translation initiation by amino acids in eukaryotic cells. Prog. Mol. Subcell Biol. 2001;26:155–184. doi: 10.1007/978-3-642-56688-2_6. [DOI] [PubMed] [Google Scholar]
- 84.Deng J., Harding H.P., Raught B., Gingras A.C., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- 85.Jiang H.Y., Wek R.C. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem. J. 2005;385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gale M., Tan S.L., Katze M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ron D. Translational control in the endoplasmic reticulum stress response. J. Clin. Invest. 2002;110:1383–1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang R., McGrath B.C., Kopp R.F., Roe M.W., Tang X., Chen G., Cavener D.R. Insulin secretion and Ca2+ dynamics in β-cells are regulated by PERK (EIF2AK3) in concert with calcineurin ∗. J. Biol. Chem. 2013;288:33824–33836. doi: 10.1074/jbc.M113.503664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kubarenko A.V., Sergiev P.V., Rodnina M.V. [GTPases of translational apparatus. Mol. Biol. (Mosk) 2005;39:746–761. [PubMed] [Google Scholar]
- 90.Kimball S.R., Fabian J.R., Pavitt G.D., Hinnebusch A.G., Jefferson L.S. Regulation of Guanine Nucleotide Exchange through Phosphorylation of Eukaryotic Initiation Factor eIF2α. J. Biol. Chem. 1998;273:12841–12845. doi: 10.1074/jbc.273.21.12841. [DOI] [PubMed] [Google Scholar]
- 91.Yoshizawa F., Kimball S.R., Jefferson L.S. Modulation of translation initiation in rat skeletal muscle and liver in response to food intake. Biochem. Biophys. Res. Commun. 1997;240:825–831. doi: 10.1006/bbrc.1997.7652. [DOI] [PubMed] [Google Scholar]
- 92.Aktas B.H., Chen T. In: Translation and its Regulation in Cancer Biology and Medicine [Internet] Parsyan A., editor. Springer Netherlands; Dordrecht: 2014. The eIF2 Complex and eIF2α; pp. 195–221. [Google Scholar]
- 93.Pelletier J., Sonenberg N. The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem. 2019;88:307–335. doi: 10.1146/annurev-biochem-013118-111042. [DOI] [PubMed] [Google Scholar]
- 94.Gingras A.-C., Raught B., Sonenberg N. In: TOR: Target of Rapamycin [Internet] Thomas G., Sabatini D.M., Hall M.N., editors. Springer; Berlin, Heidelberg: 2004. mTOR signaling to translation; pp. 169–197. (Current Topics in Microbiology and Immunology) [Google Scholar]
- 95.Korneeva N.L., Song A., Gram H., Edens M.A., Rhoads R.E. Inhibition of mitogen-activated protein kinase (MAPK)-interacting kinase (MNK) preferentially affects translation of mRNAs containing both a 5′-terminal cap and hairpin∗. J. Biol. Chem. 2016;291:3455–3467. doi: 10.1074/jbc.M115.694190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma X.M., Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 97.Ruoff R., Katsara O., Kolupaeva V. Cell type-specific control of protein synthesis and proliferation by FGF-dependent signaling to the translation repressor 4E-BP. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7545–7550. doi: 10.1073/pnas.1605451113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alain T., Morita M., Fonseca B.D., Yanagiya A., Siddiqui N., Bhat M., Zammit D., Marcus V., Metrakos P., Voyer L.A., Gandin V., Liu Y., Topisirovic I., Sonenberg N. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res. 2012;72:6468–6476. doi: 10.1158/0008-5472.CAN-12-2395. [DOI] [PubMed] [Google Scholar]
- 99.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teleman A.A., Chen Y.W., Cohen S.M. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vries R.G., Flynn A., Patel J.C., Wang X., Denton R.M., Proud C.G. Heat shock increases the association of binding protein-1 with initiation factor 4E ∗. J. Biol. Chem. 1997;272:32779–32784. doi: 10.1074/jbc.272.52.32779. [DOI] [PubMed] [Google Scholar]
- 102.Cuesta R., Laroia G., Schneider R.J. Chaperone Hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- 103.Dever T.E., Dinman J.D., Green R. Translation elongation and recoding in eukaryotes. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dever T.E., Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sokabe M., Fraser C.S. Toward a kinetic understanding of eukaryotic translation. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X., Li W., Williams M., Terada N., Alessi D.R., Proud C.G. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carlberg U., Nilsson A., Nygård O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990;191:639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- 108.Dumont-Miscopein A., Lavergne J.P., Guillot D., Sontag B., Reboud J.P. Interaction of phosphorylated elongation factor EF-2 with nucleotides and ribosomes. FEBS Lett. 1994;356:283–286. doi: 10.1016/0014-5793(94)01272-5. [DOI] [PubMed] [Google Scholar]
- 109.Ryazanov A.G., Shestakova E.A., Natapov P.G. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- 110.Ryazanov A.G., Davydova E.K. Mechanism of elongation factor 2 (EF-2) inactivation upon phosphorylation. Phosphorylated EF-2 is unable to catalyze translocation. FEBS Lett. 1989;251:187–190. doi: 10.1016/0014-5793(89)81452-8. [DOI] [PubMed] [Google Scholar]
- 111.Suzuki K., Monteggia L.M. In: Duman R.S., Krystal J.H., editors. Vol. 89. Academic Press; Cambridge, MA: 2020. The role of eEF2 kinase in the rapid antidepressant actions of ketamine; pp. 79–99. (Advances in Pharmacology [Internet]). Rapid Acting Antidepressants. [DOI] [PubMed] [Google Scholar]
- 112.Browne G.J., Proud C.G. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol. Cell Biol. 2004;24:2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Knebel A., Morrice N., Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu R., Proud C.G. Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol. Sin. 2016;37:285–294. doi: 10.1038/aps.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beckmann R.P., Mizzen L.E., Welch W.J. Interaction of hsp 70 with newly synthesized proteins: Implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- 116.Nelson R.J., Ziegelhoffer T., Nicolet C., Werner-Washburne M., Craig E.A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 117.Liu B., Han Y., Qian S.B. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R., Parker R. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell. 2017;68:808–820.e5. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Matheny T., Rao B.S., Parker R. Transcriptome-wide comparison of stress granules and P-bodies reveals that translation plays a major role in RNA partitioning. Mol. Cell Biol. 2019;39:e00313–e00319. doi: 10.1128/MCB.00313-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vandelli A., Cid Samper F., Torrent Burgas M., Sanchez de Groot N., Tartaglia G.G. The interplay between disordered regions in RNAs and proteins modulates interactions within stress granules and processing bodies. J. Mol. Biol. 2022;434:167159. doi: 10.1016/j.jmb.2021.167159. [DOI] [PubMed] [Google Scholar]
- 122.Franzmann T.M., Alberti S. Protein phase separation as a stress survival strategy. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Luo Y., Na Z., Slavoff S.A. P-Bodies: Composition, properties, and functions. Biochemistry. 2018;57:2424–2431. doi: 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Standart N., Weil D. P-Bodies: Cytosolic droplets for coordinated mRNA storage. Trends Genet. 2018;34:612–626. doi: 10.1016/j.tig.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 126.Riback J.A., Katanski C.D., Kear-Scott J.L., Pilipenko E.V., Rojek A.E., Sosnick T.R., Drummond D.A. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040.e19. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Teixeira D., Sheth U., Valencia-sanchez M.A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Franks T.M., Lykke-Andersen J. The control of mRNA decapping and P-Body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 131.Anderson P., Kedersha N. Stressful initiations. J. Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 132.Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins Tia-1 and tiar link the phosphorylation of Eif-2α to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dang Y., Kedersha N., Low W.-K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 134.Low W.-K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N.S., Merrick W.C., Romo D., Liu J.O. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 135.Mazroui R., Sukarieh R., Bordeleau M.E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mokas S., Mills J.R., Garreau C., Fournier M.J., Robert F., Arya P., Kaufman R.J., Pelletier J., Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell. 2009;20:2673–2683. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tauber D., Tauber G., Khong A., Van Treeck B., Pelletier J., Parker R. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell. 2020;180:411–426.e16. doi: 10.1016/j.cell.2019.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kimball S.R., Horetsky R.L., Ron D., Jefferson L.S., Harding H.P. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 140.Adivarahan S., Livingston N., Nicholson B., Rahman S., Wu B., Rissland O.S., Zenklusen D. Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol. Cell. 2018;72:727–738.e5. doi: 10.1016/j.molcel.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Khong A., Parker R. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol. 2018;217:4124–4140. doi: 10.1083/jcb.201806183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bley N., Lederer M., Pfalz B., Reinke C., Fuchs T., Glaß M., Möller B., Hüttelmaier S. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 2015;43:e26. doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moon S.L., Morisaki T., Khong A., Lyon K., Parker R., Stasevich T.J. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 2019;21:162–168. doi: 10.1038/s41556-018-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jobava R., Mao Y., Guan B.J., Hu D., Krokowski D., Chen C.W., Shu X.E., Chukwurah E., Wu J., Gao Z., Zagore L.L., Merrick W.C., Trifunovic A., Hsieh A.C., Valadkhan S., et al. Adaptive translational pausing is a hallmark of the cellular response to severe environmental stress. Mol. Cell. 2021;81:4191–4208.e8. doi: 10.1016/j.molcel.2021.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mateju D., Eichenberger B., Voigt F., Eglinger J., Roth G., Chao J.A. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell. 2020;183:1801–1812.e13. doi: 10.1016/j.cell.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 147.Kedersha N., Anderson P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 148.Yost H.J., Petersen R.B., Lindquist S. RNA metabolism: Strategies for regulation in the heat shock response. Trends Genet. 1990;6:223–227. doi: 10.1016/0168-9525(90)90183-7. [DOI] [PubMed] [Google Scholar]
- 149.Klemenz R., Hultmark D., Gehring W.J. Selective translation of heat shock mRNA in Drosophila melanogaster depends on sequence information in the leader. EMBO J. 1985;4:2053–2060. doi: 10.1002/j.1460-2075.1985.tb03891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McGarry T.J., Lindquist S. The preferential translation of Drosophila hsp70 mRNA requires sequences in the untranslated leader. Cell. 1985;42:903–911. doi: 10.1016/0092-8674(85)90286-7. [DOI] [PubMed] [Google Scholar]
- 151.Jang S.K., Kräusslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 153.Macejak D.G., Sarnow P. Internal initiation of translation mediated by the 5’ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 154.Bonneau A.M., Sonenberg N. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem. 1987;262:11134–11139. [PubMed] [Google Scholar]
- 155.Huang J.T., Schneider R.J. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell. 1991;65:271–280. doi: 10.1016/0092-8674(91)90161-q. [DOI] [PubMed] [Google Scholar]
- 156.Joshi-Barve S., De Benedetti A., Rhoads R.E. Preferential translation of heat shock mRNAs in HeLa cells deficient in protein synthesis initiation factors eIF-4E and eIF-4 gamma. J. Biol. Chem. 1992;267:21038–21043. [PubMed] [Google Scholar]
- 157.Rubtsova M.P., Sizova D.V., Dmitriev S.E., Ivanov D.S., Prassolov V.S., Shatsky I.N. Distinctive properties of the 5′-untranslated region of human Hsp70 mRNA∗. J. Biol. Chem. 2003;278:22350–22356. doi: 10.1074/jbc.M303213200. [DOI] [PubMed] [Google Scholar]
- 158.Vivinus S., Baulande S., van Zanten M., Campbell F., Topley P., Ellis J.H., Dessen P., Coste H. An element within the 5′ untranslated region of human Hsp70 mRNA which acts as a general enhancer of mRNA translation. Eur. J. Biochem. 2001;268:1908–1917. doi: 10.1046/j.1432-1327.2001.02064.x. [DOI] [PubMed] [Google Scholar]
- 159.Yueh A., Schneider R.J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 160.Sun J., Conn C.S., Han Y., Yeung V., Qian S.B. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J. Biol. Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Coots R.A., Liu X.M., Mao Y., Dong L., Zhou J., Wan J., Zhang X., Qian S.B. m6A Facilitates eIF4F-Independent mRNA Translation. Mol. Cell. 2017;68:504–514.e7. doi: 10.1016/j.molcel.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang X., Shu X.E., Qian S.B. O-GlcNAc modification of eIF4GI acts as a translational switch in heat shock response. Nat. Chem. Biol. 2018;14:909–916. doi: 10.1038/s41589-018-0120-6. [DOI] [PubMed] [Google Scholar]
- 165.Zhang X., Gao X., Coots R.A., Conn C.S., Liu B., Qian S.B. Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat. Struct. Mol. Biol. 2015;22:404–410. doi: 10.1038/nsmb.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zid B.M., O’Shea E.K. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014;514:117–121. doi: 10.1038/nature13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vera M., Pani B., Griffiths L.A., Muchardt C., Abbott C.M., Singer R.H., Nudler E. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. Elife. 2014;3 doi: 10.7554/eLife.03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Iserman C., Desroches Altamirano C., Jegers C., Friedrich U., Zarin T., Fritsch A.W., Mittasch M., Domingues A., Hersemann L., Jahnel M., Richter D., Guenther U.P., Hentze M.W., Moses A.M., Hyman A.A., et al. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell. 2020;181:818–831.e19. doi: 10.1016/j.cell.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 2006;6:160–170. doi: 10.1111/j.1567-1364.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- 170.Buchan J.R., Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.DiDomenico B.J., Bugaisky G.E., Lindquist S. The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell. 1982;31:593–603. doi: 10.1016/0092-8674(82)90315-4. [DOI] [PubMed] [Google Scholar]
- 172.Petersen R., Lindquist S. The Drosophila hsp70 message is rapidly degraded at normal temperatures and stabilized by heat shock. Gene. 1988;72:161–168. doi: 10.1016/0378-1119(88)90138-2. [DOI] [PubMed] [Google Scholar]
- 173.Simcox A.A., Cheney C.M., Hoffman E.P., Shearn A. A deletion of the 3’ end of the Drosophila melanogaster hsp70 gene increases stability of mutant mRNA during recovery from heat shock. Mol. Cell Biol. 1985;5:3397–3402. doi: 10.1128/mcb.5.12.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Feder J.H., Rossi J.M., Solomon J., Solomon N., Lindquist S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- 175.Kumar S., Stokes J., Singh U.P., Scissum Gunn K., Acharya A., Manne U., Mishra M. Targeting Hsp70: A possible therapy for cancer. Cancer Lett. 2016;374:156–166. doi: 10.1016/j.canlet.2016.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.DiDomenico B.J., Bugaisky G.E., Lindquist S. Heat shock and recovery are mediated by different translational mechanisms. Proc. Natl. Acad. Sci. U. S. A. 1982;79:6181–6185. doi: 10.1073/pnas.79.20.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Petersen R.B., Lindquist S. In: Post-transcriptional Control of Gene Expression. McCarthy J.E.G., Tuite M.F., editors. Springer; Berlin, Heidelberg: 1990. Differential mRNA stability: A regulatory strategy for Hsp70 synthesis; pp. 83–91. (NATO ASI Series) [Google Scholar]
- 178.Bunch T.A., Grinblat Y., Goldstein L.S. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mokdad R., Debec A., Wegnez M. Metallothionein genes in Drosophila melanogaster constitute a dual system. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2658–2662. doi: 10.1073/pnas.84.9.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Theodorakis N.G., Morimoto R.I. Posttranscriptional regulation of hsp70 expression in human cells: Effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol. Cell Biol. 1987;7:4357–4368. doi: 10.1128/mcb.7.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Petersen R.B., Lindquist S. Regulation of HSP70 synthesis by messenger RNA degradation. Cell Regul. 1989;1:135–149. doi: 10.1091/mbc.1.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhao M., Tang D., Lechpammer S., Hoffman A., Asea A., Stevenson M.A., Calderwood S.K. Double-stranded RNA-dependent protein kinase (pkr) is essential for thermotolerance, accumulation of HSP70, and stabilization of ARE-containing HSP70 mRNA during stress. J. Biol. Chem. 2002;277:44539–44547. doi: 10.1074/jbc.M208408200. [DOI] [PubMed] [Google Scholar]
- 183.Bönisch C., Temme C., Moritz B., Wahle E. Degradation of hsp70 and other mRNAs in Drosophila via the 5' 3' pathway and its regulation by heat shock. J. Biol. Chem. 2007;282:21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- 184.Deka K., Singh A., Chakraborty S., Mukhopadhyay R., Saha S. Protein arginylation regulates cellular stress response by stabilizing HSP70 and HSP40 transcripts. Cell Death Discov. 2016;2:16074. doi: 10.1038/cddiscovery.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rebeaud M.E., Mallik S., Goloubinoff P., Tawfik D.S. On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2020885118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shemesh N., Jubran J., Dror S., Simonovsky E., Basha O., Argov C., Hekselman I., Abu-Qarn M., Vinogradov E., Mauer O., Tiago T., Carra S., Ben-Zvi A., Yeger-Lotem E. The landscape of molecular chaperones across human tissues reveals a layered architecture of core and variable chaperones. Nat. Commun. 2021;12:2180. doi: 10.1038/s41467-021-22369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sweeny E.A., Shorter J. Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J. Mol. Biol. 2016;428:1870–1885. doi: 10.1016/j.jmb.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol. Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 189.Harrison A.F., Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 2017;474:1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kirstein J., Arnsburg K., Scior A., Szlachcic A., Guilbride D.L., Morimoto R.I., Bukau B., Nillegoda N.B. In vivo properties of the disaggregase function of J-proteins and Hsc70 in Caenorhabditis elegans stress and aging. Aging Cell. 2017;16:1414–1424. doi: 10.1111/acel.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nillegoda N.B., Kirstein J., Szlachcic A., Berynskyy M., Stank A., Stengel F., Arnsburg K., Gao X., Scior A., Aebersold R., Guilbride D.L., Wade R.C., Morimoto R.I., Mayer M.P., Bukau B. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524:247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gao X., Carroni M., Nussbaum-Krammer C., Mogk A., Nillegoda N.B., Szlachcic A., Guilbride D.L., Saibil H.R., Mayer M.P., Bukau B. Human Hsp70 disaggregase reverses Parkinson’s-linked α-Synuclein amyloid fibrils. Mol. Cell. 2015;59:781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Clos J., Rabindran S., Wisniewski J., Wu C. Induction temperature of human heat shock factor is reprogrammed in a Drosophila cell environment. Nature. 1993;364:252–255. doi: 10.1038/364252a0. [DOI] [PubMed] [Google Scholar]
- 194.Hentze N., Le Breton L., Wiesner J., Kempf G., Mayer M.P. Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. Elife. 2016;5 doi: 10.7554/eLife.11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hammel H.T. Regulation of internal body temperature. Annu. Rev. Physiol. 1968;30:641–710. doi: 10.1146/annurev.ph.30.030168.003233. [DOI] [PubMed] [Google Scholar]
- 196.Tomanek L. Proteomics to study adaptations in marine organisms to environmental stress. J. Proteomics. 2014;105:92–106. doi: 10.1016/j.jprot.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 197.Tomanek L. Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs. J. Exp. Biol. 2010;213:971–979. doi: 10.1242/jeb.038034. [DOI] [PubMed] [Google Scholar]
- 198.Hofmann G.E. Patterns of Hsp gene expression in ectothermic marine organisms on small to large biogeographic scales. Integr. Comp. Biol. 2005;45:247–255. doi: 10.1093/icb/45.2.247. [DOI] [PubMed] [Google Scholar]
- 199.Macario A.J., Lange M., Ahring B.K., Conway de Macario E. Stress genes and proteins in the archaea. Microbiol. Mol. Biol. Rev. 1999;63:923–967. doi: 10.1128/mmbr.63.4.923-967.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ahmed R., Duncan R.F. Translational regulation of Hsp90 mRNA. AUG-proximal 5'-untranslated region elements essential for preferential heat shock translation. J. Biol. Chem. 2004;279:49919–49930. doi: 10.1074/jbc.M404681200. [DOI] [PubMed] [Google Scholar]
- 201.Morán Luengo T., Kityk R., Mayer M.P., Rüdiger S.G.D. Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell. 2018;70:545–552.e9. doi: 10.1016/j.molcel.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 202.Nava R., Zuhl M.N. Heat acclimation-induced intracellular HSP70 in humans: A meta-analysis. Cell Stress Chaperones. 2020;25:35–45. doi: 10.1007/s12192-019-01059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chao Y.C., Merritt M., Schaefferkoetter D., Evans T.G. High-throughput quantification of protein structural change reveals potential mechanisms of temperature adaptation in Mytilus mussels. BMC Evol. Biol. 2020;20:28. doi: 10.1186/s12862-020-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Berger M.S., Emlet R.B. Heat-shock response of the upper intertidal barnacle Balanus glandula: Thermal stress and acclimation. Biol. Bull. 2007;212:232–241. doi: 10.2307/25066605. [DOI] [PubMed] [Google Scholar]
- 205.Domnauer M., Zheng F., Li L., Zhang Y., Chang C.E., Unruh J.R., Conkright-Fincham J., McCroskey S., Florens L., Zhang Y., Seidel C., Fong B., Schilling B., Sharma R., Ramanathan A., et al. Proteome plasticity in response to persistent environmental change. Mol. Cell. 2021;81:3294–3309.e12. doi: 10.1016/j.molcel.2021.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Morimoto R.I. Cell-Nonautonomous regulation of proteostasis in aging and disease. Cold Spring Harb. Perspect. Biol. 2020;12 doi: 10.1101/cshperspect.a034074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Amin V., Cumming D.V., Coffin R.S., Latchman D.S. The degree of protection provided to neuronal cells by a pre-conditioning stress correlates with the amount of heat shock protein 70 it induces and not with the similarity of the subsequent stress. Neurosci. Lett. 1995;200:85–88. doi: 10.1016/0304-3940(95)12074-e. [DOI] [PubMed] [Google Scholar]
- 208.Teranishi K.S., Stillman J.H. A cDNA microarray analysis of the response to heat stress in hepatopancreas tissue of the porcelain crab Petrolisthes cinctipes. Comp. Biochem. Physiol. Part. D Genomics Proteomics. 2007;2:53–62. doi: 10.1016/j.cbd.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 209.Currie R.W., Karmazyn M., Kloc M., Mailer K. Heat-shock response is associated with enhanced postischemic ventricular recovery. Circ. Res. 1988;63:543–549. doi: 10.1161/01.res.63.3.543. [DOI] [PubMed] [Google Scholar]
- 210.Gray C.C., Amrani M., Yacoub M.H. Heat stress proteins and myocardial protection: Experimental model or potential clinical tool? Int. J. Biochem. Cell Biol. 1999;31:559–573. doi: 10.1016/s1357-2725(99)00004-7. [DOI] [PubMed] [Google Scholar]
- 211.Alzahrani S.M., Ebert P.R. Stress pre-conditioning with temperature, UV and gamma radiation induces tolerance against phosphine toxicity. Shelden E.A., editor. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Das M., Das D.K. Molecular mechanism of preconditioning. IUBMB Life. 2008;60:199–203. doi: 10.1002/iub.31. [DOI] [PubMed] [Google Scholar]
- 213.Prahlad V., Cornelius T., Morimoto R.I. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fawcett T.W., Sylvester S.L., Sarge K.D., Morimoto R.I., Holbrook N.J. Effects of neurohormonal stress and aging on the activation of mammalian heat shock factor 1. J. Biol. Chem. 1994;269:32272–32278. [PubMed] [Google Scholar]
- 215.Hipp M.S., Kasturi P., Hartl F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- 216.Batulan Z., Shinder G.A., Minotti S., He B.P., Doroudchi M.M., Nalbantoglu J., Strong M.J., Durham H.D. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J. Neurosci. 2003;23:5789–5798. doi: 10.1523/JNEUROSCI.23-13-05789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dugger B.N., Dickson D.W. Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Penke B., Bogár F., Crul T., Sántha M., Tóth M.E., Vígh L. Heat shock proteins and autophagy pathways in neuroprotection: From molecular bases to pharmacological interventions. Int. J. Mol. Sci. 2018;19:325. doi: 10.3390/ijms19010325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gil R.S., Cox D., McAlary L., Berg T., Walker A.K., Yerbury J.J., Ooi L., Ecroyd H. Neurodegenerative disease-associated protein aggregates are poor inducers of the heat shock response in neuronal-like cells. J. Cell Sci. 2020 doi: 10.1242/jcs.243709. [DOI] [PubMed] [Google Scholar]
- 220.Hoshino T., Murao N., Namba T., Takehara M., Adachi H., Katsuno M., Sobue G., Matsushima T., Suzuki T., Mizushima T. Suppression of Alzheimer’s disease-related phenotypes by expression of heat shock protein 70 in mice. J. Neurosci. 2011;31:5225–5234. doi: 10.1523/JNEUROSCI.5478-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bobkova N.V., Garbuz D.G., Nesterova I., Medvinskaya N., Samokhin A., Alexandrova I., Yashin V., Karpov V., Kukharsky M.S., Ninkina N.N., Smirnov A.A., Nudler E., Evgen'ev M. Therapeutic effect of exogenous hsp70 in mouse models of Alzheimer’s disease. J. Alzheimers Dis. 2014;38:425–435. doi: 10.3233/JAD-130779. [DOI] [PubMed] [Google Scholar]
- 222.Evgen’ev M.B., Krasnov G.S., Nesterova I.V., Garbuz D.G., Karpov V.L., Morozov A.V., Snezhkina A.V., Samokhin A.N., Sergeev A., Kulikov A.M., Bobkova N.V. Molecular mechanisms underlying neuroprotective effect of intranasal administration of human Hsp70 in mouse model of Alzheimer’s disease. J. Alzheimers Dis. 2017;59:1415–1426. doi: 10.3233/JAD-170398. [DOI] [PubMed] [Google Scholar]
- 223.Campanella C., Pace A., Caruso Bavisotto C., Marzullo P., Marino Gammazza A., Buscemi S., Palumbo Piccionello A. Heat shock proteins in Alzheimer’s disease: Role and targeting. Int. J. Mol. Sci. 2018;19:2603. doi: 10.3390/ijms19092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kuta R., Larochelle N., Fernandez M., Pal A., Minotti S., Tibshirani M., St Louis K., Gentil B.J., Nalbantoglu J.N., Hermann A., Durham H.D. Depending on the stress, histone deacetylase inhibitors act as heat shock protein co-inducers in motor neurons and potentiate arimoclomol, exerting neuroprotection through multiple mechanisms in ALS models. Cell Stress Chaperones. 2020;25:173–191. doi: 10.1007/s12192-019-01064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 226.Webb J.L., Ravikumar B., Atkins J., Skepper J.N., Rubinsztein D.C. Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 227.Rubinsztein D.C., Codogno P., Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ghavami S., Shojaei S., Yeganeh B., Ande S.R., Jangamreddy J.R., Mehrpour M., Christoffersson J., Chaabane W., Moghadam A.R., Kashani H.H., Hashemi M., Owji A.A., Łos M.J. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog. Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 229.Huang H., Zhu J., Li Y., Zhang L., Gu J., Xie Q., Jin H., Che X., Li J., Huang C., Chen L.C., Lyu J., Gao J., Huang C. Upregulation of SQSTM1/p62 contributes to nickel-induced malignant transformation of human bronchial epithelial cells. Autophagy. 2016;12:1687–1703. doi: 10.1080/15548627.2016.1196313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wolff S., Weissman J.S., Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 231.Sato H., Das S., Singer R.H., Vera M. Imaging of DNA and RNA in living eukaryotic cells to reveal spatiotemporal dynamics of gene expression. Annu. Rev. Biochem. 2020;89:159–187. doi: 10.1146/annurev-biochem-011520-104955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tutucci E., Livingston N.M., Singer R.H., Wu B. Imaging mRNA in vivo, from birth to death. Annu. Rev. Biophys. 2018;47:85–106. doi: 10.1146/annurev-biophys-070317-033037. [DOI] [PubMed] [Google Scholar]
- 233.Pichon X., Lagha M., Mueller F., Bertrand E. A growing toolbox to image gene expression in single cells: Sensitive approaches for demanding challenges. Mol. Cell. 2018;71:468–480. doi: 10.1016/j.molcel.2018.07.022. [DOI] [PubMed] [Google Scholar]
- 234.Brandvold K.R., Morimoto R.I. The chemical biology of molecular chaperones--implications for modulation of proteostasis. J. Mol. Biol. 2015;427:2931–2947. doi: 10.1016/j.jmb.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.W., Zhou S., King D., Shen P.S., Weibezahn J., Dunn J.G., Rouskin S., Inada T., Frost A., Weissman J.S. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Massey A., Kiffin R., Cuervo A.M. Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 237.Majeski A.E., Dice J.F. Mechanisms of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 238.Morano K.A., Grant C.M., Moye-Rowley W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Snutch T.P., Baillie D.L. Alterations in the pattern of gene expression following heat shock in the nematode Caenorhabditis elegans. Can J. Biochem. Cell Biol. 1983;61:480–487. doi: 10.1139/o83-064. [DOI] [PubMed] [Google Scholar]
- 240.Candido E.P. The small heat shock proteins of the nematode Caenorhabditis elegans: Structure, regulation and biology. Prog. Mol. Subcell Biol. 2002;28:61–78. doi: 10.1007/978-3-642-56348-5_4. [DOI] [PubMed] [Google Scholar]
- 241.Velichko A.K., Markova E.N., Petrova N.V., Razin S.V., Kantidze O.L. Mechanisms of heat shock response in mammals. Cell Mol. Life Sci. 2013;70:4229–4241. doi: 10.1007/s00018-013-1348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Avila D.S., Benedetto A., Au C., Bornhorst J., Aschner M. Involvement of heat shock proteins on Mn-induced toxicity in Caenorhabditis elegans. BMC Pharmacol. Toxicol. 2016;17:54. doi: 10.1186/s40360-016-0097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Han M.J., Lee S.Y. The Escherichia coli proteome: Past, present, and future prospects. Microbiol. Mol. Biol. Rev. 2006;70:362–439. doi: 10.1128/MMBR.00036-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Mühlhofer M., Berchtold E., Stratil C.G., Csaba G., Kunold E., Bach N.C., Sieber S.A., Haslbeck M., Zimmer R., Buchner J. The heat shock response in yeast maintains protein homeostasis by chaperoning and replenishing proteins. Cell Rep. 2019;29:4593–4607.e8. doi: 10.1016/j.celrep.2019.11.109. [DOI] [PubMed] [Google Scholar]
- 245.Verghese J., Abrams J., Wang Y., Morano K.A. Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Vazquez J., Pauli D., Tissières A. Transcriptional regulation in Drosophila during heat shock: A nuclear run-on analysis. Chromosoma. 1993;102:233–248. doi: 10.1007/BF00352397. [DOI] [PubMed] [Google Scholar]
- 247.Mahat D.B., Salamanca H.H., Duarte F.M., Danko C.G., Lis J.T. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Yura T., Nagai H., Mori H. Regulation of the heat-shock response in bacteria. Annu. Rev. Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 249.McAlister L., Strausberg S., Kulaga A., Finkelstein D.B. Altered patterns of protein synthesis induced by heat shock of yeast. Curr. Genet. 1979;1:63–74. doi: 10.1007/BF00413307. [DOI] [PubMed] [Google Scholar]
- 250.Richter K., Haslbeck M., Buchner J. The heat shock response: Life on the verge of death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 251.de Benedetti A., Baglioni C. Translational regulation of the synthesis of a major heat shock protein in HeLa cells. J. Biol. Chem. 1986;261:15800–15804. [PubMed] [Google Scholar]
- 252.Yura T. Regulation of the heat shock response in Escherichia coli: History and perspectives. Genes Genet. Syst. 2019;94:103–108. doi: 10.1266/ggs.19-00005. [DOI] [PubMed] [Google Scholar]
- 253.Grossman A.D., Straus D.B., Walter W.A., Gross C.A. Sigma 32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1987;1:179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]