Abstract
The human oral cavity harbours complex microbial communities with various commensal microorganisms that play pivotal roles in maintaining host health and immunity but can elicit local and systemic diseases. The role of commensal microorganisms in SARS-CoV-2 infection and disease susceptibility and enrichment of opportunistic pathobionts in the oral cavity is poorly understood. The present study aims to understand the altered landscape of the oral microbiome and mycobiome in SARS-CoV-2 infected patients (n = 30) and its correlation with risk factors compared to non-infected individuals (n = 24) using targeted amplicon sequencing. Diminution of species richness, an elevated abundance of opportunistic pathogens (Veillonella, Acinetobacter, Klebsiella, Prevotella, Gemella, and Streptococcus) and impaired metabolic pathways were observed in the COVID-19 patients. Similarly, altered oral mycobiome with enrichment of known respiratory disease causing pathogenic fungi were observed in the infected individuals. The data further suggested that reduction in immunomodulatory microorganisms lowers the protection of individuals from SARS-CoV-2. Linear discriminant analysis identified several differentially abundant taxa associated with risk factors (ageing and co-morbidities). We also observed distinct bacterial and fungal community structures of elderly infected patients compared to the younger age group members making them highly vulnerable to SARS-CoV-2 infection and disease severity. Furthermore, we also assessed the dynamics of the oral microbiome and mycobiome in symptomatic and asymptomatic patients, host types, co-morbidities, and viral load in the augmentation of specific pathobionts. Overall, the present study demonstrates the microbiome and mycobiome profiling of the COVID-19 infected individuals, the data further suggests that the SARS-CoV-2 infection triggers the prevalence of specific pathobiont.
Keywords: SARS-CoV-2, Oral hygiene, Oral microbiome, Mycobiome, Risk factors, COVID-19
1. Introduction
Advancement in multi-omics technologies over the last decade has guided us to interrogate the complex microbial communities and their role in human health and diseases. The human oral cavity harbours the second-largest microbiome and plays a pivotal role in maintaining host health Bao et al., 2020). In contrast, it also serves as an important reservoir and direct access for various opportunistic pathogens (Streptococcus, Neisseria, Corynebacterium, Veillonella, Gemella, Haemophilus, Rothia, Porphyromonas, etc.), leading to severe periodontal and respiratory diseases (Dewhirst et al., 2010; Minty et al., 2019; Baghbani et al., 2020; Caselli et al., 2020; Lenartova et al., 2021; Soffritti et al., 2021. Respiratory viruses enter the human body via the oropharynx and nasopharynx, where they establish infections by altering the microbiome and escaping the host immunity (Bao et al., 2020). The severe acute respiratory syndrome coronavirus (SARS-CoV-2) uses the oropharynx as one of the primary sites of entry as salivary glands and oral mucosal cells express higher levels of angiotensin-converting enzyme 2 (ACE2) and transmembrane serine proteases 2 (TMPRSS2) receptors (Pascolo et al., 2020, Chen et al., 2020a, Chen et al., 2020b, Herrera et al., 2020). Supporting evidence suggests that the salivary glands of the asymptomatic patients act as a potential reservoir for SARS-CoV-2 and spread viruses through contaminated salivary droplets (Xu et al., 2020a, Xu et al., 2020b). Hence, the oral cavity is considered as a potential source for the spread of SARS-CoV-2, which influences the microbiota of the oral cavity and related body sites, especially both respiratory and gastrointestinal tracts, and has a profound effect on the host immunity (Xiang et al., 2021).
Since the outbreak of SARS-CoV-2, in-depth studies have been performed to understand the epidemiology, pathophysiology, evolution, and genomics of the virus (Fadaka et al., 2020, van Dorp et al., 2020, Moustafa and Planet, 2021). More so, within the context of the human microbiome, the relationship between SARS-CoV-2 infection and microbiome of gut and nasopharynx has been broadly described (Zuo et al., 2020, Yeoh et al., 2021, Nardelli et al., 2021, Rueca et al., 2021, Xu et al., 2020a, Xu et al., 2020b, Gupta et al., 2021). Even though the oral cavity is the entry point for the SARS-CoV-2 virus, yet our understanding of its immediate interaction with the oral microbiome and its underlying consequences has been poorly characterized (Iebba et al., 2021, Haran et al., 2021, Ward et al., 2021, Soffritti et al., 2021). Despite the low abundance of bacterial counterparts, the mycobiome has a significant impact on human health and disease (Bandara et al., 2019). The present study aims to characterize the oral microbiome and mycobiome of SARS-CoV-2 infected and non-infected individuals using targeted amplicon sequencing of 16S rRNA gene and internal transcribed spacer (ITS) region, respectively. This study demonstrates the association between the risk factors and oral microbiota (bacteriome and mycobiome) in COVID-19 patients and its implication in disease monitoring and intervention.
2. Materials and methods
2.1. Patient enrolment and sample collection
Patients with fever were tested for SARS-CoV-2 infection as a precautionary measure at Dr D. Y. Patil Medical College and Hospital, Pune (India). This study was approved by the Institutional Ethics Committee of Dr D. Y. Patil Medical College and Hospital (DYPU), Pune (India) (DYPV/EC/599/2020). Informed consent was obtained from the participants before the sample collection. We had collected data (more than 300 study participants) on the parameters which may influence the overall microbial composition in the saliva. Study participants [SARS-CoV-2 infected and non-infected (as healthy control)] which were using medications such as antibiotics, antiseptic mouth wash were excluded from the study. Data related to the medical history, co-morbidities, ongoing symptoms and anthropometric was collected at the time of enrolment. All the patients were instructed not to eat/drink for at least 3h before the sample collection. Patients were asked to collect 2–3ml of saliva inside the sterile container containing the 1ml phosphate buffer saline. Patients were supervised and assisted by the virologists and laboratory personnel from Dr D. Y. Patil Medical College and Hospital, Pune (India), for appropriate sample collection as per the biosafety norms laid down by the concerned authorities. Samples for the current study were collected between November 2020 to February 2021.
2.2. RT-PCR analysis
All the collected saliva samples were stored at − 80° C and processed for RT-PCR analysis within 24h. RT-PCR analysis was performed as per the manufacturer’s instruction (TRUPCR® SARS-CoV-2 Kit). The kit employed simultaneous detection of the E gene for the detection of the sarbecovirus (genus B-betacoronavirus (B-βCoV)) and RdRP gene for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). RNase P was used as an endogenous internal control to check the extracted RNA quality, amplification procedure and possible presence of inhibitors to avoid false-negative results. Results were determined based Cycle threshold.
2.3. DNA extraction, targeted 16S rRNA and ITS gene amplification, and sequencing
The total DNA isolation was performed using Biospin DNA/RNA extraction Kit (Prolab Supply Corp) as per the manufacturer’s instruction. Saliva samples (200 µl) were mixed with proteinase K and lysis buffer and incubated for 56 °C for 15 mins in the water bath. Further, ethanol (250 µl) was added to the tubes and thoroughly mixed. The mixture was then transferred to the spin column and centrifuged at 10,000 g for 1 min. After the washing step, DNA was eluted in the fresh tubes with elution buffer. The V4 hypervariable region of the 16S rRNA gene was selected for amplification using universal bacterial primers 515F and 806R (Bates et al., 2011) while ITS1 was amplified using ITS1F and ITS2R for fungi (Bokulich and Mills, 2013). The equimolar pooled library was sequenced on the in-house Illumina MiSeq platform using MiSeq reagent v2 kit-Illumina Inc. (2 ×250 bp chemistry). A detailed description of PCR amplification, thermal cycling conditions, library preparation, and sequencing is described in Supplementary materials.
2.4. Bioinformatics and statistical analysis
The obtained raw reads were quality checked using FastQC (Andrews, 2010) followed by pre-processing and analysis using DADA2 package v1.6.0 (Callahan et al., 2016) in R 3.6.0. The reads were quality filtered using filterAndTrim function of DADA2. The primers were removed from the reads. Chimaeras were removed using the removeBimeraDenovo function of DADA2. The taxonomy of clean amplicon sequence variants was assigned using Silva Database (silva_nr99_v138.1_train_set.fa.gz) for bacteria and UNITE database (sh_general_release_dynamic_04.02.2020.fasta) for fungi. Phyloseq v3.4.2 R package (McMurdie and Holmes, 2013) was used to generate alpha and beta diversity matrices. Alpha diversity metrics were calculated using the estimate_richness function on rarefied reads (90% of the lowest reads). Data visualizations were produced using ggplot2 (Wickham et al., 2016), Reshape2 (Wickham, 2012), vegan (Oksanen et al., 2013), ggpubr (Kassambara, 2018), and RColorBrewer (Neuwirth and Brewer, 2014), in RStudio version 1.2.1335. The fraction of ASVs shared among samples was visualized using the UpSet plot (Conway et al., 2017) R. Pairwise Wilcoxon test was used to compare the difference in alpha diversity/bacterial taxa between infected and non-infected individuals with adjusted p value. Analysis of similarities (ANOSIM) and permutational ANOVA (PERMANOVA) was performed on the infected and non-infected individuals using the Bray Curtis dissimilarity matrix to assess the difference in beta diversity. Non-metric multidimensional scaling (NMDS) was performed with Bray-Curtis dissimilarity matrix using phyloseq package. Betadispersion analysis was performed using betadisper function to test the inter-individual variation. Linear discriminant analysis Effect Size (LEfSe) was performed to determine the differentially enriched taxa and/or biomarkers between groups. The functional prediction of oral microbiota was performed using the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt2) (Douglas et al., 2020), and LEfSe analysis was performed to identify differentially enriched functions (KEGG categories) between infected and non-infected individuals. Microbiome multivariable association with linear models (MaAsLin2) was used to assess the association of covariates with microbiome data.
2.4.1. Data submission
The raw sequences were deposited into the NCBI under Bioproject (PRJNA780671).
3. Results
3.1. Subject clinical characteristics
Oral rinses were collected from 54 eligible individuals, including 24 non-infected and 30 SARS-CoV-2 infected patients. In total, 34 males (14: non-infected; 20: infected) and 20 females (10: non-infected; 10: infected) with age ranging from 15–75) were included in this study. Nearly 40% of the infected individuals had comorbid conditions (hypertension, diabetes, and chronic renal disease). The detailed clinical characteristics of study participants are presented in Table 1.
Table 1.
Demographic details of recruited subjects.
| Non-infected | Infected | |
|---|---|---|
| Total no of subjects | 24 | 30 |
| Gender | ||
| Male (%) | 14 (58) | 20 (67) |
| Female (%) | 10 (42) | 10 (33) |
| Age | ||
| Range | 15–65 | 15–75 |
| Mean/Median | 45/43.5 | 47/48.5 |
| Condition | ||
| Symptomatic | 10 | |
| Asymptomatic | 20 | |
| aComorbidity | 12 |
Comorbidity: patients having diabetes, chronic renal disease, and hypertension
3.2. Oral microbiome profiling in infected individuals
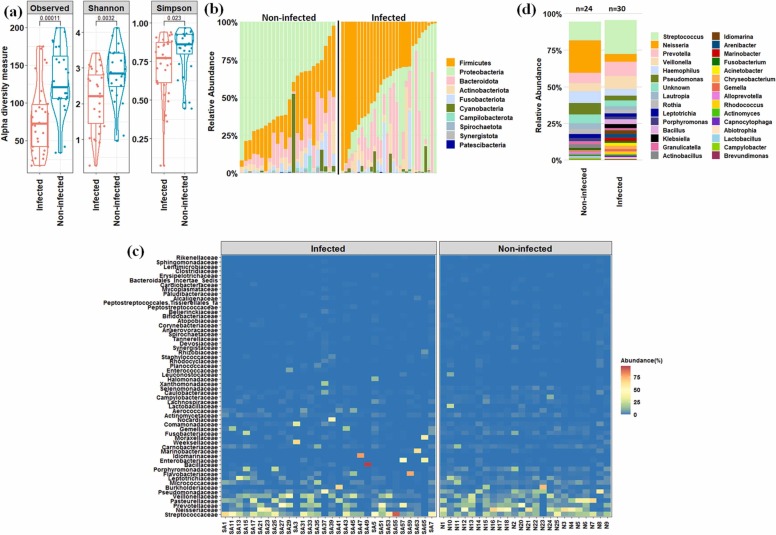
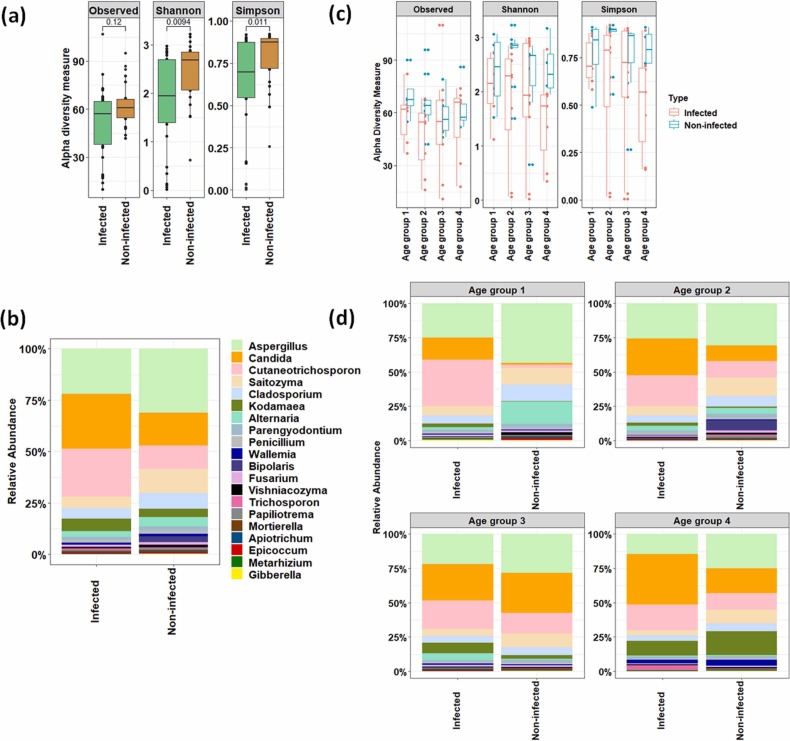
Microbiome profiling of infected and non-infected individuals and its association with host characteristics were elucidated using 16S rRNA gene-based targeted sequencing approach. Nearly 5.2 million reads were generated from 54 samples with an average of 96264 ± 17662 reads per sample. Out of the total raw reads, 4.48 million quality-filtered reads were used for the analysis. The results showed that the oral microbiome of SARS-CoV-2 infected individuals had lower alpha diversity than non-infected ones ( Fig. 1a). In total, 1312 ASVs were detected across the samples based on the rarefied reads. Out of these ASVs, 421 ASVs were found to be shared between these two states (Fig. S1). Pairwise Wilcoxon test showed a significant (p < 0.05) decrease in the number of ASVs and diminution of species richness (reduction in Shannon and Simpson indices values) in the infected individuals (Fig. 1a).
Fig. 1.
Microbial community composition in oral cavity of infected and non-infected individuals. (a) Changes in alpha diversity parameters. Pairwise Wilcoxon test was performed to assess the statistical significance (p value is mentioned). (b) Microbial community composition at phylum–level. (c) Heat-map based relative abundance distribution of major families. (d) Relative abundance of major genera between infected and non-infected groups.
Oral microbiome analysis revealed an alteration of bacterial abundance in the infected individuals compared to non-infected ones. Proteobacteria, Firmicutes, Bacteroidota, Actinobacteria, and Fusobacteriota constituted the major proportion (~96% in non-infected and ~98% in infected individuals) of the microbial community in both the states (Fig. 1b). Notably, among these dominant phyla, an abundance of Firmicutes increased significantly (p < 0.05) in infected ones, while that of Proteobacteria and Fusobacteriota reduced significantly (p < 0.05) in the same group. Moreover, a few less abundant phyla, including Campilobacteriota, Synergistota, Spirochaeota, Patescibacteria, and Desulfobacterota showed a significant reduction (p < 0.05) in their abundance pattern in infected individuals (Fig. S2). For a better taxonomic resolution and its alteration in the infected individuals, heat-map based analysis of prominent bacterial families followed by a pairwise Wilcoxon test was performed (Fig. 1c and Fig. S3). The results showed significant (p < 0.05) diminution of bacterial families such as Neisseriaceae, Pasteurellaceae, Pseudomonadaceae, Porphyromonadaceae, Leptotrichiaceae, Fusobacteriaceae, Selenomonadaceae, Gemellaceae, etc. in the infected patients. ANOSIM (p < 0.05) and PERMANOVA (p < 0.05) based statistical tests further confirmed the difference in the microbial community pattern between the two states. Furthermore, a marked difference in the abundance of various genera was observed in the infected patients than that of non-infected ones. A remarkable increase in the abundance of Streptococcus, Veillonella, Prevotella, Bacillus, Klebsiella, Idiomarina, Acinetobacter, Arenibacter, Gemella, Chryseobacterium, Capnocytophaga, etc. was observed in the infected patients. At the same time, Neisseria, Haemophilus, Pseudomonas, Lautropia, Rothia, Leptotrichia, Porphyromonas, Actinobacillus, Granulicatella, Fusobacterium, Aggregatibacter, Alloprevotella, Selenomonas, etc. decreased substantially (Fig. 1d and Fig. S4).
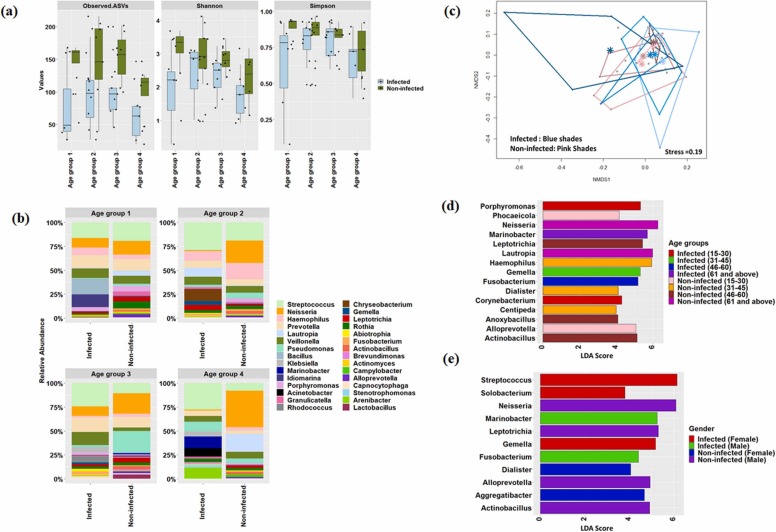
3.2.1. Differentially abundant taxa associated with host types (gender and age)
Further, we investigated alterations in the microbial community structure in the infected and non-infected individuals based on the host types; subjects were classified into four distinct age groups (age group 1:15–30 years, age group 2: 31–45 years, age group 3: 46–60, and age group 4: 61 and above) and gender (male and female). It was evident that species richness and evenness decreased with age in the infected individuals compared to non-infected ones ( Fig. 2a). The oral microbiome of infected individuals showed alteration in the abundance of various taxa in different age group category (Fig. 2b and c) which was well supported by pairwise PERMANOVA analysis (Table S1). To understand the dynamics of microbes associated with host age during SARS-CoV-2 infection, the differential abundance of microbial taxa was identified using LefSe. Distinct microbial taxa were found to be enriched (LDA score > 4.0, p < 0.05, FDR adjusted p-value of <10%) in the infected and non-infected individuals across these age groups. Corynebacterium and Porphyromonas enriched in infected individuals of age group 1, while Marinobacter was the most differentially abundant taxon in age group 4 (Fig. 2d). Similarly, LEfSe analysis was performed to assess the biomarkers associated with male and female infected individuals. The results showed that Marinobacter was significantly (p < 0.05, FDR adjusted p-value of <5%) abundant in infected males. At the same time, Gemella, Streptococcus, and Solobacterium were significantly (p < 0.05, FDR adjusted p-value of <5%) enriched in infected females (Fig. 2e). MaAsLin based analysis identified overlapping taxa as detected by lefse, however, it found few other taxa which were associated with different age groups, gender, and disease state (Table S2).
Fig. 2.
Dynamics of microbial community and its association with host age and gender. (a) Changes in alpha diversity parameters based on different age groups. (b) Microbial community composition at genera-level across the different age groups. (c) Non-metric multidimensional scaling based on a Bray-Curtis dissimilarity matrix at different age groups. Blue and pink shades represented the infected and non-infected patients. Lighter to darker shades showed age group 1–4. Centroid was also denoted in each age category. (d) Identification of differentially-abundant genera in various age groups. (e) Identification of differentially-abundant genera among infected and non-infected male and female individuals.
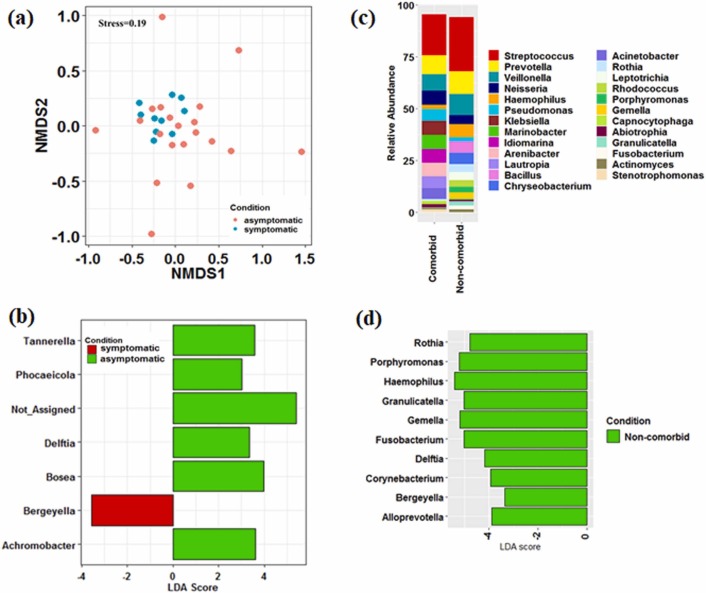
3.2.2. Dynamics of microbes associated with symptomatic and asymptomatic patients and co-morbidities
The dynamics of microbes associated with COVID-19 conditions and comorbidities were evaluated to understand the shift in the oral microbiome. NMDS based analysis showed the dispersion of asymptomatic patients while symptomatic patients cluster together. PERMANOVA analysis showed significant (p < 0.05) difference between the two conditions ( Fig. 3a). However, betadisper analysis (p < 0.05) showed that the microbial community during asymptomatic infection was more stochastic than symptomatic patients. LEfSe based analysis revealed that Bergyella was the most differentially abundant (p < 0.05) bacterial taxon in symptomatic patients (Fig. 3b). The microbiome of the asymptomatic patients was characterized by a preponderance of Tannerella, Phocaeicola, Delftia, Bosea, and Achromobacter with LDA score > 3 (Fig. 3b). In addition, the microbiome of infected patients with comorbidities showed a reduction in the abundance of Rothia, Porphyromonas, Haemophilus, Granulicatella, Gemella, Fusobacterium, Delftia, Corynebacterium, Bergeyella, and Prevotella, which constituted the significant proportion in non-comorbid patients (Fig. 3c and d). To get deeper insights into the association of microbial taxa and viral load (based on Ct values) (Mishra et al., 2021), LefSe analysis revealed that patients with high viral load (Ct value < 25) were enriched with Granulicatella, Leptotrichia, and Zoogloea (Fig. S5).
Fig. 3.
Association of microbial communities with COVID-19 conditions and co-morbidities. (a) Non-metric multidimensional scaling showed difference in the microbial community composition between symptomatic and asymptomatic infected patients. (b) Identification of differentially-abundant genera between symptomatic and asymptomatic infected patients. (c) Microbial community composition at genera-level between infected patients with or without comorbidities. (d) Identification of differentially-abundant genera in infected patients with or without comorbidities.
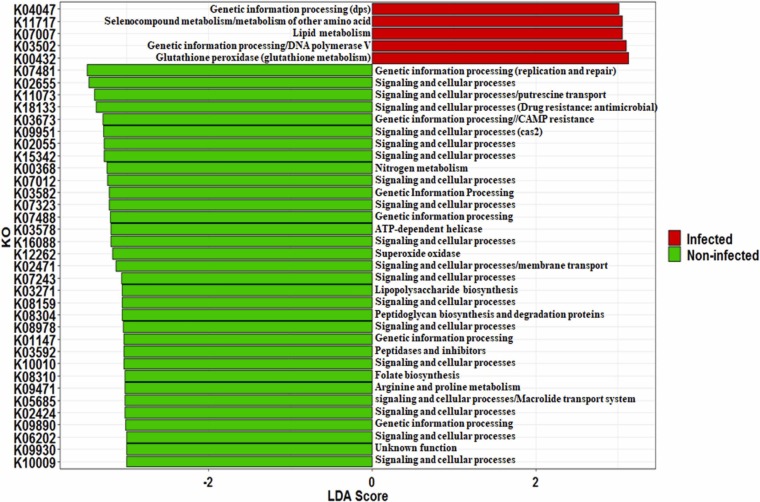
3.2.3. Predicted oral microbiome function using PICRUSt2
Functional potential of oral microbiome inferred by PICRUSt2 and differentially enriched function derived through KEGG categories were identified through LDA analysis (Fig S9 and Fig. 4). Overall, carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, replication and repair, energy metabolism, etc. were the major metabolic pathways detected in both the states (Fig S9). We observed that 169 KEGG categories showed significant (p < 0.05) differences in their abundance. Out of these 169 KEGG categories, only five were enriched (LDA score >3, p < 0.05) in infected individuals associated with lipid metabolism, glutathione peroxidase, selenocompound metabolism or other amino acids (Fig. 4). On the contrary, signaling and genetic process, replication and repair, folate biosynthesis, lipopolysaccharide biosynthesis, peptidoglycan biosynthesis and degradation proteins, arginine and proline metabolism, macrolide transport system, putrescine transport, etc. decreased significantly in the infected individuals.
Fig. 4.
Predicted microbial metabolic profiling based on PICRUSt 2 analysis. Difference in the microbial KEGG orthology was identified through LefSE and top 32 KEGG ID was presented in the figure.
3.3. Oral mycobiome profiling in infected individuals
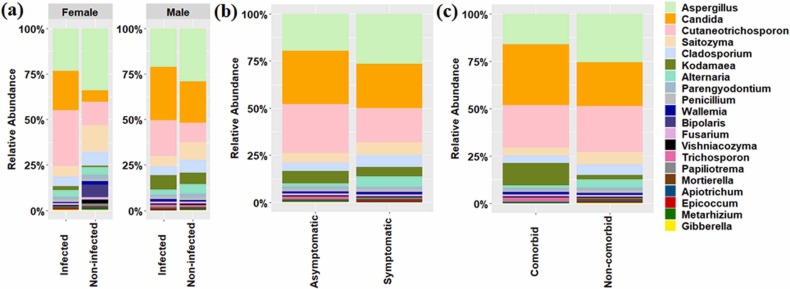
In addition to bacteriome, we have evaluated the changes in the oral mycobiome and its relation to the COVID-19. Non-bacterial components of the oral cavity are well known for their ability to cause various respiratory diseases. The results showed decreased species richness in the oral mycobiome of SARS-CoV-2 infected individuals than non-infected ones ( Fig. 5a). Mycobiome data revealed a significant (Pairwise Wilcoxon test, p. adjusted value < 0.05) decrease in the abundance of Cladosporium, Gibberella, Mortierella, Saitozyma, and Parengyodontium. However, reduction in relative abundance of Bipolaris, Aspergillus, Alternaria, and Vishniacozyma was observed in the infected patients with p.adjusted value < 10% based on pairwise Wilcoxon test (Fig. 5b). In contrast, abundance of Cutaneotrichosporon, Candida, Wallemia, Trichosporon, and Kodamaea was not found to be significantly (Pairwise Wilcoxon test, p. adjusted value > 0.05) increased in the infected patients (Fig. 5b). We further investigated the shift in the mycobiome community structure in the infected and non-infected individuals based on their host types. The alpha diversity indices showed the decrease in the species richness and evenness with age in the infected individuals compared to non-infected ones, except for age group 4, where an increase in ASVs was evident (Fig. 5c). Pairwise PERMANOVA analysis did not show a significant difference between the microbiome profile across the various age groups (Table S3). However, oral mycobiome of infected individuals showed dysbiosis with alteration in the abundance of few taxonomic groups in each age group category (Fig. 5d). The abundance of Cutaneotrichosporon, Candida, and Kodamaea increased in all the age group categories of infected patients. In contrast, few specific taxa (Alternaria in age group 3 and Trichosporon in age group 4) were enriched in particular age groups (Fig. 5d). The contrasting difference in the mycobiome community structure was well evident in the male and female infected patients compared to its non-infected counterparts in terms of few taxonomic groups ( Fig. 6a). The dynamics of fungus associated with COVID-19 conditions and comorbidities were evaluated to understand the shift in oral mycobiome of the infected patients only. A contrasting mycobiome shift was observed between the symptomatic and asymptomatic infected individuals. (Fig. 6b). Alternaria, Aspergillus, Cladosporium, and Saitozyma were predominant in the symptomatic infected patients compared to asymptomatic infected patients (Fig. 6b). However, Cutaneotrichosporon, Candida, Kodamaea, Trichosporon, and Wallemia dominated the infected patients with comorbidities (Fig. 6c).
Fig. 5.
Fungal community compositions in oral cavity of infected and non-infected individuals. (a) Changes in alpha diversity parameters in infected and non-infected individuals. (b) Microbial community composition at genera–level (top 20). (c) Changes in alpha diversity parameters across the different age groups. (d) Relative abundance of major genera across the different age groups.
Fig. 6.
Dynamics of fungal community and its association with gender, COVID-19 conditions, and co-morbidities. (a) Difference in the fungal community composition among infected and non-infected male and female individuals. (b) Relative abundance of major fungal genera between symptomatic and asymptomatic infected patients. (c) Relative abundance of major fungal genera between infected patients with or without comorbidities.
4. Discussion
Over the last decade, it has been witnessed that gut microbiota plays an essential role in maintaining human health and is also linked to diseases (Wang et al., 2017) Although the human oral cavity hosts the second-largest microbiota after the gut, its role in the pathogenesis of respiratory diseases has been understudied and requires greater attention (Bao et al., 2020). Xiang et al. (2021) observed that few of the opportunistic pathobionts of the oral cavity enter the lower respiratory and digestive tracts and colonize there. It has been reported that complex interplay among virus-host-oral microbiome during the influenza infections and presently in SARS-CoV-2 conditions at various stages of infection cycle may lead to changes in the microbial composition at local and distal organs (lung and gut), which results in secondary infections (Imai and Tanaka, 2021). Previous studies have suggested that the lung microbiota is more related to the microbiota of the oropharynx than that of the nasopharynx and gastrointestinal tract (Venkataraman et al., 2015, Segal et al., 2016). Thus, understanding the microbiome across the lung-oral axis is essential to get deeper insights into the pathogenesis, disease severity, and metabolic impairment of respiratory illness. The present study focuses on the comparative analysis of oral microbiome and mycobiome of SARS-CoV-2 infected and non-infected individuals and its correlation with host types, disease severity, and comorbidities.
The study revealed that SARS-CoV-2 infection resulted in altering bacterial species richness and enhancement of various opportunistic pathogens in the oral cavity. Our results are in line with the previous studies where the diminution in species richness and dysbiosis in the oral microbiome of SARS-CoV-2 patients or patients infected with other respiratory viruses have been observed, pointing towards the possibilities of secondary infections in the patients (Edouard et al., 2018, Iebba et al., 2021; Ma et at., 2021; Wu et al., 2021). The increased abundance of Veillonella, Acinetobacter, Klebsiella, Prevotella, Gemella, and Streptococcus have been previously reported from the oropharyngeal and nasopharyngeal microbiome of COVID-19 affected patients indicating their association with SARS-CoV-2 infection and disease severity [21, 22, 45, 46]. Veillonella has been previously identified as a differentially abundant taxon in COVID-19 patients and a cause of chronic anaerobic pneumonitis and is well represented in Bronchoalveolar lavage fluid (BALF) of COVID-19 patients (Shah et al., 2008; Ma et al., 2021; Iebba et al., 2021). Similarly, Klebsiella, Acinetobacter, and Streptococcus have been identified as indicators for ventilator-associated pneumonia or COVID-19 severity in the previous studies (Lu et al., 2014, Ma et al., 2021). The enhanced abundance of Streptococcus species leads to the upregulation of the adhesion receptors for viral entry during respiratory viral infection (Man et al., 2017). Prevotella and Gemella species have been overrepresented in COVID-19 patients and, therefore, may involve in producing proteins that can promote SARS-CoV-2 infection and disease severity (Khan and Khan, 2020, Haran et al., 2021). Most of these enriched bacteria belong to a group of periodontitis-correlated taxa, indicating the close association between periodontitis and SARS-CoV-2 infection (Ma et al., 2021). Periodontal-associated cytokines are known to involve the proliferation of pathogenic microbes in oral cavities and their contact with the lungs via altering the respiratory epithelium, thereby causing secondary infections in COVID-19 patients (Xiang et al., 2021). Interestingly, these opportunistic pathogens are enriched in BALF of COVID-19 patients suggesting the transmission of these pathobionts from the oral cavity to the lower respiratory system indicating that the oral cavity harbours such pathobionts (Bao et al., 2020, Zijie et al., 2020). SARS-CoV-2 infection provides an optimum environment for specific commensals to thrive as pathogens leading to a more complex diseased state such as those observed in co-infections. For example, cytokine storms linked with hyper inflammation during SARS-CoV-2 infection lead to notable changes in the oral microbiome landscape (Bao et al., 2020).
Immunomodulatory microorganisms (members of Bacteroidota and Fusobacteriota), which play a significant role in regulating the host immunity and health state, have been substantially decreased, leading to enhanced viral replication and disease progression, as previously discussed (Yamamoto et al., 2021, Wu et al., 2021). The members of Bacteroidota are also known to downregulate the ACE2 gene expression in the murine gut (Donati Zeppa et al., 2020, Koester et al., 2021). Similarly, Fusobacterium is involved in a defensive role against viral infections via the surface sialylation process (Nardelli et al., 2021). A decrease in Fusobacterium abundance in the oral microbiome may reduce the sialic acid metabolism, thus compromising the protection of individuals from SARS-CoV-2. Other than these bacterial taxa, reduction in Neisseria and Haemophilus may result in severe dysbiosis in the oral microbiome, leading to the enrichment of various opportunistic pathogens. The decrease in these two taxa is in concordance with the previous oral microbiome of COVID-19 patients (Ma et al., 2021). Furthermore, our study suggests a more substantial impairment in the metabolic pathways involved in genetic information processing, membrane transport, replication and repair, folate biosynthesis, etc., in the COVID-19 patients. Our observations agree with the earlier study suggesting that the members of the oropharyngeal microbiome in such patients have less potential for membrane transport of ions, lipids, sterols, peptides, proteins, and carbohydrates (Ma et al., 2021).
It has been reported that the host types (age and gender) and comorbidities influence the severity in COVID-19 patients (Gupta et al., 2021, Ma et al., 2021). Our data suggest a strong association between indigenous bacterial taxa and host types in SARS-CoV-2 infected individuals. It has been clearly understood that the oral microbiota of elderly patients shows depletion in species richness and evenness compared to other age groups, indicating the vulnerability of SARS-CoV-2 infection. This might be due to the alterations in ACE-2 expression levels, immunological profiling, oxidative stress, and mitochondrial dysfunction, which play a prominent role in the higher susceptibility of the aged population (Farshbafnadi et al., 2021, Pietrobon et al., 2020). Hence, elderly patients need to receive careful monitoring and timely medication for those with comorbidities to reduce their mortality rate. COVID-19 pandemic has affected more men than women to both mortality and severity (Gadi et al., 2020, Jin et al., 2020). Our data shows that infected male and female patients harbour distinct oral microbiota, which might influence the host immune response to infection. Based on the LefSe/MaAslin2 analysis, we could identify differences in bacterial taxa with age groups and gender, which could be used as non-invasive biomarkers for dysbiosis in SARS-Cov-2 infections or other viral respiratory diseases.
Additionally, we could explore the difference in the microbial community structure in symptomatic and asymptomatic patients and comorbid conditions and viral load. Our data reveals enrichment of pathogenic bacterial taxa in symptomatic patients responsible for inducing the expression of various virus binding receptors, thus increasing the viral replication and co-infections (Ichinohe et al., 2011, Shah, 2021). In concordance with the previous studies where viral loads are associated with enrichment of specific groups of bacterial taxa, our study identifies Leptotrichia and Granulicatella as a differentially abundant genus linked with high viral load. These two members are previously reported in higher abundance in patients with a longer COVID-19 symptom duration (Haran et al., 2021). In contrast, Rosas-Salazar et al. (2021) found the higher abundance of Granulicatella elegans in low viral load nasal samples.
Fungi, the other major group of microorganisms, play a significant role in oral and respiratory diseases (Deo and Deshmukh, 2019). Zhang et al. (2020) reported that 221 patients with laboratory-confirmed SARS-CoV-2 pneumonia had a 7.7% bacterial coinfection rate and 3.2% fungal co-infection rate. It has also been reported that fungal co-infections are common complications of viral pneumonia. Hence, they demand a detailed investigation in COVID-19 patients, as a high positive rate of fungal antigenemia has been reported in such patients (Lei et al., 2020, Zhou et al., 2020). Overall, the present study revealed the dysbiosis of oral mycobiome in the infected patients, decreasing species richness and counts. Interestingly, within the infected elderly cohort (age group 4), oral mycobiome reflected a higher fungal count, which is in line with the findings by Soffritti et al. 2021. Our study reveals the presence of various commensals fungal genera in the oral cavity of non-infected patients. Due to the SARS-COV-2 infection, a fraction of commensal fungi such as Cutaneotrichosporon, Candida, Wallemia, Trichosporon, and Kodamaea have been enriched in COVID-19 patients. It has been reported that fungi in the oral cavity are responsible for the increased inflammation and hypersensitivity due to their enzymatic and catabolic/toxic activities, which ultimately favours the increased viral load and secondary infections (Chen et al., 2020a, Chen et al., 2020b). Cutaneotrichosporon has been previously isolated from the cystic fibrosis patients responsible for the progression of respiratory diseases (van der Bruggen et al., 2018). Similarly, Trichosporon spp. is life-threatening and are associated with invasive disease in immunocompromised/immunocompetent patients (Hickey et al., 2009). These members are known to cause pulmonary deep-seated mycotic infections, hypersensitivity pneumonitis, and superficial infections (Nakajima et al., 2007). Increased abundance of Wallemia has been previously reported in allergic airway diseases in the mice models (Wheeler et al., 2016), while Kodamea is considered one of the opportunistic agents of fungemia in immunocompromised patients (Al-Sweih et al., 2011). Various species of Candida are known to cause multiple respiratory diseases such as cystic fibrosis, pneumonia, bronchiectasis, etc. (Weaver et al., 2019). Our findings are in concordance with the previous reports where COVID-19 patients carried higher numbers of Candida spp. (Chen et al., 2020a, Chen et al., 2020b, Salehi et al., 2020). This suggests that COVID-19 patients are more susceptible to Candida spp. Furthermore, our data reveals that the oral mycobiome did not show presence of mucormycosis causing fungi such as Rhizopus, Mucor, Rhizomucor, Apophysomyces, and Lichtheimia therefore it could be implied that mucormycosis is not indigenous in oral cavity at the time of admission.
Our study unequivocally revealed that SARS-CoV-2 infected patients exhibit unique microbial signatures with increased abundance of both pathogenic bacterial (Veillonella, Acinetobacter, Klebsiella, Prevotella, Gemella, and Streptococcus) and fungal (Cutaneotrichosporon, Candida, Wallemia, Trichosporon, and Kodamaea) members. Although, the current study provides a detailed investigation of microbiome and mycobiome, a sequential analysis with more subjects and immunological parameters would provide a better understanding of the virus and microbiome interactions to establish a novel perspective towards disease biology and diagnostics.
Ethical committee statement
The study was approved by the Institutional Ethical Committees of Dr. D. Y. Patil Vidyapeeth, Pune, India. Sample collection to DNA extraction was done at the Dr. D. Y. Patil Vidyapeeth, Pune, DNA samples were then transferred to the National Centre for Cell Science, Pune for further analysis.
CRediT authorship contribution statement
Conception and design of study: MS, AK, CR, RB, YS, SK, AS, Acquisition of data: AG, SB, AS, SK, AS, Analysis and/or interpretation of data: AG, SB, AS, SK, AS, Drafting the manuscript: AG, SB, AS, MS, AK, CR, RB, YS, SK, AS, Revising the manuscript critically for important intellectual content: AG, SB, AS, SK, AS.
Acknowledgement
We are grateful to the DBT/Wellcome Trust India Alliance for providing funding under the project grant (IA/E/17/1/503700). This work was also supported by the ‘Department of Biotechnology (DBT), Government of India’ (by Grant No. BT/Coord.II/01/03/2016), under the project Establishment of Centre for excellence National Centre for Microbial Resource (NCMR Grant No. BT/Coord.II/01/03/2016). We also acknowledge Dr. D. Y. Patil Vidyapeeth, Pune (India) for providing funding under the grant DPU/557(1)/2020.
Conflicts of interest/Competing interests
Authors declare no competing interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.micres.2022.127055.
Appendix A. Supplementary material
Supplementary material
.
References
- Al-Sweih N., Khan Z.U., Ahmad S., Devarajan L., Khan S., Joseph L., Chandy R. Kodamaea ohmeri as an emerging pathogen: a case report and review of the literature. Med. Mycol. 2011;49(7):766–770. doi: 10.3109/13693786.2011.572300. [DOI] [PubMed] [Google Scholar]
- Andrews S, FastQC: a quality control tool for high throughput sequence data, (2010).
- Baghbani T., Nikzad H., Azadbakht J., Izadpanah F., Kashani H.H. Dual and mutual interaction between microbiota and viral infections: a possible treat for COVID-19. Microb. Cell Fact. 2020;19(1):1–25. doi: 10.1186/s12934-020-01483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara H.M.H.N., Panduwawala C.P., Samaranayake L.P. Biodiversity of the human oral mycobiome in health and disease. Oral Dis. 2019;25(2):363–371. doi: 10.1111/odi.12899. [DOI] [PubMed] [Google Scholar]
- Bao L., Zhang C., Dong J., Zhao L., Li Y., Sun J. Oral microbiome and SARS-CoV-2: beware of lung co-infection. Front. Microbiol. 2020;11:1840. doi: 10.3389/fmicb.2020.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S.T., Berg-Lyons D., Caporaso J.G., Walters W.A., Knight R., Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5(5):908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Mills D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013;79(8):2519–2526. doi: 10.1128/AEM.03870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli E., Fabbri C., D’Accolti M., Soffritti I., Bassi C., Mazzacane S., Franchi M. Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. 2020;20:1–19. doi: 10.1186/s12866-020-01801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z., Wang S. Detection of SARS‐CoV‐2 in saliva and characterization of oral symptoms in COVID‐19 patients. Cell Prolif. 2020;53(12) doi: 10.1111/cpr.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway, J. R., Lex, A., & Gehlenborg, N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. [DOI] [PMC free article] [PubMed]
- Deo P.N., Deshmukh R. Oral microbiome: Unveiling the fundamentals. Journal of oral and maxillofacial pathology. J. Oral Maxillofac. Pathol. 2019;23(1):122. doi: 10.4103/jomfp.JOMFP_304_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F.E., Chen T., Izard J., Paster B.J., Tanner A.C., Yu W.H., Wade W.G. The human oral microbiome. J. Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati Zeppa S., Agostini D., Piccoli G., Stocchi V., Sestili P. Gut microbiota status in COVID-19: an unrecognized player? Front. Cell Infect. Microbiol. 2020;10:742. doi: 10.3389/fcimb.2020.576551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Langille M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edouard S., Million M., Bachar D., Dubourg G., Michelle C., Ninove L., Charrel R., Raoult D. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37(9):1725–1733. doi: 10.1007/s10096-018-3305-8. [DOI] [PubMed] [Google Scholar]
- Fadaka A.O., Sibuyi N.R.S., Adewale O.B., Bakare O.O., Akanbi M.O., Klein A., Meyer M. Understanding the epidemiology, pathophysiology, diagnosis and management of SARS-CoV-2. J. Int. Med. Res. 2020;48(8) doi: 10.1177/0300060520949077. 0300060520949077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshbafnadi M., Zonouzi S.K., Sabahi M., Dolatshahi M., Aarabi M.H. Aging & COVID-19 susceptibility, disease severity, and clinical outcomes: the role of entangled risk factors. Exp. Gerontol. 2021 doi: 10.1016/j.exger.2021.111507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadi N., Wu S.C., Spihlman A.P., Moulton V.R. What’s sex got to do with COVID-19? Gender-based differences in the host immune response to coronaviruses. Front. Immunol. 2020;11:2147. doi: 10.3389/fimmu.2020.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Karyakarte R., Joshi S., Das R., Jani K., Shouche Y., Sharma A. Nasopharyngeal microbiome reveals the prevalence of opportunistic pathogens in SARS-CoV-2 infected individuals and their association with host types. Microbes Infect. 2021 doi: 10.1016/j.micinf.2021.104880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran J.P., Bradley E., Zeamer A.L., Cincotta L., Salive M.C., Dutta P., Bucci V. Inflammation-type dysbiosis of the oral microbiome associates with the duration of COVID-19 symptoms and long COVID. JCI Insight. 2021;6(20) doi: 10.1172/jci.insight.152346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera D., Serrano J., Roldán S., Sanz M. Is the oral cavity relevant in SARS-CoV-2 pandemic? Clin. Oral Investig. 2020;24(8):2925–2930. doi: 10.1007/s00784-020-03413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey P.W., Sutton D.A., Fothergill A.W., Rinaldi M.G., Wickes B.L., Schmidt H.J., Walsh T.J. Trichosporon mycotoxinivorans, a novel respiratory pathogen in patients with cystic fibrosis. J. Clin. Microbiol. 2009;47(10):3091–3097. doi: 10.1128/JCM.00460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc.Natl. Acad. Sci. USA. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iebba V., Zanotta N., Campisciano G., Zerbato V., Di Bella S., Cason C., Comar M. Profiling of oral microbiota and cytokines in COVID-19 patients. Front. Microbiol. 2021:1603. doi: 10.3389/fmicb.2021.671813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Tanaka H. SARS-CoV-2 Infection and Significance of Oral Health Management in the Era of “the New Normal with COVID-19”. Int. J. Mol. Sci. 2021;22(12):6527. doi: 10.3390/ijms22126527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.M., Bai P., He W., Wu F., Liu X.F., Han D.M., Yang J.K. Gender differences in patients with COVID-19: focus on severity and mortality. Front. Public. Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, ggpubr:‘ggplot2’Based Publication Ready Plots. R package version 0.2. Retrieved September 12 (2018) 2020.
- Khan A.A., Khan Z. COVID-2019-associated overexpressed Prevotella proteins mediated host–pathogen interactions and their role in coronavirus outbreak. Bioinformatics. 2020;36(13):4065–4069. doi: 10.1093/bioinformatics/btaa285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester S.T., Li N., Lachance D.M., Morella N.M., Dey N. Variability in digestive and respiratory tract Ace2 expression is associated with the microbiome. Plos one. 2021;16(3) doi: 10.1371/journal.pone.0248730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Song Y., Shu Y., Zhao Y., Huo X., Wang H., Liu L. Fungal antigenemia in patients with severe coronavirus disease 2019 (COVID-19): the facts and challenges. J. Microbiol. Immunol. Infect. 2020;53(4):657. doi: 10.1016/j.jmii.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartova M., Tesinska B., Janatova T., Hrebicek O., Mysak J., Janata J., Najmanova L. The oral microbiome in periodontal health. Front. Cell Infect. Microbiol. 2021;11:219. doi: 10.3389/fcimb.2021.629723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Yu J., Ai Q., Liu D., Song C., Li L. Increased constituent ratios of Klebsiella sp., Acinetobacter sp., and Streptococcus sp. and a decrease in microflora diversity may be indicators of ventilator-associated pneumonia: a prospective study in the respiratory tracts of neonates. PloS one. 2014;9(2) doi: 10.1371/journal.pone.0087504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Zhang F., Zhou F., Li H., Ge W., Gan R., Huang Z. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal Transduct. Target Ther. 2021;6(1):1–11. doi: 10.1038/s41392-021-00614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W.H., de Steenhuijsen Piters W.A., Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017;15(5):259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one. 2013;8(4) doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty M., Canceil T., Serino M., Burcelin R., Tercé F., Blasco-Baque V. Oral microbiota-induced periodontitis: a new risk factor of metabolic diseases. Rev. Endocr. Metab. Disord. 2019;20(4):449–459. doi: 10.1007/s11154-019-09526-8. [DOI] [PubMed] [Google Scholar]
- Mishra, B., Ranjan, J., Purushotham, P., Saha, S., Payal, P., Kar, P., & Deshmukh, V. (2021). High proportion of low cycle threshold value as an early indicator of COVID‐19 surge. J. Med. Virol. [DOI] [PMC free article] [PubMed]
- Moustafa A.M., Planet P.J. Emerging SARS-CoV-2 diversity revealed by rapid whole-genome sequence typing. Genome Biol. Evol. 2021;13(9):evab197. doi: 10.1093/gbe/evab197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., Sugita T., Mikami Y. Granuloma associated with Trichosporon asahii infection in the lung: Unusual pathological findings and PCR detection of Trichosporon DNA. Med. Mycol. 2007;45(7):641–644. doi: 10.1080/13693780701435325. [DOI] [PubMed] [Google Scholar]
- Nardelli C., Gentile I., Setaro M., Di Domenico C., Pinchera B., Buonomo A.R., Capoluongo E. Nasopharyngeal microbiome signature in COVID-19 positive patients: can we definitively get a role to fusobacterium periodonticum? Front. Cell Infect. Microbiol. 2021;11:18. doi: 10.3389/fcimb.2021.625581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwirth, E., & Brewer, R. C. (2014). ColorBrewer palettes. R package version, 1.
- Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R. B., & Wagner, H. (2013). Community ecology package. R package version, 2(0).
- Pascolo L., Zupin L., Melato M., Tricarico P.M., Crovella S. TMPRSS2 and ACE2 coexpression in SARS-CoV-2 salivary glands infection. J. Dent. Res. 2020;99(10):1120–1121. doi: 10.1177/0022034520933589. [DOI] [PubMed] [Google Scholar]
- Pietrobon A.J., Teixeira F.M.E., Sato M.N. Immunosenescence and inflammaging: risk factors of severe COVID-19 in older people. Front. Immunol. 2020:2728. doi: 10.3389/fimmu.2020.579220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Salazar C., Kimura K.S., Shilts M.H., Strickland B.A., F Das S.R. SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome. J. Allergy Clin. Immunol. 2021;147(4):1226–1233. doi: 10.1016/j.jaci.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueca M., Fontana A., Bartolini B., Piselli P., Mazzarelli A., Copetti M., Pazienza V. Investigation of nasal/oropharyngeal microbial community of COVID-19 patients by 16S rDNA sequencing. Int. J. Environ. Res. Public Health. 2021;18(4):2174. doi: 10.3390/ijerph18042174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi M., Ahmadikia K., Mahmoudi S., Kalantari S., Jamalimoghadamsiahkali S., Izadi A., Khodavaisy S. Oropharyngeal candidiasis in hospitalised COVID‐19 patients from Iran: Species identification and antifungal susceptibility pattern. Mycoses. 2020;63(8):771–778. doi: 10.1111/myc.13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal L.N., Clemente J.C., Tsay J.C.J., Koralov S.B., Keller B.C., Wu B.G., Weiden M.D. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016;1(5):1–11. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A., Panjabi C., Nair V., Chaudhry R., Thukral S.S. Veillonella as a cause of chronic anaerobic pneumonitis. Int. J. Infect. Dis. 2008;12(6):e115–e117. doi: 10.1016/j.ijid.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Shah V., editor. Vol. 14. 2021. Letter to the Editor: Microbiota in the Respiratory System—A Possible Explanation to Age and Sex Variability in Susceptibility to SARS-CoV-2. (Microbiol. Insights). 1178636120988604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffritti I., D’Accolti M., Fabbri C., Passaro A., Manfredini R., Zuliani G., Caselli E. Oral microbiome dysbiosis is associated with symptoms severity and local immune/inflammatory response in COVID-19 patients: A cross-sectional study. Front. Microbiol. 2021;12:1397. doi: 10.3389/fmicb.2021.687513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bruggen T., Kolecka A., Theelen B., Kwakkel-van Erp J.M., Arets B., Boekhout T. Cutaneotrichosporon (Cryptococcus) cyanovorans, a basidiomycetous yeast, isolated from the airways of cystic fibrosis patients. Med. Mycol. Case Rep. 2018;22:18–20. doi: 10.1016/j.mmcr.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A., Bassis C.M., Beck J.M., Young V.B., Curtis J.L., Huffnagle G.B., Schmidt T.M. Application of a neutral community model to assess structuring of the human lung microbiome. MBio. 2015;6(1):e02284–14. doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yao M., Lv L., Ling Z., Li L. The human microbiota in health and disease. Engineering. 2017;3(1):71–82. [Google Scholar]
- Ward, D. V., Bhattarai, S., Rojas-Correa, M., Purkayastha, A., Holler, D., Da Qu, M., & Maldonado-Contreras, A. (2021). The intestinal and oral microbiomes are robust predictors of COVID-19 severity the main predictor of COVID-19-related fatality. medRxiv.
- Weaver D., Gago S., Bromley M., Bowyer P. The human lung mycobiome in chronic respiratory disease: limitations of methods and our current understanding. Curr. Fungal Infect. Rep. 2019;13(3):109–119. [Google Scholar]
- Wheeler M.L., Limon J.J., Bar A.S., Leal C.A., Gargus M., Tang J., Iliev I.D. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. 2016;19(6):865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., Chang W., Wickham M.H. Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics. Version. 2016;2(1):1–189. [Google Scholar]
- Wickham, H. (2012). reshape2: Flexibly reshape data: a reboot of the reshape package. R package version, 1(2).
- Wu Y., Cheng X., Jiang G., Tang H., Ming S., Tang L., Huang X. Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes. 2021;7(1):1–9. doi: 10.1038/s41522-021-00232-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z., Koo H., Chen Q., Zhou X., Liu Y., Simon-Soro A. Potential implications of SARS-CoV-2 oral infection in the host microbiota. J. Oral Microbiol. 2021;13(1) doi: 10.1080/20002297.2020.1853451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li Y., Gan F., Du Y., Yao Y. Salivary glands: potential reservoirs for COVID-19 asymptomatic infection. J. Dent. Res. 2020;99(8):989. doi: 10.1177/0022034520918518. -989. [DOI] [PubMed] [Google Scholar]
- Xu, R., Liu, P., Zhang, T., Wu, Q., Zeng, M., Ma, Y., & Zhang, C. (2020b). Progressive worsening of the respiratory and gut microbiome in children during the first two months of COVID-19. medRxiv.
- Yamamoto S., Saito M., Tamura A., Prawisuda D., Mizutani T., Yotsuyanagi H. The human microbiome and COVID-19: a systematic review. PloS one. 2021;16(6) doi: 10.1371/journal.pone.0253293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh Y.K., Zuo T., Lui G.C.Y., Zhang F., Liu Q., Li A.Y., Ng S.C. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70(4):698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Liu Z., Chen Y., Xiao Y., Huang X., Fan X.G. Bacterial and fungal infections in COVID-19 patients: a matter of concern. Control Hosp. Epidemiol. 2020;41(9):1124–1125. doi: 10.1017/ice.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijie, S., Yan, X., Lu, K., Wentai, M., Leisheng, S., Li, Z., & Mingkun, L. (2020). Genomic diversity of SARS-CoV-2 in Coronavirus Disease 2019 patients. Clin. Infect. Dis.
- Zuo T., Zhang F., Lui G.C., Yeoh Y.K., Li A.Y., Zhan H., Ng S.C. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159(3):944–955. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material