Summary
To date, intermediate hosts of SARS-CoV-2 remain obscure and controversial. Several studies have shown that SARS-CoV-2-related pangolin coronavirus (Pangolin-CoV) has a high sequence similarity to SARS-CoV-2 and might be the initial source of SARS-CoV-2; however, the biological characteristics of Pangolin-CoV are still largely unknown. In this study, we evaluated the pathogenicity and transmissibility of Pangolin-CoV in Syrian golden hamsters Mesocricetus auratus (Linnaeus, 1758) and compared it with SARS-CoV-2. Pangolin-CoV could effectively infect hamsters, showed similar tissue tropism to SARS-CoV-2 and replicated efficiently in the respiratory system and brain. The infected hamsters had no weight loss but had obvious viral shedding and lung pathological injury. Notably, Pangolin-CoV could transmit between hamsters by direct contact but not via aerosols, and the infected hamsters could exhale infectious viral aerosols (>1 μm). These results highlight the importance of continuous monitoring of coronaviruses in pangolins owing to the potential threat of Pangolin-CoV to human health.
Subject areas: Virology, Cell biology
Graphical abstract
Highlights
-
•
Pangolin-CoV showed similar tissue tropism to SARS-CoV-2 in hamsters
-
•
Pathogenicity and transmissibility of Pangolin-CoV were weaker than SARS-CoV-2
-
•
Pangolin-CoV could transmit between hamsters by direct contact but not via aerosols
-
•
Pangolin-CoV-infected hamsters could exhale infectious virus aerosols (>1 μm)
Virology; Cell biology.
Introduction
The pandemic of coronavirus disease-2019 (COVID-19) caused by SARS-CoV-2 poses unprecedented challenges to global health (Bull et al., 2020). As of 6 February 2022, over 392 million confirmed cases of COVID-19 and over 5.7 million deaths had been reported globally (World Health Organization, 2022). Some of the initial cases were linked to the Huanan seafood market, where live wildlife was sold (Li et al., 2020). However, the potential animal intermediate hosts involved in the transmission of SARS-CoV-2 to humans remain unknown. Lam et al., 2020 and Xiao et al., 2020 identified SARS-CoV-2-related pangolin-associated coronaviruses (Pangolin-CoV) from Malayan pangolins (Manis javanica; Desmarest, 1822), which provided new insights into the potential intermediate hosts. Pangolin-CoV played an important role in the evolution of coronaviruses. A phylogenetic analysis revealed that SARS-CoV-2 might originate from the recombination of a virus similar to Pangolin-CoV and a virus similar to a bat coronavirus (RaTG13, detected in Rhinolophus affinis) (Xiao et al., 2020; Zhou et al., 2020). In particular, the receptor-binding domain (RBD) of Pangolin-CoV shows very high sequence similarity to SARS-CoV-2, indicating a significant public health risk to humans. However, the pathogenicity and transmissibility of Pangolin-CoV remain largely unknown.
Syrian golden hamsters Mesocricetus auratus (Linnaeus, 1758; hereafter referred to as hamster) are a susceptible small animal model for studying both the pathogenicity and transmission of SARS-CoV-2 (Sia et al., 2020; Chan et al., 2020) and the evaluation of vaccines, immunotherapies, and antiviral drugs (Imai et al., 2020; Yahalom-Ronen et al., 2020; Rogers et al., 2020; Tortorici et al., 2020; Kreye et al., 2020). SARS-CoV-2 replicated efficiently in the respiratory tract of hamsters and could be transmitted between hamsters by the aerosol route. Microcomputed tomographic imaging revealed that the infected hamsters shared similar lung injury with SARS-CoV-2-infected humans (Imai et al., 2020; Simpson et al., 2020; Salehi et al., 2020). In addition, hamsters are convenient models in terms of size, cost, and husbandry requirements, hamsters can be used for large-scale studies, and obtaining statistically robust data using hamsters is easy.
In this study, we evaluated the pathogenicity and transmissibility of Pangolin-CoV (GX/P2V) in golden hamsters and compared it with a SARS-CoV-2 strain. The body weight changes, viral titers in organ tissues, lung histopathology, transmission efficiency, and neutralizing antibody titer of the infected hamsters were analyzed. In particular, we determined the concentrations and sizes of viral aerosols exhaled by Pangolin-CoV- and SARS-CoV-2-infected hamsters. These studies expand our understanding of the biological characteristics of Pangolin-CoV viruses and will aid in the Pangolin-CoV pandemic preparedness efforts.
Results
Pathogenicity comparison of Pangolin-CoV and SARS-CoV-2 in hamsters
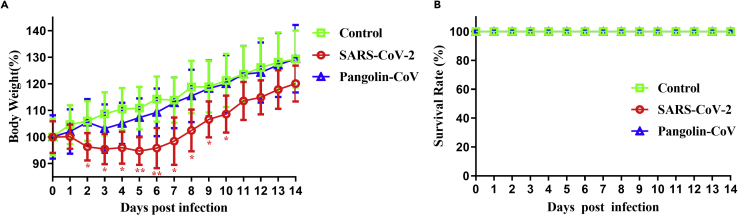
We evaluated the pathogenicities of Pangolin-CoV and SARS-CoV-2 in a hamster model. Hamsters inoculated with Pangolin-CoV showed no substantial loss of body weight (Figure 1A), whereas the body weights of hamsters infected with SARS-CoV-2 gradually decreased by approximately 5.3% during the first 5 dpi before gradually recovering to normal (Figure 1A). No hamsters died during the experiment (Figure 1B).
Figure 1.
Body weight change of Pangolin-CoV- and SARS-CoV-2-infected hamsters
Groups of five five-week-old male hamsters were intranasally inoculated with 105 TCID50 of Pangolin-CoV (GX/P2V) or SARS-CoV-2 (V34). Another five hamsters challenged with PBS were used as controls.
(A and B) The weight loss (A) and survival rates (B) of hamsters in these groups were monitored daily for 14 days. Body weight data (bars show mean ± SD) were analyzed using two-way ANOVA, and comparisons between two groups were made using the SNK method. ∗p < 0.05; ∗∗p < 0.01. The red asterisks represent the significant difference between SARS-CoV-2 and the control group. No significant differences were found between Pangolin-CoV and the control group.
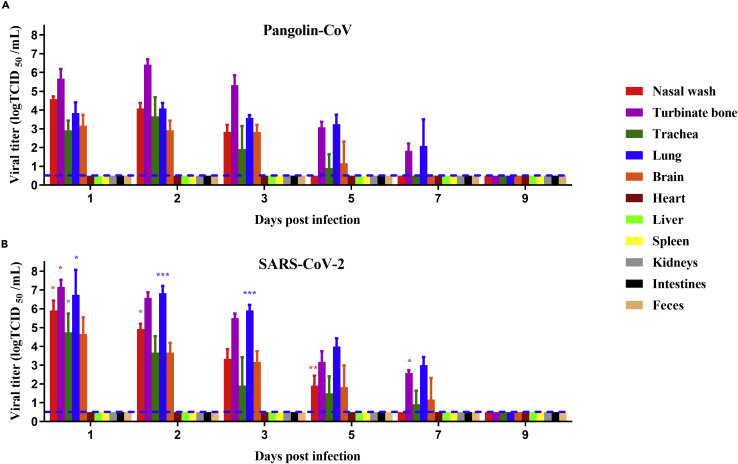
We next investigated the replication capabilities and systemic viral spreads of Pangolin-CoV and SARS-CoV-2. The two coronaviruses exhibited the same tissue tropism and could effectively replicate in the respiratory system (turbinate bones, trachea, and lung) and brain of hamsters (Figure 2). No infectious viruses were detected in feces or other tissues, including heart, liver, spleen, kidneys, and intestines. The viral titers in the respiratory system and brain peaked at one to two dpi and then gradually decreased. The replication capabilities of SARS-CoV-2 were higher than those of Pangolin-CoV. For instance, at one dpi, the viral titers in nasal wash, turbinate bones, trachea, lung, and brain of SARS-CoV-2-infected hamsters were 1.33, 1.5, 1.83, 2.9, and 1.5 log10 TCID50 higher than that of Pangolin-CoV, respectively. In addition, viral shedding in the nasal wash of Pangolin-CoV-infected hamsters lasted for 3 days, which was 2 days shorter than the viral shedding period of SARS-CoV-2-infected hamsters.
Figure 2.
Viral shedding and tissue distribution of Pangolin-CoV and SARS-CoV-2
Groups of 18 five-week-old male hamsters were intranasally inoculated with 105 TCID50 of Pangolin-CoV (GX/P2V) or SARS-CoV-2 (V34). At 1, 2, 3, 5, 7, and 9 dpi, the nasal washes and feces of three hamsters per group were collected, and then these hamsters were euthanized. Different tissue samples, including heart, liver, spleen, lung, kidney, brain, turbinate bones, intestine, trachea, and fecal samples, were homogenized in 1mL of PBS. Viral titers in the nasal washes and homogenized sample supernatants were determined by the Reed–Muench method. The results are presented as the mean ± SD, and all data were analyzed using one-way ANOVA. Comparisons between two groups were made using the SNK method. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Pangolin-CoV vs. SARS-CoV-2). The dashed lines indicate the lower limit of detection.
(A) Viral shedding and tissue distribution of Pangolin-CoV.
(B) Viral shedding and tissue distribution of SARS-CoV-2.
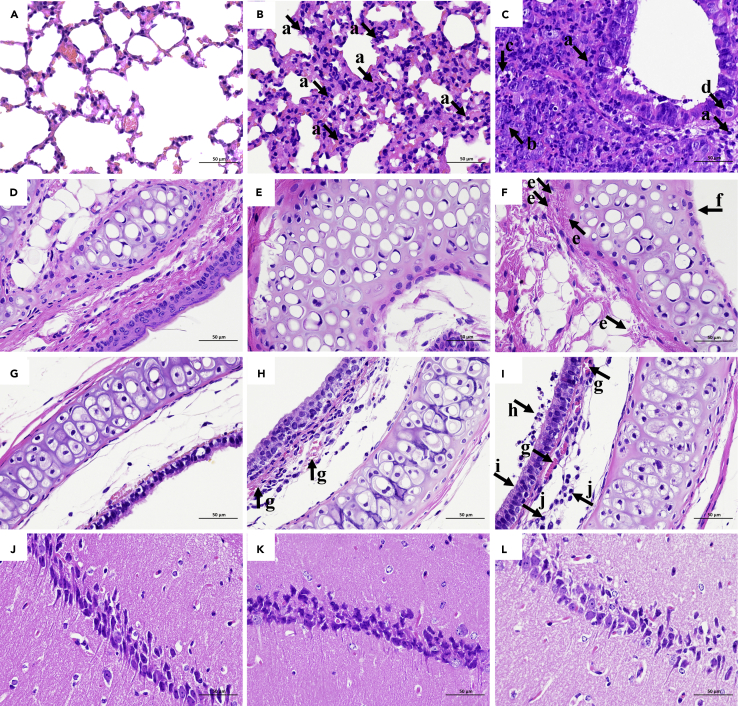
We further examined the histopathological changes, and no obvious histopathological changes were observed in the lungs, turbinate bones, trachea, and brain of the negative control group (Figures 3A, 3D, 3G, and 3J). The lungs of Pangolin-CoV-infected hamsters showed widespread alveolar wall thickening, accompanied by scattered infiltration of lymphocytes and neutrophils (Figure 3B). In contrast, the lungs of SARS-CoV-2-infected hamsters showed severe alveolar wall thickening, accompanied by the infiltration of large numbers of lymphocytes and neutrophils and a small number of macrophages (Figure 3C). A ring of inflammatory cells could be seen around the local vessels, forming vascular cuffs. There was a large amount of epithelial proliferation in the lungs with enlarged nuclei and mitotic patterns, as well as a small amount of cell necrosis and nuclear fragmentation. Pulmonary edema with eosinophilic serous exudation in the alveolar space was also observed. The turbinate bones of Pangolin-CoV-infected hamsters showed no obvious histopathological changes (Figure 3E), while the turbinate mucosa pseudostratified columnar ciliated epithelium of SARS-CoV-2-infected hamsters was completely necrotic and exfoliated, and hyperemia of the inferior vessels of the turbinate osteochondral was also observed (Figure 3F). The changes of tracheal mucosa in Pangolin-CoV-infected hamsters were not significant, but there was a small amount of hemorrhage in the submucosa (Figure 3H). In contrast, the trachea of SARS-CoV-2-infected hamsters showed marked submucosal hyperemia, accompanied by lymphocyte infiltration (Figure 3I). The mucosal surface of the trachea is covered with mucus and lymphocytes. The ciliated layer of trachea mucosa pseudostratified columnar ciliated epithelial cells was necrotic and exfoliated. No obvious histopathological changes were observed in the brain of Pangolin-CoV- and SARS-CoV-2-infected hamsters (Figures 3K and 3L). Histological analysis demonstrated that SARS-CoV-2-infected hamsters exhibited more severe histopathological changes than the Pangolin-CoV-infected hamsters.
Figure 3.
Histopathology of the tissues of hamsters inoculated with PBS, Pangolin-CoV, or SARS-CoV-2
On three dpi, (A): lungs of PBS group, (B): lungs of Pangolin-CoV group, (C): lungs of SARS-CoV-2 group, (D): turbinate bones of PBS group, (E): turbinate bones of Pangolin-CoV group, (F): turbinate bones of SARS-CoV-2 group, (G): trachea of PBS group, (H): trachea of Pangolin-CoV group, (I): trachea of SARS-CoV-2 group, (J): brain of PBS group, (K): brain of Pangolin-CoV group, (L): brain of SARS-CoV-2 group. The collected samples were fixed in formalin, embedded in paraffin and stained with hematoxylin and eosin (H & E) for pathological examination. Images were obtained at 40× magnification. Arrow a: lymphocyte and neutrophil infiltration; arrow b: epithelial proliferation with enlarged nuclei and mitotic patterns; arrow c: cell necrosis and nuclear fragmentation; arrow d: pulmonary edema with eosinophilic serous exudation in the alveolar space; arrow e: hyperemia of the inferior vessels of the turbinate osteochondral; arrow f: the mucosa pseudostratified columnar ciliated epithelium was completely necrotic and exfoliated, while the submucosa was necrotic and exfoliated; arrow g: hyperemia in the submucosa; arrow h: the mucosal surface of the trachea is covered with mucus and lymphocytes; arrow i: ciliated layer of trachea mucosa pseudostratified columnar ciliated epithelial cells was necrotic and exfoliated; arrow j: the submucosa is infiltrated with lymphocytes.
In summary, the data above indicated that Pangolin-CoV produced moderate pathogenicity in hamsters and were less virulent than SARS-CoV-2.
Varying transmissibility of Pangolin-CoV and SARS-CoV-2 in hamsters
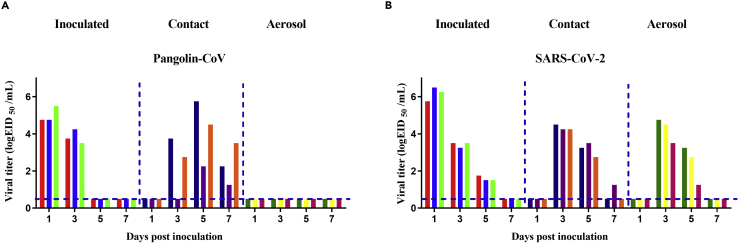
We further evaluated the transmissibility of Pangolin-CoV and SARS-CoV-2 in hamsters. Nasal washes from the inoculated, contract, and aerosol-transmission groups were collected at 2-day intervals and were titrated for virus in VeroE6 cells. Pangolin-CoV was detected in nasal washes of the naive hamsters of the contact group, but not in the nasal washes of the aerosol group (Figure 4A); therefore, Pangolin-CoV had the ability to spread via contact transmission (100%, 3/3) but not via the aerosol route in hamsters. In contrast, SARS-CoV-2 showed efficient contact transmissibility and efficient aerosol transmissibility with a transmission efficiency of 100% (3/3, Figure 4B).
Figure 4.
Transmissibility of Pangolin-CoV and SARS-CoV-2 in hamsters
For contact transmission, three hamsters were intranasally inoculated with 105 TCID50 of Pangolin-CoV or SARS-CoV-2. Three naive guinea pigs were cohoused with the three infected hamsters 24 h post-inoculation. For aerosol transmission, three hamsters were intranasally inoculated with 105 TCID50 of Pangolin-CoV (A) or SARS-CoV-2 (B). The next day, they were transferred to a wire-frame cage adjacent to another three naive hamsters. The distance between the donor hamsters and the airborne-contact hamsters was 1.8 cm. Nasal washes were collected from all hamsters for viral shedding detection every other day beginning on day 1 post-inoculation or exposure. Each color bar represents the viral titer in an individual hamster. Dashed lines indicate the lower limit of virus detection.
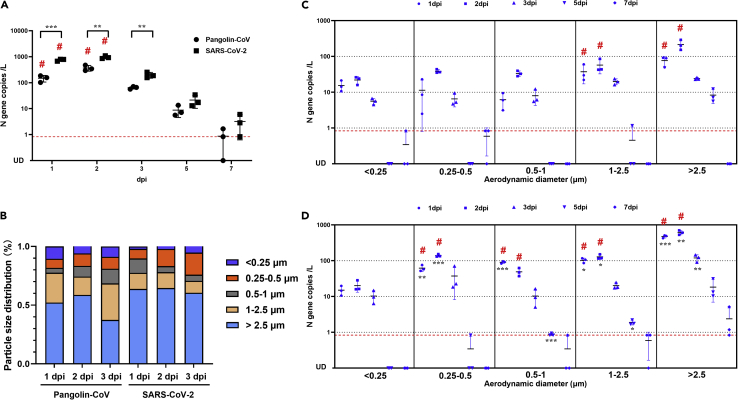
Concentrations and sizes of viral aerosols exhaled by Pangolin-CoV- and SARS-CoV-2-infected hamsters
To better understand aerosol transmissibility, aerosols exhaled by Pangolin-CoV- or SARS-CoV-2-infected hamsters were collected by a five-stage miniature cascade impactor at 1, 2, 3, 5, and 7 dpi. As shown in Figure 5A, the total viral aerosol concentrations peaked at two dpi and then gradually decreased during the duration of the infection. Viral aerosols were rarely detected 7 days after infection. In particular, infectious virus was recovered from the aerosol samples of both groups sampled at one and two dpi, indicating that Pangolin-CoV and SARS-CoV-2 were most contagious in the early stages of infection. SARS-CoV-2-infected hamsters produced significantly higher amounts of viral aerosols than the Pangolin-CoV-infected hamsters. At two dpi, the concentration of virus-laden particles exhaled by Pangolin-CoV-infected hamsters was 367.58 ± 92.43 copies per liter of air. On average, each hamster exhaled 661.64 viral particles per minute; in contrast, the concentration of virus-laden particles exhaled by SARS-CoV-2-infected hamsters was 951.67 ± 121.53 copies per liter of air. On average, each hamster exhaled 1713.01 virus particles per minute, approximately 2.59 times higher than that of Pangolin-CoV.
Figure 5.
Concentrations and sizes of viral aerosols exhaled by Pangolin-CoV- and SARS-CoV-2-infected hamsters
(A) Total viral aerosol concentrations exhaled by Pangolin-CoV- and SARS-CoV-2-infected hamsters at 1, 2, 3, 5, and 7 dpi.
(B) Size distribution of the virus-laden particles.
(C) Quantity and size distribution of virus-laden particles exhaled by Pangolin-CoV-infected hamsters.
(D) Quantity and size distribution of virus-laden particles exhaled by SARS-CoV-2-infected hamsters. Data in (A), (C), and (D) were overlaid with mean ± SD. The limit of linear range of quantification (0.833 N gene copies per liter of air) is shown with the red dotted line. # indicates that infectious virus was recovered from the aerosol samples. All data were analyzed using one-way ANOVA, and comparisons between two groups were made using the SNK method. ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001 (Pangolin-CoV vs. SARS-CoV-2).
We also measured the size distribution of the viral aerosol particles that were exhaled by the hamsters (Figure 5B), and Pangolin-CoV and SARS-CoV-2 had the same distribution tendency. Most of the viral particles were distributed in the first stage (>2.5 μm), and the fifth stage (<0.25 μm) was the least distributed. The distribution of the viral aerosol particles at levels 1 and 2 (>1 μm) accounted for more than 68% of the total viral aerosol particles.
A more detailed quantity and size distribution of virus-laden particles is shown in Figures 5C and 5D. For Pangolin-CoV, infectious virus was recovered from the first two stages (>1 μm) of aerosol samples at one dpi and two dpi, the concentrations of virus-laden particles were 76.85 ± 23.49 copies/L (>2.5 μm) and 37.44 ± 20.29 copies/L (1–2.5 μm) at 1dpi and were 215.80 ± 70.44 copies/L (>2.5 μm) and 57.48 ± 24.79 copies/L (1–2.5 μm) at two dpi, respectively. For SARS-CoV-2, infectious virus was recovered at smaller particle sizes (>0.25 μm). The concentrations of virus-laden particles were 476.76 ± 44.21 copies/L (>2.5 μm), 103.10 ± 15.91 copies/L (1–2.5 μm), 90.78 ± 6.59 copies/L (0.five to one μm) and 62.13 ± 12.60 copies/L (0.25–0.5 μm) at one dpi and 614.09 ± 85.16 copies/L (>2.5 μm), 128.48 ± 19.99 copies/L (1–2.5 μm), 49.08 ± 12.83 copies/L (0.five to one μm) and 139.79 ± 11.90 copies/L (0.25–0.5 μm) at two dpi.
Serum neutralizing antibodies of Pangolin-CoV- and SARS-CoV-2-infected hamsters showed cross-neutralizing effects
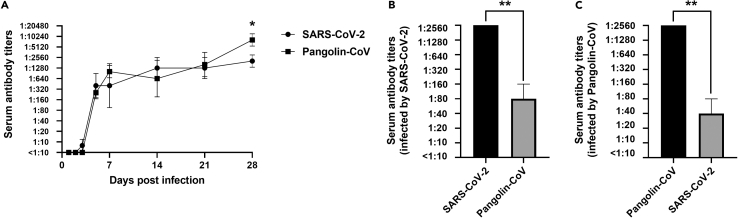
We next determined the dynamic growth of neutralizing antibody titers of Pangolin-CoV- and SARS-CoV-2-infected hamsters and asked whether the developed antibodies were resistant to each other. Serum samples were collected at 1, 2, 3, 5, 7, 14, 21, and 28 dpi, and viral neutralization titers were titrated against the inoculated virus. As shown in Figure 6A, Pangolin-CoV-infected hamsters started to develop neutralizing antibodies at five dpi, while SARS-CoV-2-infected hamsters started to develop neutralizing antibodies at three dpi, indicating that Pangolin-CoV induced a relatively later antibody response than SARS-CoV-2. However, the antibody titers induced by Pangolin-CoV were comparable to those induced by SARS-CoV-2 at 5, 7, 14, and 21 dpi and were significantly higher at 28 dpi.
Figure 6.
Dynamic growth of serum neutralizing antibody titers and the cross-neutralization effect
(A–C) Serum samples were collected from Pangolin-CoV- or SARS-CoV-2-infected hamsters at 1, 2, 3, 5, 7, 14, 21, and 28 dpi. The neutralizing antibody titers were titrated in VeroE6 cells (A). To determine the cross-neutralization titers, three antibody samples of SARS-CoV-2 (B)- or Pangolin-CoV (C)-infected hamsters with 1:2560 neutralizing titers were tested for neutralization titers with the other virus. For all panels, bars show mean ± SD. All data were analyzed using one-way ANOVA, and comparisons between two groups were made using the SNK method. ∗p < 0.05 and ∗∗p < 0.01 (SARS-CoV-2 vs. Pangolin-CoV).
Afterward, we chose three antibody samples with 1:2560 neutralizing titers per group and tested the cross-neutralization titers. As shown in Figures 6B and 6C, the neutralizing antibody induced by one coronavirus could also neutralize the other virus, but the cross-neutralization titers were significantly lower than the self-neutralization titers (p < 0.01).
Discussion
In this study, we found that Pangolin-CoV could effectively infect golden hamsters and replicate efficiently in their respiratory system and brain. However, the pathogenicity of Pangolin-CoV was relatively weak compared with that of SARS-CoV-2, and Pangolin-CoV could transmit between hamsters by direct contact but not via aerosols.
The infection begins with the binding of spike glycoprotein of both Pangolin-CoV and SARS-CoV-2 to angiotensin-converting enzyme 2 (ACE2) on the surface of host cells (Zhou et al., 2020; Hoffmann et al., 2020). Pangolin-CoV and SARS-CoV-2 exhibited the same tissue tropism in our study (Figure 2), indicating that they might have the same receptor affinity. No infectious viruses were detected in organs other than the respiratory system and brain, which is consistent with previous studies (Imai et al., 2020).
Both pangolins and hamsters are mammals; Xiao et al., 2020 reported that the Pangolin-CoV-positive pangolins they studied showed obvious clinical signs of disease, including shortness of breath, emaciation, lack of appetite, inactivity, and crying. However, a direct association between the clinical signs and active viral replication was lacking. In this study, Pangolin-CoV-infected hamsters exhibited no obvious clinical signs; thus, the clinical signs of Pangolin-CoV-positive pangolins in Xiao’s study might be caused by other reasons. In addition, SARS-CoV-2 showed higher pathogenicity than Pangolin-CoV, and the infected hamsters exhibited pronounced body weight changes, higher viral replication, and more severe pathological changes. One possible reason might be that a polybasic (furin-like) S1/S2 cleavage site was found in the spike glycoprotein of SARS-CoV-2 (Coutard et al., 2020) but was absent in Pangolin-CoV. Deletions at the S1/S1 junction of SARS-CoV-2 significantly reduced the pathogenicity of SARS-CoV-2 in hamsters (Lau et al., 2020).
However, infectious virus was recovered from the aerosol exhaled by Pangolin-CoV-infected hamsters (Figure 5A), but aerosol transmission still did not occur between the doner and naive hamsters (Figure 4A). There might be two reasons for the lack of aerosol transmission. First, Pangolin-CoV-infected hamsters produced fewer virus-laden particles than aerotransmissible SARS-CoV-2-infected hamsters (Figures 5A, 5C, and 5D). Second, the infectious viral particles exhaled by Pangolin-CoV-infected hamsters were larger in size (>1 μm) (Figures 5C and 5D). In the structure of the human respiratory tract, the commonly accepted respirable particle size range is < 5 μm (Shiu et al., 2019; Guo et al., 2020; Tellier et al., 2019). However, the structure of the hamster respiratory tract is smaller than that of humans, and the respirable particle size of hamsters might also be far less than that of humans. In this study, the infectious SARS-CoV-2 particles exhaled were recovered from aerosol particles >0.25 μm, indicating that infectious viral aerosols with a particle size range of 0.25–0.5 μm might play an important role in aerosol transmission between hamsters.
Both Pangolin-CoV and SARS-CoV-2 could induce hamsters to produce obvious neutralizing antibodies, and the neutralizing antibody induced by one coronavirus provided protective immunity against the other (Figure 6). Therefore, after proper modification, Pangolin-CoV might have the potential to be developed as a live attenuated SARS-CoV-2 vaccine or could be incorporated into other vaccines.
In summary, Pangolin-CoV showed similar infection characteristics to SARS-CoV-2, but the pathogenicity and transmissibility were relatively weaker than those of SARS-CoV-2. Notably, Pangolin-CoV could be transmitted by direct contact, and the infected hamsters could exhale infectious viral aerosols. Therefore, the public health risk of Pangolin-CoV being potential candidates for global dissemination could not be ignored. The continual monitoring of the mutation and evolution of Pangolin-CoV should be implemented in the future, and the illegal wildlife trade of pangolins should be effectively controlled.
Limitations of the study
There are two limitations of our study that need to be noted. First, the pangolin-CoV evaluated in this study was limited in quantity, and the evaluated GX/P2V strain may not represent the whole Pangolin-CoV situation. More strains should be expanded in the future. Second, in the study of systemic viral spreads in infected hamsters, the concentration of viral RNA in the tissues of hamsters was not quantified. Because if infectious viruses cannot be isolated from a tissue, even the presence of viral RNA does not indicate that the virus can effectively infect the tissue. In this respect, the viral titration results are more significant than the viral RNA concentration. However, data on viral RNA concentrations in tissues will, indeed, provide additional information to help us further understand the infectious properties of Pangolin-CoV.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 BetaCoV/Beijing/IME-BJ05-2020 | Biological Sample Library | SAMC138020 |
| Pangolin coronavirus isolate PCoV_GX-P2V | GenBank | MT072864.1 |
| Experimental models: Cell lines | ||
| VeroE6 cells | ATCC | CRL-1586 |
| Experimental models: Organisms/strains | ||
| Syrian Golden Hamster (Mesocricetus auratus) | Charles River Labs | Strain code 049 |
| Oligonucleotides | ||
| Primer for GX/P2V, Forward: AGGTGACGAGGTTAGACAAATAG | This paper | N/A |
| Primer for GX/P2V, Reverse: CCAAGCAATAACACAACCAGTAA | This paper | N/A |
| Primer for GX/P2V, Probe: FAM-5′-ACCCGGACAAACTGGTGTTATTGCT-3′-TAMRA | This paper | N/A |
| Primer for SARS-CoV-2, Forward: GGGGAACTTCTCCTGCTAGAAT | WHO | N/A |
| Primer for SARS-CoV-2, Reverse: CAGACATTTTGCTCTCAAGCTG | WHO | N/A |
| Primer for SARS-CoV-2, Probe: FAM-5′-ACCCGGACAAACTGGTGTTATTGCT-3′-TAMRA | WHO | N/A |
| Software and algorithms | ||
| PRISM | GraphPad software | Version 8 |
| Other | ||
| Miniature cascade impactors | SKC | Cat#225-370 |
| Gelatin filters | Sartorius | Cat#12602-25-ALK; Cat#12602-37-ALK |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yuwei Gao (gaoyuwei@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Viruses and culture
The SARS-CoV-2-related pangolin coronavirus, named GX/P2V, was isolated from frozen tissue samples of Malayan pangolins (M. javanica; Desmarest, 1822) which were collected between August 2017-January 2018 in Guangxi, China, as described previously (Lam et al., 2020). The SARS-CoV-2 strain, named BetaCoV/Beijing/IME-BJ05-2020 (abbreviated as V34), was isolated from COVID-19 patients in Beijing, China in 2020. Viruses were grown in VeroE6 cells and stored at −80 °C. VeroE6 cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) high glucose (Gibco), supplemented with 10% fetal bovine serum, and 100 U/mL penicillin-streptomycin. Cells were cultured at 37°C in a 5% CO2 incubator.
Hamsters
Five-week-old male Syrian golden hamsters were used in this study. They were purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd, and randomly assigned to each experimental group.
Ethics statement
All experiments with hamsters were performed in strict accordance with the guidelines of animal welfare of the World Organization for Animal Health. Experimental protocols involving hamsters were approved by the Animal Care and Use Committee of the Changchun Veterinary Institute (approval number: SMKX-20200915–11). All experiments with SARS-CoV-2 and SARS-CoV-2-related pangolin coronavirus were performed in a biosecurity level 3 laboratory.
Methods details
Pathogenicity evaluation
Groups of five five-week-old male golden hamsters were anesthetized with isoflurane and intranasally inoculated with 100 μL of GX/P2V or V34 at 105 TCID50. The weight loss and survival rates of the golden hamsters in these groups were monitored daily for 14 days. The percentages of body weight change for each group were calculated by comparing the group average weight with their initial average weight.
To assess the viral growth and pathological changes in the tissues of the infected golden hamsters, 18 golden hamsters per group were anesthetized with isoflurane and intranasally inoculated with either the GX/P2V or V34 virus at 105 EID50, while another three golden hamsters that were intranasally inoculated with PBS served as the control group. At 1, 2, 3, 5, 7 and 9 days post infection (dpi), the nasal washes and feces of three golden hamsters per group were collected, and then these golden hamsters were euthanized. The tissues, including heart, liver, spleen, lung, kidneys, brain, turbinate bones, intestines, and trachea, of these golden hamsters were removed to determine the viral titers. The feces and tissue samples were homogenized in 1 mL of PBS by Tissue Lyser (QIAGEN, Germany) and centrifuged at 12,000 rpm and 4°C. The supernatants were then collected and inoculated into VeroE6 cells. The nasal washes were directly inoculated into Vero-E6 cells. After 96 h of incubation at 37°C, the viral cytopathic effects (CPEs) were observed, and the TCID50 was determined by the Reed method. At 3 dpi, the lungs, turbinate bones, trachea, and brain of the infected golden hamsters were fixed in formalin, embedded in paraffin and stained with hematoxylin and eosin (H & E) for pathological examination.
Transmission evaluation
Six donor hamsters per group were intranasally inoculated with 105 TCID50 of either the GX/P2V or V34 virus. At 24 h post inoculation, 3 donors per group were transferred to a new cage and cohoused with three naïve hamsters for the direct contact transmission studies, and another 3 donors per group were transferred to a wire-frame cage adjacent to another 3 naïve hamsters for the airborne transmission studies. The distance between the donor hamsters and the airborne-contact hamsters was 1.8 cm, as described previously (Sia et al., 2020). Nasal washes of these hamsters were collected and titrated at 1, 3, 5, and 7 days post inoculation or exposure.
Viral aerosol emission
Five hamsters per group were intranasally inoculated with 105 TCID50 of either the GX/P2V or V34 virus. At 1, 2, 3, 5, and 7 days post infection (dpi), segregated aerosol samples in the breeding cages were collected using miniature cascade impactors (Sioutas impactor, SKC Inc., US) at a flow rate of 9.0 liters per minute for 2 hours. The aerosol particles were separated into five ranges according to their aerodynamic diameters (>2.5 μm, 1.0 to 2.5 μm, 0.5 to 1.0 μm, 0.25 to 0.50 μm and <0.25 μm) and collected on presterilized gelatin filters (Sartorius, Germany). Each filter was cut into two pieces on average after aerosol sampling. One piece was used for RNA extraction and subsequent viral nucleic acid testing, and another piece was directly inoculated into VeroE6 cells to determine whether infectious virus existed in the aerosol.
Viral nucleic acid testing
RNA was extracted using the QIAamp Viral RNA Mini kit (Qiagen, Germantown, MD, USA) and detected using the One Step PrimeScriptTM RT-PCR Kit (TaKaRa, Japan) according to the manufacturers’ protocol. Quantitative real-time PCR (Q-RT-PCR) assays were performed by using a set of primers and probes, targeting regions of the N gene [For GX/P2V, Forward: 5′-AGGTGACGAGGTTAGACAAATAG-3′; Reverse: 5′-CCAAGCAATAACACAACCAGTAA-3′; Probe: FAM-5′-ACCCGGACAAACTGGTGTTATTGCT-3′-TAMRA. For V34, Forward: 5′-GGGGAACTTCTCCTGCTAGAAT-3′; Reverse: 5′-CAGACATTTTGCTCTCAAGCTG-3′; Probe: FAM-5′-TTGCTGCTGCTTGACAGATT-3′-TAMRA], and the PCRs were run on the ABI 7500 System (ThermoFisher, Waltham, MA, USA).
Neutralizing antibody titers
Three hamsters per group were intranasally inoculated with 105 TCID50 of either the GX/P2V or V34 virus. Serum samples were collected at 1, 2, 3, 5, 7, 14, 21 and 28 dpi. 100 μL of virus (100 TCID50) was incubated with 50 μL of two-fold serial dilutions of sera in 96-well plates for 1 h at room temperature, and then 50 μL of VeroE6 cell suspension was added to the mixture. The cells were further incubated for 4 days at 37°C. The viral CPEs were observed under an inverted microscope, and virus neutralization titers were determined as the reciprocal of the highest serum dilution that completely prevented the CPE.
Quantification and statistical analysis
Quantitative data were analyzed with GraphPad Prism software (San Diego, CA, USA) using one-way or two-way analysis of variance (ANOVA) method, and statistical comparisons between two groups were tested using the Student-Newman-Keuls (SNK) method. All of the assays were run in triplicate and are representative of at least 3 separate experiments. The error bars represent the standard deviation. p-values less than 0.05 indicated significant differences.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (82150202).
Author contributions
Z.D.G., C.Z., C.M.Z. and Y.W.G. conceived the idea and designed the experiments. Z.D.G., C.Z., C.M.Z., H.C., Z.L.C., and X.Y.J. performed the animal experiments. T.C.W. and Y.G.L. completed the pathological analysis of lung tissue. J.L., Z.H.W., K.Y.M., and J.P.L. designed, analyzed, and interpreted the data. Z.D.G., Y.G.T., and Y.W.G. wrote the article.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Contributor Information
Yigang Tong, Email: tong.yigang@gmail.com.
Yuwei Gao, Email: gaoyuwei@gmail.com.
Data and code availability
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bull S., Jamrozik E., Binik A., Parker M.J. SARS-CoV-2 challenge studies: ethics and risk minimisation. J. Med. Ethics. 2020;47:e79. doi: 10.1136/medethics-2020-106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., et al. Simulation of the clinical and pathological Manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease Pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.D., Wang Z.Y., Zhang S.F., Li X., Li L., Li C., Cui Y., Fu R.B., Dong Y.Z., Chi X.Y., et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, wuhan, China, 2020. Emerg. Infect. Dis. 2020;26:1583–1591. doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sanchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.C.D., et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.Y., Jia N., Zhang Y.W., Shum M.H.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lau S.Y., Wang P., Mok B.W.Y., Zhang A.J., Chu H., Lee A.C.Y., Deng S., Chen P., Chan K.H., Song W., et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S., Lau E.H., Wong J.Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR. Am. J. Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- Shiu E.Y., Leung N.H., Cowling B.J. Controversy around airborne versus droplet transmission of respiratory viruses: implication for infection prevention. Curr. Opin. Infect. Dis. 2019;32:372–379. doi: 10.1097/QCO.0000000000000563. [DOI] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson S., Kay F.U., Abbara S., Bhalla S., Chung J.H., Chung M., Henry T.S., Kanne J.P., Kligerman S., Ko J.P., Litt H. Radiological society of north america expert consensus statement on reporting chest CT findings related to COVID-19. Endorsed by the society of thoracic radiology, the American college of radiology, and RSNA – secondary publication. J. Thorac. Imaging. 2020;35:219–227. doi: 10.1097/RTI.0000000000000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier R., Li Y., Cowling B.J., Tang J.W. Recognition of aerosol transmission of infectious agents: a commentary. BMC Infect. Dis. 2019;19:101. doi: 10.1186/s12879-019-3707-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Weekly Operational Update on COVID-19 - 8 February 2022. https://www.who.int/publications/m/item/weekly epidemiological-update-on-covid-19---8-february-2022
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., Vitner E.B., Israeli O., Milrot E., Stein D., et al. A single dose of recombinant VSV-ΔG-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11:6402. doi: 10.1038/s41467-020-20228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.