Abstract
Recently, it has been demonstrated that dysbiosis, an alteration in commensal microflora composition, is intimately involved in the onset of a variety of diseases. It is becoming increasingly evident that the composition of commensal microflora in the oral cavity is closely connected to oral diseases, such as periodontal disease, and systemic diseases, such as inflammatory bowel disease. Next-generation sequencing techniques are used as a method to examine changes in bacterial flora, but additional analytical methods to assess bacterial flora are needed to understand bacterial activity in more detail. In addition, the oral environment is unique because of the role of secretory antibodies contained in saliva in the formation of bacterial flora. The present study aimed to develop a new method for evaluating the compositional change of microbiota using flow cytometry (FCM) with specific antibodies against the bacterial surface antigen, as well as salivary antibodies. Using specific antibodies against Streptococcus mutans, a causative agent of dental caries, and human IgA, bacterial samples from human saliva were analyzed via FCM. The results showed that different profiles could be obtained depending on the oral hygiene status of the subjects. These results suggest that changes in the amount and type of antibodies that bind to oral bacteria may be an indicator for evaluating abnormalities in the oral flora. Therefore, the protocol established in this report could be applied as an evaluation method for alterations in the oral microbiota.
Keywords: Oral microbiology, Dysbiosis, Saliva, sIgA, Streptococcus mutans, LPxTG protein
Abbreviations: FCM, flow cytometry; BHI, brain heart infusion
Highlights
-
•
We aimed to develop a new method for evaluating dysbiosis using flow cytometry.
-
•
We used bacterial surface antigen-specific antibodies and salivary antibodies.
-
•
Different profiles could be obtained depending on oral hygiene status.
-
•
Changes in antibodies bound to oral bacteria may indicate oral flora abnormalities.
-
•
Our method can be used to evaluate alterations in the oral microbiota.
1. Introduction
Numerous recent studies on bacterial flora have revealed that dysbiosis, a compositional abnormality in commensal bacterial flora, is closely related to health maintenance and disease pathogenesis [1,2]. It has become increasingly evident that dysbiosis in the oral region, as well as in the intestinal tract and skin, is involved in oral and systemic diseases [3,4]. Therefore, the importance of assessing the compositional change of oral microbiota is increasing. Meta-analysis of the 16S rRNA gene using next-generation sequencers is now a standard method to evaluate dysbiosis. This method is considered effective for understanding the general profile and large-scale variation of the bacterial flora in a test sample, since it can identify the bacteria comprising the microflora at the genus and species levels from a large amount of data. More recently, to obtain a more detailed description of the behavior of each bacterium in the bacterial flora, methods such as meta-transcriptomic analysis have been developed [5]. In addition, host factors, such as the sub-mucosal immune system, are involved in microflora compositional changes, but data on these factors cannot be acquired through meta-analysis of the 16S rRNA gene. Solving the abovementioned problem, flow cytometry (FCM) can simultaneously analyze changes in bacterial flora composition and host factors [6]. FCM is widely used to identify cells by their size, internal structure, and surface molecules and to quantitatively analyze the ratio of specific cell types present in a cell population [7]. The ability of FCM to readily collect a multitude of information is useful for analyzing dysbiosis and has been applied to clinical and basic research [[8], [9], [10]]. Various immune components in the mucosal membrane play an important role in mucosal flora formation. In particular, it has been reported that secretory IgA (sIgA) binds to antigens on the surface of bacteria and eliminates them by agglutination, or that it binds to mucin on the mucosal surface and mediates bacterial colonization of the mucosal surface by binding to bacterial surface antigens [11,12]. Therefore, examining the binding state of bacteria to IgA when analyzing changes in bacterial flora is crucial to accurately understand the state of bacteria-host interactions in the mucosa. In this study, we focused on dental caries as an example of oral dysbiosis [13] and developed a method to evaluate bacterial flora using FCM by combining antibodies to detect Streptococcus mutans the causative agent of dental caries, and sIgA from saliva glands.
2. Materials and methods
2.1. Sample collection
Five dentally-healthy volunteers provided approximately 1 ml of unstimulated saliva immediately after awaking without cleaning their mouths on the previous night. Then, they cleaned their mouths in the usual manner and saliva was collected again. The saliva samples were kept on ice until analysis. Informed consent was obtained from all participants. The study was approved by the Showa University Research Ethics Review Board (#2021-003).
2.2. Bacterial culture
Escherichia coli cells were grown aerobically in Luria-Bertani medium (BD, MA, USA) at 37 °C. When appropriate, 50 μg/ml of ampicillin was added to the culture. Streptococcus mutans ATCC25175, Streptococcus sobrinus ATCC33478, Streptococcus gordonii ATCC10558, and Streptococcus oralis ATCC35037 were grown in brain heart infusion (BHI, BD) broth at 37 °C in an anaerobic chamber supplemented with 80% N2, 10% H2, and 10% CO2. If necessary, 1% sucrose was added to the BHI medium.
2.3. DNA manipulations
The wapA gene (SMU_987), except for the region that encodes the N-terminal signal peptide sequence, was amplified from the genomic DNA of S. mutans ATCC25175. The DNA fragment was amplified using LA-Taq DNA polymerase (Takara, Shiga, Japan) with the primers wapA-F (5′-TATCCCGGGGACCAAGTCACAAATTATACAAATACG-3′) and wapA-R (5′-TATCTCGAGTTAACGACGTGTTCTATAGAAATAGAC-3′). The PCR product was cloned into the SmaI/XhoI cloning site of the pGEX 4T-2 expression vector, and the obtained plasmid was transformed into E. coli BL21.
2.4. Purification of the recombinant WapA protein and polyclonal antibody production
Recombinant GST-tagged WapA was expressed in E. coli BL21 and purified using glutathione sepharose 4B (Cytiva, MA, USA), and the N-terminal GST tag was cleaved using thrombin protease. After separation on SDS-PAGE, the recombinant WapA band was excised from the SDS-PAGE gel. An antiserum against the purified recombinant WapA was raised in a rabbit by Eurofins Genomics (Tokyo, Japan). IgG was purified from the anti-WapA antiserum using a protein G column (Cytiva) according to the manufacturer's instructions.
2.5. Fluorescent labeling of antibodies
The purified anti-WapA antibody was labeled using a HiLyte Fluor 488 labeling kit (Dojindo, Kumamoto, Japan). Anti-human IgA antibodies (A80-102P) were purchased from Bethyl Laboratories (TX, USA) and labeled using a HiLyte Fluor 647 labeling kit (Dojindo).
2.6. Immunostaining
Streptococcus cells cultured in BHI broth or human saliva samples were pelleted via centrifugation, and the pellets were sonicated for 30 s at 20 W using a sonicator (Otake Works, Tokyo Japan). The samples were then washed with PBS and resuspended in PBS containing 0.25% BSA. The bacterial cell suspensions were incubated for 1 h with HiLyte Fluor 488-conjugated anti-WapA antibody (1:200) and/or HiLyte Fluor 647-conjugated anti-human IgA antibody (1:200) on ice.
2.7. Immunofluorescence microscopy
Immunostained bacterial cells were mounted on glass slides in PermaFluor (Beckmann Coulter, CA, USA), and the slides were viewed with a Carl Zeiss Axiovert 200 fluorescence microscope (Carl Zeiss, Oberkochen, Germany)
2.8. FCM analysis
FACSVerse (BD) was used for FCM analysis. Sample analysis was carried out with a flow rate of around 500–2000 events per second, and the total number of recorded particles was 50000 events. Data analysis and gating were performed using the FlowJo software (FlowJo LLC, OR, USA). To eliminate background noise, the highly positive signal at 405 nm (e-fluorescein), which is far from the target laser wavelengths of 488 nm and 647 nm, was removed by gating (Supplementary Fig. 1A and B).
2.9. Statistical analysis
The Student's t-test was used to compare the differences between the samples. The differences were considered statistically significant if p < 0.05.
3. Results and discussion
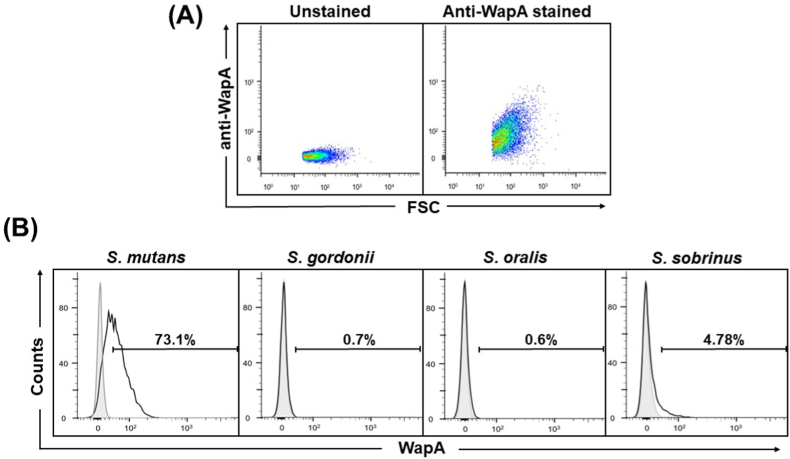
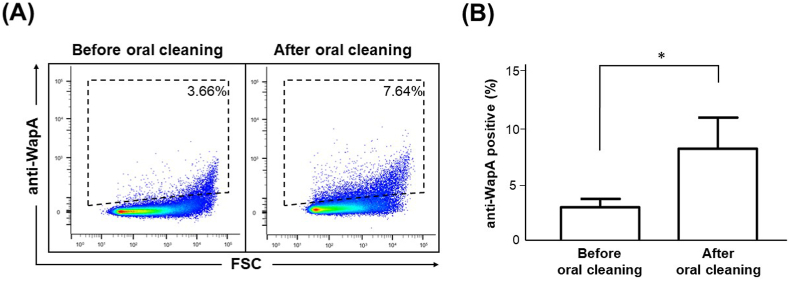
Different bacterial species possess unique surface structures, and the use of specific antibodies against them will enable the identification of bacterial species via FCM. In Gram-positive bacteria, including streptococci, many bacterial surface proteins have a characteristic peptide sequence known as the LPxTG motif, covalently anchored to the bacterial cell wall by the enzyme Sortase [14]. In S. mutans, one of the LPxTG proteins, WapA, is a major surface protein [15,16] that plays an important role in maintaining bacterial structure [17]. To specifically detect S. mutans using FCM, we generated anti-WapA rabbit polyclonal antibodies using the recombinant WapA protein as an antigen. The antibodies were fluorescently labeled. Using these antibodies, cultured S. mutans cells were stained and observed under a fluorescence microscope. As shown in Supplementary Fig. 2, binding of the fluorescently labeled antibody to the surface of S. mutans was confirmed by fluorescence microscopy. Next, we performed FCM analysis using pure S. mutans cultures stained with anti-WapA antibodies to determine whether the antibodies could be used in FCM. As shown in Fig. 1A, a significant shift was observed in S. mutans stained with anti-WapA antibodies, compared with unstained S. mutans. Next, to investigate anti-WapA antibody specificity, FCM analysis was performed on pure S. mutans cultures, as well as the closely related bacteria S. oralis, S. gordonii, and S. sobrinus treated with anti-WapA antibodies. In the case of S. mutans, a peak shift in the histogram was observed, compared with the control, but in the cases of S. oralis, S. gordonii, and S. sobrinus, almost no shift in the histogram was observed, compared with the control (Fig. 1B). These results indicate that anti-WapA antibodies can specifically detect S. mutans in FCM analysis, at least for the oral streptococci examined. Therefore, we examined whether anti-WapA antibodies detect S. mutans using FCM for saliva samples collected from the actual human oral cavity. Resting saliva was collected from healthy volunteers, and bacteria in the saliva collected through centrifugation were stained with anti-WapA antibodies and analyzed via FCM. An increase in positively stained bacteria was clearly observed in the anti-WapA antibody-stained samples, compared with the unstained samples (Supplementary Fig. 3). This indicates that anti-WapA antibodies are applicable in FCM analysis of oral samples. Next, we performed FCM bacterial analysis using saliva collected from humans before and after oral cleaning practices to determine whether anti-WapA antibodies could be used to assess human oral microbial environment. Bacteria collected from the saliva samples were stained with anti-WapA antibodies and analyzed via FCM. A density plot of typical FCM results before and after oral cleaning is shown in Fig. 2A. The proportion of WapA-positive bacteria increased significantly after oral cleaning, compared with the levels before oral cleaning (Fig. 2B). The time course before and after oral cleaning was about 1 h, and S. mutans was not considered to increase during this time. Therefore, it was thought that antibodies specific to the bacterial surface proteins recognized the bacteria more efficiently after oral cleansing. The reason for this could be that antibody binding to the bacterial surface protein is decreased in mature oral flora. In the process of biofilm formation on the tooth surface, various polysaccharide components accumulate in the bacterial flora and form a matrix during the growth of late-colonizing bacteria after the growth of early-colonizing bacteria. This biofilm matrix is known to inhibit the entry of antibodies and antimicrobial components [18]. Since biofilm formation by S. mutans prior to oral cleaning was thought to be an inhibitory factor for antibody binding, the biofilm effect on FCM analysis was investigated to simulate the characteristics of S. mutans in biofilms. S. mutans was cultured in BHI medium with or without sucrose, and the binding ability of the anti-WapA antibody was compared through FCM analysis. As a result, the binding of anti-WapA antibodies to S. mutans cultured in sucrose-containing medium was significantly decreased, compared with the sucrose-free medium (Supplementary Fig. 4A and B).
Fig. 1.
FCM detection of cultured S. mutans cells with anti-WapA antibodies. Overnight cultures of S. mutans, S. gordonii, S. oralis, and S. sobrinus were incubated with HiLyte Fluor 488-conjugated anti-WapA antibodies and analyzed using FCM (A). FCM density plots of unstained (left panel) and anti-WapA-stained (right panel) S. mutans cells. (B) Representative FCM histograms of the streptococcal cultures stained with anti-WapA antibodies. The percentage of WapA-positive bacteria (intensity greater than unstained S. mutans) is indicated in each panel.
Fig. 2.
Detection of WapA-positive bacteria from saliva samples before and after oral cleaning. Unstimulated saliva was collected before and after the subjects cleaned their mouths. The saliva samples were stained with anti-WapA antibodies and analyzed using FCM. (A) Representative FCM density plots of the saliva samples before (left panel) and after (right panel) cleaning. The percentage of WapA-positive bacteria is indicated in each panel. (B) Comparison of the amount of WapA-positive bacteria before and after oral cleaning. The data are mean ± SD from five subjects. *; P < 0.05.
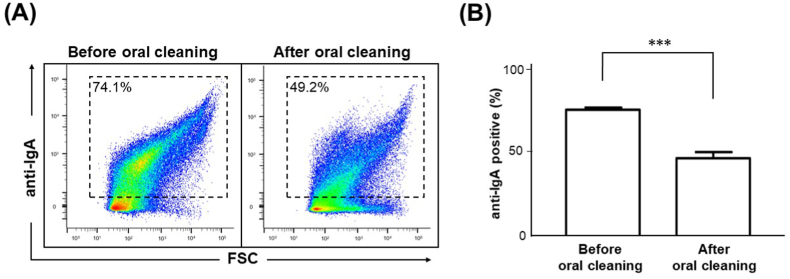
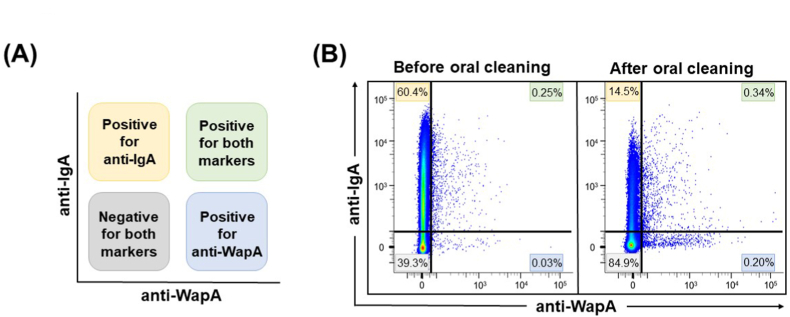
In general, mature plaque-derived bacteria, which are covered with an extracellular polysaccharide matrix, are abundant in saliva from subjects with poor oral hygiene. Therefore, it is possible that detergent cleaning removes the biofilm from the tooth surface, releasing bacteria not covered by extracellular polysaccharides into the saliva and facilitating antibody detection due to the exposure of the bacteria to WapA. Next, we examined the binding of IgA to bacteria in saliva samples. Collected saliva samples were stained with anti-human IgA antibodies and subjected to FCM analysis. A density plot of typical FCM results before and after oral cleansing is shown in Fig. 3A. The mean percentage of IgA-bound bacteria in the saliva before oral cleaning was about 75% (Fig. 3B). This result was similar to the mean value (73.6%) of the percentage of IgA-binding bacteria in saliva 24 h after tooth brushing reported in a previous study [19]. On the other hand, the ratio of IgA-bound bacteria decreased significantly after oral cleaning (Fig. 3B). It has been reported that binding to IgA is important for intestinal bacteria colonizing mucosal surfaces [12], and that salivary IgA binds to mucin on the oral mucosa surface [11]. Thus, IgA may be involved in the colonization of oral mucosal surfaces by bacteria that have resided in the oral cavity for a considerable period before oral cleaning. Next, FCM analysis was used to determine whether the simultaneous staining of saliva samples with anti-WapA and anti-IgA antibodies would provide a comprehensive assessment of qualitative oral flora alteration. A schematic diagram of FCM analysis results of double staining is shown in Fig. 4A. The FCM analysis results showed that the proportion of IgA-positive and WapA-negative bacteria (upper left) was higher before oral cleaning (Fig. 4B). In addition, the ratio of IgA-negative and WapA-positive bacteria (lower right) tended to be lower before oral cleaning (Fig. 4B). It was considered that each antibody could be applied for double staining, as well as individually. In addition, the proportion of IgA-positive and WapA-positive bacteria (upper right) increased after oral cleaning (Fig. 4B). These results suggest that this method can be used to evaluate changes in the oral microflora and present a profile before and after oral cleaning. It has been reported that salivary IgA antibodies increase in diabetic patients [20] and in food allergy model mice [21], suggesting that IgA is involved in the pathogenesis of these systemic diseases. Therefore, the FCM method which combines salivary IgA antibodies and bacteria-specific antibodies to evaluate microflora alteration would be a useful tool to evaluate the relationship between systemic diseases and oral microbial environment.
Fig. 3.
Detection of IgA-bound bacteria from saliva samples before and after oral cleaning. Unstimulated saliva was collected before and after the subjects cleaned their mouths. The saliva samples were stained with HiLyte Fluor 647-conjugated anti-human IgA antibodies and analyzed using FCM. (A) Representative FCM density plots of the saliva samples before (left panel) and after (right panel) cleaning. The percentage of IgA-bound bacteria is indicated in each panel. (B) Comparison of the amount of IgA-bound bacteria before and after oral cleaning. The data are mean ± SD from five subjects. ***; P < 0.001.
Fig. 4.
Analysis of saliva samples by double staining with anti-WapA and anti-IgA antibodies. (A) Schematic representation of the interpretation for the quadrant gates of (B). (B) Unstimulated saliva samples before and after oral cleaning were stained simultaneously with anti-WapA and anti-IgA antibodies and analyzed using FCM. Representative FCM density plots are shown. The numbers in the figure indicate the percentage of bacterial cells in each gate.
In this study, we have shown that changes in bacterial flora composition can be analyzed through FCM analysis. Our method has several advantages, such as salivary antibody binding and polysaccharide capsular coating, which can reveal the status of bacteria in the biofilm. These characteristics cannot be examined using next-generation sequencing. On the other hand, several problems were considered. For example, there are many closely related streptococcal species in the oral cavity, such as Lactococcus, as well as unknown bacterial species, and the reliability of antibody specificity is uncertain. Specificity could be improved by selecting target surface proteins. Recognizing the species that comprise the oral bacterial flora is essential for a better understanding of the state of oral health. Understanding compositional alteration in the bacterial flora using our method would be of great benefit in medical applications, including dental medicine. In the future, this technique is expected to improve by increasing the number of bacteria-detecting antibodies, which will lead to a more detailed method of evaluating dysbiosis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Hideo Kataoka (Asahi University), for helpful advice in flow cytometry.
This work was supported by JSPS KAKENHI Grants, Numbers 17K11628 to HF and 20K18542 to MY.
The funders had no role in study design, data, collection, decision to publish, or preparation of the manuscript.
The authors declare no competing financial interests.
We would like to thank Editage (www.editage.com) for English language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101269.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Das B., Nair G.B. Homeostasis and dysbiosis of the gut microbiome in health and disease. J. Biosci. 2019;44:117. doi: 10.1007/s12038-019-9926-y. [DOI] [PubMed] [Google Scholar]
- 2.Levy M., Kolodziejczyk A.A., Thaiss C.A., Elinav E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 3.Abusleme L., Morandini A.C., Hashizume-Takizawa T., Sahingur S.E. Editorial, Oral microbiome and inflammation connection to systemic health. Front. Cell. Infect. Microbiol. 2021;11:780182. doi: 10.3389/fcimb.2021.780182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solbiati J., Frias-Lopez J. Metatranscriptome of the oral microbiome in health and disease. J. Dent. Res. 2018;97:492–500. doi: 10.1177/0022034518761644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shakya M., Lo C.C., Chain P.S.G. Advances and challenges in metatranscriptomic analysis. Front. Genet. 2019;10:904. doi: 10.3389/fgene.2019.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambrecht J., Schattenberg F., Harms H., Mueller S. Characterizing microbiome dynamics - flow cytometry based workflows from pure cultures to natural communities. J. Vis. Exp. 2018;137:58033. doi: 10.3791/58033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinnon K.M. Flow cytometry: an overview. Curr. Protoc. Im. 2018;120:5. doi: 10.1002/cpim.40. 1 1-5 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Özel Duygan B.D., Hadadi N., Babu A.F., Seyfried M., van der Meer J.R. Rapid detection of microbiota cell type diversity using machine-learned classification of flow cytometry data. Commun. Biol. 2020;3:379. doi: 10.1038/s42003-020-1106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann J., Hübschmann T., Schattenberg F., Schumann J., Durek P., Riedel R., Friedrich M., Glauben R., Siegmund B., Radbruch A., Müller S., Chang H.D. High-resolution microbiota flow cytometry reveals dynamic colitis-associated changes in fecal bacterial composition. Eur. J. Immunol. 2016;46:1300–1303. doi: 10.1002/eji.201646297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch C., Harnisch F., Schröder U., Müller S. Cytometric fingerprints: evaluation of new tools for analyzing microbial community dynamics. Front. Microbiol. 2014;5:273. doi: 10.3389/fmicb.2014.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbins H.L., Proctor G.B., Yakubov G.E., Wilson S., Carpenter G.H. Concentration of salivary protective proteins within the bound oral mucosal pellicle. Oral Dis. 2014;20:707–713. doi: 10.1111/odi.12194. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson G.P., Ladinsky M.S., Yu K.B., Sanders J.G., Yoo B.B., Chou W.C., Conner M.E., Earl A.M., Knight R., Bjorkman P.J., Mazmanian S.K. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360:795–800. doi: 10.1126/science.aaq0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan L. Rebalancing the caries microbiome dysbiosis: targeted treatment and sugar alcohols. Adv. Dent. Res. 2018;29:110–116. doi: 10.1177/0022034517736498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischetti V.A. Surface proteins on Gram-positive bacteria. Microbiol. Spectr. 2019;7:10. doi: 10.1128/microbiolspec.GPP3-0012-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell M.W., Harrington D.J., Russell R.R. Identity of Streptococcus mutans surface protein antigen III and wall-associated protein antigen A. Infect. Immun. 1995;63:733–735. doi: 10.1128/iai.63.2.733-735.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han T.K., Dao M.L. Enhancement of salivary IgA response to a DNA vaccine against Streptococcus mutans wall-associated protein A in mice by plasmid-based adjuvants. J. Med. Microbiol. 2007;56:675–680. doi: 10.1099/jmm.0.47020-0. [DOI] [PubMed] [Google Scholar]
- 17.Zhu L., Kreth J., Cross S.E., Gimzewski J.K., Shi W., Qi F. Functional characterization of cell-wall-associated protein WapA in Streptococcus mutans. Microbiology (Read.) 2006;152:2395–2404. doi: 10.1099/mic.0.28883-0. [DOI] [PubMed] [Google Scholar]
- 18.Donlan R.M., Costerton J.W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simón-Soro Á., D'Auria G., Collado M.C., Džunková M., Culshaw S., Mira A. Revealing microbial recognition by specific antibodies. BMC Microbiol. 2015;15:132. doi: 10.1186/s12866-015-0456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lima-Aragão M.V., de Oliveira-Junior Jde J., Maciel M.C., Silva L.A., do Nascimento F.R., Guerra R.N. Salivary profile in diabetic patients: biochemical and immunological evaluation. BMC Res. Notes. 2016;9:103. doi: 10.1186/s13104-016-1881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsui S., Kataoka H., Tanaka J.I., Kikuchi M., Fukamachi H., Morisaki H., Matsushima H., Mishima K., Hironaka S., Takaki T., Okahashi N., Maruoka Y., Kuwata H. Dysregulation of intestinal microbiota elicited by food allergy induces IgA-mediated oral dysbiosis. Infect. Immun. 2019;88 doi: 10.1128/IAI.00741-19. e00741-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.