Abstract
Memory, defined as the storage and use of learned information in the brain, is necessary to modulate behavior and critical for animals to adapt to their environments and survive. Despite being a cornerstone of brain function, questions surrounding the molecular and cellular mechanisms of how information is encoded, stored, and recalled remain largely unanswered. One widely held theory is that an engram is formed by a group of neurons that are active during learning, which undergoes biochemical and physical changes to store information in a stable state, and that are later reactivated during recall of the memory. In the past decade, the development of engram labeling methodologies has proven useful to investigate the biology of memory at the molecular and cellular levels. Engram technology allows the study of individual memories associated with particular experiences and their evolution over time, with enough experimental resolution to discriminate between different memory processes: learning (encoding), consolidation (the passage from short-term to long-term memories), and storage (the maintenance of memory in the brain). Here, we review the current understanding of memory formation at a molecular and cellular level by focusing on insights provided using engram technology.
Keywords: engram, synapse, memory, plasticity, code, molecular mechanism
Abbreviations: AAV, adeno-associated virus; AMPAR, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor; CaM, calmodulin; CAM, cell adhesion molecule; CaMKII, CaM-dependent protein kinase II; CaMPARI, calcium-modulated photoactivatable ratiometric integrator; ChR2, channelrhodopsin-2; CREB, cAMP response element–binding protein; CS, conditioned stimulus; DG, dentate gyrus; Dnmt3a2, DNA methyltransferase 3a2; eGRASP, engram technology with GFP Reconstitution Across Synaptic Partners; FLARE, Fast Light– and Activity-Regulated Expression; IEG, immediate early gene; IgCAM, immunoglobulin superfamily cell adhesion protein; L-LTP, late phase of LTP; lncRNA, long noncoding RNA; LTP, long-term potentiation; MAPK, mitogen-activated protein kinase; NCAM, neural cell adhesion molecule; NMDAR, N-methyl-d-aspartate receptor; piRNA, Piwi-interacting RNA; PNN, perineuronal net; PRP, plasticity-related protein; PSD, postsynaptic density; Rac1, Ras-related C3 botulinum toxin substrate 1; RhoA, Ras homologous member A; tTA, tetracycline transactivator
Animals extract information from the environment through learning and modify their behavior accordingly. Memory is the ability to store and recall that knowledge (1, 2). In short, memory involves four different phenomena: encoding, consolidation, storage, and recall. Encoding is the process by which information reaching the brain through perception is written in the brain. Consolidation allows information to be selected and made stable for long-term periods. The stable storage of memory involves permanent modifications to retain the information, and recall is the process that enables the reactivation of the pertinent information upon specific and precise cues to allow the modification of behavior (3, 4, 5, 6).
In 1904, Richard Semon (7) proposed the idea of the “engram” and defined it as “the enduring though primary latent modification in the irritable substance produced by a stimulus (from an experience).” An engram, sometimes understood as a synonym for memory trace, is formed by a group of neurons that (1) become activated by a specific learning experience, (2) are modified by this experience, and (3) are reactivated by re-exposure to the same experience, inducing a change in the behavior of the animal (8). Engram cells, therefore, are at least a part of the physical place or substrate where learning leaves imprints in the brain. Sets of engram cells can be found sparse in many areas of the brain, forming an engrome, or engram complex (9).
The coding problem
To understand memory, it is necessary, but not sufficient, to describe the biological mechanisms that enable memory formation, maintenance, and expression. As well as explaining the processes required for memory, we must also explain how specific memories are formed as discrete engrams. In other words, we need to understand how specific pieces of information translate into the brain in a way that allows the animal to manage separate memories associated with concrete events. This question of how mental representations are organized in the brain is a long-standing problem. Descartes (1641) (10) proposed that the mind was organized in ideas, which are material representations of both what is presented in front of the mind and resulting from the operation of the mind itself, a thought (10, 11). The problem of mental representations, and how to operationalize it into experimental design, remains a core challenge in modern neuroscience (2, 7, 8, 12).
Early in the 1910s, before the discovery of the DNA double helix (13), the mechanism of inheritance was one of the most fascinating mysteries of biology. How is a single cell, or an embryo, able to carry the astonishingly complex information package to drive the formation of a full organism? Where in that cell is all that information stored and what is the mechanism that translates it into cells, tissues, and organs?
We understand now how biological information is precisely organized in living organisms. After the progress of Gregor Mendel (14) on understanding inheritance, the physical substrate of the information was discovered: the gene-bearing chromosomes (15). DNA was found to carry genetic traits that were trafficked between bacteria during transformation (16), and genes were discovered to be made of DNA (17). The discovery of the DNA structure (13) was the Rosetta stone that made everything else comprehensible. The structure of genes was understood (18, 19), and the genetic code was finally solved (20). The golden era of molecular genetics had finally described how biological information was written and read in living organisms. Arguably, the neurobiology of the memory field is still waiting for its golden era.
Neuroscience and experimental psychology have focused on the biology of learning and memory acquisition and retention (21, 22, 23, 24, 25, 26) but have largely sidestepped the question of the coding problem: how the specific information is written in the brain (27, 28, 29). Whilst we know a great deal about plasticity mechanisms required for learned behavior in general (which will be reviewed later), we are still far from identifying the “double helix” of memory—if one even exists. We do not have a clear idea of how long-term, specific information may be stored in the brain, into separate engrams that can be reactivated when relevant. Understanding engram organization would be the equivalent to understanding how genes are organized in the genome.
General considerations for the study of the neurobiology of memory
Taking a step back and looking at the whole picture of the neurobiology of memory can help to identify the next scientific questions. Some general considerations must first be clarified.
Information specificity: A goal in mind
Within a biological system, memory is a form of information. As discussed previously, the molecular and cellular mechanisms that underly memory function must ultimately explain how the information is specific to one experience and not others. Engram technology allows the study of how specific memories translate into neuronal changes.
Time: For now, forever
When considering how memory functions from a psychological point of view, it is crucial to consider the timescale of these processes. Any change or group of changes, whether functional or structural, that are responsible for memory should be fast enough so that learning is immediate (e.g., an unpleasant experience will immediately create aversion) and potentially permanent (this negative association will persist over time) (30). Moreover, learning is a constant process that does not cease. Therefore, the mechanisms that allow it must be constantly active and ready to function.
What memory mechanism exactly?: Call it by its name
An additional consideration in interpreting molecular studies of memory is the necessity to distinguish between the different mechanisms involved in memory function. We must clearly differentiate between the mechanisms of encoding, (re)consolidation, storage, recall, and forgetting when considering a particular molecular pathway. The lack of behavioral manifestation of a particular memory in an individual, such as an animal model, does not allow differentiation between encoding, consolidation, storage, or recall deficits (31, 32). This inherent challenge needs to be considered when investigating the precise role of molecular pathways and mechanisms involved in each of these processes. This caveat has been partly overcome by the development of engram technology.
Engram technology
In the past decade, the development of memory engram technology opened the door to identify the engram cells for a given experience, manipulate them, and study the biochemical changes that underlie engram formation and therefore, memory function.
Development of the technology
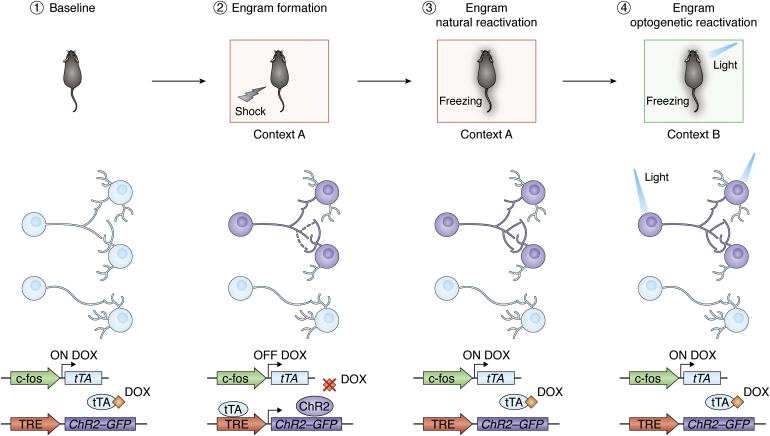
Memory engram technology involves the combination of several techniques: transgenic manipulation, optogenetics or chemogenetics, electrophysiology, and behavioral techniques (Fig. 1). It is based on the use of endogenous markers of neuronal activity, immediate early genes (IEGs) such as c-fos or mammalian activity-regulated cytoskeleton-associated protein (Arc), as drivers to target and manipulate neurons that respond to an experience—putative engram neurons. The first use of engram technology engineered viral vectors that hijacked the promoter of the IEG c-fos to drive the expression of both a fluorescent reporter (GFP) and a light-sensitive channel, channelrhodopsin-2 (ChR2) in the mouse hippocampus (33). Adding the doxycycline-inducible elements tet-ON and tet-OFF to the system (34) allows the labeling window to be reduced to target only cells that are active during a particular event, this is, the engram cells of a specific episodic memory. Finally, to evaluate memory function, mice were trained in a contextual fear conditioning paradigm, where animals learn to associate a particular contextual environment with an electric footshock. Memory reactivation was later measured by assessing the animal fear response of freezing in the same training context. Since engram cells become tagged with ChR2, they can be later artificially reactivated by light delivery resulting in memory recall. The mice, therefore, froze in a neutral context upon reactivation of the engram associated with a fearful experience (33), despite never having a negative experience in this context.
Figure 1.
Engram labeling technology. (1) The immediate early gene c-fos promoter drives the expression of tTA. Doxycycline (DOX), delivered through diet, prevents tTA-TRE element binding. (2) An electric shock is delivered when the animal is in a particular context (context A). An engram for that episodic experience is formed (purple cells). In the absence of DOX, engram neurons that are active during the encoding of that fear memory express channelrhodopsin-2 (ChR2) transgene and GFP reporter. (3) Return to context A will reactivate the engram and will induce a freezing response in the animal—behavior associated with fear. (4) The engram cells tagged with ChR2 can be optogenetically reactivated by delivering light into the brain of the animal. This artificial recall of the memory induces freezing behavior in the animal in a neutral context B. TRE, tetracycline response element; tTA, tetracycline transactivator.
What has research with engram technology taught us about how memory works?
Engram technology triggered a revolution in the memory field (8, 9). Rapidly, numerous engram studies capitalized on the ability to manipulate memories and study engram physiology, and our understanding of engram biology grew rapidly.
Activity of memory engram cells was demonstrated to be sufficient for the recall of a contextual memory associated with fear conditioning (33, 35). Later, engram cells were demonstrated to be necessary for the reactivation of the memory (36, 37). These studies described that optogenetic inhibition via inhibitory opsins prevented the elicitation of the fearful response—when silencing the activation of the engram, animals did not freeze to the context associated with a footshock. Engrams were identified and labeled in several brain areas, such as the amygdala (38, 39, 40, 41, 42, 43), dentate gyrus (DG) (33, 36, 44, 45, 46), CA1 (37), CA3 (36), cortex (42, 47, 48, 49), or nucleus accumbens (50). The connection between engram and place cells was also investigated (Box 1). Engram cell dynamics were successfully manipulated, and false memories were created in animals (45, 51, 52, 53). In an early study, Ramirez et al. (35) created a false association between a neutral context and a fearful experience. Engram technology was used to label and later artificially activate an engram for a neutral context while the animal was given a footshock. The simultaneous activation of these two engrams, one by optogenetics and one by natural contextual cues, created a false association, an artificial memory. Despite never being shocked in context A, animals were now freezing in that context—and crucially, not in an unmanipulated third context (35).
Box 1. Engram cells and place cells.
Place cells are neurons in the hippocampus that encode spatial information by tuning its firing to particular locations (291). In short, they are neurons that fire when an animal occupies a particular given location within a context. Its discovery initiated a revolution in the field and changed how the hippocampus was conceived (292).
The relation between engram cells and place cells has been the focus of several discussions that ultimately question how the hippocampus work and how it processes memories (293, 294). One view is that the activity of engram cells reflects the contextual identity of the memory, instead of specific locations (295, 296). This is, engram cells encode experience instead of place. An engram is formed by a subset of the place cells firing in the context (if the memory carries a contextual component), but it is also formed in the absence of place cells firing if the memory task does not include a spatial component (296). Another conceptual framework, not mutually exclusive, is based on the cognitive map theory (297). This theory posits that the hippocampus function as a spatial framework that links and associates the items that constitute the experience and its location on time. Following this view, engrams would act as indices that would hold the information of what parts of the memory were involved in an event (293).
The expanded engram toolbox
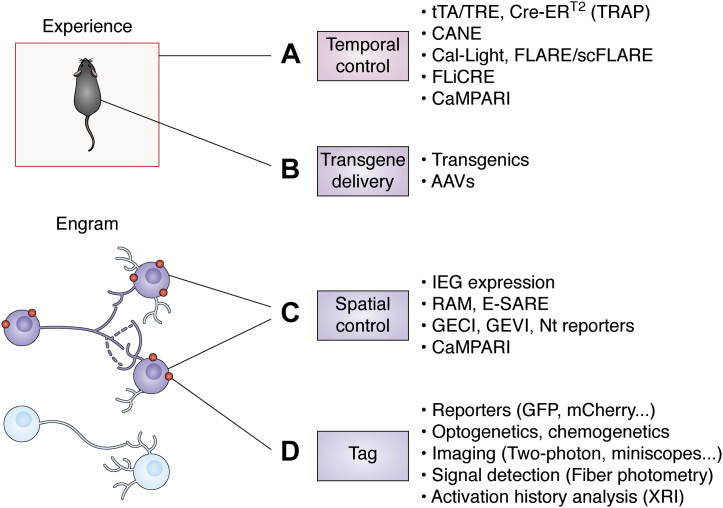
Numerous advances have been made on the technology in the last decade (Fig. 2), and a versatile and sophisticated set of tools is now available to target and study neurons activated by a given experience. Chemogenetic tools such as designer receptors exclusively activated by designer drugs have been extensively used as an alternative to optogenetics to induce both neuronal activation or neuronal inhibition in engram cells (39, 54). Compared with optogenetics, chemogenetics manipulations offer an easier alternative for sustained manipulations (55). The tamoxifen-driven inducible Cre-recombinase system (56) has been applied instead of the doxycycline-regulated system to achieve more precise temporal control (36, 48, 57). In such system, a fusion protein containing a mutated estrogen receptor and Cre recombinase (Cre-ERT2) is sequestered in the cytoplasm until tamoxifen presence induces its translocation to the nuclei leading to recombination of the desired transgene. Viral vectors can be entirely or partially substituted by the use of transgenic mice (36), in order to target bigger areas, which are logistically difficult to be reached via adeno-associated virus injections. For example, in the targeted recombination in active populations mice (48, 57, 58), the IEG c-fos promoter controls the expression of a tamoxifen-inducible Cre-ERT2 recombinase. The technology has been applied to other species via adeno-associated virus to achieve the gene delivery—that is, Fos-CreERT2 constructs (50). Engineered artificial promoters have been designed to improve the labeling specificity and efficiency. The Robust Activity Marking system (59) uses an artificial activity-induced sequence that combines the DNA sequence of the activator protein 1 and the binding motif of the neuronal-specific activity-dependent gene Npas4. The enhanced synaptic activity–regulated element system (60), on the other hand, uses a synthetic promoter that gets activated by activity-dependent transcription factors, such as cAMP response element–binding protein (CREB), myocyte enhancer factor 2, and serum response factor.
Figure 2.
The expanded engram toolbox. Engram cell tagging has been achieved with several strategies or tools that can be used individually or combined. A, temporal control allows labeling engrams responsible for encoding a particular experience and can be achieved with different strategies. The tTA/TRE or Cre-ERT2 (targeted recombination in active populations [TRAP]) genetic strategies are temporally controlled by the delivery of doxycycline or tamoxifen, respectively. The capturing activated neuronal ensembles (CANE) technology allows high temporal precision by engineering viruses to specifically infect activated neurons. Other tools (Cal-Light, Fast light–regulated and activity-regulated expression [FLARE] and its improved version single-chain FLARE [scFLARE], and fast light–regulated and calcium-regulated expression [FLiCRE]) rely on the combination of two requirements to achieve temporal control: increase in intracellular Ca2+ and delivery of light. A similar strategy based on the coincidence of activity and light is used by CaMPARI to label only activated cells. B, transgenes can be delivered by generating transgenic mice models or by the use of vectors such as adeno-associated viruses (AAVs). C, only activated cells (engram cells, purple) are tagged; thanks to several spatial control strategies. The expression of immediate early genes (IEGs) such as c-fos can be used to manipulate only activated cells. Engineered artificial promoters have been also used in the robust activity marking (RAM) or the enhanced synaptic activity–regulated element (E-SARE) systems. Intracellular Ca2+ levels are detected by genetically encoded calcium or voltage indicators (GECI and GEVI, respectively) as well as reporters that detect specific neurotransmitter (Nt) release. CaMPARI is a fluorescent indicator that responds to intracellular Ca2+ levels. D, engram cells can be tagged with reporters (e.g., GFP or mCherry fluorescent reporters). They can also be tagged with tools that will allow future manipulation by light (optogenetics) or by drugs (chemogenetics). They can be imaged with several techniques such as two-photon or head-mounted miniature microscopes (miniscopes). Their activity can be monitored by techniques that allow activity readings such as fiber photometry. Activation history over time can be investigated by the use of the expression recording island (XRI) technology. CaMPARI, calcium-modulated photoactivatable ratiometric integrator; GECI, genetically encoded calcium indicator; GEVI, genetically encoded voltage indicator; tTA, tetracycline transactivator.
Apart from changes in gene expression, other activity-triggered processes have also been used to engineer activity-dependant reporters to investigate engram cells (61). Membrane depolarization and elevation of intracellular Ca2+ can be monitored with genetically encoded voltage indicators (62) and genetically encoded calcium indicators (63), respectively. These are molecular probes that rapidly and reversibly emit fluorescent signal in response to action potentials. In particular, genetically encoded calcium indicators have been used extensively to image large populations of neuronal activity in behaving animals when combined with florescence-detecting tools such as fiber optic recording, widefield microscopy, confocal microscopy, two-photon microscopy, or head-mounted miniature microscopes (61, 64). Similarly, fluorescent neurotransmitter indicators (65) are also available to detect specific neurotransmitter release, such as glutamate (66), acetylcholine (67), or γ-aminobutyric acid (68) and monitor the response of neuronal populations. Other reporters such as calcium-modulated photoactivatable ratiometric integrator (CaMPARI) allow permanent labeling of active cells at a given time point (69). CaMPARI is a fluorescent indicator that switches from green to a red fluorescent state in the presence of both Ca2+ and a pulse of light, allowing the tagging of specific populations related to specific experiences. Although CaMPARI allows for the rapid capture of tight time windows of activity, it can only be used for identifying and studying the cells in question and does not enable the manipulation of those cells.
A similar strategy based on combining two requirements to target engram cells with higher precision has also been used to label engram cells with exogenous constructs that allow for subsequent manipulation. (1) The capturing activated neuronal ensembles (CANE) technology combines molecular tagging with engineered viruses that selectively infect activated neurons (70). (2) In the Cal-Light technology, intracellular Ca2+ binds to calcium sensors calmodulin (CaM) and M13, which then bind to each other and rescue the proteolytic activity of the tobacco etch virus protease. At the same time, light delivered by the experimenter induces the unmasking of the tobacco etch virus protease target sequence, the subsequent cleavage of tetracycline transactivator (tTA), and the expression of the reporter gene (71). (3) Fast Light– and Activity-Regulated Expression (FLARE) (72), and its improved version, single-chain FLARE (73), rely on the modification of a transcription factor to respond to both increase of intracellular Ca2+ and a pulse of light and drive the expression of a reporter gene. (4) Fast Light and Calcium-Regulated Expression (FLiCRE) technology (74) is similarly based on the induction of a reporter transcript in cells activated by intracellular Ca2+ at the time of blue light application, with higher sensitivity and temporal precision. As opposed to simply identifying or imaging neurons activated at a certain time point (such as CaMPARI), these tools allow to drive the expression of genes of interest to these cells.
Recently, a new resource has become available to investigate the history of activation of neurons over time. The expression recording island (75) is a reporter that encodes biological information of neuronal activity in the form of self-assembling protein chains. In this system, tag-labeled monomers are incorporated in a sequential order that mimics the sequence of activation states a neuron undergoes in response to experiences. The posterior analysis of such sequence reflects the involvement of individual cells in different engrams.
Overall, this powerful set of tools and its constant upgrades are approaching the field to the goal of manipulating the population of neurons associated with a particular memory with an even higher level of precision and accuracy. Because we can now artificially activate and inhibit memories, manipulate them, tag them, and study them, it is possible to address questions about the molecular and cellular mechanisms associated with memory to help us understand memory function and neurobiological correlates of memory (8).
We review here the current understanding of the neurobiology of memory, specifically at a cellular and molecular level, and with an emphasis on what we have learnt from engram studies. We discuss how these mechanisms can be interpreted to form a coherent neurobiological theory of memory that could offer verifiable explanations for each phenomenon of memory function. We classify the mechanisms as those involved in the (1) formation of an engram during learning, the (2) consolidation of an engram from short-term memory to long-term memory, and the (3) storage of the information in the brain. Finally, we also consider the role of non-neuronal cells in these processes and reflect on some open questions in the memory field. The molecular and cellular mechanisms underlying other memory phenomena such as reconsolidation (a process by which memories are susceptible to modification or updating after recall) (76, 77) or forgetting (the inability to recall a memory that was successfully learned) (78, 79) are out of the scope of this review.
Molecular and cellular mechanisms that allow the formation of the engram
Learning is the process of acquiring new information that culminates in the creation of a memory. The encoding of information for a particular event within a specific neuronal ensemble takes seconds to minutes (80). What are the changes that the neurons undergo to form the engram?
Learning and plasticity
Shortly before Semon described the concept of the engram, Santiago Ramón y Cajal (81) postulated in his Neuron Theory that neurons form a contiguous, instead of a continuous structure. They are physically separated but functionally connected by synapses, a term coined by the physiologist Charles Scott Sherrington (82). The electric impulse coming from a presynaptic neuron is transmitted through a synapse to the postsynaptic neuron, where it travels toward the axon. Going even further, Cajal predicted that synaptic strength underlies the storage of information (83). His student, Rafael Lorente de Nó, described how neurons form “multiple chains of transmission through which impulses circulate” (84), and Donald Hebb (85) described that memory lies in the strengthening of the connections between neurons that were simultaneously activated. Since then, the field has substantially advanced on our understanding of the molecular and cellular mechanisms involving memory function (21, 22, 23, 24, 26, 86, 87).
Neuronal plasticity can be defined as the mechanism that allows connections involved in a particular response to specifically strengthen, facilitating neuronal transmission between them, whereas others may weaken (88). After Hebb (85) postulated that neurons that are functionally active at the same time adapt their synapses to become linked forming an ensemble, Kandel and Tauc (89) demonstrated that learning influenced the synapses. Later, Bliss and Lomo (90) described a long-lasting modification in synaptic strength induced by electrical stimulation, also known as long-term potentiation (LTP). The opposite effect, long-term depression, was also described (91).
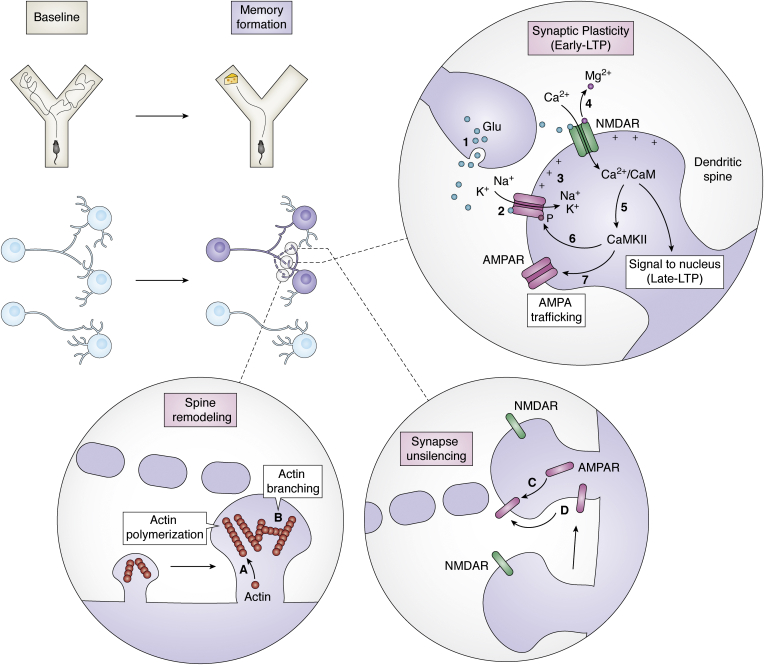
The first phase of the LTP, early-LTP (Fig. 3), lasts from seconds to a few hours and relies mainly on covalent modifications of already existing proteins (reviewed in Refs. (22, 92)). During early-LTP, glutamate is released from the presynaptic neuron and activates the α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors (AMPARs) in the postsynaptic membrane, allowing Na+ and K+ influx into the cell. N-methyl-d-aspartate receptors (NMDARs) are another type of ionotropic glutamate receptors that are sensitive to the depolarization state of the postsynaptic element. They detect coincident presynaptic and postsynaptic activation and open to permeate Ca2+ inside the postsynaptic neuron.
Figure 3.
Molecular mechanisms of learning. When an animal encodes a new memory (such as the encounter of food in a particular arm of a Y-maze, upper panel), a subset of neurons gets activated and an engram is formed (purple neurons, middle panel). Certain synapses in between engram cells (dotted lines) undergo synaptic plasticity changes (early long-term potentiation [LTP]). The (1) neurotransmitter glutamate (Glu) is released from the presynaptic neuron, Glu binds to (2) AMPARs in the postsynaptic membrane, at the level of the dendritic spine, allowing K+ and Na+ to enter the postsynaptic neuron and depolarizing it (3). The positive charges inside the postsynaptic neurons allow the release of the Mg2+ ion from the NMDARs and if the Glu release is sustained enough, it will open the channel (4). Ca2+ enters the postsynaptic neuron, activating the (5) Ca2+/calmodulin-dependent protein kinase II (CaMKII). CaMKII phosphorylates AMPARs (6) to increase their sensitivity to Glu and drives more active channels to the membrane, AMPA trafficking (7). The number and shape of dendritic spines also get modified during learning. Spine remodeling to strengthen the synapse involves cytoskeletal modification such as (A) actin polymerization and (B) actin branching. Engram cells become unsilenced by the trafficking of AMPAR to the synapses by (C) secretion from the intracellular pool and (D) diffusion from other membrane areas. AMPAR, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor; NMDAR, N-methyl-d-aspartate receptor.
This increase of Ca2+ leads to a plethora of intracellular signaling events. First, it activates the Ca2+/CaM-dependent protein kinase II (CaMKII) (92, 93, 94, 95). CaMKII is a central regulator of plasticity that increases postsynaptic responsiveness. It phosphorylates the GluA1 subunit of AMPAR, which increases single-channel conductance (96, 97, 98), and it allows more active channels in the membrane in a process known as AMPAR trafficking. AMPARs diffuse to the synapses from other membrane areas (99), as well as getting secreted from the existing intracellular pool (100), facilitating further impulse transmission.
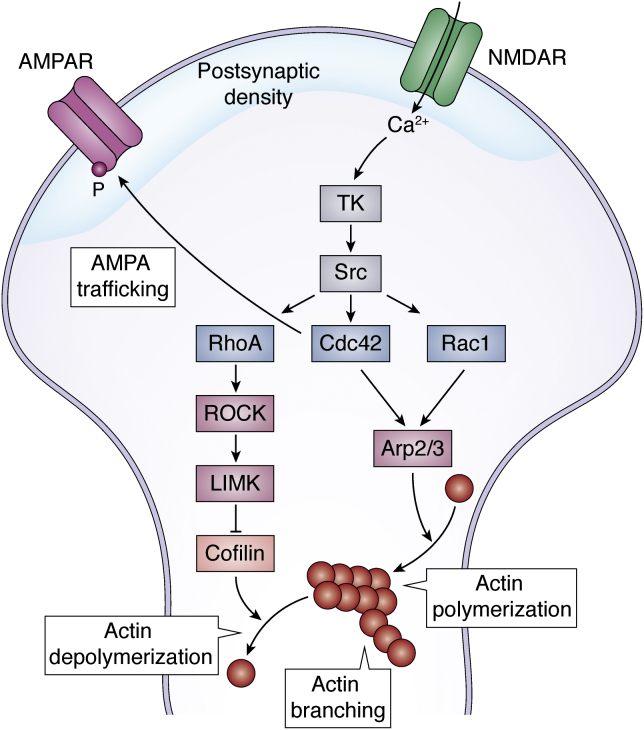
Immediate cytoskeletal modifications (Fig. 4) independent of protein synthesis are also triggered by an increase in intracellular Ca2+ (101). They contribute to enlarging the spine in the case of LTP and retracting the spine in the case of long-term depression (102, 103, 104, 105). Intracellular Ca2+ activates a transduction signal cascade mediated by tyrosine kinases and Src family kinases that in turn activate Rho GTPases and Ras GTPases (reviewed in Refs. (106, 107, 108, 109)). This family of signaling proteins includes members such as Rac1 (Ras-related C3 botulinum toxin substrate 1), Cdc42, and RhoA (Ras homologous member A), which then modulate effector molecules such as cofilin or the complex Arp2/3. Cofilin is an actin-binding protein that depolymerizes actin filaments and that is inhibited by RhoA (110, 111). Arp2/3 is an actin nucleation factor that regulates actin branching, polymerization, and recycling (112, 113). On top of their structural role, Rho GTPases also regulate AMPAR trafficking. They modulate both AMPAR insertion in the postsynaptic membrane and AMPAR stability in the membrane (109). Cdc42, for example, triggers the phosphorylation of the AMPAR subunit GluA1, which induces the insertion of AMPAR in the membrane (114). Via these signaling pathways that regulate actin polymerization and AMPAR trafficking, the synapse structure gets quickly modified in a process known as structural plasticity (103, 104, 105, 115). With the emergence of more dendritic spines, the synapse is strengthened (24, 116).
Figure 4.
Cytoskeletal modifications after learning. The increase in intracellular Ca2+ provoked by neuronal activation activates tyrosin kinases (TKs) and Src kinases. These activate members of the Rho family small GTPases: RhoA, Cdc42, and Rac1. RhoA activates ROCK and subsequently LIMK kinases, which in turn inhibits effector cofilin and eventually inhibits actin depolymerization. Rac1 and Cdc42 activate Arp2/3, an actin nucleation factor that induces actin polymerization, the elongation of actin filaments, and actin branching, and the formation of new ramifications in the actin filaments. Rho GTPases also anchor, stabilize, phosphorylate, and insert AMPARs to the postsynaptic density area of the membrane. AMPAR, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor; LIMK, LIM-domain kinases; NMDAR, N-methyl-d-aspartate receptor; Rac1, Ras-related C3 botulinum toxin substrate 1; RhoA, Ras homologous member A; ROCK, Rho-associated coiled-coil kinase.
Still much to learn
Through these processes, synaptic plasticity ultimately ties salient experiences to changes in the efficiency of the synaptic transmission between neurons, modifying the synapse accordingly for future reactivation. However, there are still significant questions opened to explain how synaptic plasticity could be understood to mediate learning, and they relate precisely to the three general considerations discussed in the introduction.
First, the goal in mind is to decipher the molecular and cellular mechanism/s that allow for one particular memory to be encoded and then ultimately escalate from that to understand how the system works for the collection of memories that a brain holds. Engram technology offers the resolution needed to understand the mechanism for a specific component of memory functionality—for a particular salient experience and not other/s.
Second, plastic changes are necessary for memory function, but they are not the only players in the game. Information in a brain is long-lasting, and yet it is constantly susceptible to modifications that allow memory updating to occur (117). Therefore, there must be an equilibrium between malleability and fidelity (118, 119). The problem of how plasticity, a mechanism based on the presence and amount of turnover-sensitive proteins, is sustained, is an old discussion in the field (120). While an engram is necessarily formed by a process of plasticity, it must be also preserved and endure across the life of an animal, by a constant process of homeostasis. It must be maintained in a state that can be reactivated during recall when it became relevant. Are plastic changes of synapses able to perform both roles, encode now and store forever?
Third, the role of plasticity in learning can now be better assessed with engram technology since it confers enough technical resolution to discriminate from its role in consolidation, recall, or storage. To illustrate this point with an example, studies based on engram technology have demonstrated that memory survives under certain types of amnesia. Though apparently lost, the memory is still retrievable by optogenetic reactivation, showing that the memory deficit is neither due to disruption of learning or storage mechanisms, but impaired recall (46, 121, 122, 123, 124, 125). It is now possible, as we discuss later, to precisely characterize the physiological process affected and call it by its name.
Creating the engrams
Engram technology has contributed to better understanding the role of synaptic and structural plasticity in the encoding of a memory.
The use of engram technology has demonstrated that engram-to-engram synapses exhibit plasticity of synaptic strength. In the hippocampus, patch-clamp recording of excitatory postsynaptic currents in engram cells upon depolarization of presynaptic input cells showed that engram to engram synapses are specifically strengthened relative to nonengram synapses, showing higher excitatory postsynaptic current amplitude, a higher spontaneous excitatory postsynaptic current amplitude, and higher ratio AMPA to NMDA (46). Structurally, engram neurons also showed an increase in dendritic spine density (46, 126, 127). This increase in plasticity between engram cells has been validated and refined by Choi et al. using a more advanced methodology. By combining engram technology with GFP Reconstitution Across Synaptic Partners (dual-eGRASP) technique (128), synapses between engram cells have been characterized. This work described that engram cells in CA1, a subarea of the hippocampus, establish more synapses with engram cells in CA3 than nonengram cells, and that the strength of memory expression correlates with the strength of the connections between these cells. Engram-to-engram cell synapses were potentiated since they showed an increase in the presynaptic transmitter release and AMPAR levels (129). Very recent studies also using dual-eGRASP from the same laboratory corroborated the increase in structural connectivity between engram cells in the auditory cortex to lateral amygdala circuit (130). Increases in structural plasticity and functional plasticity have been found specifically in engram-to-engram cells in many other circuits and areas: hippocampus to amygdala (127), cortex to amygdala (126, 131), or hippocampus to nucleus accumbens (132).
Neuronal allocation
If learning induces changes in a population of neurons that respond to an experience, then a question that must be answered is which neurons and why? In other words, what are the mechanisms that select neurons to form the engram?
One possibility is that a cue-specific afferent randomly activates a high excitable given ensemble that later becomes the engram (133). Artificial overexpression of the CREB in a subpopulation of neurons in the lateral amygdala increased preferential recruitment of these cells to the engram and enhanced auditory fear memory after training (38, 134). The transcription factor CREB, one of the best-studied genes in the memory field with a critical role in the late phase of LTP (L-LTP; see later), modulates intrinsic excitability (135, 136). Without increasing the size of the engram, CREB increases the intrinsic excitability of the neuron—the propensity to generate an action potential upon reception of the input (54). Concordantly with this hypothesis, it has been described how two experiences that occur close in time are encoded in overlapping populations, in such a way that recall of the first one will provoke the recall of the second one (137, 138). Cai et al. (137) recorded neuronal activity while the animals explored different contexts using in vivo calcium imaging, describing how the more separated in time the memories for two contexts were, the less overlapping their engrams are in the hippocampal region CA1. In aged mice, where neuronal excitability, as well as other forms of plasticity, is impaired, this overlapping phenomenon did not occur. Furthermore, the effect on overlapping and shared recall in aged mice was rescued with artificial activation of the neurons with designer receptors exclusively activated by designer drugs.
However, another study has shown that recall (and likely learning) increases engram excitability but does not increase the likelihood of memories being linked or coallocated (46, 139). Rather, it seems that increased excitability itself enhances short-term memory precision by increasing engram cell accessibility and the ability to discriminate between similar but distinguishable contexts (pattern separation), thus keeping similar experiences separated. Taken together, it seems that the overexpression of CREB influences the allocation of an engram to targeted cells and also alter the excitability state of those cells in parallel. What is the mechanism whereby natural learning decides to which neurons an engram is allocated?
While this question is largely unanswered, it is reasonable to hypothesize that the excitation state of cells at the time of learning may influence their probability of allocation to an engram. In other words, if a cell lies in an anatomical region that is innervated by a receptive field activated by a relevant perceptual experience, then its mode of excitation during that experience may influence how the engram is assembled. In addition, stable cellular properties, such as molecular subtype, location within the area (140, 141), or the age of the neuron (142), also likely determine which ones are selected to the engram. Recent studies using two-photon microscopy in engram cells have described preferential selection to neurons that receive more stable connections (143). Finally, the epigenetic stage of the cell also influences eligibility for recruitment to the engram (Box 2). Future studies that monitor neuronal activity of ensembles longitudinally, before and after engram formation, and combine this analysis with physiological and structural analysis of the allocated cells will likely generate insights into the mechanisms of engram allocation.
Box 2. Epigenetic mechanisms in memory.
The environment can influence gene expression by modulating epigenetic mechanisms (298). Therefore, it is not surprising they play a role in memory function. Learning induces a battery of epigenetic changes, including reversible modulation of both histones and DNA. These include histone methylations (299, 300), histone acetylations (301), or DNA methylations (302, 303, 304, 305) among others (reviewed in Refs. (306, 307, 308, 309)). The resolution needed to understand the mechanisms at a singular memory level has been achieved by incorporating engram labeling technology into epigenetic tools.
A study of the epigenome architecture of engram cells based on the engram technology has indeed demonstrated that memory encoding modifies chromatin structure to increase the accessibility of chromatin on enhancers that will interact with promoters during memory recall (169). This way, engram cells become epigenetically primed after encoding. Later, during memory recall, when the engram is reactivated, the primed engram cells undergo transcriptional changes. This study demonstrates that the epigenetic modifications on the engram during learning are stable and persist. These results also confirm those reported by a parallel transcriptomic and epigenetic study of activated neurons. One hour after exploration of a novel context or an artificial activation triggered by kainic acid, activated hippocampal neurons reorganized their chromatin to allow accessibility of activity-regulated genes, enhancers, and transcription factors (310).
Another interesting example of engram technology to understand epigenetic mechanisms in memory consolidation has been described by Gulmez Karaca et al. (311). Overexpression of the DNA methyltransferase 3a2 (Dnmt3a2) specifically in engram cells increased the precision of the engram reactivation during recall and strengthen memory in a context-specific manner. On the other hand, isoform Dnmt3a has been described to play a role in engram allocation: overexpression of Dnmt3a in a random sparse population of the DG increased intrinsic excitability and biased engram allocation toward them (312).
Finally, the storage of the information at a cellular level by maintaining certain epigenetic signature has been hypothesized (313, 314, 315).The epigenetic code theory explains how, by means of epigenetic reversible modification of chromosomal regions and histones, it is possible to label activity states in a permanent way in the neuron (316). Crucially, these changes can be maintained by homeostatic mechanisms. It has been proposed that, after learning and plasticity take place and the gene expression changes and structural modifications have occurred, the neurons switch from a permissive epigenetic state to a maintenance transcriptome that facilitates the long-term storage of information (317).
These studies evidence that engram cells suffer important epigenetic regulations that control cellular processes behind learning, consolidation, and storage. Engram technology, and related methodologies, will continue to help us clarify these molecular mechanisms and answer the open questions in the field.
Synaptic allocation
Another open question is how some synapses consolidate, whereas others stay available for learning-induced plasticity? This is the problem that has been commonly referred to as synaptic allocation or synaptic specificity (reviewed in Ref. (144)).
It has been suggested that synaptic specificity could be achieved thanks to the information that neurons share extrasynaptically. This noncanonical way of communication may confine plasticity mechanisms to particular synapses (21). Neurons communicate nonsynaptically via soluble factors (such as growth factors or cytokines), gap junctions (or electrical synapses) (see Ref. (145) for a review), tunneling nanotubes (see Ref. (146) for a review), or RNA-containing exosomes (see Ref. (147) for a review). Transmitted RNAs include coding RNAs (such as Arc) (148) or noncoding RNAs, particularly miRNAs and Piwi-interacting RNAs (piRNAs) (149) (see the later section). A particularly fascinating example is Arc (150), as it is trafficked interneuronally. Arc mRNA and protein appear to be self-assembled into virus-like capsids, released from neurons, and transferred into other cells. In the target cell, it is required for synaptic plasticity in the postsynaptic neuron and activity-dependent synapse formation in Drosophila (148, 151), which potentially explains how Arc could mediate synapse specificity. Since Arc expression is induced by synaptic activity, is located in the dendritic spine, and modulates local translation (152), it can be speculated that Arc plays a role in defining which synapses will be modulated by plasticity.
Either by extrasynaptic or synaptic communication, only specific synapses are formed between a given subset of engram cells after experiences. The specificity of the connections formed during learning has precisely been hypothesized to be the mechanism at the center of memory storage, as it will be discussed later (153, 154).
Molecular and cellular mechanisms that allow the consolidation of the engram
Memory consolidation is the mechanism that transforms temporary or short-term memories into stable long-term memories. It can be subdivided into two very different processes: synaptic consolidation (at the cellular level) and system consolidation (at the circuit level) (117). The first one involves biochemical and morphological changes at the synapses and in the cell, lasting from minutes to hours, with consequences even several weeks after (6, 155). On the other hand, system consolidation, which is out of the scope of this review as it does not necessitate a molecular explanation, involves the gradual reorganization of the engram from hippocampal to neocortical structures and takes from days to weeks (6, 80, 156). Overall, the final product of memory consolidation is the persistence of a modified synaptic structure and function.
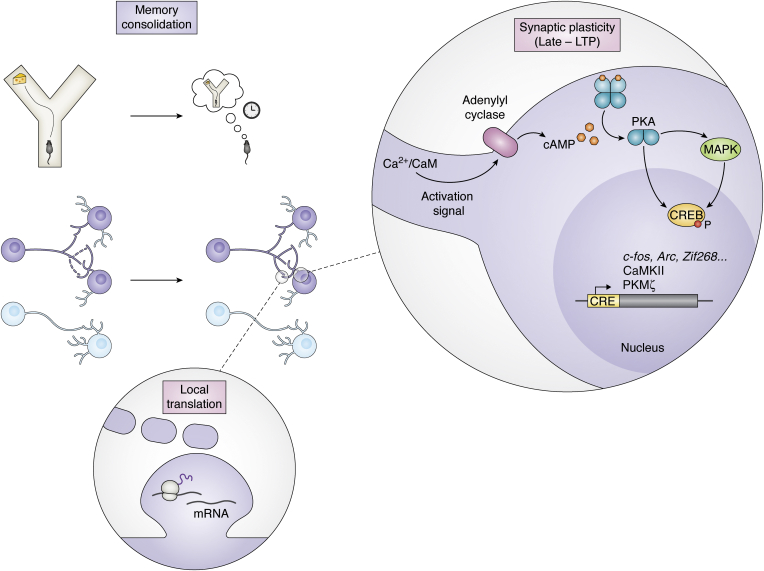
During synaptic consolidation, the synapses that encoded the information after learning go through a group of plasticity mechanisms referred as the L-LTP (Fig. 5). This phase involves (1) activation of the protein synthesis machinery in both soma and dendrites (local translation) to translate pre-existing mRNA and (2) de novo mRNA transcription (25, 157, 158, 159). The increase in intracellular Ca2+ provoked by early-LTP activates signaling cascades mediated by PKA, PKC, and mitogen-activated protein kinases (MAPKs), which then activate transcription factors. The most studied one, CREB (discussed in previous sections), is a transcription factor that initiates transcription of a group of genes containing CREB-responsive elements (160). Rapidly, induced by CREB and other transcription factors such as C/EBP (161), in a tightly regulated way, a repertoire of genes gets transcribed. These include IEGs such as Arc, Homer, c-fos or Zif268, kinases such as CaMKII and protein kinase Mzeta, and cytoskeletal proteins (see Ref. (21) for a review).
Figure 5.
Molecular mechanisms of consolidation. After the encoding of a memory (the encounter of food in a Y-maze, upper panel), memory undergoes a series of processes that allow its consolidation into long-term memories resistant to the passage of time. Engram synapses (purple, middle panel) undergo synaptic plasticity changes (late phase of long-term potentiation [or late-LTP]). Triggered by the intracellular increase in Ca2+, adenylyl cyclase is activated and the intracellular concentration of cAMP increases. The cAMP increase triggers the activation of protein kinase A (PKA) and mitogen-activated protein kinases (MAPKs). PKA translocates to the nucleus, phosphorylates the transcription factor CREB, and ultimately triggers the transcription of genes containing CREB-responsive element (CRE), such as immediate early genes (c-fos, Arc, or Zif268), kinases such as Ca2+/calmodulin-dependent protein kinase II (CaMKII) and protein kinase Mzeta (PKMζ). In the dendrites, protein synthesis occurs locally by ribosomes translating mRNAs in a localized manner in the dendritic spines.
How are these changes, occurring at the whole-cell level, able to specifically be targeted only at some synapses in the cell? A theory known as Synaptic Tagging and Capture (162, 163) has been proposed to explain this phenomenon. According to this theory, the activation of synapses during early-LTP tags the synapses and will induce the capture of plasticity-related proteins (PRPs), activated by the CaMKII–CREB pathway. Synapses in the same cell share PRPs. Therefore, a weak stimuli can induce L-LTP by tagging a synapse and then recruiting available PRPs derived from a strongly stimulated second synapse (144, 164).
The repertoire of genes
Engram technology has helped us understand how this de novo mRNA and protein synthesis modify the molecular profile of the engram synapses. Studies based on single-cell sequencing methods have characterized the transcriptomic response of engram cells (165) to identify changes associated with L-LTP. Attempts have studied engram cell transcriptomics using single-nuclei RNA-Seq, demonstrating that engram cell activation results in the expression of genes from the MAPK family, and IEG transcriptional regulators, such as Atf3, Egr1, Fosb, Homer1, and Junb (165). Further studies found a heterogeneous response within the population of engram cells, depending on the area and cell type. Even within the hippocampus, DG and CA1 neurons, as well as a subtype of inhibitory interneurons (vasoactive intestinal polypeptide positive neurons) show different profiles of gene expression after activation by learning (166). To investigate if the transcriptomic response supports reactivation during retrieval, mice were re-exposed to the same context 4 h after the first exposition. In the DG, subsets of engram cells express either (1) a characteristic early activation transcriptomic signature, (2) a late-activation signature still present 5 h after the experience, and, interestingly, (3) a particular signature that predicts that the particular engram cell will be reactivated upon exposure to the same context (166). The role of this functional response is still open to speculation.
During later consolidation into long-term memory, the transcriptomic profile of engram cells also undergoes changes. Thanks to the fact that engram cells can be permanently tagged, the mechanisms taking part in each stage of the memory process can be evaluated. Rao-Ruiz et al. (167) used RNA-Seq to demonstrate that, 1 day after training, engram cells activate a gene signature mediated mainly by CREB-induced genes. Weeks after learning, consolidated memory is known to be more dependent on cortical areas to be reactivated than on hippocampal areas (49, 156). As memory undergoes consolidation into cortical areas, engram cells, labeled in the medial prefrontal cortex 16 days after an experience is encoded, also activate specific transcriptional programs. As happens with postlearning transcriptomic profiles, consolidation transcriptomic profiles are also specific to cell type. They contain genes involved in transcriptional and translational regulation, vesicle exocytosis, transmembrane transport, dendritic spine organization, and long-range intracellular transport. Interestingly, the genes in this consolidation signature do not seem to be directly controlled by canonical transcriptional regulators, evidenced by the lack of regulatory motifs of Creb, Nfkb, Cbp, and C/ebp. Overall, this study demonstrates that activity-specific changes in engram cells occur during weeks after encoding (168). Finally, during recall, engram cells seem to exhibit a particular transcriptional program distinctive from the one associated with encoding (169), which is accompanied by an extensive chromatin reorganization, indicating epigenetic modulations are involved in the modification of the response (discussed in Box 2).
Noncoding RNAs
Gene expression can also be modulated during consolidation through noncoding RNAs. Located at the synapses, noncoding RNAs modify gene expression locally and adjust it to neuronal activity requirements. They modify mRNA stability and translation, regulate transcription and trigger epigenetic modifications (reviewed in Refs. (170, 171)). They are classified into noncoding miRNAs, piRNAs, and long noncoding RNAs (lncRNAs) (149).
miRNAs bind to complementary mRNAs preventing their translation into proteins (172). They shape the cellular response after plasticity (reviewed in Ref. (173)) by targeting transcription factors. For example, neuronal activity in Aplysia increases expression of miRNA miR-124 that modulates Creb expression inducing plasticity changes, such as dendrite morphogenesis, synaptogenesis, and glutamate receptor modulation (174). miRNAs can also modulate synaptic plasticity through the protein complex translin/trax—two dendritic proteins with RNAse activity (175). Neuronal activity triggers the RNAse activity of translin/trax, which then degrade miRNAs, releasing the silencing of PRPs (176).
A very particular subtype of noncoding RNAs, piRNAs, are small RNA molecules involved in repressing transposons (149, 177). In Aplysia neurons, the piRNA piR-F was shown to respond to the neurotransmitter serotonin, important for learning and plasticity, inducing the epigenetic silencing of transcription factor CREB2, which in turn provoked plasticity changes (178).
Finally, lncRNAs interact with RNA-binding proteins to remodel chromatin and modulate alternative splicing at the nuclear level. Locally at the synapses, lncRNAs control plasticity mechanisms by regulating mRNA stability and protein synthesis (179, 180), and its spatiotemporal regulation supports their role in synaptic specification. In an interesting example, the expression of the lncRNA BC1 has been proved inducible by neuronal activity in hippocampal primary cultures (181). Analysis of BC1 sequence allowed identification of a motif that interacts with the heterogeneous nuclear ribonucleoprotein A2 to drive BC1 exportation to the dendrite (182). Recently, an elegant study characterized a novel lncRNA, termed ADEPTR, involved in structural plasticity. ADEPTR was isolated from the synaptic fraction of hippocampal neuronal cultures treated with the secondary messenger cAMP. Using a loss-of-function analysis, knocking down of ADEPTR prevented the increase in the number of dendritic spines and in spontaneous excitatory postsynaptic currents measured by whole-cell patch clamp induced by neuronal activation in vitro. ADEPTR interacts with the actin-scaffolding regulators spectrin/ankyrin complex to drive them to the synapse and promote structural changes induced by activity (183).
The cytoskeleton
The activation of a neuron modifies the actin cytoskeleton architecture to modulate the structure, composition, and physiology of the synapses (reviewed in Refs. (115, 184)).
The cytoskeleton at the postsynaptic site is mainly formed by actin (185), and it shapes the dendritic spine, a structure that concentrates the functional components of the synapse (see the later section). Inside the spine, actin monomers (or G-actin) organize in filamentous polymers (F-actin) to form a branched network (186). F-actin polymerizes on the barbed end of the filament, whereas G-actin monomers disassemble at the other end, in a dynamic process. The velocity of polymerization and depolymerization of actin monomers, as well as their orientation and stability, determine the structure and stability of the dendritic spine, therefore modifying structural plasticity (187). Several neuropathies and disorders that lead to memory impairments have been associated with deficits in cytoskeletal function (188, 189).
Apart from being a structural component, actin modulates postsynaptic proteins in the postsynaptic density (PSD) in response to activation (190). The PSD is the area in the postsynaptic neuron located immediately opposite the presynaptic contact that organizes receptors, adhesion molecules, and signaling molecules (191). Actin is enriched at the PSD to anchor receptors via interaction with scaffolding proteins. On the other hand, actin depolymerization, in close interaction with lipids on the exterior side of the membrane, increases the motility of nonstabilized molecules at the PSD (192) and contributes to disperse AMPARs and NMDARs (193). On the presynaptic side, actin contributes to modulating the synaptic vesicle cycle and neurotransmitter release, organizing the fusion of synaptic vesicles with the active zone membrane (194).
The cytoskeleton dynamics are critical to consolidation mechanism of memories in engram cells. Engram cells express Rac1 during memory consolidation. One day after contextual fear conditioning, Rac1 expression was found in 80% of the engram population in CA1, labeled by engram technology, and was absent in nonengram cells. This expression, sustained for 10 days, induces the natural forgetting of the memory; and pharmacological inhibition of Rac1, as well as overexpression of its negative regulator α2-chimaerin, restored the manifestation of the memory, suggesting a recall deficit instead of consolidation or storage (195).
During memory consolidation, changes initiated by learning induce a more permanent and long-term adaptation, characterized by a regulation of gene expression and a structural reorganization and that culminate with an increase in synaptic plasticity and structural plasticity. Short-term memory becomes stabilized into long-term memory. But more importantly, besides which particular neurons or synapses (where) and the changes they undergo (how), an open question is what is really the substrate of the information in the brain. What is the change that grasps specific information for a certain experience in our brains?
Molecular and cellular mechanisms that allow the persistence of the engram
Learned information needs a solid system to be secured. As molecular and cellular mechanisms provide the basis for physiological mechanisms, which is the mechanism behind memory storage that maintain the information?
An early engram study provided a starting point to help clarify this question by demonstrating that L-LTP of synaptic strength is not strictly required for memory storage. Abolishing the mechanism of L-LTP by means of a protein synthesis inhibitor right after the encoding of an episodic memory did not alter its storage in the hippocampus. Injection of the protein synthesis inhibitor anysomicin induced amnesia to the mice, evidenced by the absence of freezing behavior in a context previously associated with a shock. Using optogenetics and engram-specific technology, the engram, labeled with ChR2, was reactivated and the behavioral response elicited. In the absence of L-LTP, the presence of the memory was observed (46). In the amnesic mice, the increase in engram-to-engram connectivity strength was prevented, as well as the increase in spine density between engram cells, but memory was not lost. Therefore, memory must rely on storage in other changes that are independent of L-LTP (154). This study also evidences the necessity of a fine characterization of the elements of the memory process when interpreting molecular studies of memory. A subset of studies have replicated and advanced on these results (196, 197, 198). A recent study based on genetic blockage of protein translation substantially expanded on these findings in the amygdala (198). In agreement with these findings, studies based on the use of amnesia-inducing drugs in Aplysia also demonstrated that memory does not depend on L-LTP to be stored (197). Other studies covered other types of amnesia, where memory was also hidden but not destroyed: Alzheimer’s disease (122, 123, 124) or infantile amnesia (121).
Storing as connections
If not synaptic plasticity, what then is the mechanism that retains the memory in the traces in the long term? At this point, it is important to make a distinction between plasticity mechanisms that modify synaptic weight (the strength of a particular synapse) and plasticity mechanisms that modify synaptic wiring (the flowchart diagram of connections in between neurons or structural plasticity) (23). One hypothesis is that memory is stored in the stable connectivity pattern between engram cells (153, 154), which remains unaltered in memories that can be reactivated by artificial (induced by light) but not natural recall (induced by contextual cues) (46). Molecular mechanisms involved in specifically creating and maintaining this connectivity are still to be fully understood. Two processes are particularly relevant: the formation of new connections and the activity of adhesion molecules specifying which connections must be formed.
Formation of new connections
New connections can be formed by (a) adding synapses to the system or synaptogenesis in between new or existing dendrites and/or axons or by (b) unsilencing synapses (Fig. 3).
Dendritic spines are bulbous shapes that connect to the dendrite by a neck and act as the postsynaptic component of excitatory synapses (199). They have been proposed to be relevant for memory storage since they undergo structural remodeling after plastic changes. Their remodeling, determined by the actin cytoskeleton (194), contributes to both modifying synaptic weight and synaptic wiring or connectivity. Dendritic turnover, although higher during development, also happens in adult brains (200, 201, 202). Rapidly formed after an experience, they have been suggested to act as a lasting structural ground for memory storage (203, 204, 205). Shape-wise thin-shaped spines have been associated with learning mechanisms (200, 206), whereas mushroom-shaped dendrites have been associated with memory storage since they anchor more postsynaptic molecules to their membrane (reviewed in Ref. (200)). Engram-specific manipulation demonstrated that the spine density of engram cells correlated with the temporal progression of the memories. A recent study based on eGRASP technology characterized spine morphology between engram neurons in the cortex to amygdala areas (130). Memory encoding was associated with higher spine size and that parameter correlated with the maintenance of memory (Fig. 3). After memory extinction (the targeted suppression of a learned behavioral response), spines returned to their original state (130). In the DG, engram cells showed a higher proportion of mushroom spines 5 days after encoding than engrams analyzed 24 h after encoding (169). Then hippocampal engram cells decrease their spine density as memory consolidates into a different area of the brain, the cortex, where engram neurons increase their spine density (49), which suggests that they associate with memory storage in a long term and ability to recall a memory.
New connections between engram cells might also be due to unsilencing synapses (Fig. 3). Silent synapses are synapses that are functionally silent—having NMDARs, they lack AMPA ones (207, 208). Supporting this view, engram cells have shown a higher presence of silent synapses than nonengram cells in the nucleus accumbens (209, 210), probably by means of internalizing AMPARs (211). Also supporting the role of unsilencing synapses in engram cells, overexpression of p21 kinase in engram cells increased the spine density and correlated with increased retention of memory in anisomycin-treated amnesic mice (212).
The dissociation between plasticity and storage poses the question of how L-LTP is necessary for the natural recall of memories (46). One explanation that has been proposed is that after memory is encoded, connections have been made, plasticity mechanisms distribute the synaptic weights to couple synaptic inputs with outputs to make memories accessible by recall (21). Pointing in the same direction, engram technology has shed light on the recall mechanism. Recall cues have been demonstrated to induce a rapid and transient increase in cell excitability in engram cells (139). Cellular excitability is an intrinsic cell property that allows the cell to generate an action potential in response to stimuli (213). By exposing the animals to a familiar context, the corresponding engram cells transiently upregulate excitability. Behaviorally, this temporal increase in excitability facilitates precision in context discrimination upon exposure to a context that resembles the first one and increases the efficiency in memory recall. Functionally, this effect is mediated by the internalization of Kir2.1 potassium channels (139). Concordantly with this hypothesis, it has been shown how interneurons also control engram cell reactivation, in an experience-dependent way: inhibitory interneurons in the amygdala silence engram cells to inactivate the recall of a nonrelevant memory (42).
The neural cell adhesion molecules
Molecular changes associated with memory are relevant not only for intracellular biology but also for transsynaptic interactions. Located in the presynaptic and postsynaptic membranes, cell adhesion molecules (CAMs) form bridges between cells (both from neuron to neuron and from neuron to glia) and between cells to extracellular matrix ligands. They regulate the formation of the initial contacts, stabilize the synapse and its maturation, and determine their strength (214). They physically anchor the synapses, recruit intracellular proteins to act as scaffolding, and control receptor trafficking. They also regulate molecular mechanisms involved in learning and plasticity: modulate gene transcription, activate transcription factors, regulate protein translation and regulate local protein synthesis (reviewed in Ref. (215)). Ultimately, they control neuronal morphology and network structure (216).
Many families of CAMs are involved in this complex synapse modulation: cadherins, protocadherins, immunoglobulin superfamily cell adhesion proteins (IgCAMs), integrins, and neurexins/neuroligins (217). Each of the subtypes of CAMs exhibits its particularities (see Ref. (214) for a review).
One of the best well-described adhesion molecules is the IgCAM neural cell adhesion molecule (NCAM). NCAM responds to neuronal activation upon experience (218). The homophilic interaction of NCAM triggers signal transduction pathways such as MAPK and Fyn kinases (215). These induce CREB phosphorylation (219), activation of other transcription factors such as NF-κB (220) and transcriptional changes, which culminates with LTP (221) and growth of neurites (222).
Other CAMs such as Dscam proteins (a subfamily of IgCAMs) or protocadherins (223, 224) play an important role in guiding neurons to their adequate synaptic partners. Interestingly, some of these superfamilies of CAMs exhibit a strikingly high level of diversity achieved by alternative splicing or a combination of differentially expressed isoforms (225). This high level of diversity gives them the potential to form the highly precise neural connections needed to achieve functional connectivity in the circuits formed—each neuron will find its partner (226, 227).
CAMs are necessary for the proper functioning of learning and memory, as evidenced by the multiple examples of memory affectations described by genetic or pharmacological manipulations (228). Indeed, the expression of adhesion molecules is upregulated in engram cells during consolidation. In the study discussed before, it was demonstrated that the expression of adhesion molecules such as neuroligins 1 and 3, and Ncam1 increases specifically in engram cells during memory consolidation (168). Interestingly, this modulation is mirrored by the regulation of their binding protein neurexin-1 in astrocytes (168), which may indicate crosstalk between glia and neurons to consolidate memories.
Information in a cell
Another solution for the storage problem poses that the information is stored inside the cell. Plasticity then would be responsible for quick changes that allow learning, whereas storage would happen at a nuclear level—securing long-term information (21, 229).
At the interphase of the psychology and neuroscience of memory, some authors have argued that learned information is stored in a molecular format inside the cell. Part of the motivation is the question of how organisms store integer numerical values such as memories of numbers (27, 28, 230) or conceptual representations (27, 231). In one study relevant to this perspective, Johansson et al. examined the classical eyeblink conditioning paradigm in a decerebrate ferret preparation. During one example of classical conditioning, such as the eyeblink response, an animal learns that at a particular time after a neutral cue is presented (i.e., a sound) or conditioned stimulus (CS), a second stimulus capable of provoking a response is delivered (i.e., an air puff that induces an eye blink), or unconditioned stimulus. This response is triggered by Purkinje cells in the cerebellar cortex, and it happens in a timely manner—they cease to fire and the animal blinks exactly at the time when they expect the air puff, after the sound (232). In a decerebrate ferret preparation, this learning process is electrophysiologically mimicked by direct stimulation of the relevant neurons, in substitution of the natural experience. By recording the firing of Purkinje cells with microelectrodes and stimulating the CS-carrying and unconditioned stimulus–carrying afferents (mossy and climbing fibers, respectively), the authors demonstrated that the Purkinje cells themselves were able to learn “when” after the CS they had to trigger the behavioral response. That is, the time information was somehow coded inside the Purkinje cell (233).
Studies made on the worm planaria have suggested that non-neuronal cells are also able to store information (234). Planarians have the ability to regenerate their brain after decapitation. Worms trained with a conditioning protocol where a shock was associated with light were able to retain the memory after decapitation and brain regeneration (235). More recently, similar results were obtained with more rigorous methods (236), confirming that headless worm fragments contained the memory of a familiar environment. Neoblasts (stem cells) have been hypothesized to carry the information, probably as epigenetic changes (Box 2) (237).
If memory relies on cell-autonomous processes (totally or partially), then the crucial question is where inside the cell is it encoded. Hypotheses of molecules that store memory information date back to the 1960s (238), inspired by the deciphering of the DNA code. Candidate coding substrates include DNA, RNA, epigenetic modifications (Box 2), peptides, or protein modifications (27, 229, 239, 240).
In a surprising study by Bédécarrats et al., the injection of RNA extracts from sensitized Aplysia snails enhanced the behavioral response to training in naïve animals. Donor individuals were exposed to long-term sensitization, a well-established protocol that involves the delivery of spaced tail shocks for 2 consecutive days. This training provokes a change in behavior in the animals, an enhancement in the siphon-withdrawal reflex, or sensitization. Immediately after testing the response of trained individuals, RNA from the whole nervous system was extracted and injected into receptor-naïve animals. About 24 h after the injection, recipient animals that received RNA from trained donors, but not control donors, showed sensitization. This effect is dependent on DNA methylation, since injection of the receptor animals with the DNA methyltransferase inhibitor RG-108 prevented the sensitization. This effect has been provocatively attributed to the transfer of noncoding molecules of RNA that are able to induce epigenetic modifications and prime the behavioral response to experience of receptor animals (241). However, while a modification of a behavioral response was demonstrated, this study did not investigate the transfer of steady-state RNA of trained animals, but RNA was taken from animals immediately after they recalled the experience.
Going even further, some authors have suggested that RNA may not only store information but also computationally process information; thanks to its secondary structures. In other words, RNA would not be a static reserve of experiences in the cell, but a system that can compute information, transforming different inputs into outputs (242).
Regardless of the proposed biochemical substrate, if information is stored through a molecular code inside cells, then it must be accessible in a way that is compatible (if not advantageous) with memory function. A clear physiological mechanism would be essential to handle information stored inside the cell. This postulated mechanism would translate the information inside one or more given cells to allow learning, further sustain memory progress toward consolidation, retain information for long periods, and reactivate pertinent pieces of information when these are needed during recall. On top of that, the system must assure that all this happens in an information-specific way—this is, only the relevant code-bearing cells, and not others, would be screened for that particular experience. Finally, the mechanism must be compatible with the behavioral timescale of memory—both for rapid learning and recall. It must allow immediate transit of information in and out of the cell but with a long-lasting effect, quickly writable and readable but secure enough. Although theoretically it might be feasible, substantial evidence would be necessary to validate such hypothetical mechanism.
Information outside the cell
It has been hypothesized that information could be stored at the hole pattern left by perineuronal nets (PNNs) (243). PNNs are very specialized extracellular matrix structures that surround the soma and proximal neurites of mature neurons and glia in the brain, leaving holes where synapses are established (244, 245). PNNs are initially formed right after critical periods (early developmental points when plasticity levels are high and experiences have a strong influence over the brain wiring) (246, 247). PNNs are formed by a mesh of extracellular matrix molecules, including chondroitin sulfate proteoglycans, hyaluronan, tenascin-R, and link proteins. Produced by neurons and glia (246, 248), they contribute to restricting synaptic plasticity (249, 250). The formation of PNNs and its composition are dynamic and responds to neuronal activity, not only in development (245, 249, 251) but also during adulthood (252).
The mechanism by which PNNs restrict neuronal plasticity is not fully understood. It is proposed that by forming a physical constrain around neurons, the lateral diffusion of AMPARs along the membrane is limited (244, 253). At the same time, PNNs are normally associated with inhibitory interneurons (254, 255). Deficient formation of PNNs provokes defects in inhibition over excitatory cells, which in turn increases excitatory transmission (256, 257). Surrounding excitatory neurons in the hippocampus, PNNs have also been found to respond to early life enrichment by restricting synaptic plasticity (258). Pharmacological degradation of PNNs in the hippocampus allowed plasticity modification of synapses that normally do not exhibit typical LTP (258).
The formation of PNNs has been demonstrated to be important for memory function. Injection of chondroitinase ABCh (an enzyme that degrades PNNs) in the amygdala 1 day before contextual fear conditioning decreased reinstatement of an extinguished fear memory. This is the process by which fear memories recover spontaneously after time (259, 260). These results indicate that PNNs are required at the time of learning, but not at the time of consolidation, to form memories that are later resistant to time. A similar effect occurred in the hippocampus and cortex (261, 262). Interestingly, learning or recall of recent memories were not affected by the lack of PNNs (262).
Since PNNs restrict plasticity and stabilize and protect memories from erasure (248), and because they are formed by components with a very low turnover and long life (263), they have been proposed as a mechanism to reliably store memory (243). The structure formed by PNNs at the time of learning would be then maintained and protected for longer periods to stabilize the synaptic connections that encode a particular memory. Still, much work needs to be done to reveal how the PNNs dialog with synapses and how they respond to experience.
The writers of the code?
Finally, the role of non-neuronal cell types in memory processes is crucial, though often neglected. If we understand the information as written in connections between neurons, one possibility is that immune cells are the writers that shape how the information is written. Following the hypothesis that memory is encoded in the connectivity pattern between engram cells, then the non-neuronal cells are located at an exceptional place to modify wiring plasticity and sculpt these connections (264).
Microglial cells are the patrolling immune cells in the brain parenchyma that rapidly activate and proliferate in response to insults (265). Several pathologies and disorders correlate with an impaired function of the homeostatic role of microglial cells, such as neurodegenerative diseases (Alzheimer’s disease), neurodevelopmental disorders (autism spectrum disorder), or even aging (266, 267, 268, 269). Microglial cells also respond to neuronal activation (270) and remodel the pattern of connections between neurons during development and adulthood (see Ref. (271) for a review). Engram technology was used to demonstrate that microglia engulf engram synapses, seemingly contributing to the forgetting of a specific memory. In this pioneering study, a month after an engram for a fearful memory had been formed, synapses labeled with an engram-specific marker were found within microglial cells. Ablation of microglial population with microglia-specific diphtheria toxin or inactivation of microglial phagocytic activity with the drug minocycline prevented forgetting in mice. Overall, indicating microglial control of engram persistence (272). Microglial activation provides a credible mechanism for manipulating both memory access and engram informational structure through the refinement of engram cell connectivity patterns.
Astrocytes are glial cells that govern many structural, metabolic, and homeostatic processes. Since they tile the central nervous system and interact with blood vessels, astrocytes provide metabolic support to neurons; they supply the energy necessary for the long-lasting changes that underlie memory formation and regulate neurometabolism (reviewed in Refs. (273, 274)). In particular, lactate supply from astrocytes sustains the de novo translation in the L-LTP during memory consolidation and is therefore required for long-lasting mechanisms of memory maintenance (275). Astrocytes also modulate synaptic activity by releasing molecules that signal into neurons known as gliotransmitters and by clearing out neurotransmitters from the synaptic zone (reviewed in Refs. (276, 277)). This has important implications for memory function. Artificial activation of the astrocytes in CA1 by chemogenetic tools induced spontaneous neuronal activity and de novo LTP in CA3 to CA1 synapses. Part of the mechanism that mediates the effect is NMDAR activation by d-serine secreted by astrocytes. Activated during learning through both optogenetic and chemogenetic methods in a contextual fear conditioning paradigm, astrocytes induced an enhancement in early memory retrieval (278), indicating that they modulate the formation of the task-specific engrams. Astrocytic activation during memory formation also impaired remote memory recall by disrupting hippocampal-cortical connections (279, 280), an interaction between areas that is central for the long-term consolidation of the memory (49, 156).
Oligodendrocytes are glial cells that form the protective myelin sheath around axons in the central nervous system (281, 282, 283). Neuronal activity induced by an experience triggers oligodendrocyte proliferation and myelination of active axons (284, 285). This oligodendroglial axonal support is required for memory consolidation, as evidenced by the study of mutant mice with deficits in oligodendrogenesis. By inhibiting the increase in de novo oligodendrogenesis after training without affecting existing oligodendrocytes, remote memory consolidation was impaired (285), probably by improper engram reactivation (286). Pharmacological treatment that induced myelin formation was able to prevent the forgetting of the memory over time (286), indicating a relevant role of oligodendrocytes in memory consolidation. Finally, specific transcriptomic programs associated with memory consolidation have been also described in non-neuronal cell types (168).
Precisely because they respond to neuronal activation, regulate synaptic transmission, and engulf synapses, non-neuronal cell types are uniquely suited to modulate engram cell biology. Non-neuronal cells precisely sculpt the wiring diagram and synaptic plasticity changes between engram cells that is necessary for structural and synaptic plasticity and ultimately, for memory function. Future studies that integrate engram cell labeling with glial cell biology should prove fruitful.
Conclusions and open questions
Supported by an extensive and solid base of knowledge about memory achieved by classical studies, engram studies pioneered new discoveries that have helped us clarify some of the key organizing principles of memory.
We now know that learning activates a group of neurons and induces on them a set of physical changes to form the engram. During learning, these engram cells exhibit plasticity of their synaptic strengths—the connections between them get strengthened. CAMs get redistributed, ion channels get directed to the membrane, and epigenetic mechanisms are triggered. A sparse population of neurons, and only a subset of synapses within them, exhibits these changes and get recruited to an engram, influenced by factors such as excitation state or cell type. During memory consolidation, the transition from short-term to long-term memory, a large and heterogeneous repertoire of gene expression is induced. The long-term storage of memory information is still ultimately a mystery. Some theories point to storage at connectivity patterns between engram cells—controlled by processes, such as synaptogenesis, synapse elimination, or unsilencing of synapses, whereas others point to the storage of the information through diverse molecular mechanisms inside the cell. Whole-cell plasticity mechanisms clearly contribute to information storage in engram cells but are likely insufficient to account for the full breadth of memory complexity in animals (139, 287, 288, 289, 290). Finally, non-neuronal cell types are able to modify engram function and must be accounted for in memory processes.
Despite the tremendous efforts in the field to clarify the molecular basis of memory processes, some very important questions remain unanswered:
-
•
What are the essential biochemical changes or features of engram cells that account for information specificity? How does one singular memory get recalled when it is behaviorally relevant?
-
•
How are the molecular mechanisms in memory systems not only flexible to allow immediate and quick learning but also persistent to store information during long-lasting periods? How are the systems compatible with allowing new learning involving modification of synapses while maintaining a trustworthy transcript of past experiences?
-
•
Where is the information stored for long-term? Is there such thing as a memory code? If the engram cells bear the mnemonic information, what is physically the substrate of that code? Can the experience of specific memories, and crucially not others, be transmitted as can be done with genetic information? And what is/are the associated mechanism/s that the code needs to function—where is the DNA double helix of memory information?
-
•
What is the role of traditionally less considered homeostatic processes, such as neurometabolism and thermodynamic efficiency in learning and memory?
Despite half a century of research, we are still only scratching the surface of the molecular basis for memory function. In this review, we have proposed that by keeping clearly defined questions in mind, we can find sensible ways of utilizing new technologies to experimentally test different theories of memory function. A comprehensive neurobiological account of memory requires a separation of different aspects and stages of mnemonic function and also a differentiation of enabling mechanisms (e.g., for constructing or reading engrams), from the biological substrate of information storage itself. Future research will determine which features ultimately depend on molecular explanations, and whether those mechanisms crucially work within cells or between cells.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are supported by the European Research Council, Science Foundation Ireland, the Irish Research Council, and Lister Institute of Preventive Medicine. We thank Paul Conway for proofreading and Ryan Lab members for scientific discussions.
Author contributions
C. O.-d. S. L. and T. J. R. conceptualization; C. O.-d. S. L. writing–original draft; T. J. R. writing–review & editing; T. J. R. supervision; T. J. R. funding acquisition.
Edited by Mike Shipston
Contributor Information
Clara Ortega-de San Luis, Email: ortegadc@tcd.ie.
Tomás J. Ryan, Email: tomas.ryan@tcd.ie.
References
- 1.Kandel E.R., Mack S., Jessell T.M., Schwartz J.H., Siegelbaum S.A., Hudspeth A.J. McGraw-Hill Education; New York, NY: 2013. Principles of Neural Science, Fifth Edition. [Google Scholar]
- 2.Tulving E. Episodic memory: From mind to brain. Annu. Rev. Psychol. 2002;53:1–25. doi: 10.1146/annurev.psych.53.100901.135114. [DOI] [PubMed] [Google Scholar]
- 3.Dudai Y., Roediger H.L., Tulving E. In: Science of Memory: Concepts. Roedinger H.L., Dudai Y., Fitzpatrick S.M., editors. Oxford University Press; New York, NY: 2007. Memory concepts; pp. 1–9. [Google Scholar]
- 4.McGaugh J.L. Memory - a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- 5.Schacter D.L., Tulving E. MIT Press; Cambridge, MA: 2020. Memory Systems 1994. [Google Scholar]
- 6.Squire L.R., Genzel L., Wixted J.T., Morris R.G. Memory consolidation. Cold Spring Harb. Perspect. Biol. 2015;7 doi: 10.1101/cshperspect.a021766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semon R. Engelmann W; Leipzig: 1904. Die mneme [The mneme] [Google Scholar]
- 8.Josselyn S.A., Tonegawa S. Memory engrams: Recalling the past and imagining the future. Science. 2020;367 doi: 10.1126/science.aaw4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonegawa S., Liu X., Ramirez S., Redondo R. Memory engram cells have come of age. Neuron. 2015;87:918–931. doi: 10.1016/j.neuron.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Descartes R. Oxford University Press; Oxford: 2008. Meditations on First Philosophy. [Google Scholar]
- 11.Smith K. Metaphysics Research Lab, Stanford University; Stanford, CA: 2021. Descartes’ Theory of Ideas. [Google Scholar]
- 12.Krakauer J.W., Ghazanfar A.A., Gomez-Marin A., MacIver M.A., Poeppel D. Neuroscience needs behavior: Correcting a reductionist bias. Neuron. 2017;93:480–490. doi: 10.1016/j.neuron.2016.12.041. [DOI] [PubMed] [Google Scholar]
- 13.Watson J.D., Crick F.H.C. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 14.Mendel G. Versuche uber pflanzen-hybriden. Verh. Naturforsch. Ver. Brunn. 1865;4:3–47. [Google Scholar]
- 15.Morgan T.H. Sex limited inheritance in drosophila. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- 16.Avery O.T., Macleod C.M., McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J. Exp. Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershey A.D., Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benzer S. On the topology of the genetic fine structure. Proc. Natl. Acad. Sci. U. S. A. 1959;45:1607–1620. doi: 10.1073/pnas.45.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benzer S. On the topography of the genetic fine structure. Proc. Natl. Acad. Sci. U. S. A. 1961;47:403–415. doi: 10.1073/pnas.47.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crick F.H.C., Barnett L., Brenner S., Watts-Tobin R.J. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 21.Abraham W.C., Jones O.D., Glanzman D.L. Is plasticity of synapses the mechanism of long-term memory storage? NPJ Sci. Learn. 2019;4:9. doi: 10.1038/s41539-019-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asok A., Leroy F., Rayman J.B., Kandel E.R. Molecular mechanisms of the memory trace. Trends Neurosci. 2019;42:14–22. doi: 10.1016/j.tins.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chklovskii D.B., Mel B.W., Svoboda K. Cortical rewiring and information storage. Nature. 2004;431:782–788. doi: 10.1038/nature03012. [DOI] [PubMed] [Google Scholar]
- 24.Johansen J.P., Cain C.K., Ostroff L.E., LeDoux J.E. Molecular mechanisms of fear learning and memory. Cell. 2011;147:509–524. doi: 10.1016/j.cell.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandel E.R. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 26.Kandel E.R., Dudai Y., Mayford M.R. The molecular and systems biology of memory. Cell. 2014;157:163–186. doi: 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Gallistel C.R. The coding question. Trends Cogn. Sci. 2017;21:498–508. doi: 10.1016/j.tics.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Gallistel C.R. The physical basis of memory. Cognition. 2021;213:104533. doi: 10.1016/j.cognition.2020.104533. [DOI] [PubMed] [Google Scholar]
- 29.Queenan B.N., Ryan T.J., Gazzaniga M.S., Gallistel C.R. On the research of time past: The hunt for the substrate of memory. Ann. N. Y. Acad. Sci. 2017;1396:108–125. doi: 10.1111/nyas.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallistel C.R., Matzel L.D. The neuroscience of learning: Beyond the hebbian synapse. Annu. Rev. Psychol. 2013;64:169–200. doi: 10.1146/annurev-psych-113011-143807. [DOI] [PubMed] [Google Scholar]
- 31.Josselyn S.A., Köhler S., Frankland P.W. Finding the engram. Nat. Rev. Neurosci. 2015;16:521–534. doi: 10.1038/nrn4000. [DOI] [PubMed] [Google Scholar]
- 32.Miller R.R. Failures of memory and the fate of forgotten memories. Neurobiol. Learn. Mem. 2021;181:107426. doi: 10.1016/j.nlm.2021.107426. [DOI] [PubMed] [Google Scholar]
- 33.Liu X., Ramirez S., Pang P.T., Puryear C.B., Govindarajan A., Deisseroth K., Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shockett P.E., Schatz D.G. Diverse strategies for tetracycline-regulated inducible gene expression. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5173–5176. doi: 10.1073/pnas.93.11.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez S., Liu X., Lin P.-A., Suh J., Pignatelli M., Redondo R.L., Ryan T.J., Tonegawa S. Creating a false memory in the hippocampus. Science. 2013;341:387–391. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- 36.Denny C.A., Kheirbek M.A., Alba E.L., Tanaka K.F., Brachman R.A., Laughman K.B., Tomm N.K., Turi G.F., Losonczy A., Hen R. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka K.Z., Pevzner A., Hamidi A.B., Nakazawa Y., Graham J., Wiltgen B.J. Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron. 2014;84:347–354. doi: 10.1016/j.neuron.2014.09.037. [DOI] [PubMed] [Google Scholar]
- 38.Han J.H., Kushner S.A., Yiu A.P., Hsiang H.L., Buch T., Waisman A., Bontempi B., Neve R.L., Frankland P.W., Josselyn S.A. Selective erasure of a fear memory. Science. 2009;323:1492–1496. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- 39.Kim J., Kwon J.T., Kim H.S., Josselyn S.A., Han J.H. Memory recall and modifications by activating neurons with elevated CREB. Nat. Neurosci. 2014;17:65–72. doi: 10.1038/nn.3592. [DOI] [PubMed] [Google Scholar]
- 40.Redondo R.L., Kim J., Arons A.L., Ramirez S., Liu X., Tonegawa S. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 2014;513:426–430. doi: 10.1038/nature13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reijmers L.G., Perkins B.L., Matsuo N., Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317:1230–1233. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- 42.Trouche S., Sasaki J.M., Tu T., Reijmers L.G. Fear extinction causes target-specific remodeling of perisomatic inhibitory synapses. Neuron. 2013;80:1054–1065. doi: 10.1016/j.neuron.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zelikowsky M., Hersman S., Chawla M.K., Barnes C.A., Fanselow M.S. Neuronal ensembles in amygdala, hippocampus, and prefrontal cortex track differential components of contextual fear. J. Neurosci. 2014;34:8462–8466. doi: 10.1523/JNEUROSCI.3624-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng W., Mayford M., Gage F.H. Selection of distinct populations of dentate granule cells in response to inputs as a mechanism for pattern separation in mice. Elife. 2013;2 doi: 10.7554/eLife.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez S., Liu X., MacDonald C.J., Moffa A., Zhou J., Redondo R.L., Tonegawa S. Activating positive memory engrams suppresses depression-like behaviour. Nature. 2015;522:335–339. doi: 10.1038/nature14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan T.J., Roy D.S., Pignatelli M., Arons A., Tonegawa S. Engram cells retain memory under retrograde amnesia. Science. 2015;348:1007–1013. doi: 10.1126/science.aaa5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowansage K.K., Shuman T., Dillingham B.C., Chang A., Golshani P., Mayford M. Direct reactivation of a coherent neocortical memory of context. Neuron. 2014;84:432–441. doi: 10.1016/j.neuron.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeNardo L.A., Liu C.D., Allen W.E., Adams E.L., Friedmann D., Fu L., Guenthner C.J., Tessier-Lavigne M., Luo L. Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat. Neurosci. 2019;22:460–469. doi: 10.1038/s41593-018-0318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitamura T., Ogawa S.K., Roy D.S., Okuyama T., Morrissey M.D., Smith L.M., Redondo R.L., Tonegawa S. Engrams and circuits crucial for systems consolidation of a memory. Science. 2017;356:73–78. doi: 10.1126/science.aam6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koya E., Golden S.A., Harvey B.K., Guez-Barber D.H., Berkow A., Simmons D.E., Bossert J.M., Nair S.G., Uejima J.L., Marin M.T., Mitchell T.B., Farquhar D., Ghosh S.C., Mattson B.J., Hope B.T. Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nat. Neurosci. 2009;12:1069–1073. doi: 10.1038/nn.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garner A.R., Rowland D.C., Hwang S.Y., Baumgaertel K., Roth B.L., Kentros C., Mayford M. Generation of a synthetic memory trace. Science. 2012;335:1513–1516. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohkawa N., Saitoh Y., Suzuki A., Tsujimura S., Murayama E., Kosugi S., Nishizono H., Matsuo M., Takahashi Y., Nagase M., Sugimura Y.K., Watabe A.M., Kato F., Inokuchi K. Artificial association of pre-stored information to generate a qualitatively new memory. Cell Rep. 2015;11:261–269. doi: 10.1016/j.celrep.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 53.Vetere G., Tran L.M., Moberg S., Steadman P.E., Restivo L., Morrison F.G., Ressler K.J., Josselyn S.A., Frankland P.W. Memory formation in the absence of experience. Nat. Neurosci. 2019;22:933–940. doi: 10.1038/s41593-019-0389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yiu A.P., Mercaldo V., Yan C., Richards B., Rashid A.J., Hsiang H.-L.L., Pressey J., Mahadevan V., Tran M.M., Kushner S.A., Woodin M.A., Frankland P.W., Josselyn S.A. Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron. 2014;83:722–735. doi: 10.1016/j.neuron.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 55.Vlasov K., Van Dort C.J., Solt K. Optogenetics and chemogenetics. Methods Enzymol. 2018;603:181–196. doi: 10.1016/bs.mie.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metzger D., Clifford J., Chiba H., Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guenthner C.J., Miyamichi K., Yang H.H., Heller H.C., Luo L. Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron. 2013;78:773. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allen W.E., Denardo L.A., Chen M.Z., Liu C.D., Loh K.M., Fenno L.E., Ramakrishnan C., Deisseroth K., Luo L. Thirst-associated preoptic neurons encode an aversive motivational drive. Science. 2017;1155:1149–1155. doi: 10.1126/science.aan6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sørensen A.T., Cooper Y.A., Baratta M.V., Weng F.J., Zhang Y., Ramamoorthi K., Fropf R., Laverriere E., Xue J., Young A., Schneider C., Gøtzsche C.R., Hemberg M., Yin J.C.P., Maier S.F., et al. A robust activity marking system for exploring active neuronal ensembles. Elife. 2016;5 doi: 10.7554/eLife.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawashima T., Kitamura K., Suzuki K., Nonaka M., Kamijo S., Takemoto-Kimura S., Kano M., Okuno H., Ohki K., Bito H. Functional labeling of neurons and their projections using the synthetic activity–dependent promoter E-SARE. Nat. Methods. 2013;10:889–895. doi: 10.1038/nmeth.2559. [DOI] [PubMed] [Google Scholar]
- 61.Wang W., Kim C.K., Ting A.Y. Molecular tools for imaging and recording neuronal activity. Nat. Chem. Biol. 2019;15:101–110. doi: 10.1038/s41589-018-0207-0. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y., Zou P., Cohen A.E. Voltage imaging with genetically encoded indicators. Curr. Opin. Chem. Biol. 2017;39:1–10. doi: 10.1016/j.cbpa.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mank M., Griesbeck O. Genetically encoded calcium indicators. Chem. Rev. 2008;108:1550–1564. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- 64.Kim T.H., Schnitzer M.J. Fluorescence imaging of large-scale neural ensemble dynamics. Cell. 2022;185:9–41. doi: 10.1016/j.cell.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H., Jing M., Li Y. Lighting up the brain: Genetically encoded fluorescent sensors for imaging neurotransmitters and neuromodulators. Curr. Opin. Neurobiol. 2018;50:171–178. doi: 10.1016/j.conb.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marvin J.S., Scholl B., Wilson D.E., Podgorski K., Kazemipour A., Müller J.A., Schoch S., Quiroz F.J.U., Rebola N., Bao H., Little J.P., Tkachuk A.N., Cai E., Hantman A.W., Wang S.S.H., et al. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods. 2018;15:936–939. doi: 10.1038/s41592-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jing M., Zhang P., Wang G., Feng J., Mesik L., Zeng J., Jiang H., Wang S., Looby J.C., Guagliardo N.A., Langma L.W., Lu J., Zuo Y., Talmage D.A., Role L.W., et al. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat. Biotechnol. 2018;36:726–737. doi: 10.1038/nbt.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marvin J.S., Shimoda Y., Magloire V., Leite M., Kawashima T., Jensen T.P., Kolb I., Knott E.L., Novak O., Podgorski K., Leindenheimer N.J., Rusakov D.A., Ahrens M.B., Kullmann D.M., Looger L.L. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods. 2019;16:763–770. doi: 10.1038/s41592-019-0471-2. [DOI] [PubMed] [Google Scholar]
- 69.Fosque B.F., Sun Y., Dana H., Yang C.T., Ohyama T., Tadross M.R., Patel R., Zlatic M., Kim D.S., Ahrens M.B., Jayaraman V., Looger L.L., Schreiter E.R. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 70.Sakurai K., Zhao S., Takatoh J., Rodriguez E., Lu J., Leavitt A.D., Fu M., Han B.X., Wang F. Capturing and manipulating activated neuronal ensembles with CANE delineates a hypothalamic social-fear circuit. Neuron. 2016;92:739–753. doi: 10.1016/j.neuron.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee D., Hyun J., Jung K., Hannan P., Kwon H. A calcium- and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol. 2017;35:858–863. doi: 10.1038/nbt.3902. [DOI] [PubMed] [Google Scholar]
- 72.Wang W., Wildes C.P., Pattarabanjird T., Sanchez M.I., Glober G.F., Matthews G.A., Tye K.M., Ting A.Y. A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat. Biotechnol. 2017;35:864–871. doi: 10.1038/nbt.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez M.I., Nguyen Q.A., Wang W., Soltesz I., Ting A.Y. Transcriptional readout of neuronal activity via an engineered Ca2+-activated protease. Proc. Natl. Acad. Sci. U. S. A. 2020;117:33186–33196. doi: 10.1073/pnas.2006521117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim C.K., Sanchez M.I., Hoerbelt P., Fenno L.E., Malenka R.C., Deisseroth K., Ting A.Y. A molecular calcium integrator reveals a striatal cell type driving aversion. Cell. 2020;183:2003–2019.e16. doi: 10.1016/j.cell.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Linghu C., An B., Shpokayte M., Celiker O.T., Shmoel N., Zhang C., Park W.M., Ramirez S., Boyden E.S. Recording of cellular physiological histories along optically readable self-assembling protein chains. bioRxiv. 2021 doi: 10.1101/2021.10.13.464006. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eisenberg M., Kobilo T., Berman D.E., Dudai Y. Stability of retrieved memory: Inverse correlation with trace dominance. Science. 2003;301:1102–1104. doi: 10.1126/science.1086881. [DOI] [PubMed] [Google Scholar]
- 77.Przybyslawski J., Sara S.J. Reconsolidation of memory after its reactivation. Behav. Brain Res. 1997;84:241–246. doi: 10.1016/s0166-4328(96)00153-2. [DOI] [PubMed] [Google Scholar]
- 78.Ryan T.J., Frankland P.W. Forgetting as a form of adaptive engram cell plasticity. Nat. Rev. Neurosci. 2022;23:173–186. doi: 10.1038/s41583-021-00548-3. [DOI] [PubMed] [Google Scholar]
- 79.Tulving E. Cue-dependent forgetting. Am. Sci. 1974;62:74–82. [Google Scholar]
- 80.Dudai Y. Consolidation: Fragility on the road to the engram. Neuron. 1996;17:367–370. doi: 10.1016/s0896-6273(00)80168-3. [DOI] [PubMed] [Google Scholar]
- 81.Ramon y Cajal S. Estructura de los centros nerviosos de las aves. Rev. Trim. Histol. Normal Patol. 1888;1:1–30. [Google Scholar]
- 82.Sherrington C. Yale University Press; New Haven, CT: 1906. The Integrative Action of the Nervous System. [Google Scholar]
- 83.Ramon y Cajal S. The Croonian lecture.—La fine structure des centres nerveux. Proc. R. Soc. Lond. 1894;55:444–468. [Google Scholar]
- 84.Lorente De Nó R. Analysis of the activity of the chains of internuncial neurons. J. Neurophysiol. 1938;1:207–244. [Google Scholar]
- 85.Hebb D.O. Wiley and Sons; New York, NY: 1949. The Organisation of Behaviour. [Google Scholar]
- 86.Markram H., Lübke J., Frotscher M., Sakmann B. Regulation of synaptic efficacy by coincidence of postsynaptic APs and EPSPs. Science. 1997;275:213–215. doi: 10.1126/science.275.5297.213. [DOI] [PubMed] [Google Scholar]
- 87.Markram H., Gerstner W., Sjöström P.J. Spike-timing-dependent plasticity: A comprehensive overview. Front. Synaptic Neurosci. 2012;4:2. doi: 10.3389/fnsyn.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bliss T.V.P., Collingridge G.L. A synaptic model of memory: Long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 89.Kandel E.R., Tauc L. Heterosynaptic facilitation in neurones of the abdominal ganglion of Aplysia depilans. J. Physiol. 1965;181:1–27. doi: 10.1113/jphysiol.1965.sp007742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bliss T.V., Lomo T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 1973;232:331–356. doi: 10.1113/jphysiol.1973.sp010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ito M. Cerebellar control of the vestibulo-ocular reflex--around the flocculus hypothesis. Annu. Rev. Neurosci. 1982;5:275–296. doi: 10.1146/annurev.ne.05.030182.001423. [DOI] [PubMed] [Google Scholar]
- 92.Malenka R.C., Bear M.F. LTP and LTD: An embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 93.Bayer K.U., Schulman H. CaM kinase: Still inspiring at 40. Neuron. 2019;103:380–394. doi: 10.1016/j.neuron.2019.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lisman J., Schulman H., Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 95.Lisman J., Yasuda R., Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benke T.A., Luthl A., Isaac J.T.R., Collingridge G.L. Modulation of AMPA receptor unitary conductance by synaptic activity. Nature. 1998;393:793–797. doi: 10.1038/31709. [DOI] [PubMed] [Google Scholar]
- 97.Derkach V., Barria A., Soderling T.R. Ca2+/calmodulin-kinase II enhances channel conductance of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate type glutamate receptors. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3269–3274. doi: 10.1073/pnas.96.6.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kristensen A.S., Jenkins M.A., Banke T.G., Schousboe A., Makino Y., Johnson R.C., Huganir R., Traynelis S.F. Mechanism of Ca2+/calmodulin-dependent kinase II regulation of AMPA receptor gating. Nat. Neurosci. 2011;14:727–735. doi: 10.1038/nn.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Penn A.C., Zhang C.L., Georges F., Royer L., Breillat C., Hosy E., Petersen J.D., Humeau Y., Choquet D. Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature. 2017;549:384–388. doi: 10.1038/nature23658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shirke A.M., Malinow R. Mechanisms of potentiation by calcium-calmodulin kinase II of postsynaptic sensitivity in rat hippocampal CA1 neurons. J. Neurophysiol. 1997;78:2682–2692. doi: 10.1152/jn.1997.78.5.2682. [DOI] [PubMed] [Google Scholar]
- 101.Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- 102.Matsuzaki M., Honkura N., Ellis-Davies G.C.R., Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nägerl U.V., Eberhorn N., Cambridge S.B., Bonhoeffer T. Bidirectional activity-dependent morphological plasticity in hippocampal neurons. Neuron. 2004;44:759–767. doi: 10.1016/j.neuron.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 104.Okamoto K.I., Nagai T., Miyawaki A., Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 2004;7:1104–1112. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- 105.Zhou Q., Homma K.J., Poo M.M. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 106.Carlisle H.J., Kennedy M.B. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 107.Hotulainen P., Hoogenraad C.C. Actin in dendritic spines: Connecting dynamics to function. J. Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spence E.F., Soderling S.H. Actin out: Regulation of the synaptic cytoskeleton. J. Biol. Chem. 2015;290:28613–28622. doi: 10.1074/jbc.R115.655118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang H., Ben Zablah Y., Zhang H., Jia Z. Rho signaling in synaptic plasticity, memory, and brain disorders. Front. Cell Dev. Biol. 2021;9:2673. doi: 10.3389/fcell.2021.729076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arber S., Barbayannis F.A., Hanser H., Schnelder C., Stanyon C.A., Bernards O., Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 111.Ben Zablah Y., Merovitch N., Jia Z. The role of ADF/cofilin in synaptic physiology and Alzheimer’s disease. Front. Cell Dev. Biol. 2020;8:594998. doi: 10.3389/fcell.2020.594998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goley E.D., Welch M.D. The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 113.Hotulainen P., Llano O., Smirnov S., Tanhuanpää K., Faix J., Rivera C., Lappalainen P. Defning mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J. Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hussain N.K., Thomas G.M., Luo J., Huganir R.L. Regulation of AMPA receptor subunit GluA1 surface expression by PAK3 phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E5883–E5890. doi: 10.1073/pnas.1518382112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lamprecht R., LeDoux J. Structural plasticity and memory. Nat. Rev. Neurosci. 2004;5:45–54. doi: 10.1038/nrn1301. [DOI] [PubMed] [Google Scholar]
- 116.Engert F., Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 117.Dudai Y., Eisenberg M. Rites of passage of the engram: Reconsolidation and the lingering consolidation hypothesis. Neuron. 2004;44:93–100. doi: 10.1016/j.neuron.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 118.Raman D.V., O'Leary T. Optimal plasticity for memory maintenance during ongoing synaptic change. Elife. 2021;10 doi: 10.7554/eLife.62912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Turrigiano G.G., Nelson S.B. Homeostatic plasticity in the developing nervous system. Nat. Rev. Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- 120.Crick F. Neurobiology: Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- 121.Guskjolen A., Kenney J.W., de la Parra J., Yeung B.R.A., Josselyn S.A., Frankland P.W. Recovery of “lost” infant memories in mice. Curr. Biol. 2018;28:2283–2290.e3. doi: 10.1016/j.cub.2018.05.059. [DOI] [PubMed] [Google Scholar]
- 122.Perusini J.N., Cajigas S.A., Cohensedgh O., Lim S.C., Pavlova I.P., Donaldson Z.R., Denny C.A. Optogenetic stimulation of dentate gyrus engrams restores memory in Alzheimer’s disease mice. Hippocampus. 2017;27:1110–1122. doi: 10.1002/hipo.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Poll S., Mittag M., Musacchio F., Justus L.C., Giovannetti E.A., Steffen J., Wagner J., Zohren L., Schoch S., Schmidt B., Jackson W.S., Ehninger D., Fuhrmann M. Memory trace interference impairs recall in a mouse model of Alzheimer’s disease. Nat. Neurosci. 2020;23:952–958. doi: 10.1038/s41593-020-0652-4. [DOI] [PubMed] [Google Scholar]
- 124.Roy D.S., Arons A., Mitchell T.I., Pignatelli M., Ryan T.J., Tonegawa S. Memory retrieval by activating engram cells in mouse models of early Alzheimer’s disease. Nature. 2016;531:508–512. doi: 10.1038/nature17172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao B., Sun J., Zhang X., Mo H., Niu Y., Li Q., Wang L., Zhong Y. Long-term memory is formed immediately without the need for protein synthesis-dependent consolidation in Drosophila. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-12436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim W.B., Cho J.H. Encoding of discriminative fear memory by input-specific LTP in the amygdala. Neuron. 2017;95:1129–1146.e5. doi: 10.1016/j.neuron.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 127.Kim W.B., Cho J.H. Encoding of contextual fear memory in hippocampal–amygdala circuit. Nat. Commun. 2020;11:1–22. doi: 10.1038/s41467-020-15121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Feinberg E.H., VanHoven M.K., Bendesky A., Wang G., Fetter R.D., Shen K., Bargmann C.I. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 129.Choi J.-H., Sim S.-E., Kim J., Choi D.I., Oh J., Ye S., Lee J., Kim T., Ko H.-G., Lim C.-S., Kaang B.-K. Interregional synaptic maps among engram cells underlie memory formation. Science. 2018;360:430–435. doi: 10.1126/science.aas9204. [DOI] [PubMed] [Google Scholar]
- 130.Choi D.I., Kim J., Lee H., Kim J., Sung Y., Choi J.E., Venkat S.J., Park P., Jung H., Kaang B.-K. Synaptic correlates of associative fear memory in the lateral amygdala. Neuron. 2021;109:2717–2726.e3. doi: 10.1016/j.neuron.2021.07.003. [DOI] [PubMed] [Google Scholar]
- 131.Nonaka A., Toyoda T., Miura Y., Hitora-Imamura N., Naka M., Eguchi M., Yamaguchi S., Ikegaya Y., Matsuki N., Nomura H. Synaptic plasticity associated with a memory engram in the basolateral amygdala. J. Neurosci. 2014;34:9305–9309. doi: 10.1523/JNEUROSCI.4233-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou Y., Zhu H., Liu Z., Chen X., Su X.J., Ma C., Tian Z., Huang B., Yan E., Liu X., Ma L. A ventral CA1 to nucleus accumbens core engram circuit mediates conditioned place preference for cocaine. Nat. Neurosci. 2019;22:1986–1999. doi: 10.1038/s41593-019-0524-y. [DOI] [PubMed] [Google Scholar]
- 133.Josselyn S.A., Frankland P.W. Memory allocation: Mechanisms and function. Annu. Rev. Neurosci. 2018;41:389–413. doi: 10.1146/annurev-neuro-080317-061956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Han J., Kushner S.A., Yiu A.P., Cole C.J., Matynia A., Brown R.A., Neve R.L., Guzowski J.F., Silva A.J., Josselyn S.A. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 135.Barco A., Pittenger C., Kandel E.R. CREB, memory enhancement and the treatment of memory disorders: Promises, pitfalls and prospects. Expert Opin. Ther. Targets. 2003;7:101–114. doi: 10.1517/14728222.7.1.101. [DOI] [PubMed] [Google Scholar]
- 136.Benito E., Barco A. CREB’s control of intrinsic and synaptic plasticity: Implications for CREB-dependent memory models. Trends Neurosci. 2010;33:230–240. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 137.Cai D.J., Aharoni D., Shuman T., Shobe J., Biane J., Song W., Wei B., Veshkini M., La-Vu M., Lou J., Flores S.E., Kim I., Sano Y., Zhou M., Baumgaertel K., et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature. 2016;534:115–118. doi: 10.1038/nature17955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rashid A.J., Yan C., Mercaldo V., Hsiang H.-L.L., Park S., Cole C.J., De Cristofaro A., Yu J., Ramakrishnan C., Lee S.Y., Deisseroth K., Frankland P., Josselyin S.A. Competition between engrams influences fear memory formation and recall. Science. 2016;353:383–387. doi: 10.1126/science.aaf0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pignatelli M., Ryan T.J., Roy D.S., Lovett C., Smith L.M., Muralidhar S., Tonegawa S. Engram cell excitability state determines the efficacy of memory retrieval. Neuron. 2019;101:274–284.e5. doi: 10.1016/j.neuron.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 140.Chawla M.K., Penner M.R., Olson K.M., Sutherland V.L., Mittelman-Smith M.A., Barnes C.A. Spatial behavior and seizure-induced changes in c-fos mRNA expression in young and old rats. Neurobiol. Aging. 2013;34:1184–1198. doi: 10.1016/j.neurobiolaging.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Erwin S.R., Sun W., Copeland M., Lindo S., Spruston N., Cembrowski M.S. A sparse, spatially biased subtype of mature granule cell dominates recruitment in hippocampal-associated behaviors. Cell Rep. 2020;31:107551. doi: 10.1016/j.celrep.2020.107551. [DOI] [PubMed] [Google Scholar]
- 142.Kee N., Teixeira C.C.M., Wang A.H.A., Frankland P.W. Preferential incorporation of adult-generated granule cells into spatial memory networks in the dentate gyrus. Nat. Neurosci. 2007;10:355–362. doi: 10.1038/nn1847. [DOI] [PubMed] [Google Scholar]
- 143.Castello-Waldow T.P., Weston G., Ulivi A.F., Chenani A., Loewenstein Y., Chen A., Attardo A. Hippocampal neurons with stable excitatory connectivity become part of neuronal representations. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rogerson T., Cai D.J., Frank A., Sano Y., Shobe J., Lopez-Aranda M.F., Silva A.J. Synaptic tagging during memory allocation. Nat. Rev. Neurosci. 2014;15:157–169. doi: 10.1038/nrn3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Söhl G., Maxeiner S., Willecke K. Expression and functions of neuronal gap junctions. Nat. Rev. Neurosci. 2005;6:191–200. doi: 10.1038/nrn1627. [DOI] [PubMed] [Google Scholar]
- 146.Ariazi J., Benowitz A., De Biasi V., Den Boer M.L., Cherqui S., Cui H., Douillet N., Eugenin E.A., Favre D., Goodman S., Gousset K., Hanein D., Israel D.I., Kimura S., Kirkpatrick R.B., et al. Tunneling nanotubes and gap junctions–their role in long-range intercellular communication during development, health, and disease conditions. Front. Mol. Neurosci. 2017;10:333. doi: 10.3389/fnmol.2017.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Smalheiser N.R. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol. Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ashley J., Cordy B., Lucia D., Fradkin L.G., Budnik V., Thomson T. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell. 2018;172:262–274.e11. doi: 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Landry C.D., Kandel E.R., Rajasethupathy P. New mechanisms in memory storage: piRNAs and epigenetics. Trends Neurosci. 2013;36:535–542. doi: 10.1016/j.tins.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 150.Shepherd J.D., Bear M.F. New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pastuzyn E.D., Day C.E., Kearns R.B., Kyrke-Smith M., Taibi A.V., McCormick J., Yoder N., Belnap D.M., Erlendsson S., Morado D.R., Briggs J.A.G., Feschotte C., Shepherd J.D. The neuronal gene Arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer. Cell. 2018;172:275–288.e18. doi: 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Farris S., Lewandowski G., Cox C.D., Steward O. Selective localization of Arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J. Neurosci. 2014;34:4481–4493. doi: 10.1523/JNEUROSCI.4944-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ryan T.J., Ortega-de San Luis C., Pezzoli M., Sen S. Engram cell connectivity: An evolving substrate for information storage. Curr. Opin. Neurobiol. 2021;67:215–227. doi: 10.1016/j.conb.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 154.Tonegawa S., Pignatelli M., Roy D.S., Ryan T.J. Memory engram storage and retrieval. Curr. Opin. Neurobiol. 2015;35:101–109. doi: 10.1016/j.conb.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 155.Staubli U., Lynch G. Stable hippocampal long-term potentiation elicited by “theta” pattern stimulation. Brain Res. 1987;435:227–234. doi: 10.1016/0006-8993(87)91605-2. [DOI] [PubMed] [Google Scholar]
- 156.Tonegawa S., Morrissey M.D., Kitamura T. The role of engram cells in the systems consolidation of memory. Nat. Rev. Neurosci. 2018;19:485–498. doi: 10.1038/s41583-018-0031-2. [DOI] [PubMed] [Google Scholar]
- 157.Raymond C.R., Thompson V.L., Tate W.P., Abraham W.C. Metabotropic glutamate receptors trigger homosynaptic protein synthesis to prolong long-term potentiation. J. Neurosci. 2000;20:969–976. doi: 10.1523/JNEUROSCI.20-03-00969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Steward O. mRNA localization in neurons: A multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 159.Sutton M.A., Schuman E.M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 160.Dash P.K., Hochner B., Kandel E.R. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 161.Alberini C.M., Ghirardl M., Metz R., Kandel E.R. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 162.Frey U., Morris R.G.M. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 163.Frey U., Morris R.G. Weak before strong: Dissociating synaptic tagging and plasticity-factor accounts of late-LTP. Neuropharmacology. 1998;37:545–552. doi: 10.1016/s0028-3908(98)00040-9. [DOI] [PubMed] [Google Scholar]
- 164.Martin K.C., Kosik K.S. Synaptic tagging — who’s it? Nat. Rev. Neurosci. 2002;3:813–820. doi: 10.1038/nrn942. [DOI] [PubMed] [Google Scholar]
- 165.Lacar B., Linker S.B., Jaeger B.N., Krishnaswami S.R., Barron J.J., Kelder M.J.E., Parylak S.L., Paquola A.C.M., Venepally P., Novotny M., O’Connor C., Fitzpatrick C., Erwin J.A., Hsu J.Y., Husband D., et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jaeger B.N., Linker S.B., Parylak S.L., Barron J.J., Gallina I.S., Saavedra C.D., Fitzpatrick C., Lim C.K., Schafer S.T., Lacar B., Jessberger S., Gage F.H. A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nat. Commun. 2018;9:3084. doi: 10.1038/s41467-018-05418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rao-Ruiz P., Couey J.J., Marcelo I.M., Bouwkamp C.G., Slump D.E., Matos M.R., van der Loo R.J., Martins G.J., van den Hout M., van IJcken W.F., Costa R.M., van den Oever M.C., Kushner S.A. Engram-specific transcriptome profiling of contextual memory consolidation. Nat. Commun. 2019;10:2232. doi: 10.1038/s41467-019-09960-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen M.B., Jiang X., Quake S.R., Südhof T.C. Persistent transcriptional programmes are associated with remote memory. Nature. 2020;587:437–442. doi: 10.1038/s41586-020-2905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Marco A., Meharena H.S., Dileep V., Raju R.M., Davila-Velderrain J., Zhang A.L., Adaikkan C., Young J.Z., Gao F., Kellis M., Tsai L.H. Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci. 2020;23:1606–1617. doi: 10.1038/s41593-020-00717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mercer T.R., Dinger M.E., Mariani J., Kosik K.S., Mehler M.F., Mattick J.S. Noncoding RNAs in long-term memory formation. Neuroscientist. 2007;14:434–445. doi: 10.1177/1073858408319187. [DOI] [PubMed] [Google Scholar]
- 171.Smalheiser N.R. The RNA-centred view of the synapse: Non-coding RNAs and synaptic plasticity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130504. doi: 10.1098/rstb.2013.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 173.Bredy T.W., Lin Q., Wei W., Baker-Andresen D., Mattick J.S. MicroRNA regulation of neural plasticity and memory. Neurobiol. Learn. Mem. 2011;96:89–94. doi: 10.1016/j.nlm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 174.Rajasethupathy P., Fiumara F., Sheridan R., Betel D., Puthanveettil S.V., Russo J.J., Sander C., Tuschl T., Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Finkenstadt P.M., Kang W.-S., Jeon M., Taira E., Tang W., Baraban J.M. Somatodendritic localization of translin, a component of the translin/trax RNA binding complex. J. Neurochem. 2000;75:1754–1762. doi: 10.1046/j.1471-4159.2000.0751754.x. [DOI] [PubMed] [Google Scholar]
- 176.Park A.J., Havekes R., Fu X., Hansen R., Tudor J.C., Peixoto L., Li Z., Wu Y.C., Poplawski S.G., Baraban J.M., Abel T. Learning induces the translin/trax RNase complex to express activin receptors for persistent memory. Elife. 2017;6 doi: 10.7554/eLife.27872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ishizu H., Siomi H., Siomi M.C. Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rajasethupathy P., Antonov I., Sheridan R., Frey S., Sander C., Tuschl T., Kandel E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Grinman E., Espadas I., Puthanveettil S.V. Emerging roles for long noncoding RNAs in learning, memory and associated disorders. Neurobiol. Learn. Mem. 2019;163:107034. doi: 10.1016/j.nlm.2019.107034. [DOI] [PubMed] [Google Scholar]
- 180.Liau W.-S., Samaddar S., Banerjee S., Bredy T.W. On the functional relevance of spatiotemporally-specific patterns of experience-dependent long noncoding RNA expression in the brain. RNA Biol. 2021;18:1025–1036. doi: 10.1080/15476286.2020.1868165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Muslimov I.A., Banker G., Brosius J., Tiedge H. Activity-dependent regulation of dendritic BC1 RNA in hippocampal neurons in culture. J. Cell Biol. 1998;141:1601–1611. doi: 10.1083/jcb.141.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Muslimov I.A., Iacoangeli A., Brosius J., Tiedge H. Spatial codes in dendritic BC1 RNA. J. Cell Biol. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grinman E., Nakahata Y., Avchalumov Y., Espadas I., Swarnkar S., Yasuda R., Puthanveettil S.V. Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Basu S., Lamprecht R. The role of actin cytoskeleton in dendritic spines in the maintenance of long-term memory. Front. Mol. Neurosci. 2018;11:143. doi: 10.3389/fnmol.2018.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Landis D.M.D., Reese T.S. Cytoplasmic organization in cerebellar dendritic spines. J. Cell Biol. 1983;97:1169–1178. doi: 10.1083/jcb.97.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Korobova F., Svitkina T.M. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol. Biol. Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Star E.N., Kwiatkowski D.J., Murthy V.N. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat. Neurosci. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- 188.Kommaddi R.P., Das D., Karunakaran S., Nanguneri S., Bapat D., Ray A., Shaw E., Bennett D.A., Nair D., Ravindranath V. Aβ mediates F-actin disassembly in dendritic spines leading to cognitive deficits in Alzheimer’s disease. J. Neurosci. 2018;38:1085–1099. doi: 10.1523/JNEUROSCI.2127-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pelucchi S., Stringhi R., Marcello E. Dendritic spines in Alzheimer’s disease: How the actin cytoskeleton contributes to synaptic failure. Int. J. Mol. Sci. 2020;21:908. doi: 10.3390/ijms21030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kuriu T., Inoue A., Bito H., Sobue K., Okabe S. Differential control of postsynaptic density scaffolds via actin-dependent and -independent mechanisms. J. Neurosci. 2006;26:7693–7706. doi: 10.1523/JNEUROSCI.0522-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sheng M., Kim M.J. Postsynaptic signaling and plasticity mechanisms. Science. 2002;298:776–780. doi: 10.1126/science.1075333. [DOI] [PubMed] [Google Scholar]
- 192.Renner M., Choquet D., Triller A. Control of the postsynaptic membrane viscosity. J. Neurosci. 2009;29:2926–2937. doi: 10.1523/JNEUROSCI.4445-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Allison D.W., Gelfand V.I., Spector I., Craig A.M. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: Differential attachment of NMDA versus AMPA receptors. J. Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cingolani L.A., Goda Y. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- 195.Lv L., Liu Y., Xie J., Wu Y., Zhao J., Li Q., Zhong Y. Interplay between α2-chimaerin and Rac1 activity determines dynamic maintenance of long-term memory. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-13236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Abdou K., Shehata M., Choko K., Nishizono H., Matsuo M., Muramatsu S., Inokuchi K. Synapse-specific representation of the identity of overlapping memory engrams. Science. 2018;360:1227–1231. doi: 10.1126/science.aat3810. [DOI] [PubMed] [Google Scholar]
- 197.Chen S., Cai D., Pearce K., Sun P.Y.W., Roberts A.C., Glanzman D.L. Reinstatement of long-term memory following erasure of its behavioral and synaptic expression in Aplysia. Elife. 2014;3 doi: 10.7554/eLife.03896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Shrestha P., Ayata P., Herrero-Vidal P., Longo F., Gastone A., LeDoux J.E., Heintz N., Klann E. Cell-type-specific drug-inducible protein synthesis inhibition demonstrates that memory consolidation requires rapid neuronal translation. Nat. Neurosci. 2020;23:281–292. doi: 10.1038/s41593-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Harris K.M. Structure, development, and plasticity of dendritic spines. Curr. Opin. Neurobiol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- 200.Bourne J., Harris K.M. Do thin spines learn to be mushroom spines that remember? Curr. Opin. Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 201.Holtmaat A.J.G.D., Trachtenberg J.T., Wilbrecht L., Shepherd G.M., Zhang X., Knott G.W., Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 202.Yu X., Zuo Y. Spine plasticity in the motor cortex. Curr. Opin. Neurobiol. 2011;21:169–174. doi: 10.1016/j.conb.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Attardo A., Fitzgerald J.E., Schnitzer M.J. Impermanence of dendritic spines in live adult CA1 hippocampus. Nature. 2015;523:592–596. doi: 10.1038/nature14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yang G., Pan F., Gan W.-B. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yuste R., Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu. Rev. Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- 206.Xu T., Yu X., Perlik A.J., Tobin W.F., Zweig J.A., Tennant K., Jones T., Zuo Y. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Isaac J.T., Nicoll R.A., Malenka R.C. Evidence for silent synapses: Implications for the expression of LTP. Neuron. 1995;15:427–434. doi: 10.1016/0896-6273(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 208.Liao D., Hessler N.A., Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- 209.Koya E., Cruz F.C., Ator R., Golden S.A., Hoffman A.F., Lupica C.R., Hope B.T. Silent synapses in selectively activated nucleus accumbens neurons following cocaine sensitization. Nat. Neurosci. 2012;15:1556–1562. doi: 10.1038/nn.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Whitaker L.R., Carneiro de Oliveira P.E., McPherson K.B., Fallon R.V., Planeta C.S., Bonci A., Hope B.T. Associative learning drives the formation of silent synapses in neuronal ensembles of the nucleus accumbens. Biol. Psychiatry. 2016;80:246–256. doi: 10.1016/j.biopsych.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Whitaker L.R., Hope B.T. Chasing the addicted engram: Identifying functional alterations in Fos-expressing neuronal ensembles that mediate drug-related learned behavior. Learn. Mem. 2018;25:455–460. doi: 10.1101/lm.046698.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Roy D.S., Muralidhar S., Smith L.M., Tonegawa S. Silent memory engrams as the basis for retrograde amnesia. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E9972–E9979. doi: 10.1073/pnas.1714248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hille B. Ionic channels in excitable membranes. Current problems and biophysical approaches. Biophys. J. 1978;22:283–294. doi: 10.1016/S0006-3495(78)85489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Togashi H., Sakisaka T., Takai Y. Cell adhesion molecules in the central nervous system. Cell Adhes. Migr. 2009;3:29–35. doi: 10.4161/cam.3.1.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kozlova I., Sah S., Keable R., Leshchyns'ka I., Janitz M., Sytnyk V. Cell adhesion molecules and protein synthesis regulation in neurons. Front. Mol. Neurosci. 2020;13:592126. doi: 10.3389/fnmol.2020.592126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Martin K.C., Kandel E.R. Cell adhesion molecules, CREB, and the formation of new synaptic connections. Neuron. 1996;17:567–570. doi: 10.1016/s0896-6273(00)80188-9. [DOI] [PubMed] [Google Scholar]
- 217.Shapiro L., Love J., Colman D.R. Adhesion molecules in the nervous system: Structural insights into function and diversity. Annu. Rev. Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- 218.Sandi C., Merino J.J., Isabel Cordero M., Kruyt N.D., Murphy K.J., Regan C.M. Modulation of hippocampal NCAM polysialylation and spatial memory consolidation by fear conditioning. Biol. Psychiatry. 2003;54:599–607. doi: 10.1016/s0006-3223(03)00182-3. [DOI] [PubMed] [Google Scholar]
- 219.Schmid R.S., Graff R.D., Schaller M.D., Chen S., Schachner M., Hemperly J.J., Maness P.F. NCAM stimulates the Ras-MAPK pathway and CREB phosphorylation in neuronal cells. J. Neurobiol. 1999;38:542–558. [PubMed] [Google Scholar]
- 220.Krushel L.A., Cunningham B.A., Edelman G.M., Crossin K.L. NF-κB activity is induced by neural cell adhesion molecule binding to neurons and astrocytes. J. Biol. Chem. 1999;274:2432–2439. doi: 10.1074/jbc.274.4.2432. [DOI] [PubMed] [Google Scholar]
- 221.Lüthi A., Laurent J.-P., Figurovt A., Mullert D., Schachnert M. Hippocampal long-term potentiation and neural cell adhesion molecules L1 and NCAM. Nature. 1994;372:777–779. doi: 10.1038/372777a0. [DOI] [PubMed] [Google Scholar]
- 222.Bodrikov V., Sytnyk V., Leshchyns'ka I., den Hertog J., Schachner M. NCAM induces CaMKIIα-mediated RPTPα phosphorylation to enhance its catalytic activity and neurite outgrowth. J. Cell Biol. 2008;182:1185–1200. doi: 10.1083/jcb.200803045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lefebvre J.L. Neuronal territory formation by the atypical cadherins and clustered protocadherins. Semin. Cell Dev. Biol. 2017;69:111–121. doi: 10.1016/j.semcdb.2017.07.040. [DOI] [PubMed] [Google Scholar]
- 224.Yamagata M., Sanes J.R. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- 225.Sanes J.R., Zipursky S.L. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell. 2020;181:536–556. doi: 10.1016/j.cell.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 226.Agi E., Kulkarni A., Hiesinger P.R. Neuronal strategies for meeting the right partner during brain wiring. Curr. Opin. Neurobiol. 2020;63:1–8. doi: 10.1016/j.conb.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hassan B.A., Hiesinger P.R. Beyond molecular codes: Simple rules to wire complex brains. Cell. 2015;163:285–291. doi: 10.1016/j.cell.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Benson D. Making memories stick: Cell-adhesion molecules in synaptic plasticity. Trends Cell Biol. 2000;10:473–482. doi: 10.1016/s0962-8924(00)01838-9. [DOI] [PubMed] [Google Scholar]
- 229.Gold A.R., Glanzman D.L. The central importance of nuclear mechanisms in the storage of memory. Biochem. Biophys. Res. Commun. 2021;564:103–113. doi: 10.1016/j.bbrc.2021.04.125. [DOI] [PubMed] [Google Scholar]
- 230.Langille J.J., Gallistel C.R. Locating the engram: Should we look for plastic synapses or information-storing molecules? Neurobiol. Learn. Mem. 2020;169:107164. doi: 10.1016/j.nlm.2020.107164. [DOI] [PubMed] [Google Scholar]
- 231.Prasada S. The physical basis of conceptual representation – an addendum to. Cognition. 2021;214:104751. doi: 10.1016/j.cognition.2021.104751. [DOI] [PubMed] [Google Scholar]
- 232.Jirenhed D.A., Rasmussen A., Johansson F., Hesslow G. Learned response sequences in cerebellar Purkinje cells. Proc. Natl. Acad. Sci. U. S. A. 2017;114:6127–6132. doi: 10.1073/pnas.1621132114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Johansson F., Jirenhed D.-A., Rasmussen A., Zucca R., Hesslow G. Memory trace and timing mechanism localized to cerebellar Purkinje cells. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14930–14934. doi: 10.1073/pnas.1415371111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Baluška F., Levin M. On having no head: Cognition throughout biological systems. Front. Psychol. 2016;7:902. doi: 10.3389/fpsyg.2016.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.McConnell J.V., Jacobson A.L., Kimble D.P. The effects of regeneration upon retention of a conditioned response in the planarian. J. Comp. Physiol. Psychol. 1959;52:1–5. doi: 10.1037/h0048028. [DOI] [PubMed] [Google Scholar]
- 236.Shomrat T., Levin M. An automated training paradigm reveals long-term memory in planarians and its persistence through head regeneration. J. Exp. Biol. 2013;216:3799–3810. doi: 10.1242/jeb.087809. [DOI] [PubMed] [Google Scholar]
- 237.Blackiston D.J., Shomrat T., Levin M. The stability of memories during brain remodeling: A perspective. Commun. Integr. Biol. 2015;8 doi: 10.1080/19420889.2015.1073424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Chapouthier G. From the search for a molecular code of memory to the role of neurotransmitter: A historical perspective. Neural Plast. 2004;11:151–158. doi: 10.1155/NP.2004.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gershman S.J., Balbi P.E.M., Gallistel C.R., Gunawardena J. Reconsidering the evidence for learning in single cells. Elife. 2021;10 doi: 10.7554/eLife.61907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hydén H. Biochemical aspects of learning and memory. Nord Med. 1967;77:205–209. [PubMed] [Google Scholar]
- 241.Bédécarrats A., Chen S., Pearce K., Cai D., Glanzman D.L. RNA from trained Aplysia can induce an epigenetic engram for long-term sensitization in untrained Aplysia. eNeuro. 2018;5 doi: 10.1523/ENEURO.0038-18.2018. ENEURO.0038-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Akhlaghpour H. An RNA-based theory of natural universal computation. J. Theor. Biol. 2022;537:110984. doi: 10.1016/j.jtbi.2021.110984. [DOI] [PubMed] [Google Scholar]
- 243.Tsien R.Y. Very long-term memories may be stored in the pattern of holes in the perineuronal net. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12456–12461. doi: 10.1073/pnas.1310158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Frischknecht R., Gundelfinger E.D. The brain’s extracellular matrix and its role in synaptic plasticity. Adv. Exp. Med. Biol. 2012;970:153–171. doi: 10.1007/978-3-7091-0932-8_7. [DOI] [PubMed] [Google Scholar]
- 245.Wang D., Fawcett J. The perineuronal net and the control of cns plasticity. Cell Tissue Res. 2012;349:147–160. doi: 10.1007/s00441-012-1375-y. [DOI] [PubMed] [Google Scholar]
- 246.Carulli D., Rhodes K.E., Brown D.J., Bonnert T.P., Pollack S.J., Oliver K., Strata P., Fawcett J.W. Composition of perineuronal nets in the adult rat cerebellum and the cellular origin of their components. J. Comp. Neurol. 2006;494:559–577. doi: 10.1002/cne.20822. [DOI] [PubMed] [Google Scholar]
- 247.Cisneros-Franco J.M., Voss P., Thomas M.E., de Villers-Sidani E. Critical periods of brain development. Handb. Clin. Neurol. 2020;173:75–88. doi: 10.1016/B978-0-444-64150-2.00009-5. [DOI] [PubMed] [Google Scholar]
- 248.Carulli D., Verhaagen J. An extracellular perspective on cns maturation: Perineuronal nets and the control of plasticity. Int. J. Mol. Sci. 2021;22:1–26. doi: 10.3390/ijms22052434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Carulli D., Pizzorusso T., Kwok J.C.F., Putignano E., Poli A., Forostyak S., Andrews M.R., Deepa S.S., Glant T.T., Fawcett J.W. Animals lacking link protein have attenuated perineuronal nets and persistent plasticity. Brain. 2010;133:2331–2347. doi: 10.1093/brain/awq145. [DOI] [PubMed] [Google Scholar]
- 250.Pizzorusso T., Medini P., Berardi N., Chierzi S., Fawcett J.W., Maffei L. Reactivation of ocular dominance plasticity in the adult visual cortex. Science. 2002;298:1248–1251. doi: 10.1126/science.1072699. [DOI] [PubMed] [Google Scholar]
- 251.Zaremba S., Guimaraes A., Kalb R.G., Hockfield S. Characterization of an activity-dependent, neuronal surface proteoglycan identified with monoclonal antibody Cat-301. Neuron. 1989;2:1207–1219. doi: 10.1016/0896-6273(89)90305-x. [DOI] [PubMed] [Google Scholar]
- 252.Sale A., Maya Vetencourt J.F., Medini P., Cenni M.C., Baroncelli L., De Pasquale R., Maffei L. Environmental enrichment in adulthood promotes amblyopia recovery through a reduction of intracortical inhibition. Nat. Neurosci. 2007;10:679–681. doi: 10.1038/nn1899. [DOI] [PubMed] [Google Scholar]
- 253.Frischknecht R., Heine M., Perrais D., Seidenbecher C.I., Choquet D., Gundelfinger E.D. Brain extracellular matrix affects AMPA receptor lateral mobility and short-term synaptic plasticity. Nat. Neurosci. 2009;12:897–904. doi: 10.1038/nn.2338. [DOI] [PubMed] [Google Scholar]
- 254.Celio M.R., Spreafico R., De Biasi S., Vitellaro-Zuccarello L. Perineuronal nets: Past and present. Trends Neurosci. 1998;21:510–515. doi: 10.1016/s0166-2236(98)01298-3. [DOI] [PubMed] [Google Scholar]
- 255.Härtig W., Derouiche A., Welt K., Brauer K., Grosche J., Mäder M., Reichenbach A., Brückner G. Cortical neurons immunoreactive for the potassium channel Kv3.1b subunit are predominantly surrounded by perineuronal nets presumed as a buffering system for cations. Brain Res. 1999;842:15–29. doi: 10.1016/s0006-8993(99)01784-9. [DOI] [PubMed] [Google Scholar]
- 256.Bukalo O., Schachner M., Dityatev A. Hippocampal metaplasticity induced by deficiency in the extracellular matrix glycoprotein tenascin-R. J. Neurosci. 2007;27:6019–6028. doi: 10.1523/JNEUROSCI.1022-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Nikonenko A., Schmidt S., Skibo G., Brückner G., Schachner M. Tenascin-R-deficient mice show structural alterations of symmetric perisomatic synapses in the CA1 region of the hippocampus. J. Comp. Neurol. 2003;456:338–349. doi: 10.1002/cne.10537. [DOI] [PubMed] [Google Scholar]
- 258.Carstens K.E., Phillips M.L., Pozzo-Miller L., Weinberg R.J., Dudek S.M. Perineuronal nets suppress plasticity of excitatory synapses on CA2 pyramidal neurons. J. Neurosci. 2016;36:6312–6320. doi: 10.1523/JNEUROSCI.0245-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Gogolla N., Caroni P., Lüthi A., Herry C. Perineuronal nets protect fear memories from erasure. Science. 2009;325:1258–1261. doi: 10.1126/science.1174146. [DOI] [PubMed] [Google Scholar]
- 260.Pizzorusso T. Erasing fear memories. Science. 2009;325:1214–1215. doi: 10.1126/science.1179697. [DOI] [PubMed] [Google Scholar]
- 261.Hylin M.J., Orsi S.A., Moore A.N., Dash P.K. Disruption of the perineuronal net in the hippocampus or medial prefrontal cortex impairs fear conditioning. Learn. Mem. 2013;20:267–273. doi: 10.1101/lm.030197.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Thompson E.H., Lensjø K.K., Wigestrand M.B., Malthe-Sørenssen A., Hafting T., Fyhn M. Removal of perineuronal nets disrupts recall of a remote fear memory. Proc. Natl. Acad. Sci. U. S. A. 2018;115:607–612. doi: 10.1073/pnas.1713530115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Toyama B.H., Hetzer M.W. Protein homeostasis: Live long, won’t prosper. Nat. Rev. Mol. Cell Biol. 2013;14:55–61. doi: 10.1038/nrm3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hong S., Dissing-Olesen L., Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr. Opin. Neurobiol. 2016;36:128–134. doi: 10.1016/j.conb.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kreutzberg G.W. Microglia: A sensor for pathological events in the CNS. Trends Neurosci. 1996;19:312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- 266.Bachiller S., Jiménez-Ferrer I., Paulus A., Yang Y., Swanberg M., Deierborg T., Boza-Serrano A. Microglia in neurological diseases: A road map to brain-disease dependent-inflammatory response. Front. Cell. Neurosci. 2018;12:488. doi: 10.3389/fncel.2018.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Li Q., Barres B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018;18:225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 268.Salter M.W., Stevens B. Microglia emerge as central players in brain disease. Nat. Med. 2017;23:1018–1027. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- 269.Wolf S.A., Boddeke H.W.G.M., Kettenmann H. Microglia in physiology and disease. Annu. Rev. Physiol. 2017;79:619–643. doi: 10.1146/annurev-physiol-022516-034406. [DOI] [PubMed] [Google Scholar]
- 270.Tremblay M.Ě., Lowery R.L., Majewska A.K. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Frost J.L., Schafer D.P. Microglia: Architects of the developing nervous system. Trends Cell Biol. 2016;26:587–597. doi: 10.1016/j.tcb.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wang C., Yue H., Hu Z., Shen Y., Li J., Wang X., Wang L., Sun B., Shi P., Wang L., Gu Y. Microglia mediate forgetting via complement- dependent synaptic elimination. Science. 2020;367:688–694. doi: 10.1126/science.aaz2288. [DOI] [PubMed] [Google Scholar]
- 273.Alberini C.M., Cruz E., Descalzi G., Bessières B., Gao V. Astrocyte glycogen and lactate: New insights into learning and memory mechanisms. Glia. 2018;66:1244–1262. doi: 10.1002/glia.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Sofroniew M.V., Vinters H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Descalzi G., Gao V., Steinman M.Q., Suzuki A., Alberini C.M. Lactate from astrocytes fuels learning-induced mRNA translation in excitatory and inhibitory neurons. Commun. Biol. 2019;2:1–11. doi: 10.1038/s42003-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Adamsky A., Goshen I. Astrocytes in memory function: Pioneering findings and future directions. Neuroscience. 2018;370:14–26. doi: 10.1016/j.neuroscience.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 277.Allen N.J., Eroglu C. Cell biology of astrocyte-synapse interactions. Neuron. 2017;96:697–708. doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Adamsky A., Kol A., Kreisel T., Doron A., Ozeri-Engelhard N., Melcer T., Refaeli R., Horn H., Regev L., Groysman M., London M., Goshen I. Astrocytic activation generates de novo neuronal potentiation and memory enhancement. Cell. 2018;174:59–71.e14. doi: 10.1016/j.cell.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 279.Kol A., Goshen I. The memory orchestra: The role of astrocytes and oligodendrocytes in parallel to neurons. Curr. Opin. Neurobiol. 2021;67:131–137. doi: 10.1016/j.conb.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Kol A., Adamsky A., Groysman M., Kreisel T., London M., Goshen I. Astrocytes contribute to remote memory formation by modulating hippocampal–cortical communication during learning. Nat. Neurosci. 2020;23:1229–1239. doi: 10.1038/s41593-020-0679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Fünfschilling U., Supplie L.M., Mahad D., Boretius S., Saab A.S., Edgar J., Brinkmann B.G., Kassmann C.M., Tzvetanova I.D., Möbius W., Diaz F., Meijer D., Suter U., Hamprecht B., Sereda M.W., et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 2012;485:517–521. doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Griffiths I., Klugmann M., Anderson T., Yool D., Thomson C., Schwab M.H., Schneider A., Zimmermann F., McCulloch M., Nadon N., Nave K.A. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998;280:1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- 283.Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P.E., Griffiths I.R., Navel K.A. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 284.Gibson E.M., Purger D., Mount C.W., Goldstein A.K., Lin G.L., Wood L.S., Inema I., Miller S.E., Bieri G., Zuchero J.B., Barres B.A., Woo P.J., Vogel H., Monje M. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344:1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Steadman P.E., Xia F., Ahmed M., Mocle A.J., Penning A.R.A., Geraghty A.C., Steenland H.W., Monje M., Josselyn S.A., Frankland P.W. Disruption of oligodendrogenesis impairs memory consolidation in adult mice. Neuron. 2020;105:150–164.e6. doi: 10.1016/j.neuron.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Pan S., Mayoral S.R., Choi H.S., Chan J.R., Kheirbek M.A. Preservation of a remote fear memory requires new myelin formation. Nat. Neurosci. 2020;23:487–499. doi: 10.1038/s41593-019-0582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Disterhoft J.F., Oh M.M. Learning, aging and intrinsic neuronal plasticity. Trends Neurosci. 2006;29:587–599. doi: 10.1016/j.tins.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 288.Lisman J., Cooper K., Sehgal M., Silva A.J. Memory formation depends on both synapse-specific modifications of synaptic strength and cell-specific increases in excitability. Nat. Neurosci. 2018;21:309–314. doi: 10.1038/s41593-018-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Mozzachiodi R., Byrne J.H. More than synaptic plasticity: Role of nonsynaptic plasticity in learning and memory. Trends Neurosci. 2010;33:17–26. doi: 10.1016/j.tins.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Titley H.K., Brunel N., Hansel C. Toward a neurocentric view of learning. Neuron. 2017;95:19–32. doi: 10.1016/j.neuron.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.O'Keefe J., Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- 292.Moser E.I., Moser M.B., McNaughton B.L. Spatial representation in the hippocampal formation: A history. Nat. Neurosci. 2017;20:1448–1464. doi: 10.1038/nn.4653. [DOI] [PubMed] [Google Scholar]
- 293.Goode T.D., Tanaka K.Z., Sahay A., McHugh T.J. An integrated index: Engrams, place cells, and hippocampal memory. Neuron. 2020;107:805–820. doi: 10.1016/j.neuron.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Tanaka K.Z. Heterogeneous representations in the hippocampus. Neurosci. Res. 2021;165:1–5. doi: 10.1016/j.neures.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 295.Hainmueller T., Bartos M. Dentate gyrus circuits for encoding, retrieval and discrimination of episodic memories. Nat. Rev. Neurosci. 2020;21:153–168. doi: 10.1038/s41583-019-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Tanaka K.Z., He H., Tomar A., Niisato K., Huang A.J.Y., McHugh T.J. The hippocampal engram maps experience but not place. Science. 2018;361:392–397. doi: 10.1126/science.aat5397. [DOI] [PubMed] [Google Scholar]
- 297.O’keefe J., Nadel L. Oxford University Press; Oxford: 1978. The Hippocampus as a Cognitive Map. [Google Scholar]
- 298.Sweatt J., Meaney M., Nestler E., Akbarian S. Academic Press; London: 2012. Epigenetic Regulation in the Nervous System: Basic Mechanisms and Clinical Impact. [Google Scholar]
- 299.Gupta-Agarwal S., Franklin A.V., DeRamus T., Wheelock M., Davis R.L., McMahon L.L., Lubin F.D. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J. Neurosci. 2012;32:5440–5453. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Oliveira A.M.M. DNA methylation: A permissive mark in memory formation and maintenance. Learn. Mem. 2016;23:587–593. doi: 10.1101/lm.042739.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Levenson J.M., O'Riordan K.J., Brown K.D., Trinh M.A., Molfese D.L., Sweatt J.D. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 302.Day J.J., Childs D., Guzman-Karlsson M.C., Kibe M., Moulden J., Song E., Tahir A., Sweatt J.D. DNA methylation regulates associative reward learning. Nat. Neurosci. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Guo N., Soden M.E., Herber C., Kim M.T.W., Besnard A., Lin P., Ma X., Cepko C.L., Zweifel L.S., Sahay A. Dentate granule cell recruitment of feedforward inhibition governs engram maintenance and remote memory generalization. Nat. Med. 2018;24:438–449. doi: 10.1038/nm.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Miller C.A., Sweatt J.D. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 305.Miller C.A., Gavin C.F., White J.A., Parrish R.R., Honasoge A., Yancey C.R., Rivera I.M., Rubio M.D., Rumbaugh G., Sweatt J.D. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Campbell R.R., Wood M.A. How the epigenome integrates information and reshapes the synapse. Nat. Rev. Neurosci. 2019;20:133–147. doi: 10.1038/s41583-019-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Gräff J., Tsai L.-H. Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 308.Sweatt J.D. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Zovkic I.B., Guzman-Karlsson M.C., Sweatt J.D. Epigenetic regulation of memory formation and maintenance. Learn. Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Fernandez-Albert J., Lipinski M., Lopez-Cascales M.T., Rowley M.J., Martin-Gonzalez A.M., del Blanco B., Corces V.G., Barco A. Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 2019;22:1718–1730. doi: 10.1038/s41593-019-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Gulmez Karaca K., Kupke J., Brito D.V.C., Zeuch B., Thome C., Weichenhan D., Lutsik P., Plass C., Oliveira A.M.M. Neuronal ensemble-specific DNA methylation strengthens engram stability. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-14498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Odell S.C., Taki F., Klein S.L., Chen R.J., Levine O.B., Skelly M.J., Nabila A., Brindley E., Gal Toth J., Dündar F., Sheridan C.K., Fetcho R.N., Alonso A., Liston C., Landau D.A., et al. Epigenomically bistable regions across neuron-specific genes govern neuron eligibility to a coding ensemble in the Hippocampus. Cell Rep. 2020;31:107789. doi: 10.1016/j.celrep.2020.107789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Aoued H.S., Sannigrahi S., Doshi N., Morrison F.G., Linsenbaum H., Hunter S.C., Walum H., Baman J., Yao B., Jin P., Ressler K.J., Dias B.G. Reversing behavioral, neuroanatomical, and germline influences of intergenerational stress. Biol. Psychiatry. 2019;85:248–256. doi: 10.1016/j.biopsych.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Dias B.G., Ressler K.J. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Gräff J., Mansuy I.M. Epigenetic codes in cognition and behaviour. Behav. Brain Res. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 316.Day J.J., Sweatt J.D. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kyrke-Smith M., Williams J.M. Bridging synaptic and epigenetic maintenance mechanisms of the engram. Front. Mol. Neurosci. 2018;11:369. doi: 10.3389/fnmol.2018.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]