Abstract
Aims
The HFA‐PEFF score was developed to optimize diagnosis and to aid in early recognition of heart failure (HF) with preserved ejection fraction (HFpEF) in patients who present with HF‐like symptoms. Recognizing early‐HFpEF phenogroups is essential to better understand progression towards overt HFpEF and pave the way for early intervention and treatment. Whether the HFA‐PEFF domain scores can identify ‘early‐HFpEF’ phenogroups remains unknown. The aims of this pilot study are to (i) identify distinct phenogroups by cluster analysis of HFA‐PEFF domain scores in subjects that present with HF‐like symptoms and (ii) study whether these phenogroups may be associated with distinct blood proteome profiles.
Methods and results
Subjects referred to the Cardiology Centers of the Netherlands, location Utrecht, with non‐acute possibly cardiac‐related symptoms (such as dyspnoea or fatigue) were prospectively enrolled in the HELPFul cohort (N = 507) and were included in the current analysis. Inclusion criteria for this study were (i) age ≥ 45 years and (ii) a left ventricular ejection fraction (LVEF) ≥ 50%, in the absence of a history of HF, coronary artery disease, congenital heart disease, or any previous cardiac interventions. Multinominal‐based clustering with latent class model using the HFA‐PEFF domain scores (functional, structural, and biomarker scores) as input was used to detect distinct phenotypic clusters. For each bootstrapping run, the 92 Olink proteins were analysed for their association with the identified phenogroups. Four distinct phenogroups were identified in the current analysis (validated by bootstrapping 1000×): (i) no left ventricular diastolic dysfunction (no LVDD, N = 102); (ii) LVDD with functional left ventricular (LV) abnormalities (N = 204); (iii) LVDD with functional and structural LV abnormalities (N = 204); and (iv) LVDD with functional and structural LV abnormalities and elevated BNP (N = 107). The HFA‐PEFF total score risk categories significantly differed between the phenogroups (P < 0.001), with an increase of the HFA‐PEFF score from Phenogroup 1 to 4 (low/intermediate/high HFA‐PEFF risk score: Phenogroup 1: 88%/12%/0%; Phenogroup 2: 9%/91%/0%; Phenogroup 3: 0%/92%/8%; Phenogroup 4: 5%/83%/12%). Thirty‐two out of the 92 Olink protein biomarkers significantly differed among the phenogroups. The top eight biomarkers—N‐terminal prohormone brain natriuretic peptide, growth differentiation factor‐15, matrix metalloproteinase‐2, osteoprotegerin, tissue inhibitor of metalloproteinase‐4, chitinase‐3‐like protein 1, insulin‐like growth factor‐binding protein 2, and insulin‐like growth factor‐binding protein 7—are mainly involved in inflammation and extracellular matrix remodelling, which are currently proposed key processes in HFpEF pathophysiology.
Conclusions
This study identified distinct phenogroups by using the HFA‐PEFF domain scores in ambulant subjects referred for HF‐like symptoms. The newly identified phenogroups accompanied by their circulating biomarkers profile might aid in a better understanding of the pathophysiological processes involved during the early stages of the HFpEF syndrome.
Keywords: HFpEF, Left ventricular diastolic dysfunction, biomarkers, HFA‐PEFF
Background
Heart failure with preserved ejection fraction (HFpEF) is a heterogeneous clinical syndrome that is associated with a poor quality of life, high mortality rates, and significant healthcare‐related costs. 1 , 2 Recently, the HFA‐PEFF diagnostic algorithm was developed to optimize diagnosis and aid in the early recognition of this syndrome in patients who present with heart failure (HF)‐like symptoms. 3 However, whether the HFA‐PEFF domain scores can identify ‘early‐HFpEF’ phenogroups remains unknown. Recognizing early‐HFpEF phenogroups is essential to better understand progression towards overt HFpEF and pave the way for early treatment.
Aims
The aims of this pilot study are to (i) identify distinct phenogroups by cluster analysis of HFA‐PEFF domain scores in subjects that present with HF‐like symptoms and (ii) study whether these phenogroups may be associated with distinct blood proteome profiles.
Methods
Consecutive participants (n = 507) of the previously described HELPFul observational cohort 4 were included in this study. In summary, the HELPFul cohort is a single‐centre [Cardiology Centers of the Netherlands (CCN), location Utrecht] prospective case‐cohort study designed to better understand early HFpEF and its progression towards overt HFpEF. The CCN cardiology outpatient clinic is positioned between the general practitioner and the hospital. It is intended to allow fast cardiac screening in subjects with non‐acute potential cardiac‐related symptoms such as dyspnoea or fatigue. 4 The HELPFul study population therefore provides a unique possibility to study biomarkers and risk factors in patients that have not yet developed (overt) left ventricular diastolic dysfunction (LVDD) or HFpEF or are still in the early stages of these conditions. 4 Inclusion criteria for this study were (i) age ≥ 45 years, (ii) signed informed consent, and (iii) a left ventricular ejection fraction (LVEF) ≥ 50%, in the absence of a medical history of HF (hospitalization), coronary artery disease, congenital heart disease, or any previous cardiac interventions. As a result, subjects with HF‐like symptoms and structural/functional/biomarkers abnormalities in line with recently published HFA‐PEFF score but without a medical history of HFpEF diagnosis are among others included in current study. 3
At baseline visit, history taking, physical examination, laboratory measurements, and transthoracic echocardiography were performed as part of routine clinical care. For this study, baseline plasma samples were analysed for 92 protein biomarkers using the Olink Proseek Multiplex cardiovascular panel III (CVDIII) as described previously. 5 Missing clinical data (total missing <2% with <10% missing per variable) were imputed using factor analysis for mixed data (missMDA v1.17). Subsequently, the structural, functional, and biomarker HFA‐PEFF domain scores were calculated (maximum score of 2 for each domain). 3 Multinominal‐based clustering with latent class model using the domain scores as categorical input was performed with Rmixmod v2.1.5. Four phenogroups were identified based on the Bayesian information criterion. The clustering was validated by bootstrapping (n = 1000) with boot‐package v1.3–25. The statistical significance of the difference in clinical characteristics among the phenogroups was estimated using the Kruskal–Wallis rank‐sum test and Mann–Whitney U test, or ANOVA and t‐test for continuous variables, and χ 2 test or Fisher's exact test for categorical variables, where appropriate. For each bootstrapping run, the 92 Olink proteins were analysed for their association with the four phenogroups using the Kruskal–Wallis rank‐sum test (Figure 1 ). All analyses were carried out with the R software (Version 4.0.4).
Figure 1.
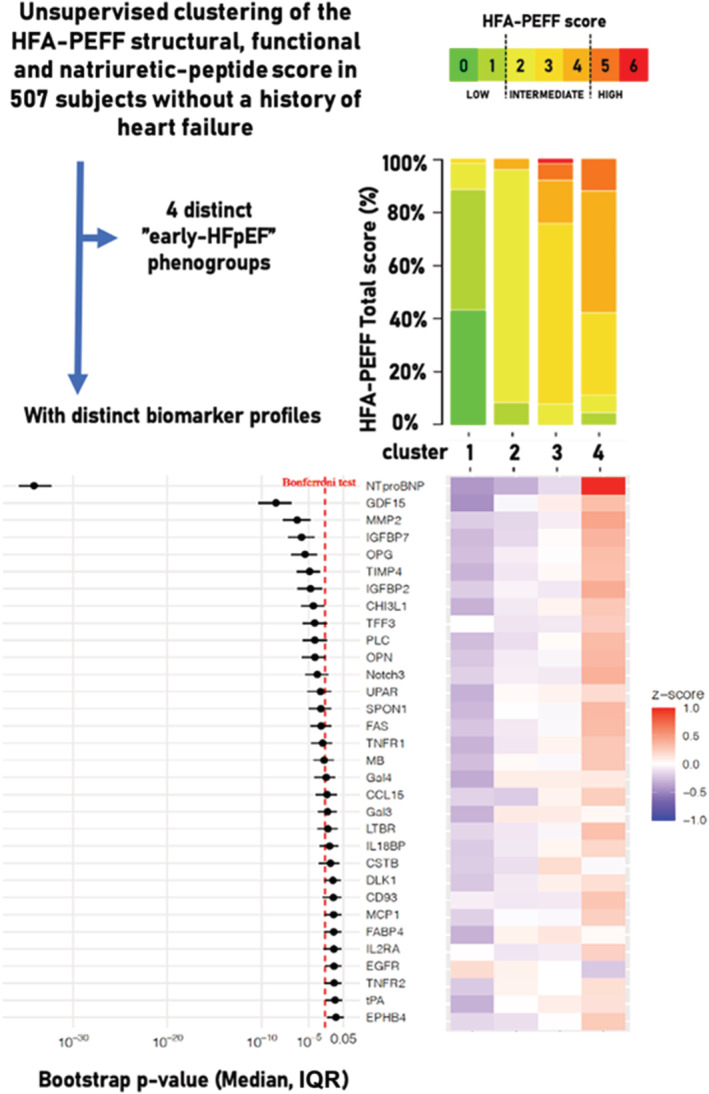
Top panel: Multinominal‐based clustering with latent class model using the HFA‐PEFF domain scores as categorical input revealed four distinct phenogroups with significant difference between the HFA‐PEFF total score risk categories (P < 0.001, after applying Bonferroni's correction). Left bottom panel: Bootstrapping (1000×) results [P‐value and interquartile range (IQR)] of the Olink proteins for their association with the four phenogroups using the Kruskal–Wallis rank‐sum test. Biomarkers of which the upper interquartile range (IQR) limit of the bootstrapping results were significantly (P < 0.05) associated with the clusters are shown (vertical red dotted line indicates the P‐value cut‐off after Bonferroni's correction: 0.05/92). Right bottom panel: Heatmap of the mean value of z‐scores of these Olink proteins in each cluster. CCL15, C–C motif chemokine 15; CD93, complement component C1q receptor; CHI3L1, chitinase‐3‐like protein 1; CSTB, cystatin‐B; DLK‐1, protein delta homologue 1; EGFR, epidermal growth factor receptor; EPHB4, ephrin type‐B receptor 4; FABP4, fatty acid‐binding protein 4; FAS, tumour necrosis factor receptor superfamily member 6; Gal‐3, galectin‐3; Gal‐4, galectin‐4; GDF‐15, growth differentiation factor‐15; IGFBP‐2, insulin‐like growth factor‐binding protein 2; IGFBP‐7, insulin‐like growth factor‐binding protein 7; IL‐18BP, interleukin‐18‐binding protein; IL2‐RA, interleukin‐2 receptor subunit alpha; LTBR, lymphotoxin‐beta receptor; MB, myoglobin; MCP‐1, monocyte chemotactic protein 1; MMP‐2, matrix metalloproteinase‐2; Notch3, neurogenic locus notch homologue protein 3; NT‐proBNP, N‐terminal prohormone brain natriuretic peptide; OPG, osteoprotegerin; OPN, osteopontin; PLC, perlecan; SPON1, spondin‐1; TFF3, trefoil factor 3; TIMP4, tissue inhibitor of metalloproteinase‐4; TNF‐R1, tumour necrosis factor receptor 1; TNF‐R2, tumour necrosis factor receptor 2; t‐PA, tissue‐type plasminogen activator; U‐PAR, urokinase plasminogen activator surface receptor.
Results
Compared with the other clusters, subjects in Phenogroup 1 were relatively young and had a normal left ventricular (LV) function; subjects in Phenogroup 2 were characterized by functional (diastolic) LV abnormalities but normal LV structure; Phenogroup 3 by both structural and functional LV abnormalities, normal BNP plasma levels, and a higher prevalence of hypertension; and Phenogroup 4 by elevated BNP levels (mostly) accompanied by structural and functional LV abnormalities (Table 1 ). The HFA‐PEFF total score risk categories significantly differed between the phenogroups (P < 0.001, Bonferroni's correction), with an increase of the HFA‐PEFF score from Phenogroup 1 to 4 (low/intermediate/high HFA‐PEFF risk score: Phenogroup 1: 88%/12%/0%; Phenogroup 2: 9%/91%/0%; Phenogroup 3: 0%/92%/8%; Phenogroup 4: 5%/83%/12%). Prevalence of sex, medical history of atrial fibrillation, LVEF, creatinine levels, and body mass index did not significantly differ between the four phenogroups (Table 1 ). Thirty‐two out of the 92 Olink protein biomarkers significantly differed among clusters (Figure 1 ; proteins with an upper interquartile range limit of P‐value in bootstrapping < 0.05 are shown, with a P‐value < 0.05 for the top eight after applying Bonferroni's correction). The top eight biomarkers—N‐terminal prohormone brain natriuretic peptide, growth differentiation factor‐15, matrix metalloproteinase‐2, insulin‐like growth factor‐binding protein 2, insulin‐like growth factor‐binding protein 7, osteoprotegerin, tissue inhibitor of metalloproteinase‐4, and chitinase‐3‐like protein 1—included biomarkers that have been previously associated with HFpEF and/or LVDD and are mainly involved in inflammation and extracellular matrix remodelling. 6 , 7
Table 1.
Baseline clinical characteristics stratified for the four identified phenogroups
| Total (N = 507) |
Cluster 1 No LVDD (N = 102) |
Cluster 2 LVDD with functional LV abnormalities (N = 94) |
Cluster 3 LVDD with functional and structural LV abnormalities (N = 204) |
Cluster 4 LVDD with functional and structural LV abnormalities and elevated BNP (N = 107) |
P‐value | |
|---|---|---|---|---|---|---|
| Age, years | 62.9 ± 9.5 | 56.3 ± 8.1¥£† | 62.5 ± 8.0§† | 64.0 ± 8.8§† | 67.5 ± 9.8§¥£ | <0.001 |
| Female, n % | 344 (67.9%) | 64 (62.7%) | 62 (66.0%) | 146 (71.6%) | 72 (67.3%) | 0.443 |
| BMI, kg/m2 | 26.6 [24.0–29.6] | 26.3 [24.4–28.9] | 26.0 [23.8–29.6] | 27.2 [23.9–30.1] | 26.6 [23.4–29.5] | 0.562 |
| HR, b.p.m. | 72 ± 11 | 70 ± 10 | 73 ± 10 | 74 ± 12† | 69 ± 11£ | 0.002 |
| SBP, mmHg | 148 ± 20 | 139 ± 19¥£† | 148 ± 18§ | 152 ± 19§ | 151 ± 23§ | <0.001 |
| DBP, mmHg | 87 ± 11 | 83 ± 10¥£ | 87 ± 9§ | 89 ± 10§ | 88 ± 12 | <0.001 |
| Medication, n % | ||||||
| Beta‐blocker | 81 (16.0%) | 8 (7.8%) | 13 (13.8%) | 39 (19.1%) | 21 (19.6%) | 0.048 |
| ACEi/ARB | 118 (23.3%) | 13 (12.7%)£ | 18 (19.1%) | 61 (29.9%)§ | 26 (24.3%) | 0.006 |
| Loop diuretic | 15 (3.0%) | 2 (2.0%) | 0 (0.0%) | 6 (2.9%) | 7 (6.5%) | 0.046 |
| MRA | 4 (0.8%) | 1 (1.0%) | 0 (0.0%) | 2 (1.0%) | 1 (0.9%) | 0.999 |
| Medical history, n % | ||||||
| AF | 15 (3.0%) | 4 (3.9%) | 1 (1.1%) | 7 (3.4%) | 3 (2.8%) | 0.643 |
| Hypertension | 298 (58.8%) | 37 (36.3%)¥£† | 51 (54.3%)§£ | 145 (71.1%)§¥ | 65 (60.7%)§ | <0.001 |
| DM | 41 (8.1%) | 5 (4.9%) | 8 (8.5%) | 20 (9.8%) | 8 (7.5%) | 0.517 |
| COPD | 57 (11.2%) | 10 (9.8%) | 11 (11.7%) | 23 (11.3%) | 13 (12.1%) | 0.956 |
| Blood assessment | ||||||
| BNP, pg/mL | 19.4 [10.0–36.6] | 13.5 [10.0–20.4]† | 14.9 [10.0–23.9]† | 15.8 [10.0–24.5]† | 46.4 [40.0–60.0]§¥£ | <0.001 |
| Creatinine, μmol/L | 66.1 [60.7–74.8] | 65.4 [60.9–74.1] | 67.7 [60.4–74.9] | 66.5 [61.5–73.9] | 66.2 [60.0–76.9] | 0.979 |
| CRP, μmol/L | 1.5 [0.7–3.3] | 1.6 [0.7–3.7] | 1.4 [0.7–3.2] | 1.5 [0.7–3.5] | 1.5 [0.7–3.2] | 0.841 |
| Echocardiography | ||||||
| LVEF, % | 67.6 ± 7 | 66.8 ± 6.8 | 68.9 ± 7.8 | 67.5 ± 6.9 | 67.4 ± 7.2 | 0.183 |
| LVEDD, mm | 44.7 ± 5.1 | 44.8 ± 4.6 | 46.4 ± 4.4£ | 44.0 ± 5.5¥ | 44.5 ± 4.8 | 0.003 |
| LVESD, mm | 27.6 ± 4.2 | 28.1 ± 3.7 | 27.9 ± 4.1 | 27.4 ± 4.5 | 27.4 ± 4.3 | 0.414 |
| LAVI, mL/m2 | 24.4 [19.8–30.5] | 22.9 [19.1–27.1]£† | 21.3 [17.9–24.6]£† | 27.1 [20.8–33.4]§¥ | 27.0 [20.5–32.6]§¥ | <0.001 |
| LVMI, g/m2 | 71.8 [61.3–85.1] | 64.8 [56.2–75.2]£† | 68.6 [61.8–77.3]£ | 77.6 [65.1–88.9]§¥ | 70.6 [62.0–87.3]§ | <0.001 |
| RWT | 0.42 [0.37–0.47] | 0.41 [0.35–0.46]¥£ | 0.37 [0.34–0.39]§£† | 0.45 [0.41–0.50]§¥† | 0.43 [0.37–0.47]¥£ | <0.001 |
| e′ septal, cm/s | 7.0 ± 1.9 | 8.9 ± 1.4¥£† | 6.6 ± 1.7§ | 6.5 ± 1.6§ | 6.7 ± 1.9§ | <0.001 |
| e′ lateral, cm/s | 8.7 ± 2.4 | 11.4 ± 1.5¥£† | 8.1 ± 2.1§ | 8.0 ± 1.9§ | 8.2 ± 2.4§ | <0.001 |
| E/e′ | 9.0 [7.9–10.3] | 7.5 [6.3–8.3]¥£† | 9.1 [8.0–10.6]§ | 9.7 [8.6–11.0]§ | 9.4 [8.3–10.7]§ | <0.001 |
| PASP > 35, mmHg | 7 (1.4%) | 0 (0.0%) | 2 (2.1%) | 3 (1.5%) | 2 (1.9%) | 0.586 |
| HFA‐PEFF score | ||||||
| Functional, minor/major | 50 (10%)/342 (67%) | 0 (0%)/0 (0%) | 8 (9%)/86 (91%) | 26 (13%)/178 (87%) | 16 (15%)/78 (73%) | <0.001 |
| Structural, minor/major | 267 (53%)/75 (15%) | 48 (47%)/9 (9%) | 0 (0%)/0 (0%) | 157 (77%)/47 (23%) | 62 (58%)/19 (18%) | <0.001 |
| Biomarker, minor/major | 107 (21%)/24 (5%) | 0 (0%)/3 (3%) | 0 (0%)/4 (4%) | 0 (0%)/17 (8%) | 107 (100%)/0 (0%) | <0.001 |
| HFA‐total score | <0.001 | |||||
| Low (<2) | 103 (20%) | 90 (88%) | 8 (9%) | 0 (0.0%) | 5 (5%) | |
| Intermediate (2–4) | 374 (74%) | 12 (12%) | 86 (91%) | 187 (92%) | 89 (83%) | |
| High (>4) | 30 (6%) | 0 (0%) | 0 (0.0%) | 17 (8%) | 13 (12%) | |
ACEi, angiotensin‐converting enzyme inhibitor; AF, atrial fibrillation; ARB, angiotensin‐receptor blocker; BMI, body mass index; BNP, B‐type natriuretic peptide; COPD, chronic obstructive pulmonary disease; CRP, C‐reactive protein; DBP, diastolic blood pressure; DM, diabetes mellitus; HR, heart rate; LAVI, left atrial volume index; LVDD, left ventricular diastolic dysfunction; LVEDD, left ventricular end‐diastolic diameter; LVEF, left ventricular ejection fraction; LVMI, left ventricular mass index; LVSED, left ventricular end‐systolic diameter; MRA, mineralocorticoid receptor antagonist; PASP, pulmonary artery systolic pressure; RWT, relative wall thickness; SBP, systolic blood pressure.
Values are shown as mean ± SD, or median [IQR], or counts (percentage). A significant difference (P < 0.05 after Bonferroni's adjustment) compared with Cluster 1, 2, 3, or 4 is indicated with symbols §, ¥, £, and †, respectively.
Conclusions
This is the first study revealing distinct phenogroups by using the HFA‐PEFF domain scores in ambulant subjects referred for HF‐like symptoms. While it is unlikely that individual circulating biomarkers will have diagnostic value to detect ‘early HFpEF’, 7 the newly identified phenogroups accompanied by their circulating biomarkers profile might aid in a better understanding of the pathophysiological processes involved during the early stages of the heterogeneous HFpEF syndrome. In addition, this information might help to identify those individuals who progress from LVDD towards overt HFpEF and possibly could benefit from early treatment in the future.
Certain study limitations have to be addressed, including the case‐cohort cross‐sectional design, non‐fasting blood samples, the lack of information on global longitudinal strain (which was therefore not used for the calculation of the functional HFA‐PEFF score), and potential under‐detection of LVDD because no exercise echocardiography or invasive haemodynamic stress testing was performed. 8 Moreover, it is unclear whether the biomarkers are a primary cause or effect of the phenogroups, and whether the biomarker profiles itself are (indirectly) driven by elevated BNP levels, which needs to be determined in longitudinal studies with sequential biobanking. The current approach's strength is the usage of easy to assess, widely available diagnostic parameters that are currently being used in cardiology and HFpEF clinics. 3 Follow‐up of clinical and biomarker data with serial (exercise) echocardiographies—along with validation in similar cohorts—is required to prove the added value of the currently identified phenogroups in predicting new‐onset HFpEF and its progression.
Conflict of interest
None declared.
Funding
We acknowledge the support of the Netherlands Cardiovascular Research Initiative: an initiative with support of the Dutch Heart Foundation, CVON2015‐RECONNECT, CVON2016‐Early HFPEF, and CVON 2017‐ShePREDICTS. Additionally, J.W.J.B. is supported by a ZonMw VIDI grant.
Henkens, M. T. H. M. , van Ommen, A.‐M. , Remmelzwaal, S. , Valstar, G. B. , Wang, P. , Verdonschot, J. A. J. , Hazebroek, M. R. , Hofstra, L. , van Empel, V. P. M. , Beulens, J. W. J. , den Ruijter, H. M. , and Heymans, S. R. B. (2022) The HFA‐PEFF score identifies ‘early‐HFpEF’ phenogroups associated with distinct biomarker profiles. ESC Heart Failure, 9: 2032–2036. 10.1002/ehf2.13861.
References
- 1. Ziaeian B, Fonarow GC. Epidemiology and aetiology of heart failure. Nat Rev Cardiol 2016; 13: 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dunlay SM, Roger VL, Redfield MM. Epidemiology of heart failure with preserved ejection fraction. Nat Rev Cardiol 2017; 14: 591–602. [DOI] [PubMed] [Google Scholar]
- 3. Pieske B, Tschope C, de Boer RA, Fraser AG, Anker SD, Donal E, Edelmann F, Fu M, Guazzi M, Lam CSP, Lancellotti P, Melenovsky V, Morris DA, Nagel E, Pieske‐Kraigher E, Ponikowski P, Solomon SD, Vasan RS, Rutten FH, Voors AA, Ruschitzka F, Paulus WJ, Seferovic P, Filippatos G. How to diagnose heart failure with preserved ejection fraction: the HFA‐PEFF diagnostic algorithm: a consensus recommendation from the Heart Failure Association (HFA) of the European Society of Cardiology (ESC). Eur Heart J 2019; 40: 3297–3317. [DOI] [PubMed] [Google Scholar]
- 4. Valstar GB, Bots SH, Groepenhoff F, Gohar A, Rutten FH, Leiner T, Cramer MJM, Teske AJ, Suciadi LP, Menken R, Pasterkamp G, Asselbergs FW, Hofstra L, Bots ML, den Ruijter HM. Discovery of biomarkers for the presence and progression of left ventricular diastolic dysfunction and HEart faiLure with Preserved ejection Fraction in patients at risk for cardiovascular disease: rationale and design of the HELPFul case‐cohort study in a Dutch cardiology outpatient clinic. BMJ Open 2019; 9: e028408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferreira JP, Verdonschot J, Collier T, Wang P, Pizard A, Bar C, Bjorkman J, Boccanelli A, Butler J, Clark A, Cleland JG, Delles C, Diez J, Girerd N, Gonzalez A, Hazebroek M, Huby AC, Jukema W, Latini R, Leenders J, Levy D, Mebazaa A, Mischak H, Pinet F, Rossignol P, Sattar N, Sever P, Staessen JA, Thum T, Vodovar N, Zhang ZY, Heymans S, Zannad F. Proteomic bioprofiles and mechanistic pathways of progression to heart failure. Circ Heart Fail 2019; 12: e005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mishra S, Kass DA. Cellular and molecular pathobiology of heart failure with preserved ejection fraction. Nat Rev Cardiol 2021; 18: 400–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henkens M, Remmelzwaal S, Robinson EL, van Ballegooijen AJ, Barandiaran Aizpurua A, Verdonschot JAJ, Raafs AG, Weerts J, Hazebroek MR, Sanders‐van Wijk S, Handoko ML, den Ruijter HM, Lam CSP, de Boer RA, Paulus WJ, van Empel VPM, Vos R, Brunner‐La Rocca HP, Beulens JWJ, Heymans SRB. Risk of bias in studies investigating novel diagnostic biomarkers for heart failure with preserved ejection fraction. A systematic review. Eur J Heart Fail 2020; 22: 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reddy YNV, Carter RE, Obokata M, Redfield MM, Borlaug BA. A simple, evidence‐based approach to help guide diagnosis of heart failure with preserved ejection fraction. Circulation 2018; 138: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]


