Highlights
-
•
Ultrasonication was successfully used for production of apple seeds protein nanoparticles.
-
•
Apple seed protein nanoparticles were characterisation for various structural and functional properties.
-
•
Protein nanoparticles showed enhanced techno-functional properties.
-
•
Apple seed protein nanoparticles can be efficiently used as functional ingredient in food industry.
Keywords: Apple seed protein, Ultrasoication, Nanotechnology, Hydrophobicity, Functional properties
Abstract
In this study, protein was extracted from the apple seed flour using alkali-acid precipitation method. The main objective of this study was to evaluate the impact of ultrasonication on structural and techno-functional properties of apple seed protein. Both native (N-protein) and ultra-sonicated protein (US-protein) were characterized for size, zeta potential, structure, protein pattern, crystallinity, thermal stability and functional properties. The results revealed that the hydrodynamic diameter of N-protein and US-protein was 1.2 µm and 484 nm while zeta potential was −11 and −19 mV, respectively. Fourier transform infrared-spectroscopy and X-ray diffraction analysis showed change in the conformational characteristics and functional groups of proteins after nano-reduction. SEM revealed change in the surface morphology of protein molecule upon ultrasonication. Differential scanning calorimetry showed decreased denaturation temperature for US-protein compared to N-protein . SDS-PAGE depicted no change in protein pattern upon ultrasonication. Ultrasonicated protein exhibited increased functional properties like emulsification, foaming, hydrophobicity and oil absorbing properties and hence can be efficiently used as functional ingredient in food and nutraceutical industry.
1. Introduction
The demand for proteins is increasing globally due to the consumer’s awareness towards functional foods that provide health benefit beyond basic nutrition [1]. Presently, both plants and animals are a good source of protein however, animal protein is not preferred because of the high cost of production, ethical, cultural, and dietary choices [2]. Therefore, there is an urgent need to shift towards plant-based protein alternatives that are cost-effective, safe and have a positive effect on health [3]. Also, growing trend of veganism because of the sustainability concerns have increased the demand for plant-based protein alternatives that are eco-friendly and more economical. For this purpose, utilisation of agro-waste to produce plant-based protein is the innovative approach which will enhance the marketable worth to this agro-waste and could be a better substitute to animal-derived proteins for satisfying the demands of the consumer. Apple is an important fruit rich in fibre, vitamins, and minerals. During its processing apple seeds are discarded as waste therefore, damaging the local environment apart from acquiring economic loss [4]. Apple seeds are reported to have a good protein content (49.5 g/100 g) which comprises of essential amino acids and hence can be a novel and alternative source over animal protein for food and nutraceutical applications [5]. Besides this apple seed flour has been reported to have different functional and nutraceutical properties which include antioxidant, anti-diabetic, anti-obesity, and antimicrobial [5]. Also, functional properties are an important characteristic feature which gives knowledge about the application of proteins in food industry. As a result, many technologies based on physical, chemical, and enzymatic methods have been proposed to improve protein bioactivity and functionality. Ultrasonication which is a considered as a green technology has been found as an effective alternate to enhance the physcio-chemical properties of various seed protein [2], [6].
Hence, in this study ultrasonication has been used to reduce the size of the protein . Ultrasonication is a physical technique that uses high-intensity sound waves of frequency >16 kHz and can be used to produce nanoparticles [2]. Ultrasonication causes unfolding of the protein structure which enhances the functionality of protein. It has been demonstrated that ultra-sonication could effectively modify the functional and structural properties of proteins such as millet protein, walnut meal protein and pea protein [2], [7], [8], [9].
Therefore, the main purpose of this study was to prepare apple protein nanoparticles using green technology i.e. Ultra-sonication and evaluate its impact on the techno-functional properties andprotein structure. It is hypothesized that apple seed protein nanoparticles will offer enhanced functional properties and could be better exploited in food systems. Furthermore, we expect that this study will pave the way to offer an innovative approach for utilizing the protein from waste with increased techno-functional property which can find its use in food industry.
2. Materials
Apple seeds were procured from the FIL Industries, Pvt. Ltd., Srinagar and were manually cleaned in the laboratory. Apple seed flour was obtained by manually removing the seed coat and collecting the inner white cotyledon. All chemicals and reagents used in this work were procured from Sigma Aldrich Pvt Ltd unless otherwise mentioned.
2.1. Protein extraction and estimation
Extraction of protein from defatted apple seed flour was done using alkali solubilization and acid precipitation method following the method of Malik et al. [10] with some modifications. Briefly, defatted apple seed flour was dissolved in five volumes of distilled water and pH was raised to 10.5 and stirred for 2 h for complete solubilisation of proteins. The mixture was then centrifuged at 3500 rpm for 10 min and the supernatant obtained was precipitated at iso-electric pH 4.5 using 1 N HCl. Precipitated protein was re-centrifuged, and protein pellet was recovered and freeze-dried. The freeze dried sample was stored at −20 °C till further analysis. The protein content in the freeze dried sample was estimated to be 95.4 ± 0.54% on dry weight basis (AOAC 1997). The nitrogen conversion factor was 6.25.
2.2. Preparation of apple seed protein nanoparticles
Apple protein was dissolved in sodium phosphate buffer (pH 7.2) and then subjected to probe ultra-sonication treatment for 30 min. The sonication was done at frequency of 40 kHz at 5 sec lag time to avoid overheating. Also, ice bath was used during sonication so that the temperature remains constant (20–30 °C). The sonicated solution was then freeze dried and stored at −20 °C till further analysis.
2.3. Particle size distribution (PSD), polydispersity index (PI) and zeta potential (ZP)
The PSD, PI and ZP of native (N-protein) and ultrasonicated (US-protein) apple seed protein were determined using a dynamic light scattering (DLS) (Anton Paar, Australia). Briefly, the dispersion of 0.01% protein samples was prepared in deionised water and sonicated at 20 kHz for 20 min using sonication bath (model: LTUSB) followed by sample analysis using DLS.
2.4. Protein pattern
Protein profile was visualized using SDS-PAGE following the method of Gani et al. [11]. The protein sample was prepared by adding 0.3 g of sample to 2.7 mL of 5% (w/v) SDS solution. The mixture was vortexed for 2 min and the homogenate was incubated at 85 °C for 1 h. The samples were centrifuged at 17,000g for 20 min at room temperature (26–28 °C) using a centrifuge (Eppendrof, centrifuge, 5418 R, Hamburg, Germany). Protein concentration in the supernatant was determined according to Biuret method . Solubilized samples were mixed with the sample buffer at a ratio of 1:1 (v/v) and boiled for 3 min. Samples (15 µg protein) were loaded onto polyacrylamide gels comprising a 15% running gel and a 4% stacking gel and subjected to electrophoresis at a constant current of 15 mA/gel using a Mini Protein III unit (Bio-Rad Laboratories, Inc., Richmond, CA, USA). After electrophoresis, staining of the gel was carried out with 0.02% (w/v) Coomassie Blue R-250 in 50% (v/v) methanol and 7.5% (v/v) acetic acid and destained with 50% (v/v) methanol and 7.5% (v/v) acetic acid. The protein standards (Biostep™ prestained protein marker) with protein markers ranging from 11 kDa to 180 kDa were used to estimate the molecular weight of the proteins.
2.5. Scanning electron microscopy (SEM)
Microstructure of protein samples (N-protein and US-protein) were analysed using scanning electron microscopy (Hitachi S-300H-Tokyo, Japan). The sample was placed on an adhesive tape attached to aluminium specimen stub and covered with gold. After gold-palladium coating, the samples were visualised at high vacuum.
2.6. X-ray diffraction analysis
X-ray diffractometer (Rigaku Smart lab) was used to examine the crystallographic structure of N-protein and US-protein following the method of Jhan et al. [2]. It was operated at 40 kV voltage, 30 mA current, and radiation (λ) of 1.5418 Å. The samples were scanned from 0⁰–25⁰ (2θ) at the rate of 3.0/min with a step width of 0.02°.
2.7. Structural analysis
The structural changes in protein samples were determined by Fourier transform infrared spectroscopy (CARY 630, Agilent Technologies, USA) at wavelength ranging from 500 to 4000 cm−1 by taking regular 32 scans at a resolution of 4 cm−1.
2.8. Differential scanning calorimetry (DSC) for thermal analysis
The protein samples (N-protein and US-protein) were evaluated for thermal properties using differential scanning calorimeter (DSC-1 STARe, Mettler-Toledo) following the method of Manzoor et al. [5]. The 5 mg of sample was sealed in platinum pans and scanned at the rate of 10 °C/min from 20–250 °C, and an empty platinum pan was used as a reference. The flow rate of nitrogen gas was 40 mL/min to deliver the inert atmosphere. The thermal curves were recorded.
2.9. Functional properties
2.9.1. Protein solubility profile
Protein solubility was determined following the method of Klompong et al. [12] with some modification. Briefly, 100 mg of protein sample was dispersed in 20 mL of distilled water and pH was adjusted from 2 to 11 using either 6 N HCl or 6 N NaOH. The mixture was stirred for 60 min at room temperature and centrifuged at 11,000g for 15 min. Protein contents in the supernatant were determined using the Biuret method while total protein content in the sample was determined after solubilizing the sample in 0.5 N NaOH. Protein solubility was calculated as follows:
Solubility (%) = (protein content in supernatant/total protein content in sample) × 10.
2.9.2. Surface hydrophobicity
Fluorescence spectroscopy was used to evaluate surface hydrophobicity of N-protein and US-protein using ANS (8-anilinonaphthalene-1-sulfonic acid) fluorescence probe following the method of Malik and Saini. [10]. Briefly, 4 mL of N-protein and US-protein solution (1 mg/mL) prepared in sodium phosphate buffer (0.01 M, pH 7.2) were mixed with 20 µl of freshly prepared ANS. The mixture was incubated for 20 min at room temperature in dark. Spectrofluorometer (Cary Eclipse, Alginate technologies, Malaysia) was used to measure fluorescence intensity with an excitation wavelength of 360 nm (slit width 5 nm) and emission wavelength between 400 and 600 nm (slit width 5 nm).
2.9.3. Foaming properties
Foaming properties of the protein samples were determined following the protocol of Gaoa et al. [13]. Sample of 1 g was dissolved in 25 mL deionised water. The mixture was then blended using homogenizer (WiseTis, HG-15A, Wisd Instruments, Korea) at 10,000 rpm for 5 min which resulted in production of foam. Foaming capacity (FC) was calculated using the following equation.
Where, V1 is the volume of sample after homogenization and Vo is the volume of sample before homogenization.
However foaming stability (FS) was evaluated using the equation.
FS = Volume after standing of the foam (mL)/Volume before standing of the foam (mL) × 100.
2.9.4. Emulsifying properties
Emulsifying activity (EA) and emulsion stability (ES) were determined by method of Malik and Saini [14] with some modification. Protein solution (2%) was homogenized with soyabean oil at 1:1 (v/v) ratio. The emulsions were centrifuged for 10 min at 3000 rpm. The height of emulsified layer and that of the total contents in the tube was measured. The emulsifying activity (EA) was calculated as:
EA (%) = (Height of emulsified layer/Height of total content in the tube) × 100.
Emulsion stability was determined by heating the emulsion at 80 °C for 30 min using a hot water bath followed by centrifugation at 3000 rpm for 10 min. Emulsion stability was calculated as:
ES (%) = (Height of emulsified layer/Height of total content in the tube) × 100.
2.9.5. Water (WAC) and oil absorption capacity (OAC)
The WAC and OAC of samples were done following the method of Deng et al. [1]. Briefly, 0.2 g of sample on dry basis was mixed with 10 mL of deionised water. The solution was centrifuged for 10 min at 6000g and the supernatant was decanted. The WAC was calculated as:
Where, W1 represents saturated weight of sample and W2 represents the dry weight of the sample.
For evaluating the OAC, 0.2 g of protein sample was dissolved in 10 g of soya bean oil and stirred for 30 min at ambient temperature. The mixture was then centrifuged at 6000g for 10 min and the unabsorbed oil was decanted and weighed. The OAC was calculated as:
Where, O1 represents oil saturated weight of sample and O2 represents the dry weight of the sample.
2.10. Statistical analysis
Experiments were carried out in triplicate using three different lots of samples and data was presented as mean ± standard deviation. For pair comparison, T-test was used, and significance of difference was established at p < 0.05. Statistical analysis was performed using the Statistical Package for Social Science (SPSS 11.0 for windows, SPSS Inc., Chicago, IL, USA).
3. Results and discussion
3.1. Particle size distribution, polydispersity index and zeta potential
The size of the polymer is an important characteristic feature to understand its physicochemical and functional properties. The average particle size of N-protein and US-protein was found to be 1.2 ± 0.04 µm and 484 ± 0.3 nm respectively as shown in Table 1. The results depicted a reduction in the size of particles upon the ultrasonication treatment. The reduction may be because of the unfolding of the protein structure stimulated by the ultra-sonication procedure, or it may be due to cavitation triggered by the breakdown of microbubbles which rupture and propagate in the mixture as a sound wave [15]. Disaggregation of soluble aggregates is also reported to decrease the size of protein upon ultrasonication [16]. The emulsion stability and foam stability were enhanced by the use of US-protein (p < 0.05), therefore conforming the impact of particle size on functional properties (Table 2). The cavitation moderately cleaves the intermolecular hydrophobic sites of protein and therefore reducing the particle size and increasing the surface area. Jhan et al. [2] also reported a decrease in size of foxtail millet protein using ultra-sonication treatment (20 kHz) for 15 and 30 min. The polydispersity index measures the particle distribution in a solution and was found to be 0.5 for N-protein representing broader range of size distribution and 0.2 for US-protein representing the narrow range of size distribution.
Table 1.
DLS analysis of native protein (N-protein) and ultrasonicated protein (US-protein).
| N-protein | US-protein | |
|---|---|---|
| Particle Size | 1.2 ± 0.04 a µm | 484 ± 0.3b nm |
| Polydispersity index (PDI) | 0.5 ± 0.03 a | 0.27 ± 0.04b |
| Zeta potential (mV) |
−11 ± 0.01 a | −19 ± 0.03b |
Results are represented as mean±standard deviation, n=3 and values in the same row with same superscripts are not significantly different (p>0.05).
Table 2.
Thermal properties of native (N-protein) and ultrasonicated (US-protein) protein.
| Sample | Td (°C) | ΔH |
|---|---|---|
| N-protein | 75 ± 0.5a | 178.13 ± 0.2a |
| US-protein | 63 ± 0.6b | 166.75 ± 0.1b |
Results are represented as mean±standard deviation, n=3 and values in the same row with same superscripts are not significantly different (p>0.05).
Zeta potential (ZP) signifies the net charge on a particle surface and gives knowledge about attractive or repulsive forces between particles to predict the stability of colloidal systems [17], [18]. The higher value of zeta potential indicates better stability of colloidal system [16]. The ZP value of N-protein and US-protein was found to be −11 and −19 mV. Cui et al. [16] reported the increase in zeta potential of protein from tiger nut upon ultrasonication. The US-protein had strong negative surface charge than the N-protein depicting better colloidal stability upon ultrasonication. Change in zeta potential value may be because of the disruption of protein structure by ultrasonication process, resulting in exposure of polar groups [16]. As a result, electrostatic repulsion between protein particles was enhanced and hence enhanced the stability of US-protein.
3.2. Protein pattern
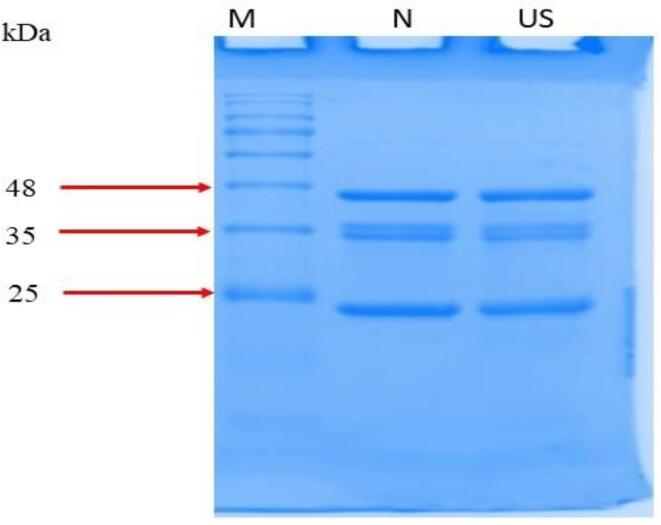
The electrophoretic protein profile of native and ultrasonicated apple seed protein is shown in Fig. 1. Major protein bands were identified at 25, 35 and 48 kDa in both native and ultrasonicated proteins. The results indicate that ultrasonication did not break the peptide bonds and therefore, had no impact on molecular weight of the protein. Similar results were reported by Du et al. [19] for ultrasonic treated pumpkin seed protein and Meng et al. [20] for ultrasonic treated whey protein. Hence, ultrasonication could be used a technique to improve the functional properties of apple seed proteins without having any impact on its primary structure.
Fig. 1.
SDS-PAGE native protein (N-protein) and ultrasonicated protein (US-protein).
3.3. Structural properties using ATR-FTIR
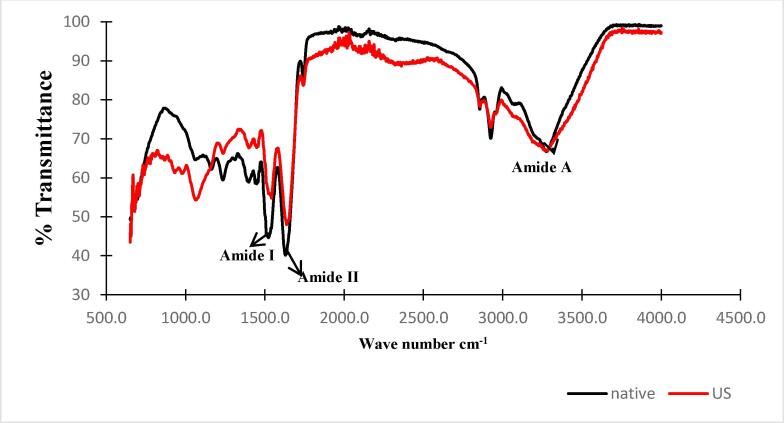
The secondary structure change of N-protein and US-protein upon ultrasonication treatment was evaluated using FTIR analysis as shown in Fig. 2 Both the spectra showed strong band ranging from 3100 to 3300 cm−1 which corresponds to the occurrence of amide-A. The amide-A is due to the stretching of N–H bonds of protein signifying aggregation and unfolding of the protein. The peaks from 1600 to 1750 cm−1 and 1490–1550 cm−1 represents amide I and amide II regions of proteins which occur because of C O stretching and combination of C-N and N–H bending vibrations, that plays an important role in secondary protein structure such as α-helix, β-sheet, β-turn, and random coil. The US-protein showed increased intensity at amide I and amide II however, decreased intensity at the amide A zone in comparison to N-protein signifying a modification of secondary protein structure. Similar results of ultrasonicated samples were reported by Tang et al [6]. Also, minor positional shift in the ultrasonicated sample might be because of the alteration in the random coil part of the β-sheet. The increase in intensity also suggested a decrease in α-helical content of protein after ultrasonication treatment. Moreover, stretching vibration of C O of amide group, bending of in-plane N–H and stretching modes of C–N are based on the absorption in amide-I region and are indications of presence of interactions, which led to the formation of secondary structure elements [21]. Jhan et al. [2] reported similar results of fox millet protein samples upon nano-reduction using ultra-sonication. Therefore, increase in transmittance of US-protein indicates changes in the secondary structure of protein due to ultrasonication.
Fig. 2.
FTIR graph of native protein (N-protein) and ultrasonicated protein (US-protein).
3.4. X-ray diffraction (XRD) analysis
The basic crystallographic diffraction pattern of N-protein and US-protein are represented in Fig. 3. XRD determines the size of the crystal and probable conformational changes in the structure of the protein. The N-protein showed characteristic peaks at Bragg angle values of around 8.4° and 19.94° and US-protein showed a major peak at 4.4° and a minor peak at 19.58°, with lowered intensity. The peak around 10° represent crystalline region I however, the peak around 2θ = 20° represents the crystalline region II and the β-sheet of protein structure [2]. Low intensities of this peak in the US-protein clearly show a lack of crystallinity or orderly arrangement in the structure that is attributed to the change in protein structure because of the cavitation phenomenon of ultrasound waves [2]. Furthermore, there is a direct correlation between particle size and diffraction intensity, low diffraction intensity specifies smaller crystal size of proteins [22]. Kaushik et al. [23] reported a similar single peak for flax seed protein. Mir et al. [24] and Malik et al. [21] also observed similar peaks at a diffraction angle of 10° and 20° for quinoa, sunflower protein isolates respectively.
Fig. 3.
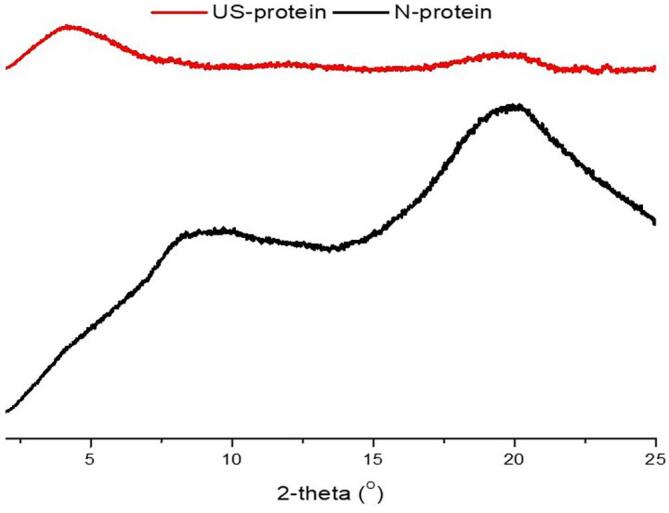
X-ray diffraction pattern of native protein (N-protein) and ultrasonicated protein (US-protein).
3.5. Morphological analysis
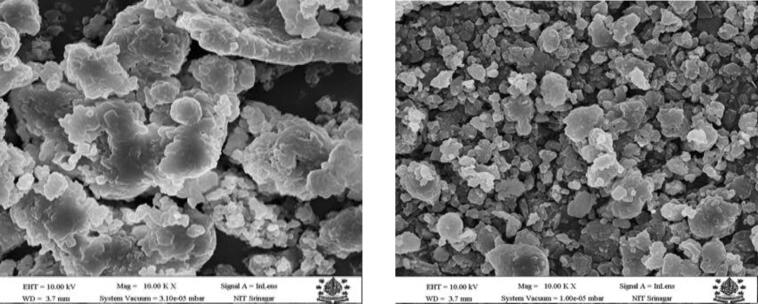
The difference in surface morphology of N-protein and US-protein was visualized using scanning electron microscopy as shown in Fig. 4. The N-protein micrographs displayed hollow spongy tangled structure, smooth surface, with particles accumulated together. However, the micrograph of US-protein showed polyhedral shapes with slackened protein granules. Ultrasonication has resulted in marked reduction in size of US-protein compared to N-protein (Fig. 4). The distortion of protein upon ultra-sonication may be because of the denaturation and unfolding of protein structure due to cavitation forces resulting in change of morphological properties of protein [24]. Du et al. [25] reported the similar morphological change in pumpkin seed protein upon sonication.
Fig. 4.
SEM micrograph of native protein (N-protein) and ultrasonicated protein (US-protein).
3.6. Differential scanning calorimeter
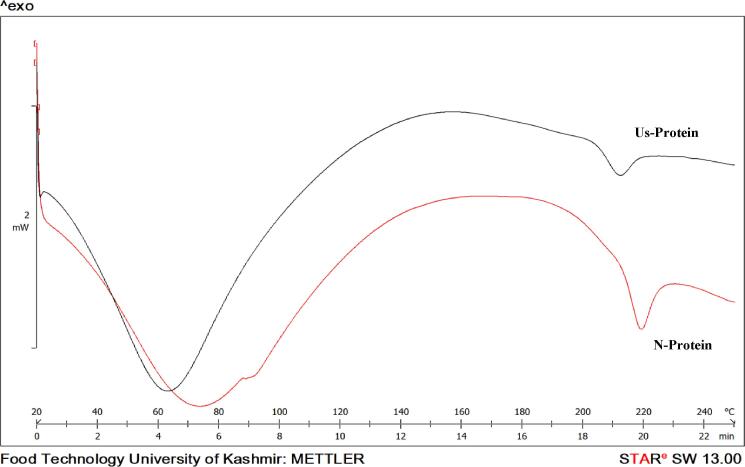
The transition temperature (Td) and enthalpy (ΔH) of denaturation for N-protein and US-protein are presented in Table 2. The N-protein showed a denaturation temperature at 75 °C which shifted to 63 °C in case of US-protein (Fig. 5). The enthalpy of denaturation for N-protein was 178.13 J/g which decreased to 166.75 for US-protein (Table 2). Td commonly indicates the thermal stability of a protein, while ΔH represents the extent of ordered structure of a protein [6]. The decrease in Td and ΔH indicated breakdown of non-covalent bonds between protein molecules upon ultrasonication and decrease in ordered structure of US-protein. It was also evident from SDS-PAGE results (Fig. 1) as no difference was observed in the band pattern of N-protein and US-protein indicating no breakdown of covalent bonds. This change in the thermal behaviour of US-protein upon ultrasonication is because of the conformational changes in structure of protein as indicated by FTIR results (Fig. 2). Similar results were reported in case of millet protein in which the thermal properties of protein decreased upon ultra-sonication treatment [2]. Furthermore, ΔH which is indicative of crystallinity and presence of double-helical structure subsequently decreased from 178 J/g to 166.75 J/g indicating loss of crystallinity as provided by XRD spectroscopy results.
Fig. 5.
Thermograph of native protein (N-protein) and ultrasonicated protein (US-protein).
3.7. Functional properties
3.7.1. Surface hydrophobicity
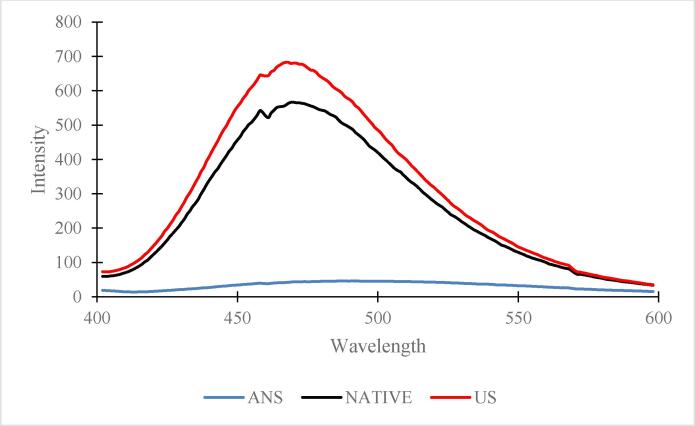
Surface hydrophobicity of protein is a structural characteristic and is important for protein stability and conformation which in turn impacts functionality of protein [26]. The surface hydrophobicity of N-protein and US-protein in term of ANS fluorescence is shown in Fig. 6 The US-protein showed higher value of hydrophobicity in comparison to N-protein. Increase in hydrophobicity of US-protein upon sonication is attributed to unfolding of protein structure due to acoustic cavitation caused by the ultra-sonication resulting in exposure of hydrophobic groups of protein available for ANS fluorescent probe [27]. Greater the exposure of hydrophobic sites to solvent upon protein unfolding greater will be the fluorescence intensity. These findings are also supported by decrease in protein solubility in US-protein (Fig. 7). Similar hydrophobicity results were reported in previous literature for wheat germ, whey protein isolates upon sonication which is attributed to increased emulsification and foaming properties [28], [29].
Fig. 6.
Surface hydrophobicity of native protein (N-protein) and ultrasonicated protein (US-protein).
Fig. 7.
Solubility pattern of native protein (N-protein) and ultrasonicated protein (US-protein)with respect to pH.
3.7.2. Protein solubility
Solubility is an important parameter for evaluating the application of protein as a functional ingredient in different food systems. The solubility of N-protein and US-protein was evaluated at different pH values ranging from 2 to 10 as shown in Fig. 7. Results depicted that the ultrasonication treatment generally decreased the solubility of protein at all pH values. The solubility of N-protein and US-protein samples decreased in the pH range of 4 showing minimum values of 5.5 and 6% respectively, which is near to the isoelectric point (pH 4.5). The decreased solubility may be due to the exposure of hydrophobic domains to the aqueous environment resulting in hydrophobic-hydrophobic interactions (Fig. 6).However, solubility gradually increased as the pH of the solutions increased from 4 to 10 showing value of solubility 86.7.61% and 64.3% for N-protein and US-protein. In general, high solubility at alkaline pH was attributed to negative charge which may have dissociated protein aggregates due to electrostatic repulsion. Nevertheless, some studies have shown an increase in protein solubility due to ultrasonication treatment [16].
3.7.3. Foaming properties
Foaming properties such as foam capacity and foam stability is an important property for the processing of the foods. Foam formation is governed by transportation, penetration and reorganization of the molecules on the air/water interface. It is mainly dependent upon the size, hydrophobicity and structural flexibility of protein molecules [26]. The foaming capacity of N-protein was 60 ± 2.4 % and US-protein showed increased foaming capacity of 72.5 ± 2.0 % (p < 0.05) as shown in Table 3. It has been reported that the exposed hydrophobic groups contribute to enhanced interaction at the air–water interface. Additionally, particle size in nano-range might have enhanced adsorption rate of protein at the air–water interfaces [31]. Increased foaming property of soy protein and α-lactalbumin upon ultrasonication has been reported in the literature [32], [33]. In addition, ultra-sonication involves cavitation phenomenon resulting in partially unfolded protein structure and contributed to improvements in foaming properties. Similar increase in foam stability was also found in ultra-sonicated sample (61.5 ± 2.0 %) in comparison to native protein (49.5 ± 2.0 %). Jambrak et al. [33] reported same trend of foaming properties for ultrasonically treated soy protein and wheat gluten [34]. The increase in surface hydrophobicity values may also be responsible for improving the foaming capacity and stability which might be due to the exposure of hydrophobic groups to accelerated adsorption at the gas/water interface [35].
Table 3.
Functional properties.
| Functional property | N-Protein | Us-protein |
|---|---|---|
| Water absorption capacity (g/g) | 6.9 ± 0.7a | 5.3 ± 0.1b |
| Oil absorption capacity (g/g) | 2.6 ± 0.2a | 3.9 ± 0.2b |
| Foaming capacity (%) | 60 ± 2.4a | 72.5 ± 2.0b |
| Foaming stability (%) | 49.5 ± 2.0a | 61.5 ± 2.0b |
| Emulsifying activity (%) | 77 ± 1.6a | 89 ± 1.2b |
| Emulsifying stability (%) | 58 ± 1.2a | 69 ± 2.2b |
Results are mean ± S.D, n=3 and values in the same row with same are not significantly different (p>0.05).
3.7.4. Emulsifying properties
Emulsifying property is defined as the capability of proteins to adsorb at oil–water interface. EA represents the ability of proteins to be adsorbed at the interface of water and oil, while ES is the capacity of proteins to stay at oil–water interface after emulsion storage or heating. The emulsifying properties of N-protein and US-protein are presented in Table 3. The emulsion activity was found to be 77 ± 1.6 % and 89 ± 1.2 % for N-protein and US-protein. Results revealed highest emulsifying activity of ultrasonicated protein particles in comparison to native. The probable mechanism of increased emulsification activity might be due to rapid adsorption of the proteins on the surface of oil droplets upon ultrasonication treatment leading to increase in capability of protein to decrease the interfacial tension [16]. However, the emulsion stability was found to be 58 ± 1.2 % and 69 ± 2.2 % for N-protein and US-protein. Results reveal that ultrasonication has positive role in enhancing the emulsifying property of proteins which is ascribed to charge, solubility and hydrophobicity.
3.7.5. Water absorption capacity and oil absorption capacity
Water absorption capability (WAC) indicates interaction between water and protein molecules that partially determines its behaviour in different complex food systems. The US-protein showed decrease in WAC compared to N-protein (p < 0.05) (Table 2). The decrease in WAC of US-protein may be due to the unfolding of the polypeptide chain leading to exposure of hydrophobic groups consequently decreasing protein solubility (Fig. 7) in addition to WAC [27]. Nevertheless, several studies indicated increase in WAC of protein isolate upon ultrasonication [30], [36].
The US-protein showed higher oil absorption capacity (OAC) compared to N-protein (p < 0.05). This can be attributed to more exposure of hydrophobic domains in US-protein as indicated by increased hydrophobicity (Fig. 6). The collapse of the cavitation bubbles near the protein molecules caused conformational changes and unfolding of the polypeptide chains in the protein structure resulting in exposure of non-polar hydrophobic sites which increases binding of the oil molecules to the protein molecules.
4. Conclusion
The study provides valuable knowledge about utilising the agro waste such as apple seeds for exploration of protein that can be used as an ingredient in functional food industry. The protein nanoparticle was produced using green technology i.e. ultrasonication. The thermal stability decreased upon ultra-sonication which specifies denaturation of proteins. Furthermore, this study can give new hope for the exploration of the plant-based protein as functional constituent in food system which can help to decrease the existing food issues of the population. It was concluded that nano-reduction using ultra-sonication positively enhanced the techno-functional properties of protein and hence can be efficiently used as functional ingredient in food and nutraceutical industry. Moreover, the developed nanostructures can pave way as a possible wall material for control release of bioactive ingredients into food systems.
Declaration of Competing Interest
The authors have no conflict of interest.
Acknowledgement
Dr. Asir Gani is thankful to Department of Biotechnology, GOI (DBT-RA/2021/January/N/860) for the financial support.
References
- 1.Denga Y., Huanga L., Zhanga C., Xiea P., Chenga J., Wanga X., Lia S. Physicochemical and functional properties of Chinese quince seed protein isolate. Food Chem. 2019;283:539–548. doi: 10.1016/j.foodchem.2019.01.083. [DOI] [PubMed] [Google Scholar]
- 2.Jhan F., Gani A., Noor N., Shah A. Nano reduction of millet proteins: Effect on structural and functional properties. ACS Food Sci. Technol. 2021;8:1418–1427. [Google Scholar]
- 3.Ahnen R.T., Jonnalagadda S.S., Slavin J.L. Role of plant protein in nutrition, wellness, and health. Nutr. Rev. 2019;7:735–747. doi: 10.1093/nutrit/nuz028. [DOI] [PubMed] [Google Scholar]
- 4.Freitas L.C., Barbosa J.R., da Costa A.L.C., Bezerra F.W.F., Pinto R.H.H., Carvalho Junior R.N.d. From waste to sustainable industry: How can agro-industrial wastes help in the development of new products? Elsevier. 2021;169:105466. [Google Scholar]
- 5.Manzoor M., Singh J., Gani A. Characterization of apple (Malus domestica) seed flour 443 for its structural and nutraceutical potential. LWT. 2021;112138:444. doi: 10.1016/J.LWT.2021.112138. [DOI] [Google Scholar]
- 6.S-Q. Tang, Q-H. Du, Z. Fu .Ultrasonic treatment on physicochemical properties of water-soluble protein from Moringa oleifera seed, Ultrason. Sonochem., 71 (2021) 105357. [DOI] [PMC free article] [PubMed]
- 7.Hu H., Wu J., Li-Chan E.C.Y., Zhu L., Zhang F., Xu X., Fan G., Wang L., Huang X., Pan S. Pan Effects of ultrasound on structural and physical properties of soy protein isolate (SPI) dispersions. Food Hydrocoll. 2013;30(2):647–655. [Google Scholar]
- 8.Y. Di , X. Li , X. Chang , R. Gu , X. Duan , F. Liu , X. Liu , Y. Wang, Impact of germination on structural, functional properties and in vitro protein digestibility of sesame (Sesamum indicum L.) protein. LWT 154 (2022) 112651.
- 9.Nazaria B., Mohammadifar M.A., Shojaee-Aliabadia S., Feizollahia E., Mirmoghtadaiea L. Effect of ultrasound treatments on functional properties and structure of millet protein concentrate. Ultrason. Sonochem. 2018;4:383–388. doi: 10.1016/j.ultsonch.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Malik M.A., Saini C.S. Improvement of functional properties of sunflower protein isolates near isoelectric point: Application of heat treatment. LWT-Food Sci. Technol. 2018;98:411–417. [Google Scholar]
- 11.Gani A., Benjakul S., Nuthong P. Effect of virgin coconut oil on properties of surimi gel. J Food Sci. Technol. 2018;55:496–505. doi: 10.1007/s13197-017-2958-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klompong V., Benjakul S., Kantachote D., Shahidi F. Antioxidative activity and functional properties of protein hydrolysate of yellow stripe trevally (Selaroides leptolepis) as influenced by the degree of hydrolysis and enzyme type. Food Chem. 2007;102:1317–1327. [Google Scholar]
- 13.Gaoa L.L., Lia Y.Q., Wang Z.-S., Suna G.-J., Qic X.-M., H-Z, Mod, Physicochemical characteristics and functionality of tree peony (Paeonia suffruticosa Andr.) seed protein. Food Chem. 2018;240:980–988. doi: 10.1016/j.foodchem.2017.07.124. [DOI] [PubMed] [Google Scholar]
- 14.Malik M.A., Sharma H.K., Saini C.S. High intensity ultrasound treatment of protein isolate extracted from dephenolized sunflower meal: Effect on physicochemical and functional properties. Ultrason. Sonochem. 2017;39:511–519. doi: 10.1016/j.ultsonch.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 15.Jianga S., Dinga J., Andradea J., Rababahb T.M., Almajwalc A., Abulmeatyc M.M., Fenga H. Modifying the physicochemical properties of pea protein by pH-shifting and ultrasound combined treatments. Ultrason. Sonochem. 2017;38:835–842. doi: 10.1016/j.ultsonch.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 16.Cui Q., Wang L., Wang G., Zhang A., Wang X., Jiang L. Ultrasonication effects on physicochemical and emulsifying properties of Cyperus esculentus seed (tiger nut) proteins. LWT. 2021;142:110979. [Google Scholar]
- 17.Gani A., Ashraf Z.U., Shah A., Noor N., Gani A. Encapsulation of vitamin D3 into β-glucan matrix using the supercritical carbon dioxide. ACS Food Sci. Technol. 2021;1(10):1880–1887. [Google Scholar]
- 18.Ashraf Z.U., Shah A., Gani A., Gani A., Masoodi F.A., Noor N. N, Nanoreduction as a technology to exploit β-Glucan from cereal and fungal sources for enhancing its nutraceutical potential. Carbohydr. Polym. 2021;258 doi: 10.1016/j.carbpol.2021.117664. [DOI] [PubMed] [Google Scholar]
- 19.Du H., Zhang J., Wang S., Manyande A., Wang J. Effect of high-intensity ultrasonic treatment on the physicochemical, structural, rheological, behavioral, and foaming properties of pumpkin (Cucurbita moschata Duch.)-seed protein isolates. LWT - Food Sci. Technol. 2022;155 [Google Scholar]
- 20.Y. Meng., Z. Liang., C. Zhang., S. Hao., H. Han., P. Du., A. Li., H. Shao., C. Li., L. Liu. Ultrasonic modification of whey protein isolate: Implications for the structural and functional properties, LWT - Food Sci. Technol. 152 (2021) 112272.
- 21.Malik M.A., Saini C.S. Rheological and structural properties of protein isolates extracted from dephenolized sunflower meal: Effect of high intensity ultrasound. Food Hydrocoll. 2018;81:229–241. [Google Scholar]
- 22.Cao Y., Tan H.M. Study on crystal structures of enzyme hydrolyzed cellulosic materials by X-ray diffraction. Enzyme Microb. Technol. 2005;36:314–317. [Google Scholar]
- 23.Kaushika P., Dowlinga K., McKnight S., Barrow C.J., Wang B., Adhikaric B. Preparation, characterization and functional properties of flax seed protein isolate. Food Chem. 2016;197:212–220. doi: 10.1016/j.foodchem.2015.09.106. [DOI] [PubMed] [Google Scholar]
- 24.Mir N.A., Riar C.S., Singh S. Improvement in the functional properties of quinoa (Chenopodium quinoa) protein isolates after the application of controlled heat treatment: Effect on structural properties. Food Struct. 2021;28 doi: 10.1016/J.FOOSTR.2021.100189. [DOI] [Google Scholar]
- 25.Shah A., ul Ashraf Z., Gani A., Masoodi F.A., Gani A. A Gani - β-Glucan from mushrooms and dates as a wall material for targeted delivery of model bioactive compound: Nutraceutical profiling and bioavailability. Ultrason. Sonochem. 2022;82:105884. doi: 10.1016/j.ultsonch.2021.105884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik M.A., Sharma H.K., Saini C.S. High intensity ultrasound treatment of protein isolate extracted from dephenolized sunflower meal: Effect on physicochemical and functional properties. Ultrason. Sonochem. 2017;39:511–519. doi: 10.1016/j.ultsonch.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C., Ma H., Yu X., Liu B., Yagoub Ael G., Pan Z. Pretreatment of defatted wheat germ proteins (by-products of flour mill industry) using ultrasonic horn and bath reactors: effect on structure and preparation of ACE-inhibitory peptides. Ultrason. Sonochem. 2013;20:1390–1400. doi: 10.1016/j.ultsonch.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Jiang L., Wang J., Li Y., Wang Z., Liang J., Wang R., Chen Y., Ma W., Qi B., Zhang M. Effects of ultrasound on the structure and physical properties of black bean protein isolates. Food Res. Int. 2014;62:595–601. [Google Scholar]
- 29.Huang L., Ding X., Li Y., Ma H. The aggregation, structures and emulsifying properties of soybean protein isolate induced by ultrasound and acid. Food Chem. 2019;279:114–119. doi: 10.1016/j.foodchem.2018.11.147. [DOI] [PubMed] [Google Scholar]
- 30.Hu H., Li-Chan E.C., Wan L., Tian M., Pan S. The effect of high intensity ultrasonic pre-treatment on the properties of soybean protein isolate gel induced by calcium sulfate. Food Hydrocoll. 2012;32:303–311. [Google Scholar]
- 31.Zhang H., Claver I.P., Zhu K.X., Zhou H. The effect of ultrasound on the functional properties of wheat gluten. Molecules. 2011;16:4231–4240. [Google Scholar]
- 32.Damodaran S. Protein stabilization of emulsions and foams. J. Food Sci. 2005;70:R54–R66. [Google Scholar]
- 33.Morales R., Martínez K.D., Pizones Ruiz-Henestrosa V.M., Pilosof A.M.R. Pilosof, Modification of foaming properties of soy protein isolate by high ultrasound intensity: particle size effect. Ultrason. Sonochem. 2015;26:48–55. doi: 10.1016/j.ultsonch.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Jambrak A.R., Mason T.J., Lelas V., Krešić G. Ultrasonic effect on physicochemical and functional properties of α-lactalbumin. LWT-Food Sci. Technol. 2010;43:254–262. [Google Scholar]
- 35.Golly M.K., Ma H., Yuqing D., Wu P., Dabbour M., Sarpong F., Farooq M. Enzymolysis of walnut (Juglans regia L.) meal protein: ultrasonication-assisted alkaline pretreatment impact on kinetics and thermodynamics. J. Food Biochem. 2019;43:12948. doi: 10.1111/jfbc.12948. [DOI] [PubMed] [Google Scholar]
- 36.Biswas B., Sit N. Effect of ultrasonication on functional properties of tamarind seed protein isolates. J Food Sci Technol. 2020;57:2070–2078. doi: 10.1007/s13197-020-04241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]