Summary
Background
In HIV infection, even under long-term antiretroviral therapy (ART), up to 20% of HIV-infected individuals fail to restore CD4+ T cell counts to the levels similar to those of healthy controls. The mechanisms of poor CD4+ T cell reconstitution on suppressive ART are not fully understood.
Methods
Here, we tested the hypothesis that lipopolysaccharide (LPS) from bacteria enriched in the plasma from immune non-responders (INRs) contributes to blunted CD4+ T cell recovery on suppressive ART in HIV. We characterized plasma microbiome in HIV INRs (aviremic, CD4+ T cell counts < 350 cells/μl), immune responders (IRs, CD4+ T cell counts > 500 cells/μl), and healthy controls. Next, we analyzed the structure of the lipid A domain of three bacterial species identified by mass spectrometry (MS) and evaluated the LPS function through LPS induced proinflammatory responses and CD4+ T cell apoptosis in PBMCs. In comparison, we also evaluated plasma levels of proinflammatory cytokine and chemokine patterns in these three groups. At last, to study the causality of microbiome-blunted CD4+ T cell recovery in HIV, B6 mice were intraperitoneally (i.p.) injected with heat-killed Burkholderia fungorum, Serratia marcescens, or Phyllobacterium myrsinacearum, twice per week for total of eight weeks.
Findings
INRs exhibited elevated plasma levels of total microbial translocation compared to the IRs and healthy controls. The most enriched bacteria were Burkholderia and Serratia in INRs and were Phyllobacterium in IRs. Further, unlike P. myrsinacearum LPS, B. fungorum and S. marcescens LPS induced proinflammatory responses and CD4+ T cell apoptosis in PBMCs, and gene profiles of bacteria-mediated cell activation pathways in THP-1 cells in vitro. Notably, LPS structural analysis by mass spectrometry revealed that lipid A from P. myrsinacearum exhibited a divergent structure consistent with weak toll-like receptor (TLR) 4 agonism, similar to the biological profile of probiotic bacteria. In contrast, lipid A from B. fungorum and S. marcescens showed structures more consistent with canonical TLR4 agonists stemming from proinflammatory bacterial strains. Finally, intraperitoneal (i.p.) injection of inactivated B. fungorum and S. marcescens but not P. myrsinacearum resulted in cell apoptosis in mesenteric lymph nodes of C57BL/6 mice in vivo.
Interpretation
These results suggest that the microbial products are causally associated with INR phenotype. In summary, variation in blood microbial LPS immunogenicity may contribute to immune reconstitution in response to suppressive ART. Collectively, this work is consistent with immunologically silencing microbiome being causal and targetable with therapy in HIV.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID; R01 AI128864, Jiang) (NIAID; P30 AI027767, Saag/Health), the Medical Research Service at the Ralph H. Johnson VA Medical Center (merit grant VA CSRD MERIT I01 CX-002422, Jiang), and the National Institute of Aging (R21 AG074331, Scott). The SCOPE cohort was supported by the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763, Gandhi) and the CFAR Network of Integrated Clinical Systems (R24 AI067039, Saag). The National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001450 (the pilot grant, Jiang). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Keywords: HIV, Immune non-responders, Immune responders, Lipopolysaccharide, Lipid A
Abbreviations: ART, antiretroviral therapy; INRs, immune non-responders; IRs, immune responders; i.p., intraperitoneal; LPS, lipopolysaccharide; TLR, toll-like receptor; ASVs, amplicon sequence variants; sCD14, soluble CD14; MD2, myeloid differentiation factor 2; MS, mass spectrometry; APC, antigen-presenting cells; PBS, phosphate-buffered saline; m/z, mass-to-charge ratio; PBMC, peripheral blood mononuclear cells; FDR, Benjamini and Hochberg false-discovery rate; GO, gene ontology; IL, Interleukin; GM-CSF, Granulocyte-macrophage colony-stimulating factor; IFN-γ, Interferon-γ; IP-10, Interferon gamma-induced protein 10; MCP, Monocyte chemoattractant protein; MDC, Macrophage-derived chemokine; MIP, Macrophage inflammatory protein; TARC, Thymus and activation-regulated chemokine; TNF, Tumor necrosis factor; VEGF, Vascular endothelial growth factor
Research in context.
Evidence before this study
In HIV disease, B. fungorum was found to colonize duodenum in patients with low CD4+ T cell counts but neither in patients with normal CD4+ T cell counts nor in HIV-negative controls. Furthermore, the enrichment of B. fungorum in the gut was inversely correlated with CD4+ T cell counts. These results suggest a link between microbiome and CD4+ T cell counts in HIV disease.
Added value of this study
In the current study, both quantitative and qualitative plasma microbial translocation differed in INRs compared to IRs and healthy individuals. The enriched plasma bacteria from INRs belonged to proinflammatory bacterial strains, whereas the enriched plasma bacteria in IRs belonged to non-inflammatory bacterial strains. Structural analysis of LPS from P. myrsinacearum, mainly enriched in IRs, showed a structure with low predicted TLR4 stimulatory properties. LPS from B. fungorum and S. marcescens, mainly enriched in INRs, showed a lipid A structure closely related to the canonical TLR4 agonist, E. coli lipid A. Further, INR-enriched microbial LPS induced proinflammatory responses, CD4+ T cell apoptosis, and CD4+ T cell dysfunction, whereas IR-enriched microbial LPS did not exhibit such pathogenic activity.
Implications of all evidence available
Our findings reveal variation of circulating microbial LPS contributes to chronic inflammation and immune reconstitution failure in HIV patients on ART. In the future, a therapeutic strategy targeting anti-bacterial peptides, inhibitors for pathogenesis mediated by the inflammatory bacterial strain LPS, or changing diet, together with ART, could improve CD4+ T cell recovery and reduce chronic immune activation and inflammation, morbidity, and mortality in HIV.
Alt-text: Unlabelled box
Introduction
In HIV infection, circulating CD4+ T cell counts predict disease progression regardless of antiretroviral therapy (ART).1 Even under long-term suppressive ART, up to 20% of HIV-infected individuals fail to restore CD4+ T cell counts to the levels similar to those of healthy controls2; increased complications, morbidity, and mortality are observed among these patients.3, 4, 5, 6, 7, 8, 9 Despite long-term suppressive ART, HIV immunologic responders (IRs) are defined by aviremic, ART-treaeted, and peripheral CD4+ T cell counts above 500 cells/μl10; HIV immunologic non-responders (INRs) are defined by aviremic, ART-treaeted, CD4+ T cell counts lower than 350 cells/μl.11 The mechanisms of poor CD4+ T cell reconstitution on suppressive ART are not fully understood, which include insufficient thymic output, lymph node fibrosis, persistent microbial translocation and inflammation, residual virus replication, and autoantibody induced antibody-dependent cellular cytotoxicity.10,12, 13, 14
It is well established that HIV infection alters the mucosal microbiota, elevates gut permeability, and increases microbial translocation. However, ART does not fully restore gut microbial dysbiosis. The altered gut microbiota is associated with chronic inflammation in the gut mucosa, peripheral CD4+ T cell counts, and mortality in HIV-infected individuals.15 Increased relative abundance of Prevotella and Proteobacteria and decreased relative abundance of Bacteroides were observed in the gut microbiota from the HIV patients compared to controls.16 Among HIV-infected individuals with CD4+ T cell counts lower than 200 cells/μl, reduced enteric phylogenetic diversity and increased specific bacteria (i.e., Enterobacteriaceae) are observed compared to patients with normal CD4+ T cell counts.17 These studies indicate that the altered microbiome may play a role in the immunological outcome of ART; however, associations do not indicate causality. The distinction between beneficial and harmful microbial communities and the functional mechanisms underlying their effects in ART outcomes in HIV are poorly understood.
Many studies have analyzed microbiome in stools or in samples from other mucosal sites that may not represent systemic microbiome and may not play a major role in systemic immune reconstitution from ART. HIV infection is associated with a compromised mucosal epithelial barrier, which allows the translocation of bacteria or microbial products into the circulation. Thus, bacterial fragments or whole bacteria can appear in the blood through translocation and influence host immune system.18 Recently, blood microbiome has been applied to investigate the microbiome-host interaction in disease pathogenesis19, 20, 21; we designed a quality-filtering pipeline to exclude amplicon sequence variants (ASVs) of contaminations and artifacts in the human plasma microbiome.22 In this study, we identified the translocated microbiome in INRs and analyzed their effects on CD4+ T cells' recovery from ART.
Methods
Subjects
This study was conducted using plasma samples from 71 ART-treated HIV individuals on suppressive ART, including 31 IRs and 40 INRs, and 60 healthy individuals. All HIV individuals had ART for at least eight years. Samples were received from the University of California, San Francisco (UCSF, SCOPE cohort), University of Alabama at Birmingham (UAB), and Medical University of South Carolina (MUSC). The clinical characteristics are shown in Table 1 and Supplementary Table 1. This study was approved by each participating institutional review board. All recruited participants for this study provided written consent.
Table 1.
Demographical clinical characteristics of all study participants.
| Value for groupa |
||||
|---|---|---|---|---|
| HIV- | HIV+, IRs | HIV+, INRs | p value (two HIV+ groups)c | |
| Number of participants | 60 | 31 | 40 | |
| Sex (Male/Female) | 42/18 | 23/8 | 39/1 | |
| Age (yr) | 37.5 | 46 | 52 | 0.0011 |
| (31-55.75) | (35-53) | (45.25-60.75) | ||
| Race (AA/W/Others)b | 15/38/7 | 17/14/0 | 4/30/2006 | NA |
| CD4+ T cell counts (Cells/μl) | 707 | 253 | <0.0001 | |
| (545-958) | (212.5-310.8) | |||
| Nadir CD4+ T cell counts | 207 | 59.5 | <0.0001 | |
| (50-353) | (23-128.8) | |||
| Viral load pre-ARTd | 37300 | 57307 | 0.78 | |
| (31336-119535) | (48816-60202) | |||
| Time from diagnosis to ART (year)d | 7 | 10 | 0.196 | |
| (4-12) | (6-14) | |||
| Duration of ARTd | 5 | 7.5 | 0.0006 | |
| (4-6) | (5-9.75) | |||
| Duration of undetectable virusd | 4 | 7 | 0.0023 | |
| (3-6) | (4-8) | |||
| Viral blipsd | 1 | 1 | 0.4 | |
| (0-4) | (0-2) | |||
Data are medians (interquartile ranges).
AA: African American; W: White; Others: Mixed Ethnicity/Multiracial and Hispanic/Latino.
Fisher's exact test or Mann-Whitney's U test (unpaired) were used.
Descriptive statistics have removed unknown or unavailable data (NA), details see Supplementary Table 1.
Plasma microbial 16S rDNA isolation, sequencing, data processing and analysis
The plasma microbial 16S rDNA isolation, circulating qPCR bacterial 16S rDNA detection, and sequencing were performed according to our previously published method.23 Bacteria 16S rDNA amplicons covering variable regions V3 to V4 were amplified using primers (515F and 806R with Illumina adapters and eight base-pair dual indices). All samples were run concurrently to avoid batch-to-batch variation. The PCR reaction was incubated at 94°C for 3 min, the 35 cycles at 94°C for 30 s, 53°C for 40 s and 72°C for 60 s, and followed by a final elongation step at 72°C for 5 min. PCR products were checked on a 2% agarose gel to assess the amplification. Multiple samples were pooled together in equal concentrations and purified using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA). The purified PCR products were used to prepare the DNA library according to the Illumina TruSeq DNA library preparation protocol. 16S rDNA libraries were sequenced at MR DNA (Shallowater, TX, USA) on a MiSeq platform using a v2 2 × 250 base-pair kit (Illumina, Inc). QIIME2 (https://qiime2.org/) was applied to demultiplex the data generated by Illumina MiSeq sequencing into paired forward and reverse FASTQ. Demultiplexed sequences were processed using the DADA2 (version 1.8)24 analysis pipeline in the R (https://www.r-project.org/, version 3.5.0) environment. Amplicon sequence variants (ASV) tables and different levels of taxonomic tables were imported into the phyloseq package (version 3.7)25 for statistical analysis. A user-defined filter pipeline was performed to remove the low abundance, low prevalence, and potential contaminants and artifacts from the plasma microbiome. Differential abundance testing between groups was compared by nonparametric Mann-Whitney's U tests at the ASV level. P-values were adjusted for multiple comparisons by the Benjamin-Hochberg false discovery rate (FDR). Comparison analysis was performed using R.
Bacterial culture and LPS extraction
B. fungorum (strain: CCUG 31961, ATCC, Manassas, VA) was grown in trypticase soy broth at 30°C in a shaker incubator for 24-48 h. S. marcescens (strain: PCI 1107, ATCC) and P. myrsinacearum (strain: NCIB 12127, ATCC) were grown in nutrient broth at 26°C in a shaker incubator for 48-72 h. The bacteria species selection was based on the following criteria: 1. The species should be contained in the predicted species using 16S sequencing; 2. The bacteria species selected were commercially available and reported in human microbiome. After centrifugation of culture media at 5000 rpm for 30 min, sedimented bacteria were harvested. The LPS was isolated using LPS Extraction Kit (Boca Scientific, Dedham, MA) according to the manufacturer's instructions. The concentration of LPS was determined using LAL Chromogenic Endotoxin Quantitation Kit (Thermo Fisher, Waltham, MA).
mRNA sequencing
THP-1 cells (5 × 105 cells per well) were seeded in 2 ml RPMI-1640 medium containing 10% FBS (culture medium) onto the 12 well plate. The final concentration of 2 ng/ml LPS isolated from B. fungorum, S. marcescens, and P. myrsinacearum were added to each well. After 24 h culture, total RNA was extracted from the bacteria treated THP-1 cells using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's protocol, the RNA purity was determined by a Nanodrop 2000 (Thermo Fisher). The mRNA libraries construction and sequencing were performed at Novogene (Durham, NC). The library was quantified using a Qubit 2.0 fluorometer (Thermo Fisher) and real-time quantitative PCR (TaqMan Probe), and the average length was detected by an Agilent 2100. Next, the libraries were sequenced on a HiSeq 2000 system (Illumina, San Diego, CA). The quality of the reads was checked using FastQC. The reference genome GENCODE v38 (GRCh38.p13) and corresponding gene model annotation files were downloaded from GENCODE (gencodegenes.org). STAR (v2.7)26 was applied using the default parameters to build the indexes of the reference genome and align the paired-end clean reads to the reference genome. Transcripts per kilobase million (TPM) were calculated based on the length of the gene and read count mapped to the gene. Differential expression between two groups was identified by the R Bioconductor package DESeq2 (v1.24.0).27 The resulting p-values were adjusted using Benjamini and Hochberg's approach for controlling the FDR. Genes with an adjusted p-value < 0.05 were assigned as differentially expressed.
Plasma levels of antibodies against specific bacterial antigens
The B. fungorum, S. marcescens, and P. myrsinacearum (1 × 1010/ml) were sonicated for 90 s on ice. The bacteria lysates were spun at 13,000 × g at 4°C for 20 min, and the bacteria lysates were treated using DNase I (5 µg/ml) in 37°C for 30 min. High binding microtiter plates were coated with LPS (2 µg/100 µl/well) or 1:200 diluted bacteria lysates at 4°C overnight. Microwells were washed three times with fresh prepared 1 x PBST wash buffer (1x PBS, 0.1% Tween 20). The plates were blocked using 1 x PBST, containing 3% BSA for 120 min at 37°C. After washing, diluted plasma was added to each well for 1 h at room temperature. After four times washing, horseradish peroxidase-labeled goat anti-human IgG (KPL, Gaithersburg, MD) was added at a 1:5000 dilution in PBS containing 3% BSA and incubated for 60 min at room temperature. After four times washing, 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) substrate solutions were used to detect binding. Absorbance was measured at 405 nm emission within 30 min.
Bacterial lipid A structural analysis
Bacterial lipid A structures were analyzed in the Department of Microbial Pathogenesis, University of Maryland School of Dentistry. Cultures of B. fungorum, S. marcescens, and P. myrsinacearum were plated overnight, and three colonies used to inoculate a 5 mL overnight culture of the respective nutrient broths (agar plates and broth per ATCC specifications) without antibiotics. Liquid cultures were grown aerobically. Bacterial pellets were prepared and a colony-equivalent placed on a MALDI target plate for direct lipid A analysis and structural characterization using our previously described method.28,29 MALDI-MS and MS/MS were performed in negative ion mode on a timsTOF Flex instrument with norharmane matrix.
Levels of cytokines and chemokines in plasma in vivo and cell culture supernatant in vitro
The isolated PBMCs from healthy individuals were cultured into 96-well plates at 2 × 105 cells/well in culture medium, consisting of RPMI-1640 supplemented with 10% fetal bovine serum (FBS, vol/vol) and 50 μg/ml penicillin/streptomycin. These cells were stimulated using LPS isolated from B. fungorum, S. marcescens, and P. myrsinacearum. LPS from E.coli 055:B5 was used as a positive control with a final concentration of 2 ng/ml. After 48 h, the cell culture supernatant was collected. The levels of the following 33 cytokines or chemokines were measured from the cell culture supernatant or human plasma from IRs, INRs, and healthy controls using Human Th17/Cytokine/Chemokine/ProInflam kit (Meso Scale Diagnostics, Rockville, Maryland) according to the manufacturer's instructions.
Mice
C57BL/6 mice were purchased from the Jackson Laboratories (Bar Harbor, ME) and housed at the MUSC animal facility. All animal studies were approved by MUSC Institutional Animal Care and Use Committee (IACUC). Mice were injected with PBS, heat-killed B. fungorum, S. marcescens, or P. myrsinacearum (5 mice for each group) twice a week for total of eight weeks by intraperitoneal (i.p.) route. The heat-killed bacteria were given 5*107 CFU/mice/time. The mice were sacrificed on day three after the last injection. For cell death and caspase detection, cells from mesenteric lymph nodes were digested, passed through a 40µm filter, and stained using Muse MultiCaspase Kit (Luminex Corporate, Austin) according to the manufacturer's protocol. The cells were analyzed using a MUSE flow cytometer (Luminex Corporate). For the T cell function study, mouse spleen cells were collected using the same method as described for cells from mesenteric lymph nodes. Next, spleen cells were stained for surface markers and intracellular cytokines using standard flow cytometric protocols. The following antibodies (Miltenyi biotec, Bergisch Gladbach, Germany) were used for cell staining: anti-CD3 (17A2), anti-CD4 (REA604), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-IFN-γ (XMG1.2), anti-IL-2 (REA665), anti-TNF-α (REA636). Cells were stimulated in complete RPMI-1640 + 10% FBS with leukocyte activation cocktail (BD, San Jose, CA) at 2 µl/mL. After being cultured at 37°C for 16 h, cells were collected and washed with PBS. 50 μL aqua blue (Life Technologies, Carlsbad, CA) was used at 4°C for 20 min to exclude dead cells, and cells were stained with surface markers and intracellular markers. Cells were analyzed using a BD FACSverse flow cytometer (BD) and data were analyzed by FlowJo software (Version 10.0.8).
Plasma LPS measurement
Fresh blood samples or endotoxin-free water (a negative control, Catalog number: W50-640, LONZA, Walkersville, MD, USA) in EDTA-containing tubes (BD, San Jose, CA, USA) were centrifuged at 800 g for 15 min, which was followed by transferring the samples to new centrifuge tubes (Catalog number: 352098, BD). Plasma and water controls were placed in aliquots and stored at −80°C. We avoided repeated freezing and thawing. Plasma LPS levels were measured according to our previous publication.23 Endpoint chromogenic limulus amebocyte lysate assays kit (Lonza, Basel, Switzerland) was used in the detection according to the manufacturer's protocol. Samples were 1:10 diluted with endotoxin-free water and subsequently heated to 85°C for 15 min to inactivate inhibitory proteins. LPS levels were calculated based on the standards. The background was subtracted using corresponding sample controls without adding chromogenic substrate.
Statistical analysis
One-way ANOVA tests were used to compare differences among more than two groups. Comparisons of individual gene expression in RNA-seq data were tested using the Wald test in DESeq2 package; p values were corrected using the Benjamini-Hochberg method. Comparison analysis was performed using R (version 3.3.1) or GraphPad Prism 8, and p ≤ 0.05 was considered statistically significant.
Ethics approval and consent to participate
This study was approved by Medical University of South Carolina institutional review boards (IBC-2019-00745). All participants provided written informed consent. All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Medical University of South Carolina (IACUC-2019-00858).
Role of the funding source
No entity other than the authors listed played any role in the design of the study; the collection, analysis, or interpretation of the data; writing of the report; or in the decision to submit the paper for publication.
Results
Increased systemic microbial translocation in HIV INRs
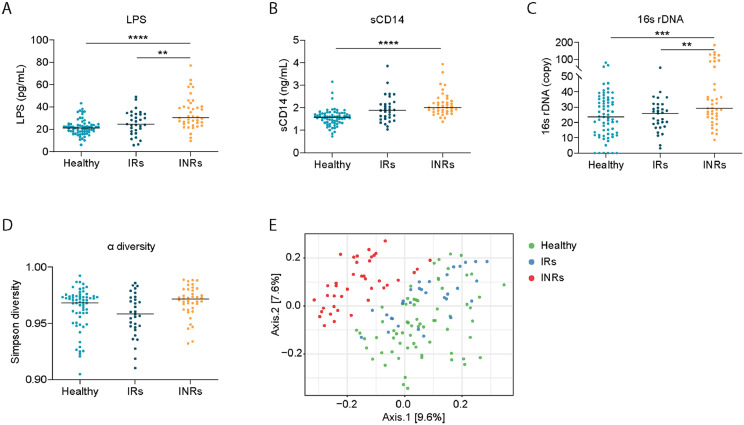
Plasma levels of LPS, total bacterial 16s rDNA, and soluble CD14 (sCD14) are proven markers of systemic microbial translocation, were evaluated in 31 HIV IRs, 40 HIV INRs, and 68 healthy controls. As expected, plasma levels of LPS, sCD14, and total bacterial 16s rDNA were significantly increased in INRs compared to healthy controls; but no difference was observed between IRs and healthy controls (Figure 1A–C).
Figure 1.
Systemic microbial translocation and distinct circulating microbial profiles in INRs. Plasma levels of LPS (A), sCD14 (B), and bacterial 16S rDNA (C) from the three study groups. (D) The Gini Simpson diversity index (α-diversity) was used to compare the diversity of the plasma-circulating microbial community within INRs and IRs or healthy controls. (E) PCoA was conducted based on the unweighted UniFrac distance to determine the beta diversity of the plasma microbial community. One-way ANOVA was used, followed by Tukey's post hoc test. The statistical significance of the beta diversity was tested using the Multivariate Welch t-test. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Distinct circulating microbiome in HIV INRs
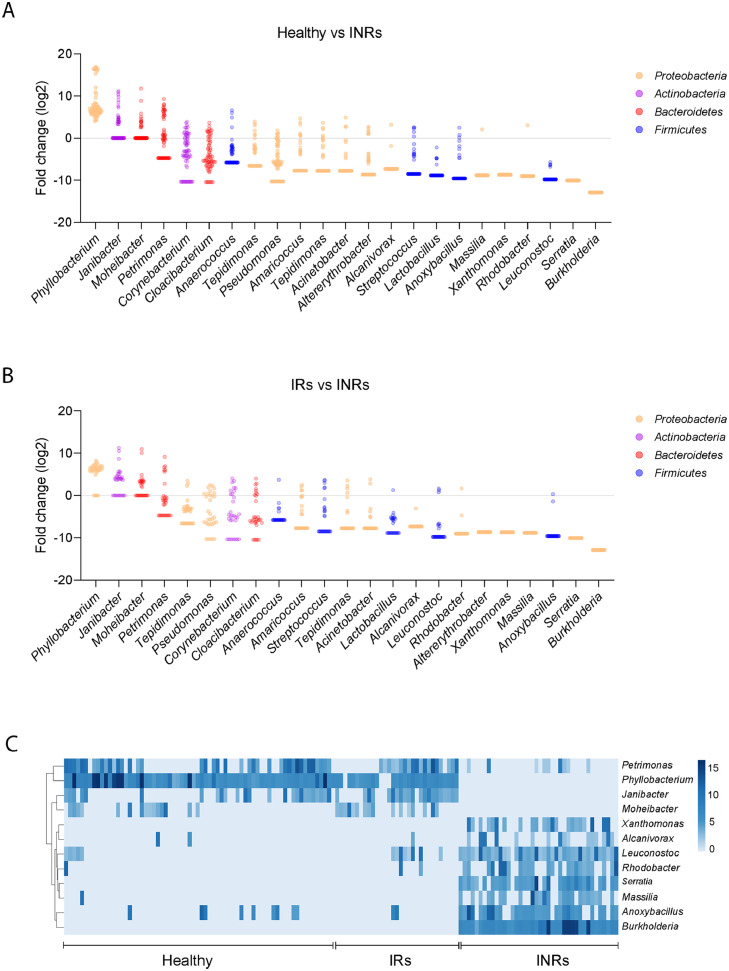
The plasma microbiome is known to contribute to immune perturbations and disease pathogenesis.23,30 To study the role of the plasma microbiome in immune failure in HIV+ individuals under suppressive ART, we performed microbial 16S rRNA sequencing. It has been reported that contamination and false-positive microbial DNA signals may generate noise during sequencing.31 Here, we focused on an extremely low-biomass microbiome sample (blood) and presented the identification of atypical human microbiome signals, it is critical to remove the background and artifacts from sequencing data to obtain accurate results from plasma microbiome analysis. After carefully removing potential contaminants using blank extractions according to our previous methods,32 there was no difference in Gini Simpson (alpha-diversity) index among the three study groups (Figure 1D). However, unweighted UniFrac phylogenetic distance analysis showed that INRs had a significantly altered circulating microbiome community compared to the other two groups, whereas similar communities were found between IRs and healthy controls (Figure 1E). After excluding the ASVs with a low prevalence across samples,32 Actinobacteria and Bacteroidetes phylum were predominantly decreased in INRs compared to IRs and healthy controls (Figure 2A). The top increased phylum in INRs compared to IRs were Proteobacteria and Firmicutes (Figure 2B). At the class level, both IRs and INRs had increased Gammaproteobacteria and decreased Alphaproteobacteria compared to healthy controls (Figure S1A). The abundance of Alphaproteobacteria and Bacteroidia was increased in healthy controls and less in IRs. The abundance of Bacteroidia was almost absent in INRs (Figure S1A). After adjusting p-values for multiple comparisons (Benjamini and Hochberg false-discovery rate [FDR]), 12 ASVs at the genus level showed significant differences in INRs relative to IRs and healthy controls, including four decreased taxa (Phyllobacterium, Janibacter, Moheibacter, and Petrimonas) and eight increased taxa (Burkholderia, Serratia, Xanthomonas, Massilia, Alcanivorax, Leuconostoc, Anoxybacillus, and Rhodobacter) (Figure 2C).
Figure 2.
Differentially abundant bacterial taxa from circulating microbiome. Dots represent ASVs assigned to the indicated bacterial (column label). The y-axis indicates the relative abundance of ASVs in healthy individuals or IRs compared to INRs. Bacterial ASVs with significant differences between healthy control versus INRs (A) and IRs versus INRs (B) were graphed by log2 fold-change (y-axis). Different colors show various genus bacteria and their affiliated phylum bacteria. (C) Heatmap showing the relative abundance of the 12 bacteria that differed the most (y-axis) by sample (x-axis) when INRs were compared with IRs or healthy controls. The gradient key indicates percent abundance.
To verify the enrichment of translocated bacterial antigens in INRs, we evaluated plasma levels of antibodies against bacterial lysate or LPS extracted from Phyllobacterium, Burkholderia, and Serratia (Figure S1B). Consistently, plasma levels of IgGs against Phyllobacterium myrsinacearum LPS were increased in healthy controls and IRs compared to those in INRs (Figure S1C). However, the plasma levels of IgGs against LPS from Burkholderia fungorum or Serratia marcescens were similar among the three groups (Figure S1C). Nonetheless, plasma levels of IgGs against B. fungorum and S. marcescens bacterial lysates were increased in INRs compared to IRs and healthy controls (Figure S1D). In terms of IgA, plasma levels of IgA against all three bacterial lysates were decreased in INRs compared to healthy controls (Figure S1E).
Variation in LPS structure from blood predominant microbiome in INRs and IRs
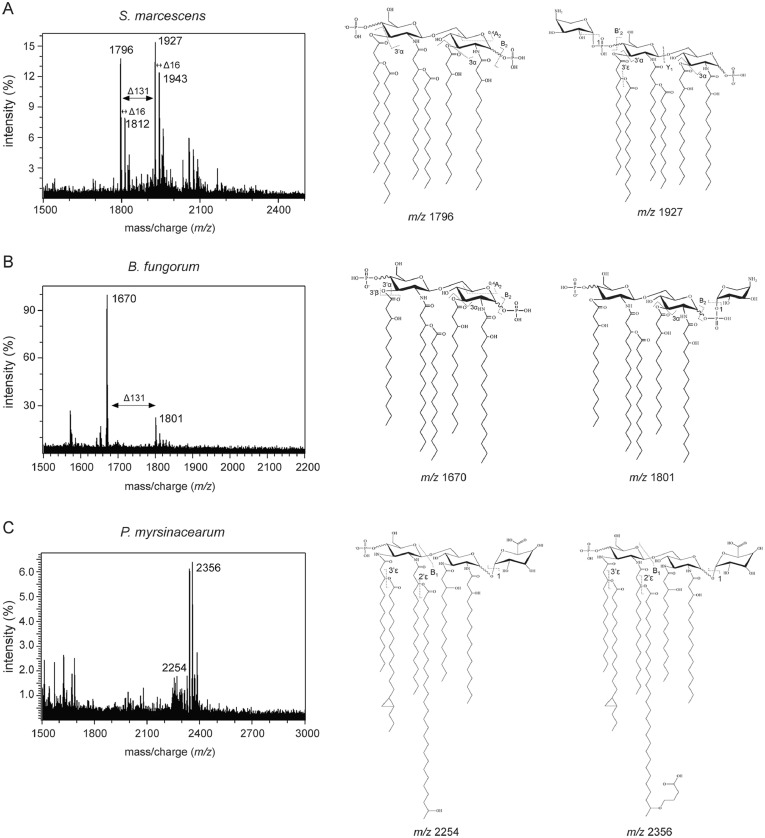
LPS is the major component of the Gram-negative bacterial outer membrane and is highly antigenic with conserved structural molecular motifs. The lipid A domain of LPS is responsible for immune signaling through the myeloid differentiation factor 2 (MD2)/TLR4 coreceptor complex.33 Structural changes in lipid A impact TLR4 recognition and downstream immune responses.34 We utilized mass spectrometry (MS) to analyze the structure of the lipid A domain of three bacterial species identified in this study.35 Notably, the lipid A from S. marcescens presented two predominant peaks at mass-to-charge ratio (m/z) of 1,796 and 1,927, which was structurally similar to E. coli lipid A,33 a potent agonist of MD2/TLR4, which is the most common immunostimulatory lipid A structure. However, important diversifications from the canonical E. coli lipid A structure was observed as hydroxylation and aminoarabinose modification of the terminal phosphate moiety of lipid A from S. marcescens (Figure 3A). Lipid A extracted from B. fungorum presented predominant peaks at m/z of 1,670 and 1,801. The lipid A observed from B. fungorum showed longer primary acyl chains at the 2- and 2’- positions and lacking the typical 3’-acyl-oxo-acyl chain. Aminoarabinose modification was also found in lipid A from B. fungorum. Aminoarabinose modification of lipid A has been linked to the susceptibility to antibiotics (e.g., Colistin resistance)36 (Figure 3B). In contrast, lipid A from P. myrsinacearum showed an extremely elongated 2’-acyl-oxo-acyl modification (28 carbon) similar to those seen in other environmental bacterial species (e.g., Agrobacterium),37 which had a predominant peak at a m/z of 2,356 (Figure 3C). These elongated, exotic lipid A structures have been predicted to be far too long to sit in the binding pocket of MD2 and expected to fail in stimulating TLR4.38
Figure 3.
Determination of lipid A structures. (A–C) MALDI-MS and MS/MS analysis of lipid A from S. marcescens (A), B. fungorum (B), and P. myrsinacearum (C). Representative major structures with predicted mass of the deprotonated ion are shown. Evidence of hydroxylation shown as delta m/z 16 and aminoarabinose as delta m/z 131.
Gene signatures of myeloid cell responses to the bacterial LPS enriched in IRs and INRs
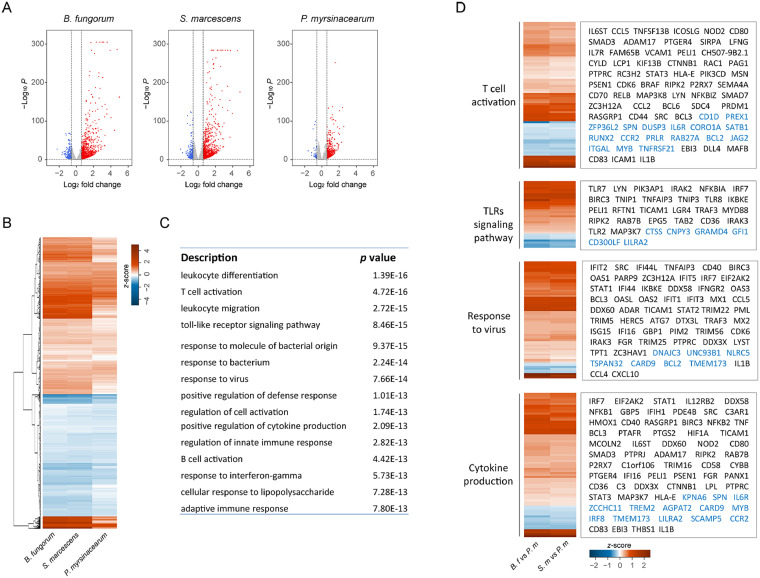
In order to understand the consequences of the structural differences between the enriched bacterial LPS from two patient populations, we first analyzed RNA gene expression profiles in a human myeloid cell line THP-1 cells, which express high levels of TLR4. RNA-seq transcriptome was analyzed in THP-1 cells treated with LPS from three bacteria enriched in IRs and INRs in vitro. In general, B. fungorum and S. marcescens LPS induced a more robust cell activation compared to P. myrsinacearum. Compared to unstimulated THP-1 cells, a total of 5711 and 6275 gene expression was regulated by B. fungorum and S. marcescens, respectively (FDR adjusted p < 0.05). In contrast, 4866 genes were regulated by P. myrsinacearum (FDR adjusted p < 0.05) (Figure 4A). B. fungorum and S. marcescens induced a similar gene expression pattern compared to P. myrsinacearum (Figure 4B). Further, gene ontology (GO) is used to characterize the function of regulated genes; the top functional entities were “leukocyte differentiation, activation, and migration”, “response to bacterium and TLR signaling pathways”, “response to virus and regulation of defense response”, and “cell activation and cytokine production” represented cellular responses to B. fungorum or S. marcescens compared to those in P. myrsinacearum treated cells (Figure 4C). Although all three tested bacteria induced the gene expression related to the TLR response, B. fungorum and S. marcescens induced greater TLR-related gene expression compared to those responses to P. myrsinacearum. Moreover, B. fungorum and S. marcescens increased TLR2, TLR7, TLR8 expression and upregulated genes of cytokines when compared with P. myrsinacearum (Figure 4D).
Figure 4.
Gene expression profiles of THP-1 cells after stimulation with different bacterial LPS. (A) Volcano plots of differential expression genes (DEGs, threshold fold change > 1.5 and FDR < 0.05) in THP-1 cells stimulated with LPS from each bacterium compared with unstimulated THP-1 cells. Red dots indicate upregulated DEGs and blue dots indicate downregulated DEGs. The horizontal and vertical dark green lines indicate fold-change (log2) thresholds and adjusted p-value (log10). (B) Heatmap of fold changes (log2) in gene expression for B. fungorum, S. marcescens, or P. myrsinacearum stimulated THP-1 cells compared to unstimulated THP-1 cells. (C) The most altered GO pathways enriched in coherently changed genes in THP-1 cells stimulated by B. fungorum or S. marcescens versus P. myrsinacearum. (D) The expression of various genes encoding products in selected pathways showing increased (red) or decreased (blue) expression in THP-1 cells treated with B. fungorum (B. f) or S. marcescens (S. m) compared to P. myrsinacearum (P. m).
LPS from B. fungorum and S. marcescens but not P. myrsinacearum induce CD4+ T cell apoptosis and proinflammatory responses
To study the causality of microbiome-blunted CD4+ T cell recovery in HIV, we evaluated CD4+ T cell apoptosis in response to LPS from B. fungorum, S. marcescens, and P. myrsinacearum in peripheral blood mononuclear cells (PBMC) or in isolated CD4+ T cells from healthy subjects in vitro. LPS from B. fungorum and S. marcescens increased CD4+ T cell apoptosis in PBMCs (Figure S2A) but not in purified CD4+ T cells (Figure S2B). In general, healthy human CD4+ T cells express low to undetectable levels of toll-like receptors (TLRs) and do not respond to most TLR ligands directly.39,40 These results suggest LPS from INR enriched bacteria induces CD4+ T cell apoptosis indirectly.
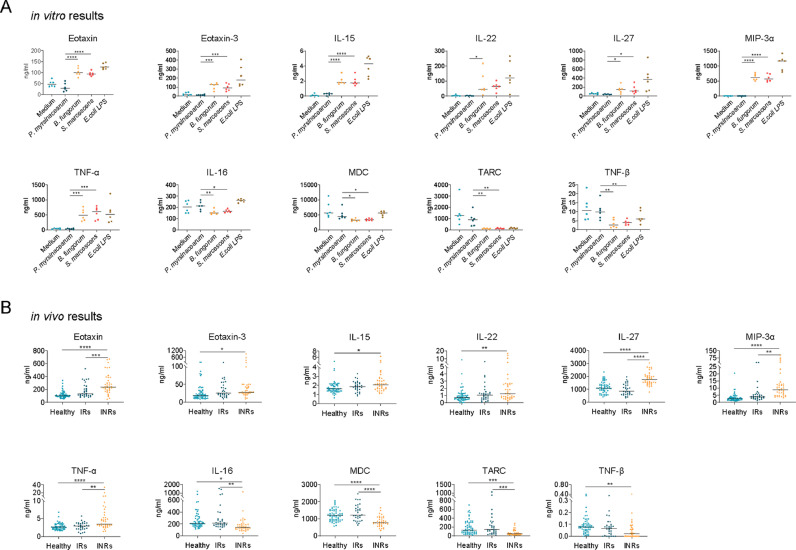
Given that LPS from B. fungorum and S. marcescens induced CD4+ T cell apoptosis indirectly, we evaluated cytokine and chemokine production in PBMC culture supernatants after culturing with LPS from three bacteria and LPS from E. coli 055:B5 as a positive control. LPS from B. fungorum and S. marcescens showed a proinflammatory response like pattern similar to LPS from E coli, including increased production of Eotaxin, Eotaxin-3, IL-15, IL-22, IL-27, MIP-3α, TNF-α, IFN-γ, IL-10, IL-12/IL-23p40, IL-12p70, IL-1α, IL-1β, IL-4, IL-6, IL-7, IL-8, MCP-1, MIP-1α, MIP-1β, and VEGF (Figures 5A, S3A), and decreased production of IL-16, MDC, TARC, TNF-β, IL-5, and IP-10 (Figures 5A, S3B), compared to those treated with P. myrsinacearum LPS.
Figure 5.
Plasma cytokines in INRs in vivo were consistent with B. fungorum and S. marcescens-induced cytokines in vitro. (A) The extracted LPS from B. fungorum, S. marcescens, and P. myrsinacearum were used to stimulate peripheral blood mononuclear cells (PBMC) from healthy subjects; LPS from E. coli 055:B5 was chosen as a positive control. The LPS induced multiplex cytokines in the cell culture supernatants were measured. (B) Plasma levels of cytokines in INRs compared with IRs or healthy controls. One-way ANOVA followed by Tukey's post hoc test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To further understand the link between the plasma microbiome and its-mediated proinflammatory responses in INRs in vivo, we evaluated plasma levels of proinflammatory cytokine and chemokine patterns. Although a more complicated pattern was observed in humans in vivo compared to the results in vitro (Figures 5B, S4), many cytokine or chemokine patterns in INRs in vivo were consistent with those in vitro in PBMCs in response to LPS of B. fungorum and S. marcescens (Figure 5A, B); plasma levels of Eotaxin, Eotaxin-3, IL-15, IL-22, IL-27, MIP-3α, and TNF-α were increased in INRs compared to those in IRs or healthy controls (Figure 5A, B), and levels of IL-16, MDC, TARC, and TNF-β were decreased in INRs compared to levels in IRs or healthy controls (Figure 5A, B).
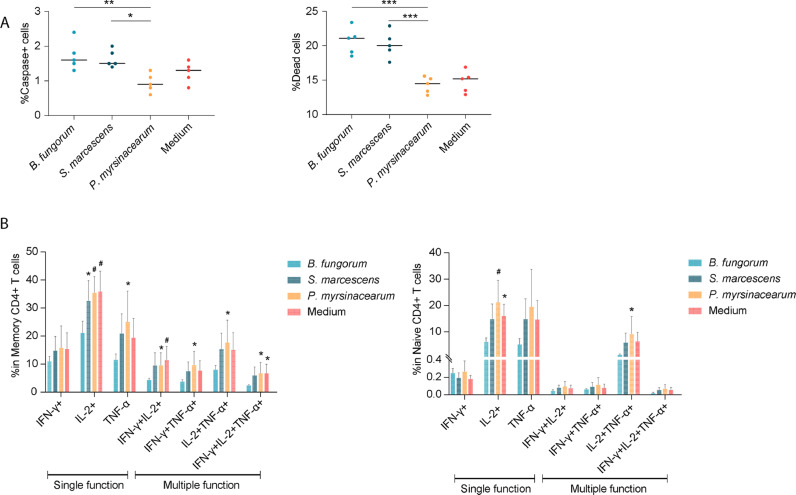
B. fungorum and S. marcescens induce intestinal lymph node cell apoptosis and impair CD4+ T cell function in mice
To further verify the effects of B. fungorum and S. marcescens on CD4+ T cell recovery in INRs, B6 mice were intraperitoneally (i.p.) injected with phosphate-buffered saline (PBS), heat-killed B. fungorum, S. marcescens, or P. myrsinacearum, twice per week for total of eight weeks. LPS structure is highly sensitive to temperature and growth condition changes resulting in unique structural characteristics.41,42 Thus, we injected mice with whole inactivated bacteria but not purified LPS. Notably, B. fungorum and S. marcescens administration resulted in cell apoptosis from mesenteric lymph nodes compared to those with injection of PBS or P. myrsinacearum (Figure 6A). Next, to evaluate CD4+ T cell function after i.p. injection of bacteria in the spleen cells from mice, we conducted a standard T cell functional assay for cytokine production in response to a leukocyte activation cocktail. Note, injection of B. fungorum impaired CD4+ T cell function compared to the controls (Figure 6B, C). These results suggested that B. fungorum and S. marcescens induced cell apoptosis from intestinal lymph nodes, and B. fungorum further impaired CD4+ T cell function.
Figure 6.
B. fungorum and S. marcescens, the bacteria enriched in INRs, impaired CD4+ T cell function. 6-week-old healthy C57BL/6 mice were injected with PBS, heat-killed B. fungorum, S. marcescens, or P. myrsinacearum twice a week for eight weeks by i.p. route (n = 5 per group). The mice were sacrificed on day three after the last injection (day 1). (A) The percentage of apoptotic cells and dead cells in the mesenteric lymph nodes. One-way ANOVA followed by Tukey's post hoc test, *p < 0.05, **p < 0.01. (B, C) The portion of functional memory CD4+ T cells (B) and functional naïve CD4+ T cells (C) from spleen. The spleen CD4+ T cells were stimulated using a leukocyte activation cocktail after 18 h and measured cytokine production using flow cytometry. One-way ANOVA followed by Dunnett's multiple comparisons test. *p < 0.05, #p < 0.01.
Discussion
In the current study, both quantitative and qualitative plasma microbial translocation differed in INRs compared to IRs and healthy individuals. Further, INR-enriched microbial LPS induced proinflammatory responses, CD4+ T cell apoptosis, and CD4+ T cell dysfunction, whereas IR-enriched microbial LPS did not exhibit such pathogenic activity. Structural analysis of LPS from P. myrsinacearum, mainly enriched in IRs, showed a structure with low predicted TLR4 stimulatory properties. LPS from B. fungorum and S. marcescens, mainly enriched in INRs, showed a lipid A structure closely related to the canonical TLR4 agonist, E. coli lipid A.
Results from previous studies on plasma levels of microbial translocation in INRs compared to IRs in HIV show either significantly increased or slightly increased.10,43 Consistently, we found that INRs have increased plasma levels of total microbial or microbial product translocation compared to IRs and healthy controls, suggesting a compromised mucosal barrier in INRs. Next, not only the total levels of microbial translocation but also the microbial composition differed in INRs versus IRs. Gut microbial dysbiosis has been associated with poor CD4+ T cell recovery in HIV-1 infected individuals on ART.44 A previous study found sexual preference has been shown to affect gut microbiome diversity, gut microbiota in MSM had higher Shannon diversity than non-MSM among HIV+ individuals.45 In our study, the alpha diversity of plasma microbiome in IRs tends to be lower than in INRs, although did not achieve significance. The current study reports a unique finding in the INRs, i.e., the consensus loss of colonization resistance to exogenous Burkholderia or Serratia. The enrichment of Burkholderia and Serratia in INRs were further confirmed by bacterial antigen-specific IgG in plasma. In contrast, IgAs of B. fungorum and S. marcescens were decreased in INRs. HIV infection is associated with impaired IgA production.46 The impaired IgA production specific to the enriched bacteria in INRs may stem from reduced CD4+ T cell counts and function and its mediated impairment of antibody class switch recombination to IgA,47 and may contribute to reduced mucosal surface protection against bacteria.48
Inter-species differences in LPS structure are associated with alterations of immunoregulatory properties.49 A recent study reveals that Bacterioides LPS, enriched in gut microbiome from a population with low prevalence of early-onset autoimmune diseases, is structurally distinct from E. coli LPS, and inhibits innate immune signaling and endotoxin tolerance; early colonization by immunologically silencing microbiota (i.e., B. dorei) may contribute to early immune education and low prevalence of autoimmune diseases.49 Moreover, B. dorei previously associated with type 1 diabetes pathogenesis.50 Notably, the lipid A domain of LPS is the minimal structure sufficient for TLR4 recognition of Gram-negative bacteria; TLR4 is expressed on the surface of many types of immune cells (i.e., myeloid cells). The recognition of LPS by TLR4 activates an intracellular signaling cascade and mediates proinflammatory responses, which plays a role in both innate and adaptive immune responses as well as autoimmune disease pathogenesis.51, 52, 53, 54 Besides binding to TLR4, the lipid A of LPS can directly bind to human caspase-4 and mouse homolog caspase-11 (caspase-4/11) with high specificity and affinity, which is next to activate caspase-4/11 then triggers cell pyroptosis and IL-1β/18 release.55,56
In the current study, the lipid A from B. fungorum and S. marcescens exhibited stimulatory properties in monocytes. In contrast, the lipid A from P. myrsinacearum exhibited weak stimulation of monocytes, which is consistent with the predicted profile of LPS from a beneficial bacterial species (Figure 4).49 Moreover, the enriched bacterial LPS from INRs and IRs was evaluated for their activities of inducing innate immune responses and apoptosis and function of CD4+ T cells in vitro. INR-enriched bacterial LPS showed largely proinflammatory responses and pathologic strain for inducing CD4+ T cell apoptosis and dysfunction. The gut microbial LPS with different degree of acylation on the lipid A was found related to the level of inflammation in HIV, indicating a link between proinflammatory LPS and inflammation.57 Indeed, INRs displayed increased spontaneous cell activation in both monocytes and DCs compared with those in cells from IRs.58 HIV-associated monocyte activation and chronic inflammation, reflected by elevated levels of proinflammatory cytokines and monocyte dysfunction, play a critical role in disease pathogenesis despite ART.59 Previous studies have shown that antiretroviral drug combinations affect the gut microbiome, immune activation and microbial translocation.60 In this study, there was no difference of ART regimens in IRs and INRs. However, the relative small sample size prevents us to draw further conclusions. Moreover, the majority of CD4+ T cells are depleted from the gut lymphoid tissues during HIV/SIV infection, regardless of the route of exposure.61 In this study, B. fungorum and S. marcescens LPS had extensive abilities to promote proinflammatory responses by human monocytes; i.p. injection of inactivated B. fungorum and S. marcescens, resulted in intestinal lymph node cell apoptosis in B6 mice. Both bacteria and bacterial LPS promoted cell apoptosis in vitro and in vivo. In general, human CD4+ T cells express low to undetectable TLRs and do not directly respond to bacterial products.39,40 We found that B. fungorum and S. marcescens increased apoptotic CD4+ T cells in PBMCs but not in isolated CD4+ T cells, suggesting that the accessory cells in PBMCs (e.g., monocytes) or cytokines mediated CD4+ T cell apoptosis in response to bacteria.
Previous studies show that TLR4-mediated responses contributed to persistent cell activation in response to LPS in vivo in HIV.62 However, cells may respond differently to different bacterial LPS as LPS from different strains of bacteria has shown different immunoregulatory properties.63 A previous study found that the initial binding of Burkholderia spp to the cell surface receptors triggered the proinflammatory signaling, which facilitated bacterial invasion into host epithelial cells.64 Our in vitro results indicated that LPS from B. fungorum and S. marcescens, but not from P. myrsinacearum, induced robust proinflammatory chemokine and cytokine responses. Notably, the overall plasma cytokine and chemokine patterns in INRs in vivo were consistent with those in B. fungorum and S. marcescens LPS-treated PBMCs in vitro. Intriguingly, RNAseq revealed that the gene profiles in THP-1 cells treated with LPS from B. fungorum and S. marcescens predicted T cell activation and viral responses. Those findings indicate that circulation translocation of LPS from proinflammatory bacterial strains (i.e., B. fungorum and S. marcescens) might contribute to persistent elevated cell activation and inflammation in INRs. Proinflammatory cytokines (e.g., TNF-α) have been shown to increase HIV infection.65, 66, 67, 68 Given that B. fungorum and S. marcescens LPS induced proinflammatory cytokines in vitro, their LPS-mediated inflammation may play a role in HIV infection or latency, which deserves further investigations. In contrast, LPS from P. myrsinacearum induced limited inflammation and thus prevented persistent monocyte activation, inflammation, and CD4+ T cell death. In this study, the LPS structure of a representative species was assigned and the resulting proinflammatory profile analyzed for the three implicated genera. Lipid A structural diversity can be observed at an intra-genus level and refinement of the species identity and respective lipid A structural assignments are worth future investigations. Furthermore, it is critical for future studies to determine the mechanism of translocated pathogens or microbial antigens-mediated remodeling of immune responses and disease progression in HIV.69
The limitation in this study includes: 1) the source of the enriched microbial translocation in INRs is not clear, comparing the stool and oral microbiota with blood microbiota would be essential to understand the origin of microbiota in blood. Thus, it would be important to study microbiome from these paired samples from different sites in the future. 2) We tested one species in each genus. However, the other strains within the same genus were not tested and may contribute to immune reconstruction. Furthermore, besides LPS, other microbial products or antigens may play a role in immune failure in HIV. 3) Using myeloid cells line THP-1 cells to analyze LPS-regulated gene expression instead of using primarily myeloid cells from patients. 4) No information of sexual preference and BMI in each individual, which were found as major confounders to affect the gut microbiome in HIV+ individuals.45,70 We also do not have information of gastrointestinal disease, liver disease, and medications of prophylactic antibiotics, probiotics, and steroids, which may impact gut microbiome and microbial translocation.
Conclusions
The present study analyzed the systemic translocated microbiome associated with CD4+ T cell counts in ART-treated HIV-infected subjects. We found that different structures of lipid A domain from plasma enriched bacterial LPS in INRs and IRs; unlike LPS of IR-enriched bacteria, LPS of INR-enriched bacteria activated monocytes to produce proinflammatory cytokines or chemokines and increased CD4+ T cell apoptosis. The enriched plasma bacteria from INRs belonged to proinflammatory bacterial strains, whereas the enriched plasma bacteria in IRs belonged to non-inflammatory bacterial strains. We conclude that variation of circulating microbial LPS contributes to chronic inflammation and immune reconstitution failure in HIV patients on ART. In the future, a therapeutic strategy targeting anti-bacterial peptides,71 inhibitors for inflammatory bacterial strain LPS-mediated pathogenesis, or changing diet, together with ART, could improve CD4+ T cell recovery and reduce chronic immune activation and inflammation, morbidity, and mortality in HIV.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We would like to thank John D. Dinolfo, Ph.D. from MUSC for English Language editing. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID; R01 AI128864, Jiang) (NIAID; P30 AI027767, Saag/Health), the Medical Research Service at the Ralph H. Johnson VA Medical Center (merit grant VA CSRD MERIT I01 CX-002422, Jiang), and the National Institute of Aging (R21 AG074331, Scott). The SCOPE cohort was supported by the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763, Gandhi) and the CFAR Network of Integrated Clinical Systems (R24 AI067039, Saag). The National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001450 (the pilot grant, Jiang).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104037.
Contributor Information
Alison Scott, Email: aliscott@umaryland.edu.
Wei Jiang, Email: jianw@musc.edu, wei.jiang1@va.gov.
Appendix. Supplementary materials
References
- 1.Gazzola L., Tincati C., Bellistri G.M., Monforte A., Marchetti G. The absence of CD4+ T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: clinical risk, immunological gaps, and therapeutic options. Clin Infect Dis. 2009;48(3):328–337. doi: 10.1086/595851. [DOI] [PubMed] [Google Scholar]
- 2.Robbins G.K., Spritzler J.G., Chan E.S., et al. Incomplete reconstitution of T cell subsets on combination antiretroviral therapy in the AIDS clinical trials group protocol 384. Clin Infect Dis. 2009;48(3):350–361. doi: 10.1086/595888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewden C., Chene G., Morlat P., et al. HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J Acquir Immune Defic Syndr. 2007;46(1):72–77. doi: 10.1097/QAI.0b013e318134257a. [DOI] [PubMed] [Google Scholar]
- 4.Baker J.V., Peng G., Rapkin J., et al. CD4+ count and risk of non-AIDS diseases following initial treatment for HIV infection. AIDS. 2008;22(7):841–848. doi: 10.1097/QAD.0b013e3282f7cb76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutierrez F., Padilla S., Masia M., et al. Patients' characteristics and clinical implications of suboptimal CD4 T-cell gains after 1 year of successful antiretroviral therapy. Curr HIV Res. 2008;6(2):100–107. doi: 10.2174/157016208783885038. [DOI] [PubMed] [Google Scholar]
- 6.Lapadula G., Chatenoud L., Gori A., et al. Risk of severe non AIDS events is increased among patients unable to increase their CD4+ T-cell counts >200+/mul despite effective HAART. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0124741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battegay M., Nüesch R., Hirschel B., Kaufmann G.R. Immunological recovery and antiretroviral therapy in HIV-1 infection. Lancet Infect Dis. 2006;6(5):280–287. doi: 10.1016/S1473-3099(06)70463-7. [DOI] [PubMed] [Google Scholar]
- 8.Baker J.V., Peng G., Rapkin J., et al. CD4+ count and risk of non-AIDS diseases following initial treatment for HIV infection. AIDS. 2008;22(7):841. doi: 10.1097/QAD.0b013e3282f7cb76. (London, England) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapadula G., Cozzi-Lepri A., Marchetti G., et al. Risk of clinical progression among patients with immunological nonresponse despite virological suppression after combination antiretroviral treatment. AIDS. 2013;27(5):769–779. doi: 10.1097/QAD.0b013e32835cb747. [DOI] [PubMed] [Google Scholar]
- 10.Lederman M.M., Calabrese L., Funderburg N.T., et al. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. J Infect Dis. 2011;204(8):1217–1226. doi: 10.1093/infdis/jir507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley C.F., Kitchen C.M., Hunt P.W., et al. Incomplete peripheral CD4+ cell count restoration in HIV-infected patients receiving long-term antiretroviral treatment. Clin Infect Dis. 2009;48(6):787–794. doi: 10.1086/597093. an official publication of the Infectious Diseases Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz A., Alos L., Leon A., et al. Factors associated with collagen deposition in lymphoid tissue in long-term treated HIV-infected patients. AIDS. 2010;24(13):2029–2039. doi: 10.1097/QAD.0b013e32833c3268. [DOI] [PubMed] [Google Scholar]
- 13.Hunt P.W., Martin J.N., Sinclair E., et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187(10):1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 14.Teixeira L., Valdez H., McCune J.M., et al. Poor CD4 T cell restoration after suppression of HIV-1 replication may reflect lower thymic function. AIDS. 2001;15(14):1749–1756. doi: 10.1097/00002030-200109280-00002. [DOI] [PubMed] [Google Scholar]
- 15.Shenoy M.K., Fadrosh D.W., Lin D.L., et al. Gut microbiota in HIV–pneumonia patients is related to peripheral CD4 counts, lung microbiota, and in vitro macrophage dysfunction. Microbiome. 2019;7(1):1–16. doi: 10.1186/s40168-019-0651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur U.S., Shet A., Rajnala N., et al. High Abundance of genus Prevotella in the gut of perinatally HIV-infected children is associated with IP-10 levels despite therapy. Sci Rep. 2018;8(1):1–16. doi: 10.1038/s41598-018-35877-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monaco C.L., Gootenberg D.B., Zhao G., et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19(3):311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchetti G., Tincati C., Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26(1):2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poore G.D., Kopylova E., Zhu Q., et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puri P., Liangpunsakul S., Christensen J.E., et al. The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology. 2018;67(4):1284–1302. doi: 10.1002/hep.29623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schierwagen R., Alvarez-Silva C., Madsen M.S.A., et al. Circulating microbiome in blood of different circulatory compartments. Gut. 2019;68(3):578–580. doi: 10.1136/gutjnl-2018-316227. [DOI] [PubMed] [Google Scholar]
- 22.Luo Z., Alekseyenko A.V., Ogunrinde E., et al. Rigorous plasma microbiome analysis method enables disease association discovery in clinic. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.613268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Z., Li M., Wu Y., et al. Systemic translocation of Staphylococcus drives autoantibody production in HIV disease. Microbiome. 2019;7(1):25. doi: 10.1186/s40168-019-0646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorensen M., Chandler C.E., Gardner F.M., et al. Rapid microbial identification and colistin resistance detection via MALDI-TOF MS using a novel on-target extraction of membrane lipids. Sci Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-78401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H., Chandler C.E., Jackson S.N., et al. On-tissue derivatization of lipopolysaccharide for detection of lipid A using MALDI-MSI. Anal Chem. 2020;92(20):13667–13671. doi: 10.1021/acs.analchem.0c02566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogunrinde E., Zhou Z., Luo Z., et al. A link between plasma microbial translocation, microbiome, and autoantibody development in first-degree relatives of systemic lupus erythematosus patients. Arthritis Rheumatol. 2019;71(11):1858–1868. doi: 10.1002/art.40935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fricker A.M., Podlesny D., Fricke W.F. What is new and relevant for sequencing-based microbiome research? A mini-review. J Adv Res. 2019;19:105–112. doi: 10.1016/j.jare.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Z., Alekseyenko A., Ogunrinde E., et al. Rigorous plasma microbiome analysis method enables disease association discovery in clinic. Front Microbiol. 2020;11:3350. doi: 10.3389/fmicb.2020.613268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vatanen T., Kostic A.D., d'Hennezel E., et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajjar A.M., Ernst R.K., Tsai J.H., Wilson C.B., Miller S.I. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3(4):354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- 35.Scott A.J., Post J.M., Lerner R., et al. Host-based lipid inflammation drives pathogenesis in Francisella infection. Proc Natl Acad Sci. 2017;114(47):12596–12601. doi: 10.1073/pnas.1712887114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panta P.R., Kumar S., Stafford C.F., et al. A DedA family membrane protein is required for Burkholderia thailandensis colistin resistance. Front Microbiol. 2019;10:2532. doi: 10.3389/fmicb.2019.02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Castro C., Molinaro A., Lanzetta R., Silipo A., Parrilli M. Lipopolysaccharide structures from Agrobacterium and Rhizobiaceae species. Carbohydr Res. 2008;343(12):1924–1933. doi: 10.1016/j.carres.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 38.Scott A.J., Oyler B.L., Goodlett D.R., Ernst R.K. Lipid A structural modifications in extreme conditions and identification of unique modifying enzymes to define the Toll-like receptor 4 structure-activity relationship. Biochim Biophys Acta (BBA) Mol Cell Biol Lipids. 2017;1862(11):1439–1450. doi: 10.1016/j.bbalip.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornung V., Rothenfusser S., Britsch S., et al. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168(9):4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 40.Funderburg N., Luciano A.A., Jiang W., Rodriguez B., Sieg S.F., Lederman M.M. Toll-like receptor ligands induce human T cell activation and death, a model for HIV pathogenesis. PLoS One. 2008;3(4):e1915. doi: 10.1371/journal.pone.0001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar G.S., Jagannadham M.V., Ray M.K. Low-temperature-induced changes in composition and fluidity of lipopolysaccharides in the antarctic psychrotrophic bacterium Pseudomonas syringae. J Bacteriol. 2002;184(23):6746–6749. doi: 10.1128/JB.184.23.6746-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtis M.A., Percival R.S., Devine D., et al. Temperature-dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defenses. Infect Immun. 2011;79(3):1187–1193. doi: 10.1128/IAI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchetti G., Bellistrì G.M., Borghi E., et al. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS. 2008;22(15):2035–2038. doi: 10.1097/QAD.0b013e3283112d29. [DOI] [PubMed] [Google Scholar]
- 44.Lu W., Feng Y., Jing F., et al. Association between gut microbiota and cd4 recovery in HIV-1 infected patients. Front Microbiol. 2018;9:1451. doi: 10.3389/fmicb.2018.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelpi M., Vestad B., Hansen S.H., et al. Impact of human immunodeficiency virus–related gut microbiota alterations on metabolic comorbid conditions. Clin Infect Dis. 2020;71(8):e359–ee67. doi: 10.1093/cid/ciz1235. [DOI] [PubMed] [Google Scholar]
- 46.Belec L., Meillet D., Gaillard O., et al. Decreased cervicovaginal production of both IgA1 and IgA2 subclasses in women with AIDS. Clin Exp Immunol. 1995;101(1):100–106. doi: 10.1111/j.1365-2249.1995.tb02284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiao X., He B., Chiu A., Knowles D.M., Chadburn A., Cerutti A. Human immunodeficiency virus 1 Nef suppresses CD40-dependent immunoglobulin class switching in bystander B cells. Nat Immunol. 2006;7(3):302–310. doi: 10.1038/ni1302. [DOI] [PubMed] [Google Scholar]
- 48.Macpherson A.J., Geuking M.B., McCoy K.D. Homeland security: IgA immunity at the frontiers of the body. Trends Immunol. 2012;33(4):160–167. doi: 10.1016/j.it.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Vatanen T., Kostic A.D., d'Hennezel E., et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis-Richardson A.G., Triplett E.W. A model for the role of gut bacteria in the development of autoimmunity for type 1 diabetes. Diabetologia. 2015;58(7):1386–1393. doi: 10.1007/s00125-015-3614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Umiker B.R., Andersson S., Fernandez L., et al. Dosage of X-linked Toll-like receptor 8 determines gender differences in the development of systemic lupus erythematosus. Eur J Immunol. 2014;44(5):1503–1516. doi: 10.1002/eji.201344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong C.K., Wong P.T., Tam L.S., Li E.K., Chen D.P., Lam C.W. Activation profile of Toll-like receptors of peripheral blood lymphocytes in patients with systemic lupus erythematosus. Clin Exp Immunol. 2010;159(1):11–22. doi: 10.1111/j.1365-2249.2009.04036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thibault D.L., Chu A.D., Graham K.L., et al. IRF9 and STAT1 are required for IgG autoantibody production and B cell expression of TLR7 in mice. J Clin Investig. 2008;118(4):1417–1426. doi: 10.1172/JCI30065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwasaki A., Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327(5963):291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagar J.A., Powell D.A., Aachoui Y., Ernst R.K., Miao E.A. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341(6151):1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi J., Zhao Y., Wang Y., et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514(7521):187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 57.Storm-Larsen C., Stiksrud B., Eriksen C., et al. Microbial translocation revisited: targeting the endotoxic potential of gut microbes in HIV-infected individuals. AIDS. 2019;33(4):645–653. doi: 10.1097/QAD.0000000000002087. [DOI] [PubMed] [Google Scholar]
- 58.Stiksrud B., Aass H.C., Lorvik K.B., Ueland T., Trøseid M., Dyrhol-Riise A.M. Activated dendritic cells and monocytes in HIV immunological nonresponders: HIV-induced interferon-inducible protein-10 correlates with low future CD4+ recovery. AIDS. 2019;33(7):1117. doi: 10.1097/QAD.0000000000002173. (London, England) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deeks S.G. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinto-Cardoso S., Klatt N.R., Reyes-Terán G. Impact of antiretroviral drugs on the microbiome: unknown answers to important questions. Currt Opin HIV AIDS. 2018;13(1):53. doi: 10.1097/COH.0000000000000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veazey R.S. Intestinal CD4 Depletion in HIV /SIV Infection. Curr Immunol Rev. 2019;15(1):76–91. doi: 10.2174/1573395514666180605083448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L., Luo Z., Sieg S.F., et al. Plasmacytoid dendritic cells mediate synergistic effects of HIV and lipopolysaccharide on CD27+ IgD–memory B cell apoptosis. J Virol. 2014;88(19):11430–11441. doi: 10.1128/JVI.00682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vatanen T., Kostic A.D., d'Hennezel E., et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(6):1551. doi: 10.1016/j.cell.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 64.Gillette D.D., Shah P.A., Cremer T., et al. Analysis of human bronchial epithelial cell proinflammatory response to Burkholderia cenocepacia infection: inability to secrete il-1β. J Biol Chem. 2013;288(6):3691–3695. doi: 10.1074/jbc.C112.430298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duh E.J., Maury W.J., Folks T.M., Fauci A.S., Rabson A.B. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci USA. 1989;86(15):5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Folks T.M., Clouse K.A., Justement J., et al. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci USA. 1989;86(7):2365–2368. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffin G.E., Leung K., Folks T.M., Kunkel S., Nabel G.J. Activation of HIV gene expression during monocyte differentiation by induction of NF-kappa B. Nature. 1989;339(6219):70–73. doi: 10.1038/339070a0. [DOI] [PubMed] [Google Scholar]
- 68.Osborn L., Kunkel S., Nabel G.J. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc Natl Acad Sci USA. 1989;86(7):2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brenchley J.M., Price D.A., Schacker T.W., et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 70.Turnbaugh P.J., Hamady M., Yatsunenko T., et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansson M.E., Hansson G.C. Microbiology. Keeping bacteria at a distance. Science. 2011;334(6053):182–183. doi: 10.1126/science.1213909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.