Abstract
Face masks do not completely prevent transmission of respiratory infections, but masked individuals are likely to inhale fewer infectious particles. If smaller infectious doses tend to yield milder infections, yet ultimately induce similar levels of immunity, then masking could reduce the prevalence of severe disease even if the total number of infections is unaffected. It has been suggested that this effect of masking is analogous to the pre-vaccination practice of variolation for smallpox, whereby susceptible individuals were intentionally infected with small doses of live virus (and often acquired immunity without severe disease). We present a simple epidemiological model in which mask-induced variolation causes milder infections, potentially with lower transmission rate and/or different duration. We derive relationships between the effectiveness of mask-induced variolation and important epidemiological metrics (the basic reproduction number and initial epidemic growth rate, and the peak prevalence, attack rate and equilibrium prevalence of severe infections). We illustrate our results using parameter estimates for the original SARS-CoV-2 wild-type virus, as well as the Alpha, Delta and Omicron variants. Our results suggest that if variolation is a genuine side-effect of masking, then the importance of face masks as a tool for reducing healthcare burdens from COVID-19 may be under-appreciated.
Keywords: infectious diseases, COVID-19, variolation, face masks, SIR model, differential equations
1. Introduction
Early in the COVID-19 pandemic, face masking was discouraged outside healthcare settings [1,2], and inadequate supplies of masks created significant challenges for healthcare workers [3–5]. By 3 April 2020 appreciation of the potential value of masking had increased sufficiently for the US Centers for Disease Control and Prevention (CDC) to recommend the wearing of face coverings in public [6]. Similar recommendations were made in Canada on 6 April 2020 [7] and in the UK on 11 May 2020 [8]. Over the course of the pandemic, evidence that masking is an effective tool to reduce community transmission of SARS-CoV-2 has continued to accumulate [9–16].
An additional potential benefit of masking—even in situations where it fails to prevent transmission—was proposed in the summer of 2020 by Gandhi & Rutherford [17] and Gandhi et al. [18]. They noted that if transmission occurs in the presence of face masks, then the infecting viral inoculum is likely to be smaller than is typical when masks are not worn. If smaller inocula tend to lead to less severe infections, then—even if masks fail to block transmission completely—masking could reduce morbidity and mortality from COVID-19 and boost the level of herd immunity in the population.
The notion that promoting mild infections could be an effective disease control strategy has a long history. In the eighteenth century, it was common to infect children with smallpox intentionally—a process known as variolation—by administering a small inoculum of smallpox virus taken from an infected person [19–24]. While SARS-CoV-2 infections are never intentional, Gandhi & Rutherford [17] refer to SARS-CoV-2 transmission via small inocula that penetrate masks as variolation. In this paper, we explore the potential benefits of SARS-CoV-2 variolation using a mathematical model.
We begin in §2 by reviewing evidence that supports the hypothesis that face masking may promote SARS-CoV-2 variolation. In subsequent sections, we present and analyse a simple model that allows us to investigate potential effects of variolation induced by face masking.
2. Variolation as a side effect of face masking
Face masks can reduce the probability of transmission either through outward or inward filtration. Outward filtration occurs when droplets containing viral inoculum are captured when leaving the mouth or nose of an infected person [25]. Inward filtration occurs when viral particles are prevented from entering someone’s nose or mouth.
Gandhi & Rutherford [17] suggested that people who are infected with SARS-CoV-2 when wearing a face mask might experience less severe illness, because the size of the viral inoculum they receive is reduced by inward filtration. Assuming that individuals infected in this way still develop lasting immunity, they can be considered variolated. Variolation in this sense might not reduce the total proportion of the population infected, but would nevertheless benefit the population by increasing the proportion of infections that are mild or asymptomatic [17].
The variolation hypothesis, as originally formulated by Gandhi & Rutherford [17], depends on three key assumptions:
Reduced inocula: individuals who are infected while wearing a mask receive a smaller viral inoculum than if they had not been masked.
Reduced severity: smaller viral inocula tend to yield infections with milder symptoms, i.e. there is a positive dose–response relationship.
Acquired immunity: mild infections still provide long-lasting natural immunity to the disease.
Reduced inocula is plausible because masks are known to filter droplets that may contain virus particles [26].
Reduced severity from smaller inocula is indicated by several lines of evidence. The dose–response relationship is supported by an investigation in Madrid that found that ‘distinct sizes of viral inoculum at the time of exposure’ could explain different illness courses in three clusters of SARS-CoV-2 infection (infecteds in different clusters developed COVID-19 with different severity, and the only clear difference among the clusters was the degree of SARS-CoV-2 exposure) [27]. In addition, disproportionately severe disease outcomes were documented following a choir rehearsal in the state of Washington in March 2020 [28]; the observed severity of illness among attendees is consistent with a positive dose–response relationship because infected individuals likely spew many more virus particles when singing, compared to simply talking at a social gathering. Finally, an experimental study exposed hamsters to SARS-CoV-2 in a laboratory and found that COVID-19 symptoms were much less severe in infected animals that were shielded by a surgical mask partition [29].
Immunity after mild infection is suggested by a 2020 study in the New York City area, where 13.7% of a sample of healthcare personnel (HCP) were found to have antibodies to SARS-CoV-2 [30]; HCP were not asked if they were symptomatic, but 5% (11%) of those who tested positive for SARS-CoV-2 antibodies reported low (medium) likelihood of SARS-CoV-2 exposure. Another study found that 15% of those with SARS-CoV-2 antibodies reported never having COVID-19 symptoms [31]. Other investigations have revealed that antibodies to SARS-CoV-2 were present in individuals five months after asymptomatic or mild infections [32,33]. Thus, asymptomatic and mild SARS-CoV-2 infections likely do provide at least some protection against reinfection.
3. Model
We investigate the potential effects of facemask-induced variolation by expanding the standard Susceptible-Infectious-Removed (SIR) model [34,35].
We assume that all members of the population are identical—and that the population is homogeneously mixed—so we do not distinguish between situations in which poor facemasks are worn universally by everyone versus situations in which better masks are worn by a subset of the population; thus, masks are implicitly assumed to be distributed to individuals at random.
We capture the variolating effect of facemasks by imagining that adherence to facemasking causes a proportion of infections to be mild, and we therefore refer to as the probability of mild infection. Mild infections might be shorter and/or less transmissible (however, the effective infectious period for severe infections could, in practice, be shorter than for mild infections, because severe cases are likely to be isolated quickly). Thus, three distinct effects—causing mild infections, reducing transmission rate and potentially shortening infectious periods—contribute to the overall effectiveness of mask-induced variolation. We allow for the possibility that immunity decays [36], but assume the duration of immunity is the same following mild or severe infections.
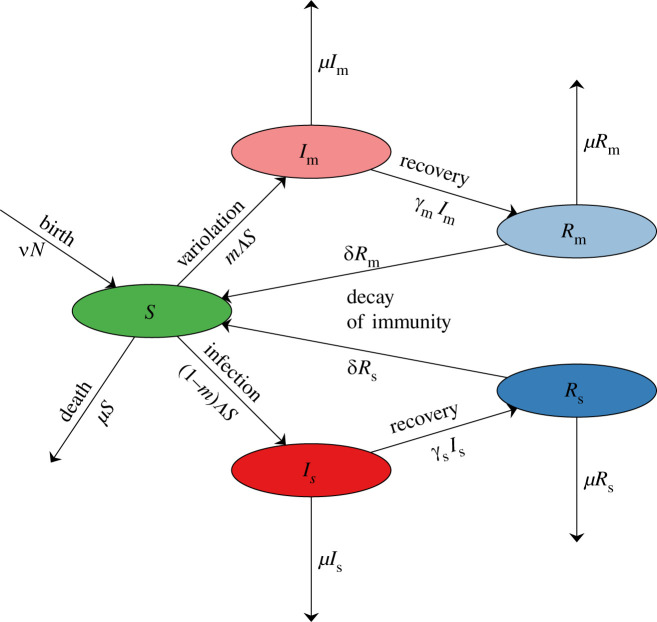
Figure 1 presents a flow chart for the model, which we represent formally as a system of ordinary differential equations (ODEs),
| 3.1a |
| 3.1b |
| 3.1c |
| 3.1d |
| 3.1e |
where the force of infection is
| 3.1f |
and the total population size is
| 3.1g |
Figure 1.
Flow chart for the model defined by equation (3.1). The state variables are defined in table 1, and the parameters are defined in table 3.
All the state variables are listed in table 1. and do not appear in the first three lines of equation (3.1) and can be ignored for the purposes of dynamical analysis. We retain equations (3.1d,e) for convenience, to keep track of the numbers of immune individuals who had mild versus severe infections.
Table 1.
State variables for the model specified in figure 1 and equation (3.1). All variables represent numbers of individuals.
| state variable | meaning |
|---|---|
| S | susceptible to infection |
| mildly infectious (variolated) | |
| severely infectious | |
| removed, after mild infection | |
| removed, after severe infection |
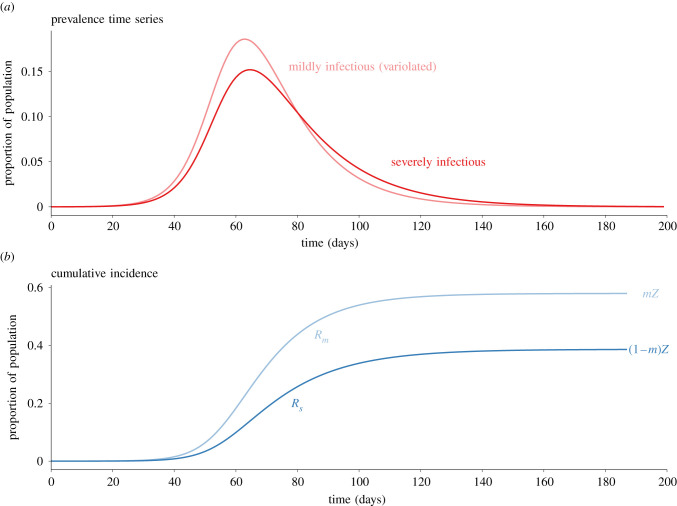
Estimated values of parameters associated with COVID-19 are listed in table 2. We use these estimates to set default values for the parameters of our model, which are listed in table 3. Since we ignore the latent stage in our model, we use the estimated mean generation interval for COVID-19 in place of the mean infectious period in our model (cf. [49,52]). We ignore disease-induced mortality, but we (more than) compensate for this by assuming for simplicity that the death rate from other causes (μ) is equal to the birth rate (ν); thus, births are balanced by deaths and the population size (N) is constant. Figure 2 shows prevalence and cumulative incidence time series, obtained by solving equation (3.1) numerically with the default parameters.
Table 2.
Parameter estimates (and 95% confidence intervals) for COVID-19; these estimates are examples from published studies and are not based on a systematic review of the literature. WT refers to the wild-type virus. The latent period (time from exposure to infectiousness) is extremely difficult to estimate, so the incubation period (time from exposure to appearance of symptoms) is often used as a proxy. The incubation period for the Alpha variant was estimated to be [37]. is expressed in terms of the model parameters in equation (4.1). for the Alpha, Delta and Omicron variants were approximated using estimates of the increase in transmissibility for each successive variant, i.e. [38], [39–41] and [42].
| parameter | meaning | estimate | references |
|---|---|---|---|
| Tlat | mean latent period | 3.7 [3.3–3.9] days | WT [43] |
| 4.0 [3.5–4.4] days | Delta [44] | ||
| Tinc | mean incubation period | 6.4 [4.9–8.5] days | WT [45] |
| 4.2 [3.2–5.6] days | Alpha [37] | ||
| 5.8 [5.2–6.4] days | Delta [44] | ||
| ∼3 days | Omicron [46] | ||
| Tinf,m | mean infectious period for mild infections | 9 [6–11] days | [47] |
| Tinf,s | mean infectious period for severe infections | 14 [8–20] days | [48] |
| Tgen,m | mean generation interval for mild infections | Tlat + Tinf,m days | [49, equation (4.1)] |
| Tgen,s | mean generation interval for severe infections | Tlat + Tinf,s days | [49, equation (4.1)] |
| Timm | mean immune period | 7 [6–8] months | [32,33,50] |
| basic reproduction number | 3 [2.1–4.6] | WT [38] | |
| 4.5 [3.15–6.9] | Alpha [51] | ||
| 6.75 [4.7–10.4] | Delta [39–41] | ||
| 28.4 [9.9–94.6] | Omicron [42] |
Table 3.
Parameters of the model described in figure 1 and equation (3.1). The probability with which masking causes infections to be mild () is unknown. Our default value is chosen to be substantial so that for illustrative graphs constructed with fixed (figures 5, 6 in §5) the effect of mask-induced variolation is non-negligible. The recovery rates can be interpreted as the rates of ‘recovery or death’ since we do not explicitly model disease-induced mortality (cf. final paragraph of §3). The death rate μ refers to mortality from causes other than the focal disease. Note that mild illness is assumed to be associated with mild infectiousness. All birth rates were estimated for the years 2015–2020. We use a default latent period of Tlat = 3.7 days for all variants. The generation interval for an SEIR model is Tlat + Tinf [49, eqn (4.1)]. Setting γx = 1/Tgen,x in our model yields dynamics more similar to an SEIR version (cf. [49,52]), so it is a better approximation of the real world than an SIR version with 1/γ taken to be the observed mean infectious period. The transmission rate for severe infections () is set for each variant using equation (4.1) with and the associated estimate listed in table 2. After specifying , we then set .
| parameter | meaning | expression or default value |
|---|---|---|
| probability that an infected individual develops mild illness | 0.6 | |
| transmission rate from mildly infectious individuals | equation (4.1) | |
| transmission rate from severely infectious individuals | equation (4.1) | |
| ratio of transmission rates | 1/2 | |
| recovery rate from mild infections | 1/Tgen,m | |
| recovery rate from severe infections | 1/Tgen,s | |
| ν | per capita annual birth rate [53] | 0.0105 (Canada) |
| 0.0115 (UK) | ||
| 0.012 (USA) | ||
| μ | per capita annual death rate | ν |
| δ | rate of decay of immunity | 1/Timm |
Figure 2.
Solution curves for the model (3.1) with the parameter values specified in table 3 (birth rate for Canada). The initial conditions were . Panel (b) shows the proportions in the removed compartments (approximately cumulative incidence in those compartments), which converge to the indicated values derived in equation (4.18).
4. Analysis
4.1. Basic reproduction number ()
The contribution to from infectors who are mildly infectious is the probability of mild infection (), times the transmission rate from mildly infectious individuals (), times the expected time that a mildly infectious individual is infectious (). There is a similar contribution from severely infectious individuals, hence
| 4.1 |
A formal calculation, applying the method of [54] to equation (3.1), yields the same expression.
4.2. Dimensionless stage durations
The mean time spent in the mildly infectious state, as a fraction of the mean lifetime, is
| 4.2a |
Similarly, for severe infections, we have
| 4.2b |
The mean across both types of infections is
| 4.2c |
It is worth noting that
| 4.3 |
The mean duration of immunity, as a fraction of the mean lifetime, is
| 4.4 |
4.3. Equilibria and stability
The system described by equation (3.1) always has a disease free equilibrium (DFE), at which S = N and all other compartments are empty. In addition, if then there is an endemic equilibrium (EE) given by
| 4.5a |
| 4.5b |
| 4.5c |
In the limit of permanent immunity (δ → 0, η → ∞), the final factors in equations (4.5b,c) simplify to 1. Since , decay of immunity (η < ∞) necessarily increases equilibrium prevalence. Theorem 2 of [54] establishes that is the boundary between local stability and instability of the DFE. In fact, the model (3.1) is a special case of the class of SIR models with multiple parallel infectious stages considered by Korobeinikov [55]. Consequently, theorem 1 of [55] establishes that the DFE is globally asymptotically stable (GAS) if , and that the EE is GAS if .
4.4. Initial growth rate (r)
The initial exponential growth rate of an epidemic beginning near the DFE of equation (3.1) is the largest eigenvalue of the Jacobian derivative of the vector field at the DFE. This Jacobian has a first column (− δ − μ, 0, 0), so −(δ + μ) is an eigenvalue. The other two eigenvalues are determined by the submatrix
| 4.6 |
If we write, for convenience,
| 4.7 |
then the larger of the two eigenvalues of (4.6) is
| 4.8 |
Note that, as written, the discriminant in equation (4.8) is manifestly positive if , but it can also be written1
| 4.9 |
which is manifestly positive if . In the limit of equal mild and severe infectious periods, the initial growth rate is simply
| 4.10 |
For any initial growth rate r (equation (4.8)), the initial doubling time, i.e. the time required for prevalence to double during the exponential growth phase, is
| 4.11 |
4.5. Final size
If there is no source of new susceptibles (ν = μ = δ = 0), we can derive a final size relation based on equation (3.1). Following [56,57], we look for a constant of the motion that is the sum of log(S/N) and a linear combination of the other state variables. Solving for coefficients that cause the time-derivative of this expression to vanish, we find that
is a constant of the motion. In particular, F(0) = F(∞); consequently, since as t → 0 and as t → ∞ (and noting that the coefficient of the final term in equation (4.12) is ) we have
| 4.13 |
Writing Z = 1 − S(∞)/N and noting that
| 4.14 |
we obtain the standard final size relation [34,57],
| 4.15 |
With the same approach, but insisting that the coefficient of vanishes, we find another constant of the motion
| 4.16 |
Considering the limits t → 0 and t → ∞ yields
| 4.17 |
From this expression and equations (4.14) and (4.15), we therefore have exact expressions for the mild and severe attack rates,
| 4.18 |
Solving equation (4.15) for Z [57], we can write
| 4.19 |
where W0 is the principal branch of Lambert’s W function [58]. In equation (4.19), we have emphasized the dependence of on (including the dependence of on , equation (4.1)), since we are especially interested in understanding how an increase in the proportion of cases that are mild influences the expected number of severe infections.
4.6. Peak prevalence
Another way of writing equation (4.12) is
| 4.20 |
which does not depend explicitly on the probability that an infection is mild (there is implicit dependence on through ; equation (4.1)). In the limit that mild and severe infections have the same reproduction number (), the left-hand side of equation (4.20) reduces to the total prevalence,
| 4.21 |
and equation (4.20) agrees exactly with the formula for the phase portrait I(S) of the standard SIR model. The right-hand side of equation (4.20) is maximized at , so inserting into the equation (4.20) yields the peak prevalence formula for the standard SIR model,
| 4.22 |
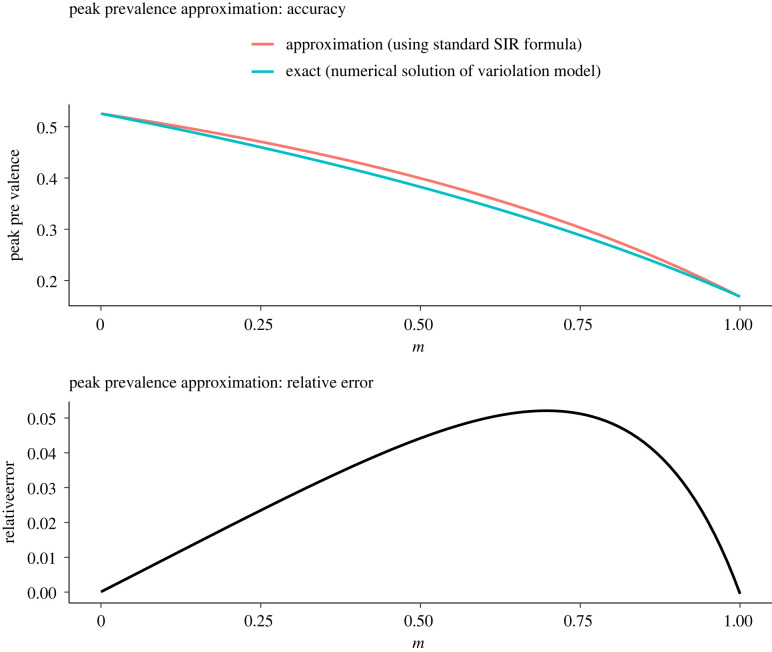
This formula will provide a good approximation to the peak total prevalence in our model (equation (3.1)) to the extent that the left-hand side of equation (4.20) approximates total prevalence near its peak. Figure 3 compares total prevalence (equation (4.21)) with the left-hand side of equation (4.20) for the full range of possible mask-induced increase in the probability of mild infection () with other parameters fixed at the values specified in tables 2 and 3. The figure indicates that approximating peak prevalence with the left-hand side of equation (4.20) is reasonable (the maximum relative error is 5.5%).
Figure 3.
Accuracy and relative error of the peak total prevalence approximated using the formula for the basic SIR model (equation (4.22)), as a function of the probability that an infection is mild (). Parameters other than are set to the values specified in tables 2 and 3. For each , the value of is computed using equation (4.1). The relative error is , where TRUE refers to the peak total prevalence obtained by numerically solving equation (3.1), and SIR refers to equation (4.22). The maximum relative error of the approximation is 5.5%, for (). In the extreme of perfect variolation (), , whereas in the extreme of completely ineffective variolation (), .
If we approximate the susceptible population S at peak total prevalence by , then inserting into equation (4.20) provides an estimate of how prevalence at its peak is partitioned between mild and severe cases. Of course, we can approximate the value of S at peak total prevalence to any desired accuracy by solving equation (3.1) numerically; with that approach, we can use equation (4.20) to determine, to any desired accuracy, the partitioning of peak prevalence between mild and severe cases.
In practice, the peak prevalence of severe cases is what is most important from the point of view of stress on health care systems, and it is simplest to find the peak of directly from numerical solutions of equation (3.1). However, it is important to bear in mind that is the number of individuals who are still contributing to transmission dynamics but are suffering from disease that is so severe that they will need substantial healthcare. does not include people who have already been isolated in hospitals or other settings (and can therefore be considered unable to cause further infections). Thus, is a measure of upcoming healthcare demand, as opposed to the current burden on the system. The peak healthcare burden can be estimated roughly by multiplying the peak of by TH/Tgen,s, where TH is the mean length of stay in hospital and is the mean time spent in the compartment of our model (cf. tables 2 and 3).
5. Illustration of results for COVID-19
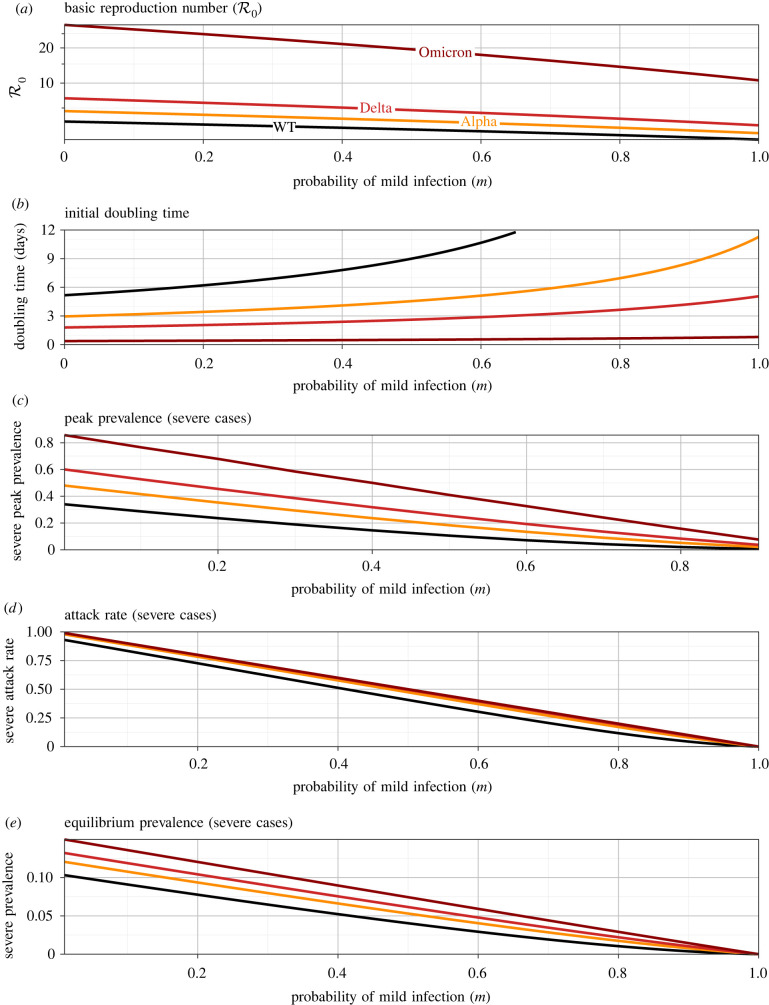
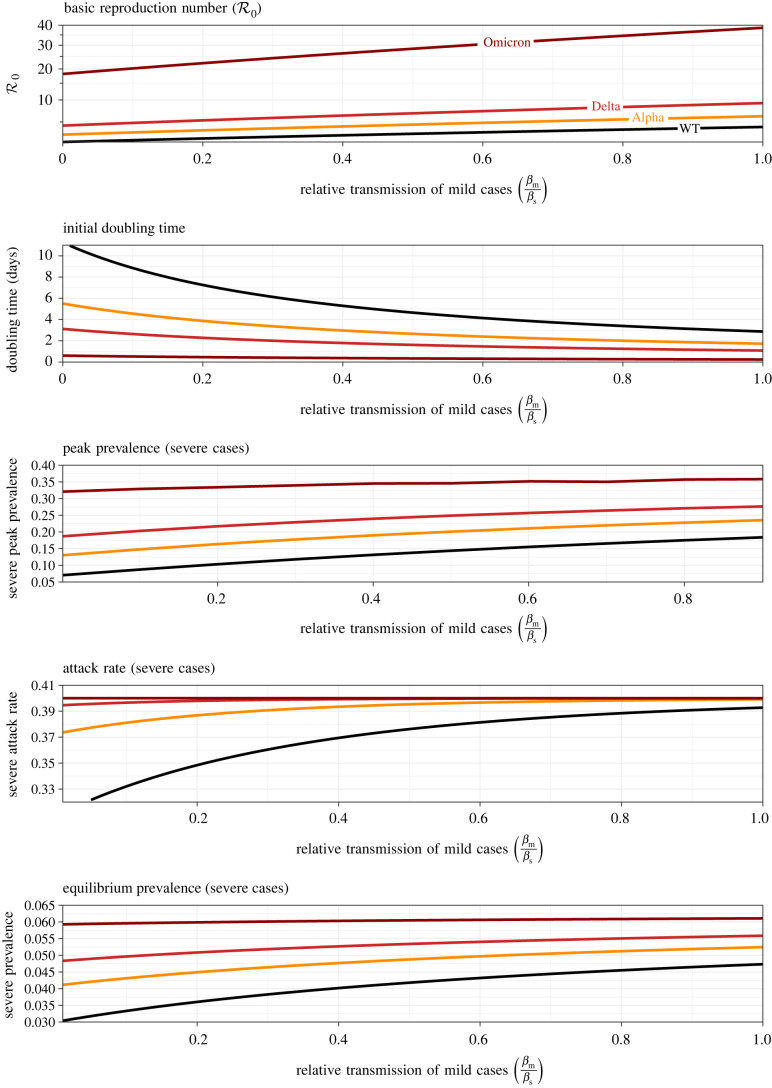
Figure 4 shows how several important epidemiological risk metrics depend on the variolating effect of masking, if it acts principally by increasing the probability of mild infection (). Each panel shows three curves, associated with the estimated basic reproduction number for the original wild-type SARS-CoV-2 virus (WT, ; [38]), the Alpha variant that began to spread in late 2020 (; [51]), the Delta variant that emerged in the spring of 2021 (; [39–41]) and the Omicron variant that emerged in late 2021 (; [42]). for each variant is assumed to take the observed value in the limit of no variolating effect () and then to decrease according to equation (4.1). All other parameters are fixed at the default values listed in tables 2 and 3; in particular, we assume that the transmission rates of mild and severe infections (, ) are independent of their probability of occurrence (, ).
Figure 4.
Epidemiological risk metrics as a function of the probability, , that an infection is mild (as a result of mask-induced variolation). In each panel, results are shown for three situations, corresponding to the original SARS-CoV-2 wild-type, and the Alpha, Delta and Omicron variants (for which , respectively; cf. table 2). (a) Basic reproduction number (equation (4.1)); the estimated values of for each variant are assumed to be associated with ineffective variolation (). All other parameters are set to the default values indicated in table 3 (the Canadian birth rate is assumed). (b) Doubling time T2 (equation (4.11)); for the original wild-type (WT), the curve extends far above the vertical maximum of the plotted graph (T2 → 65 days as ). (c) Peak prevalence of severe cases (§4.6). (d) Expected final proportion of the population that will have experienced a severe illness (equation (4.19)). (e) Equilibrium prevalence of severe cases (equation (4.5c)).
Figure 4b shows that the initial growth rate of the epidemic is strongly dependent on the probability , and is more sensitive to if the variant is more transmissible. As is increased from 0 to 1, the initial doubling time increases from 5.1 to 65 days for WT (upper limit not shown on the graph), 2.9 to 11 days for Alpha, 1.8 to 5.0 days for Delta and 0.37 to 0.79 days for Omicron. (Note that these are estimates of expected doubling times in a completely susceptible population, whereas the doubling times actually observed when the later variants emerged were in populations that already had substantial levels of immunity from previous infections and vaccination.)
Figure 4c–e shows that risk measures related to severe infections decline substantially with , as expected since severe illness is completely eliminated in the limit of perfect variolation (; all cases mild). Variolation has relatively greater effect (on prevalence of severe illness) for more transmissible variants.
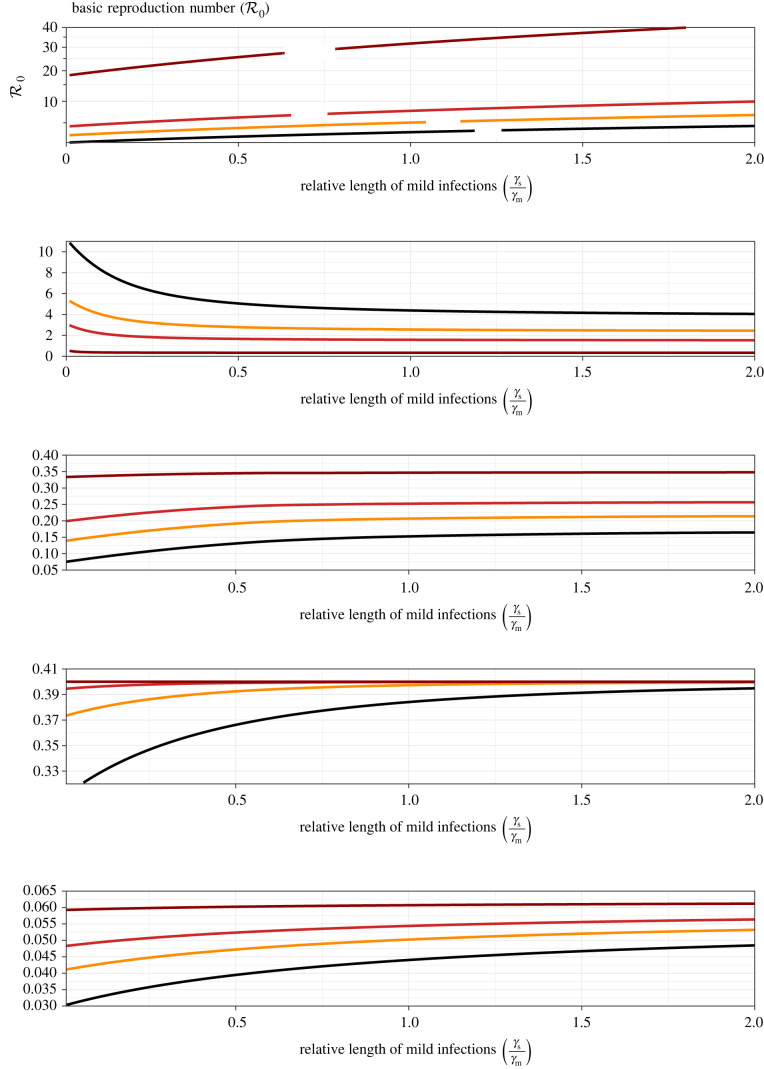
Figures 5 and 6 show how the same risk metrics depend on the relative transmissibility of mild infections () and the relative length of mild infections (). The horizontal scale in figure 5 ends at 1 because it is implausible that mild infections are intrinsically more transmissible than severe infections. By contrast, the horizontal scale in figure 6 extends beyond 1 because it is plausible that the time during which an infection can be transmitted is shorter for severe infections (e.g. severe cases are likely to be isolated sooner and more stringently). The similarities between figures 5 and 6 can be attributed to the fact that increasing or decreasing has a similar effect on (equation (4.1)).
Figure 5.
Epidemiological risk metrics as a function of the relative transmissibility of mild infections (). The panels correspond to those in figure 4, but the vertical axis ranges are different. The graphs were generated by varying while fixing all other parameters at their default values. The estimated values of (listed in table 2) are associated with (the default ratio listed in table 3).
Figure 6.
Epidemiological risk metrics as a function of the relative length of mild infections (). The panels correspond to those in figures 4 and 5, but with different vertical axis ranges. The graphs were generated by varying while fixing all other parameters at their default values. The estimated values of (listed in table 2) are associated with the ratio of generation intervals listed in table 3 ().
6. Discussion
We have explored the potential role of mask-induced variolation in reducing the impact of COVID-19 (or other directly transmitted infectious diseases). Our approach has been to analyse a highly idealized mathematical model (figure 1 and equation (3.1)) that captures the key mechanisms that we wished to investigate, namely the potential for masking to cause a proportion of infections to be mild, reduce the probability of infection upon contact and/or change the infectious period. Because our model does not attempt to be realistic in detail, conclusions we draw are qualitative only. However, the simplicity of the model has made it possible to derive results analytically, so qualitative conclusions can easily be drawn for pathogens with reproduction numbers or generation intervals that are different from those of SARS-CoV-2.
6.1. Summary of results
Our main results are analytical formulae that show how the model parameters—including the probability of mask-induced mild infection (), the relative transmissibility of mild infections () and the relative length of mild infections ()—influence the initial epidemic doubling time (equation (4.11)), the peak prevalence of severe infections (§4.6), the attack rate for severe infections (equation (4.19)) and the equilibrium prevalence of severe infections (equation (4.5c)). These results are illustrated for parameters that are representative of four variants of SARS-CoV-2 in figures 4–6.
-
—
Figure 4 shows that if masking primarily influences the probability that an infection is mild (), then more effective masking strongly affects transmission (reducing ) and lengthens the doubling time substantially, especially for less transmissible variants. In addition, the peak prevalence of severe cases is strongly affected (and the effect on peak prevalence is substantially greater for more transmissible variants). The expected number of severe cases during the initial wave of infections is also strongly dependent on , but is not sensitive to transmissibility over the range of observed for SARS-CoV-2 variants. The equilibrium prevalence of severe cases also declines with (equations (4.1) and (4.5c)).
-
—
Figures 5 and 6 show the effects of greater transmissibility of mild infections (increasing ), and longer infectious periods of mild cases (increasing ), respectively. The effects are similar, as might be expected given that (equation (4.1)).
Overall, increasing the effectiveness of mask-induced variolation—whether by increasing the probability that an infection will be mild, reducing the transmissibility of mild infections or reducing the length of mild infections—has the potential to drastically impact disease control, by slowing spread and reducing the magnitude of the epidemic peak (‘flattening the curve’ [59,60]), reducing the number of severe cases in the initial wave and reducing the prevalence of severe cases at equilibrium.
6.2. Limitations
The principal limitation of our analysis is that the hypothetical variolating effect of facemasks is unproven. We do not know that masking does tend to induce milder infections nor, if so, that variolated individuals attain a similar level of immunity as those who are infected in the absence of masks. Comments [61] and responses [62–64] to Gandhi & Rutherford’s initial article [17] make clear that further experimental and observational research is required. It is also worth noting that even if mask-induced variolation were very effective, promoting it could ‘implicitly encourage reckless behaviour’ [63] and consequently could present additional challenges for public health messaging.
6.3. Conclusion
Beyond qualitative conclusions, we are not likely to be able to make more powerful inferences without experimental studies that convincingly quantify the magnitudes of the effects that induce variolation from masking. If such experimental data do become available—and support the hypothesis that masking induces a substantial variolating effect—it will then be worth expanding our simple model (3.1) to include explicit latent periods, hospitalization, age and social structure (e.g. schools, workplaces) and heterogeneities in adherence to masking and other control measures. With appropriate data and more realistic models, we may be able to make quantitative inferences that could usefully inform policy decisions.
In the context of the highly transmissible Delta and Omicron variants [39–42], and the potential evolution of new SARS-CoV-2 variants that are even more transmissible and/or more successfully evade existing vaccines [65], a better understanding of the effectiveness of masking in promoting variolation could be of great value. At the time of writing, vaccines for children under 5 are not yet approved [66,67], but approval is expected soon [68,69]. While vaccine availability for people of all ages is imperative, substantial vaccine hesitancy [70] and breakthrough infections among the vaccinated [71–74], make achieving herd immunity through vaccination an unattainable target at present. If that situation persists, potential mask-induced variolation could contribute to COVID-19 control as we transition to endemicity.
Endnote
The equivalence of the expressions for the discriminant in (4.8) and (4.9) can be verified directly, or by recognizing that the results must be invariant to changing the labelling of mild and severe: m → s, s → m, m → (1 − m).
Contributor Information
Zachary Levine, Email: zachary.levine@weizmann.ac.il.
David J. D. Earn, Email: earn@math.mcmaster.ca.
Data accessibility
This article has no additional data.
Authors' contributions
Z.L.: formal analysis, funding acquisition, investigation, resources, software, visualization, writing—original draft, writing—review and editing; D.E.: conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing—original draft, writing—review and editing.
Both authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
Both authors received funding from the Natural Sciences and Engineering Research Council of Canada (NSERC): Z.L. was supported by an Undergraduate Student Research Award, and DJDE was supported by a Discovery Grant and an Emerging Infectious Diseases Modelling Initiative grant.
References
- 1.Dr Vivek Murthy, U S Surgeon General. 2020 The best way to protect yourself and your community is with everyday preventive actions, like staying home when you are sick and washing hands with soap and water, to help slow the spread of respiratory illness. Get your FluShot- fewer flu patients = more resources for COVID19. (accessed 15 July 2021). See https://twitter.com/Surgeon_General/status/1233726563881029632.
- 2.World Health Organization. 2020 Advice on the use of masks in the context of COVID-19: interim guidance, 6 April 2020. (accessed 15 July 2021). See https://apps.who.int/iris/handle/10665/331693.
- 3.Khazan O. 2020. Why we are running out of masks. See https://www.theatlantic.com/health/archive/2020/04/why-were-running-out-of-masks-in-the-coronavirus-crisis/609757/.
- 4.Sun L. 2020. Face mask shortage prompts CDC to loosen coronavirus guidance. Washington Post. See https://www.washingtonpost.com/health/2020/03/10/face-mask-shortage-prompts-cdc-loosen-coronavirus-guidance/.
- 5.Dyer E. 2020. The great PPE panic: how the pandemic caught Canada with its stockpiles down. See https://www.cbc.ca/news/politics/ppe-pandemic-covid-coronavirus-masks-1.5645120.
- 6.NPR. 2020 CDC Now Recommends Americans Consider Wearing Cloth Face Coverings In Public. (accessed 15 July 2021). See https://www.npr.org/sections/coronavirus-live-updates/2020/04/03/826219824/president-trump-says-cdc-now-recommends-americans-wear-cloth-masks-in-public.
- 7.Britneff B. 2020 Coronavirus: wearing non-medical masks will help others but not you, Tam says—National — Globalnews.ca. Global News. See https://globalnews.ca/news/6782934/coronavirus-non-medical-masks-community-covid-19/.
- 8.So E, Baker H. 2020. Mask or no mask? A look at UK’s policy over time. Centre for Research in the Arts, Social Sciences, and Humanities. See https://www.crassh.cam.ac.uk/blog/mask-or-no-mask-a-look-at-uks-policy-over-time/.
- 9.Stutt ROJH, Retkute R, Bradley M, Gilligan CA, Colvin J. 2020. A modelling framework to assess the likely effectiveness of facemasks in combination with lock-down in managing the COVID-19 pandemic. Proc. R. Soc. A 476, 20200376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aydin O, Emon B, Cheng S, Hong L, Chamorro LP, Saif MTA. 2020. Performance of fabrics for home-made masks against the spread of COVID-19 through droplets: a quantitative mechanistic study. Extreme Mech. Lett. 40, 100924. ( 10.1016/j.eml.2020.100924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks JT, Butler JC. 2021. Effectiveness of mask wearing to control community spread of SARS-CoV-2. JAMA 325, 998-999. ( 10.1001/jama.2021.1505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu DK, et al. 2020. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet 395, 1973-1987. ( 10.1016/S0140-6736(20)31142-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer EP, Fischer MC, Grass D, Henrion I, Warren WS, Westman E. 2020. Low-cost measurement of face mask efficacy for filtering expelled droplets during speech. Sci. Adv. 6, eabd3083. ( 10.1126/sciadv.abd3083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard J, et al. 2021. An evidence review of face masks against COVID-19. Proc. Natl Acad. Sci. USA 118, e2014564118. ( 10.1073/pnas.2014564118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrejko K, et al. 2022. SARS-CoV-2 incidence in K-12 school districts with mask-required versus mask-optional policies - Arkansas, August - October 2021. Morb. Mortal. Wkly Rep. (MMWR) 71, 384-389. ( 10.15585/mmwr.mm7110e1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donovan C, et al. 2022. Effectiveness of face mask or respirator use in indoor public settings for prevention of SARS-CoV-2 Infection- California, February - December 2021. Clin. Transl. Med. 71, 212-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandhi M, Rutherford GW. 2020. Facial masking for Covid-19: potential for variolation as we await a vaccine. N. Engl. J. Med. 383, e101. ( 10.1056/NEJMp2026913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi M, Beyrer C, Goosby E. 2020. Masks do more than protect others during COVID-19: reducing the inoculum of SARS-CoV-2 to protect the wearer. J. Gen. Intern. Med. 35, 3063-3066. ( 10.1007/s11606-020-06067-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krylova O, Earn DJD. 2020. Patterns of smallpox mortality in London, England, over three centuries. PLoS Biol. 18, e3000506. ( 10.1371/journal.pbio.3000506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss RA, Esparza J. 2015. The prevention and eradication of smallpox: a commentary on Sloane (1755) An account of inoculation. Phil. Trans. R. Soc. B 370, 20140378. ( 10.1098/rstb.2014.0378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Razzell P. 2011. The decline of adult smallpox in eighteenth-century London: a commentary. Econ. History Rev. 64, 1315-1335. ( 10.1111/j.1468-0289.2011.00620.x) [DOI] [PubMed] [Google Scholar]
- 22.Hopkins DR. 2002. The greatest killer: smallpox in history, with a new introduction. Chicago, IL: University of Chicago Press. [Google Scholar]
- 23.Geddes AM. 2006. The history of smallpox. Clin. Dermatol. 24, 152-157. ( 10.1016/j.clindermatol.2005.11.009) [DOI] [PubMed] [Google Scholar]
- 24.Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID, Organization WH. 1988. Smallpox and its eradication. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 25.Leung NHL, et al. 2020. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 26, 676-680. ( 10.1038/s41591-020-0843-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sande M, Teunis P, Sabel R. 2008. Professional and home-made face masks reduce exposure to respiratory infections among the general population. PLoS ONE 3, e2618. ( 10.1371/journal.pone.0002618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guallar MP, Meirino R, Donat-Vargas C, Corral O, Jouve N, Soriano V. 2020. Inoculum at the time of SARS-CoV-2 exposure and risk of disease severity. Int. J. Infect. Dis. 97, 290-292. ( 10.1016/j.ijid.2020.06.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamner L. 2020. High SARS-CoV-2 attack rate following exposure at a choir practice Skagit County, Washington, 3 2020. Morb. Mortal. Wkly Rep. 69, 606-608. ( 10.15585/mmwr.mm6919e6) [DOI] [PubMed] [Google Scholar]
- 29.Chan JFW, et al. 2020. Surgical mask partition reduces the risk of noncontact transmission in a Golden Syrian Hamster Model for Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 71, 2139-2149. ( 10.1093/cid/ciaa644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moscola J, Sembajwe G, Jarrett M, Farber B, Chang T, McGinn T, Davidson KW. 2020. Prevalence of SARS-CoV-2 antibodies in health care personnel in the New York City Area. JAMA 324, 893. ( 10.1001/jama.2020.14765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steensels D, Oris E, Coninx L, Nuyens D, Delforge ML, Vermeersch P, Heylen L. 2020. Hospital-wide SARS-CoV-2 antibody screening in 3056 staff in a Tertiary Center in Belgium. JAMA 324, 195. ( 10.1001/jama.2020.11160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds CJ, et al. 2020. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 5, eabf3698. ( 10.1126/sciimmunol.abf3698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wajnberg A, et al. 2020. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370, 1227-1230. ( 10.1126/science.abd7728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kermack W, McKendrick AG. 1927. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. A 115, 700-721. [Google Scholar]
- 35.Anderson RM, May RM. 1991. Infectious diseases of humans: dynamics and control. Oxford, UK: Oxford University Press. [Google Scholar]
- 36.Dushoff J, Plotkin JB, Levin SA, Earn DJD. 2004. Dynamical resonance can account for seasonality of influenza epidemics. Proc. Natl Acad. Sci. USA 101, 16 915-16 916. ( 10.1073/pnas.0407293101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homma Y, Katsuta T, Oka H, Inoue K, Toyoshima C, Iwaki H, Yamashita Y, Shinomiya H. 2021. The incubation period of the SARS-CoV-2 B1.1.7 variant is shorter than that of other strains. J. Infect. 83, 15-17. ( 10.1016/j.jinf.2021.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SW, Bolker BM, Champredon D, Earn DJD, Li M, Weitz JS, Grenfell BT, Dushoff J. 2020. Reconciling early-outbreak estimates of the basic reproductive number and its uncertainty: framework and applications to the novel coronavirus (SARS-CoV-2) outbreak. J. R. Soc. Interface 17, 20200144. ( 10.1098/rsif.2020.0144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H et al. 2021. Herd immunity induced by COVID-19 vaccination programs to suppress epidemics caused by SARS-CoV-2 wild type and variants in China. medRxiv. See https://www.medrxiv.org/content/10.1101/2021.07.23.21261013v1.
- 40.Dagpunar J. 2021. Interim estimates of increased transmissibility, growth rate, and reproduction number of the Covid-19 B.1.617.2 variant of concern in the United Kingdom. medRxiv. See https://www.medrxiv.org/content/10.1101/2021.06.03.21258293v1.
- 41.Challen R, et al. 2021. Early epidemiological signatures of novel SARS-CoV-2 variants: establishment of B.1.617.2 in England. medRxiv. See https://www.medrxiv.org/content/10.1101/2021.06.05.21258365v1.
- 42.Nishiura H, Ito K, Anzai A, Kobayashi T, Piantham C, Rodriguez-Morales AJ. 2022. Relative reproduction number of SARS-CoV-2 Omicron (B.1.1.529) compared with Delta variant in South Africa. J. Clin. Med. 11, 30. ( 10.3390/jcm11010030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J. 2020. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 368, 489-493. ( 10.1126/science.abb3221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang M, et al. 2021. Transmission dynamics and epidemiological characteristics of Delta variant infections in China. medRxiv. 2021.08.12.21261991. See https://www.medrxiv.org/content/10.1101/2021.08.12.21261991v1.
- 45.Bui LV, et al. 2020. Estimation of the incubation period of SARS-CoV-2 in Vietnam. PLoS ONE 15, e0243889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jansen L. 2021. Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) Variant Cluster in Nebraska, November to December 2021. MMWR Morbidity and Mortality Weekly Report. 70. See https://www.cdc.gov/mmwr/volumes/70/wr/mm705152e3.htm.
- 47.Sakurai A, Sasaki T, Kato S, Hayashi M, Tsuzuki SI, Ishihara T, Iwata M, Morise Z, Doi Y. 2020. Natural History of Asymptomatic SARS-CoV-2 Infection. N. Engl. J. Med. 383, 885-886. ( 10.1056/NEJMc2013020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voinsky I, Baristaite G, Gurwitz D. 2020. Effects of age and sex on recovery from COVID-19: analysis of 5769 Israeli patients. J. Infect. 81, e103. ( 10.1016/j.jinf.2020.05.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krylova O, Earn DJD. 2013. Effects of the infectious period distribution on predicted transitions in childhood disease dynamics. J. R. Soc. Interface 10, 20130098. ( 10.1098/rsif.2013.0098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dan JM, et al. 2021. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063. ( 10.1126/science.abf4063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Washington NL, et al. 2021. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 184, 2587-2594.e7. ( 10.1016/j.cell.2021.03.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Champredon D, Dushoff J, Earn DJD. 2018. Equivalence of the Erlang SEIR epidemic model and the renewal equation. SIAM J. Appl. Math. 78, 3258-3278. ( 10.1137/18M1186411) [DOI] [Google Scholar]
- 53.United Nations and Department of Economic and Social Affairs and Population Division. 2019 World Population Prospects Volume 1. United Nations.
- 54.van den Driessche P, Watmough J. 2002. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 180, 29-48. ( 10.1016/S0025-5564(02)00108-6) [DOI] [PubMed] [Google Scholar]
- 55.Korobeinikov A. 2009. Global properties of SIR and SEIR epidemic models with multiple parallel infectious stages. Bull. Math. Biol. 71, 75-83. ( 10.1007/s11538-008-9352-z) [DOI] [PubMed] [Google Scholar]
- 56.Anderson D, Watson R. 1980. On the spread of a disease with gamma distributed latent and infectious periods. Biometrika 67, 191-198. ( 10.1093/biomet/67.1.191) [DOI] [Google Scholar]
- 57.Ma J, Earn DJD. 2006. Generality of the final size formula for an epidemic of a newly invading infectious disease. Bull. Math. Biol. 68, 679-702. ( 10.1007/s11538-005-9047-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corless RM, Gonnet GH, Hare DEG, Jeffrey DJ, Knuth DE. 1996. On the Lambert W function. Adv. Comput. Math. 5, 329-359. ( 10.1007/BF02124750) [DOI] [Google Scholar]
- 59.The Economist. 2020. Covid 19 is now in 50 countries, and things will get worse. See https://www.economist.com/briefing/2020/02/29/covid-19-is-now-in-50-countries-and-things-will-get-worse.
- 60. Centers for Disease Control and Prevention. 2007 Interim pre-pandemic planning guidance: community strategy for pandemic influenza mitigation in the United States: early, targeted, layered use of nonpharmaceutical interventions. See https://stacks.cdc.gov/view/cdc/11425 .
- 61.Klompas M, Morris CA, Sinclair J, Pearson M, Shenoy ES. 2020. Universal masking in hospitals in the Covid-19 Era. N. Engl. J. Med. 382, e63. ( 10.1056/NEJMp2006372) [DOI] [PubMed] [Google Scholar]
- 62.Rasmussen A, Escandón K, Popescu S. 2020. Facial Masking for Covid-19. N. Engl. J. Med. 383, 2092. ( 10.1056/NEJMc2030886) [DOI] [PubMed] [Google Scholar]
- 63.Brosseau L. 2020. Facial Masking for Covid-19. N. Engl. J. Med. See https://www.nejm.org/doi/full/10.1056/NEJMc2030886.
- 64.Gandhi M. 2020. Facial Masking for Covid-19. N. Engl. J. Med. See https://www.nejm.org/doi/full/10.1056/NEJMc2030886.
- 65.Otto SP, et al. 2021. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 31, R918-R929. ( 10.1016/j.cub.2021.06.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Centers for Disease Control and Prevention. 2022 COVID-19 Vaccines for Children and Teens. See https://www.cdc.gov/coronavirus/2019-ncov/vaccines/recommendations/children-teens.html. [PubMed]
- 67.Public Health Agency of Canada. Vaccines for children: COVID-19. 2021. Last Modified: 28 January 2022. See https://www.canada.ca/en/public-health/services/vaccination-children/covid-19.html.
- 68.Johnson C. 2022. What to know about the coronavirus vaccine for children younger than 5. See https://www.washingtonpost.com/health/2022/01/11/covid-vaccine-kids-under-5/.
- 69.Bever L. 2022 Pfizer and BioNTech seek emergency use authorization of the first coronavirus vaccine for children under 5. See https://www.washingtonpost.com/health/2022/02/01/coronavirus-vaccine-under-5-pfizer/.
- 70.Bonyton S. 2021 Nearly 20% of Canadians still hesitant or refusing to get COVID-19 vaccine: poll. (accessed 7 September 2021). See https://globalnews.ca/news/7960345/covid-canada-vaccine-hesitancy-poll/.
- 71.Public Health Ontario. 2021 Confirmed Cases of COVID-19 Following Vaccination in Ontario: 14 December 2020 to 21 August 2021. (accessed 7 September 2021). See https://www.publichealthontario.ca/-/media/documents/ncov/epi/covid-19-epi-confirmed-cases-post-vaccination.pdf?la=en.
- 72.Zhou R, et al. 2022. Vaccine-breakthrough infection by the SARS-CoV-2 omicron variant elicits broadly cross-reactive immune responses. Clin. Transl. Med. 12, 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen J, Wang R, Gilby NB, Wei GW. 2022. Omicron Variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J. Chem. Inf. Model. 62, 412-422. ( 10.1021/acs.jcim.1c01451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dimeglio C, Migueres M, Mansuy JM, Saivin S, Miedougé M, Chapuy-Regaud S. 2022. Antibody titers and breakthrough infections with Omicron SARS-CoV-2. J. Infect. 84, e13-e15. ( 10.1016/j.jinf.2022.01.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.