Abstract
The IncC plasmid R55, initially described in the 1970s and isolated from Klebsiella pneumoniae, confers nonenzymatic chloramphenicol resistance. The gene coding for this resistance was cloned and sequenced and shows 95 to 97% nucleotide identity with the recently reported floR gene from Salmonella enterica serovar Typhimurium DT104 and from Escherichia coli animal isolates, respectively, conferring cross-resistance to florfenicol.
Resistance to chloramphenicol (CHL) has been reported to be mainly due to the production of inactivating enzymes, the CHL acetyl transferases (CATs) (11). Nonenzymatic CHL resistance, however, was described in the late 1970s and early 1980s for plasmids of different incompatibility groups from gram-negative bacteria, such as the IncP-1 plasmid R26 from Pseudomonas aeruginosa and the IncC plasmid R55 from Klebsiella pneumoniae (10, 11). The cml gene of plasmid R26 conferring nonenzymatic CHL resistance was reported in 1986 by Dorman et al. (9) and codes for a putative efflux pump related to the more recently described CmlA protein of the P. aeruginosa In4 integron of transposon Tn1696 (3). Nonenzymatic CHL resistance has gained importance with the spread of multidrug-resistant Salmonella enterica serovar Typhimurium DT104 world-wide epidemic strains, which harbor on their chromosome an antibiotic resistance gene cluster comprising a nonenzymatic CHL resistance gene conferring cross-resistance to florfenicol (FFC) (1, 2, 4, 5).
FFC is a fluorinated analog of CHL approved in Europe for use against pasteurellosis in cattle since January 1995. Previous studies have shown that FFC is active against CHL-resistant strains producing either CATs (6) or nonenzymatic CHL resistance mediated by the CmlA efflux pump (3). The gene conferring cross-resistance to FFC has been named floR, floSt, flo, or cmlA-like (1, 2, 4, 5, 15) and is closely related (97% identity) to the pp-flo gene described in 1996 from a transferable R plasmid of the fish pathogen Pasteurella piscicida (13), recently renamed Photobacterium damselae subsp. piscicida. Their deduced amino acid sequences show about 47% identity to that of the CmlA protein. All of the gene products are assumed to belong to the 12 transmembrane segments family of export proteins of the major facilitator superfamily reviewed by Paulsen et al. (14). FFC resistance conferred by the floR gene has also recently been reported in Escherichia coli strains isolated from cattle and poultry (7, 12, 15) and in S. enterica serovar Agona strains isolated from poultry (8).
In the present study we analyzed nonenzymatic CHL resistance, with particular attention to possible FFC cross-resistance, conferred by the IncC plasmid R55 (150 kb; Tra+ Ap Cm Gm Su), among the first to have been described as conferring nonenzymatic CHL resistance in the 1970s (10, 11). Interestingly, this plasmid, isolated from K. pneumoniae, has been reported to encode enzymatic CHL resistance as well, namely, a type I CAT (11).
R55 FFC resistance and detection of the floR gene.
E. coli strain K-12 BM14 (pro met azi) carrying plasmid R55 was verified as FFC resistant. Antibiograms and the MICs of FFC were determined as described previously (1, 2). FFC disks and the drug itself were purchased from Schering-Plough Animal Health (Kenilworth, N.J.). E. coli BM14 carrying plasmid R55 showed resistance to FFC (MIC, 32 μg/ml) to the same extent as S. enterica serovars Typhimurium DT104 and Agona (8) and previously described E. coli strains (7). PCR was performed on the extracted plasmid DNA using internal primers of the floR gene, cml01 and cml15, as described previously (1, 2, 8). An amplification fragment of the expected size (496 bp) was obtained (data not shown). Nucleotide sequencing of the fragment revealed 95% identity with the floR nucleotide sequence of S. enterica serovar Typhimurium DT104 (data not shown), thus indicating that plasmid R55 carries a floR gene variant conferring resistance to FFC. Southern blot hybridization of plasmid R55 digested by SacI, BamHI, and BglI using a floR probe produced and labeled as described previously (1, 2, 7, 8) revealed bands of 12, 6, and 3 kb, respectively (not shown), and thus confirmed the presence of a floR gene variant on this plasmid.
R55 floR gene variant and flanking regions.
The floR-carrying 6-kb BamHI fragment of plasmid R55 was cloned in plasmid pGEM-7Zf (Ampr) (Promega, Charbonnieres, France) and sequenced. Briefly, BamHI-digested fragments of plasmid R55 were ligated into plasmid pGEM-7Zf. Competent E. coli JM109 cells were transformed with the recombinant plasmids. Selection of transformants was done using Luria-Bertani agar plates supplemented with ampicillin (100 μg/ml) and FFC (10 μg/ml). Positive clones were confirmed by floR PCR on the extracted plasmids. One pGEM-7Zf plasmid clone carrying the 6-kb BamHI insert was kept and named plasmid pSR511. The FFC and CHL MICs for E. coli JM109 carrying plasmid pSR511 were 32 and 128 μg/ml, respectively. Those for E. coli JM109 without the plasmid were 4 μg/ml for both antibiotics. DNA sequencing of the insert was performed by Génome Express (Grenoble, France).
Comparative sequence analysis showed that the R55 floR gene variant was 95 and 97% identical to previously reported floR genes of S. enterica serovar Typhimurium DT104 and E. coli animal isolates, respectively (data not shown). The deduced amino acid sequence of the R55 floR gene variant showed 97 and 98% identity to that of floR of S. enterica serovar Typhimurium DT104 and E. coli, respectively. Amino acid changes occured principally in the sixth transmembrane segment of the protein (data not shown).
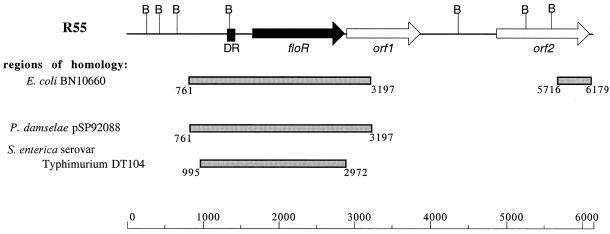
A database search for homologies revealed that the flanking regions of the R55 floR gene partly matched those of previously described E. coli and S. enterica serovar Typhimurium DT104 floR genes and also those of the pp-flo gene of P. damselae subsp. piscicida (Fig. 1). In all cases, the upstream region of floR with its putative promoter region, and a stretch of 99 bp which is repeated in S. enterica serovar Typhimurium DT104 downstream of the floR gene, appears conserved. Also, two open reading frames were detected downstream of floR of plasmid R55 (Fig. 1). The deduced amino acid sequence of orf1 shows homology with those of transcriptional regulators of the LysR family, the closest homolog being that of S. enterica serovar Typhimurium (accession number AAK02052) with 67% amino acid identity (data not shown). The C-terminal end of the deduced amino acid sequence of orf2 is identical to that of the putative transposases found up- and downstream of the floR gene of E. coli (Fig. 1) (7).
FIG. 1.
Structural organization of the 6,179-bp floR locus of plasmid R55. Regions which exhibit homology to the floR-carrying E. coli BN10660 plasmid (GenBank accession no. AF231986), to the pp-flo-carrying P. damselae plasmid pSP92088 (GenBank accession no. D37826), and to the S. enterica serovar Typhimurium DT104 antibiotic resistance gene cluster (GenBank accession no. AF071555) are indicated. The numbers of the homologous segments refer to their positions within the sequence of the plasmid R55 floR locus. The extent and the direction of transcription of the floR, orf1, and orf2 reading frames are marked by arrows. The solid box upstream of floR (DR) indicates the 99-bp direct repeat. The BglI restriction sites are abbreviated as B. The distance scale below the map of the floR locus is given in basepairs.
In conclusion, this study showed that the nonenzymatic CHL resistance described in the late 1970s and mediated by the IncC plasmid R55, initially isolated from K. pneumoniae, is conferred by a floR gene variant which confers FFC cross-resistance to the same extent as previously described floR genes. From a historical point of view, this is thus the first example of FFC resistance, long before its description in P. damselae subsp. piscicida (13), S. enterica serovars Typhimurium DT104 and Agona (1, 2, 4, 5, 8), or E. coli of animal origin (7, 12, 15).
Nucleotide sequence accession number.
The sequence of the floR-containing BamHI fragment from plasmid R55 has been deposited in GenBank under accession no. AF332662.
Acknowledgments
We thank P. Courvalin for providing E. coli strain BM14 and plasmid R55. We also thank M. A. Arcangioli for preliminary studies and C. Mouline for expert technical assistance.
REFERENCES
- 1.Arcangioli M A, Leroy-Sétrin S, Martel J L, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimuriumDT104. FEMS Microbiol Lett. 1999;174:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 2.Arcangioli M A, Leroy-Sétrin S, Martel J L, Chaslus-Dancla E. Evolution of chloramphenicol resistance, with emergence of cross-resistance to florfenicol, in bovine SalmonellaTyphimurium strains implicates definitive phage type (DT) 104. J Med Microbiol. 2000;49:103–110. doi: 10.1099/0022-1317-49-1-103. [DOI] [PubMed] [Google Scholar]
- 3.Bissonnette L, Champetier S, Buisson J P, Roy P H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolton L F, Kelley L C, Lee M D, Fedorka-Cray P J, Maurer J J. Detection of multidrug-resistant Salmonella entericaserotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348–1351. doi: 10.1128/jcm.37.5.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimuriumDT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon M, Harford S, Davies J. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol and some fluorinated derivatives. J Antimicrob Chemother. 1990;26:307–317. doi: 10.1093/jac/26.3.307. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert A, Baucheron S, Flaujac G, Schwarz S, Kehrenberg C, Martel J L, Chaslus-Dancla E. Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coliisolated from cattle. Antimicrob Agents Chemother. 2000;44:2858–2860. doi: 10.1128/aac.44.10.2858-2860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloeckaert A, Sidi Boumedine K, Flaujac G, Imberechts H, D'hooghe I, Chaslus-Dancla E. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. entericaserovar Agona. Antimicrob Agents Chemother. 2000;44:1359–1361. doi: 10.1128/aac.44.5.1359-1361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman C J, Foster T J, Shaw W V. Nucleotide sequence of the R26 chloramphenicol resistance determinant and identification of its gene product. Gene. 1986;41:349–353. doi: 10.1016/0378-1119(86)90119-8. [DOI] [PubMed] [Google Scholar]
- 10.Gaffney D F, Cundliffe E, Foster T J. Chloramphenicol resistance that does not involve chloramphenicol acetyltransferase encoded by plasmids from Gram-negative bacteria. J Gen Microbiol. 1981;125:113–121. doi: 10.1099/00221287-125-1-113. [DOI] [PubMed] [Google Scholar]
- 11.Gaffney D F, Foster T J. Chloramphenicol acetyltransferases determined by R plasmids from Gram-negative bacteria. J Gen Microbiol. 1978;109:351–358. doi: 10.1099/00221287-125-1-113. [DOI] [PubMed] [Google Scholar]
- 12.Keyes K, Hudson C, Maurer J J, Thayer S, White D G, Lee M D. Detection of florfenicol resistance genes in Escherichia coliisolated from sick chickens. Antimicrob Agents Chemother. 2000;44:421–424. doi: 10.1128/aac.44.2.421-424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E H, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida. Microbiol Immunol. 1996;40:665–669. doi: 10.1111/j.1348-0421.1996.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 14.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White D G, Hudson C, Maurer J J, Ayers S, Zhao S, Lee M D, Bolton L, Foley T, Sherwood J. Characterization of chloramphenicol and florfenicol resistance in Escherichia coliassociated with bovine diarrhea. J Clin Microbiol. 2000;38:4593–4598. doi: 10.1128/jcm.38.12.4593-4598.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]