Abstract
The gene encoding a β-lactamase of Prevotella intermedia was cloned and sequenced. This gene, called cfxA2, shared 98% identity with cfxA, the structural gene of a β-lactamase previously described in Bacteroides vulgatus. The deduced protein sequence had a K272E substitution. CfxA2 had the characteristics of class A, group 2e β-lactamases.
Prevotella intermedia, a black-pigmented gram-negative anaerobic rod that is a member of the family Bacteroidaceae, is associated with periodontal diseases and infections of dental origin (19). Several studies have reported increasing resistance to antibiotics in gram-negative anaerobes, especially to β-lactam antibiotics, mostly by the production of β-lactamases (3, 4, 8, 13, 14). Only a few genes encoding β-lactamases have been cloned and sequenced in members of the family Bacteroidaceae (Bacteroides fragilis, Bacteroides vulgatus, and Bacteroides uniformis) but not in Prevotella and Porphyromonas species (12, 16, 18, 20). A preliminary work concluded that P. intermedia and Prevotella buccae were the predominant β-lactamase-producing species among anaerobic gram-negative rods isolated from periodontal pockets, with 35 and 42% of these, respectively, β-lactamase-positive strains (9). Biochemical characterization of these β-lactamases is difficult because of fastidious bacterial growth and weak enzymatic activity. The purpose of this work was to clone the β-lactamase gene of a strain of P. intermedia for biochemical and genetic analysis.
P. intermedia NI-1187 was a clinical isolate obtained from the subgingival flora of a male adult patient suffering from periodontitis. The wild-type strain showed resistance to penicillin, amoxycillin, tetracycline, and erythromycin and susceptibility to the amoxycillin-clavulanic acid combination. Isoelectric focusing experiments performed with sonified crude extracts of P. intermedia did not allow us to visualize the β-lactamase. All of the media and compounds used have been previously described (9). Sequences were determined from both strands of DNA with an Applied Biosystems sequencer (Eurogentec, Herstal, Belgium). Deduced protein sequences and sequence alignments were performed with the National Center for Biotechnology Information, Infobiogen, and ExPaSy suite of programs, and β-lactamase relatedness was investigated by comparison with the GenBank-EMBL-DDBJ databases.
For cloning experiments, chromosomal DNA from P. intermedia NI-1187 was obtained with conventional phenol-chloroform extraction methods, restricted with EcoRI, ligated in pZErO-2-Kan, and transferred by electroporation in Escherichia coli Top 10 (InvitroGen, Leek, The Netherlands) (12, 16, 20). A bank of approximately 106 recombinant clones was obtained on kanamycin selective plates and yielded about 10 colonies of β-lactamase-producing E. coli transformants on ampicillin selective plates (40 μg/ml). Plasmids from E. coli NI-14 transformants presented a 15-kb DNA EcoRI insert. After subcloning was done, one clone of E. coli, NI-141, was chosen for genetic analysis. It harbored the pNCE-3 plasmid with an EcoRI/PstI 4.9-kb insert. The MIC determination was suggestive of a class A, group 2e β-lactamase (Table 1) (2, 7). This phenotype, with penicillinase and cephalosporinase properties and characteristic low resistance to cefotaxime and cefpirome, is a common feature of P. intermedia strains producing β-lactamases (4, 9, 23).
TABLE 1.
MICs of 10 β-lactam antibiotics alone in E. coli Top 10 host strains and alone and in combination with β-lactamase inhibitors (clavulanic acid and tazobactam) in E. coli NI-141 cloned with the cfxA2 β-lactamase gene of an oral strain of P. intermedia (DNA insert, 4.9-kb)
| Substrate | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| E. coli Top 10 (antibiotic alone) |
E. coli NI-141 coding for CfxA2 β-lactamase
|
|||
| Antibiotic alone | Combination with clavulanic acida | Combination with tazobactamb | ||
| Amoxycillin | 4c | 1,024 | 16 | 16 |
| Ticarcillin | 4 | 256 | 8 | 4 |
| Cephalothin | 4 | 32 | 8 | 8 |
| Cefuroxime | 8 | 64 | 8 | 8 |
| Cefoxitin | 4 | 4 | 4 | 4 |
| Cefotaxime | 0.06 | 1 | 0.125 | 0.125 |
| Ceftazidime | 0.5 | 2 | NDd | ND |
| Aztreonam | 0.125 | 0.125 | 0.125 | 0.125 |
| Cefpirome | 0.03 | 1 | 0.125 | 0.125 |
| Imipenem | 0.06 | 0.125 | ND | ND |
Clavulanic acid at a fixed concentration of 2 μg/ml.
Tazobactam at a fixed concentration of 4 μg/ml.
Amoxycillin and clavulanic acid, MIC <2 μg/ml.
ND, not done.
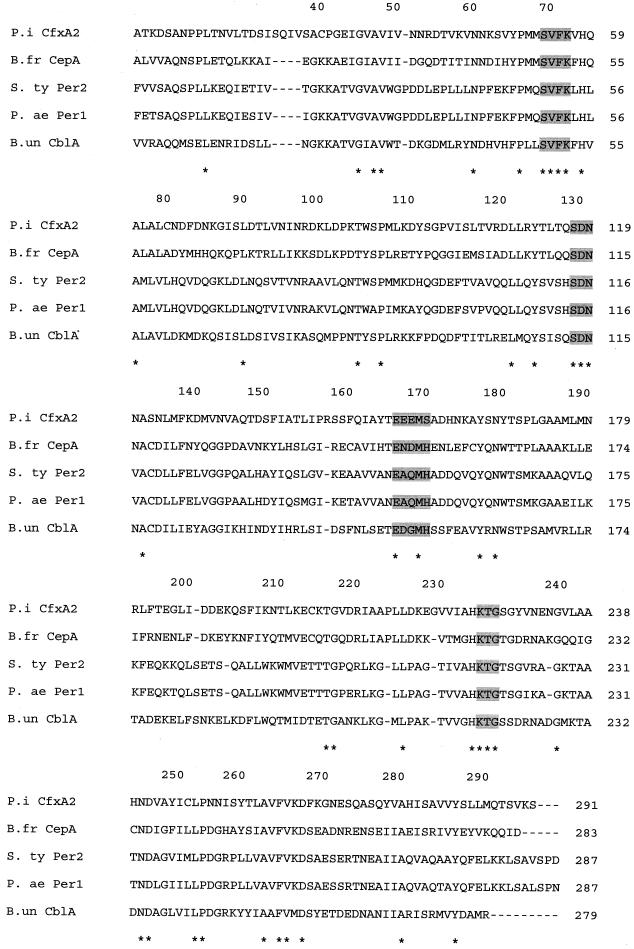
The 4.9-kb insert was sequenced and assigned GenBank accession no. AF118110. The β-lactamase gene of P. intermedia NI-1187 shared 98% identity with cfxA, the structural gene of a class A β-lactamase previously characterized in a cefoxitin-resistant B. vulgatus CLA-341 strain, and was provisionally called cfxA2 (16). The deduced protein sequence called CfxA2 contained 321 amino acids with a K272E substitution (Fig. 1). The flanking sequences revealed transposition genes. The left flanking region contained mobA and the partial sequence of a gene related to TnpA (98 and 40% homology, respectively) previously described on the Bacteroides mobilizable transposon Tn4555 (11, 22). The right flanking region shared 27% homology with mobC and bfmC, associated with the pathogenicity island of enterotoxigenic B. fragilis strains (10).
FIG. 1.
Multiple sequence alignment of the amino acid sequences of CfxA2 and related β-lactamases. Amino acids that are identical in all five sequences are marked with asterisks. The amino acid numbering for the class A β-lactamases is used (2). The main conserved amino acid motifs of class A β-lactamases are shaded. Abbreviations: P.i, P. intermedia NI-1187; B.fr, B. fragilis (17); S. ty, S. enterica serovar Typhimurium (4); P. ae, P. aeruginosa (14); B.un, B. uniformis (19). CfxA2 differs from CfxA of B. vulgatus by a K272E substitution (16).
For purification purposes, two sets of primers were designed in order to produce the CfxA2 protein with a C- (Set 1, 5′- AAAAAACCATGGAAAAAAACAGAAAAAAACAAATC G-3′ and 5′-AAAAAACTCGAGAGATTTTACTGAAGTTTGCATTAATAAAGAATATAC-3′) and an N-terminal (Set 2, 5′-GGGATCCGAAAAAAACAGAAAAAAACAAATC-3′ and 5′-CGAATCCTTAAGATTTTACTGAAGTTAG-3′) histidine tag. After PCR amplification of cfxA2, including the promotor region, cfxA2 was inserted into pET28 and cloned into E. coli BL21(DE3) (TA cloning kit; Novagen, Madison, Wis.) (17, 21). The transformed bacteria were grown in 1 liter of Luria-Bertani broth (kanamycin [50 μg/ml]) for 5 h at 37°C, followed by IPTG (isopropyl-β-d-thiogalactopyranoside) (0.3 mM) induction for 4 h at 30°C, centrifugation, and disruption by two passages through a French pressure cell. The tagged proteins were then purified by affinity chromatography through nickel-coated Sepharose beads with an imidazole elution buffer (20 to 500 mM, pH 7.4) according to the manufacturer's recommendations (HiTrap chelating column and GradiFrac; Pharmacia Biotec, Uppsala, Sweden). β-Lactamase activity was monitored with the chromogenic nitrocefin (482 nm) (Uvikon 820 spectrophotometer; Kontron Instruments, Zürich, Switzerland). The C-terminally tagged CfxA2 protein from clone NI-124 (33 to 35 kDa) was obtained in a pure but inactive form and was resistant to thrombin hydrolysis (21). The N-terminally tagged protein (clone NI-142) was highly purified in an active form. After an affinity purification step, the β-lactamase was purified about 50-fold compared to crude homogenate supernatant. Active fractions were pooled, extensively dialyzed, thrombin treated, and stored at −70°C until use. Protein concentrations were determined by the method of Bradford, with bovine serum albumin used as a standard (protein assay; Bio-Rad Laboratories GmbH, Munich, Germany). Repeated isoelectric focusing experiments with crude extracts were necessary to visualize a discrete reactive band with a pI of 5.4. The pI reported for Bacteroides species was 5.8, but CfxA2 differed from CfxA by a glutamic acid (acid) replacing a lysine (base) in position 272 (12, 16). The hydrolysis of β-lactams was monitored at 37°C in sodium phosphate buffer (0.05 M; pH 7.0) with 20 μg of β-lactamase in a 500-μl reaction mixture. Kinetic parameters were estimated for at least three different assays, and substrate inhibition was confirmed with antibiotic concentrations above 50 μM (23). The apparent Km and relative Vmax values were calculated from Eadie-Hofstee plots (Table 2), with Vmax values relative to that of benzylpenicillin, which was set as 100 as previously described (1, 17). The kinetic parameters of CfxA2 are characterized by Km values ranging from 12 to 38 μM for all of the β-lactams tested, except for cefoxitin, which was not hydrolyzed. The inhibitory kinetic parameters (Ki) of CfxA2 with cefazolin as a substrate were as follows: cefoxitin, 10 nM, and clavulanic acid, 200 nM. Inhibitors were preincubated with the enzyme for 10 min at 37°C before the rate of cefazolin inhibition was tested.
TABLE 2.
Steady-state kinetic parameters of highly purified CfxA2 β-lactamase cloned in E. coli NI-142
| Substrate | Relative Vmaxa | Km (μM) | Relative Vmax/Km |
|---|---|---|---|
| Benzylpenicillin | 100 | 20.7 | 4.8 |
| Ampicillin | 160 | 38.0 | 4.2 |
| Cefazolin | 300 | 12.3 | 24.4 |
| Cefuroxime | 1,500 | 60.6 | 24.8 |
| Cefotaxime | 600 | 12.9 | 46.5 |
| Cephalothin | 40 | 24.7 | 1.6 |
| Cefoxitin | <0.01 |
Values relative to that of benzylpenicillin, which was set at 100.
In order to determine whether the K272E substitution affects the kinetic parameters towards representative β-lactam substrates, and particularly resistance to cefoxitin (6), we compared the kinetic properties of the original CfxA (B. vulgatus CLA-341 [kindly provided by J. C. Smith]) and CfxA2 with substitutions (B. vulgatus NI-2869 [a clinical laboratory strain]) produced in wild-type B. vulgatus strains. For comparison, (i) wild-type β-lactamase genes were sequenced for identification purposes (PCR amplification); (ii) susceptibility profiles and MICs were determined, and (iii) kinetic parameters were calculated from partially purified β-lactamase crude extracts as previously described (23). B. vulgatus CLA-341 was resistant to benzylpenicillin, amoxycillin, cefoxitin, and moxalactam and susceptible to the amoxycillin-clavulanic acid combination, piperacillin, and imipenem. No synergy was observed between cefoxitin and amoxycillin-clavulanic acid. In comparison, B. vulgatus NI-2869 was susceptible to moxalactam and showed decreased susceptibility to cefoxitin. Amoxycillin, amoxycillin-clavulanic acid, cefoxitin, and cefuroxime MICs were as follows: 256, 0.125, 256, and 256 μg/ml (CLA-341) and 4, 0.94, 12, and 1 μg/ml (NI-2869), respectively. Comparison of CfxA and CfxA2 kinetic parameters showed that the K272E substitution has no significant influence on their catalytic properties towards benzylpenicillin, ampicillin, cefotaxime, cephalothin (hydrolyzed), and cefoxitin (not hydrolyzed) but increases CfxA2 affinity for cefazolin about 10-fold (Table 3). The high level of resistance of B. vulgatus CLA-341 towards cefoxitin should be attributed to another resistance mechanism than CfxA production, such as a porin mutation (16).
TABLE 3.
Comparison of kinetic parameters of partially purified CfxA and CfxA2 β-lactamases obtained from wild-type strains (CfxA, B. vulgatus CLA-341; CfxA2, B. vulgatus NI-2869)
| Substrate | Relative Vmaxa
|
Km (μM)
|
||
|---|---|---|---|---|
| CfxA | CfxA2 | CfxA | CfxA2 | |
| Benzylpenicillin | 100 | 100 | 30.0 | 20.0 |
| Ampicillin | 35 | 130 | 40.0 | 40.0 |
| Cefazolin | 740 | 170 | 133.3 | 10.2 |
| Cefotaxime | 90 | 670 | 40.0 | 8.0 |
| Cefoxitin | <0.01 | <0.01 | ||
Values relative to that of benzylpenicillin, which was set at 100.
In order to determine the location of enzymatic activity, cultures of E. coli NI-142 were fractionated according to the osmotic-shock method (16). Cell membranes were finally treated with 2% polyoxyethylene 10–tridecyl ether, a detergent which does not inhibit enzyme activity (Emulphogene-BC-720; Sigma, St. Louis, Mo.). Cefazolin (50 μM) was used as a substrate, and no enzymatic activity could be detected in extracellular, periplasmic, or cytoplasmic fractions, while 52% of enzymatic activity was recovered in the pellet after the French press treatment. Eighty-three percent of this membrane activity could be solubilized after detergent treatment. These results in E. coli suggest a cytoplasmic membrane localization of CfxA β-lactamases. However, interference of the N-terminal histidine tag cannot be excluded (16).
Phylogenetic analysis revealed homologies with other β-lactamases of anaerobes (CfxA of B. vulgatus, >99%; CepA of B. fragilis, 35%; CblA of B. uniformis, 28%) and with extended-spectrum β-lactamases of aerobic rods (Per-1 of Pseudomonas aeruginosa, 27%; Per-2 of Salmonella enterica serovar Typhimurium, 28%) (Fig. 1) (5, 6, 15, 16, 18, 20). From genetic data, Nordmann and Naas proposed a novel subgroup of class A β-lactamases for CfxA and Per-1 (15). Per-2 and CfxA2 would be new members of this subgroup, but CfxA (B. vulgatus and P. intermedia) is not an extended-spectrum β-lactamase. Additional work is necessary to characterize the mobilization genes associated with cfxA2 and its spread among Prevotella species.
Acknowledgments
We thank C. J. Smith, Department of Microbiology and Immunology, East Carolina University, who sent us Bacteroides vulgatus CLA-341, and A. Galmiche, INSERM 452, Faculté de Médecine de Nice, Nice, France, for helpful assistance in protein purification.
REFERENCES
- 1.Ahamed J, Kundu M. Molecular characterization of the SHV-11 β-lactamase of Shigella dysenteriae. Antimicrob Agents Chemother. 1999;43:2081–2083. doi: 10.1128/aac.43.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P, Coulson A F W, Frere J, Ghuysen J, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andres M T, Chung W O, Roberts M C, Fierro J F. Antimicrobial susceptibilities of Porphyromonas gingivalis, Prevotella intermedia, and Prevotella nigrescens spp. isolated in Spain. Antimicrob Agents Chemother. 1998;42:3022–3023. doi: 10.1128/aac.42.11.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appelbaum P C, Spangler S K, Jacobs M R. β-Lactamase production and susceptibilities to amoxycillin, amoxycillin-clavulanate, ticarcillin, ticarcillin-clavulanate, cefoxitin, imipenem, and metronidazole of 320 non-Bacteroides fragilis Bacteroides isolates and 129 fusobacteria from 28 U.S. centers. Antimicrob Agents Chemother. 1990;34:1546–1550. doi: 10.1128/aac.34.8.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Stemplinger I, Jungwirth R, Pangold P, Amann S, Akalin E, Ang O, Bal C, Casellas J M. Characterization of β-lactamase gene bla-PER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob Agents Chemother. 1996;40:616–620. doi: 10.1128/aac.40.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouthors A T, Dagoneau-Blanchard N, Naas T, Nordmann P, Jarlier V, Sougakoff W. Role of residues 104, 164, 166, 238 and 240 in the substrate profile of PER-1 beta-lactamase hydrolyzing third-generation cephalosporins. Biochem J. 1998;15:1443–1449. doi: 10.1042/bj3301443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards R, Thirlwell D, Greenwood D. Changes in beta-lactam antibiotic susceptibility and beta-lactamase production of clinical isolates of Bacteroides and Prevotella species over a 9 year period. J Antimicrob Chemother. 1996;37:636–638. doi: 10.1093/jac/37.3.636. [DOI] [PubMed] [Google Scholar]
- 9.Fosse T, Madinier I, Hitzig C, Charbit Y. Prevalence of β-lactamase producing strains among 149 anaerobic Gram negative rods isolated from periodontal pockets. Oral Microbiol Immunol. 1999;14:352–357. doi: 10.1034/j.1399-302x.1999.140604.x. [DOI] [PubMed] [Google Scholar]
- 10.Franco A A, Cheng R K, Chung G T, Wu S, Oh H B, Sears C L. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J Bacteriol. 1999;181:6623–6633. doi: 10.1128/jb.181.21.6623-6633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guiney D J, Hasegawa P. Transfer of conjugal elements in oral black-pigmented Bacteroides (Prevotella) spp. involves DNA rearrangements. J Bacteriol. 1992;174:4853–4855. doi: 10.1128/jb.174.14.4853-4855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedberg M, Lindqvist L, Bergman T, Nord C E. Purification and characterization of a new β-lactamase from Bacteroides uniformis. Antimicrob Agents Chemother. 1995;39:1458–1461. doi: 10.1128/aac.39.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Könonen E, Saarela M, Kanervo A, Karjalainen J, Asikainen S, Jousimies-Somer H. β-Lactamase production and benzylpenicillin susceptibility among different ribotypes of Prevotella melaninogenica simultaneously colonizing the oral cavity. Clin Infect Dis. 1995;20(Suppl. 2):S364–S366. doi: 10.1093/clinids/20.supplement_2.s364. [DOI] [PubMed] [Google Scholar]
- 14.Medeiros A A. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl. 1):S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 15.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker A C, Smith C J. Genetic and biochemical analysis of a novel Ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob Agents Chemother. 1993;37:1028–1036. doi: 10.1128/aac.37.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Naas T, Guibert M, El Bachir C, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodgers M B, Parker A C, Smith C J. Cloning and characterization of the endogenous cephalosporinase gene, CEP-A, from Bacteroides fragilis reveals a new subgroup of Ambler class A β-lactamases. Antimicrob Agents Chemother. 1993;37:2391–2400. doi: 10.1128/aac.37.11.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah H N, Gharbia S E. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the Bacteroides. Int J Syst Bacteriol. 1995;40:5426. doi: 10.1099/00207713-40-2-205. [DOI] [PubMed] [Google Scholar]
- 20.Smith C J, Bennett T K, Parker A C. Molecular and genetic analysis of the Bacteroides uniformis cephalosporinase gene, CBL-A, encoding the species-specific β-lactamase. Antimicrob Agents Chemother. 1994;38:1711–1715. doi: 10.1128/aac.38.8.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 22.Tribble G D, Parker A C, Smith C J. Transposition genes of the Bacteroides mobilizable transposon Tn4555: role of a novel targeting gene. Mol Microbiol. 1999;34:385–394. doi: 10.1046/j.1365-2958.1999.01616.x. [DOI] [PubMed] [Google Scholar]
- 23.Valle G, Quiros L M, Andrés M T, Fierro J F. A β-lactamase belonging to group 2e from oral clinical isolates of Prevotella intermedia. FEMS Microbiol Lett. 1998;158:191–194. doi: 10.1111/j.1574-6968.1998.tb12819.x. [DOI] [PubMed] [Google Scholar]